Summary
Microglia utilize their phagocytic activity to prune redundant synapses and refine neural circuits during precise developmental periods. However, the neuronal signals that control this phagocytic clockwork remain largely undefined. Here, we showed that neuronal signal regulatory protein alpha (SIRPα) was a permissive cue for microglial phagocytosis in the developing murine retina. Removal of neuronal, but not microglial, SIRPα reduced microglial phagocytosis, increased synaptic number, and impaired circuit function. Conversely, prolonging neuronal SIRPα expression extended developmental microglial phagocytosis. These outcomes depended on the interaction of presynaptic SIRPα with postsynaptic CD47. Global CD47 deficiency modestly increased microglial phagocytosis, while CD47 overexpression reduced it. This effect was rescued by co-expression of neuronal SIRPα or co-deletion of neuronal SIRPα and CD47. These data indicate that neuronal SIRPα regulated microglial phagocytosis by limiting access of microglial SIRPα to neuronal CD47. This discovery may aid our understanding of synapse loss in neurological diseases.
Keywords: microglia, SIRPα, synapse refinement, retina
eTOC blurb
Temporal regulation of microglia phagocytosis is central to nervous system development, but the underlying mechanisms for this regulation remain poorly understood. Jiang et al. reveal that neuronal, but not microglial, signal regulatory protein alpha (SIRPα) is necessary for microglia phagocytosis and synapse refinement during development. To achieve this, neuronal SIRPα functions as a decoy receptor to prevent microglial SIRPα-CD47 interaction.
Graphical Abstract
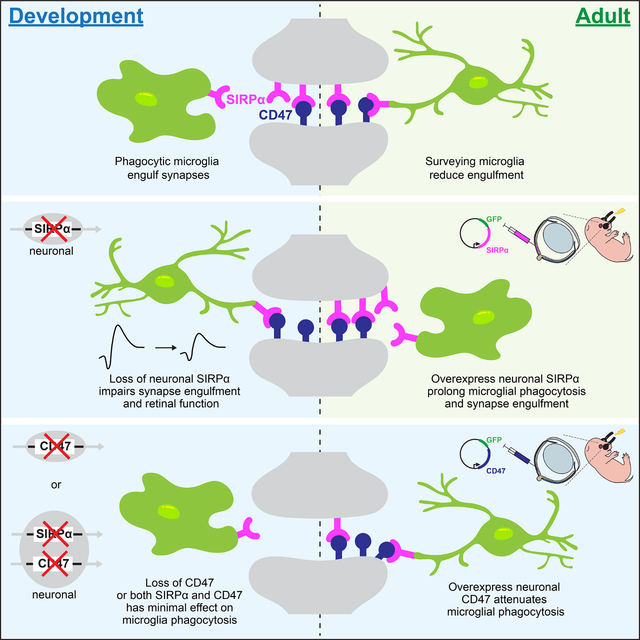
Introduction
Microglia are resident central nervous system (CNS) immune cells that display defined windows of heightened phagocytosis which align precisely with periods of neuron growth and remodeling (Silverman and Wong, 2018; Wilton et al., 2019; Wu et al., 2015). Microglia are highly phagocytic during neuron refinement and become less phagocytic as neurons mature, suggesting that the CNS experiences a “critical period” of heightened microglial phagocytosis that is tightly controlled (Bessis et al., 2007; Perry et al., 2010; Schafer et al., 2014; Schafer et al., 2012; Sierra et al., 2013; Tremblay et al., 2010). The importance of understanding microglia phagocytosis regulation is underscored by the large number of neurodegenerative disorders in which dysregulated microglia phagocytosis is implicated (Estes and McAllister, 2015; Hong et al., 2016; Lall et al., 2021; Lui et al., 2016; Perry et al., 2010; Salter and Stevens, 2017; Sellgren et al., 2019; Vasek et al., 2016; Werneburg et al., 2020). Several microglia cell-surface receptors have been identified that regulate phagocytosis over development (Fu et al., 2014; Gardai et al., 2005; Lui et al., 2016; Oldenborg et al., 2001; Oldenborg et al., 2000), but whether neuron-derived cues can also be instructive remains largely unknown.
Previously, we uncovered a candidate engulfment pathway controlled by signal regulatory protein alpha (SIRPα) (Jiang et al., 2020). In the periphery, professional phagocytes can express SIRPα, and binding of its receptor CD47 on host cells serves as a “don’t eat me signal” that reduces phagocytosis (Gardai et al., 2005; Oldenborg et al., 2001; Oldenborg et al., 2000). Various cancers have exploited this pathway by upregulating CD47 to avoid immune detection (Chao et al., 2011; Chao et al., 2012; Majeti et al., 2009; Weiskopf et al., 2016; Willingham et al., 2012; Zhao et al., 2016). However, our view of these interactions is expanding with the observation that cancer cells also express SIRPα, and that SIRPα downregulation can likewise enhance cancer immune evasion (Chen et al., 2004; Qin et al., 2006; Takahashi, 2018; Wu et al., 2000; Yan et al., 2004; Yao et al., 2017). Thus, the relative levels of SIRPα in phagocytic and non-phagocytic cells appear important for modulating immune outcomes.
Complementary roles for diverse cellular sources of SIRPα may also extend to the CNS. SIRPα is present on both neurons and microglia (Barclay and Brown, 2006; Chuang and Lagenaur, 1990; Jiang et al., 1999; Kharitonenkov et al., 1997; Mi et al., 2000; van Beek et al., 2005), while high amounts of CD47 are present on neurons throughout development. Surprisingly, microglia are highly phagocytic during this period despite the presence of inhibitory CD47 (Lehrman et al., 2018). Whole-body deletion of CD47 can increase neural refinement by microglia in an activity-dependent manner (Lehrman et al., 2018). In parallel, SIRPα has also been shown to regulate activity-dependent synapse maturation (Nagappan-Chettiar et al., 2018; Toth et al., 2013; Umemori and Sanes, 2008). Whether or how microglia- or neuron-derived SIRPα differentially contribute to these outcomes remains unclear. Further, the molecular pathways that permit microglia phagocytic activity during development despite high amounts of inhibitory CD47 protein are unknown.
Here we used the murine retina to identify neuronal SIRPα as an unexpected permissive cue for developmental microglia phagocytosis. Neurons produced the bulk of SIRPα in temporal alignment with heightened microglial phagocytosis. SIRPα was located presynaptically, where it colocalized with postsynaptic CD47. We found that neuronal, but not microglial, SIRPα was required for developmental microglia phagocytosis and synaptic refinement. Conversely, prolonging neuronal SIRPα expression extended the window in which microglia were highly phagocytic. We further showed that these outcomes depended on the interaction of neuronal SIRPα with CD47. Increasing neuronal CD47 alone reduced microglial phagocytosis, while SIRPα and CD47 co-expression in neurons was sufficient to restore microglia phagocytosis. These data identified SIRPα as a critical neuron-derived cue that instructed both the timing and degree of microglia phagocytosis through modulating the accessibility of inhibitory CD47 over development.
Results
Retinal neuron refinement coincided with heightened microglia phagocytosis
As in the brain, developmental refinement of the retina’s diverse neuron types occurs during the first two postnatal weeks when neurites become restricted to two synapse layers (Kim et al., 2010; Wong and Ghosh, 2002) (Figure 1A–B). The inner plexiform layer (IPL) appears first (~P2), followed by the outer plexiform layer (OPL) at P5. By P14, neurons have largely adopted their adult morphologies. To examine microglia during this period, we used Cx3cr1GFP/+ reporter mice in which microglia are selectively labeled with GFP (Jung et al., 2000). We found that the location and number of retinal microglia coincided precisely with synapse refinement (Figure 1C, Figure S1A). At P9, 97% of microglia were present in retinal synapse layers (Figure S1A). Because OPL synapses between presynaptic photoreceptors and postsynaptic horizontal and bipolar cells are particularly large and highly ordered, we focused on this region for our analyses. We found that high levels of microglia phagocytosis accompanied OPL synapse refinement. At P9, microglia adopted a morphology characteristic of active engulfment, with shorter process length, larger somas, and more phagocytic cups (round-shaped invaginations associated with phagocytosis(Swanson, 2008)) compared to time points prior to and after P9 (Figure 1D–H, Figure S1B). Consistent with heightened phagocytosis, microglia in P9 retina displayed increased expression of the lysosomal membrane marker CD68 (Figure 1I). As refinement ended at P14, microglia adopted a morphology characteristic of more mature microglia with reduced engulfment. This included increased ramification, smaller somas, decreased CD68 protein expression, and fewer numbers of phagocytic cups (Figure 1E–G, 1J, Figure S1C). Together, these data showed that elevations in microglia phagocytic activity temporally and spatially aligned with retinal neuron refinement. As neurons matured and refinement concluded, microglial became ramified and lysosomal content declined, consistent with a decrease in phagocytic function.
Figure 1. Retinal neuron refinement coincided with heightened microglia phagocytosis.
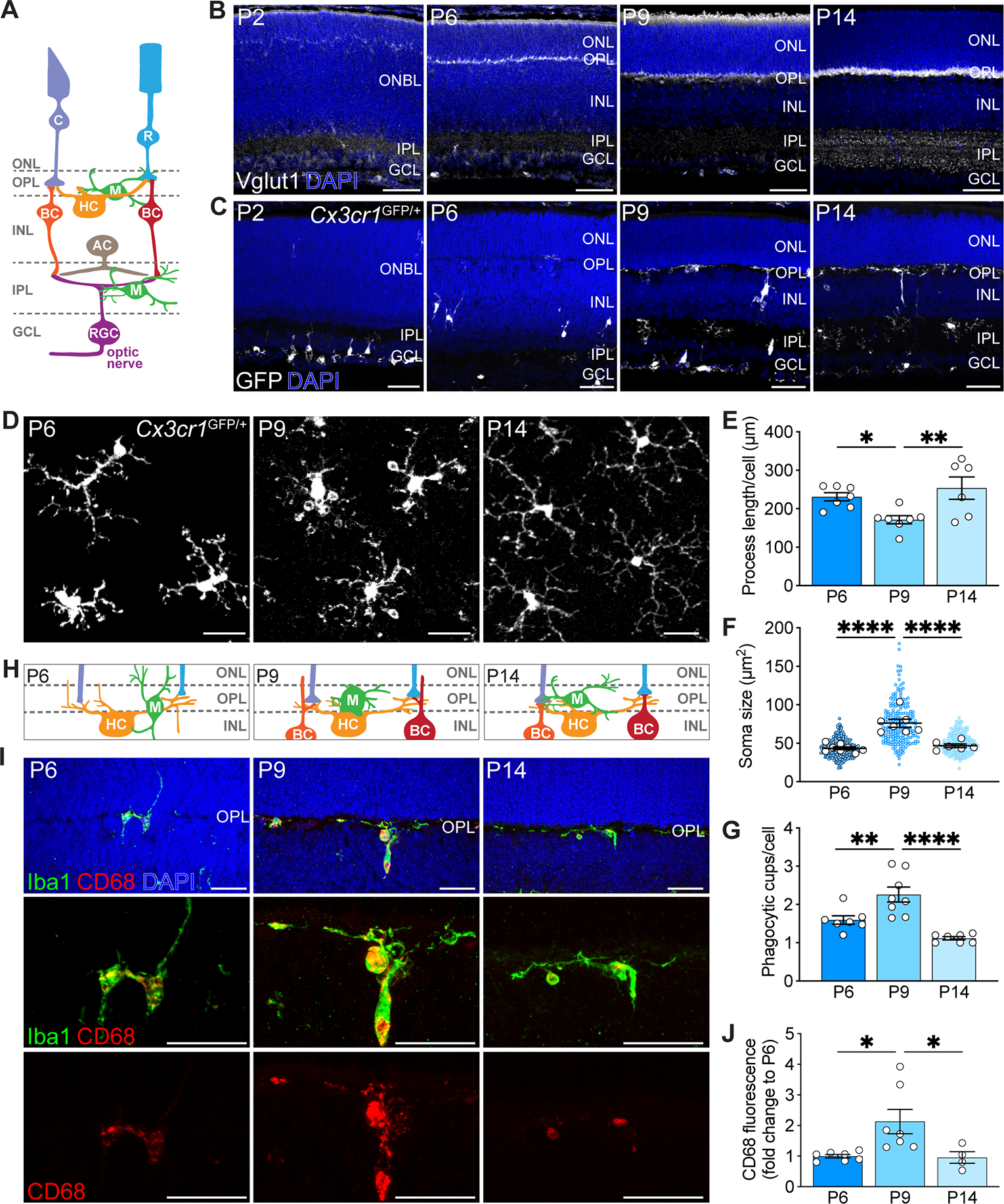
(A) Schematic of adult retina. Rods (R) and cones (C) in the outer nuclear layer (ONL) synapse onto bipolar cells (BC) and horizontal cells (HC) in the inner nuclear layer (INL), forming a thin synaptic band called outer plexiform layer (OPL). Bipolar cells and amacrine cells (AC) relay signals to retinal ganglion cells (RGC) in the inner plexiform layer (IPL). RGCs reside in the ganglion cell layer (GCL), and their axons form the optic nerve which projects to the brain. Microglia (M) occupy the synaptic layers.
