Abstract
For a long time, myelin was thought to be restricted to excitatory neurons, and studies on dysmyelination focused primarily on excitatory cells. Recent evidence showed that axons of inhibitory neurons in the neocortex are also myelinated, but the role of myelin on inhibitory circuits remains unknown. Here we studied the impact of mild hypomyelination on both excitatory and inhibitory connectivity in the primary auditory cortex (A1) with well‐characterized mouse models of hypomyelination due to loss of oligodendrocyte ErbB receptor signaling. Using laser‐scanning photostimulation, we found that mice with mild hypomyelination have reduced functional inhibitory connections to A1 L2/3 neurons without changes in excitatory connections, resulting in altered excitatory/inhibitory balance. These effects are not associated with altered expression of GABAergic and glutamatergic synaptic components, but with reduced density of parvalbumin‐positive (PV+) neurons, axons, and synaptic terminals, which reflect reduced PV expression by interneurons rather than PV+ neuronal loss. While immunostaining shows that hypomyelination occurs in both PV+ and PV− axons, there is a strong correlation between MBP and PV expression, suggesting that myelination influences PV expression. Together, the results indicate that mild hypomyelination impacts A1 neuronal networks, reducing inhibitory activity, and shifting networks towards excitation.
Keywords: auditory cortex, circuit, E/I balance, inhibition, myelin basic protein, oligodendrocytes
Main Point
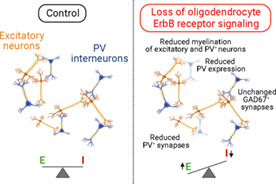
Loss of oligodendrocyte ErbB receptor signaling leads to A1 hypomyelination, reduced PV expression and density of PV+ neurons and synapses, hypoconnectivity of inhibitory circuits and shift of the E/I balance towards excitation.
1. INTRODUCTION
It has long been known that myelination increases conduction velocity of action potentials and provides metabolic support to axons (Foster et al., 2019; Monje, 2018). Therefore, myelination is critical for the normal patterns of neural circuits' activation and synchronization, as well as for normal physiological, cognitive, and behavioral performance. Given the abundant presence of myelin in the mammalian central nervous system, its disruption can cause severe information processing defects, and has been associated with neurodegenerative and psychiatric disorders (Du & Ongur, 2013; Gibson et al., 2018; Meuth et al., 2010).
Whereas myelin was thought to be restricted primarily to excitatory neurons, it has recently been appreciated that in the neocortex, myelin is also present on inhibitory neurons (Micheva et al., 2016). EM‐based studies indicate that axon collaterals of inhibitory neurons, especially those of a large proportion of parvalbumin‐positive (PV+) cells, are myelinated (Micheva et al., 2016; Peters & Proskauer, 1980; Tamas et al., 1997). PV+ cell axons constitute about half of all myelinated axons in somatosensory cortical layer 2/3 (L2/3), and a quarter of myelinated axons in L4, and myelination of GABAergic axons is enriched in MBP expression (Micheva et al., 2016). However, the impact of myelin defects on inhibitory circuits and neurons remains unknown.
Most insights into the impact of myelin on the function of neural circuits have been obtained by analyzing the consequences of severe demyelination, which can affect regions throughout the brain, including the cerebral cortex. For example, cuprizone‐induced demyelination promotes hyper‐ and depolarizing shifts of the resting membrane potential of auditory thalamocortical pathway neurons and reduction in action potential firing frequency of primary auditory cortex (A1) neurons (Ghaffarian et al., 2016). Furthermore, focal demyelination in A1 permanently disrupts its tonotopic organization (Cerina et al., 2017) and auditory frequency‐specific responses in the medial geniculate body (Narayanan et al., 2018). Whether these effects were mediated by myelin in excitatory or inhibitory neurons is unknown.
Moreover, in recent years there has been an increased appreciation that myelination is not an all‐or‐none process, but rather a continuum based on subtle differences in myelin thickness of the density of myelin segments (Monje, 2018). Furthermore, we now know that central nervous system (CNS) myelin thickness and density is influenced by experience, that is, myelin is negatively influenced by deprivation (Liu et al., 2012; Makinodan et al., 2012) and increased by neuronal activity and novelty (Gibson et al., 2014; McKenzie et al., 2014). Additionally, a recent study showed that while both excitatory and PV+ inhibitory neurons undergo homeostatic myelin remodeling under normal vision, monocular deprivation only induces adaptive myelin remodeling in PV+ interneurons (Yang et al., 2020). However, much less is known about how subtle changes in myelin alters the balance between excitation and inhibition.
To gain insights into the impact of hypomyelination on inhibitory and excitatory neuronal network function, we studied A1 in mice with mild hypomyelination caused by loss of oligodendrocyte ErbB receptor signaling, either by oligodendrocyte specific expression of dominant‐negative ErbB4 (Roy et al., 2007), or oligodendrocyte‐specific inducible ErbB3 knock‐out (Makinodan et al., 2012). To evaluate whether hypomyelination alters intracortical neural circuits in A1, we used laser‐scanning photostimulation (LSPS) to optically probe functional intracortical circuits to L2/3 neurons. We found that hypomyelination leads to a reduction in inhibitory but not excitatory circuits and a consequent alteration in the excitatory/inhibitory balance of inputs to L2/3 neurons. The functional alterations are not associated with changes in gene expression of molecules involved in GABAergic and glutamatergic synaptic function, but rather with a reduced density of PV+ neurons and lower number of myelinated PV+ axons in A1. Remarkably, the reduced number of PV+ neurons is not due to neuronal loss, but rather to a reduction in PV expression by interneurons. These results show that subtle defects in myelination can lead to large changes in gene expression and function of PV interneurons which result in large‐scale changes in network function in the neocortex.
2. MATERIALS AND METHODS
2.1. Animals
All animal procedures were carried out with prior approval from the University of Michigan and the University of Maryland Committees on Use and Care of Animals in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals. Mice were housed in an Association for Assessment and Accreditation of Laboratory Animal Care–accredited facility in the University of Michigan. Mice were kept in a light‐ (12 h on/off) and temperature‐ (21–23°C) controlled environment and were fed with a standard chow diet (5LOD, LabDiet).
Transgenic mice expressing a dominant‐negative ErbB4 receptor under control of the CNPase promoter (CNP‐DN‐ErbB4) (Chen et al., 2006) were crossed to wild type FVB/N J mice (JAX® mice, stock # 001800). The experimental mice were those hemizygous for CNP‐DN‐ErbB4 or wild type littermates. To evaluate the role of NRG‐1/ErbB3 signaling in the hypomyelination and expression of PV in the A1, mice expressing inducible cre recombinase under the control of the proteolipid protein promoter (PLP/creERT) were crossed to ErbB3flox/flox mice (Makinodan et al., 2012). Floxed allele recombination was induced by intraperitoneal injection of tamoxifen (Sigma, St. Louis, MO) dissolved in corn oil (10 mg/mL), by the time the peripheral myelination is known to be established (from P6). For recombination between P6 and P30, mice were treated with a dose of 33 mg/kg/day. From P30 to P56, tamoxifen dose was 100 mg/kg body weight, every other day. This regimen was used because PLP expressing oligodendrocytes differentiate and mature over time until adulthood. The experimental mice were those homozygous for ErbB3 flox expressing creERT, and homozygous ErbB3 flox littermates without cre recombinase expression. All mice were treated with tamoxifen, regardless of genotype. Experiments were performed in 2‐month age mice.
The genotypes of mice were confirmed by PCR detection of the transgenes in tail‐derived DNA from the CNP‐DN‐ErbB4, wild type, PLP/creERT and ErbB3fl/fl mice at weaning and at the end of experiments. The following pairs of primers were used: CNP‐DN‐ErbB4: F: 5′ TGCTGAAGGAATGGTGTGC 3′; R: 5′ CTTGTCGTCATCGTCTTTG 3′; PLP/CreERT: F: 5′ GATGTAGCCAGCAGCATGTC 3; R: 5′ ACTATATCCGTAACCTGGAT 3′ and ErbB3‐flox: F: 5′ CCAACCCTTCTCCTCAGATAGG 3′; R: 5′ TGTTTGTGAAATGTGGACTTTACC 3′ and R: 5′ GGCAGGCATGTTGACTTCACTTGT 3′. Expression of the DN‐ErbB4 FLAG transgene was also validated by RT‐qPCR in the primary auditory cortex, using the following pair of primers: F: 5′ GAGCCTTGAGAAGATTCTTG 3′; R: 5′ TGTCGTCATCGTCTTTGTAG 3′.
2.2. Dissection of the primary auditory cortex
Male mice were anesthetized with isoflurane (Fluoriso, Vet One) and euthanized by decapitation. Brain was collected and the primary auditory cortex (A1) fragments were micro dissected (thickness: 1.0 mm) from an area located laterally to the border of the hippocampus, according to the coordinates from Paxinos and Franklin Mouse Brain Atlas (2.06 mm posterior to bregma, 3.5 mm lateral to the midline and 2.0 mm dorsoventrally). Tissues were immediately frozen in dry ice for posterior processing for RT‐qPCR (n = 7–12/group) or Western blotting (n = 6/group) analyses. Tissue harvesting was performed between 8 am and 10 am.
2.3. Real‐time quantitative RT‐PCR
Total RNA was isolated from the A1 samples using RNA extraction kit, Qiazol Reagent (RNeasy mini kit; Qiagen, Germany), and DNase treatment was performed (RNase‐free; Qiagen). The complementary DNA was synthesized using iScript cDNA synthesis kit (Bio‐Rad, #1708891), according to the manufacturers' protocol. Quantitative RT‐PCR was performed on a CFX‐96 Bio‐Rad reverse transcription polymerase chain reaction detection system (Hercules, CA) using iTaq Universal SYBR® Green supermix (Bio‐Rad, # 172‐5121) and primer pairs were synthesized by IDT (Coralville, IA). All samples and standard curves were run in triplicate. Water instead of complementary DNA was used as a negative control. The mRNA expression in transgenic versus control mice was determined by a comparative cycle threshold (Ct) method and relative gene copy number was calculated as normalized gene expression, defined as described previously (Stankovic & Corfas, 2003). Ribosomal protein L19 (RPL19) and GAPDH were used as housekeeping genes. The following specific oligo primers were used for the target genes: RPL19, F: 5′ACCTGGATGAGAAGGATGAG 3′; R: 5′ACCTTCAGGTACAGGCTGTG 3′; GAPDH, F: 5′ TCACTGCCACCCAGAAGA 3′; R: 5′ GACGGACACATTGGGGGTAG 3′; MBP, F: 5′ATCCAAGTACCTGGCCACAG 3′; R: 5′CCTGTCACCGCTAAAGAAGC 3′; GAD65, F: 5′ CATTGATAAGTGTTTGGAGCTAGCA 3′; R: 5′ GTGCGCAAACTAGGAGGTACAA 3′; GAD67, F: 5′ TCGATTTTTCAACCAGCTCTCTACT 3′; R: 5′ GTGCAATTTCATATGTGAACATATT 3′; VGAT, F: 5′ TCCTGGTCATCGCTTACTGTCTC 3′; R: 5′ CGTCGATGTAGAACTTCACCTTCTC 3′; GABARα1, F: 5′ CCCCGGCTTGGCAACTA 3′; R: 5′ TGGTTTTGTCTCAGGCTTGAC 3′; VGLUT1, F: 5′ TCGCTACATCATCGCCATC 3′; R: 5′ GTTGTGCTGTTGTTGACCAT 3′; VGLUT2, F: 5′ CTGCGATACTGCTCACCTCTA 3′; R: 5′ GCCAACCTACTCCTCTCCAA 3′; GRIA1, F: 5′ GCTATTCCTACCGACTTGA 3′; R: 5′ CCACATCTGCTCTTCCATA 3′; GRIA2, F: 5′ CCTCATCATCATCTCCTCCTAC 3′; R: 5′ GAGCCAGAGTCTAATGTTCCA 3′; GRIA3, F: 5′ TCTAAGCCTGAGCAATGTG 3′; R: 5′ CCTTCTCTGTATGTAGCGTAAT 3′ and GRIA4, F: 5′ GCATACCTTGACCTCCTTCTG 3′; R: 5′ GCACGAACTGGCTCTCTC 3′, PARVALBUMIN: F: 5′ GCAAGATTGGGGTTGAAGAA 3′; R: 5′ GTGTCCGATTGGTACAGCCT 3′, VIP: F: 5′ CTGGCCTCTCTTTGGACCAC 3′; R: 5′ ACGGCATCAGAGTGTCGTTT 3′, SOMATOSTATIN: F: 5′ CCAACTCGAACCCAGCAATG 3′; R: 5′ TCAGAGGTCTGGCTAGGACA 3′ and ErbB3: F: 5′ TTGCCTACAGGAACGCTTACCCG 3′; R: 5′ ACCCCCCAAAACCGCAGAATC 3″. Changes in mRNA expression were calculated as relative expression (arbitrary units) respective to the wild‐type group.
