Abstract
Background:
A key goal of precision medicine is to disaggregate common, complex diseases into discrete molecular subtypes. Rare coding variants in the low-density lipoprotein receptor gene (LDLR) are identified in 1–2% of coronary artery disease (CAD) patients, defining a molecular subtype with risk driven by hypercholesterolemia.
Methods:
To search for additional subtypes, we compared the frequency of rare, predicted loss-of-function and damaging missense variants aggregated within a given gene in 41,081 CAD cases versus 217,115 controls.
Results:
Rare variants in LDLR were most strongly associated with CAD, present in 1% of cases and associated with 4.4-fold increased CAD risk. A second subtype was characterized by variants in endothelial nitric oxide synthase gene (NOS3), a key enzyme regulating vascular tone, endothelial function, and platelet aggregation. A rare predicted loss-of-function or damaging missense variants in NOS3 was present in 0.6% of cases and associated with 2.42-fold increased risk of CAD (95%CI 1.80 to 3.26; p= 5.5 × 10−9). These variants were associated with higher systolic blood pressure (+ 3.25 mm Hg; 95%CI 1.86 to 4.65; p= 5.0 × 10−6) and increased risk of hypertension (adjusted odds ratio 1.31; 95%CI 1.14 to 1.51; p = 0.0002) but not circulating cholesterol concentrations, suggesting that – beyond lipid pathways – nitric oxide synthesis is a key nonlipid driver of CAD risk.
Conclusions:
Beyond LDLR, we identified an additional nonlipid molecular subtype of CAD characterized by rare variants in the NOS3 gene.
Keywords: rare variant association study, NOS3, coronary artery disease, precision medicine
Introduction
Careful study of patients with a specific molecular defect can provide generalizable insights into disease biology and – in some cases – enable targeted therapies, as recently demonstrated for genetically defined subtypes of severe obesity and congestive heart failure1,2.
For coronary artery disease, loss-of-function variants in the gene encoding the low-density lipoprotein receptor (LDLR) are the prototypical molecular subtype3. This condition – known as familial hypercholesterolemia – is characterized by impaired hepatic clearance of LDL cholesterol from the circulation. Although patients with rare LDLR variants account for only 1–2% of patients with coronary artery disease4–8, recognition of this subtype is nonetheless important. Of particular value is identifying individuals prior to disease onset, given recent evidence that early initiation of statin therapy in patients with familial hypercholesterolemia can largely offset the natural history of accelerated atherosclerosis9.
Rare variant association studies – recently enabled by large-scale gene sequencing efforts – provide an opportunity to identify new subtypes for a given disease. Because any individual rare variant is not observed with adequate frequency to test for an association with a given trait, variants are grouped into sets with aggregate frequencies compared between cases and controls10,11. One principled strategy aggregates putative loss-of-function variants in each gene (‘pLoF’), with the potential additional inclusion of very rare missense variants predicted to be damaging by computational algorithms (‘pLoF+missense’)4,12–14, as described in Online Methods.
For coronary artery disease, rare variants in at least ten genes have been shown to impact coronary artery disease risk, all related to lipid pathways15. We set out to test the hypothesis that rare variant association analyses might allow for the identification of damaging variants in nonlipid genes – feature additional novel molecular subtypes – that impact the risk of coronary artery disease. To this end, we aggregated gene sequencing data from 41,081 cases and 217,115 controls from four independent datasets.
Methods
To minimize the possibility of unintentionally sharing information that can be used to reidentify private information, the human genetic data used in this study are available at the database of Genotypes and Phenotypes (dbGaP) and can be accessed through the accession number listed for each study in the Data Supplement. The UK Biobank data with the full summary statistics generated in this study can be applied through the UK Biobank Access Management System. This research was approved by the Mass General Brigham institutional review board (protocol 2013P001840) and was performed under UK Biobank application #7089. For all the study samples used in this study, written informed consent was received from participants prior to inclusion in the study. Full description of methods is provided in the Data Supplement.
Results
To test the hypothesis that rare genetic variants in a given gene might enable identification of molecular subtypes of coronary artery disease, we studied gene sequencing data from 41,081 cases and 217,115 controls from four independent datasets. Across the four cohorts analyzed, the mean age at the time of coronary artery disease onset was 53 years and 51.9% were male (Table 1 and Supplemental Table I–V). The Myocardial Infarction Genetics ExSeq (MIGen ExSeq) study and WGSeq (MIGen WGSeq) included a range of ancestries – 40% European, 2% East Asian, 49% South Asian, and 7% African – while the majority of participants in the UK Biobank 13K and 200K studies16–18 were of European ancestry (Table 1 and Supplemental Figure I).
Table 1.
Coronary artery disease cases versus control datasets
| MIGen ExSeq | MIGen WGSeq | UK Biobank 13K | UK Biobank 200K | |
|---|---|---|---|---|
| N Cases | N Controls | 24,097 | 30,354 | 2,369 | 4,218 | 6,446 | 5,932 | 8,169 | 176,611 |
| Age of cases, years, mean (SD) | 50.9 (10.4) | 48.3 (6.4) | 50.5 (7.9) | 62.3 (7.6) |
| Sex, Male, n (%) | 38,850 (73%) | 2,944 (45%) | 8,099 (65%) | 83,612 (45%) |
| Ancestry, n (%) | ||||
| African | 3087 (6%) | 1,298 (20%) | 128 (1%) | 3,061 (1.7%) |
| East Asian | 5 (0%) | 1,289 (20%) | 23 (0.2%) | 622 (0.3%) |
| European | 21,413 (39%) | 3,081 (47%) | 11,698 (94.5%) | 173,060 (93.7%) |
| Other | 81 (0.1%) | 919 (14%) | 214 (1.7%) | 3,995 (2.2%) |
| South Asian | 29,865 (55%) | 0 (0%) | 315 (2.5%) | 4,042 (2.2%) |
SD, standard deviation.
ASSOCIATION OF LDLR VARIANTS AND RISK OF CORONARY ARTERY DISEASE
As expected, variants in LDLR – known to cause the familial hypercholesterolemia subtype – were most strongly associated with coronary artery disease using either of the two variant aggregation strategies (Figure 1 and Figure 2). Aggregated across all four datasets using the ‘pLoF+missense’ strategy, a rare variant in LDLR was noted in 0.91% of cases versus 0.34% of controls, corresponding to an adjusted odds ratio of 4.39 (95%CI 3.44 to 5.60; p = 1.7 × 10−32). As in previous studies7,8, this association was somewhat stronger among carriers of inactivating variants (LOFTEE predicated high confidence variants, adjusted odds ratio 6.58, 95%CI 3.76 to 11.50, p-value = 4.1 × 10−11) as compared to those previously annotated as pathogenic in the ClinVar database (adjusted odds ratio 3.80, p-value = 5.2 × 10–20, p-value for heterogeneity = 0.09), or missense variants predicted to be damaging by five prediction algorithms (adjusted odds ratio 2.65, p-value = 1.0 × 10−21, p-value for heterogeneity = 0.003 when compared the the LOFTEE variants).
