Abstract
New antimalarial compounds with novel mechanisms of action are urgently needed to combat the recent rise in antimalarial drug resistance. Phenotypic high-throughput screens have proven to be a successful method for identifying new compounds, however, do not provide mechanistic information about the molecular target(s) responsible for antimalarial action. Current and emerging target identification methods such as in vitro resistance generation, metabolomics screening, chemoproteomic approaches and biophysical assays measuring protein stability across the whole proteome have successfully identified novel drug targets. This review provides an overview of these techniques, comparing their strengths and weaknesses and how they can be utilised for antimalarial target identification.
Keywords: Target identification, Plasmodium falciparum, Antimalarial, CETSA, Chemoproteomics
Graphical abstract
Highlights
-
•
Novel antimalarial compounds with unknown mechanisms of action require new methods of target identification.
-
•
Many methods have complementary strengths and weaknesses.
-
•
IVIEWGA and systems biology approaches have traditionally been effective target ID methods.
-
•
Emerging proteomics-based techniques offer new options for target ID approaches.
-
•
Direct small molecule-protein interactions can now be identified across a whole proteome.
1. The need for new antimalarial compounds with novel mechanisms of action
Malaria remains one of the most prevalent infectious diseases throughout the world, with more than 40% of the world's population living in malaria endemic regions (World Health Organization, 2021). In 2020, there were over 200 million cases and over 600 thousand deaths caused by malaria, predominantly due to two Plasmodium species, P. falciparum and P. vivax (World Health Organization, 2021). Of the two, P. falciparum is responsible for over 90% of human infections and represents a significant global health burden. These cases disproportionately affect developing countries, with nineteen African countries along with India carrying 85% of the malaria burden. Furthermore, the majority of deaths occur in children under 5 years of age. Efforts in malaria prevention have had mixed success. The use of insecticide treated bed nets has been effective in reducing parasite prevalence and child mortality, and has been a core aspect of malaria control programs (Pryce et al., 2018). However, the development of a vaccine has had limited success, with the recently approved RTS,S vaccine (Mosquirix™) demonstrating limited efficacy, reducing clinical malaria episodes by 26–38% in children (Morrison, 2015). In the absence of a highly efficacious vaccine, management of malaria currently relies on small molecule antimalarial drugs that can quickly and effectively treat active malaria infection, prevent new infections or remove quiescent liver-stage parasites.
Currently, artemisinin-based combination therapies (ACTs) are the front-line treatment for P. falciparum infections. These combine fast acting artemisinin derivatives with complementary antimalarials that act via a different mechanism of action (Heller and Roepe, 2019). Combination therapies provided a solution to the development of resistance to previously-used antimalarials such as chloroquine, sulfadoxine and pyrimethamine throughout Africa (D'Alessandro and Buttiens, 2001). However, the development of artemisinin resistance throughout South East Asia over the previous decade threatens the effectiveness of ACTs (Dondorp et al., 2009). Therefore, the development of new antimalarials with novel mechanisms of action is necessary to further reduce malaria-associated mortality and proceed towards complete eradication (Anthony et al., 2012; Phillips et al., 2017).
Many new compounds with promising antimalarial potential have recently been discovered using phenotypic high throughput screens. Phenotypic screens test libraries of compounds against whole cells in order to determine those compounds with antiparasitic activity, via a phenotypic readout such as parasite survival (Hovlid and Winzeler, 2016). These screens have been a valuable tool for the identification of new classes of antimalarials as they do not require prior knowledge of a validated biological target (Hovlid and Winzeler, 2016; Katsuno et al., 2015) and have identified many novel antiplasmodial chemotypes (Gamo et al., 2010). However, the unknown mechanism of action of these novel ‘hit’ compounds has limited further development, as structure-based drug design strategies are not a viable option without a known target (Anderson, 2003; Drinkwater and McGowan, 2014). There are examples of compounds that have progressed through the drug development pipeline without a known target, however, understanding the mechanism of action of novel drug candidates can substantially enhance their development and subsequent utilisation (Tse et al., 2019).
This review provides an overview of current and emerging target identification methods for novel compounds, used in the investigation of both the mode and mechanism of action (MoA). These include genomic and systems biology approaches such as in vitro resistance selection and metabolomics screening, along with mass-spectrometry based approaches to detect drug-target binding, including chemoproteomic approaches and biophysical assays. We have highlighted studies that demonstrate how these techniques can provide insight into novel mechanisms of action and compared some of the strengths and limitations of each technique (Table 1).
Table 1.
Strengths and weaknesses of target identification techniques.* = Recent literature examples of target ID methods. Listed examples are not exhaustive.
| Technique | Strengths | Weaknesses | Examples* (Plasmodium) |
|---|---|---|---|
| In vitro resistance selection and whole genome sequencing |
|
|
|
| Untargeted multi-omics approaches |
|
|
|
| Affinity-based probe chemoproteomics |
|
|
|
| CETSA proteomics |
|
|
|
| Limited proteolysis mass spectrometry |
|
|
|
2. Drug target identification methods
2.1. In vitro evolution and whole genome sequencing (IVIEWGA)
A commonly used method for the identification of unknown targets involves in vitro selection of resistance to a novel compound coupled with whole genome sequencing (IVIEWGA) (Cowell and Winzeler, 2019a; Xie et al., 2018). In this method, parasites are exposed to low concentrations of antiparasitic compounds until resistant parasites are selected. The resistant clones can then be sequenced via whole genome sequencing to identify genetic mutations or copy number amplifications that confer resistance (Cowell et al., 2018; Xie et al., 2018). These mutations typically confer resistance by reducing the ability of the drug to bind to its target or by developing a compensatory mechanism through which the parasite can overcome the drug pressure (Cowell et al., 2018). Successful IVIEWGA approaches, experimental design, and the role that it has played in antimalarial drug discovery have previously been reviewed in detail (Cowell and Winzeler, 2019b; Flannery et al., 2013; Luth et al., 2018; Nzila and Mwai, 2010; Okombo et al., 2021).
IVIEWGA has been used to explore the MoA of many novel antimalarial chemotypes. This is perhaps most evident in the mapping of the P. falciparum druggable genome (Cowell and Winzeler, 2018). Cowell et al. selected for in vitro resistance against 37 chemically distinct small molecules, identifying 35 genes thought to be either a drug-resistance determinant or an actual target. A number of these were new drug targets, including farnesyltransferase, dipeptidyl-aminopeptidase 1 and the aminophospholipid transporting P-type ATPase. IVIEWGA has also successfully identified a number of new targets for compounds now in clinical development, such as DDD107498 and DSM265, which target translation elongation factor 2 (eEF2), and dihydroorotate dehydrogenase (PfDHODH), respectively, highlighting the power of IVIEWGA as an untargeted approach for target identification (Baragana et al., 2015; Cowell and Winzeler, 2019a; White et al., 2019).
A primary advantage of IVIEWGA is that identified mutations have demonstrable phenotypic implications for parasite survival, in contrast with other target identification methods that identify direct target-protein interactions. Furthermore, identified resistance mechanisms could provide insight into the potential for resistance to develop in the field if these compounds are used clinically. However, IVIEWGA also has a number of limitations. The identification of mutations via whole genome sequencing must be validated, either by reintroduction of the mutation into wild type parasites, or an extensive mechanistic investigation to confirm the mutation identified is responsible for the resistant phenotype observed.
In vitro resistance generation can induce non-specific, multi drug resistance mechanisms that provide limited information about the specific target of the test compound. For example, four months of continuous culturing of P. falciparum in the presence of sub-lethal concentrations of the imidazolopiperazine KAF156 (ganaplacide), resulted in the selection of six resistant clones (Kuhen et al., 2014; LaMonte et al., 2016; Meister et al., 2011). Upon sequencing, mutations in one specific gene, P. falciparum cyclic amine resistance locus (PfCARL), were found to confer resistance (Kuhen et al., 2014). The function of PfCARL is unknown, although homology models have suggested a potential role in protein trafficking (Jonikas et al., 2009). Interestingly, mutations in PfCARL also conferred resistance to structurally unrelated compounds (LaMonte et al., 2016; Magistrado et al., 2016) and the resistance-causing mutations were not present in an obvious catalytic site, suggesting that PfCARL is likely a general resistance determinant rather than a molecular target. A recent study has localised the imidazolopiperazine GNF179 to the endoplasmic reticulum and demonstrated inhibition of protein transport in P. falciparum as well as the expansion of the ER following imidazolopiperazine treatment (LaMonte et al., 2020). However, the molecular target of the imidazolopiperazines has not yet been identified and further studies using chemoproteomic or biophysical methods may be required to identify a direct target.
Other examples of non-specific resistance mechanisms include an increased copy number of Pf multi-drug resistance 1 transporter (PfMDR1). PfMDR1 confers multi-drug resistance via enhanced drug efflux, and therefore would provide little insight into a potential target (Blasco et al., 2017; Cowell and Winzeler, 2018). PfATP4 mutations are another resistance mechanism which have been shown to confer resistance to multiple, chemically distinct classes of compounds including the spiroindolones, pyrazoles and the dihydroisoquinolones (Flannery et al., 2015; Spillman and Kirk, 2015). While PfATP4 is an important drug target for the clinical candidate cipargamin (Rottmann et al., 2010), its propensity for conferring resistance suggests that it could have a compensatory function and therefore provides limited insight into the binding target of cipargamin.