(B) Generation of retinal synaptic layers. Vglut1-labeled inner retina synapses (white) were present at P2. At P5-P6, Vglut1+ photoreceptor terminals were visible in the OPL. At P9, both layers continued to be refined. Synaptogenesis largely completed by P14. Scale bars, 50 μm.
(C) Microglia (white) migration to the synaptic layers. Scale bars, 50 μm.
(D) Representative wholemount images of P6, P9, and P14 OPL microglia in Cx3cr1GFP/+ mice. Scale bars, 25 μm.
(E-G) Developmental time course of wildtype (WT) microglial morphology. Quantifications of process length (E), process endpoints (F), and number of phagocytic cups per microglia (G). n=7 for P6, n=7 for P9, n=6 for P14. Data were compared using one-way ANOVA with posthoc Bonferroni correction.
(H) Schematic of OPL synaptogenesis.
(I) Representative retinal cross-sections showing WT P6, P9, and P14 Iba1+ OPL microglia (green), CD68+ lysosomes (red), and merge (yellow). Scale bars, 25 μm.
(J) Quantification depicting the percentages of P6, P9, and P14 WT CD68+ microglia. n=7 for P6, n=7 for P9, and n=4 for P14. Data were compared using one-way ANOVA with posthoc Bonferroni correction.
Data from (E) to (J) were pooled from two independent experiments. All data are shown as the mean ± SEM. *p<0.05, **p<0.01, ****p<0.0001. See also Figure S1.
Neuronal SIRPα was enriched during periods of peak microglia phagocytosis
Through a screen for laminar-restricted molecules in the retina, we previously uncovered SIRPα as a candidate regulator (Jiang et al., 2020). We documented significant SIRPα expression in both inner and outer retina synapse layers using a beta-galactosidase reporter line (Jiang et al., 2020), validating a prior report (Mi et al., 2000). To determine whether SIRPα was at the right place and time to modulate microglia activity in the retina, we mapped the histological distribution of SIRPα over development. SIRPα first appeared as each retinal synapse layer emerged, and its expression increased as synapses refined (P9-14), coinciding with a high degree of microglial phagocytosis (Figure 2A, Figure S2A). At the conclusion of refinement at P14, SIRPα protein levels declined, though some SIRPα remained in the OPL (Figure 2A, Figure S2B). In microglia, SIRPα was present at low but detectable amounts, showing dim co-staining with the microglia marker Iba1 (Figure 2B). However, the bulk of SIRPα signal was localized to retinal synapse layers (Figure 2C). We further confirmed SIRPα localization at synapses by staining with pre- and postsynaptic neuronal markers. We found that SIRPα colocalized with presynaptic cone and rod terminal markers (mCAR and PSD95) but not with postsynaptic horizontal cell and cone bipolar cell terminals (Calbindin and SCGN, Figure 2D–E, Figure S2C–D). Together, these results demonstrated that, as in the brain (Lehrman et al., 2018; Toth et al., 2013), SIRPα was found in both neurons and microglia in the retina during neuron refinement but that the majority of SIRPα was associated with synapses. Further, the amount of neuronal SIRPα was highest when microglia were most phagocytic. Thus, neuronal SIRPα is in the right place at the right time to impact microglial phagocytosis.
Figure 2. Neuronal SIRPα was enriched during periods of peak microglia phagocytosis.
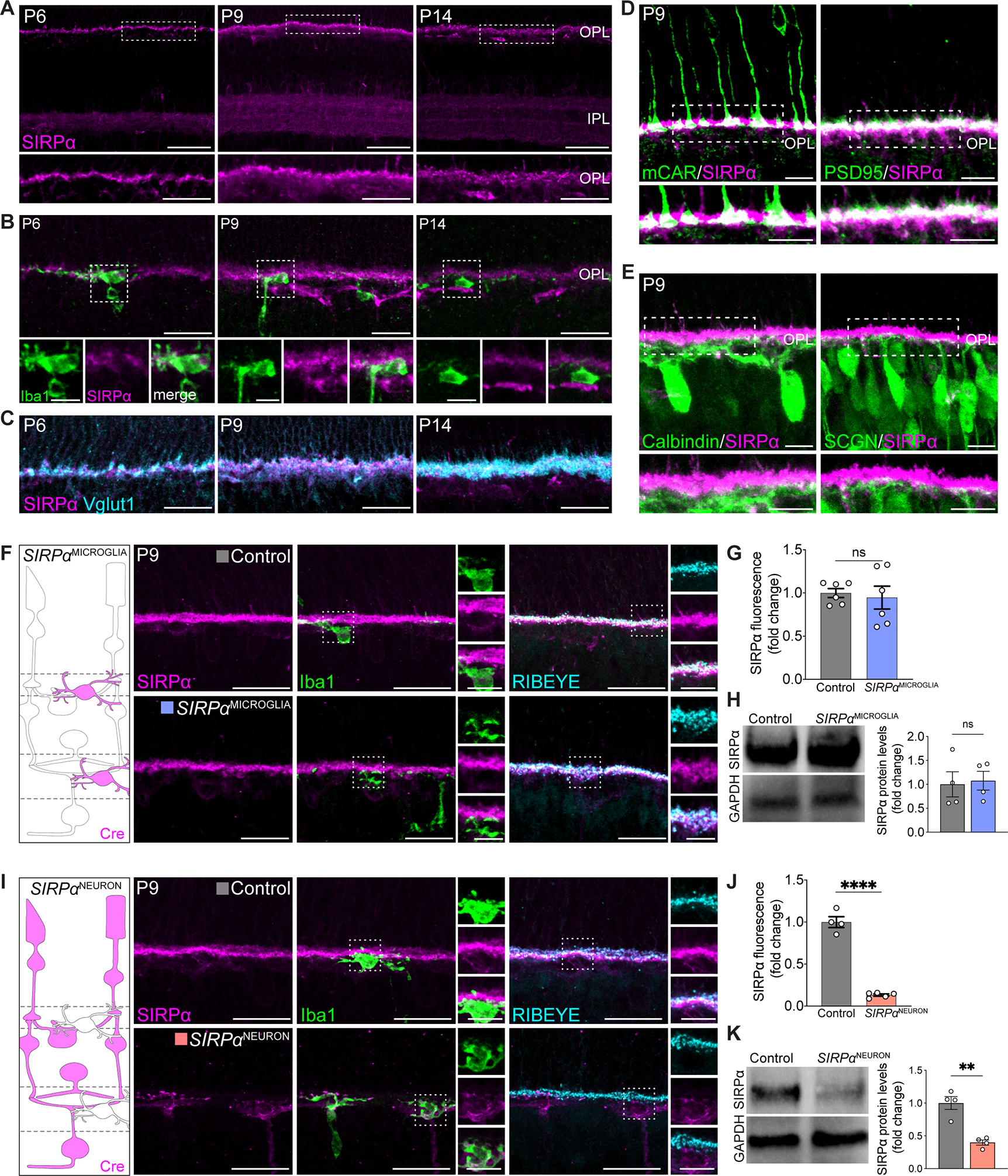
(A) Representative images showing P6, P9, and P14 WT SIRPα staining (magenta) in the synaptic layers. Scale bars, 50 μm (top) and 25 μm (bottom). See also Figure S2A–B.
(B) Representative images showing little SIRPα signal in Iba1+ microglia (green). Scale bars, 25 μm and 10 μm (insets). See also Figure S2C.
(C) Representative images showing colocalization of SIRPα (magenta) and Vglut1+ photoreceptor terminals (cyan) in the OPL. Scale bars, 25 μm. See also Figure S2C.
(D) Representative images showing colocalization of SIRPα (magenta) with cone (mCAR) and rod (PSD95) terminals (green). Scale bars, 10 μm. See also Figure S2D.
(E) Representative images showing SIRPα (magenta) with horizontal cell (Calbindin) and cone bipolar cell (SCGN) terminals (green). Scale bars, 10 μm. See also Figure S2D.
(F) Schematic of microglial SIRPα deficiency model (SIRPαMICROGLIA). Example images showing staining of SIRPα (magenta), microglia (Iba1, green), and OPL synapses (RIBEYE, cyan) in this model at P9. Scale bars, 25 μm and 10 μm (insets). See also Figure S2F.
(G) Levels of SIRPα fluorescence in OPL in SIRPαMICROGLIA relative to controls, n=6 per group. Data were compared using an unpaired t-test.
(H) Representative immunoblot image and quantification of SIRPα in whole retina from P9 WT and SIRPαMICROGLIA mice. n=3 per group. Data were compared using an unpaired t-test.
(I) Schematic of neuronal SIRPα deficiency model (SIRPαNEURON). Example images showing staining of SIRPα (magenta), microglia (Iba1, green), and OPL synapses (RIBEYE, cyan) in this model at P9. Scale bars, 25 μm and 10 μm (insets). See Figure S2F.
(J) Levels of SIRPα fluorescence in OPL in SIRPαNEURON mice relative to controls, n=4 control and 5 SIRPαNEURON. Data were compared using an unpaired t-test.
(K) Representative immunoblot image and quantification of SIRPα in whole retina from P9 WT and SIRPαNEURON. n=3 per group. Data were compared using an unpaired t-test.