Figure 1.
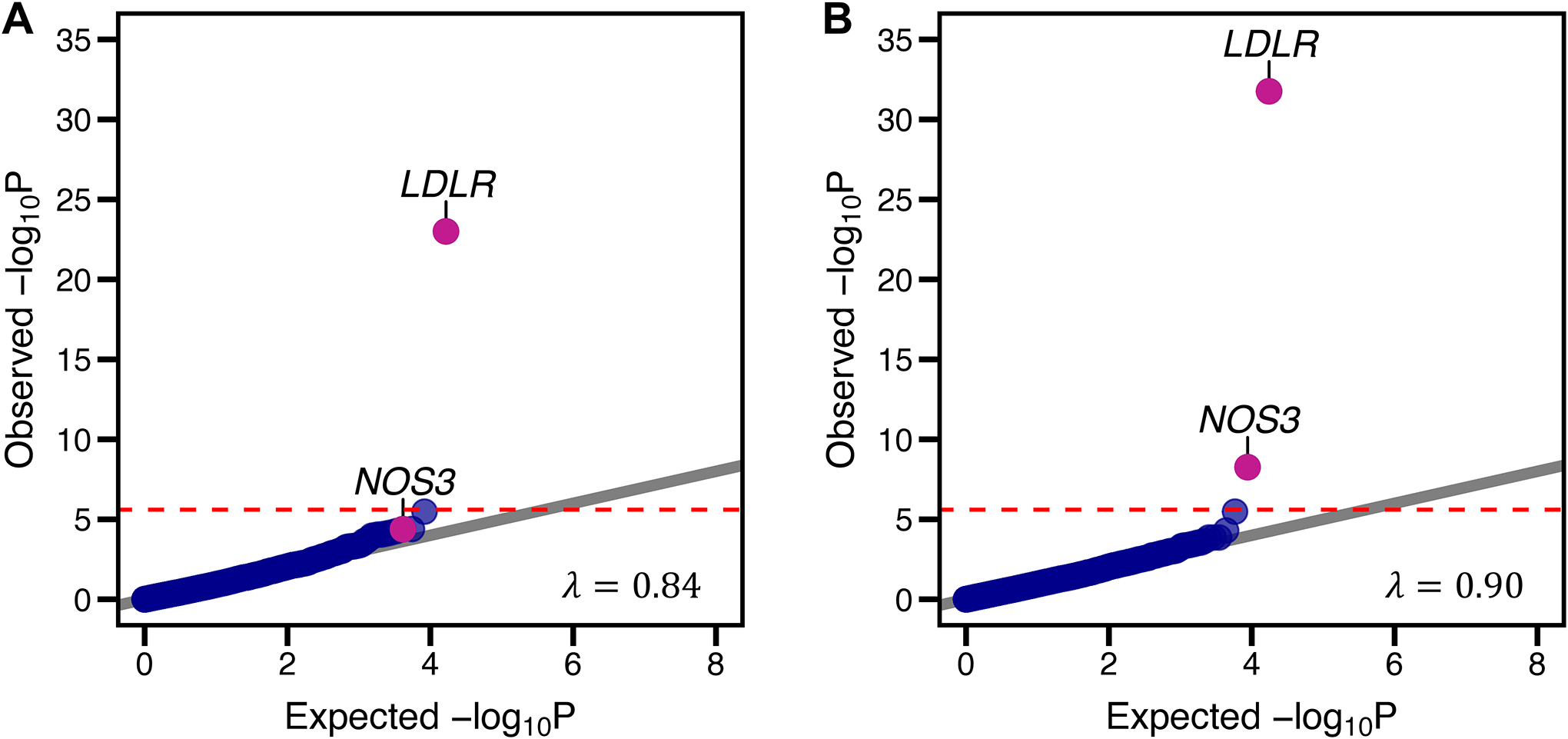
Association of predicted loss-of-function variants and risk of coronary artery disease. Rare DNA variants predicted to lead to loss-of-function, disrupt mRNA splicing, or annotated as pathogenic or likely pathogenic within the ClinVar database were aggregated within each gene (‘pLoF’ strategy). Panel A) is a quantile-quantile plot of observed versus expected p-value distributions observed using this strategy. A second variant annotation strategy (‘pLoF+missense’) additionally included ultra-rare missense variants predicted to be damaging by each of five computational prediction algorithms within each gene. Panel B) is a quantile-quantile plot of observed versus expected p-value distributions of this mask. The horizontal line represents the Bonferroni-corrected p-value threshold of 1.25 × 10−6, assuming 20,000 genes tested and two rare variant grouping masks used. λ refers to the genomic inflation factor, with values significantly higher than 1 suggestive of inadequate control for population stratification.
LDLR – low-density lipoprotein receptor; NOS3 – endothelial nitric oxide synthase (NOS3).
Figure 2. Association of rare variants in the LDLR gene and risk of coronary artery disease.