Whilst IVIEWGA has proven very effective at identifying the mechanisms of action and/or resistance of promising drug candidates, it should also be noted that this approach preferentially identifies targets that are inherently prone to developing resistance, which could be detrimental to future clinical usage. For example, resistance to atovaquone can be rapidly selected for in vitro (Musset et al., 2007; Phillips et al., 2015), and resistance to atovaquone rapidly emerged in the field shortly after it was first introduced as a monotherapy (Looareesuwan et al., 1996). Therefore, new strategies to expedite the early prediction of parasite resistance to new antimalarial compounds are of significant interest to the antimalarial drug discovery community. One such strategy is the minimum inoculation of resistance (MIR) parameter, which describes the potential for selection of resistance to a given candidate compound at a given concentration (Duffey et al., 2021). Obtaining an MIR involves exposing a range of parasite inocula (105 to 109 parasites) to constant drug pressure to determine the minimum number of parasites required for selection of a resistant population. The MIR parameter allows for the identification of compounds for which resistance can easily be selected. These compounds can then be deprioritised in preference for compounds which may be less susceptible to the emergence of resistance in the field. The MIR parameter is impacted by both the concentration of compound used and the parasite strain tested. Therefore, the use of the multi-drug resistant P. falciparum Dd2-B2 line is recommended and it is important to generate a MIR at multiple concentrations (Duffey et al., 2021). The MIR and speed of resistance selection can provide insight into the MoA of new compounds. For example, compounds for which resistance is difficult to select could possess multiple parasite targets, or have a fast onset of action.
Furthermore, ‘irresistible’ compound classes; where the specific MoA of the compound prevents the selection of resistant parasites via in vitro evolution methods, are becoming of increasing interest to the antimalarial drug discovery community (Yang et al., 2021). These compounds differ from ‘targetless’ compound classes, where resistant parasites can be obtained in vitro however, a target cannot be determined from IVIEWGA alone. Irresistible compounds are less likely to result in drug resistance when used clinically and often have a fast onset of action (Cowell and Winzeler, 2018). Therefore, the Medicines for Malaria Venture (MMV) have highlighted the development of ‘resistance-proof’ chemical scaffolds as a priority for future antimalarials (Burrows et al., 2017). Other target identification methods may be necessary for these compound classes. However, the use of mutagenic agents such as ethyl methanesulfonate to increase the likelihood of selecting resistant parasites has seen success in generating resistance-causing mutations for compounds where traditional IVIEWGA experimental designs have failed (Gisselberg et al., 2018). Furthermore, hypermutator parasite lines that possess mutations in the DNA polymerase delta, which ablates their proofreading activity, have been shown to increase the rate of DNA mutation (Honma et al., 2014). These lines have been successfully used to identify putative targets for the antimalarial Salinipostin A, after traditional IVIEWGA approaches failed to identify resistance-causing mutations (Yoo et al., 2020). While hyper-mutating strategies can increase the odds of identifying resistance-causing mutations, extensive validation of identified mutations is necessary to ensure they are responsible for the resistance phenotype, and are directly related to the compound's mechanism of action.
2.2. Systems biology multi-omics approaches
Untargeted omics techniques, such as metabolomics, proteomics and transcriptomics, can provide an untargeted overview of cellular systems by the simultaneous detection and quantification of small molecules, proteins and transcript levels in a biological system (Dunn et al., 2012; Scalbert et al., 2009). By comparing drug-treated samples to untreated controls, perturbations in cellular processes resulting from drug treatment can be identified. This can provide insight into the MoA of novel compounds and in some limited instances, identify specific targets.
Of the different omics techniques, untargeted metabolomics is best positioned for identifying direct effects of drugs on parasite metabolism, whereas transcriptomics and proteomics are more likely to detect downstream or secondary responses. Because of this, untargeted metabolomics approaches have been successfully applied to confirm identified antimalarial targets, with studies into the MoA of atovaquone providing a good example of how untargeted metabolomics can demonstrate a specific MoA (Allman et al., 2016; Dickerman et al., 2016). Atovaquone is a direct inhibitor of the bc1 complex, an essential component of the mitochondrial electron transport chain (Fry and Pudney, 1992). Inhibition of the bc1 complex impairs de novo pyrimidine synthesis through the indirect inhibition of PfDHODH and this produces a clear metabolic signature, with a rapid and robust accumulation of the precursors for pyrimidine synthesis, N-carbamoyl-L-aspartate and dihydroorotate, detected following atovaquone treatment (Allman et al., 2016). This clear metabolic signature is consistently seen with other inhibitors of pyrimidine synthesis, such as DSM265, and has been used to identify PfDHODH as a target of MMV020439 and MMV007571 (Coteron et al., 2011; Creek et al., 2016; Dickerman et al., 2016).
Metabolomics can also be used to characterise the MoA of novel compounds with higher throughput than other target identification methods, by comparing the metabolic profile of test compounds with those of known compounds. Two studies into the MMV Malaria Box were able to predict a substantial number of target pathways for compounds with previously unknown MoA (Allman et al., 2016; Creek et al., 2016). One study simultaneously investigated the mode of action of 90 antimalarial compounds, with over half of the tested compounds demonstrating significant metabolic perturbations. Of these compounds, nearly one quarter caused an increase in N-carbamoyl-L-aspartate and dihydroorotate, indicating that they inhibit pyrimidine synthesis, while 13 compounds possessed a metabolic signature similar to artemisinin and 6 shared a metabolic signature with chloroquine. This allows for the quick characterisation of novel compounds, providing rapid insight into their potential MoA. However, it is important to consider that while similar metabolic profiles could indicate similar MoA, they do not necessarily have the same biological target. For instance, it was hypothesised that those compounds that clustered with artemisinin may share its potent, fast acting profile, and therefore the observed metabolic perturbation was likely due to rapidly mediated cell death, rather than an indicator of a specific drug target or pathway (Creek et al., 2016).
A limitation of typical metabolomics workflows is the fact that they only detect small molecules and therefore, are not useful for compounds which do not interfere with parasite metabolism. Metabolomics has been combined with other ‘omics’ approaches, such as transcriptomics or proteomics to overcome this limitation (Birrell et al., 2020; Giannangelo et al., 2020; van Brummelen et al., 2009). These complimentary approaches allow for a greater range of biomolecules to be detected, providing more scope for the identification of changes that can provide insight into the MoA. For instance, a recent investigation into synthetic ozonide antimalarials combined metabolomics, peptidomics and proteomics to identify the MoA of the ozonides in P. falciparum. In this study, global metabolomics and peptidomics analysis identified an initial impairment of the haemoglobin digestion pathway, with depletion of short haemoglobin-derived peptides and an accumulation of long chain haemoglobin-derived peptides identified through these complimentary techniques (Giannangelo et al., 2020). Subsequent proteomics and metabolomics performed at later time points identified secondary perturbations in pyrimidine biosynthesis, the Kennedy pathway, translation regulation and the ubiquitin-proteasome system, providing a comprehensive analysis of both primary and secondary mechanisms of ozonide-induced parasite death.
Another limitation of typical metabolomics-based approaches is that while they often provide valuable mechanistic information, they often identify a target pathway or signature rather than the specific drug target, and they cannot measure direct drug-target binding. Systems biology-based approaches to investigate compound mechanisms can also be confounded by many experimental design factors, including compound concentration, duration of incubation and parasite lifecycle stage. Designing experiments with multiple distinct time points and concentrations can assist with identifying drug-specific perturbations. Nevertheless, untargeted metabolomics, proteomics and transcriptomics approaches provide useful and unbiased characterisations of the impact of novel drug candidates on parasite biochemistry, enabling the generation or confirmation of hypotheses regarding their MoA.
2.3. Chemoproteomic approaches to target identification
Small molecule affinity-based probes have been widely used for the identification of novel drug targets and new biological pathways (Heal et al., 2011; Su et al., 2013). Affinity-based probes require functionalisation of the compound of interest with chemical handles that allow the compound to be enriched, along with its bound protein target(s) (Kawatani and Osada, 2014). Recently, novel antimalarial compounds with unknown MoAs have been functionalised to allow for the identification of new drug targets (Ismail et al., 2016; Paquet et al., 2017). The study into MMV390048 is an example where an affinity-based probe was used to successfully identify P. falciparum phosphatidylinositol 4-kinase (PfPI4K) as a new target for a novel antimalarial compound (Paquet et al., 2017). An active analogue, MMV666845, was covalently immobilised on Sepharose beads and used as bait to capture protein targets from a cell lysate. To distinguish between specific and non-specific binding, this pull-down approach was also performed in the presence of the active parent compound MMV390048. The ‘free’ MMV390048 outcompetes the probe MMV666845 for the protein target, resulting in less enrichment of the target protein compared to non-specific binding proteins. This allows the target to be distinguished from those proteins with non-specific binding. This approach identified PfPI4K as the only specifically-bound protein and was supported by in vitro generation of MMV390048-resistant parasites, which identified mutations in PfPI4K.
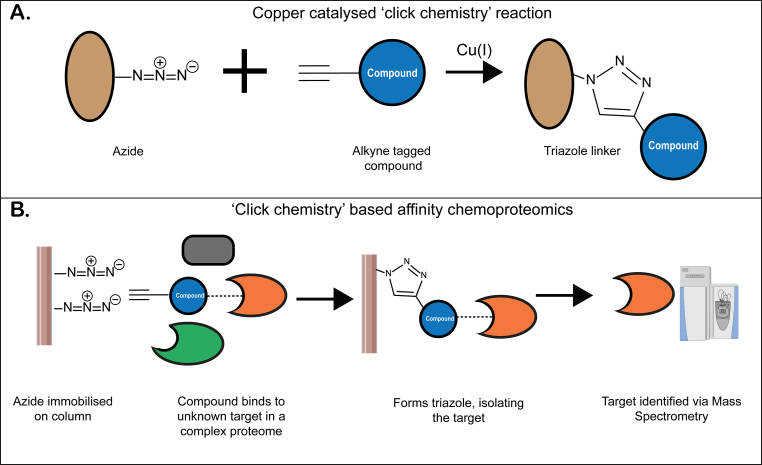
A convenient form of affinity-based probes are those based on copper catalysed ‘click chemistry’ reactions (Speers and Cravatt, 2004). ‘Clickable’ azide and alkyne functional groups can react, forming stable triazole conjugates (Fig. 1. A). These ‘clickable’ functional groups can be synthetically introduced onto the compound of interest and, after exposing these functionalised probe compounds to either live cells or a cell lysate, they can be reacted with the reciprocal functional group which is typically immobilised on beads. This reaction forms a covalently-linked triazole conjugate, allowing for the probe to be pulled out of the complex proteome, along with any interacting protein(s) that can subsequently be detected via mass spectrometry (Fig. 1. B). ‘Clickable’ probes have been used to identify protein targets for endoperoxide antimalarials including artemisinin and synthetic ozonide compounds (Ismail et al., 2016; Jourdan et al., 2019; Siddiqui et al., 2022; Wang et al., 2015). These studies identified proteins from a range of pathways including haemoglobin digestion, antioxidant defence systems, glycolysis, nucleic acid and protein biosynthesis, demonstrating the expected promiscuous nature of artemisinin binding within the parasite as a result of the radical chemistry involved with the endoperoxide bond cleavage.