2.4. Western blot analysis
Total protein from A1 was extracted using RIPA buffer (#R0278, Sigma Aldrich) and protease inhibitor cocktail kit (#78410, ThermoFisher Scientific). Homogenates were centrifuged at 4°C and 14,000 g for 15 min. Aliquots of the lysates containing 10 mg of protein were denatured in Laemmli buffer and β‐mercaptoethanol (Bio‐Rad) at 95°C for 5 min. After electrophoresis (Mini‐protean TGX gel, #456‐1086, Bio‐Rad), samples were blotted onto nitrocellulose membranes (Immobilon‐PSQ, #ISEQ00010, Merk Millipore). Nonspecific binding was prevented by immersing the membranes in a blocking buffer (5% BSA in Tris‐buffered saline ‐Tween 20, TBS‐T) for 60 min at room temperature. The membranes were then exposed overnight to the primary antibodies: mouse anti‐GAPDH (1:3000, MA5‐15738, RRID: AB_10977387, ThermoFisher Scientific) and rat anti‐MBP (1: 1000, MAB386, RRID: AB_94975, Millipore, Germany). The blots were rinsed in TBS‐T and then incubated with horseradish peroxidase‐conjugated anti‐mouse antibody (1:4000, SC‐516102, RRID: AB_2687626, Santa Cruz) or anti‐rat antibody (1: 4000, #7077, RRID: AB_10694715, Cell Signaling) for 1 h at room temperature. Antibody–antigen complexes were visualized by detecting enhanced chemiluminescence using a Pierce ECL detection system (#32209, ThermoFisher Scientific) and digital images with Chemi Doc Touch Image System (Bio‐Rad). Expression of MBP was normalized to the expression of GAPDH. Data were analyzed as relative expressions (arbitrary units) respective to wild type group.
2.5. Immunostaining and image processing
To assess the pattern of expression of the MBP (6–8/group) or PV (8–12/group) in A1, PFC and M1 CNP‐DN‐ErbB4, PLP/creERT:ErbB3fl/fl or their respective littermate controls (wild types or ErbB3fl/fl mice), were anesthetized using isoflurane (Fluriso, Vet One) and transcardially perfused with saline, followed by 4% formaldehyde in 0.1 M phosphate buffer (PBS). Brains were dissected, post‐fixed in the same fixative for 1 h, placed in PBS containing 30% sucrose and sectioned on a cryostat (30‐mm sections, 4 series) in the frontal plane. For MBP staining, brain coronal sections were rinsed with PBS and nonspecific binding was prevented by immersing the sections in blocking buffer (PBS, 5% normal horse serum and 0.3% Triton X‐100) for 1 h at room temperature. The sections were incubated overnight at 37°C with primary antibody rat anti‐MBP (1:1000, MAB386, RRID: AB_94975, Millipore, Germany) in PBS, 1% normal horse and 0.3% Triton X‐100 solution. After rinses, sections were incubated for 1 h with the chicken anti‐rat AlexaFluor 647 secondary antibody (1:400, #A‐21472, RRID: AB_2535875, Thermo Fisher Scientific). For PV and GAD67 staining, brain coronal sections were rinsed with TBS and nonspecific binding was prevented by immersing the sections in blocking buffer (TBS, 5% normal goat serum and 0.2% Triton X‐100) for 1 h at room temperature. The sections were incubated overnight at 4°C with primary antibody rabbit anti‐PV (1:1000, PV25, RRID: AB_10000344, Swant) or mouse anti‐GAD67 (1:1000, MAB5406, RRID: AB_2278725, Millipore). After rinses, sections were incubated for 1 h with the goat anti‐rabbit AlexaFluor 488 (1:400, #A‐11094, RRID: AB_221544, Thermo Fisher Scientific) or goat anti‐mouse AlexaFluor 594 (1:400, A‐11005, RRID: AB_2534073, ThermoFisher Scientific) secondary antibodies. Finally, the sections were mounted on superfrost microscope slides (Fisher Scientific) and coversliped with Fluoro‐Gel II with DAPI mounting medium (#17985–50, Electron Microscopy Sciences). For double‐labeling, tissue has been stained for PV, followed by MBP, as described above. For PV and Vicia villosa agglutinin (VVA) co‐staining, mice (n = 6–7) were perfused as described above. PV staining was performed as above and then sections were incubated at 4°C overnight with biotinylated VVA (1:5000, #B‐1235, RRID: AB_2336855, Vector Laboratories). After rinsing, sections were incubated for an additional 2 h with Alexa 594 streptavidin‐conjugated antibody (1:500, #S32356, RRID: N/A, Thermo Fisher Scientific). Sections were imaged after rinsing.
Series of systematically selected brain sections (30‐mm thick, every 120‐mm) representing the A1 starting on 2.06 mm posterior to bregma and ending on bregma – 3.5, the PFC (starting on 2.22 mm anterior to bregma and ending on 1.54 mm anterior to bregma) and the M1 (starting on 2.15 mm anterior to bregma and ending on 1.98 mm anterior to bregma) were acquired using a Leica SP8 confocal laser scanning microscope and 10×, 20×, or 63× oil‐immersion lens. The immunoreactive structures were excited using lasers with the excitation and barrier filters set for the fluorochromes used (magenta for MBP, green for PV, red for GAD67 or VVA, and blue for DAPI). Quantification of PV and MBP immunofluorescence signal, axonal area of coverage, or the number of PV positive cells were performed in 10 different fields of the A1 per mouse, and 6 different fields of the PFC and M1 per mouse, using the Image J software (FIJI version, NIH). Signal intensity was calculated by converting each frame into grayscale using the plugging for measurement of pixel intensity. The same image was then binarized for measurement of the area covered by PV or MBP pixels. Binarized images had their thresholds homogeneously adjusted. Threshold value was set based on the average of auto‐threshold values obtained in the control group. Histograms indicating the number of pixels of PV or MBP staining within the same area of each image were recorded and expressed as Integrated Density. Averaged Integrated Density of the fields analyzed per mouse was used to compare the levels of PV and MBP expression between groups. All immunofluorescence images shown are representative of one mouse from each group.
Quantification of myelinated PV+ axons was performed in A1 by obtaining z‐stack images (1064 × 1064 pixels) at 63× magnification (oil immersion) with 1× digital zoom at a step size of 0.5 μm. Every 5 stacks were sampled across A1, based on DAPI nuclear labeling. Immunofluorescence colocalization of MBP and PV was manually counted by a blind investigator in 5 stacks per field, 4–5 fields per mouse using ImageJ (FIJI). Co‐localization was defined as the circumferential bordering of a PV‐labeled axon by an MBP+ myelin signal (Stedehouder et al., 2017; Stedehouder et al., 2019).
To estimate the area of total MBP, non‐PV associated MBP, and PV associated MBP signal, we performed immunostaining for PV and MBP and image acquisition as described above. Using the FIJI software, images were binarized and we generated a mask for the PV signal and subtracted it from the double labeled image, thus removing all the MBP signaling that would be associated with the PV. Quantification of the area covered by total MBP, by non‐PV associated MBP and by PV associated MBP in A1 was performed in 5 images/mouse, in a 63× magnification.
Quantification of PV‐VVA+ cells has been performed manually using the FIJI cell counter plugin in 3–4 A1 sections/mouse. For quantification of GAD67+ and PV+ perisomatic puncta, brain slices were stained for GABA‐synthetic enzyme GAD67, and PV as described above. To count the number of perisomatic puncta, we randomly acquired three non‐overlapping images per mouse from a single confocal plane of the A1, using a 63× oil immersion objective lens. Quantification was performed by a blind investigator using the FIJI puncta analyzer plugin (Li et al., 2017; Ueno et al., 2013). Briefly, in RGB images containing both red and green channels a circular area (ROI) was traced enclosing the soma and 2 μm away from the GAD67 signal. GAD67+ and PV+ puncta with 0.5–2.0 μm2 area sizes were counted as the number of synaptic terminals. To minimize the fusion of puncta, the same threshold of each image was adjusted manually for each color channel. The puncta analyzer plugin first quantifies the number of puncta in the red channel (GAD67), then the number of puncta in the green channel (PV) and estimates the number of positive puncta for both channels (colocalized). The perimeter of each ROI enclosing the cell was also determined. The number of puncta surrounding 15–20 pyramidal neurons distributed in three sections/mouse were quantified and the result represents the average of positive puncta/100 μm.
2.6. In vitro Laser‐Scanning Photostimulation (LSPS)
LSPS experiments were performed as previously described (Meng et al., 2015; Meng et al., 2017; Meng et al., 2020).
2.7. Slice preparation
Mice were deeply anesthetized with isoflurane (Halocarbon). A block of brain containing the A1 and the medial geniculate nucleus (MGN) was removed and thalamocortical slices (500 μm thick) were cut on a vibrating microtome (Leica) in ice‐cold ACSF containing (in mM): 130 NaCl, 3 KCl, 1.25 KH2PO4, 20 NaHCO3, 10 glucose, 1.3 MgSO4, 2.5 CaCl2 (pH 7.35–7.4, in 95%O2–5%CO2). The cutting angle was ~15° from the horizontal plane (lateral raises) and A1 was identified as described previously (Meng et al., 2015; Meng et al., 2017; Meng et al., 2020). Slices were incubated for 1 h in ACSF at 30°C and then kept at room temperature. Slices were held in a chamber on a fixed‐stage microscope (Olympus BX51) for recording and superfused (2–4 ml/min) with high‐Mg2+ ACSF recording solution at room temperature to reduce spontaneous activity in the slice. The recording solution contained (in mM): 124 NaCl, 5 KCl, 1.23 NaH2PO4, 26 NaHCO3, 10 glucose, 4 MgCl2, 4 CaCl2. The location of the recording site in A1 was identified by landmarks (Meng et al., 2015; Meng et al., 2017; Meng et al., 2020).
2.8. Electrophysiology
Whole‐cell recordings from L2/3 cells were performed with a patch clamp amplifier (Multiclamp 700B, Axon Instruments) using pipettes with input resistance of 4–9 MΩ. Data acquisition was performed with National Instruments AD boards and custom software (Ephus) (Suter et al., 2010), which was written in MATLAB (Mathworks) and adapted to our setup. Voltages were corrected for an estimated junction potential of 10 mV. Electrodes were filled with (in mM): 115 cesium methanesulfonate (CsCH3SO3), 5 NaF, 10 EGTA, 10 HEPES, 15 CsCl, 3.5 MgATP, 3 QX‐314 (pH 7.25, 300 mOsm). Cesium and QX314 block most intrinsic active conductances and thus make the cells electrotonically compact. Series resistances were typically 20–25 MΩ. Photostimulation: 0.5–1 mM caged glutamate (N‐[6‐nitro‐7‐coumarinylmethyl]‐L‐glutamate; Ncm‐Glu) (Muralidharan et al., 2016) is added to the ACSF. This compound has no effect on neuronal activity without UV light (Muralidharan et al., 2016). UV laser light (500 mW, 355 nm, 1 ms pulses, 100 kHz repetition rate, DPSS) was split by a 33% beam splitter (CVI Melles Griot), attenuated by a Pockels cell (Conoptics), gated with a laser shutter (NM Laser), and coupled into a microscope via scan mirrors (Cambridge Technology) and a dichroic mirror. The laser beam in LSPS enters the slice axially through the objective (Olympus 10·, 0.3NA/water) and has a diameter of <20 μm. Laser power at the sample is <25 mW. Laser power was constant between slices and recording days. We typically stimulated up to 30 × 25 sites spaced 40 μm apart, enabling us to probe areas of 1 mm2; such dense sampling reduces the influence of potential spontaneous events. Repeated stimulation yielded essentially identical maps. Stimuli were applied at 1 Hz. Analysis was performed essentially as described previously with custom software written in MATLAB (Meng et al., 2015; Meng et al., 2017; Meng et al., 2020). To detect monosynaptically evoked postsynaptic currents (PSCs), we detected PSCs with onsets in an approximately 50‐ms window after the stimulation. This window was chosen based on the observed spiking latency under our recording conditions (Meng et al., 2015; Meng et al., 2017; Meng et al., 2020). Our recordings were performed at room temperature and in high‐Mg2+ solution to reduce the probability of polysynaptic inputs. We measured both peak amplitude and transferred charge; transferred charge was measured by integrating the PSC. While the transferred charge might include contributions from multiple events, our prior studies showed a strong correlation between these measures (Meng et al., 2015; Meng et al., 2017; Meng et al., 2020). Traces containing a short‐latency (<8 ms) “direct” response were discarded from the analysis (black patches in color‐coded maps) as were traces that contained longer latency inward currents of long duration (>50 ms) (Figure 3b). The short‐latency currents could sometimes be seen in locations surrounding (<100 μm) areas that gave a “direct” response. Occasionally some of the “direct” responses contained evoked synaptic responses that we did not separate out, which leads to an underestimation of local short‐range connections. Cells that did not show any large (>100 pA) direct responses were excluded from the analysis as these could be astrocytes. It is likely that the observed PSCs at each stimulus location represent the activity of multiple presynaptic cells.
FIGURE 3.