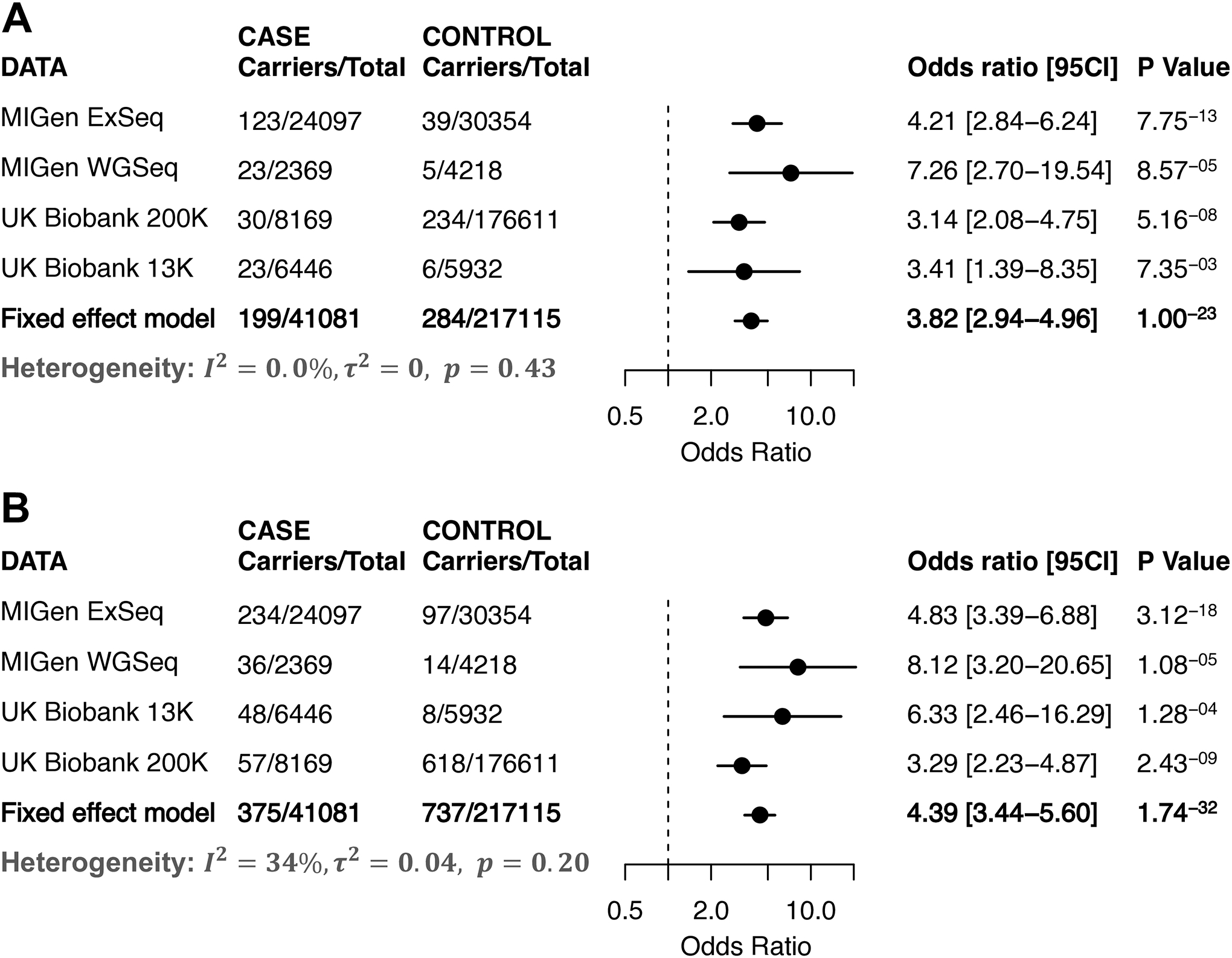
Forest plots including carrier count across cases and controls within four studies for the gene encoding the low-density lipoprotein receptor (LDLR). Panel A) are the results for the counts of variants predicted to lead to loss-of-function, disrupt mRNA splicing, or annotated as pathogenic or likely pathogenic within the ClinVar database. Panel B) is the results from a second variant annotation strategy that additionally included ultra-rare missense variants predicted to be damaging by each of five computational prediction algorithms. The meta-analysis was performed by a fixed-effects meta-analysis model based on the effect size estimated from a Firth logistic regression analysis in each of the four studies. The bar in both plots presents 95% confidence interval.
Consistent with hypercholesterolemia as the driving physiology, estimated untreated LDL cholesterol concentrations in UK Biobank 200K participants were significantly higher in carriers of LDLR variants identified using the ‘pLoF+missense’ strategy versus noncarriers – mean 182 versus 145 mg/dl respectively (adjusted difference +37 mg/dl; 95%CI 34.71 to 39.79; p= 2.91 × 10−181). Importantly, our estimate of a 4.4-fold increased risk for coronary artery disease may have been attenuated by differential treatment of carriers with risk-reducing therapies in clinical practice. Taking the UK Biobank datasets as an example, for those people without diagnosed coronary artery disease, 40% (247 of 618) of LDLR variant carriers reported treatment with lipid-lowering medications as compared to 17% (30,023 of 175,993) of non-carriers.
NOS3 VARIANTS, HYPERTENSION, AND RISK OF CORONARY ARTERY DISEASE
Rare variants in the gene encoding endothelial nitric oxide synthase 3 (NOS3) were identified as a second driver of coronary artery disease risk (Figure 1 and Figure 3). Using the ‘pLoF+missense’ strategy, a NOS3 variant was present in 0.59% of cases versus 0.41% of controls, corresponding to an adjusted odds ratio of 2.42 (95%CI 1.80 to 3.26; p = 5.5 × 10−9). This association was consistently driven by variants identified using the ‘pLoF’ strategy (adjusted odds ratio 2.30, 95%CI 1.54 to 3.42, p-value = 4.1 × 10−5), as well as by the additional missense variants predicted to be damaging by five prediction algorithms added using the ‘pLoF+missense’ strategy (adjusted odds ratio 1.51, 95%CI 1.24 to 1.84, p-value = 4.9 × 10−5). Consistent with a known role of this pathway in the regulation of vascular tone, higher systolic blood pressure (+ 3.25 mm Hg; 95%CI 1.86 to 4.65; p= 5.0 × 10−6) and increased risk of hypertension (adjusted odds ratio 1.31; 95%CI 1.14 to 1.51; p = 0.0002) were noted among 850 carriers of a NOS3 variant in the UK Biobank 200K dataset as compared to 173,697 non-carriers with blood pressure trait data available, but without a significant association with LDL cholesterol, HDL cholesterol, total cholesterol or triglycerides (Supplemental Table VI, Figure 3B, and Supplemental Table VII). A similar result of sensitivity analysis (adjusted odds ratio 2.41, 95%CI 1.78 to 3.25, p-value = 9.5 × 10−9) for the NOS3 gene by partitioning the sample by European and non-European ancestry reassured the robustness of the association results discovered in this study (Supplemental Figure II).
Figure 3. Association of rare variants in the NOS3 gene and risk of coronary artery disease and hypertension.