Data from (H) and (K) were obtained from one experiment. (G) and (J) were pooled from two independent experiments. All data are presented as the mean ± SEM. **p<0.01, ****p<0.0001, ns, not significant. See also Figures S2–3.
SIRPα can be cleaved and secreted such that its histological localization may not necessarily reflect its primary cellular source (Nagappan-Chettiar et al., 2018; Toth et al., 2013). Accordingly, we sought to determine the cellular source of SIRPα over development and performed single-molecule fluorescent in situ hybridization (smFISH) for Sirpα mRNA (Figure S2E). We found that early in development (P2), Sirpα was present in both neurons and microglia, and this pattern persisted throughout refinement. From P14, Sirpα signal appears largely restricted to neurons. To confirm and extend these findings, we genetically assessed which cells produce SIRPα by selectively eliminating SIRPα in microglia or neurons. To achieve this, we crossed conditional SIRPαF/F mice (Skarnes et al., 2011) to either a yolk sac-derived erythro-myeloid progenitor Cre line TNFRSF11ACre (Maeda et al., 2012), which in the brain are largely comprised of microglia (Jordao et al., 2019) or a retina neuron-specific Cre line Six3Cre (Furuta et al., 2000). We termed these mouse lines SIRPαMICROGLIA and SIRPαNEURON, respectively, and confirmed their specificity (Figure S2F). We found that in the absence of microglia-derived SIRPα, total SIRPα protein levels were unaffected (Figure 2F–H). In contrast, in the absence of neuron-derived SIRPα, protein levels were significantly decreased. We observed a marked decrease of SIRPα immunofluorescent signal at synapses and in total protein levels (Figure 2I–K). Low levels of microglia-localized SIRPα protein remained visible in SIRPαNEURON mice, while neuron-associated SIRPα was unaltered in SIRPαMICROGLIA mice. To independently confirm these results, we utilized the microglia depletion model Cx3cr1CreER; Rosa26iDTR (Zhao et al., 2019). This model resulted in 96% microglia depletion at P8 (Figure S2G). Consistent with our results in SIRPαMICROGLIA mice, synaptic SIRPα was intact in Cx3cr1CreER; Rosa26iDTR animals, showing comparable staining and localization to that in controls (Figure S2H). Together, these data indicate that neurons are responsible for producing nearly all synapse-associated SIRPα and the majority of total SIRPα during neuron refinement.
Microglia phagocytosis was impaired in neuronal SIRPα-deficient mice
Given that neurons produced a high amount of SIRPα, we sought to determine the relative roles of neuron- and microglia-derived SIRPα in modulating microglia activity. To assess this, we examined microglia at P9 in SIRPαNEURON and SIRPαMICROGLIA mice using seven independent measures. These included more general morphological features (soma size, process length, and process endpoint number) and phagocytic machinery markers (CD68 and prevalence of phagocytic cups). As expected at P9, microglia in control animals displayed shorter and less branched neurites, large somas, high amounts of the lysosomal marker CD68, and a high number of phagocytic cups. Hallmarks of microglia phagocytosis were largely absent in SIRPαNEURON mice during this period. SIRPαNEURON microglia were highly ramified at P9 with long, extensive processes resulting in a significant increase in total process endpoints and length relative to controls (Figure 3A–C). Microglia in SIRPαNEURON mice also had smaller somas (Figure 3D), and CD68 was drastically reduced (Figure 3E–F). In addition, we imaged and reconstructed individual microglia at P9 and assessed the average percent volume of CD68 within each cell (Figure 3G). SIRPαNEURON mice displayed a significantly decreased volume of CD68 within microglia compared to controls (Figure 3H). Assessment of selected phagocytic pathway genes by qPCR using retinal RNA was largely consistent with our immunostaining data, showing reductions in SIRPαNEURON mice (Figure S3A). Next, we quantified phagocytic cups. Significantly fewer microglia in SIRPαNEURON mice displayed phagocytic cups, and those with cups contained half the number per cell compared to controls (Figure 3I–J, Figure S3B). To further determine whether microglial phagocytic capacity was altered in the absence of neuronal SIRPα, we electroporated GFP plasmids into the retina at P0 and assessed internalized GFP within microglia at P9. This method only transfected dividing cells, which consisted primarily of photoreceptors at this age (Figure S3C) (Matsuda and Cepko, 2004). Because microglia are born embryonically outside the retina, they are not affected (Gomez Perdiguero et al., 2015; Mass et al., 2016). Consistent with previous results, microglial internalization of GFP+ photoreceptor was significantly lower in SIRPαNEURON mice when compared to controls (Figure 3K–L). Finally, we generated SIRPαNEURON; Cx3cr1GFP/+, SIRPαMICROGLIA; Cx3cr1GFP/+, and SIRPαF/F; Cx3cr1GFP/+ mice by crossing the cell type-specific knockouts of SIRPα with Cx3cr1GFP animals, in which microglia express GFP. We confirmed that most GFP+ cells in this line were CD11b+ and CD45low, consistent with their microglial identity (Figure S3D) (Ford et al., 1995). We then performed functional phagocytosis assays by measuring the engulfment of pHrodo-red-conjugated yeast particles in retina explants followed by dissociation and flow cytometry. This pH-sensitive dye conjugate only fluoresces upon lysosomal acidification allowing measures of phagocytosis in individual microglia (Miksa et al., 2009; Wang et al., 2021b). We found that significantly fewer microglia from SIRPαNEURON; Cx3cr1GFP/+ retinas engulfed labeled particles relative to controls (Figure 3M, Figure S3E). Microglia phagocytosis is thus impaired in neuronal SIRPα-deficient mice.
Figure 3. Microglia phagocytosis was impaired in neuronal SIRPα-deficient mice.
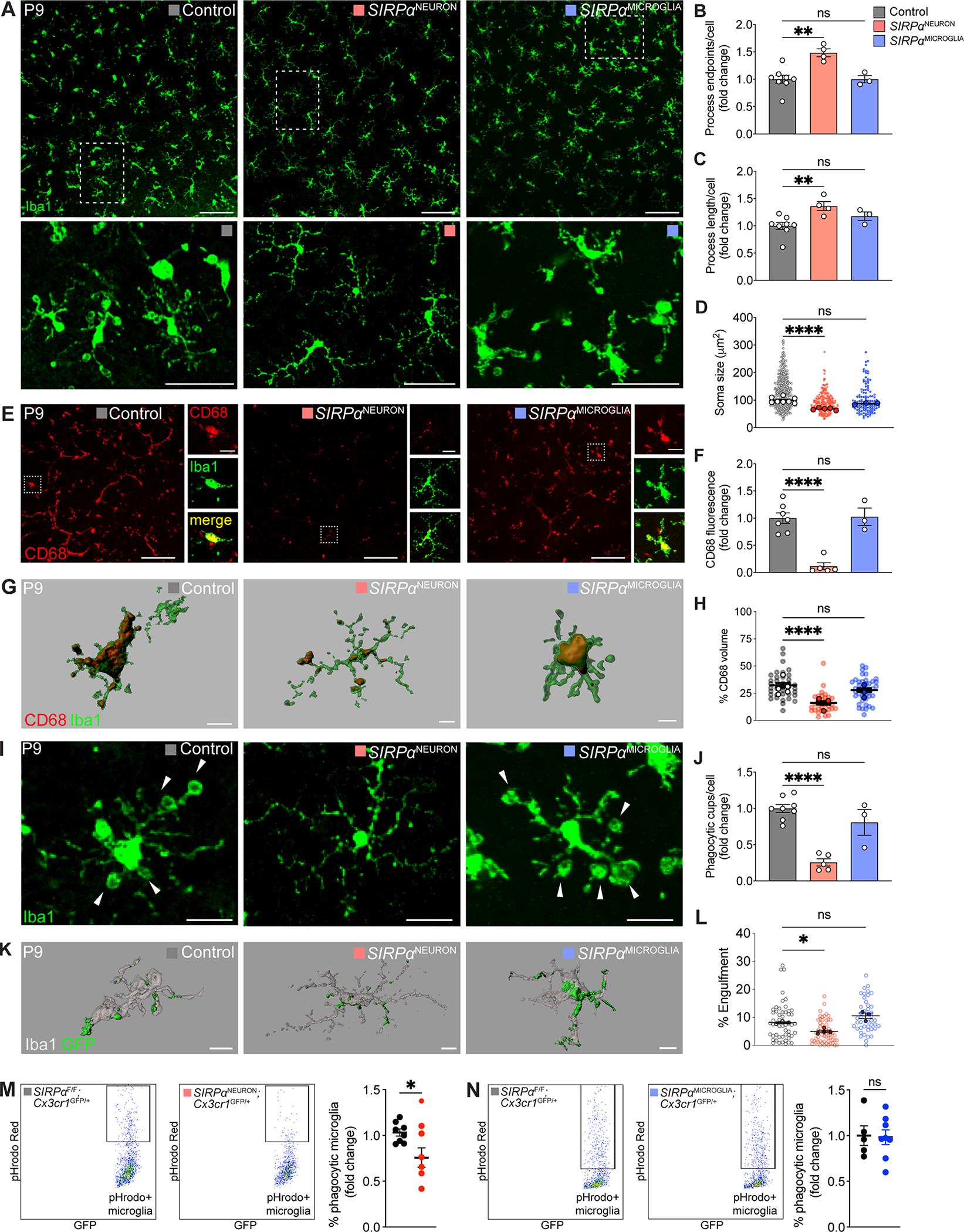
(A) Representative images of control, SIRPαNEURON, and SIRPαMICROGLIA OPL microglia at P9. Scale bars, 100 μm (top) and 50 μm (below).
(B-D) Quantifications of microglia process endpoints (B), process length (C), and soma size (D) in P9 control, SIRPαNEURON, and SIRPαMICROGLIA mice. n=8 control, 4 SIRPαNEURON, and 3 SIRPαMICROGLIA mice, one-way ANOVA with posthoc Bonferroni correction.
(E-F) Representative images showing the lysosomal marker CD68 in microglia in P9 control, SIRPαNEURON, and SIRPαMICROGLIA mice. Scale bars, 100 μm and 20 μm (insets). (F) Bar graphs depicting the levels of CD68 staining in control, SIRPαNEURON, and SIRPαMICROGLIA animals. n=8 control, 4 SIRPαNEURON, and 3 SIRPαMICROGLIA, one-way ANOVA with posthoc Bonferroni correction.
(G-H) Representative 3D reconstructions of control, SIRPαNEURON, and SIRPαMICROGLIA microglia (green) with internalized CD68+ lysosomes (red). Scale bars, 10 μm. (H) Graph showing percent volume of CD68+ lysosome in microglia from P9 SIRPαNEURON and SIRPαMICROGLIA mice relative to control. n=8 control, 4 SIRPαNEURON, 3 SIRPαMICROGLIA mice, one-way ANOVA with posthoc Bonferroni correction.
(I-J) Representative images of phagocytic cups (arrowheads) in control, SIRPαNEURON, and SIRPαMICROGLIA microglia (green). Scale bars, 20 μm. The graphs depict the number of phagocytic cups per microglia (I). Data were compared using two-way ANOVA with posthoc Bonferroni correction. See also Figure S3B.