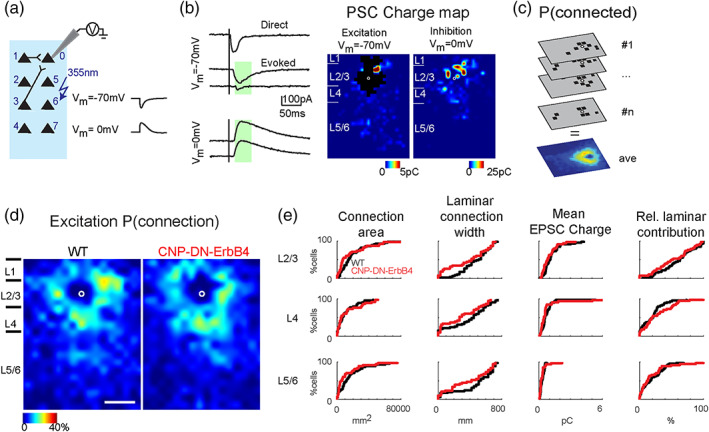
Excitatory circuits to L2/3 neurons are normal in A1 of CNP‐DN‐ErbB4 mice. (a) Schematic diagram shows how whole‐cell voltage‐clamp combined with LSPS experiments are performed. (b) Whole‐cell voltage‐clamp recordings at holding potential of ‐70 mV (top) and 0 mV (bottom). Left, shows the traces obtained with photostimulation at different locations. Solid lines indicate time of photostimulation. Green shaded area is the analysis window. It started at 8 ms and ended at 50 ms after laser onset. Right, example of excitatory and inhibitory maps for a L2/3 cell. Pseudocolor indicates the PSC charge at each stimulation location. Black area indicates where direct responses are. White circle marks the soma location. Horizontal bars indicate layer borders and are 100 μm long. (c) Cartoon illustrating how the connection probability (P[connection]) map is calculated. All the input maps contain 0 (gray, the area has no connection to the recorded cell) and 1 (black square, the area has monosynaptic connection to the recorded cell) and are aligned to soma (white circles). The P (connection) map is calculated by averaging all the input maps along the z‐axis. (d) Spatial connection probability of excitatory connections for WT (left) and CNP‐DN‐ErbB4 (right) mice, n = 36 cells for WT and n = 32 cells for CNP‐DN‐ErbB4. The border bars are the averaged borders across all the cells in each group. Scale bar is 200 μm long. (e) Cumulative distribution functions (CDFs) of LSPS data for excitatory connections. Total area, laminar connection width, mean charge, and relative laminar charge contribution from L2/3, L4, and L5/6 to L2/3 neurons of WT (black) and CNP‐DN‐ErbB4 (red) mice. All p > .05 (Rank Sum test).
Stimulus locations that showed PSC were deemed connected and we derived binary connection maps. We aligned connection maps for L2/3 cells in the population and averaged connection maps to derive a spatial connection probability map (Figure 3c). In these maps the value at each stimulus location indicates the fraction of L2/3 cells that received input from these stimulus locations. Layer boundaries were determined from the infrared pictures. We derived laminar measures on the single cell level to compare the cell populations. Input area is calculated as the area within each layer that gave rise to PSCs. Mean charge is the average charge of PSCs from each stimulus location in each layer. Intralaminar integration distance is the extent in the rostro‐caudal direction that encompasses connected stimulus locations in each layer. We calculated an excitation to inhibition index (“E/I balance”) in each layer for measures of input area and strength as (E – I)/(E + I), for example, (AreaE – AreaI)/(AreaE + AreaI), resulting in a number that varied between −1 and 1 with 1 indicating dominant excitation and −1 indicating dominant inhibition. For visualization, we plotted the data both as cumulative distribution functions (CDFs, Figures 3 and 4) as well as histograms (Figures [Link], [Link]). In the histograms, we fitted the distribution with a smoothing distribution using the matlab function histfit (with dist = “kernel”).
Spatial connection probability maps show the average connection pattern in each group. To compare the large‐scale connectivity between cells in each group we calculated the spatial correlation of the binary connection maps in each group by calculating the pairwise cross‐correlations (Meng et al., 2020).
2.9. Statistical analysis
Results are expressed as means ± SEM and data were analyzed using MATLAB or GraphPad Prism 9 software. Results of qRT‐PCR, Western blots and immunofluorescence experiments were analyzed by unpaired two‐tailed Student's t‐test if the variables passed a normality test or by Mann–Whitney non‐parametric test if not. Cumulative distribution functions (CDFs) for LSPS results were analyzed with Rank Sum test. To evaluate the correlation between PV and MBP mRNA expression a Pearson's correlation test was performed. Histograms for excitatory and inhibitory connections and excitatory/inhibitory balance were fit using Kernel function. Differences were accepted as significant at P < .05.
3. RESULTS
3.1. Disruption of oligodendrocyte ErbB signaling results in reduced myelin basic protein and mRNA levels in A1
We previously showed that expression of a dominant‐negative ErbB4 receptor in cells of the oligodendrocyte lineage under the control of the CNPase promoter (CNP‐DN‐ErbB4) results in mice with reduced myelin thickness in optic nerve, corpus callosum and prefrontal cortex axons (Roy et al., 2007) as well as thinner myelin and reduced levels of MBP protein and mRNA levels in peripheral nerves (Chen et al., 2006). DN‐ErbB4, which in these mice is only expressed in myelinating glia, is a truncated ErbB4 receptor, and its expression abolishes the signaling of NRG1 receptors ErbB2, 3 and 4 (Chen et al., 2006; Prevot et al., 2003; Rio et al., 1997) without affecting signaling by ErbB1, the receptor for EGF (Prevot et al., 2003). We now find that DN‐ErbB4 expression in oligodendrocytes leads to similar alterations in A1 myelination. Quantitative immunofluorescence showed that the intensity of MBP in A1 is markedly reduced in adult mutant mice compared to their wild type (WT) littermates (Figure 1a,b). Western blot analysis (Figure 1c,d) showed a reduction in MBP protein in A1 of CNP‐DN‐ErbB4 mice, and quantitative RT‐PCR showed a similar reduction in MBP mRNA levels (Figure 1e). Furthermore, quantification of the MBP immunofluorescence density showed that the area of MBP signal is reduced in the mutant, consistent with a reduction in the number of MBP+ processes in A1 (Figure 1f). Together, these results indicate that loss of oligodendrocyte ErbB receptor signaling leads to A1 hypomyelination.
FIGURE 1.
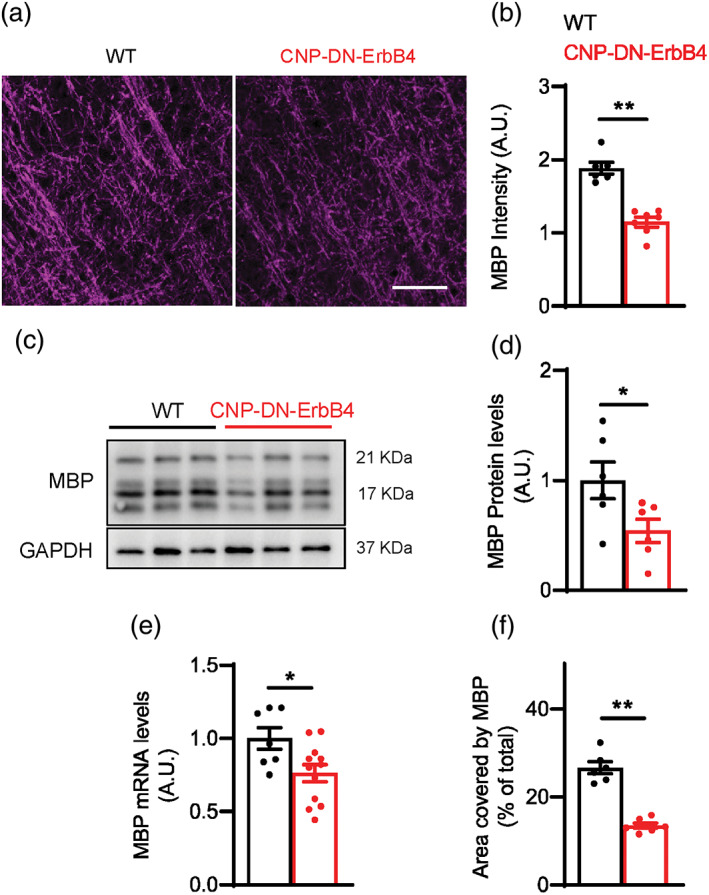
Loss of oligodendrocyte ErbB receptor signaling leads to a reduction in MBP protein and mRNA expression levels in A1. (a) Representative photomicrographs of primary auditory cortex of WT and CNP‐DN‐ErbB4 mice showing the expression of myelin basic protein (MBP) (magenta). Scale bar: 20 μm. (b) Quantification of MBP staining intensity (WT: black; CNP‐DN‐ErbB4: red) (n = 6–7 mice per genotype; **p = .001). (c) Representative MBP and GAPDH Western blots of A1 samples from WT and CNP‐DN‐ErbB4 mice. (d) Quantification of MBP protein expression normalized by GAPDH in A1 of WT and CNP‐DN‐ErbB4 mice (n = 6; p = .0413). (e) MBP mRNA levels in A1 of WT and CNP‐DN‐ErbB4 mice (n = 7–12; p = .0226). (f) Quantification of the percent of area covered by MBP (WT: black; CNP‐DN‐ErbB4: red) (n = 6–7 mice per genotype; **p ˂ .001) (A.U.: arbitrary units). Unpaired two‐tailed Student's t test was performed. Data are expressed as mean ± SEM.
3.2. Hypomyelination does not alter sensitivity of cortical neurons to photostimulation
We next examined the impact of hypomyelination on A1 intracortical neural circuits using laser‐scanning photostimulation (LSPS) using caged glutamate (Meng et al., 2015; Meng et al., 2017; Meng et al., 2020) in brain slices of adult mice. We first tested if oligodendrocyte DN‐ErbB4 expression affects the ability of A1 neurons to fire action potentials to photo released glutamate by performing cell‐attached patch recordings from cells in L2/3 and L4 (N = 27 L4 and N = 38 L2/3 neurons in WT, N = 20 L4 and N = 36 L2/3 neurons in CNP‐DN‐ErbB4) (Figure 2a). Short UV laser pulses (1 ms) to focally release glutamate were targeted to multiple stimulus spots covering the extent of A1 (Figure 2b). Stimulation close to the cell body led to action potentials (Figure 2b). There were no differences between the genotypes in the area around targeted neurons where action potentials could be generated (Figure 2b,c, L2/3: p = .14; L4: p = .62) or in the number of evoked action potentials (Figure 2c, L2/3: p = .29; L4: p = .74). Thus, the spatial resolution of LSPS is not affected by hypomyelination.
FIGURE 2.
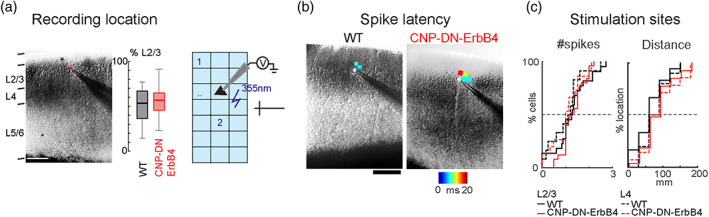
Sensitivity to photo released glutamate in layer 2/3 neurons is normal in A1 of CNP‐DN‐ErbB4 mice. (a) Left, Infrared image of cortical field with patch pipette on a. Scale bar is 200 μm. Cortical layers are identified based on the DIC image. Right, Position of recorded cells within Layer 2/3. Plotted are the relative positions within Layer 2/3 for WT (black) and CNP‐DN‐ErbB4 (red) mice. 0 refers to the border with layer 4 and 100 refers to the border with Layer 1. (b) Left, Graphic illustration on how cell attach LSPS experiments were performed. The cortical fields were divided into approximately 30 by 25 grids. UV laser targets stimulation sites in a grid in a pseudorandom pattern to make sure that two nearby locations will not be stimulated sequentially to avoid adaptation. Cells under laser activation sites could be activated and generate action potentials (APs). Right, Cell attached recordings on Layer 2/3 and 4 cells show areas that evoke action potentials. Maps show first spike latencies encoded by color and overlaid on infrared images. (c) Cumulative distributions of number of spikes (Left) and distances from locations that resulted in APs to the soma of L2/3 (solid) and L4 (dashed) cells for both WT (black) and CNP‐DN‐ErbB4 (red) mice.
3.3. Hypomyelination does not alter excitatory connections to L2/3 neurons
To map and quantify functional excitatory inputs to L2/3 neurons, we performed whole‐cell recording combined with LSPS while cells were held at a holding potential of –70 mV. All recorded cells were from similar laminar positions (Figure 2a, p = .062). We targeted the laser pulse to multiple (600–900) stimulus locations spanning across the cortical extent and recorded the resulting membrane currents (Figure 3a). Targeting the cell body and the proximal dendrites of the cell under study caused large amplitude and short‐latency (<8 ms) currents (Figure 3b), reflecting direct activation of glutamate receptors on the neuron. These “direct” currents were excluded from the analysis. Targeting other sites could result in longer‐latency (>8 ms) currents, which under our recording conditions, reflect monosynaptically evoked postsynaptic currents (PSCs) (Meng et al., 2015; Meng et al., 2017; Meng et al., 2020) (Figure 3b).
We mapped 68 L2/3 cells (36 cells in 14 WT mice, 32 cells in 11 CNP‐DN‐ErbB4 mice) and generated spatial connection maps for each cell which indicated which stimulus location resulted in an evoked PSC. We then aligned and averaged all individual spatial connection maps to the soma position of the individual cells (Figure 3c). By averaging the individual connection maps, we obtained the spatial connection probability map for excitatory inputs where each value (represented as color in the maps) denotes the fraction of neurons in the population that receives input from a particular location (Figure 3d). Qualitatively, a comparison of the excitatory connection probability maps from WT and in CNP‐DN‐ErbB4 animals showed no differences (Figure 3d).