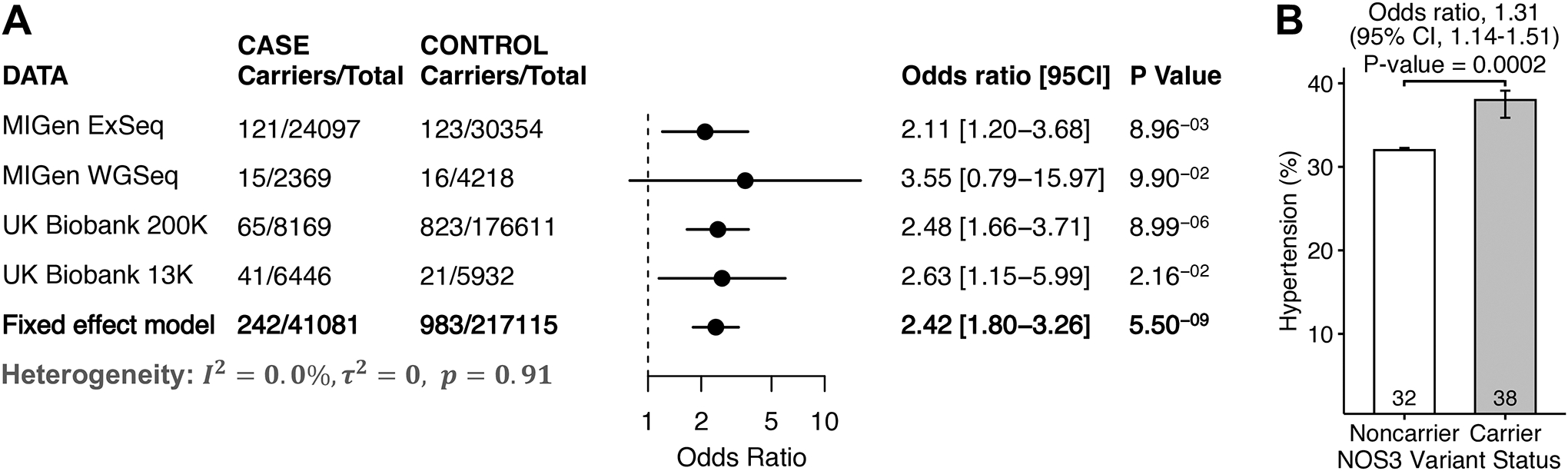
Panel A) is a forest plot including carrier counts across cases and controls within four studies for the gene encoding endothelial nitric oxide synthase (NOS3). Variants included those predicted to lead to loss-of-function, disrupt mRNA splicing, or annotated as pathogenic or likely pathogenic within the ClinVar database, and ultra-rare missense variants predicted to be damaging by each of five computational prediction algorithms (‘pLoF+missense). The meta-analysis was performed by a fixed-effects meta-analysis model based on the effect size estimated from a Firth logistic regression analysis in each of the four studies. The bar in the plot presents 95% confidence interval. Panel B) is the proportion of individuals from the UK Biobank 200K dataset who had been diagnosed with hypertension in carriers versus noncarriers of NOS3 rare variants. The error bar in the bar plot represents the standard error.
Nitric oxide produced by NOS3 acts as a signaling molecule to activate soluble guanylyl cyclase via a heterodimeric receptor encoded by the GUCY1A3 and GUCY1B3 genes19. In an exploratory analysis across all four datasets using the ‘pLoF+missense’ strategy, we observe nominally significant associations for these two additional genes with risk of coronary artery disease, adjusted odds ratios 1.75 (95%CI 1.16 to 2.64; p = 0.007) and 2.31 (95%CI 1.29 to 4.12; p = 0.005) respectively, Supplemental Figure III. As noted for NOS3, carriers of variants in either GUCY1A3 or GUCY1B3 also had increased risk of hypertension, adjusted odds ratios of 1.39 (95%CI 1.14 to 1.69; p = 0.001) and 1.53 (95%CI 1.15 to 2.03; p = 0.004) respectively. A post hoc pathway analysis that aggregated variants in any of three genes – NOS3, GUCY1A3, and GUCY1B3 – using the ‘pLoF+missense’ strategy noted a variant in 1.05% of cases versus 0.80% of controls, corresponding to an adjusted odds ratio of 2.19 for CAD disease risk; 95% CI 1.76 to 2.74; p = 4.5 × 10−12, Supplemental Table VII.
Discussion
By comparing the frequency of rare DNA variants within the coding sequence of a given gene in 41,081 coronary artery disease cases versus 217,115 controls, we identify one more subtype distinct from LDL cholesterol pathways. 0.6% of patients with coronary artery disease inherit an abnormality in nitric oxide production – associated with increased risk of hypertension.
Our identification of rare LDLR variants as the most strongly associated with coronary artery disease – present in 1% of affected individuals – confirms prior results and provides a useful positive control for the overall analytic framework. Previous studies have similarly noted an LDLR variant prevalence of 1–2% among patients afflicted by coronary artery disease, corresponding to a three- to five-fold increased risk4–8. Importantly, individuals have increased risk of coronary artery disease even when compared to those with similarly elevated LDL cholesterol levels – likely reflecting increased lifelong exposure – but remain underdiagnosed and undertreated within current practice8,20.
The second molecular subtype relates to perturbation of the nitric oxide pathway, present in 0.6% of coronary artery disease cases and associated with 2.42-fold increased risk of coronary artery disease. This is consistent with impairment of endothelial function and nitric oxide production as the earliest derangement in coronary atherosclerosis21,22. Two additional lines of genetic support for the involvement of this pathway include prior association of a rare, loss-of-function variant in GUCY1A3 with coronary artery disease in a large family, and common variant association studies that linked noncoding regulatory variants near NOS3 and GUCY1A3 with increases in risk of coronary disease23–25. Beyond an impact on vascular tone, previous studies have additionally linked deficiency of platelet-derived nitric oxide with arterial thrombosis26,27. Whether individuals who inherit a defect in nitric oxide signaling might derive selective benefit in treatment or prevention of coronary artery disease from pharmacologic upregulation of the pathway – already possible using several existing classes of medication – remains uncertain28,29.
Despite our careful analysis of over 40,000 coronary artery disease cases, our analysis likely remained underpowered. To that end, we agree with recent recommendations that analysis of at least 250,000 afflicted individuals will be required to adequately test the hypothesis of a gene-disease relationship for the majority of genes30. Importantly, these sample sizes have become increasingly tractable in recent years with the advent of sequencing of large and ancestrally-diverse populations12,31–33. We anticipate that these future analyses will confirm that a subset of the most strongly associated – but subthreshold – genes are drivers of risk for coronary artery disease. As an example, carriers of variants in the ZNF687 gene tended to have increased risk of coronary artery disease using both the ‘pLoF’ and ‘pLoF+missense’ strategies, ranking 2nd and 14th among the studied genes respectively (Figure 1A and Supplemental Table VIII). Interesting, rare variants in this gene have previously been linked with Paget disease of bone, with preliminary evidence of accelerated cardiovascular disease in several familial and sporadic cases34,35. The fourth most strongly associated gene using the ‘pLoF+missense’ strategy (LPIN2) plays a role in lipid metabolism, and the loss of function of this gene leads to lipodystrophy and increased susceptibility to atherosclerosis in a mouse model36,37. The eleventh gene (PANX1) has been reported to have a role in cardiac response to ischemia and regulation of regulate blood pressure38 (Supplemental Table VIII).