(K-L) Representative 3D reconstructions of control, SIRPαNEURON, and SIRPαMICROGLIA microglia (gray) with internalized GFP+ neuronal material (green). Scale bars, 10 μm. (L) Graph showing percent volume of GFP-labeled neuronal material in microglia from P9 SIRPαNEURON, and SIRPαMICROGLIA mice relative to control. n=3 control, 4 SIRPαNEURON, 3 SIRPαMICROGLIA mice. Data were compared using one-way ANOVA with posthoc Bonferroni correction.
(M-N) Flow cytometry gating and quantification of microglial phagocytosis of pHrodo-red-conjugated yeast particles in (M) SIRPαNEURON; Cx3cr1GFP/+ (n=20) and SIRPαF/F; Cx3cr1GFP/+ (n=16) retinas as well as (N) SIRPαMICROGLIA; Cx3cr1GFP/+ (n=16) and SIRPαF/F; Cx3cr1GFP/+ (n-12) retinas at P9. *p<0.05, unpaired t-test. See also Figure S3D–E.
Data from (B) to (J) were obtained from one experiment. Data in (L) to (N) were pooled from three independent experiments. All data are presented as the mean ± SEM. *p<0.05, **p<0.01, ****p<0.0001, ns, not significant. See also Figure S3.
By marked contrast, microglia phagocytic activity was largely unaffected in SIRPαMICROGLIA retina. Microglial morphology in these animals was indistinguishable from that of P9 controls, and cells displayed comparable numbers of total process endpoints, length, and soma size (Figure 3A–D). In addition, CD68 staining and the internalized volume of CD68 within 3D-reconstructed microglia were similar to P9 controls (Figure 3E–H), as were the percentage of microglia with phagocytic cups and the number of cups per cell (Figure 3I–J, Figure S3B). Finally, microglia from SIRPαMICROGLIA; Cx3cr1GFP/+ retinas internalized GFP-labeled neuronal material (Figure 3K–L) and pHrodo-red-conjugated yeast (Figure 3N) at amounts similar to those of control microglia. Together these results suggest that neuronal, but not microglial SIRPα, is required to modulate microglia phagocytosis during development.
Neuronal SIRPα was required for synapse refinement and circuit function in the retina
To investigate whether decreased phagocytosis could alter synapse refinement outcomes, we next assayed synapses in neuronal and microglial SIRPα knockouts. We utilized the helpful organizational features of the OPL. Individual synapses in this layer can be quantified using the ribbon marker RIBEYE due to their large size and laminar arrangement (Samuel et al., 2011; Sarin et al., 2018). In SIRPαNEURON retina, decreased microglia phagocytosis was associated with an increase in both RIBEYE fluorescence intensity and the total number of RIBEYE+ synapses. In contrast, synapses were largely unaffected in SIRPαMICROGLIA mice (Figure 4A–F). To assess whether these alterations in synapse number affected visual function, we recorded electroretinograms (ERGs). We found that SIRPαNEURON, but not SIRPαMICROGLIA mice, showed decreased scotopic a-wave amplitudes, which report directly on photoreceptor function (Figure 4G–L). These data indicate that neuronal SIRPα-dependent microglia phagocytosis directly influences synapse refinement and circuit function.
Figure 4. Neuronal SIRPα was required for synapse refinement and circuit function in the retina.
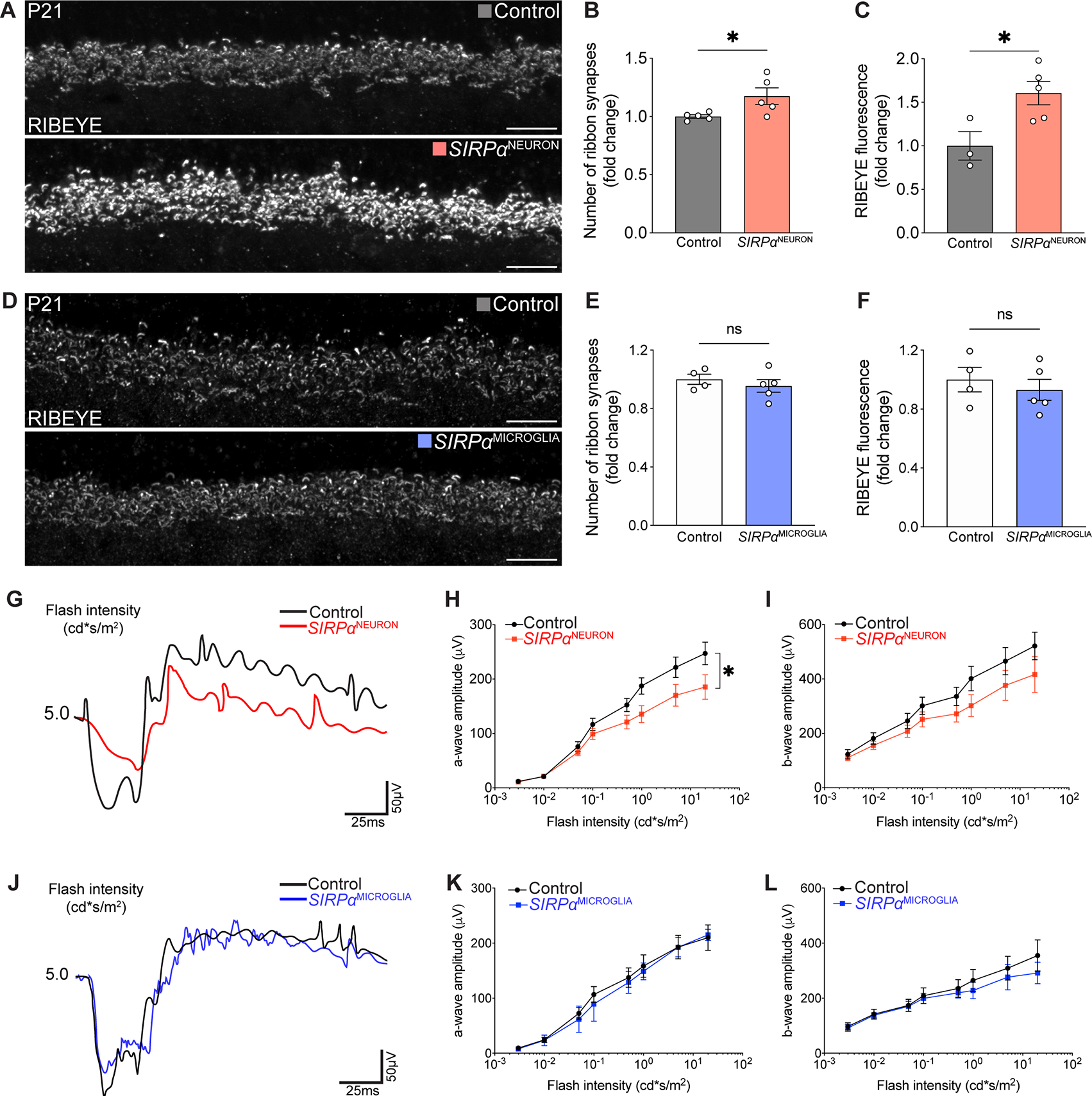
(A) Representative images of RIBEYE+ OPL ribbon synapses in control and SIRPαNEURON retinas. Scale bars, 10 μm.
(B-C) Graphs depicting the number of OPL ribbon synapses (B) and RIBEYE intensity (C) in P9 SIRPαNEURON mice relative to controls. n=5 per group, unpaired t-test.
(D) Representative images of RIBEYE-labeled OPL ribbon synapses in control and SIRPαMICROGLIA retinas. Scale bars, 10 μm.
(E-F) Graphs depicting the number of OPL ribbon synapses (E) and RIBEYE intensity (F) at P9 in SIRPαMICROGLIA mice relative to controls. n>4 per group, unpaired t-test.
(G) Representative traces of scotopic recording from control and SIRPαNEURON mice.
(H and I) Quantifications of the amplitudes of the scotopic a-wave and b-wave in control and SIRPαNEURON mice. n=7 per group, paired t-test.
(J) Representative traces of scotopic recording from control and SIRPαMICROGLIA mice.
(K and L) Quantifications of the amplitudes of the scotopic a-wave and b-wave in control and SIRPαMICROGLIA mice. n=7 per group, paired t-test.
Data were obtained from two to three independent experiments. All data are presented as the mean ± SEM. *p<0.05, ns, not significant.
Prolonging neuronal SIRPα expression extended microglial phagocytosis
To test whether neuronal SIRPα alone was sufficient to define when and where microglia were phagocytic, we used a gain-of-function approach in which we introduced SIRPα by electroporating plasmid DNA in the retina at P0. We again confirmed that neurons, but not microglia, expressed plasmid DNA following electroporation and that this method successfully increased the amount of neuronal SIRPα (Figure 5A, Figure S4A). To test the hypothesis that neuronal SIRPα can define the window in which microglia are phagocytic, we assessed microglial morphology and CD68 levels at P21 when microglia phagocytosis was low, cells were correspondingly ramified, and CD68 was reduced. Expression of SIRPα and GFP (but not GFP alone) resulted in a significant increase in markers of microglial phagocytosis (Figure 5B, Figure S4B). These cells displayed shorter processes and larger somas and showed significantly increased levels of CD68 globally and in individual microglia (Figure 5C–F). Next, we asked whether neuronal SIRPα acted as a local cue to affect microglia phagocytosis. We took advantage of the fact that electroporation targets the retina regionally (Matsuda and Cepko, 2004), generating patches of high neuronal SIRPα expression adjacent to control un-transfected regions that contain wildtype SIRPα expression. Notably, changes in microglial morphology and CD68 expression were restricted precisely to regions in which SIRPα was overexpressed, and adjacent un-transfected regions showed normal, ramified microglia that did not differ significantly from GFP-only transfected controls (Figure 5G–K). Thus, neuronal SIRPα appears sufficient to instruct both the timing and location of microglial phagocytic activity.
Figure 5. Prolonging neuronal SIRPα expression extended microglia phagocytosis.