Since spatial averaging can obscure differences in functional connections, we quantified the laminar changes of the connection properties for each cell as in prior studies (Meng et al., 2015; Meng et al., 2017; Meng et al., 2020). For each neuron, we quantified the total area in each layer where stimulation resulted in a response. The total area within each layer where stimulation can evoke EPSCs in L2/3 neurons was similar in all layers between genotypes (Figures 3e and S1, L2/3: p = .13; L4: p = .96; L5/6: p = .12). The thalamocortical slices are cut such that the macroscale rostro‐caudally oriented tonotopic map is preserved in the slice plane. Thus, the spatial extent of functional integration along the slice is a proxy for the integration along the tonotopic axis. To assess this laminar connection width, we calculated the distance in each layer that covers 80% of the input sites. We found that the laminar connection width is similar between genotypes indicating that the functional integration across the tonotopic axis was unchanged in CNP‐DN‐ErbB4 mice (Figures 3e and S1, L2/3: p = .11; L4: p = .14; L5/6: p = .14). Circuit changes can result from both connection probability and input strength. Thus, we measured the average charge of the evoked EPSC and the relative EPSC charge contribution from each layer to L2/3 cells (Figure 3e). Evoked EPSCs and relative contribution from all layers were not different between genotypes (Figure 3e, S1, evoked EPSC, L2/3: p = .15; L4: p = .16; L5/6: p = .14; relative contribution, L2/3: p = .22; L4: p = .21; L5/6: p = .52). Together, these results indicate that hypomyelination does not affect the spatial pattern of functional excitatory connectivity to L2/3 neurons. To complement the cumulative distribution functions (CDFs) in Figure 3e, we also plotted histograms of the data (Figure S1). Distributions for excitatory inputs from L2/3, L4 and L5/6 to L2/3 neurons were overlapping, confirming that there are no differences in excitatory inputs between WT and CNP‐DN‐ErbB4 mice.
3.4. Hypomyelination leads to reduced inhibitory connectivity to L2/3 neurons
To map and quantify functional inhibitory input to L2/3 neurons we held cells at 0 mV and repeated the LSPS. Qualitative comparison of the resulting connection probability maps from WT and CNP‐DN‐ErbB4 animals showed an apparent reduction in inputs from L4 and from within L2/3 (Figure 4a). We confirmed this observation by quantifying the total area within each layer where stimulation evoked IPSCs in the recorded L2/3 neurons. We found that total input from L2/3, L4 and L5/6 was reduced in CNP‐DN‐ErbB4 animals (Figures 4b and S2, L2/3: p = 5.1 × 10–3; L4: p = 4.5 × 10–3; L5/6: p = 3.7 × 10–3). Moreover, the laminar connection width was smaller in all layers of CNP‐DN‐ErbB4 mice than in cells from WT (Figures 4b and S2, L2/3: p = 8.5 × 10–3; L4: p = 2.3 × 10–3; L5/6: p = 2.9 × 10–3). This indicates that cells from CNP‐DN‐ErbB4 mice receive inputs from a more restricted area than in cells from WTs. The average charge of the evoked IPSC within L2/3 in CNP‐DN‐ErbB4 animals was reduced (L2/3: p = .02; L4: p = .23; L5/6: p = .36) but the relative contribution from L2/3 is increased (Figures 4c and S2, L2/3: p = .031; L4: p = .11; L5/6: p = .021). These results suggest that hypomyelination causes a spatial hypoconnectivity of inhibitory connections. As expected from the decreased inhibitory connectivity in face of unchanged excitatory connectivity, calculating the balance between excitation and inhibition showed that this balance is shifted towards excitation (Figures 4d and S3, EI charge: L2/3: p = .901; L4: p = 2.4 × 10–3; L5/6: p = .012; EI peak: L2/3: p = .32; L4: p = 6.2 × 10–4; L5/6: p = 6.9 × 10–3; EI Area: L2/3: p = .52; L4: p = 7.6 × 10–4; L5/6: p = .011). Histograms of the inhibitory inputs from L2/3, L4 and L5/6 to L2/3 neurons were not overlapping (Figure S2), confirming the conclusion that inhibitory connections are reduced in the CNP‐DN‐erbB4 mice, and that the excitatory/inhibitory balance is shifted towards excitation (Figure S3).
FIGURE 4.
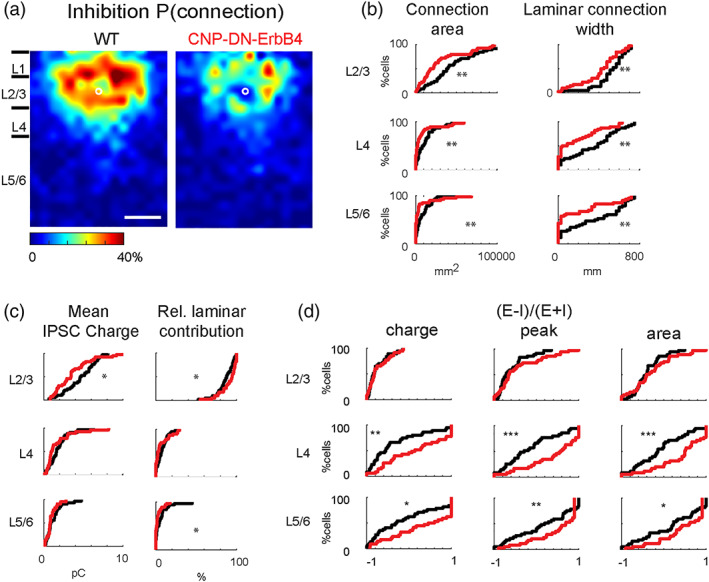
Reduced connectivity of inhibitory circuits to L2/3 neurons in A1 of CNP‐DN‐ErbB4 mice. (a) Spatial connection probability of inhibitory connections for WT (left) and CNP‐DN‐ErbB4 (right) mice. The border bars are the averaged borders across all the cells in each group. Scale bar is 200 μm long. (b) CDF plots of LSPS data for inhibitory connections. Connection area and laminar connection width of inhibitory inputs from L2/3 (p = 5.1 10–3, p = 8.5 10–3), L4 (p = 4.5 10–3, p = 4.5 10–3), and L5/6 (p = 3.7 10–3, p = 2.9 10–3) to L2/3 neurons of WT (black) and CNP‐DN‐ErbB4 (red) mice (Rank Sum test). (c) CDF plots of LSPS data for inhibitory connections. Mean charge (left) and relative laminar charge contribution of inhibitory inputs from L2/3 (p = .02, p = .031), L4 (p = .23, p = .11), and L5/6 (p = .36, p = .021) to L2/3 neurons of WT (black) and CNP‐DN‐ErbB4 (red) mice. (Rank Sum test). Loss of myelin reduces the inhibitory inputs from all layers. (d) CDF plots of excitatory/inhibitory ([E – I]/[E + I]) balance for inputs from L2/3 (all p > .05), L4 (p = 2.4 10–3, p = 6.2 10–4, p = 7.6 10–4, and L5/6 (p = .012, p = 6.9 10–3, p = .011) based on charge (left), peak (middle) and area (right) (Rank Sum test).
Individual cells can vary in their inputs and functional circuit diversity can emerge through development (Meng et al., 2020). We thus investigated the functional spatial diversity of the circuits impinging on L2/3 cells by calculating the similarity (spatial correlation) between connection maps within each population (Meng et al., 2020). We found that the circuit similarity is increased for both excitatory and inhibitory circuits in cells from CNP‐DN‐ErbB4 mice (Figure 5, Excitatory correlation: p = 3.5 × 10−3; Inhibitory correlation: 7.4 × 10−24). Thus, loss of oligodendrocyte ErbB signaling leads to a hypoconnectivity of inhibitory circuits and reorganized excitatory circuits.
FIGURE 5.
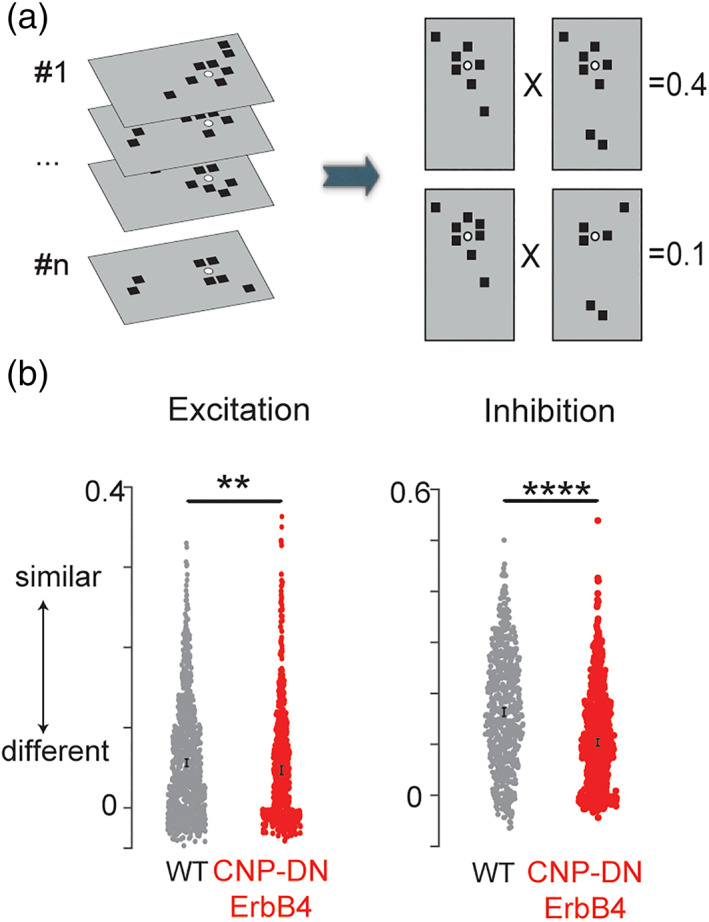
Loss of myelin increased the diversity of excitatory and inhibitory functional circuits. (a) Graphical representation of calculation of pairwise correlation between functional connection maps. Each black square represents the area that has monosynaptic connection to the recorded cell. Each connection map will be first vectorized and pairwise correlation between all the vectors will be calculated. (b) The mean and 95% bootstrap confidence interval for 10,000 resampled means of pairwise correlations of both excitatory (left) and inhibitory (right) maps for WT (black) and CNP‐DN‐ErbB4 (red) mice. Lack of myelin reduced the pairwise correlation of both excitatory and inhibitory maps, **p < .01, ****p < .0001.
3.5. Hypomyelination does not alter expression of inhibitory or excitatory synaptic components but reduces PV mRNA levels and PV + cells number in the auditory cortex
Our functional studies show changes in inhibitory circuits. Since ErbB receptor signaling has been reported to regulate neurotransmitter receptor expression in a diverse set of cells (Gerecke et al., 2001), we tested if the levels of mRNA for several genes central to inhibitory and excitatory neurotransmission are altered in A1 in CNP‐DN‐ErbB4 mice. Quantitative RT‐PCR analysis revealed normal mRNA levels for molecules associated with GABAergic neurotransmission (GAD65, GAD67, VGAT and GABARα1) (Figure 6a), and glutamatergic neurotransmission (VGLUT1, VGLUT2, GRIA1, GRIA2, GRIA3, and GRIA4) (Figure 6b). Thus, the observed circuit differences do not appear to result from alteration in expression of components of GABAergic or glutamatergic synapses.
FIGURE 6.
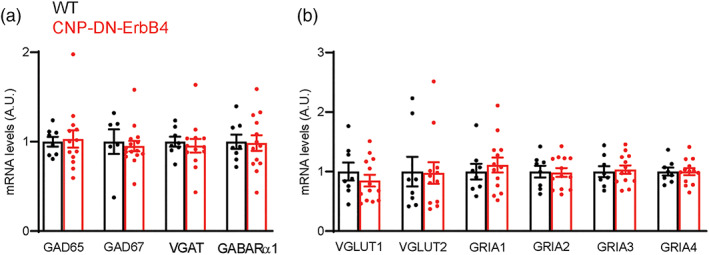
mRNA levels for excitatory and inhibitory synaptic proteins is normal in A1 of CNP‐DN‐ErbB4 mice. mRNA expression for synaptic proteins in A1 of WT (black) and CNP‐DN‐ErbB4 (red) male mice were measured by real‐time RT‐PCR. (a) GABAergic markers: GAD65 (p = .9574), GAD67 (p = .8027), VGAT (p = .7639) and GABARα1 (p = .4925). (b) Glutamatergic markers: VGLUT1 (p = .1843), VGLUT2 (p = .2377), GRIA1 (p = .8244), GRIA2 (p = .7100), GRIA3 (p = .5533) and GRIA4 (p = 6197). Unpaired two‐tailed Student's t test was performed. Data are expressed as mean ± SEM. No differences were found between genotypes (n = 8–12).
We then wondered if the observed reduction in functional connections could be due to alterations in specific types of inhibitory neurons. To test this scenario, we first measured the mRNA levels of genes that mark the three main populations of inhibitory interneurons, that is, parvalbumin (PV), vasoactive intestinal peptide (VIP) and somatostatin (SST). Only PV mRNA levels were reduced in A1 of CNP‐DN‐ErbB4 mice (Figure 7a). We found a strong correlation between the levels of PV and MBP mRNA in A1 (p = .0064) (Figure 7b), suggesting that key aspects of PV neurons are linked to myelination. Consistent with the reduction in PV mRNA, PV immunostaining showed that the density of A1 PV+ neuron cell bodies is reduced in CNP‐DN‐ErbB4 mice (Figure 7c,d). Moreover, intensity of PV immunostaining in the neuropil is also lower, suggesting that the number of PV+ axons is reduced as well (Figure 7e–h).
FIGURE 7.