We note that, we used the weight of 0.75 for variants identified by the SpliceAI algorithm suggested by the developers of this tool39. However, the results for the NOS3 variants associated with coronary artery disease were largely unaffected by this choice of weighting, with odds ratios ranging from 2.26 to 2.55 for weight ranging from 0.5 to 1 using the ‘pLoF+missense’ strategy. In each case the strength of statistical association was below the Bonferroni-corrected p-value of 1.25 × 10−6, Supplemental Table IX.
This study also has several limitations which may guide our future improvements. First, although we were able to gather a large number of CAD cases and controls, the power for studying rare variant association is still not sufficient, with our results consistent with other recent large-scale sequencing studies12,40. Second, computational predictions of a given variant’s impact on protein function remain imperfect as compared to functional assays, which may have resulted in reduced statistical power41,42. Third, additional work is needed to build a rare variant analysis framework that additionally considers impact on related traits, such as circulation lipids or blood pressure to improve statistical power43,44.
In conclusion, we analyze gene sequencing data from 258,196 individuals and identify two molecular subtypes of coronary artery disease based on rare DNA variants in the LDLR and NOS3 genes that confer significantly increased risk.
Supplementary Material
Acknowledgments:
We are grateful to study participants for their contribution to this research.
Sources of Funding:
Newly generated sequencing data in the Myocardial Infarction Genetics ExSeq, the VIRGO and TAICHI participants of the Myocardial Infarction Genetics WGSeq study, and the UK Biobank 13K datasets was supported by the National Human Genome Research Institute (NHGRI) Center for Common Disease Genetics program under award number 5UM1HG008895 (principal investigators Dr. Lander, Gabriel, Kathiresan). Whole genome sequencing (WGS) for the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung and Blood Institute (NHLBI). WGS for “NHLBI TOPMed: Multi-Ethnic Study of Atherosclerosis (MESA)” (phs001416.v1.p1) was performed at the Broad Institute of MIT and Harvard (3U54HG003067-13S1). Centralized read mapping and genotype calling, along with variant quality metrics and filtering were provided by the TOPMed Informatics Research Center (3R01HL-117626-02S1). Phenotype harmonization, data management, sample-identity quality control, and general study coordination, were provided by the TOPMed Data Coordinating Center (3R01HL-120393-02S1). MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts 75N92020D00001, HHSN268201500003I, N01-HC-95159, 75N92020D00005, N01-HC-95160, 75N92020D00002, N01-HC-95161, 75N92020D00003, N01-HC-95162, 75N92020D00006, N01-HC-95163, 75N92020D00004, N01-HC-95164, 75N92020D00007, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-000040, UL1-TR-001079, UL1-TR-001420. This study was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR001881, the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The academic coordinating centre for the BRAVE study was supported by grants from the British Heart Foundation, Health Data Research UK, UK National Institute for Health Research, and UK Research and Innovation (Medical Research Council). Professor John Danesh holds a British Heart Foundation Professorship and a National Institute for Health and Care Research (NIHR) Senior Investigator Award. This work was also supported by core funding from the: UK Medical Research Council (MR/L003120/1), British Heart Foundation (RG/13/13/30194; RG/18/13/33946) and NIHR Cambridge Biomedical Research Centre (BRC-1215-20014). The views expressed are those of the author(s) and not necessarily those of the NIHR or the Department of Health and Social Care. This work was also supported by Health Data Research UK, which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation and Wellcome. A.V.K. was supported by grant 1K08HG010155 from the National Human Genome Research Institute, a Hassenfeld Scholar Award from Massachusetts General Hospital, a Merkin Institute Fellowship from the Broad Institute of MIT and Harvard. Dr. Danesh is supported by a British Heart Foundation Personal Chair and a National Institute for Health Research Senior Investigator Award. The funding sources had no role in the design, conduct, or analysis of the study or in the decision to submit the manuscript for publication.
Disclosures:
A.V.K. has served as a scientific advisor to Sanofi, Amgen, Maze Therapeutics, Navitor Pharmaceuticals, Sarepta Therapeutics, Verve Therapeutics, Veritas International, Color Health, Third Rock Ventures, and Columbia University (NIH); received speaking fees from Illumina, MedGenome, Amgen, and the Novartis Institute for Biomedical Research; and received sponsored research agreements from the Novartis Institute for Biomedical Research and IBM Research. Dr. Lander serves on the Board of Directors for Codiak; serves on the Scientific Advisory Board of F-Prime Capital Partners and Third Rock Ventures; serves on the Board of Directors of the Innocence Project, Count Me In, and Biden Cancer Initiative; and serves on the Board of Trustees for the Parker Institute for Cancer Immunotherapy. S.K. is an employee of Verve Therapeutics, holds equity in Verve Therapeutics and Maze Therapeutics, and has served as a consultant for Acceleron, Eli Lilly, Novartis, Merck, Novo Nordisk, Novo Ventures, Ionis, Alnylam, Aegerion, Haug Partners, Noble Insights, Leerink Partners, Bayer Healthcare, Illumina, Color Health, MedGenome, Quest and Medscape. All the other authors have declared that no conflict of interest exists. John Danesh reports grants, personal fees and non-financial support from Merck Sharp & Dohme (MSD), grants, personal fees and non-financial support from Novartis, grants from Pfizer and grants from AstraZeneca outside the submitted work. John Danesh sits on the International Cardiovascular and Metabolic Advisory Board for Novartis (since 2010); the Steering Committee of UK Biobank (since 2011); the MRC International Advisory Group (ING) member, London (since 2013); the MRC High Throughput Science ‘Omics Panel Member, London (since 2013); the Scientific Advisory Committee for Sanofi (since 2013); the International Cardiovascular and Metabolism Research and Development Portfolio Committee for Novartis; and the Astra Zeneca Genomics Advisory Board (2018). Adam Butterworth reports institutional grants from AstraZeneca, Bayer, Biogen, BioMarin, Bioverativ, Novartis, Regeneron and Sanofi.