Fig. 1.
A. Copper catalysed click chemistry reaction between azide and alkyne moieties. B. Theoretical chemoproteomic pull down. A novel compound modified with an alkyne moiety interacts with an unknown target in a complex proteome. The alkyne interacts with an immobilised azide, forming a covalent triazole conjugate which allows the target protein to be isolated and subsequently identified via mass spectrometry.
A limitation of affinity-based probes such as MMV666845, is that the interactions with their target proteins are non-covalent and therefore can be transient. One approach to overcome this challenge is the use of bifunctional probes that can covalently bind to target proteins through photoactivatable functional groups such as phenyl azides or diazirines, as well as possessing a chemoreactive group suitable for enrichment. A novel, non-covalently binding antimalarial, albitiazolium, was investigated with photoactivatable bifunctional probes which could covalently bond to target proteins to aid in the determination of specific drug-protein interactions (Penarete-Vargas et al., 2014). These bifunctional probes showed good specificity, identifying eleven proteins as interactors and providing a shortlist of promising targets for further validation. This demonstrated the scope for development of advanced probes that improve the reliability and versatility of this approach for compounds that don't act through covalent binding.
These studies demonstrated how chemoproteomic approaches can identify binding targets for antimalarial compounds with unknown MoAs, in an untargeted fashion, however, a number of factors limit their use. Firstly, this approach requires the time-consuming synthesis of specifically designed chemical probes with the desired functionalities. This is further complicated by the need for good knowledge of the structure activity relationships of the compound of interest, to ensure that the probe modifications don't interfere with compound activity or target specificity. Therefore, it is not suitable for high throughput screening applications and likely will only be applicable for compounds showing significant promise. Secondly, approaches that are dependent on affinity to identify target proteins can be susceptible to identifying non-specific interactions and co-purified proteins (Gingras et al., 2007). While this can potentially confound the results, sufficient negative controls, particularly with competitive binders, can ameliorate this. Furthermore, the identification of co-purified proteins can provide context for the biological role of the identified target. This can be particularly important in the context of P. falciparum, where a significant proportion of the genome has no functional annotation.
Databases of commonly identified contaminants, such as the Contaminant Repository for Affinity Purification Mass Spectrometry Data (CRAPome) can help to identify non-specific protein binding for Homo sapiens and Saccharomyces cerevisiae data (Mellacheruvu et al., 2013). However, a similar P. falciparum database is not currently available, placing more importance on having sufficient negative controls and replicates to help discern non-specific interactions from real targets. This is highlighted by the recent success of a chemoproteomic approach to directly compare the alkylation profiles of endoperoxide antimalarial probes, where the use of advanced mass spectrometry techniques and extensive negative controls allowed for the high confidence identification of new alkylation targets for endoperoxide antimalarials (Siddiqui et al., 2022).
Overall, these studies demonstrate the power of chemoproteomics as an untargeted technique for target identification, and highlights the importance of direct approaches that can distinguish between molecular targets and indirect resistance mechanisms, provided strategies to distinguish between target binding and non-specific binding are employed effectively.
2.4. Protein stability target identification
When proteins are heated they denature, unfold and form insoluble aggregates (Varela et al., 2019). However, the preferential binding of a ligand to a target protein can make that protein more resistant to denaturation, resulting in a higher proportion of ligand-bound protein remaining in its soluble form following a thermal challenge. After a thermal challenge, soluble proteins can be separated from denatured aggregates by centrifugation and ligand-stabilised proteins can be identified by an apparent increase in abundance compared to a negative control. This concept has been applied to a number of targeted thermal shift style assays utilising Western blot or fluorescent detection methods (Martinez Molina et al., 2013; Martinez et al., 2018). However, these methods typically require either purified protein, or antibodies directed at a suspected target, and therefore are more suitable for the validation of suspected targets rather than the identification of unknown MoAs (Martinez et al., 2018; McNulty et al., 2018; Shaw et al., 2018). The recent incorporation of quantitative proteomics techniques (Franken et al., 2015; Martinez Molina et al., 2013) has allowed these thermal shift assays to be simultaneously performed across whole proteomes, therefore making it possible to identify previously unknown protein targets from a complex proteome. These whole proteome approaches are typically referred to as Cellular Thermal Shift Assays (CETSA).
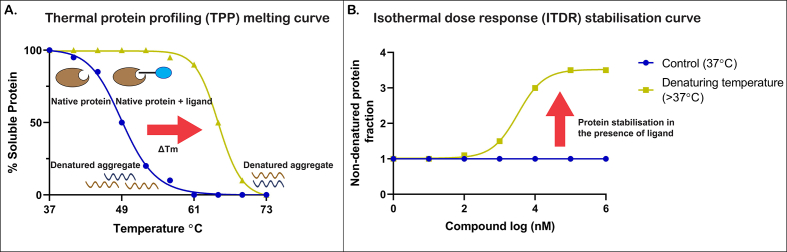
CETSA has two main variants: the thermal proteome profiling (TPP) or ‘melt curve’ approach, whereby the temperature of the heat challenge is varied in the presence of a consistent concentration of compound (Franken et al., 2015), and the isothermal dose response (ITDR), which employs increasing compound concentrations with a single temperature (Fig. 2) (Dziekan et al., 2019; Franken et al., 2015). In TPP CETSA the thermal melting temperature (Tm) increases in proteins bound by the ligand, resulting in an increase in the amount of soluble protein remaining following heating. In ITDR CETSA, as the concentration of ligand is increased, more protein is protected from the thermal challenge, resulting in an increase in soluble protein proportional to the increase in ligand.
Fig. 2.
A. Example melting curve for thermal protein profiling (TPP) CETSA. ΔTm represents the increased thermal melting temperature of the target protein in the presence of the ligand. B. Example stabilisation curve for an isothermal dose response (ITDR) CETSA. Red arrow represents the increased stability of the target protein in the presence of increasing drug concentrations following a heat challenge. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Recently, a TPP CETSA proteomics study in P. falciparum was performed in order to identify binding targets for quinine (QN) and mefloquine (MQ), two antimalarials with previously unknown MoAs. QN was found to selectively stabilise the purine nucleoside phosphorylase (PfPNP) in a dose-dependent manner in both lysate and intact cells. Stabilisation of PfPNP was also observed in lysate samples following MQ treatment, albeit to a lesser extent (Dziekan et al., 2019). This assay was supported by other biophysical assays and cocrystal structures in order to confirm the binding of quinine to PfPNP. It was also demonstrated that MQ can bind to PfPNP, however, the therapeutic relevance could be limited due to lower potency.
TPP approaches to target identification have also been applied to other apicomplexan parasites, including Toxoplasma gondii. The calcium dependent protein kinase 1 (CDPK1) was identified as a target of a novel antiparasitic compound termed ENH1 although, follow up validation studies did identify ENH1 polypharmacology, making it difficult to discern the primary mechanism of ENH1 activity (Herneisen et al., 2020). ITDR style CETSA approaches have also been recently used to identify previously unknown targets of the pan-kinase inhibitor staurosporine in mammalian cells (Ball et al., 2020). By focusing on increasing sample replicates, rather than having multiple temperatures, Ball et al.'s isothermal shift assay (iTSA) identified 51 new kinase targets for staurosporine, a significant improvement on previously performed TPP CETSA studies (Savitski et al., 2014). The iTSA method also reduced the total number of samples required for each compound tested, ultimately reducing analysis time and increasing throughput, which could make mass spectrometry-based thermal shift approaches more amenable to routine application.
Untargeted CETSA proteomics can identify potential protein binding targets from cell lysates and whole cell samples, making it a powerful technique for identifying targets of compounds with unknown MoAs. CETSA is also advantaged by the fact that it does not rely on the time-consuming synthesis of specific probes or generation of resistant parasites and therefore can be performed relatively rapidly (Dziekan et al., 2020). However, it is important to consider the possibility that off-target binding can occur at high concentrations. Therefore, subsequent validation with targeted methods is necessary to confirm the binding and the biological relevance of identified targets. Current data analysis techniques for CETSA proteomics have also been thought to result in high levels of false negative identifications, largely due to the fact that they typically rely on a significant change in a single parameter such as melting point or protein abundance (Childs et al., 2019). New data analysis techniques such as the nonparametric analysis of response curves (NPARC) could help improve target identification for TPP CETSA (Childs et al., 2019). NPARC takes advantage of functional data analysis and nonlinear regression, rather than relying on changes in a single parameter, to identify stabilised proteins with non-canonical melting profiles, reducing the number of false-negative identifications and increasing the theoretical coverage of TPP CETSA. Current CETSA proteomics approaches also have difficulty identifying membrane proteins due to the detergent-free cell extraction procedures used (Franken et al., 2015; Rawlings, 2016). While there are some studies exploring the use of detergents in CETSA protocols (Franken et al., 2015), detergents are expected to alter protein structure and stability, and more development is required before membrane proteins can be reliably identified. Finally, there are examples of compounds that do not induce any changes in thermal stability in their validated targets, highlighting how CETSA proteomics may not be appropriate for all compounds (Becher et al., 2016; Savitski et al., 2014). Despite this, CETSA proteomics can be a powerful tool for the target identification of novel compounds with unknown MoAs, particularly when combined with targeted validation assays.