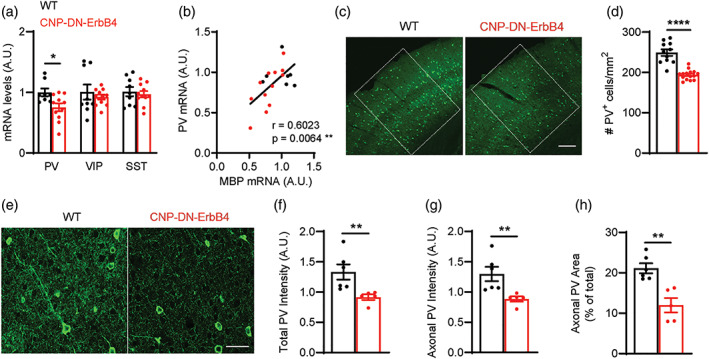
Loss of oligodendrocyte ErbB signaling leads to reduced A1 PV+ cell density and PV mRNA levels. (a) mRNA levels for parvalbumin (PV), vasoactive intestinal peptide (VIP) and somatostatin (SST) in the A1 of WT (black) and CNP‐DN‐ErbB4 (red) mice. PV mRNA levels are reduced in A1 of CNP‐DN‐ErbB4 mice (n = 8–11; p = .0290) whereas no changes are observed in VIP (p = .51) and STT (p = .6301) mRNA. (b) PV mRNA levels correlate with MBP mRNA levels in A1 (WT: black dots; CNP‐DN‐ErbB4: red dots; Pearson's correlation r = 0.6023; p = .0064). (c) Representative photomicrographs of A1 sections from WT and CNP‐DN‐ErbB4 mice showing PV+ cells (green). Scale bar: 100 μm. (d) A1 PV+ cell density is reduced in CNP‐DN‐ErbB4 mice (n = 10–14; p < .0001). (e) Representative photomicrographs of A1 sections from WT and CNP‐DN‐ErbB4 mice showing PV+ neurons and axons (green). Scale bar: 20 μm. (f) Quantification of total PV staining intensity (cell bodies + axons) in WT (black) and CNP‐DN‐ErbB4 (red) mice (n = 5–6, p = .0043). (g) Quantification of axonal PV staining intensity (total PV intensity – cell body PV intensity) in WT (black) and CNP‐DN‐ErbB4 (red) mice (n = 5–6, p = .0043). (h) Quantification of the area covered by PV+ axons in WT (black) and CNP‐DN‐ErbB4 (red) mice (n = 5–6; p = .0087) (A.U.: arbitrary units). Unpaired two‐tailed Student's t test or Mann–Whitney test were performed. Data are expressed as mean ± SEM.
To determine if the reduced A1 PV+ neuron density in CNP‐DN‐ERrB4 mice is due to neuronal loss or altered PV expression, we co‐labeled A1 sections with PV antibodies and VVA, a lectin that preferentially binds to the extracellular matrix enwrapping PV neurons (Drake et al., 1991; Luth et al., 1992) (Figure 8a). PV+ and VVA+ cells were identified by confocal microscopy. Quantitative analysis showed that, whereas PV+ neuron density is significantly reduced in CNP‐DN‐ErbB4 mice, VVA+ neuron density remains unchanged (Figure 8b,c). Importantly, there was a clear impact on the pattern of PV and VVA double labeled neurons. While 84% of VVA+ cells also expressed PV in controls, only 67% did in CNP‐DN‐ErbB4 mice (Figure 8d). In contrast, the proportion of PV+ cells that were also VVA+ was not altered by the hypomyelination (87.8% in controls and 86% in CNP‐DN‐ErbB4 mice) (Figure 8e). These results show that the reduction in A1 PV+ neurons is due to reduced PV expression by some interneurons rather than their loss. Together, these results suggest that A1 hypomyelination affects PV neurons, and that myelination and PV expression are linked.
FIGURE 8.
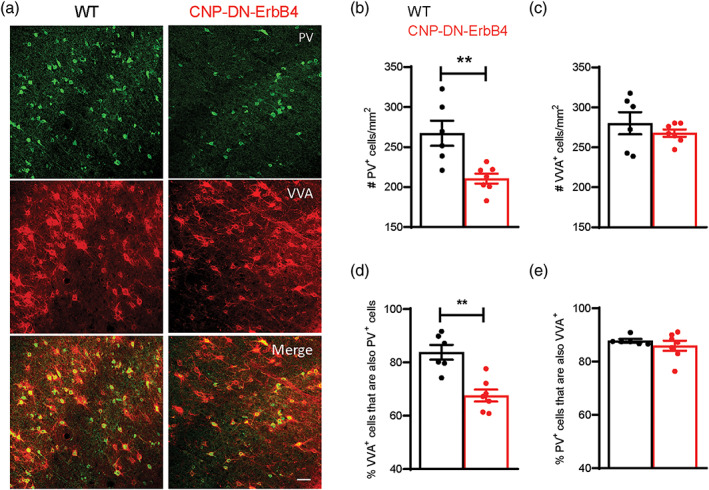
Hypomyelination leads to reduced density of PV+ neurons but normal density of VVA+ cells in A1. (a) Representative photomicrographs of A1 sections from WT and CNP‐DN‐ErbB4 mice showing PV+ (green) and VVA+ (red) immunostaining. Scale bar: 50 μm. (b) A1 PV+ cell density is reduced in CNP‐DN‐ErbB4 mice (n = 6–7; p = .0058). (c) A1 VVA+ cell density is not altered in CNP‐DN‐ErbB4 mice (n = 6–7; p = .60). (d) The fraction of VVA+ neurons also expressing PV is reduced in CNP‐DN‐erbB4 mice (67.5%) in comparison to WT (84%) (n = 6–7; p = .0023). (e) The fraction of PV+ neurons also expressing VVA is similar between groups (WT: 87.8%; CNP‐DN‐ErbB4: 86% (n = 6–7; p = .73). Mann–Whitney test was performed. Data are expressed as mean ± SEM.
3.6. Loss of oligodendrocyte ErbB signaling due to expression of DN‐ErbB4 reduces the number of myelinated PV + axons in the A1
To determine if A1 PV+ interneurons are hypomyelinated in CNP‐DN‐ErbB4, we quantified the density of PV+ axonal segments that are ensheated by MBP using immunofluorescence and confocal imaging as done previously by others (Stedehouder et al., 2017; Stedehouder et al., 2019) (Figure 9a,b). We found a 57% reduction in the number of PV+/MBP+ axonal segments in A1 of CNP‐DN‐ErbB4 mice (Figure 9a,c). Furthermore, we observed a strong correlation between the intensity of PV and MBP signal in A1 axons (r = 0.6021, p‐value .0001) (Figure 9d), and between the area covered by PV+ and MBP+ in the neuropil (r = 0.6373, p < .0001) (Figure 9e), supporting the notion that myelination of axons of PV+ neurons are negatively impacted by disruption of ErbB receptor signaling and might explain the changes in E/I balance in the A1 of CNP‐DN‐ErbB4 mice.
FIGURE 9.
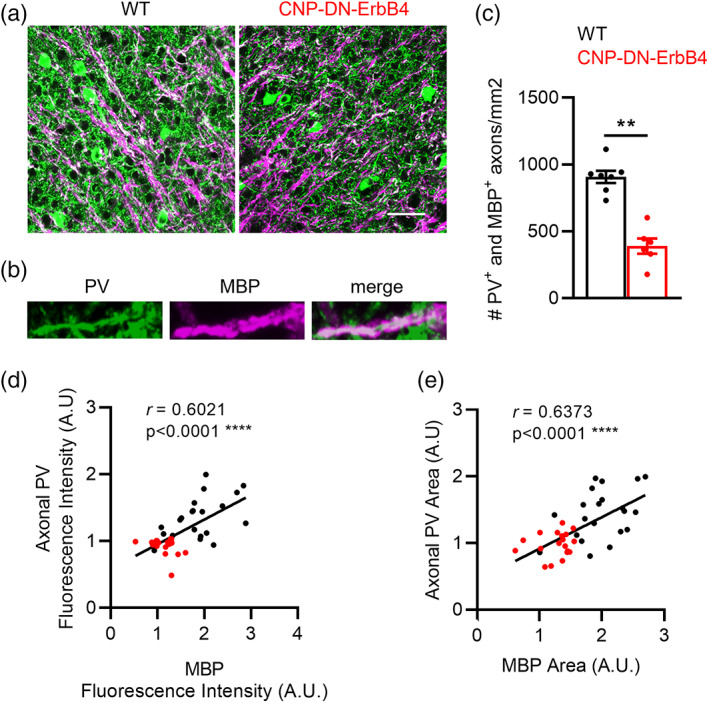
Loss of oligodendrocyte ErbB signaling leads to reduced number of A1 PV+ axons associated with MBP staining, reduced intensity of PV axonal immunostaining, and the area of PV+ axons. The intensity and area of PV+ axons correlate with the intensity and area of their associated MBP immunostaining. (a) Representative photomicrographs of A1 sections from WT and CNP‐DN‐ErbB4 mice showing PV+ (green) and MBP+ (magenta) immunostaining. Scale bar: 20 μm. (b) Representative photomicrographs of 2× zoom images from the 63× images shown in panel. (c) Density of PV‐MBP double‐labeled axons is reduced in A1 of CNP‐DN‐ErbB4 mice (n = 6–7; p = .0012). (d) A1 axonal PV fluorescence intensity correlates with MBP fluorescence intensity (WT: black dots, CNP‐DN‐erbB4: red dots; Pearson's correlation r = 0.6021; p < .0001). (e) A1 axonal PV area correlates with MBP area (WT: black dots, CNP‐DN‐erbB4: red dots; Pearson's correlation r = 0.6373; p < .0001). Mann–Whitney test was performed. Data are expressed as mean ± SEM.
3.7. Loss of oligodendrocyte ErbB signaling leads to reduction in MBP associated with both PV + and PV − axons
Since cortical myelin is present on axons of both excitatory neurons and PV+ interneurons (Micheva et al., 2016; Peters & Proskauer, 1980; Tamas et al., 1997), it was important to determine if loss of oligodendrocyte ErbB signaling affects these axons differentially. To this end, we used quantitative immunofluorescence on a new set of samples (Figure 10). As we found earlier (Figure 1), the area covered by MBP immunoreactivity was reduced in A1 of CNP‐DN‐ErbB4 mice (Figure 10a,b). To identify the portion of the image reflecting MBP associated with PV‐ axons, we created a mask for all the PV+ pixels and subtracted it from the MBP image (Figure 10a). The area of MBP immunofluorescence associated with PV+ axons was calculated as the difference between total MBP area and the MBP not associated with PV signal. Both portions of the MBP signal were reduced in CNP‐DN‐ErbB4 (Figure 10c,d), suggesting that ErbB receptor signaling regulates myelination of both PV+ and PV‐ axons, the latter most probably representing axons from excitatory neurons. The area of MBP associated with PV+ pixels was reduced to a larger extent, most probably reflecting the reduced abundance of PV+ axons in CNP‐DN‐ErbB4. However, we cannot rule out the possibility that myelination of PV+ axons in A1 has a stronger dependence on ErbB signaling than excitatory axons.
FIGURE 10.
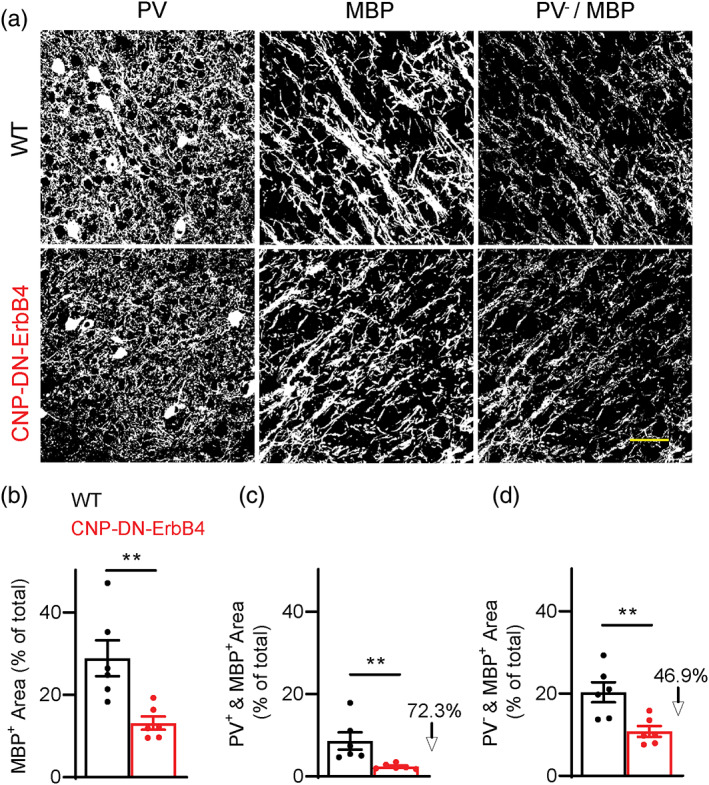
Loss of oligodendrocyte ErbB signaling leads to reduction in A1 MBP immunostaining both associated and not associated with PV immunostaining. (a) Representative photomicrographs of A1 sections from WT and CNP‐DN‐ErbB4 mice showing the area with PV immunostaining (left), the area with MBP immunostaining (middle) and the MBP+ area not associated with PV staining (right). Scale bar: 20 μm. (b) Percent area covered by total MBP signal in A1 of WT (black) and CNP‐DN‐ErbB4 (red) mice (n = 6; p = .0043). (c) Percent area covered by MBP signal associated with PV signal (PV+ & MBP+) in A1 of WT (black) and CNP‐DN‐ErbB4 (red) mice (n = 6; p = .0022). (d) Percent area covered by MBP signal not associated with PV signal (PV− & MBP+) in A1 of WT (black) and CNP‐DN‐ErbB4 (red) mice (n = 6; p = .0087). Mann–Whitney test was performed. Data are expressed as mean ± SEM.