Nonstandard Abbreviations and Acronyms
- CAD
coronary artery disease
- CI
confidence interval
- LOFTEE
Loss-Of-Function Transcript Effect Estimator
- MIGen ExSeq
Myocardial Infarction Genetics exome sequencing study
- MIGen WGSeq
Myocardial Infarction Genetics whole genome sequencing study
- pLoF
predicted to be loss-of-function
Footnotes
Supplemental Materials:
References:
- 1.Kuhnen P, Clement K, Wiegand S, Blankenstein O, Gottesdiener K, Martini LL, Mai K, Blume-Peytavi U, Gruters A, Krude H. Proopiomelanocortin Deficiency Treated with a Melanocortin-4 Receptor Agonist. N Engl J Med. 2016;375:240–246. [DOI] [PubMed] [Google Scholar]
- 2.Maurer MS, Schwartz JH, Gundapaneni B, Elliott PM, Merlini G, Waddington-Cruz M, Kristen AV, Grogan M, Witteles R, Damy T, et al. Tafamidis Treatment for Patients with Transthyretin Amyloid Cardiomyopathy. N Engl J Med. 2018;379:1007–1016. [DOI] [PubMed] [Google Scholar]
- 3.Lehrman MA, Schneider WJ, Sudhof TC, Brown MS, Goldstein JL, Russell DW. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.NHLBI Exome Sequencing Project, Do R, Stitziel NO, Won H-H, Jørgensen AB, Duga S, Angelica Merlini P, Kiezun A, Farrall M, Goel A, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khera AV, Chaffin M, Zekavat SM, Collins RL, Roselli C, Natarajan P, Lichtman JH, D’Onofrio G, Mattera J, Dreyer R, et al. Whole-Genome Sequencing to Characterize Monogenic and Polygenic Contributions in Patients Hospitalized With Early-Onset Myocardial Infarction. Circulation. 2019;139:1593–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benn M, Watts GF, Tybjaerg-Hansen A, Nordestgaard BG. Mutations causative of familial hypercholesterolaemia: screening of 98 098 individuals from the Copenhagen General Population Study estimated a prevalence of 1 in 217. Eur Heart J. 2016;37:1384–1394. [DOI] [PubMed] [Google Scholar]
- 7.Abul-Husn NS, Manickam K, Jones LK, Wright EA, Hartzel DN, Gonzaga-Jauregui C, O’Dushlaine C, Leader JB, Kirchner HL, Lindbuchler DM, et al. Genetic identification of familial hypercholesterolemia within a single U.S. health care system. Science. 2016;354. [DOI] [PubMed] [Google Scholar]
- 8.Khera AV, Won HH, Peloso GM, Lawson KS, Bartz TM, Deng X, van Leeuwen EM, Natarajan P, Emdin CA, Bick AG, et al. Diagnostic Yield and Clinical Utility of Sequencing Familial Hypercholesterolemia Genes in Patients With Severe Hypercholesterolemia. J Am Coll Cardiol. 2016;67:2578–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luirink IK, Wiegman A, Kusters DM, Hof MH, Groothoff JW, de Groot E, Kastelein JJP, Hutten BA. 20-Year Follow-up of Statins in Children with Familial Hypercholesterolemia. N Engl J Med. 2019;381:1547–1556. [DOI] [PubMed] [Google Scholar]
- 10.Povysil G, Petrovski S, Hostyk J, Aggarwal V, Allen AS, Goldstein DB. Rare-variant collapsing analyses for complex traits: guidelines and applications. Nat Rev Genet. 2019. [DOI] [PubMed] [Google Scholar]
- 11.Zuk O, Schaffner SF, Samocha K, Do R, Hechter E, Kathiresan S, Daly MJ, Neale BM, Sunyaev SR, Lander ES. Searching for missing heritability: Designing rare variant association studies. Proc Natl Acad Sci. 2014;111:E455–E464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Backman JD, Li AH, Marcketta A, Sun D, Mbatchou J, Kessler MD, Benner C, Liu D, Locke AE, Balasubramanian S, et al. Exome sequencing and analysis of 454,787 UK Biobank participants. Nature. 2021;599:628–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akbari P, Gilani A, Sosina O, Kosmicki JA, Khrimian L, Fang Y-Y, Persaud T, Garcia V, Sun D, Li A, et al. Sequencing of 640,000 exomes identifies GPR75 variants associated with protection from obesity. Science. 2021;373:eabf8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khera AV, Won H-H, Peloso GM, O’Dushlaine C, Liu D, Stitziel NO, Natarajan P, Nomura A, Emdin CA, Gupta N, et al. Association of Rare and Common Variation in the Lipoprotein Lipase Gene With Coronary Artery Disease. JAMA. 2017;317:937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khera AV, Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet. 2017;18:331–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fahed AC, Wang M, Homburger JR, Patel AP, Bick AG, Neben CL, Lai C, Brockman D, Philippakis A, Ellinor PT, et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat Commun. 2020;11:3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szustakowski JD, Balasubramanian S, Sasson A, Khalid S, Bronson PG, Kvikstad E, Wong E, Liu D, Davis JW, Haefliger C, et al. Advancing Human Genetics Research and Drug Discovery through Exome Sequencing of the UK Biobank. Genetic and Genomic Medicine; 2020. [DOI] [PubMed] [Google Scholar]
- 19.Farah C, Michel LYM, Balligand J-L. Nitric oxide signalling in cardiovascular health and disease. Nat Rev Cardiol. 2018;15:292–316. [DOI] [PubMed] [Google Scholar]
- 20.Nordestgaard BG, Chapman MJ, Humphries SE, Ginsberg HN, Masana L, Descamps OS, Wiklund O, Hegele RA, Raal FJ, Defesche JC, et al. Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease: consensus statement of the European Atherosclerosis Society. Eur Heart J. 2013;34:3478–90a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludmer PL, Selwyn AP, Shook TL, Wayne RR, Mudge GH, Alexander RW, Ganz P. Paradoxical Vasoconstriction Induced by Acetylcholine in Atherosclerotic Coronary Arteries. N Engl J Med. 1986;315:1046–1051. [DOI] [PubMed] [Google Scholar]
- 22.Russell R Atherosclerosis — An Inflammatory Disease. N Engl J Med. 1999:12. [Google Scholar]
- 23.Erdmann J, Stark K, Esslinger UB, Rumpf PM, Koesling D, de Wit C, Kaiser FJ, Braunholz D, Medack A, Fischer M, et al. Dysfunctional nitric oxide signalling increases risk of myocardial infarction. Nature. 2013;504:432–436. [DOI] [PubMed] [Google Scholar]
- 24.Emdin CA, Khera AV, Klarin D, Natarajan P, Zekavat SM, Nomura A, Haas M, Aragam K, Ardissino D, Wilson JG, et al. Phenotypic Consequences of a Genetic Predisposition to Enhanced Nitric Oxide Signaling. Circulation. 2018;137:222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freedman JE, Loscalzo J, Barnard MR, Alpert C, Keaney JF, Michelson AD. Nitric oxide released from activated platelets inhibits platelet recruitment. J Clin Invest. 1997;100:350–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freedman Jane E., Ting Brian, Hankin Beth, Loscalzo Joseph, Keaney John F., Vita Joseph A. Impaired Platelet Production of Nitric Oxide Predicts Presence of Acute Coronary Syndromes. Circulation. 1998;98:1481–1486. [DOI] [PubMed] [Google Scholar]
- 28.Steinhorn BS, Loscalzo J, Michel T. Nitroglycerin and Nitric Oxide — A Rondo of Themes in Cardiovascular Therapeutics. Http://DxDoiOrg/101056/NEJMsr1503311. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall KT, Kessler T, Buring JE, Passow D, Sesso HD, Zee RYL, Ridker PM, Chasman DI, Schunkert H. Genetic variation at the coronary artery disease risk locus GUCY1A3 modifies cardiovascular disease prevention effects of aspirin. Eur Heart J. 2019;40:3385–3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.ICDA Recommendations and White Paper.pdf. n.d.