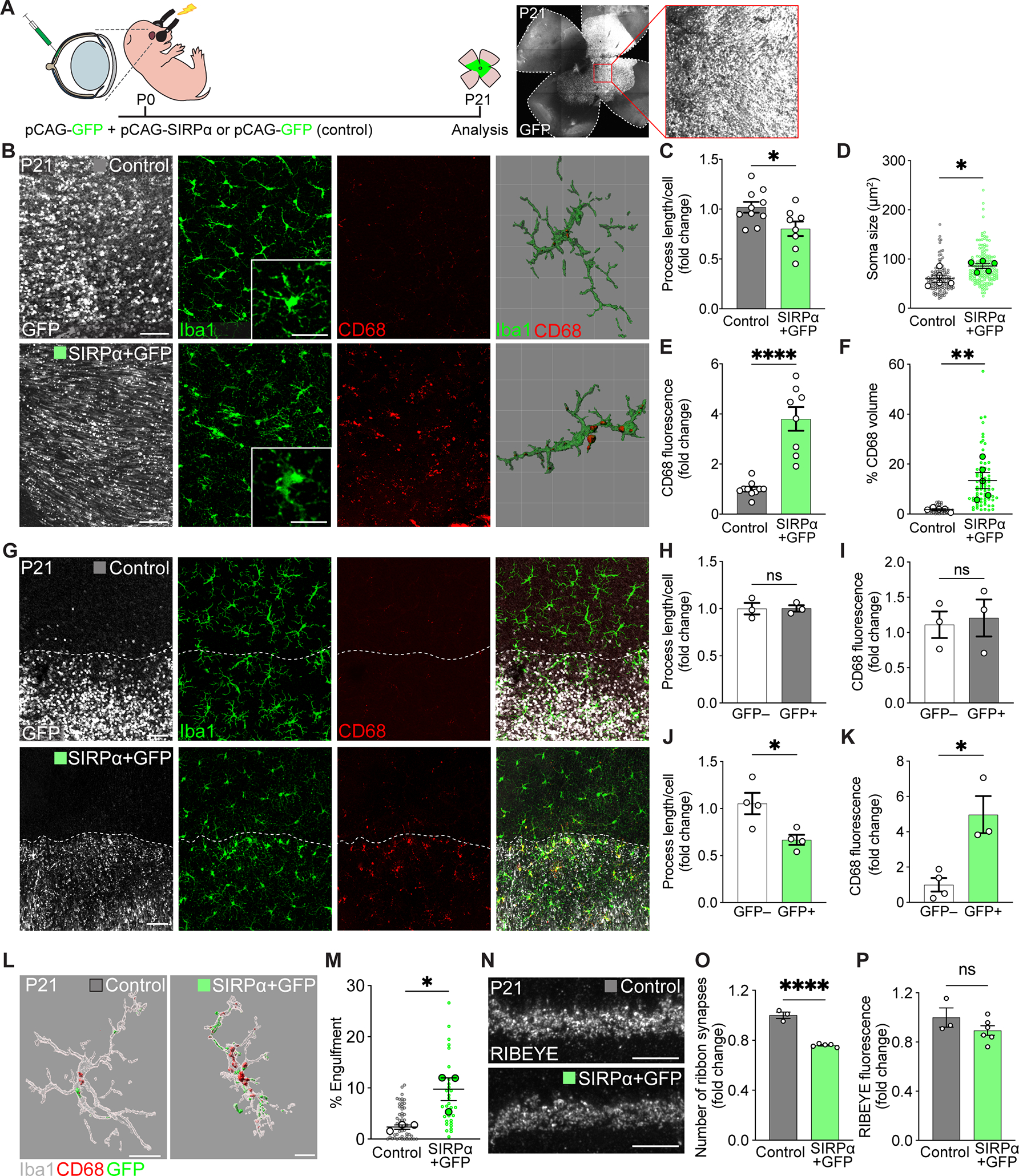
(A) Schematic illustration of in vivo electroporation. See also Figure S4A.
(B) Representative confocal and 3D reconstructed images of GFP-expressing cells (white), Iba1+ microglia (green), and CD68+ lysosomes (red) in control (GFP only) and SIRPα+GFP retinas at P21, viewed in wholemount. Scale bars, 50 μm and 25 μm (insets). See also Figure S4B.
(C-D) Quantifications of microglial morphology, including process length (C) and soma size (D), in control and SIRPα+GFP groups. n=10 control, 8 SIRPα+GFP mice, unpaired t-test. (E-F) Quantification of the levels of CD68 staining (E) and internalized CD68+ lysosome volume (F) in SIRPα+GFP versus control groups. n=10 control, 8 SIRPα+GFP mice, unpaired t-test.
(G) Representative confocal images showing borders of the electroporated retinal patch (GFP, white, border indicated by the dotted line), microglia (Iba1, green) morphology, and the levels of CD68 staining (red) in control and SIRPα+GFP regions. Scale bars, 50 μm.
(H-I) Quantifications of microglia process length (H) and CD68 staining levels (I) inside and outside GFP control transfected regions. n=3 per group, unpaired t-test.
(J-K) Quantifications of microglia process length (J) and CD68 staining levels (K) inside and outside SIRPα+GFP transfected regions. n=4 per group, unpaired t-test.
(L-M) Representative 3D-reconstructed images of P21 Iba1+ microglia (gray), internalized GFP-labeled neuronal material (green), and CD68+ lysosomes (red) in control and SIRPα+GFP regions (L), and graph showing percent volume of GFP+ material in microglia from these groups (M). Scale bars, 20 μm. n=3 per group, unpaired t-test.
(N) Representative images of RIBEYE-labeled OPL ribbon synapses in control and SIRPα+GFP groups. Scale bars, 10 μm.
(O-P) Graphs depicting the number of OPL ribbon synapses (O) and RIBEYE intensity (P) in P21 control and SIRPα+GFP groups. n=3 control and 5 SIRPα+GFP mice, unpaired t-test. Data were pooled from at least three independent experiments. All data are presented as the mean ± SEM. *p<0.05, **p<0.01, ****p<0.0001, ns, not significant. See also Figure S4.
To determine whether prolonging microglia phagocytosis beyond the normal developmental window impacted neuronal refinement, we assayed synaptic engulfment and synapse density in SIRPα electroporated retinas and respective controls. We quantified the volume of engulfed GFP+ neuronal material through 3D reconstruction of individual microglia in SIRPα+GFP and control (GFP only) transfected retinas. Microglia in SIRPα+GFP patches showed significantly increased engulfed neural material relative to those in controls (Figure 5L–M, Figure S4C). Increased engulfment was also associated with decreased synapse numbers, as the total number of RIBEYE+ synapses was significantly lower in SIRPα+GFP regions relative to controls (Figure 5N–P). Together, these data suggest that neuronal SIRPα acts as a locally restricted cue that determines microglial phagocytosis and is sufficient to extend the developmental window in which neuronal material is engulfed by microglia.
Neuronal SIRPα is juxtaposed with CD47 at synapses during development
In the periphery, SIRPα is found on phagocytes and serves to limit engulfment through recognition of its only known ligand CD47, which has been characterized as a “don’t eat me” signal (Ishikawa-Sekigami et al., 2006; Kojima et al., 2016; Willingham et al., 2012). To elucidate the cellular mechanisms through which neuronal SIRPα may impact microglia function, we first determined where and when CD47 was present in the retina. Immunostaining for CD47 revealed that it was localized to synapse layers as refinement initiated at P2 and increased as refinement progressed (Figure 6A, Figure S5A). Notably, high CD47 protein levels were present in both synapse layers at P9 during the peak of microglia-mediated neuron remodeling, and CD47 was further increased in these regions in adults. We confirmed CD47 localization at synapses by staining with pre- and postsynaptic protein markers in the OPL. Little CD47 colocalized with pre-synaptic markers (Vglut1 and PSD95). Instead, the bulk of CD47 signal overlapped with postsynaptic markers (Calbindin and SCGN), with a particular enrichment at horizontal cell terminals (Figure 6B, Figure S5B). We then performed smFISH to determine the cells responsible for CD47 mRNA production. Co-staining with cell type-specific markers confirmed high expression in postsynaptic horizontal cells (Figure 6C). Signal was also present in the INL and GCL but was largely absent from microglia (Figure 6C, Figure S5C). Together, these data suggest that CD47 is localized postsynaptically in the outer retina and that high levels of this inhibitory cue are present during peak periods of microglia phagocytosis.
Figure 6. Neuronal SIRPα was juxtaposed with CD47 at synapses during development.
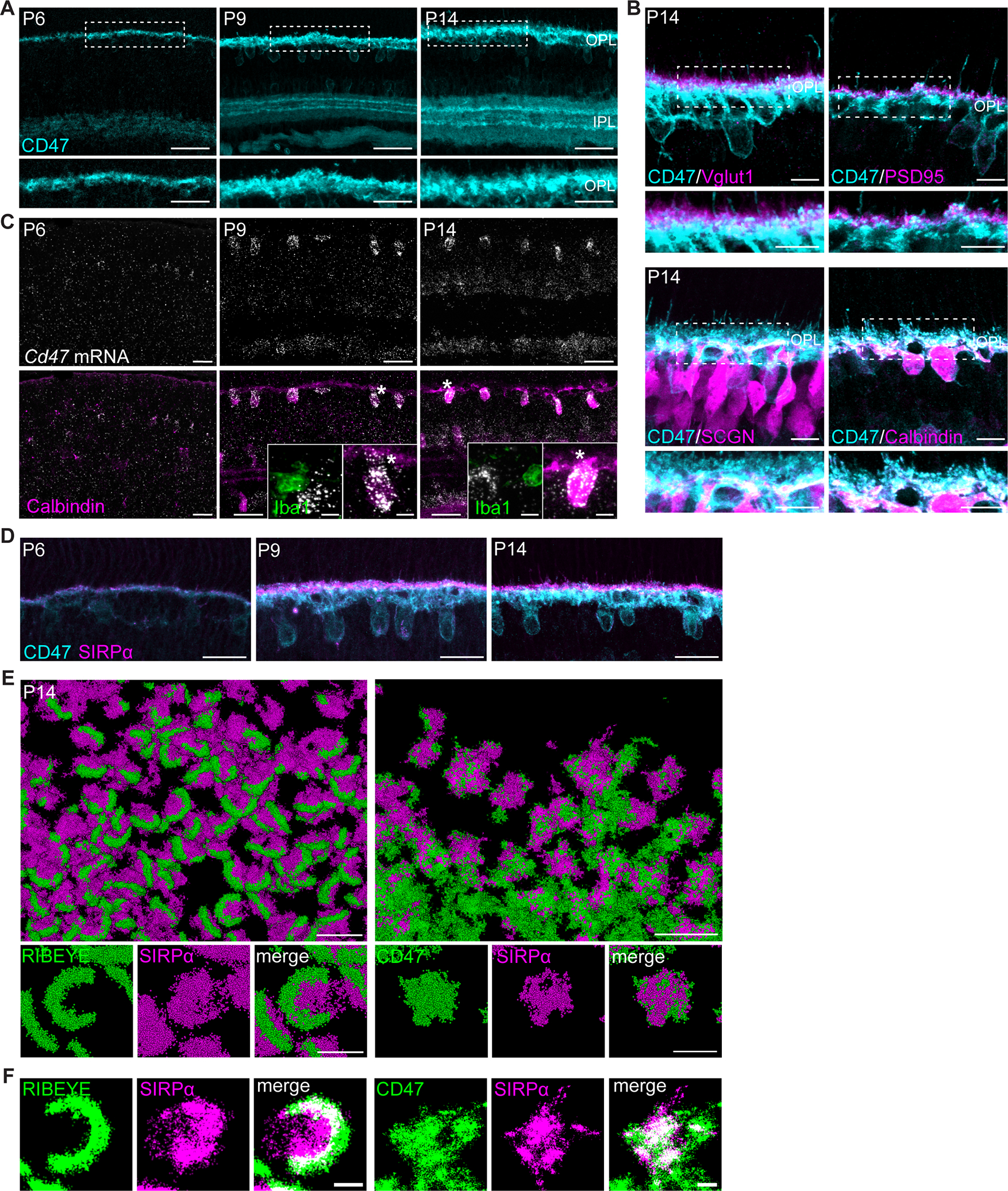
(A) Representative images showing P6, P9, and P14 WT CD47 staining (cyan) in retinal synaptic layers. Scale bars, 50 μm (top) and 25 μm (bottom). See also Figure S5A.