3.8. Loss of oligodendrocyte ErbB signaling leads to reduced density of perisomatic PV+ synaptic terminals, without altering the density of GAD67+ terminals in A1
To assess changes in GABAergic circuits in mice with hypomyelination due to loss of oligodendrocyte ErbB signaling we used GAD67 and PV antibodies to label inhibitory nerve terminals in the A1 of WT and CNP‐DN‐ErbB4 mice. To avoid confounding signals from PV+ axons, we did not quantify the number of puncta in the neuropil, but rather the number of puncta surrounding the soma of pyramidal neurons as done previously by (Bragina et al., 2011; Li et al., 2017; Sommeijer & Levelt, 2012; Ueno et al., 2013). The density of perisomatic GAD67+ puncta was not altered by hypomyelination and loss of ErbB signaling. However, CNP‐DN‐ErbB4 mice showed a reduced density of total PV+ puncta, and reduced density of PV+ puncta colocalized with GAD67 (Figure 11a,b). The proportion of GAD67+ puncta that are also PV+ was significantly reduced (by 19%) in the A1 of CNP‐DN‐ErbB4 mice, compared with controls (Figure 11c). Accordingly, the proportion of GAD67+ puncta that is not PV+ was significantly increased (by 13%) in CNP‐DN‐ErbB4 mice (Figure 11d).
FIGURE 11.
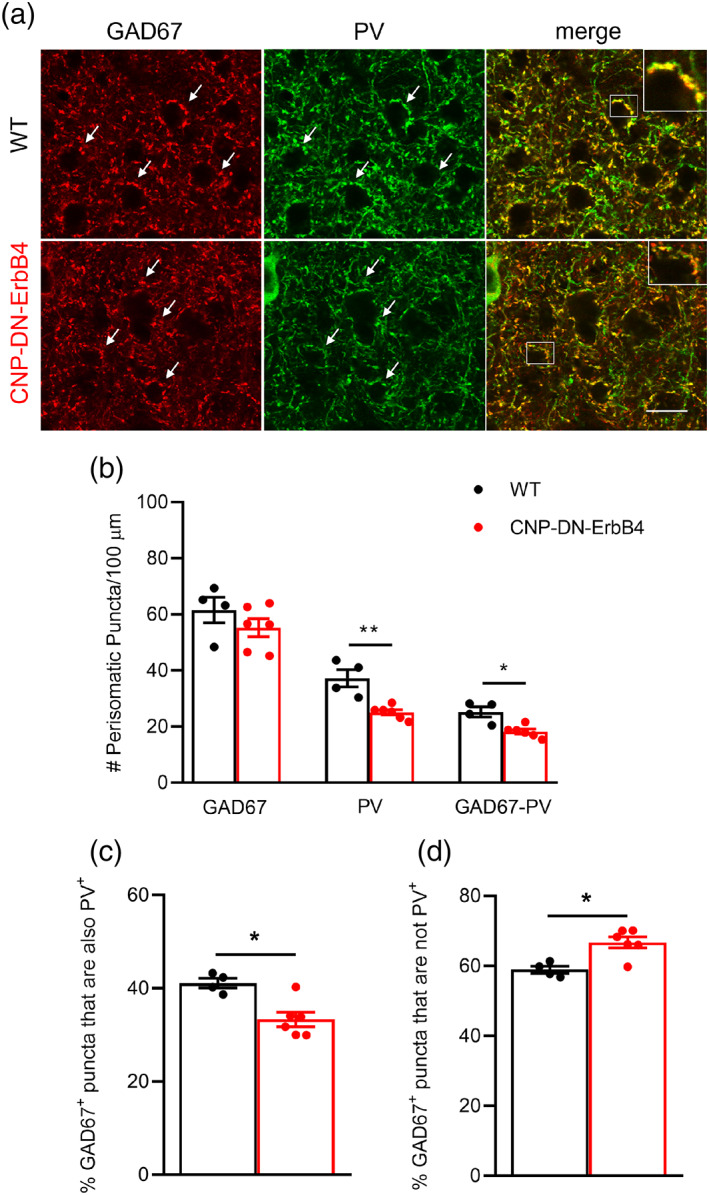
Loss of oligodendrocyte ErbB signaling leads to reduction in PV+, but not GAD67+ synaptic terminals in A1. (a) Representative photomicrographs of A1 sections from WT and CNP‐DN‐ErbB4 mice showing the GAD67 (red), the PV (green) and colocalized GAD67‐PV (merge) perisomatic puncta. Scale bar: 10 μm. (b) Number of GAD67+, PV+ and GAD67+‐PV+ perisomatic puncta/100 μm in A1 of WT (black) and CNP‐DN‐ErbB4 (red) mice (n = 4–6; **p = .0095; *p = .0190). (c) Percent % GAD67+ puncta that are also PV+ in A1 of WT (black) and CNP‐DN‐ErbB4 (red) mice (n = 4–6; p = .0381). (d) Percent % GAD67+ puncta that are not PV+ in A1 of WT (black) and CNP‐DN‐ErbB4 (red) mice (n = 4–6; p = .0381). Mann–Whitney test was performed. Data are expressed as mean ± SEM.
3.9. Oligodendrocyte‐specific ErbB3 KO has the same effects on A1 myelination and PV+ neuron number as oligodendrocyte DN‐ErbB4 expression
To test if the effects of hypomyelination on A1 interneurons occur in a different mouse model, we used a line in which we induce loss of oligodendrocyte ErbB signaling by a distinct approach, that is, inducible oligodendrocyte specific knockout of ErbB3, that is, mice homozygous for an ErbB3 floxed allele (ErbB3fl/fl) (Qu et al., 2006) and expressing the tamoxifen‐inducible Cre recombinase (CreERT) under the control of the PLP1 promoter (PLP/creERT) (Doerflinger et al., 2003). We previously showed that these mice have CNS hypomyelination comparable to that seen in the CNP‐DN‐ErbB4 mice in other CNS regions (Makinodan et al., 2012), consistent with the overlapping expression of CNPase and Plp1 in mature oligodendrocytes (Marques et al., 2016).
Tamoxifen‐induced oligodendrocyte ErbB3 KO led to a 28% reduction in A1 ErbB3 mRNA levels in the adult (Figure 12a), consistent with loss of ErbB3 in oligodendrocytes, but not in the other cells expressing this receptor, including microglia, astrocytes, tanycytes, and some neurons (Gerecke et al., 2001). Since PV+ neurons do not ErbB3 receptors (Mayer et al., 2018; Que et al., 2021; Yau et al., 2003) and Plp1 is expressed in myelinating cells (Marques et al., 2016), it is extremely unlikely that PV ErbB receptor signaling was altered in these mice. Oligodendrocyte ErbB3 KO led to a reduction in A1 MBP mRNA levels, indicative of hypomyelination (Figure 12a). Like in CNP‐DN‐ErbB4 mice, oligodendrocyte ErbB3 KO also led to a reduction in A1 PV mRNA levels without affecting VIP and SST expression (Figure 12b). In this model we again found a strong correlation between PV and MBP mRNA levels in A1 (r = 0.7787, p = .01; Figure 12c). The number of PV+ cells in the A1 of PLP‐ErbB3 KO mice was also significantly reduced respectively to controls (Figure 12d,e).
FIGURE 12.
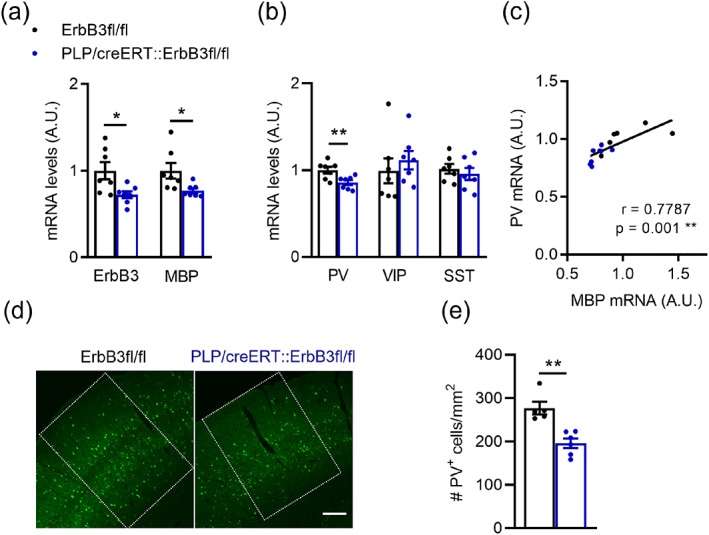
Loss of oligodendrocyte ErbB signaling by ErbB3 KO leads to reduced A1 MBP and PV mRNA levels and PV+ cell density. (a) Reduced ErbB3 (n = 7; p = .0230) and MBP (n = 7; p = .0175) mRNA levels in A1 after oligodendrocyte‐specific ErbB3 KO (PLP/creERT::ErbB3fl/fl). (b) Relative mRNA expression for parvalbumin (PV), vasoactive intestinal peptide (VIP) and somatostatin (SST) in the A1 of WT (black) and PLP/creERT::ErbB3fl/fl (blue) mice. PV mRNA levels are reduced in A1 of PLP/creERT::ErbB3fl/fl mice (n = 6–7; p = .0093). No changes are observed in VIP (p = .3176) and STT (p = .62) mRNA expression. (c) A1 PV mRNA levels correlate with MBP mRNA levels (WT: black dots, PLP/creERT::ErbB3fl/fl: blue dots; Pearson's correlation r = 0.7787; p = .001). (d) Representative photomicrographs of A1 sections from WT and PLP/creERT::ErbB3fl/fl mice showing PV+ cells (green). Scale bar: 100 μm. (e) A1 PV+ cell density is reduced in PLP/creERT::ErbB3fl/fl mice (n = 5–6; p = .0043). Mann–Whitney test was performed. Data are expressed as mean ± SEM.
Finally, to test if the impact of loss of oligodendrocyte ErbB receptor signaling on PV+ neurons is specific to A1, we quantified the number of PV+ cells in two other brain areas, the medial prefrontal cortex (PFC) and the primary motor cortex (M1), for both mutant lines and their respective wild type littermates. Consistent with the findings from A1, both CNP‐DN‐ErbB4 (Figure 13a–d) and PLP‐ErbB3 KO mice (Figure 13e–h) showed reduced number of PV cells in the PFC and M1, compared to their littermate controls. These data indicate that hypomyelination leads to alterations in the number of PV‐expressing neurons in the whole brain.
FIGURE 13.
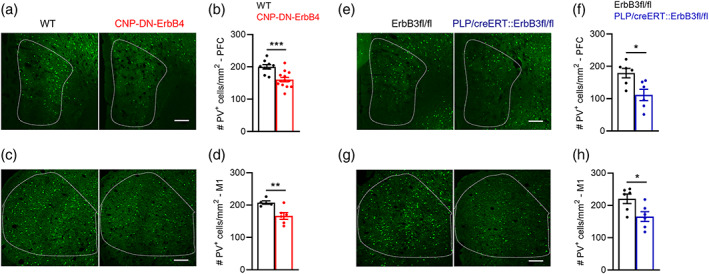
Loss of oligodendrocyte ErbB signaling by expression of DN‐ErbB4 or ErbB3 KO leads to reduced PV cell density in other brain areas such as prefrontal cortex and primary motor cortex. (a) Representative photomicrographs of prefrontal cortex (PFC) sections from WT and CNP‐DN‐ErbB4 mice showing PV+ cells (green). Scale bar: 100 μm. (b) PFC PV+ cell density is reduced in PFC of CNP‐DN‐ErbB4 mice (n = 9–13; p = .0009). (c) Representative photomicrographs of primary motor cortex (M1) sections from WT and CNP‐DN‐ErbB4 mice showing PV+ cells (green). Scale bar: 100 μm. (d) PFC PV+ cell density is reduced in M1 of CNP‐DN‐ErbB4 mice (n = 5–6; p = .0083). (e) Representative photomicrographs of prefrontal cortex (PFC) sections from WT and PLP/creERT::ErbB3fl/fl mice showing PV+ cells (green). Scale bar: 100 μm. (f) PFC PV+ cell density is reduced in PFC of PLP/creERT::ErbB3fl/fl mice (n = 6; p = .0411). (g) Representative photomicrographs of primary motor cortex (M1) sections from WT and PLP/creERT::ErbB3fl/fl mice showing PV+ cells (green). Scale bar: 100 μm. (h) PFC PV+ cell density is reduced in M1 of PLP/creERT::ErbB3fl/fl (n = 6; p = .0260). Mann–Whitney test was performed. Data are expressed as mean ± SEM.
4. DISCUSSION
Our results indicate that hypomyelination leads to altered excitatory/inhibitory balance in A1 cortical circuits, resulting in a shift towards excitation. Importantly, the mutant mice do not have visible seizures or overt symptoms related to hyper‐excitation, indicating that the changes in excitatory/inhibitory balance are mild. The observed functional changes correlate with a reduced density of PV+ neurons, which, as illustrated by the preservation of VVA+ neuron density and the number of GAD67+ terminals, reflects a loss of PV expression by some of the interneurons rather than the loss of the inhibitory neurons themselves. PV expression levels are regulated by neuronal activity (Dehorter et al., 2015; Favuzzi et al., 2017; Lagler et al., 2016; Stedehouder et al., 2018), and it has been proposed that myelin regulates PV+ neurons metabolism as well as improves the energy efficiency of signal propagation (Lee et al., 2012; Micheva et al., 2016; Rinholm et al., 2011). Thus, our results suggest that hypomyelination causes PV+ neuron hypoactivity, leading to the observed reduction in PV expression and inhibitory hypoconnectivity.
The tight correlation between the levels of MBP and PV expression point to a powerful regulation of PV+ neuron function and gene expression by myelin. The observation that PV+ neuron density is also reduced in the prefrontal and motor cortices suggest that the impact of hypomyelination on PV+ neuron function is likely to occur in the whole brain. While our results suggest a direct relationship between the changes in PV neurons and the hypomyelination, we cannot rule out that changes in ErbBR signaling alters expression of oligodendrocyte‐derived factors that participate in the regulation of inhibitory neuron function.