- 31.Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, Whitbourne S, Deen J, Shannon C, Humphries D, et al. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–223. [DOI] [PubMed] [Google Scholar]
- 32.Nagai A, Hirata M, Kamatani Y, Muto K, Matsuda K, Kiyohara Y, Ninomiya T, Tamakoshi A, Yamagata Z, Mushiroda T, et al. Overview of the BioBank Japan Project: Study design and profile. J Epidemiol. 2017;27:S2–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beesley LJ, Salvatore M, Fritsche LG, Pandit A, Rao A, Brummett C, Willer CJ, Lisabeth LD, Mukherjee B. The emerging landscape of health research based on biobanks linked to electronic health records: Existing resources, statistical challenges, and potential opportunities. Stat Med. 2020;39:773–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gianfrancesco F, Rendina D, Merlotti D, Esposito T, Amyere M, Formicola D, Muscariello R, De Filippo G, Strazzullo P, Nuti R, et al. Giant cell tumor occurring in familial Paget’s disease of bone: Report of clinical characteristics and linkage analysis of a large pedigree. J Bone Miner Res. 2013;28:341–350. [DOI] [PubMed] [Google Scholar]
- 35.Divisato G, Formicola D, Esposito T, Merlotti D, Pazzaglia L, Del Fattore A, Siris E, Orcel P, Brown JP, Nuti R, et al. ZNF687 Mutations in Severe Paget Disease of Bone Associated with Giant Cell Tumor. Am J Hum Genet. 2016;98:275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reue K, Xu P, Wang X-P, Slavin BG. Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene. J Lipid Res. 2000;41:1067–1076. [PubMed] [Google Scholar]
- 37.Dwyer JR, Donkor J, Zhang P, Csaki LS, Vergnes L, Lee JM, Dewald J, Brindley DN, Atti E, Tetradis S, et al. Mouse lipin-1 and lipin-2 cooperate to maintain glycerolipid homeostasis in liver and aging cerebellum. Proc Natl Acad Sci. 2012;109:E2486–E2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, He L, Wu D, Chen L, Jiang Z. Pannexin-1 channels and their emerging functions in cardiovascular diseases. Acta Biochim Biophys Sin. 2015;47:391–396. [DOI] [PubMed] [Google Scholar]
- 39.Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, Kosmicki JA, Arbelaez J, Cui W, Schwartz GB, et al. Predicting Splicing from Primary Sequence with Deep Learning. Cell. 2019;176:535–548.e24. [DOI] [PubMed] [Google Scholar]
- 40.Broad Genomics Platform, DiscovEHR Collaboration, CHARGE, LuCamp, ProDiGY, GoT2D, ESP, SIGMA-T2D, T2D-GENES, AMP-T2D-GENES, et al. Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature. 2019. [Google Scholar]
- 41.Wang M, Lee-Kim VS, Atri DS, Elowe NH, Yu J, Garvie CW, Won H-H, Hadaya JE, MacDonald BT, Trindade K, et al. Rare, Damaging DNA Variants in CORIN and Risk of Coronary Artery Disease: Insights From Functional Genomics and Large-Scale Sequencing Analyses. Circ Genomic Precis Med. 2021;14:e003399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stitziel NO, Khera AV, Wang X, Bierhals AJ, Vourakis AC, Sperry AE, Natarajan P, Klarin D, Emdin CA, Zekavat SM, et al. ANGPTL3 Deficiency and Protection Against Coronary Artery Disease. J Am Coll Cardiol. 2017;69:2054–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peloso GM, Nomura A, Khera AV, Chaffin M, Won H-H, Ardissino D, Danesh J, Schunkert H, Wilson JG, Samani N, et al. Rare Protein-Truncating Variants in APOB, Lower Low-Density Lipoprotein Cholesterol, and Protection Against Coronary Heart Disease. Circ Genomic Precis Med. 2019;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nioi P, Sigurdsson A, Thorleifsson G, Helgason H, Agustsdottir AB, Norddahl GL, Helgadottir A, Magnusdottir A, Jonasdottir A, Gretarsdottir S, et al. Variant ASGR1 Associated with a Reduced Risk of Coronary Artery Disease. N Engl J Med. 2016;374:2131–2141. [DOI] [PubMed] [Google Scholar]
- 45.Assimes TL, Lee I-T, Juang J-M, Guo X, Wang T-D, Kim ET, Lee W-J, Absher D, Chiu Y-F, Hsu C-C, et al. Genetics of Coronary Artery Disease in Taiwan: A Cardiometabochip Study by the Taichi Consortium. PLOS ONE. 2016;11:e0138014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy‐Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. From FastQ Data to High‐Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr Protoc Bioinforma. 2013;43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Jang W, et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-Variant Association Analysis: Study Designs and Statistical Tests. Am J Hum Genet. 2014;95:5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park J-H, Gail MH, Weinberg CR, Carroll RJ, Chung CC, Wang Z, Chanock SJ, Fraumeni JF, Chatterjee N. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc Natl Acad Sci. 2011;108:18026–18031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dey R, Schmidt EM, Abecasis GR, Lee S. A Fast and Accurate Algorithm to Test for Binary Phenotypes and Its Application to PheWAS. Am J Hum Genet. 2017;101:37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. [DOI] [PubMed] [Google Scholar]
- 53.Han B, Eskin E. Random-Effects Model Aimed at Discovering Associations in Meta-Analysis of Genome-wide Association Studies. Am J Hum Genet. 2011;88:586–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patel AP, Wang M, Fahed AC, Mason-Suares H, Brockman D, Pelletier R, Amr S, Machini K, Hawley M, Witkowski L, et al. Association of Rare Pathogenic DNA Variants for Familial Hypercholesterolemia, Hereditary Breast and Ovarian Cancer Syndrome, and Lynch Syndrome With Disease Risk in Adults According to Family History. JAMA Netw Open. 2020;3:e203959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–2935. [DOI] [PubMed] [Google Scholar]