Denaturation methods other than heating have also been used to identify drug targets by measuring changes in protein stability. Stability of Proteins from Rates of Oxidation (SPROX) is a method that induces protein denaturation with hydrogen peroxide in order to test protein stability and its subsequent stabilisation by ligands in a similar way to TPP-CETSA (Strickland et al., 2013). SPROX has recently been applied to antimalarial drug target discovery. Performed in concert with TPP-CETSA, SPROX identified the essential Plasmodium protein chaperonin TRiC/CCT as a target of the antihistamine clemastine (Lu et al., 2020).
Other methods, such as limited proteolysis coupled mass spectrometry (LiP-MS) and Drug Affinity Responsive Target Stability (DARTS) use limited proteolytic digestion to identify proteins stabilised by the ligand in question. These methods rely on applying proteases with broad specificity, such as proteinase K, to either whole proteomes or purified proteins for a short period of time (Lomenick et al., 2009; Schopper et al., 2017). The limited exposure of the proteases ensures that proteolysis is directed by protein structure, with exposed proteolysis sites digested first. When a ligand binds to a target site, it prevents proteolysis of the target site and results in a differential peptide pattern which can be detected via MS. This approach has previously been used to demonstrate subtle changes in secondary protein structures, structural changes induced by allostery and identify new small molecule-protein interactions (Feng et al., 2014; Geiger et al., 2016). LiP-MS has been previously validated with the antifungal drug cerulenin, successfully identifying its known target, Fatty Acid Synthase 2 (FAS2) from over 2500 identified proteins (Piazza et al., 2018). LiP-MS has historically been limited by the difficulty of detecting LiP-MS sites against a complex peptide background (Suh et al., 2007). However, LiP-MS and DARTS have the potential to provide another proteome-wide target identification method for novel antimalarial compounds with the ability to provide unique insight that is complementary to currently established methods.
3. Prospects and challenges
Each of the untargeted techniques detailed in this review have their own strengths and limitations that can affect how they can be applied. IVIEWGA and untargeted metabolomics approaches both benefit from identified mechanisms having clear, phenotypic implications for the parasite. However, neither method directly measures target engagement and therefore both identified resistance mechanisms and metabolic perturbations could be indirectly related to the compound's target. In contrast, chemoproteomic and CETSA proteomic approaches can identify specific drug-protein interactions, however, require validation to confirm that identified ‘hits’ have biological relevance. The ease of application of these methods can also be a limiting factor. IVIEWGA is time consuming, often requiring multiple months of continuous parasite culture and can be particularly challenging for compounds with pleiotropic effects or a fast onset of action. Traditional affinity-based chemoproteomic approaches require the synthesis of specific probe compounds, which requires medicinal chemistry expertise and can also be a time-consuming process.
It should be noted that all of the methods discussed in this review take advantage of ‘omic’ style approaches to identify specific targets from within a whole genome or proteome. While the ability to investigate the whole genome/proteome is undoubtedly powerful, it can be limited by the time and cost associated with performing large experiments reliant on sophisticated instrumentation such as mass spectrometers. Whilst the cost-efficiency of these omics approaches has improved in recent years, this barrier can encourage the design of experiments with limited samples, reducing the number of concentrations, time points or controls tested. In turn, this can increase the likelihood of false positive target identifications from these untargeted methods. All the methods discussed in this review are susceptible to false-positive ‘hits’ being identified and this highlights the crucial need for extensive target validation following the identification of promising targets. Approaches that can directly measure compound–target binding, such as surface plasmon resonance (SPR) based assays, targeted CETSA approaches and x-ray crystallography can all provide confidence in direct compound–target binding (Maveyraud and Mourey, 2020; Molina et al., 2013; Schneider et al., 2015). Furthermore, using multiple target identification methods in parallel can also provide confidence in targets that are consistently identified. Successful target identification programs have often combined multiple approaches, with complimentary techniques confirming both target engagement and the phenotypic relevance of identified targets (Milne et al., 2022). For example, the plasmepsin inhibitors WM4, WM5 and WM382 utilised IVIEWGA to originally identify plasmepsin X as a potential target, and demonstrated that WM382 specifically bound to both plasmepsin IX and X through targeted chemoproteomic and CETSA assays (Favuzza et al., 2020). However, one caveat is that the level of expertise required to perform multiple untargeted identification techniques for a single target identification program can be restrictive, and therefore, collaboration is critical (Cowell and Winzeler, 2018).
It should also be noted that these techniques have typically been applied to investigate compounds with a blood stage phenotype, and have seen limited use in the sexual and liver stages. The recent development of high-throughput screens for both liver stage and transmission inhibitors will provide new lead compounds which will require target identification methods that have been optimised to those stages of the lifecycle (Delves et al., 2018; Swann et al., 2016). Techniques that rely on biophysical changes, such as CETSA proteomics and LiP-MS, should be easily transferable to other parasite stages. However, further optimisation of sample preparation procedures is likely necessary.
4. Concluding remarks
The use of target identification methods greatly assists the development of new antimalarial compounds identified in phenotypic high throughput screens. Currently-used techniques such as IVIEWGA and untargeted metabolomics profiling have been successful in providing insight into the MoA of multiple compounds currently in preclinical development, fast-tracking their progress through the antimalarial development pipeline. The emergence of affinity-based chemoproteomics, CETSA and SPROX proteomics, and the potential for approaches such as LiP-MS to be used for target identification, can improve the options available for drug discovery programs to identify new targets.
There remains a number of outstanding questions regarding the scope for further development of these techniques. They are yet to be applied in typically understudied stages of the Plasmodium lifecycle and there are still opportunities to improve their ability to delineate off target or indirect effects from biologically relevant targets directly involved in the MoA. Simplifying these methods to allow for their incorporation into high throughput screening approaches and enabling earlier access for drug discovery projects may be an important area of development. The early identification of biologically relevant targets can assist with fast compound development and the triaging of promising candidate compounds early in the drug discovery process. In the context of P. falciparum drug target identification, the development of a database of common off-target or non-specific binding proteins will assist in distinguishing between off target or indirect effects and finally, the emergence of new methods such as LiP-MS, DARTS and SPROX open up exciting new approaches to antimalarial target identification that will provide alternate avenues for identifying new biologically relevant targets.
Overall, it is worth noting the importance of the combined use of multiple techniques in parallel to overcome the individual weaknesses of each method, and the subsequent validation of novel targets is crucial for the advancement of new compounds that are urgently needed to combat growing antimalarial drug resistance, and progress towards malaria elimination.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Darren Creek reports financial support was provided by National Health and Medical Research Council.
Acknowledgements
The authors acknowledge financial support from NHMRC project grant #APP1163235 and fellowship #APP1148700.