(B) Representative images showing the juxtaposition of CD47 (cyan) with photoreceptor terminals (Vglut1 and PSD95, magenta) as well as colocalization with cone bipolar cell (SCGN) and horizontal cell (Calbindin) terminals (magenta). Scale bars, 10 μm. See also Figure S5B.
(C) Representative images of smFISH for Cd47 mRNA(white) combined with IHC for horizontal cell marker Calbindin (magenta) and microglia marker Iba1 (green). Scale bars, 25 μm and 5 μm (insets). See also Figure S5C.
(D) Representative images showing CD47 colocalization with SIRPα at P6, P9, and P14 in WT retinas. Scale bars, 25 μm.
(E-F) Images showing examples of CD47 colocalization with SIRPα (right) and RIBEYE colocalization with SIRPα (left) in P14 retina using Stochastic Optical Reconstruction Microscopy (STORM). In (F), co-localization between SIRPα and CD47 is depicted in white. Scale bars, 2 μm (top) and 500 nm (bottom).
See also Figure S5.
We next sought to examine the structural localization of neuronal SIRPα relative to CD47. We first co-labeled both proteins in the OPL over development and examined their structural arrangement via confocal microscopy. We found that at P6, P9, and P14, SIRPα and CD47 were concentrated in the OPL and were closely associated (Figure 6D). To examine this arrangement in more detail, we performed stochastic optical reconstruction microscopy optimized for tissue imaging (Albrecht et al., 2022). Dual-color RAIN-STORM imaging confirmed that SIRPα expression was predominantly associated with RIBEYE-labeled ribbon synapses and CD47 colocalized with SIRPα at synapses (Figure 6E–F). These data indicate that neuronal SIRPα overlaps with its binding partner CD47 at synapses during development.
Neuronal SIRPα promotes microglia phagocytosis by interacting with CD47
We next investigated whether neuronal SIRPα instructed microglial phagocytosis through its interaction with CD47. To begin, we asked if CD47 itself regulated developmental microglia phagocytosis. Microglia in CD47 null mice showed a small but significant change in the number of process endpoints (Figure 7A–B). However, microglia process length, soma size, CD68 protein levels, and phagocytic cups did not differ significantly from that of controls (Figure 7C–D, Figure S6A). Based on these modest effects, we considered whether neuronal SIRPα limits inhibitory CD47 signaling to microglia during development. This model predicted that removing both neuronal CD47 and SIRPα together would restore phagocytosis that is limited by removal of neuronal SIRPα alone. To test this, we generated SIRPαF/F; CD47F/F; Six3Cre mice we termed SIRPαNEURON; CD47NEURON in which both SIRPα and CD47 were removed only in neurons. We found that microglia in SIRPαNEURON; CD47NEURON mice showed similar morphologies (Figure 7E–G), comparable CD68 expression (Figure 7H), and indistinguishable numbers and prevalence of phagocytic cups relative to controls (Figure S6B).
Figure 7. Neuronal SIRPα and CD47 functioned together to limit microglial phagocytosis.
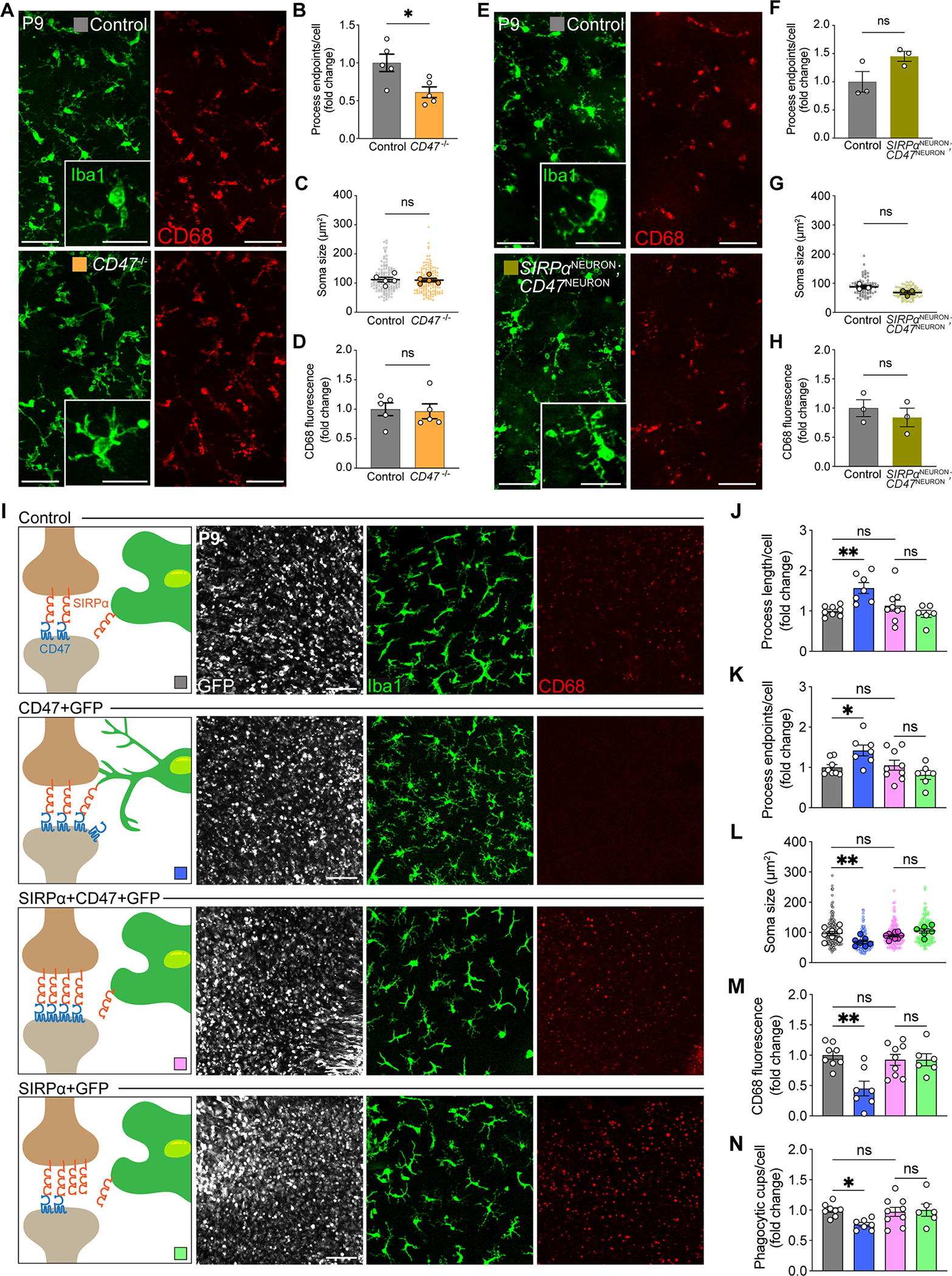
(A) Representative images of Iba1+ microglia (green) and CD68+ lysosomes (red) in control and CD47 knockout mice. Scale bars, 50 μm and 25 μm (insets).
(B-D) Quantifications of microglial morphology and levels of activation, including process endpoints (B), soma size (C), and levels of CD68 staining (D). n=5 per group, unpaired t-test. See also Figure S6A.
(E) Representative images of Iba1+ microglia (green) and CD68+ lysosomes (red) in control and SIRPα/CD47 neuron-specific double knockout mice (SIRPαNEURON; CD47NEURON). Scale bars, 50 μm and 25 μm (insets).
(F-H) Quantifications of microglial morphology and levels of activation, including process endpoints (F), soma size (G), and levels of CD68 staining (H). n=3 per group, unpaired t-test. See also Figure S6B.
(I) Representative confocal images of GFP-expressing cells (white), Iba1-labeled microglia (green), and CD68+ lysosomes (red) in control (GFP only), CD47+GFP, SIRPα+CD47+GFP, and SIRPα+GFP retinas, viewed in wholemount. Scale bars, 50 μm.
(J-N) Quantifications of microglial morphology and CD68 levels, including process length (J), process endpoints (K), soma size (L), levels of CD68 staining (M), and phagocytic cups per cell (N). n=8 control, 9 SIRPα+CD47+GFP, 7 CD47+GFP, and 6 SIRPα+GFP mice, one-way ANOVA with posthoc Bonferroni correction.
Data from (F) to (H) were obtained from one experiment. All other data were pooled from two to three independent experiments. All data are presented as the mean ± SEM. *p<0.05, **p<0.01, ns, not significant. See also Figure S6.
Our model also predicted that increasing CD47 during development may limit microglia phagocytosis, while increasing CD47 and neuronal SIRPα together would restore microglial phagocytosis (Figure 7I). To examine this, we overexpressed CD47 via electroporation at P0 and assayed microglial morphology and CD68 at P9 when microglia were highly phagocytic. Microglia in CD47+GFP patches appeared significantly more ramified relative to controls, with increased process length, process endpoints, and reduced soma size (Figure 7J–L). This was accompanied by decreased CD68 and a reduced number of phagocytic cups per cell (Figure 7M–N). The inhibitory effect of increasing neuronal CD47 on microglial phagocytic features was mitigated by co-elevating neuronal SIRPα (Figure 7I–N). Microglia in co-transfected regions displayed less ramified morphology, and the number of process endpoints, process length, and soma size were all indistinguishable from that in GFP control regions or regions in which SIRPα+GFP was transfected (Figure 7I–L). In addition, CD68 was unaltered, as was the number of phagocytic cups per cell (Figure 7M–N).
Finally, our model predicted important roles for microglia SIRPα in sensing neuronal CD47-mediated inhibition. In line with this idea, we found that microglia SIRPα is required for neuronal CD47-mediated phagocytosis inhibition. CD47 overexpression limited microglia engulfment in controls but had no effect in SIRPαMICROGLIA mice, and microglia displayed similar morphology and comparable CD68 expression (Figure S6C). We validated the critical role of neuronal SIRPα in these interactions and confirmed that genetic models did not cause baseline alterations in microglia function by restoring neuronal SIRPα in SIRPαNEURON animals via electroporation. Re-introduction of neuronal SIRPα at P0 significantly restored soma size and CD68 expression in SIRPαNEURON animals (Figure S6D). Together, these results suggest that neuronal SIRPα promotes microglia phagocytosis in development by limiting the accessibility of neuronal CD47 to microglia SIRPα.