- 56.The International Consortium of Blood Pressure (ICBP) 1000G Analyses, The CHD Exome+ Consortium, The ExomeBP Consortium, The T2D-GENES Consortium, The GoT2DGenes Consortium, The Cohorts for Heart and Ageing Research in Genome Epidemiology (CHARGE) BP Exome Consortium, The International Genomics of Blood Pressure (iGEN-BP) Consortium, The UK Biobank CardioMetabolic Consortium BP working group, Warren HR, Evangelou E, et al. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet. 2017;49:403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 58.Atherosclerosis, Thrombosis, and Vascular Biology Italian Study Group. No Evidence of Association Between Prothrombotic Gene Polymorphisms and the Development of Acute Myocardial Infarction at a Young Age. Circulation. 2003;107:1117–1122. [DOI] [PubMed] [Google Scholar]
- 59.Cardiology Research Group, Chowdhury R, Alam DS, Fakir II, Adnan SD, Naheed A, Tasmin I, Monower MM, Hossain F, Hossain FM, et al. The Bangladesh Risk of Acute Vascular Events (BRAVE) Study: objectives and design. Eur J Epidemiol. 2015;30:577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Do R, Stitziel NO, Won HH, Jorgensen AB, Duga S, Angelica Merlini P, Kiezun A, Farrall M, Goel A, Zuk O, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann H-E, et al. Genomewide Association Analysis of Coronary Artery Disease. N Engl J Med. 2007;357:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Myocardial Infarction Genetics and CARDIoGRAM Exome Consortia Investigators. Coding Variation in ANGPTL4, LPL, and SVEP1 and the Risk of Coronary Disease. N Engl J Med. 2016;374:1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, et al. A Common Allele on Chromosome 9 Associated with Coronary Heart Disease 2007;316:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clarke R, Peden JF, Hopewell JC, Kyriakou T, Goel A, Heath SC, Parish S, Barlera S, Franzosi MG, Rust S, et al. Genetic Variants Associated with Lp(a) Lipoprotein Level and Coronary Disease. N Engl J Med. 2009;361:2518–2528. [DOI] [PubMed] [Google Scholar]
- 65.Saleheen D, Zaidi M, Rasheed A, Ahmad U, Hakeem A, Murtaza M, Kayani W, Faruqui A, Kundi A, Zaman KS, et al. The Pakistan Risk of Myocardial Infarction Study: a resource for the study of genetic, lifestyle and other determinants of myocardial infarction in South Asia. Eur J Epidemiol. 2009;24:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saleheen D, Natarajan P, Armean IM, Zhao W, Rasheed A, Khetarpal SA, Won H-H, Karczewski KJ, O’Donnell-Luria AH, Samocha KE, et al. Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity. Nature. 2017;544:235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sentí M, Tomás M, Marrugat J, Elosua R. Paraoxonase1–192 Polymorphism Modulates the Nonfatal Myocardial Infarction Risk Associated With Decreased HDLs. Arterioscler Thromb Vasc Biol. 2001;21:415–420. [DOI] [PubMed] [Google Scholar]
- 68.The TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute. Loss-of-Function Mutations in APOC3, Triglycerides, and Coronary Disease. N Engl J Med. 2014;371:22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seidelmann SB, Feofanova E, Yu B, Franceschini N, Claggett B, Kuokkanen M, Puolijoki H, Ebeling T, Perola M, Salomaa V, et al. Genetic Variants in SGLT1, Glucose Tolerance, and Cardiometabolic Risk. J Am Coll Cardiol. 2018;72:1763–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ArXiv13033997 Q-Bio. 2013. [Google Scholar]
- 71.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, Kling DE, Gauthier LD, Levy-Moonshine A, Roazen D, et al. Scaling accurate genetic variant discovery to tens of thousands of samples. Genomics; 2017. [Google Scholar]
- 73.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen W-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li H Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bailey JA. Segmental Duplications: Organization and Impact Within the Current Human Genome Project Assembly. Genome Res. 2001;11:1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bild DE. Multi-Ethnic Study of Atherosclerosis: Objectives and Design. Am J Epidemiol. 2002;156:871–881. [DOI] [PubMed] [Google Scholar]
- 78.Lichtman JH, Lorenze NP, D’Onofrio G, Spertus JA, Lindau ST, Morgan TM, Herrin J, Bueno H, Mattera JA, Ridker PM, et al. Variation in Recovery: Role of Gender on Outcomes of Young AMI Patients (VIRGO) Study Design. Circ Cardiovasc Qual Outcomes. 2010;3:684–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao H, Sun Z, Wang J, Huang H, Kocher J-P, Wang L. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics. 2014;30:1006–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM. Control of Confounding of Genetic Associations in Stratified Populations. Am J Hum Genet. 2003;72:1492–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Libiger O, Schork NJ. A Method for Inferring an Individual’s Genetic Ancestry and Degree of Admixture Associated with Six Major Continental Populations. Front Genet. 2013;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, Fernandez Banet J, Billis K, García Girón C, Hourlier T, et al. The Ensembl gene annotation system. Database. 2016;2016:baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Exome Aggregation Consortium, Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Khera AV, Won HH, Peloso GM, O’Dushlaine C, Liu D, Stitziel NO, Natarajan P, Nomura A, Emdin CA, Gupta N, et al. Association of Rare and Common Variation in the Lipoprotein Lipase Gene With Coronary Artery Disease. JAMA. 2017;317:937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum Mutat. 2016;37:235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function n.d.:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