The observation that loss of oligodendrocyte ErbBR signaling alters excitatory/inhibitory balance in the cortex are similar to those made in mice with KO of the GABAA receptor γ2 subunit in NG2+ cells, which include oligodendrocyte precursor cells (Benamer et al., 2020). This study showed that barrel‐cortex fast spiking interneurons in the GABAAR γ2 subunit KO mice present with severe myelin and axonal defects, as well as alterations in their function, leading to excitation‐inhibition imbalance. Together, these observations suggest that communication between PV+ neurons and cells of the oligodendrocyte lineage are complex and could involve several signaling molecules and processes.
The finding that hypomyelination leads to reduced PV+ neuron density but normal density of VVA+ cells is remarkably similar to what has been reported for several mutant mice that display autism spectrum disorder (ASD) endophenotypes, that is, PV+/−, Shank1−/− and Shank3B−/− mice (Filice et al., 2016). Interestingly, myelin defects have been found in ASD patients (Graciarena et al., 2018; Steinman & Mankuta, 2019), which also display auditory processing deficits (Jones et al., 2020; Rotschafer, 2021; Srinivasan et al., 2020). Together, these observations raise the possibility that defects in myelination might contribute to some aspects of ASD by their negative impact on inhibitory networks.
The LSPS experiments show a significant spatial hypoconnectivity of inhibitory connections to L2/3 neurons in mice which hypomyelination, that is, individual L2/3 neurons in the mutant mice receive inhibitory inputs from fewer locations, especially from L2/3 and L4. However, individual inhibitory connections are of comparable strength, indicating that hypomyelination causes a loss but not a weakening of connections. This is consistent with the observation that some GAD67+ synapses lost PV expression, suggesting they are inactive, but others appear to express normal levels of both GAD67 and PV. In contrast, excitatory connectivity in A1 appears to be normal. However, we cannot exclude the possibility that the effects of hypomyelination on excitatory axons are subtle and not resolved by our LSPS method. For example, hypomyelination could lead to increased conduction times in excitatory axons without loss of connections and cells, but since LSPS probes relatively short‐range connections, such changes might be too small to be detected. Moreover, LSPS cannot reveal connections from close‐by neurons.
The specific effects of myelin on inhibitory circuits suggest that myelin might play multiple roles beyond controlling timing of axonal conduction. Our results suggest that hypomyelination affects axons of PV+ cells, which are fast‐spiking neurons with metabolic demands higher than those of excitatory axons (Hu et al., 2014). In fact, the number of myelinated PV axons in A1 is reduced in the mutant mice, and the axonal PV fluorescence intensity and area strongly correlate with MBP fluorescence intensity and area. The reduced density of PV+ synaptic terminals without changes in overall GAD67+ terminals reinforce the link between hypomyelination due to loss of oligodendrocyte ErbBR signaling and the physiology of parvalbumin interneurons. Of note, no alteration of expression of markers for two other important populations of inhibitory interneurons, the VIP and somatostatin, was found in A1 of mice with hypomyelination. This agrees with previous reports showing no myelin in VIP+ neurons and a rare myelination of somatostatin+ neurons in the somatosensory cortex (Micheva et al., 2016).
Our findings of altered A1 circuits suggests that hypomyelination might lead to degraded fidelity of sound representation. This is consistent with the observation that focal demyelination of A1 L4 alters auditory frequency‐specific responses (Narayanan et al., 2018) and abolishes tonotopic organization (Cerina et al., 2017). Since A1 neurons that encode the fast fluctuation of acoustic stimuli tend to receive balanced and concurrent excitation and inhibition (Bendor, 2015; Wang et al., 2008), our observation of altered inhibitory circuits in CNP‐DN‐ErbB4 mice are consistent with the functional deficits after hypomyelination. Thus, hypomyelination could play a role in the development of cognitive deficits. Indeed, we previously showed that CNP‐DN‐ErbB4 mice have behavioral phenotypes consistent with psychiatric disorders (Roy et al., 2007) and interneuron hypomyelination was reported in rat models of schizophrenia (Maas et al., 2020; Stedehouder & Kushner, 2017). Hence, our results showing reduced density of PV+ cells and altered network and excitatory/inhibitory balance suggest that the absence of myelin might lead to circuit changes that manifest as impaired cognitive functions. Furthermore, the observation that the density of VVA+ neurons and GAD67+ terminal in A1 is normal in the mutant mice indicates that the PV neurons affected by the hypomyelination remain and likely have normal connectivity, raising the possibility that, if myelination could be restored, they could regain their normal activity patterns.
CONFLICT OF INTEREST
Gabriel Corfas is a scientific founder of Decibel Therapeutics, has an equity interest in and has received compensation for consulting. The company was not involved in this study.
Supporting information
Figure S1 Excitatory circuits to L2/3 neurons are unchanged in the A1 of CNP‐DN‐ErbB4 mice. Histograms of excitatory connections (up left), laminar connection width (up right), mean charge (bottom left) and relative laminar charge (bottom right) of excitatory inputs from L2/3, L4 and L5/6 to L2/3 neurons of WT (black) and CNP‐DN‐ErbB4 (red) mice. The black and red lines are the histogram fit with kernel function for WT (black) and CNP‐DN‐ErbB4 (red) mice. The data depicted in these graphs is the same as in Figure 3e.
Figure S2 Inhibitory circuits to L2/3 neurons are altered in the A1 of CNP‐DN‐ErbB4 mice. Histograms of inhibitory connections (up left), laminar connection width (up right), mean charge (bottom left) and relative laminar charge (bottom right) of inhibitory inputs from L2/3, L4 and L5/6 to L2/3 neurons of WT (black) and CNP‐DN‐ErbB4 (red) mice. The black and red lines are the histogram fit with kernel function for WT (black) and CNP‐DN‐ErbB4 (red) mice. The data depicted in these graphs is the same as in Figure 4b,c.
Figure S3 CNP‐DN‐ErbB4 mice show a shift of the excitatory/inhibitory balance towards excitation. Histograms of excitatory/inhibitory balance showing the mean charge, peak and area of inputs from L2/3, L4 and L5/6 to L2/3 neurons of WT (black) and CNP‐DN‐ErbB4 (red) mice. The black and red lines are the histogram fit with kernel function for WT (black) and CNP‐DN‐ErbB4 (red) mice. The data depicted in these graphs is the same as in Figure 4d.
ACKNOWLEDGMENTS
This work was supported in part by NIH/NIDCD R01DC018500 (Gabriel Corfas) and NIH/NIDCD R01DC009607 (Patrick O. Kanold). We thank Ilhan Onder for helping with MATLAB and Michael Hayes for helping with mouse colony management.
Borges, B. C. , Meng, X. , Long, P. , Kanold, P. O. , & Corfas, G. (2023). Loss of oligodendrocyte ErbB receptor signaling leads to hypomyelination, reduced density of parvalbumin‐expressing interneurons, and inhibitory function in the auditory cortex. Glia, 71(2), 187–204. 10.1002/glia.24266
Beatriz C. Borges and Xiangying Meng contributed equally to this study.
Patrick O. Kanold and Gabriel Corfas are co‐senior authors.
Funding information National Institute on Deafness and Other Communication Disorders, Grant/Award Numbers: R01DC009607, R01DC018500
Contributor Information
Patrick O. Kanold, Email: pkanold@jhu.edu.
Gabriel Corfas, Email: corfas@med.umich.edu.
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in the manuscript. Original blots and images used for calculations, Matlab codes, and maps into 3D matrices used in electrophysiology experiments will be uploaded to Dryad.
REFERENCES
- Benamer, N. , Vidal, M. , Balia, M. , & Angulo, M. C. (2020). Myelination of parvalbumin interneurons shapes the function of cortical sensory inhibitory circuits. Nature Communications, 11(1), 5151. 10.1038/s41467-020-18984-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendor, D. (2015). The role of inhibition in a computational model of an auditory cortical neuron during the encoding of temporal information. PLoS Computational Biology, 11(4), e1004197. 10.1371/journal.pcbi.1004197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragina, L. , Fattorini, G. , Giovedi, S. , Melone, M. , Bosco, F. , Benfenati, F. , & Conti, F. (2011). Analysis of synaptotagmin, SV2, and Rab3 expression in cortical glutamatergic and GABAergic axon terminals. Frontiers in Cellular Neuroscience, 5, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerina, M. , Narayanan, V. , Gobel, K. , Bittner, S. , Ruck, T. , Meuth, P. , & Meuth, S. G. (2017). The quality of cortical network function recovery depends on localization and degree of axonal demyelination. Brain, Behavior, and Immunity, 59, 103–117. 10.1016/j.bbi.2016.08.014 [DOI] [PubMed] [Google Scholar]
- Chen, S. , Velardez, M. O. , Warot, X. , Yu, Z. X. , Miller, S. J. , Cros, D. , & Corfas, G. (2006). Neuregulin 1‐erbB signaling is necessary for normal myelination and sensory function. The Journal of Neuroscience, 26(12), 3079–3086. 10.1523/jneurosci.3785-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehorter, N. , Ciceri, G. , Bartolini, G. , Lim, L. , del Pino, I. , & Marin, O. (2015). Tuning of fast‐spiking interneuron properties by an activity‐dependent transcriptional switch. Science, 349(6253), 1216–1220. 10.1126/science.aab3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger, N. H. , Macklin, W. B. , & Popko, B. (2003). Inducible site‐specific recombination in myelinating cells. Genesis, 35(1), 63–72. 10.1002/gene.10154 [DOI] [PubMed] [Google Scholar]
- Drake, C. T. , Mulligan, K. A. , Wimpey, T. L. , Hendrickson, A. , & Chavkin, C. (1991). Characterization of Vicia villosa agglutinin‐labeled GABAergic interneurons in the hippocampal formation and in acutely dissociated hippocampus. Brain Research, 554(1–2), 176–185. 10.1016/0006-8993(91)90186-y [DOI] [PubMed] [Google Scholar]
- Du, F. , & Ongur, D. (2013). Probing myelin and axon abnormalities separately in psychiatric disorders using MRI techniques. Frontiers in Integrative Neuroscience, 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favuzzi, E. , Marques‐Smith, A. , Deogracias, R. , Winterflood, C. M. , Sanchez‐Aguilera, A. , Mantoan, L. , & Rico, B. (2017). Activity‐dependent gating of parvalbumin interneuron function by the perineuronal net protein brevican. Neuron, 95(3), 639–655, e610. 10.1016/j.neuron.2017.06.028 [DOI] [PubMed] [Google Scholar]
- Filice, F. , Vorckel, K. J. , Sungur, A. O. , Wohr, M. , & Schwaller, B. (2016). Reduction in parvalbumin expression not loss of the parvalbumin‐expressing GABA interneuron subpopulation in genetic parvalbumin and shank mouse models of autism. Molecular Brain, 9, 10. 10.1186/s13041-016-0192-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, A. Y. , Bujalka, H. , & Emery, B. (2019). Axoglial interactions in myelin plasticity: Evaluating the relationship between neuronal activity and oligodendrocyte dynamics. Glia, 67(11), 2038–2049. 10.1002/glia.23629 [DOI] [PubMed] [Google Scholar]
- Gerecke, K. M. , Wyss, J. M. , Karavanova, I. , Buonanno, A. , & Carroll, S. L. (2001). ErbB transmembrane tyrosine kinase receptors are differentially expressed throughout the adult rat central nervous system. The Journal of Comparative Neurology, 433(1), 86–100. 10.1002/cne.1127 [DOI] [PubMed] [Google Scholar]
- Ghaffarian, N. , Mesgari, M. , Cerina, M. , Gobel, K. , Budde, T. , Speckmann, E. J. , & Gorji, A. (2016). Thalamocortical‐auditory network alterations following cuprizone‐induced demyelination. Journal of Neuroinflammation, 13(1), 160. 10.1186/s12974-016-0629-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, E. M. , Geraghty, A. C. , & Monje, M. (2018). Bad wrap: Myelin and myelin plasticity in health and disease. Developmental Neurobiology, 78(2), 123–135. 10.1002/dneu.22541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson, E. M. , Purger, D. , Mount, C. W. , Goldstein, A. K. , Lin, G. L. , Wood, L. S. , & Monje, M. (2014). Neuronal activity promotes oligodendrogenesis and adaptive myelination in the mammalian brain. Science, 344(6183), 1252304. 10.1126/science.1252304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graciarena, M. , Seiffe, A. , Nait‐Oumesmar, B. , & Depino, A. M. (2018). Hypomyelination and oligodendroglial alterations in a mouse model of autism spectrum disorder. Frontiers in Cellular Neuroscience, 12, 517. 10.3389/fncel.2018.00517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, H. , Gan, J. , & Jonas, P. (2014). Interneurons. Fast‐spiking, parvalbumin(+) GABAergic interneurons: From cellular design to microcircuit function. Science, 345(6196), 1255263. 10.1126/science.1255263 [DOI] [PubMed] [Google Scholar]