Discussion
Microglia display defined windows of phagocytosis, with high engulfment during neural refinement that is restricted over time. Signals that limit phagocytosis as neurons mature remain largely unknown. Using the murine retina, we showed that neurons use the membrane glycoprotein SIRPα to tune the levels and timing of microglia phagocytosis. SIRPα localized to both neurons and microglia, and its expression correlated with peak developmental pruning. Using cell type-specific deletion models, we showed that while microglia-derived SIRPα is dispensable, neuron-derived SIRPα is required for elevated microglial phagocytosis during development. Deletion of neuronal SIRPα dampened microglia phagocytosis and increased retinal synapse number, while prolonging neuronal SIRPα extended the window of heightened microglial phagocytosis and reduced synapse number. Interactions between neuronal SIRPα and its binding partner CD47 drove these outcomes. The phagocytic inducing effects of prolonging neuronal SIRPα in development were restored by co-expression of neuronal CD47. Conversely, the phagocytic reducing effects of increasing neuronal CD47 were counteracted by increasing neuronal SIRPα. Finally, co-deletion of neuronal SIRPα and CD47 restored microglia phagocytosis. These results indicate that neuronal SIRPα permits microglia phagocytosis by limiting the accessibility of neuronal CD47. These results define unappreciated roles for cell type-specific SIRPα in modulating synapse engulfment.
The nervous system limits microglia engulfment to developmental periods in which neuron remodeling occurs to ensure proper circuit outcomes. Our data indicate that neuronal SIRPα is sufficient to instruct the timing of microglia phagocytosis. In support of this idea, removing neuronal SIRPα caused microglia to adopt a homeostatic morphology in development. Conversely, SIRPα overexpression was sufficient to sustain microglia phagocytosis in normally homeostatic periods. These data raise important questions regarding cause and effect. Does neuronal SIRPα influence synapse-specific decisions that alter global microglial phagocytic capacity, or does microglia phagocytic capacity fundamentally rely on the amount of CD47-SIPRα signaling? Our data cannot rule out the former possibility but most strongly support the latter. Neuronal CD47 was sufficient to rescue the effects of increasing SIRPα on microglia phagocytosis. This suggests an indirect “decoy receptor” mechanism whereby interactions between presynaptic neuronal SIRPα and postsynaptic CD47 influence phagocytosis by modulating the ability of microglia SIRPα to detect neuronal CD47. In further support of this idea, neuron-independent measures of microglia engulfment using labeled yeast particles confirmed a reduction in microglia phagocytic capacity in neuronal SIRPα mutants. Direct signaling mechanisms may also contribute. For example, neuronal SIRPα-dependent synapse loss may affect microglia, or neuronal SIRPα could bind directly to putative microglia CD47. While we did not detect measurable CD47 in microglia, CD47 has been documented at low levels on peripheral phagocytes (Doucey et al., 2004; Hayes et al., 2020). Further, while CD47 lacks a substantial cytoplasmic signaling domain (Brooke et al., 2004; Matozaki et al., 2009), it is possible that SIRPα-dependent lateral CD47 interactions with other binding partners could play important roles.
In addition to temporal alignment with neuron remodeling, microglia activity is also spatially restricted. This is particularly easy to appreciate in the laminated retina, where most microglia processes are found within synaptic regions (Li et al., 2019; Rashid et al., 2019). How local neuron-derived cues spatially restrict microglia activity was unknown. We assessed the spatial relationship between neuronal SIRPα and local microglia phagocytosis using electroporation to create restricted regions of neuronal SIRPα manipulation. Neuronal SIRPα was sufficient to locally instruct microglia activity only in the regions in which it was present. These results have a few implications. First, they help explain how microglia phagocytosis can proceed during development despite high anti-phagocytic CD47 levels by locally controlling the degree to which microglia can detect this “don’t eat me” cue. Second, they suggest that even though SIRPα can be cleaved and secreted (Toth et al., 2013), it does not appear to diffuse broadly beyond the neurons from which it is derived. Third, they suggest that despite the ability of microglia to migrate and dynamically survey diverse neural regions, movements might be limited such that the bulk of signaling occurs locally. It will be informative to determine how neuronal SIRPα influences the rate of microglia environmental sampling in real time.
This study raises important questions about the impact of local activity-dependent synapse refinement and microglia engulfment. Several cues that target synapses for removal appear to be regulated by activity. These include the complement proteins C1q and C3 (Burger et al., 2020; Schafer et al., 2012), TREM2 (triggering receptor expressed on myeloid cells 2) and its adaptor DAP12 (Filipello et al., 2018; Roumier et al., 2004), major histocompatibility complex (MHC) class I molecules (Datwani et al., 2009; Huh et al., 2000), and fractalkine and its receptor (Gunner et al., 2019; Paolicelli et al., 2011; Rogers et al., 2011). In line with these ideas, SIRPα can directly contribute to synapse maturation in an activity-dependent manner, while CD47 can serve as an activity-dependent “don’t eat me” cue that modifies microglia-mediated synapse pruning (Lehrman et al., 2018; Toth et al., 2013). These models imply that the amounts of these synapse-associated proteins vary from synapse to synapse in a way that is predictive of whether a particular synapse will be removed or maintained. Our results may help shed may light on these questions. Using STORM-microscopy, we found that nearly all synapses in the OPL contained both CD47 and SIRPα, and amounts did not appreciably differ from synapse to synapse. While we cannot rule out that minor differences in SIRPα may influence whether a synapse is lost or maintained, these results are more consistent with the idea that CD47-SIRPα signaling at a local population level rather than at single synapses impacts microglia phagocytic activity.
How might our results be viewed in the context of SIRPα and CD47 whole-body knockout experiments in the brain? Lehrman et al. show limited microglia morphological changes but enhanced microglia engulfment when either SIRPα or CD47 is removed from all cells, resulting in a ~20–30% decrease in synapse number in the dorsal lateral geniculate nucleus (Lehrman et al., 2018). Similarly, we observed minor morphological changes in CD47 global knockouts with limited but measurable impacts on phagocytosis. Three factors might contribute to the observed brain outcomes in whole-body SIRPα and CD47 knockouts. First, global deletion may obscure the cell subtype-specific contribution of SIRPα on neurons and microglia. Our model predicts that neuronal and microglial SIRPα have opposing roles. The former is required to promote microglia phagocytosis by temporally limiting microglia SIRPα access to CD47, while the latter is required to limit microglia phagocytosis when neuronal SIRPα decreases, exposing CD47. Consistent with this idea, loss of microglial SIRPα worsens outcomes in a mouse model of Alzheimer’s disease (Ding et al., 2021). Second, it is possible that the cellular mechanisms through which SIRPα and CD47 signal may differ between the retina and retinorecipient areas in the brain. However, we view this as unlikely given that: 1) a large body of evidence suggests that retina microglia are structurally, functionally, and developmentally analogous to those in the brain (Anderson et al., 2019; Burger et al., 2020; Hooks and Chen, 2007; Hume et al., 1983; O’Koren et al., 2019; Punal et al., 2019; Schafer et al., 2012; Silverman and Wong, 2018; Stevens et al., 2007; Umpierre and Wu, 2021; Wang et al., 2016; Werneburg et al., 2017), and 2) the expression of these proteins in neurons and microglia are temporally and structurally conserved in the retina and the brain (Adams et al., 1998; Comu et al., 1997; Jiang et al., 2020; Mi et al., 2000). Studies aimed at addressing CD47 and SIRPα cell-specific signaling in the dLGN and other brain regions may aid in resolving these questions.
Finally, our results have potential implications for neurodegenerative diseases. Microglia reactivation is increasingly implicated in the pathogenesis of a large number of both retina and brain diseases and injuries, including diabetic retinopathy, Alzheimer’s disease, frontal temporal dementia, demyelinating diseases, and psychiatric diseases (Altmann and Schmidt, 2018; Estes and McAllister, 2015; Hong et al., 2016; Kinuthia et al., 2020; Lall et al., 2021; Lui et al., 2016; Perry et al., 2010; Salter and Stevens, 2017; Sellgren et al., 2019; Vasek et al., 2016; Werneburg et al., 2020). Might neuronal SIRPα and CD47 be involved in these outcomes? Our model predicts that the answer might depend on the timing of intervention and the regional amounts of neuronal SIRPα, microglial SIRPα, and CD47. For example, in a disease-affected region in which high amounts of neuronal SIRPα and CD47 are present, decreasing neuronal SIRPα may be sufficient to reduce microglia activity and improve neural outcomes. In contrast, for diseases of excess connectivity (e.g. autism), elevating neuronal SIRPα in otherwise low SIRPα regions may be sufficient to locally induce microglia phagocytosis. Similarly, reduced CD47 expression has been documented in patients with multiple sclerosis (Han et al., 2012; Koning et al., 2007), and experimental models suggest that CD47-SIRPα signaling plays dual roles in this disease (Azcutia et al., 2017; Han et al., 2012; Wang et al., 2021a). Given these results, understanding the regional, neuron-subtype, and synapse-specific consequences of CD47-SIRPα signaling may provide new therapeutic opportunities for precisely intervening in neurological disease progression.
Limitations of the study
In this study, we demonstrated that neuronal SIRPα promotes microglia phagocytosis because it prevents CD47 from accessing microglial SIRPα. Removing SIRPα from neurons, but not microglia, reduced microglial phagocytosis and increased synapse number. However, it is not clear if neuronal SIRPα alters other non-phagocytic microglia functions. Future work will also be needed to determine whether neuronal SIRPα is required in adulthood to promote microglia phagocytosis in normal or disease conditions. Our data also showed the SIRPα-CD47 signaling is sufficient to alter microglia state during development despite the presumed presence of a variety of pro-engulfment cues, but precisely how microglia reconcile conflicting “eat me” and “don’t eat me” cues was not determined.
STAR METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Melanie Samuel (melanie.samuel@bcm.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
SIRPαF/F mice were kindly provided by Dr. Beth Steven, Boston Children’s Hospital. To broadly delete SIRPα from retinal neurons, SIRPαF/F mice were crossed to Six3Cre mice (Furuta et al., 2000), referred here as SIRPαNEURON mice. To delete SIRPα in microglia, SIRPαF/F mice were crossed to TNFRSF11ACre mice (Maeda et al., 2012) to generate animals referred here as SIRPαMICROGLIA mice. TNFRSF11ACre is expressed in and targets yolk sac-derived erythro-myeloid progenitors (Jordao et al., 2019), which in the brain are comprised of microglia. For these lines, SIRPαF/F littermates were used as controls. To deplete microglia, Cx3cr1CreER mice (Yona et al., 2013) were crossed to ROSA26iDTR mice (Buch et al., 2005) to generate animals referred here as Cx3cr1CreER; Rosa26iDTR mice. C57BL/6 mice, Cx3Cr1GFP/+ mice, CD47F/F, and CD47−/− mice were obtained from Jackson Labs. SIRPαNEURON; CD47NEURON double knockouts were generated by crossing SIRPαF/F and CD47F/F mice to Six3Cre mice. For this line, SIRPαF/F; CD47F/F littermates were used as controls. All mice were used at the ages specified in the experimental procedures outlined below, and a mixture of male and female mice were used. Experiments were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the NIH under protocols approved by the BCM Institutional Animal Care and Use Committee.
METHOD DETAILS
Microglia depletion
For microglia ablation experiments, the ROSA26iDTR line (Buch et al., 2005) was crossed to the Cx3cr1CreER line (Yona et al., 2013) to generate animals referred here as Cx3cr1CreER; Rosa26iDTR. To ablate microglia for longer periods of time and prevent repopulation of microglia, we administered tamoxifen and diphtheria toxin as previously described (Punal et al., 2019). In brief, 100 μg of tamoxifen was administered via intraperitoneal (IP) injection to neonatal pups at P1, P5, and P7, and single doses of 80 ng of Diphtheria toxin were administered at P4, P6, and P8. Depletion (~96% compared to control) was confirmed by staining with the microglial marker Iba1 at P8 (Figure S2G).