Abbreviations
- CETSA
Cellular thermal shift assay
- E.coli
Escherichia coli
- ITDR
Isothermal dose response
- IVIEWGA
In vitro evolution and whole genome sequencing
- LiP-MS
Limited proteolysis mass spectrometry
- MoA
Mechanism of Action
- MS
Mass spectrometry
- Pf
Plasmodium falciparum
- PfATP4
Plasmodium falciparum P-type ATPase 4
- PfCARL
Plasmodium falciparum cyclic amine resistance locus
- PfDHODH
Plasmodium falciparum dihydroorotate dehydrogenase
- PfMDR1
Plasmodium falciparum multidrug resistance 1
- PfPI4K
Plasmodium falciparum phosphatidylinositol 4-kinase
- PfPNP
Plasmodium falciparum purine nucleoside phosphorylase
- TPP
Thermal proteome profiling
References
- Allman E.L., Painter H.J., Samra J., Carrasquilla M., Llinas M. Metabolomic profiling of the malaria Box reveals antimalarial target pathways. Antimicrob. Agents Chemother. 2016;60:6635–6649. doi: 10.1128/AAC.01224-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A.C. The process of structure-based drug design. Chem. Biol. 2003;10:787–797. doi: 10.1016/j.chembiol.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Anthony M.P., Burrows J.N., Duparc S., Moehrle J.J., Wells T.N. The global pipeline of new medicines for the control and elimination of malaria. Malar. J. 2012;11:316. doi: 10.1186/1475-2875-11-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball K.A., Webb K.J., Coleman S.J., Cozzolino K.A., Jacobsen J., Jones K.R., Stowell M.H.B., Old W.M. An isothermal shift assay for proteome scale drug-target identification. Commun Biol. 2020;3:75. doi: 10.1038/s42003-020-0795-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baragana B., Hallyburton I., Lee M.C., Norcross N.R., Grimaldi R., Otto T.D., Proto W.R., Blagborough A.M., Meister S., Wirjanata G., Ruecker A., Upton L.M., Abraham T.S., Almeida M.J., Pradhan A., Porzelle A., Luksch T., Martinez M.S., Luksch T., Bolscher J.M., Woodland A., Norval S., Zuccotto F., Thomas J., Simeons F., Stojanovski L., Osuna-Cabello M., Brock P.M., Churcher T.S., Sala K.A., Zakutansky S.E., Jimenez-Diaz M.B., Sanz L.M., Riley J., Basak R., Campbell M., Avery V.M., Sauerwein R.W., Dechering K.J., Noviyanti R., Campo B., Frearson J.A., Angulo-Barturen I., Ferrer-Bazaga S., Gamo F.J., Wyatt P.G., Leroy D., Siegl P., Delves M.J., Kyle D.E., Wittlin S., Marfurt J., Price R.N., Sinden R.E., Winzeler E.A., Charman S.A., Bebrevska L., Gray D.W., Campbell S., Fairlamb A.H., Willis P.A., Rayner J.C., Fidock D.A., Read K.D., Gilbert I.H. A novel multiple-stage antimalarial agent that inhibits protein synthesis. Nature. 2015;522:315–320. doi: 10.1038/nature14451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher I., Werner T., Doce C., Zaal E.A., Togel I., Khan C.A., Rueger A., Muelbaier M., Salzer E., Berkers C.R., Fitzpatrick P.F., Bantscheff M., Savitski M.M. Thermal profiling reveals phenylalanine hydroxylase as an off-target of panobinostat. Nat. Chem. Biol. 2016;12:908–910. doi: 10.1038/nchembio.2185. [DOI] [PubMed] [Google Scholar]
- Birrell G.W., Challis M.P., De Paoli A., Anderson D., Devine S.M., Heffernan G.D., Jacobus D.P., Edstein M.D., Siddiqui G., Creek D.J. Multi-omic characterization of the mode of action of a potent new antimalarial compound, JPC-3210, against Plasmodium falciparum. Mol. Cell. Proteomics. 2020;19:308–325. doi: 10.1074/mcp.RA119.001797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco B., Leroy D., Fidock D.A. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat. Med. 2017;23:917–928. doi: 10.1038/nm.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows J.N., Duparc S., Gutteridge W.E., Hooft van Huijsduijnen R., Kaszubska W., Macintyre F., Mazzuri S., Mohrle J.J., Wells T.N.C. New developments in anti-malarial target candidate and product profiles. Malar. J. 2017;16:26. doi: 10.1186/s12936-016-1675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs D., Bach K., Franken H., Anders S., Kurzawa N., Bantscheff M., Savitski M.M., Huber W. Nonparametric analysis of thermal proteome profiles reveals novel drug-binding proteins. Mol. Cell. Proteomics. 2019;18:2506–2515. doi: 10.1074/mcp.TIR119.001481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements R.L., Streva V., Dumoulin P., Huang W., Owens E., Raj D.K., Burleigh B., Llinas M., Winzeler E.A., Zhang Q., Dvorin J.D. A novel antiparasitic compound kills ring-stage Plasmodium falciparum and retains activity against artemisinin-resistant parasites. J. Infect. Dis. 2020;221:956–962. doi: 10.1093/infdis/jiz534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coteron J.M., Marco M., Esquivias J., Deng X., White K.L., White J., Koltun M., El Mazouni F., Kokkonda S., Katneni K., Bhamidipati R., Shackleford D.M., Angulo-Barturen I., Ferrer S.B., Jimenez-Diaz M.B., Gamo F.J., Goldsmith E.J., Charman W.N., Bathurst I., Floyd D., Matthews D., Burrows J.N., Rathod P.K., Charman S.A., Phillips M.A. Structure-guided lead optimization of triazolopyrimidine-ring substituents identifies potent Plasmodium falciparum dihydroorotate dehydrogenase inhibitors with clinical candidate potential. J. Med. Chem. 2011;54:5540–5561. doi: 10.1021/jm200592f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell A., Winzeler E. Exploration of the Plasmodium falciparum resistome and druggable genome reveals new mechanisms of drug resistance and antimalarial targets. Microbiol. Insights. 2018;11 doi: 10.1177/1178636118808529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell A.N., Istvan E.S., Lukens A.K., Gomez-Lorenzo M.G., Vanaerschot M., Sakata-Kato T., Flannery E.L., Magistrado P., Owen E., Abraham M., LaMonte G., Painter H.J., Williams R.M., Franco V., Linares M., Arriaga I., Bopp S., Corey V.C., Gnadig N.F., Coburn-Flynn O., Reimer C., Gupta P., Murithi J.M., Moura P.A., Fuchs O., Sasaki E., Kim S.W., Teng C.H., Wang L.T., Akidil A., Adjalley S., Willis P.A., Siegel D., Tanaseichuk O., Zhong Y., Zhou Y., Llinas M., Ottilie S., Gamo F.J., Lee M.C.S., Goldberg D.E., Fidock D.A., Wirth D.F., Winzeler E.A. Mapping the malaria parasite druggable genome by using in vitro evolution and chemogenomics. Science. 2018;359:191–199. doi: 10.1126/science.aan4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell A.N., Winzeler E.A. Advances in omics-based methods to identify novel targets for malaria and other parasitic protozoan infections. Genome Med. 2019;11:63. doi: 10.1186/s13073-019-0673-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell A.N., Winzeler E.A. Advances in omics-based methods to identify novel targets for malaria and other parasitic protozoan infections. Genome Med. 2019;11:63. doi: 10.1186/s13073-019-0673-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creek D.J., Barrett M.P. Determination of antiprotozoal drug mechanisms by metabolomics approaches. Parasitology. 2014;141:83–92. doi: 10.1017/S0031182013000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creek D.J., Chua H.H., Cobbold S.A., Nijagal B., MacRae J.I., Dickerman B.K., Gilson P.R., Ralph S.A., McConville M.J. Metabolomics-based screening of the malaria Box reveals both novel and established mechanisms of action. Antimicrob. Agents Chemother. 2016;60:6650–6663. doi: 10.1128/AAC.01226-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alessandro U., Buttiens H. History and importance of antimalarial drug resistance. Trop. Med. Int. Health. 2001;6:845–848. doi: 10.1046/j.1365-3156.2001.00819.x. [DOI] [PubMed] [Google Scholar]
- Delves M.J., Miguel-Blanco C., Matthews H., Molina I., Ruecker A., Yahiya S., Straschil U., Abraham M., Leon M.L., Fischer O.J., Rueda-Zubiaurre A., Brandt J.R., Cortes A., Barnard A., Fuchter M.J., Calderon F., Winzeler E.A., Sinden R.E., Herreros E., Gamo F.J., Baum J. A high throughput screen for next-generation leads targeting malaria parasite transmission. Nat. Commun. 2018;9:3805. doi: 10.1038/s41467-018-05777-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerman B.K., Elsworth B., Cobbold S.A., Nie C.Q., McConville M.J., Crabb B.S., Gilson P.R. Identification of inhibitors that dually target the new permeability pathway and dihydroorotate dehydrogenase in the blood stage of Plasmodium falciparum. Sci. Rep. 2016;6 doi: 10.1038/srep37502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp A.M., Nosten F., Yi P., Das D., Phyo A.P., Tarning J., Lwin K.M., Ariey F., Hanpithakpong W., Lee S.J., Ringwald P., Silamut K., Imwong M., Chotivanich K., Lim P., Herdman T., An S.S., Yeung S., Singhasivanon P., Day N.P., Lindegardh N., Socheat D., White N.J. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorjsuren D., Eastman R.T., Wicht K.J., Jansen D., Talley D.C., Sigmon B.A., Zakharov A.V., Roncal N., Girvin A.T., Antonova-Koch Y., Will P.M., Shah P., Sun H., Klumpp-Thomas C., Mok S., Yeo T., Meister S., Marugan J.J., Ross L.S., Xu X., Maloney D.J., Jadhav A., Mott B.T., Sciotti R.J., Winzeler E.A., Waters N.C., Campbell R.F., Huang W., Simeonov A., Fidock D.A. Chemoprotective antimalarials identified through quantitative high-throughput screening of Plasmodium blood and liver stage parasites. Sci. Rep. 2021;11:2121. doi: 10.1038/s41598-021-81486-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drinkwater N., McGowan S. From crystal to compound: structure-based antimalarial drug discovery. Biochem. J. 2014;461:349–369. doi: 10.1042/BJ20140240. [DOI] [PubMed] [Google Scholar]