- Jones, M. K. , Kraus, N. , Bonacina, S. , Nicol, T. , Otto‐Meyer, S. , & Roberts, M. Y. (2020). Auditory processing differences in toddlers with autism spectrum disorder. Journal of Speech, Language, and Hearing Research, 63(5), 1608–1617. 10.1044/2020_JSLHR-19-00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagler, M. , Ozdemir, A. T. , Lagoun, S. , Malagon‐Vina, H. , Borhegyi, Z. , Hauer, R. , & Klausberger, T. (2016). Divisions of identified parvalbumin‐expressing basket cells during working memory‐guided decision making. Neuron, 91(6), 1390–1401. 10.1016/j.neuron.2016.08.010 [DOI] [PubMed] [Google Scholar]
- Lee, Y. , Morrison, B. M. , Li, Y. , Lengacher, S. , Farah, M. H. , Hoffman, P. N. , & Rothstein, J. D. (2012). Oligodendroglia metabolically support axons and contribute to neurodegeneration. Nature, 487(7408), 443–448. 10.1038/nature11314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Han, W. , Pelkey, K. A. , Duan, J. , Mao, X. , Wang, Y. X. , & Lu, W. (2017). Molecular dissection of neuroligin 2 and Slitrk3 reveals an essential framework for GABAergic synapse development. Neuron, 96(4), 808–826. 10.1016/j.neuron.2017.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Dietz, K. , DeLoyht, J. M. , Pedre, X. , Kelkar, D. , Kaur, J. , & Casaccia, P. (2012). Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nature Neuroscience, 15(12), 1621–1623. 10.1038/nn.3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luth, H. J. , Fischer, J. , & Celio, M. R. (1992). Soybean lectin binding neurons in the visual cortex of the rat contain parvalbumin and are covered by glial nets. Journal of Neurocytology, 21(3), 211–221. 10.1007/BF01194979 [DOI] [PubMed] [Google Scholar]
- Maas, D. A. , Eijsink, V. D. , Spoelder, M. , van Hulten, J. A. , De Weerd, P. , Homberg, J. R. , & Martens, G. J. M. (2020). Interneuron hypomyelination is associated with cognitive inflexibility in a rat model of schizophrenia. Nature Communications, 11(1), 2329. 10.1038/s41467-020-16218-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinodan, M. , Rosen, K. M. , Ito, S. , & Corfas, G. (2012). A critical period for social experience‐dependent oligodendrocyte maturation and myelination. Science, 337(6100), 1357–1360. 10.1126/science.1220845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques, S. , Zeisel, A. , Codeluppi, S. , van Bruggen, D. , Mendanha Falcao, A. , Xiao, L. , & Castelo‐Branco, G. (2016). Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science, 352(6291), 1326–1329. 10.1126/science.aaf6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, C. , Hafemeister, C. , Bandler, R. C. , Machold, R. , Batista Brito, R. , Jaglin, X. , & Satija, R. (2018). Developmental diversification of cortical inhibitory interneurons. Nature, 555(7697), 457–462. 10.1038/nature25999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie, I. A. , Ohayon, D. , Li, H. , de Faria, J. P. , Emery, B. , Tohyama, K. , & Richardson, W. D. (2014). Motor skill learning requires active central myelination. Science, 346(6207), 318–322. 10.1126/science.1254960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X. , Kao, J. P. , Lee, H. K. , & Kanold, P. O. (2015). Visual deprivation causes refinement of intracortical circuits in the auditory cortex. Cell Reports, 12(6), 955–964. 10.1016/j.celrep.2015.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X. , Solarana, K. , Bowen, Z. , Liu, J. , Nagode, D. A. , Sheikh, A. , & Kanold, P. O. (2020). Transient subgranular hyperconnectivity to L2/3 and enhanced pairwise correlations during the critical period in the mouse auditory cortex. Cerebral Cortex, 30(3), 1914–1930. 10.1093/cercor/bhz213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, X. , Winkowski, D. E. , Kao, J. P. Y. , & Kanold, P. O. (2017). Sublaminar subdivision of mouse auditory cortex layer 2/3 based on functional translaminar connections. The Journal of Neuroscience, 37(42), 10200–10214. 10.1523/JNEUROSCI.1361-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuth, S. G. , Bittner, S. , Ulzheimer, J. C. , Kleinschnitz, C. , Kieseier, B. C. , & Wiendl, H. (2010). Therapeutic approaches to multiple sclerosis: An update on failed, interrupted, or inconclusive trials of neuroprotective and alternative treatment strategies. BioDrugs, 24(5), 317–330. 10.2165/11537190-000000000-00000 [DOI] [PubMed] [Google Scholar]
- Micheva, K. D. , Wolman, D. , Mensh, B. D. , Pax, E. , Buchanan, J. , Smith, S. J. , & Bock, D. D. (2016). A large fraction of neocortical myelin ensheathes axons of local inhibitory neurons. eLife, 5, 1–29. 10.7554/eLife.15784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje, M. (2018). Myelin plasticity and nervous system function. Annual Review of Neuroscience, 41, 61–76. 10.1146/annurev-neuro-080317-061853 [DOI] [PubMed] [Google Scholar]
- Muralidharan, S. , Dirda, N. D. , Katz, E. J. , Tang, C. M. , Bandyopadhyay, S. , Kanold, P. O. , & Kao, J. P. (2016). NCM, a photolabile group for preparation of caged molecules: Synthesis and biological application. PLoS One, 11(10), e0163937. 10.1371/journal.pone.0163937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan, V. , Cerina, M. , Gobel, K. , Meuth, P. , Herrmann, A. M. , Fernandez‐Orth, J. , & Pape, H. C. (2018). Impairment of frequency‐specific responses associated with altered electrical activity patterns in auditory thalamus following focal and general demyelination. Experimental Neurology, 309, 54–66. 10.1016/j.expneurol.2018.07.010 [DOI] [PubMed] [Google Scholar]
- Peters, A. , & Proskauer, C. C. (1980). Synaptic relationships between a multipolar stellate cell and a pyramidal neuron in the rat visual cortex. A combined Golgi‐electron microscope study. Journal of Neurocytology, 9(2), 163–183. 10.1007/BF01205156 [DOI] [PubMed] [Google Scholar]
- Prevot, V. , Rio, C. , Cho, G. J. , Lomniczi, A. , Heger, S. , Neville, C. M. , & Corfas, G. (2003). Normal female sexual development requires neuregulin‐erbB receptor signaling in hypothalamic astrocytes. The Journal of Neuroscience, 23(1), 230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, S. , Rinehart, C. , Wu, H. H. , Wang, S. E. , Carter, B. , Xin, H. , & Arteaga, C. L. (2006). Gene targeting of ErbB3 using a Cre‐mediated unidirectional DNA inversion strategy. Genesis, 44(10), 477–486. 10.1002/dvg.20243 [DOI] [PubMed] [Google Scholar]
- Que, L. , Lukacsovich, D. , Luo, W. , & Foldy, C. (2021). Transcriptional and morphological profiling of parvalbumin interneuron subpopulations in the mouse hippocampus. Nature Communications, 12(1), 108. 10.1038/s41467-020-20328-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinholm, J. E. , Hamilton, N. B. , Kessaris, N. , Richardson, W. D. , Bergersen, L. H. , & Attwell, D. (2011). Regulation of oligodendrocyte development and myelination by glucose and lactate. The Journal of Neuroscience, 31(2), 538–548. 10.1523/JNEUROSCI.3516-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio, C. , Rieff, H. I. , Qi, P. , Khurana, T. S. , & Corfas, G. (1997). Neuregulin and erbB receptors play a critical role in neuronal migration. Neuron, 19(1), 39–50. 10.1016/s0896-6273(00)80346-3 [DOI] [PubMed] [Google Scholar]
- Rotschafer, S. E. (2021). Auditory discrimination in autism spectrum disorder. Frontiers in Neuroscience, 15, 651209. 10.3389/fnins.2021.651209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, K. , Murtie, J. C. , El‐Khodor, B. F. , Edgar, N. , Sardi, S. P. , Hooks, B. M. , & Corfas, G. (2007). Loss of erbB signaling in oligodendrocytes alters myelin and dopaminergic function, a potential mechanism for neuropsychiatric disorders. Proceedings of the National Academy of Sciences of the United States of America, 104(19), 8131–8136. 10.1073/pnas.0702157104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommeijer, J. P. , & Levelt, C. N. (2012). Synaptotagmin‐2 is a reliable marker for parvalbumin positive inhibitory boutons in the mouse visual cortex. PLoS One, 7(4), e35323. 10.1371/journal.pone.0035323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, V. , Udayakumar, N. , & Anandan, K. (2020). Influence of primary auditory cortex in the characterization of autism Spectrum in young adults using brain connectivity parameters and deep belief networks: An fMRI study. Current Medical Imaging, 16(9), 1059–1073. 10.2174/1573405615666191111142039 [DOI] [PubMed] [Google Scholar]
- Stankovic, K. M. , & Corfas, G. (2003). Real‐time quantitative RT‐PCR for low‐abundance transcripts in the inner ear: Analysis of neurotrophic factor expression. Hearing Research, 185(1–2), 97–108. 10.1016/s0378-5955(03)00298-3 [DOI] [PubMed] [Google Scholar]
- Stedehouder, J. , Brizee, D. , Shpak, G. , & Kushner, S. A. (2018). Activity‐dependent myelination of parvalbumin interneurons mediated by axonal morphological plasticity. The Journal of Neuroscience, 38(15), 3631–3642. 10.1523/JNEUROSCI.0074-18.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedehouder, J. , Brizee, D. , Slotman, J. A. , Pascual‐Garcia, M. , Leyrer, M. L. , Bouwen, B. L. , & Kushner, S. A. (2019). Local axonal morphology guides the topography of interneuron myelination in mouse and human neocortex. eLife, 8, 1–28. 10.7554/eLife.48615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedehouder, J. , Couey, J. J. , Brizee, D. , Hosseini, B. , Slotman, J. A. , Dirven, C. M. F. , & Kushner, S. A. (2017). Fast‐spiking parvalbumin interneurons are frequently myelinated in the cerebral cortex of mice and humans. Cerebral Cortex, 27(10), 5001–5013. 10.1093/cercor/bhx203 [DOI] [PubMed] [Google Scholar]
- Stedehouder, J. , & Kushner, S. A. (2017). Myelination of parvalbumin interneurons: A parsimonious locus of pathophysiological convergence in schizophrenia. Molecular Psychiatry, 22(1), 4–12. 10.1038/mp.2016.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman, G. , & Mankuta, D. (2019). Molecular biology of autism's etiology ‐ an alternative mechanism. Medical Hypotheses, 130, 109272. 10.1016/j.mehy.2019.109272 [DOI] [PubMed] [Google Scholar]
- Suter, B. A. , O'Connor, T. , Iyer, V. , Petreanu, L. T. , Hooks, B. M. , Kiritani, T. , Svoboda, K. , & Shepherd, G. M. (2010). Ephus: multipurpose data acquisition software for neuroscience experiments. Frontiers in Neural Circuits, 4, 100. 10.3389/fncir.2010.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamas, G. , Buhl, E. H. , & Somogyi, P. (1997). Fast IPSPs elicited via multiple synaptic release sites by different types of GABAergic neurone in the cat visual cortex. The Journal of Physiology, 500(3), 715–738. 10.1113/jphysiol.1997.sp022054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno, H. , Shoshi, C. , Suemitsu, S. , Usui, S. , Sujiura, H. , & Okamoto, M. (2013). Somatosensory and visual deprivation each decrease the density of parvalbumin neurons and their synapse terminals in the prefrontal cortex and hippocampus of mice. Acta Medica Okayama, 67(3), 135–143. 10.18926/AMO/50406 [DOI] [PubMed] [Google Scholar]
- Wang, X. , Lu, T. , Bendor, D. , & Bartlett, E. (2008). Neural coding of temporal information in auditory thalamus and cortex. Neuroscience, 154(1), 294–303. 10.1016/j.neuroscience.2008.03.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. M. , Michel, K. , Jokhi, V. , Nedivi, E. , & Arlotta, P. (2020). Neuron class‐specific responses govern adaptive myelin remodeling in the neocortex. Science, 370(6523), 1–10. 10.1126/science.abd2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau, H. J. , Wang, H. F. , Lai, C. , & Liu, F. C. (2003). Neural development of the neuregulin receptor ErbB4 in the cerebral cortex and the hippocampus: Preferential expression by interneurons tangentially migrating from the ganglionic eminences. Cerebral Cortex, 13(3), 252–264. 10.1093/cercor/13.3.252 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Excitatory circuits to L2/3 neurons are unchanged in the A1 of CNP‐DN‐ErbB4 mice. Histograms of excitatory connections (up left), laminar connection width (up right), mean charge (bottom left) and relative laminar charge (bottom right) of excitatory inputs from L2/3, L4 and L5/6 to L2/3 neurons of WT (black) and CNP‐DN‐ErbB4 (red) mice. The black and red lines are the histogram fit with kernel function for WT (black) and CNP‐DN‐ErbB4 (red) mice. The data depicted in these graphs is the same as in Figure 3e.
Figure S2 Inhibitory circuits to L2/3 neurons are altered in the A1 of CNP‐DN‐ErbB4 mice. Histograms of inhibitory connections (up left), laminar connection width (up right), mean charge (bottom left) and relative laminar charge (bottom right) of inhibitory inputs from L2/3, L4 and L5/6 to L2/3 neurons of WT (black) and CNP‐DN‐ErbB4 (red) mice. The black and red lines are the histogram fit with kernel function for WT (black) and CNP‐DN‐ErbB4 (red) mice. The data depicted in these graphs is the same as in Figure 4b,c.
Figure S3 CNP‐DN‐ErbB4 mice show a shift of the excitatory/inhibitory balance towards excitation. Histograms of excitatory/inhibitory balance showing the mean charge, peak and area of inputs from L2/3, L4 and L5/6 to L2/3 neurons of WT (black) and CNP‐DN‐ErbB4 (red) mice. The black and red lines are the histogram fit with kernel function for WT (black) and CNP‐DN‐ErbB4 (red) mice. The data depicted in these graphs is the same as in Figure 4d.
Data Availability Statement
All data generated or analyzed during this study are included in the manuscript. Original blots and images used for calculations, Matlab codes, and maps into 3D matrices used in electrophysiology experiments will be uploaded to Dryad.