- Duffey M., Blasco B., Burrows J.N., Wells T.N.C., Fidock D.A., Leroy D. Assessing risks of Plasmodium falciparum resistance to select next-generation antimalarials. Trends Parasitol. 2021;37:709–721. doi: 10.1016/j.pt.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W.B., Erban A., Weber R.J.M., Creek D.J., Brown M., Breitling R., Hankemeier T., Goodacre R., Neumann S., Kopka J., Viant M.R. Mass appeal: metabolite identification in mass spectrometry-focused untargeted metabolomics. Metabolomics. 2012;9:44–66. [Google Scholar]
- Dziekan J.M., Wirjanata G., Dai L., Go K.D., Yu H., Lim Y.T., Chen L., Wang L.C., Puspita B., Prabhu N., Sobota R.M., Nordlund P., Bozdech Z. Cellular thermal shift assay for the identification of drug-target interactions in the Plasmodium falciparum proteome. Nat. Protoc. 2020;15:1881–1921. doi: 10.1038/s41596-020-0310-z. [DOI] [PubMed] [Google Scholar]
- Dziekan J.M., Yu H., Chen D., Dai L., Wirjanata G., Larsson A., Prabhu N., Sobota R.M., Bozdech Z., Nordlund P. Identifying purine nucleoside phosphorylase as the target of quinine using cellular thermal shift assay. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aau3174. [DOI] [PubMed] [Google Scholar]
- Favuzza P., de Lera Ruiz M., Thompson J.K., Triglia T., Ngo A., Steel R.W.J., Vavrek M., Christensen J., Healer J., Boyce C., Guo Z., Hu M., Khan T., Murgolo N., Zhao L., Penington J.S., Reaksudsan K., Jarman K., Dietrich M.H., Richardson L., Guo K.Y., Lopaticki S., Tham W.H., Rottmann M., Papenfuss T., Robbins J.A., Boddey J.A., Sleebs B.E., Sabroux H.J., McCauley J.A., Olsen D.B., Cowman A.F. Dual plasmepsin-targeting antimalarial agents disrupt multiple stages of the malaria parasite life cycle. Cell Host Microbe. 2020;27:642–658. doi: 10.1016/j.chom.2020.02.005. e612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., De Franceschi G., Kahraman A., Soste M., Melnik A., Boersema P.J., de Laureto P.P., Nikolaev Y., Oliveira A.P., Picotti P. Global analysis of protein structural changes in complex proteomes. Nat. Biotechnol. 2014;32:1036–1044. doi: 10.1038/nbt.2999. [DOI] [PubMed] [Google Scholar]
- Flannery E.L., Fidock D.A., Winzeler E.A. Using genetic methods to define the targets of compounds with antimalarial activity. J. Med. Chem. 2013;56:7761–7771. doi: 10.1021/jm400325j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannery E.L., McNamara C.W., Kim S.W., Kato T.S., Li F., Teng C.H., Gagaring K., Manary M.J., Barboa R., Meister S., Kuhen K., Vinetz J.M., Chatterjee A.K., Winzeler E.A. Mutations in the P-type cation-transporter ATPase 4, PfATP4, mediate resistance to both aminopyrazole and spiroindolone antimalarials. ACS Chem. Biol. 2015;10:413–420. doi: 10.1021/cb500616x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken H., Mathieson T., Childs D., Sweetman G.M., Werner T., Togel I., Doce C., Gade S., Bantscheff M., Drewes G., Reinhard F.B., Huber W., Savitski M.M. Thermal proteome profiling for unbiased identification of direct and indirect drug targets using multiplexed quantitative mass spectrometry. Nat. Protoc. 2015;10:1567–1593. doi: 10.1038/nprot.2015.101. [DOI] [PubMed] [Google Scholar]
- Fry M., Pudney M. Site of action of the antimalarial hydroxynaphthoquinone, 2-[trans-4-(4'-chlorophenyl) cyclohexyl]-3-hydroxy-1,4-naphthoquinone (566C80) Biochem. Pharmacol. 1992;43:1545–1553. doi: 10.1016/0006-2952(92)90213-3. [DOI] [PubMed] [Google Scholar]
- Gamo F.J., Sanz L.M., Vidal J., de Cozar C., Alvarez E., Lavandera J.L., Vanderwall D.E., Green D.V., Kumar V., Hasan S., Brown J.R., Peishoff C.E., Cardon L.R., Garcia-Bustos J.F. Thousands of chemical starting points for antimalarial lead identification. Nature. 2010;465:305–310. doi: 10.1038/nature09107. [DOI] [PubMed] [Google Scholar]
- Geiger R., Rieckmann J.C., Wolf T., Basso C., Feng Y., Fuhrer T., Kogadeeva M., Picotti P., Meissner F., Mann M., Zamboni N., Sallusto F., Lanzavecchia A. L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell. 2016;167:829–842 e813. doi: 10.1016/j.cell.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannangelo C., Siddiqui G., De Paoli A., Anderson B.M., Edgington-Mitchell L.E., Charman S.A., Creek D.J. System-wide biochemical analysis reveals ozonide antimalarials initially act by disrupting Plasmodium falciparum haemoglobin digestion. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.C., Gstaiger M., Raught B., Aebersold R. Analysis of protein complexes using mass spectrometry. Nat. Rev. Mol. Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- Gisselberg J.E., Herrera Z., Orchard L.M., Llinás M., Yeh E. Specific inhibition of the bifunctional farnesyl/geranylgeranyl diphosphate Synthase in malaria parasites via a new small-molecule binding site. Cell Chem Biol. 2018;25:185–193. doi: 10.1016/j.chembiol.2017.11.010. e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heal W.P., Dang T.H., Tate E.W. Activity-based probes: discovering new biology and new drug targets. Chem. Soc. Rev. 2011;40:246–257. doi: 10.1039/c0cs00004c. [DOI] [PubMed] [Google Scholar]
- Heller L.E., Roepe P.D. Artemisinin-based antimalarial drug therapy: molecular pharmacology and evolving resistance. Trav. Med. Infect. Dis. 2019;4:89. doi: 10.3390/tropicalmed4020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herneisen A.L., Sidik S.M., Markus B.M., Drewry D.H., Zuercher W.J., Lourido S. Identifying the target of an antiparasitic compound in Toxoplasma using thermal proteome profiling. ACS Chem. Biol. 2020;15:1801–1807. doi: 10.1021/acschembio.0c00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma H., Hirai M., Nakamura S., Hakimi H., Kawazu S., Palacpac N.M., Hisaeda H., Matsuoka H., Kawai S., Endo H., Yasunaga T., Ohashi J., Mita T., Horii T., Furusawa M., Tanabe K. Generation of rodent malaria parasites with a high mutation rate by destructing proofreading activity of DNA polymerase δ. DNA Res. 2014;21:439–446. doi: 10.1093/dnares/dsu009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovlid M.L., Winzeler E.A. Phenotypic screens in antimalarial drug discovery. Trends Parasitol. 2016;32:697–707. doi: 10.1016/j.pt.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail H.M., Barton V., Phanchana M., Charoensutthivarakul S., Wong M.H., Hemingway J., Biagini G.A., O'Neill P.M., Ward S.A. Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. Proc. Natl. Acad. Sci. U. S. A. 2016;113:2080–2085. doi: 10.1073/pnas.1600459113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas M.C., Collins S.R., Denic V., Oh E., Quan E.M., Schmid V., Weibezahn J., Schwappach B., Walter P., Weissman J.S., Schuldiner M. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdan J., Walz A., Matile H., Schmidt A., Wu J., Wang X., Dong Y., Vennerstrom J.L., Schmidt R.S., Wittlin S., Maser P. Stochastic protein alkylation by antimalarial peroxides. ACS Infect. Dis. 2019;5:2067–2075. doi: 10.1021/acsinfecdis.9b00264. [DOI] [PubMed] [Google Scholar]
- Katsuno K., Burrows J.N., Duncan K., Hooft van Huijsduijnen R., Kaneko T., Kita K., Mowbray C.E., Schmatz D., Warner P., Slingsby B.T. Hit and lead criteria in drug discovery for infectious diseases of the developing world. Nat. Rev. Drug Discov. 2015;14:751–758. doi: 10.1038/nrd4683. [DOI] [PubMed] [Google Scholar]
- Kawatani M., Osada H. Affinity-based target identification for bioactive small molecules. Medchemcomm. 2014;5:277–287. [Google Scholar]
- Krishnan K., Ziniel P., Li H., Huang X., Hupalo D., Gombakomba N., Guerrero S.M., Dotrang T., Lu X., Caridha D., Sternberg A.R., Hughes E., Sun W., Bargieri D.Y., Roepe P.D., Sciotti R.J., Wilkerson M.D., Dalgard C.L., Tawa G.J., Wang A.Q., Xu X., Zheng W., Sanderson P.E., Huang W., Williamson K.C. Torin 2 derivative, NCATS-SM3710, has potent multistage antimalarial activity through inhibition of P. falciparum phosphatidylinositol 4-kinase (PfPI4KIIIβ) ACS Pharmacol Transl Sci. 2020;3:948–964. doi: 10.1021/acsptsci.0c00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhen K.L., Chatterjee A.K., Rottmann M., Gagaring K., Borboa R., Buenviaje J., Chen Z., Francek C., Wu T., Nagle A., Barnes S.W., Plouffe D., Lee M.C., Fidock D.A., Graumans W., van de Vegte-Bolmer M., van Gemert G.J., Wirjanata G., Sebayang B., Marfurt J., Russell B., Suwanarusk R., Price R.N., Nosten F., Tungtaeng A., Gettayacamin M., Sattabongkot J., Taylor J., Walker J.R., Tully D., Patra K.P., Flannery E.L., Vinetz J.M., Renia L., Sauerwein R.W., Winzeler E.A., Glynne R.J., Diagana T.T. KAF156 is an antimalarial clinical candidate with potential for use in prophylaxis, treatment, and prevention of disease transmission. Antimicrob. Agents Chemother. 2014;58:5060–5067. doi: 10.1128/AAC.02727-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMonte G., Lim M.Y., Wree M., Reimer C., Nachon M., Corey V., Gedeck P., Plouffe D., Du A., Figueroa N., Yeung B., Bifani P., Winzeler E.A. Mutations in the Plasmodium falciparum cyclic amine resistance Locus (PfCARL) confer multidrug resistance. mBio. 2016;7 doi: 10.1128/mBio.00696-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaMonte G.M., Rocamora F., Marapana D.S., Gnadig N.F., Ottilie S., Luth M.R., Worgall T.S., Goldgof G.M., Mohunlal R., Santha Kumar T.R., Thompson J.K., Vigil E., Yang J., Hutson D., Johnson T., Huang J., Williams R.M., Zou B.Y., Cheung A.L., Kumar P., Egan T.J., Lee M.C.S., Siegel D., Cowman A.F., Fidock D.A., Winzeler E.A. Pan-active imidazolopiperazine antimalarials target the Plasmodium falciparum intracellular secretory pathway. Nat. Commun. 2020;11:1780. doi: 10.1038/s41467-020-15440-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawong A., Gahalawat S., Okombo J., Striepen J., Yeo T., Mok S., Deni I., Bridgford J.L., Niederstrasser H., Zhou A., Posner B., Wittlin S., Gamo F.J., Crespo B., Churchyard A., Baum J., Mittal N., Winzeler E., Laleu B., Palmer M.J., Charman S.A., Fidock D.A., Ready J.M., Phillips M.A. Novel antimalarial tetrazoles and amides active against the hemoglobin degradation pathway in Plasmodium falciparum. J. Med. Chem. 2021;64:2739–2761. doi: 10.1021/acs.jmedchem.0c02022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomenick B., Hao R., Jonai N., Chin R.M., Aghajan M., Warburton S., Wang J., Wu R.P., Gomez F., Loo J.A., Wohlschlegel J.A., Vondriska T.M., Pelletier J., Herschman H.R., Clardy J., Clarke C.F., Huang J. Target identification using drug affinity responsive target stability (DARTS) Proc. Natl. Acad. Sci. U. S. A. 2009;106:21984–21989. doi: 10.1073/pnas.0910040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looareesuwan S., Viravan C., Webster H.K., Kyle D.E., Hutchinson D.B., Canfield C.J. Clinical studies of atovaquone, alone or in combination with other antimalarial drugs, for treatment of acute uncomplicated malaria in Thailand. Am. J. Trop. Med. Hyg. 1996;54:62–66. doi: 10.4269/ajtmh.1996.54.62. [DOI] [PubMed] [Google Scholar]
- Lu K.Y., Quan B., Sylvester K., Srivastava T., Fitzgerald M.C., Derbyshire E.R. Plasmodium chaperonin TRiC/CCT identified as a target of the antihistamine clemastine using parallel chemoproteomic strategy. Proc. Natl. Acad. Sci. U. S. A. 2020;117:5810–5817. doi: 10.1073/pnas.1913525117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luth M.R., Gupta P., Ottilie S., Winzeler E.A. Using in vitro evolution and whole genome analysis to discover next generation targets for antimalarial drug discovery. ACS Infect. Dis. 2018;4:301–314. doi: 10.1021/acsinfecdis.7b00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistrado P.A., Corey V.C., Lukens A.K., LaMonte G., Sasaki E., Meister S., Wree M., Winzeler E., Wirth D.F. Plasmodium falciparum cyclic amine resistance Locus (PfCARL), a resistance mechanism for two distinct compound classes. ACS Infect. Dis. 2016;2:816–826. doi: 10.1021/acsinfecdis.6b00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Molina D., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C., Sreekumar L., Cao Y., Nordlund P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- Martinez N.J., Asawa R.R., Cyr M.G., Zakharov A., Urban D.J., Roth J.S., Wallgren E., Klumpp-Thomas C., Coussens N.P., Rai G., Yang S.M., Hall M.D., Marugan J.J., Simeonov A., Henderson M.J. A widely-applicable high-throughput cellular thermal shift assay (CETSA) using split Nano Luciferase. Sci. Rep. 2018;8:9472. doi: 10.1038/s41598-018-27834-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maveyraud L., Mourey L. Protein X-ray crystallography and drug discovery. Molecules. 2020;25:1030. doi: 10.3390/molecules25051030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNulty D.E., Bonnette W.G., Qi H., Wang L., Ho T.F., Waszkiewicz A., Kallal L.A., Nagarajan R.P., Stern M., Quinn A.M., Creasy C.L., Su D.S., Graves A.P., Annan R.S., Sweitzer S.M., Holbert M.A. A high-throughput dose-response cellular thermal shift assay for rapid screening of drug target engagement in living cells, exemplified using SMYD3 and Ido1. SLAS Discov. 2018;23:34–46. doi: 10.1177/2472555217732014. [DOI] [PubMed] [Google Scholar]
- Meister S., Plouffe D.M., Kuhen K.L., Bonamy G.M., Wu T., Barnes S.W., Bopp S.E., Borboa R., Bright A.T., Che J., Cohen S., Dharia N.V., Gagaring K., Gettayacamin M., Gordon P., Groessl T., Kato N., Lee M.C., McNamara C.W., Fidock D.A., Nagle A., Nam T.G., Richmond W., Roland J., Rottmann M., Zhou B., Froissard P., Glynne R.J., Mazier D., Sattabongkot J., Schultz P.G., Tuntland T., Walker J.R., Zhou Y., Chatterjee A., Diagana T.T., Winzeler E.A. Imaging of Plasmodium liver stages to drive next-generation antimalarial drug discovery. Science. 2011;334:1372–1377. doi: 10.1126/science.1211936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D., Wright Z., Couzens A.L., Lambert J.P., St-Denis N.A., Li T., Miteva Y.V., Hauri S., Sardiu M.E., Low T.Y., Halim V.A., Bagshaw R.D., Hubner N.C., Al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W.H., Goudreault M., Lin Z.Y., Badillo B.G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A.J., Choi H., Gstaiger M., Mohammed S., Cristea I.M., Bennett K.L., Washburn M.P., Raught B., Ewing R.M., Gingras A.C., Nesvizhskii A.I. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne R., Wiedemar N., Corpas-Lopez V., Moynihan E., Wall R.J., Dawson A., Robinson D.A., Shepherd S.M., Smith R.J., Hallyburton I., Post J.M., Dowers K., Torrie L.S., Gilbert I.H., Baragaña B., Patterson S., Wyllie S. Toolkit of approaches to support target-focused drug discovery for Plasmodium falciparum lysyl tRNA synthetase. ACS Infect. Dis. 2022;8:1962–1974. doi: 10.1021/acsinfecdis.2c00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina D.M., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C., Sreekumar L., Cao Y., Nordlund P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 2013;341:84–87. doi: 10.1126/science.1233606. [DOI] [PubMed] [Google Scholar]
- Morrison C. Landmark green light for Mosquirix malaria vaccine. Nat. Biotechnol. 2015;33:1015–1016. doi: 10.1038/nbt1015-1015. [DOI] [PubMed] [Google Scholar]
- Musset L., Le Bras J., Clain J. Parallel evolution of adaptive mutations in Plasmodium falciparum mitochondrial DNA during atovaquone-proguanil treatment. Mol. Biol. Evol. 2007;24:1582–1585. doi: 10.1093/molbev/msm087. [DOI] [PubMed] [Google Scholar]
- Nzila A., Mwai L. In vitro selection of Plasmodium falciparum drug-resistant parasite lines. J. Antimicrob. Chemother. 2010;65:390–398. doi: 10.1093/jac/dkp449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okombo J., Kanai M., Deni I., Fidock D.A. Genomic and genetic approaches to studying antimalarial drug resistance and Plasmodium biology. Trends Parasitol. 2021;37:476–492. doi: 10.1016/j.pt.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet T., Le Manach C., Cabrera D.G., Younis Y., Henrich P.P., Abraham T.S., Lee M.C.S., Basak R., Ghidelli-Disse S., Lafuente-Monasterio M.J., Bantscheff M., Ruecker A., Blagborough A.M., Zakutansky S.E., Zeeman A.M., White K.L., Shackleford D.M., Mannila J., Morizzi J., Scheurer C., Angulo-Barturen I., Martinez M.S., Ferrer S., Sanz L.M., Gamo F.J., Reader J., Botha M., Dechering K.J., Sauerwein R.W., Tungtaeng A., Vanachayangkul P., Lim C.S., Burrows J., Witty M.J., Marsh K.C., Bodenreider C., Rochford R., Solapure S.M., Jimenez-Diaz M.B., Wittlin S., Charman S.A., Donini C., Campo B., Birkholtz L.M., Hanson K.K., Drewes G., Kocken C.H.M., Delves M.J., Leroy D., Fidock D.A., Waterson D., Street L.J., Chibale K. Antimalarial efficacy of MMV390048, an inhibitor of Plasmodium phosphatidylinositol 4-kinase. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aad9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penarete-Vargas D.M., Boisson A., Urbach S., Chantelauze H., Peyrottes S., Fraisse L., Vial H.J. A chemical proteomics approach for the search of pharmacological targets of the antimalarial clinical candidate albitiazolium in Plasmodium falciparum using photocrosslinking and click chemistry. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M.A., Burrows J.N., Manyando C., van Huijsduijnen R.H., Van Voorhis W.C., Wells T.N.C. Malaria. Nat. Rev. Dis. Prim. 2017;3 doi: 10.1038/nrdp.2017.50. [DOI] [PubMed] [Google Scholar]
- Phillips M.A., Lotharius J., Marsh K., White J., Dayan A., White K.L., Njoroge J.W., El Mazouni F., Lao Y., Kokkonda S., Tomchick D.R., Deng X., Laird T., Bhatia S.N., March S., Ng C.L., Fidock D.A., Wittlin S., Lafuente-Monasterio M., Benito F.J., Alonso L.M., Martinez M.S., Jimenez-Diaz M.B., Bazaga S.F., Angulo-Barturen I., Haselden J.N., Louttit J., Cui Y., Sridhar A., Zeeman A.M., Kocken C., Sauerwein R., Dechering K., Avery V.M., Duffy S., Delves M., Sinden R., Ruecker A., Wickham K.S., Rochford R., Gahagen J., Iyer L., Riccio E., Mirsalis J., Bathhurst I., Rueckle T., Ding X., Campo B., Leroy D., Rogers M.J., Rathod P.K., Burrows J.N., Charman S.A. A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria. Sci. Transl. Med. 2015;7:296ra111. doi: 10.1126/scitranslmed.aaa6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza I., Beaton N., Bruderer R., Knobloch T., Barbisan C., Chandat L., Sudau A., Siepe I., Rinner O., de Souza N., Picotti P., Reiter L. A machine learning-based chemoproteomic approach to identify drug targets and binding sites in complex proteomes. Nat. Commun. 2020;11:4200. doi: 10.1038/s41467-020-18071-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza I., Kochanowski K., Cappelletti V., Fuhrer T., Noor E., Sauer U., Picotti P. A map of protein-metabolite interactions reveals principles of chemical communication. Cell. 2018;172:358–372 e323. doi: 10.1016/j.cell.2017.12.006. [DOI] [PubMed] [Google Scholar]
- Pryce J., Richardson M., Lengeler C. Insecticide-treated nets for preventing malaria. Cochrane Database Syst. Rev. 2018;11:CD000363. doi: 10.1002/14651858.CD000363.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings A.E. Membrane proteins: always an insoluble problem? Biochem. Soc. Trans. 2016;44:790–795. doi: 10.1042/BST20160025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reader J., van der Watt M.E., Taylor D., Le Manach C., Mittal N., Ottilie S., Theron A., Moyo P., Erlank E., Nardini L., Venter N., Lauterbach S., Bezuidenhout B., Horatscheck A., van Heerden A., Spillman N.J., Cowell A.N., Connacher J., Opperman D., Orchard L.M., Llinas M., Istvan E.S., Goldberg D.E., Boyle G.A., Calvo D., Mancama D., Coetzer T.L., Winzeler E.A., Duffy J., Koekemoer L.L., Basarab G., Chibale K., Birkholtz L.M. Multistage and transmission-blocking targeted antimalarials discovered from the open-source MMV Pandemic Response Box. Nat. Commun. 2021;12:269. doi: 10.1038/s41467-020-20629-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottmann M., McNamara C., Yeung B.K., Lee M.C., Zou B., Russell B., Seitz P., Plouffe D.M., Dharia N.V., Tan J., Cohen S.B., Spencer K.R., Gonzalez-Paez G.E., Lakshminarayana S.B., Goh A., Suwanarusk R., Jegla T., Schmitt E.K., Beck H.P., Brun R., Nosten F., Renia L., Dartois V., Keller T.H., Fidock D.A., Winzeler E.A., Diagana T.T. Spiroindolones, a potent compound class for the treatment of malaria. Science. 2010;329:1175–1180. doi: 10.1126/science.1193225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitski M.M., Reinhard F.B., Franken H., Werner T., Savitski M.F., Eberhard D., Martinez Molina D., Jafari R., Dovega R.B., Klaeger S., Kuster B., Nordlund P., Bantscheff M., Drewes G. Tracking cancer drugs in living cells by thermal profiling of the proteome. Science. 2014;346 doi: 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- Scalbert A., Brennan L., Fiehn O., Hankemeier T., Kristal B.S., van Ommen B., Pujos-Guillot E., Verheij E., Wishart D., Wopereis S. Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics. 2009;5:435–458. doi: 10.1007/s11306-009-0168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.S., Bhargav A.G., Perez J.G., Wadajkar A.S., Winkles J.A., Woodworth G.F., Kim A.J. Surface plasmon resonance as a high throughput method to evaluate specific and non-specific binding of nanotherapeutics. J. Contr. Release. 2015;219:331–344. doi: 10.1016/j.jconrel.2015.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopper S., Kahraman A., Leuenberger P., Feng Y., Piazza I., Muller O., Boersema P.J., Picotti P. Measuring protein structural changes on a proteome-wide scale using limited proteolysis-coupled mass spectrometry. Nat. Protoc. 2017;12:2391–2410. doi: 10.1038/nprot.2017.100. [DOI] [PubMed] [Google Scholar]
- Shaw J., Leveridge M., Norling C., Karen J., Molina D.M., O'Neill D., Dowling J.E., Davey P., Cowan S., Dabrowski M., Main M., Gianni D. Determining direct binders of the androgen receptor using a high-throughput cellular thermal shift assay. Sci. Rep. 2018;8:163. doi: 10.1038/s41598-017-18650-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui G., Giannangelo C., De Paoli A., Schuh A.K., Heimsch K.C., Anderson D., Brown T.G., MacRaild C.A., Wu J., Wang X., Dong Y., Vennerstrom J.L., Becker K., Creek D.J. Peroxide antimalarial drugs target redox homeostasis in Plasmodium falciparum infected red blood cells. ACS Infect. Dis. 2022;8:210–226. doi: 10.1021/acsinfecdis.1c00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speers A.E., Cravatt B.F. Profiling enzyme activities in vivo using click chemistry methods. Chem. Biol. 2004;11:535–546. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Spillman N.J., Kirk K. The malaria parasite cation ATPase PfATP4 and its role in the mechanism of action of a new arsenal of antimalarial drugs. Int J Parasitol Drugs Drug Resist. 2015;5:149–162. doi: 10.1016/j.ijpddr.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland E.C., Geer M.A., Tran D.T., Adhikari J., West G.M., DeArmond P.D., Xu Y., Fitzgerald M.C. Thermodynamic analysis of protein-ligand binding interactions in complex biological mixtures using the stability of proteins from rates of oxidation. Nat. Protoc. 2013;8:148–161. doi: 10.1038/nprot.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Ge J., Zhu B., Zheng Y.G., Zhu Q., Yao S.Q. Target identification of biologically active small molecules via in situ methods. Curr. Opin. Chem. Biol. 2013;17:768–775. doi: 10.1016/j.cbpa.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Suh M.J., Pourshahian S., Limbach P.A. Developing limited proteolysis and mass spectrometry for the characterization of ribosome topography. J. Am. Soc. Mass Spectrom. 2007;18:1304–1317. doi: 10.1016/j.jasms.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swann J., Corey V., Scherer C.A., Kato N., Comer E., Maetani M., Antonova-Koch Y., Reimer C., Gagaring K., Ibanez M., Plouffe D., Zeeman A.M., Kocken C.H., McNamara C.W., Schreiber S.L., Campo B., Winzeler E.A., Meister S. High-throughput luciferase-based assay for the discovery of therapeutics that prevent malaria. ACS Infect. Dis. 2016;2:281–293. doi: 10.1021/acsinfecdis.5b00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse E.G., Korsik M., Todd M.H. The past, present and future of anti-malarial medicines. Malar. J. 2019;18:93. doi: 10.1186/s12936-019-2724-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brummelen A.C., Olszewski K.L., Wilinski D., Llinas M., Louw A.I., Birkholtz L.M. Co-inhibition of Plasmodium falciparum S-adenosylmethionine decarboxylase/ornithine decarboxylase reveals perturbation-specific compensatory mechanisms by transcriptome, proteome, and metabolome analyses. J. Biol. Chem. 2009;284:4635–4646. doi: 10.1074/jbc.M807085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanaerschot M., Murithi J.M., Pasaje C.F.A., Ghidelli-Disse S., Dwomoh L., Bird M., Spottiswoode N., Mittal N., Arendse L.B., Owen E.S., Wicht K.J., Siciliano G., Bosche M., Yeo T., Kumar T.R.S., Mok S., Carpenter E.F., Giddins M.J., Sanz O., Ottilie S., Alano P., Chibale K., Llinas M., Uhlemann A.C., Delves M., Tobin A.B., Doerig C., Winzeler E.A., Lee M.C.S., Niles J.C., Fidock D.A. Inhibition of resistance-refractory P. falciparum kinase PKG delivers prophylactic, blood stage, and transmission-blocking antiplasmodial activity. Cell Chem Biol. 2020;27:806–816 e808. doi: 10.1016/j.chembiol.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela A.E., England K.A., Cavagnero S. Kinetic trapping in protein folding. Protein Eng. Des. Sel. 2019;32:103–108. doi: 10.1093/protein/gzz018. [DOI] [PubMed] [Google Scholar]
- Wang J., Zhang C.J., Chia W.N., Loh C.C., Li Z., Lee Y.M., He Y., Yuan L.X., Lim T.K., Liu M., Liew C.X., Lee Y.Q., Zhang J., Lu N., Lim C.T., Hua Z.C., Liu B., Shen H.M., Tan K.S., Lin Q. Haem-activated promiscuous targeting of artemisinin in Plasmodium falciparum. Nat. Commun. 2015;6 doi: 10.1038/ncomms10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J., Dhingra S.K., Deng X., El Mazouni F., Lee M.C.S., Afanador G.A., Lawong A., Tomchick D.R., Ng C.L., Bath J., Rathod P.K., Fidock D.A., Phillips M.A. Identification and mechanistic understanding of dihydroorotate dehydrogenase point mutations in Plasmodium falciparum that confer in vitro resistance to the clinical candidate DSM265. ACS Infect. Dis. 2019;5:90–101. doi: 10.1021/acsinfecdis.8b00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; Geneva: 2021. World Malaria Report 2021. [Google Scholar]
- Xie S.C., Gillett D.L., Spillman N.J., Tsu C., Luth M.R., Ottilie S., Duffy S., Gould A.E., Hales P., Seager B.A., Charron C.L., Bruzzese F., Yang X., Zhao X., Huang S.C., Hutton C.A., Burrows J.N., Winzeler E.A., Avery V.M., Dick L.R., Tilley L. Target validation and identification of novel boronate inhibitors of the Plasmodium falciparum proteasome. J. Med. Chem. 2018;61:10053–10066. doi: 10.1021/acs.jmedchem.8b01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Ottilie S., Istvan E.S., Godinez-Macias K.P., Lukens A.K., Baragaña B., Campo B., Walpole C., Niles J.C., Chibale K., Dechering K.J., Llinás M., Lee M.C.S., Kato N., Wyllie S., McNamara C.W., Gamo F.J., Burrows J., Fidock D.A., Goldberg D.E., Gilbert I.H., Wirth D.F., Winzeler E.A. MalDA, accelerating malaria drug discovery. Trends Parasitol. 2021;37:493–507. doi: 10.1016/j.pt.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo E., Schulze C.J., Stokes B.H., Onguka O., Yeo T., Mok S., Gnädig N.F., Zhou Y., Kurita K., Foe I.T., Terrell S.M., Boucher M.J., Cieplak P., Kumpornsin K., Lee M.C.S., Linington R.G., Long J.Z., Uhlemann A.C., Weerapana E., Fidock D.A., Bogyo M. The antimalarial natural product Salinipostin A identifies essential α/β serine hydrolases involved in lipid metabolism in P. falciparum parasites. Cell Chem Biol. 2020;27:143–157. doi: 10.1016/j.chembiol.2020.01.001. e145. [DOI] [PMC free article] [PubMed] [Google Scholar]