Immunohistochemistry
Immunohistochemistry was performed as previously described (Jiang et al., 2020). Briefly, eyes were harvested from mice at P2, P6, P9, P14, P21, and 14 weeks and fixed in 4% PFA for 45 min at room temperature. For cross-section analysis, eye cups were dissected, and the cornea and lens were removed. Following cryoprotection in 30% sucrose, eyes were embedded in OCT compound (VWR) and sectioned at 20 μm thickness. Cryosections were incubated with blocking buffer (3% normal donkey serum and 0.3% Triton X-100 in PBS) for 1 h, and then with primary antibodies diluted in blocking buffer overnight at 4°C. After washing, secondary antibodies were applied and incubated for 1 h at room temperature. Slides were then washed again and mounted with Vectashield (Vector Labs). For whole-mount preparations, the retinas were removed from the eye cups and blocked with a 10% normal donkey serum and 0.5% Triton X-100 solution in PBS for 1 h before proceeding with incubation with primary antibodies diluted in blocking buffer for 5 days followed by washes and staining with secondary antibodies for 3 days at 4°C. All images were acquired using an Olympus Fluoview FV1200 confocal microscope and processed using FIJI.
RNAscope
RNAscope single-molecule fluorescence RNA in situ hybridization (smFISH) was performed on 20 μm sections of retina collected as described for immunohistochemistry using Probe-Mm-SIRPα (837091) and Probe-Mm-CD47-C2 (515461-C2, ACD-bio). RNAscope fluorescent multiplex assays were performed according to the manufacturer’s instructions (ACD-bio) with the following modifications. Tissue samples were dehydrated using an ethanol gradient of 10%, 30%, 50%, 70%, and 100% (3 min each), and the boiling time in target retrieval solution was modified to 5 min. After RNAscope, slides were co-stained with Iba1, Calbindin, RBPMS, and AP2 to visualize microglia, horizontal cells, ganglion cells and amacrine cells, respectively.
Quantitative Real-Time PCR
Total RNA was isolated from whole retinas of P9 control and SIRPαNEURON animals using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. 100ng of RNA was used to generate cDNA by reverse transcription using the iScript Reverse Transcription Supermix (BioRad). qRT-PCR was subsequently performed using the iTaq Universal SYBR Green Supermix (BioRad) on a CFX384 Touch Real-Time PCR Detection System with primer sequences listed in Key Resources Table.
Key resources table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Ibal | Wako | Cat#019-19741; RRID: AB_839504 |
| Goat polyclonal anti-Ibal | Abcam | Cat#ab5076; RRID: AB_2224402 |
| Rat monoclonal anti-CD68 | Bio-Rad | Cat#MCA1957; RRID:AB_322219 |
| Rat monoclonal anti-SIRPα | BD Biosciences | Cat#552371; RRID:AB 394371 |
| Rabbit polyclonal anti-SIRPα | Thermo Fisher | Cat#PA5-19869; RRID:AB 11155968 |
| Rat monoclonal anti-CD47 | BD Biosciences | Cat#555297; RRID:AB 395713 |
| Goat polyclonal anti-CD47 | R&D Systems | Cat#AF1866; RRID:AB 2074942 |
| Chicken polyclonal anti-GFP | Abcam | Cat#ab13970; RRID:AB 300798 |
| Rabbit polyclonal anti-Vglut1 | Synaptic Systems | Cat#135302; RRID:AB 887877 |
| Rabbit polyclonal anti-mouse cone arrestin (mCAR) | Millipore | Cat#AB15282; RRID:AB 11210270 |
| Goat polyclonal anti-PSD95 | Abcam | Cat#ab12093; RRID:AB 298846 |
| Rabbit polyclonal anti-RIBEYE | Synaptic Systems | Cat#192103; RRID:AB_2086775 |
| Rabbit polyclonal anti-secretagogin (SCGN) | BioVendor Laboratory Medicine | Cat#RD181120100; RRID:AB_2034060 |
| Chicken polyclonal anti-Calbindin D-28k | Novus biologicals | Cat#NBP2-50028; N/A |
| Guinea pig polyclonal anti-RBPMS | PhosphoSolutions | Cat#1832-RBPMS; RRID: AB 2395389 |
| Mouse monoclonal anti-AP2 alpha (3B5) | Developmental Studies Hybridoma Bank | Cat#3b5; RRID: AB 528084 |
| Rabbit polyclonal anti-GAPDH | MIllipore | Cat#ABS16; RRID:AB 10806772 |
| Donkey anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor 488 | Jackson ImmunoResearch Labs | Cat#711-545-152; RRID:AB_2313584 |
| Donkey anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor 594 | Jackson ImmunoResearch Labs | Cat#711-585-152; RRID:AB_2340621 |
| Donkey anti-rabbit IgG (H+L) secondary antibody, Alexa Fluor 647 | Jackson ImmunoResearch Labs | Cat#711-605-152; RRID:AB_2492288 |
| Donkey anti-rat IgG (H+L) secondary antibody, Alexa Fluor 488 | Jackson ImmunoResearch Labs | Cat#712-545-150; RRID:AB_2340683 |
| Donkey anti-rat IgG (H+L) secondary antibody, Alexa Fluor 594 | Jackson ImmunoResearch Labs | Cat#712-585-150; RRID:AB_2340688 |
| Donkey anti-rat IgG (H+L) secondary antibody, Alexa Fluor 647 | Jackson ImmunoResearch Labs | Cat#712-605-150; RRID:AB_2340693 |
| Donkey anti-goat IgG (H+L) secondary antibody, Alexa Fluor 488 | Jackson ImmunoResearch Labs | Cat#705-545-003; RRID:AB_2340428 |
| Donkey anti-goat IgG (H+L) secondary antibody, Alexa Fluor 594 | Jackson ImmunoResearch Labs | Cat#705-585-004; RRID:AB_2340432 |
| Donkey anti-goat IgG (H+L) secondary antibody, Alexa Fluor 647 | Jackson ImmunoResearch Labs | Cat#705-605-005; RRID:AB_2340436 |
| Donkey anti-chicken IgY (IgG) (H+L) secondary antibody, Alexa Fluor 488 | Jackson ImmunoResearch Labs | Cat#703-545-155; RRID:AB_2340375 |
| Donkey anti-mouse IgG (H+L) secondary antibody, Alexa Fluor 488 | Jackson ImmunoResearch Labs | Cat#715-545-150; RRID:AB_2340846 |
| Donkey anti-mouse IgG (H+L) secondary antibody, Alexa Fluor 594 | Jackson ImmunoResearch Labs | Cat#715-585-150; RRID:AB_2340854 |
| Critical Commercial Assays | ||
| Plasmid Plus Midi Kit | QIAGEN | Cat#12943 |
| RNAscope Multiplex Fluorescent Reagent Kit v2 Assay | ACD-Biotechne | Cat#323100 |
| RNeasy Mini Kit | QIAGEN | Cat#74104 |
| iScript Reverse Transcription Supermix | BioRad | Cat#170-8840 |
| iTaq Universal SYBR Green Supermix | BioRad | Cat#172-5124 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Cx3cr1+/eGFP: B6.129P-Cx3cr1tm1Litt/J | The Jackson Laboratory | JAX stock #005582 |
| Mouse: Sirpatm1b(EUCOMM)Hmgu | Gift from Beth Stevens (Boston Children’s Hospital) | N/A |
| Mouse: Cd47tm1Fpl | The Jackson Laboratory | JAX stock #003173 |
| Mouse: Six3-Cre | The Jackson Laboratory | Furuta et al., 2000; JAX stock #019755 |
| Mouse: Tnfrsf11a-Cre | Gift from Frederic Geissmann (Memorial Sloan Kettering Cancer Center) | N/A |
| Mouse: Tnfrsf11a-Cre | Gift from Frederic Geissmann (Memorial Sloan Kettering Cancer Center) | N/A |
| Mouse: Cx3cr1tm2.1(cre/ERT2)Jung |
The Jackson Laboratory | Yona et al., 2013; JAX stock #020940 |
| Mouse: ROSA26iDTR | The Jackson Laboratory | Buch et al., 2005; JAX stock #003173 |
| Recombinant DNA | ||
| Plasmid: pCAG-SIRPα | This paper | N/A |
| Plasmid: pCAG-CD47 | This paper | N/A |
| Plasmid: pCAG-GFP (in pcDNA3.1) | Gift from Elizabeth Zuniga-Sanchez, Matsuda and Cepko, 2007 | N/A |
| Plasmid: pCAG in pcDNA3.1 | Gift from Elizabeth Zuniga-Sanchez | N/A |
| Plasmid: Sirpa mouse tagged ORF clone | ORIGENE | Cat# MG208194 |
| Plasmid: Cd47 mouse tagged ORF clone | ORIGENE | Cat# MG204706 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Diphtheria toxin | Sigma | Cat#D0564 |
| Tamoxifen | Sigma | Cat#T5648 |
| 10X PBS | VWR | Cat#101175-842 |
| Normal Donkey Serum | Jackson ImmunoResearch Labs | Cat#017-000-121 |
| Triton X-100 | EMD | Cat#TX1568-1 |
| Vectashield | Vector Labs | Cat#H-1000-10 |
| FastGreen Dye | Sigma | Cat#F7252-5G |
| RNAscope Probe Mm-Sirpa-C1 | ACD-Biotechne | Cat#837091 |
| RNAscope Probe Mm-Cd47-C2 | ACD-Biotechne | Cat#515461-C2 |
| pHrodo Red Zymosan Bioparticles Conjugate for Phagocytosis | Thermo Fisher | Cat#017-000-121 |
| DMEM/F12 (1:1) | Gibco | Cat#11039-021 |
| B27 (50X) | Gibco | Cat#17504-044 |
| BDNF | Invitrogen | Cat#PHC7074 |
| Dnase I | Sigma | Cat#D4527 |
| Papain | Worthington | Cat#LS003126 |
| MEM | Gibco | Cat#11090 |
| Bovine serum albumin (BSA) | Sigam | Cat#A-9418 |
| Ovomucoid | Worthington | Cat#LS003087 |
| L-cysteine | Sigma | Cat#C1276 |
| 2.5% Phenylephrine Hydrochloride Ophthalmic Solution | Akorn | NDC: 17478-201-15 |
| 1% Tropicamide Ophthalmic Solution | Akorn | NDC: 17478-101-12 |
| Gonak 3ypromellose ophthalmic demulcent solution | Akorn | NDC: 17478-064-12 |
| cOmplete protease inhibitor | Roche | Cat#04693124001 |
| Phosphatase inhibitor I | Calbiochem | Cat#524624 |
| Phosphatase inhibitor II | Calbiochem | Cat#524625 |
| Oligonucleotides | ||
| qRT-PCR primers | ||
| β actin | Fwd – TGAGAGGGAAATCGTGCGTG Rev – TCGTTGCCAATAGTGATGACCTG |
Anderson et al., 2019 |
| Cx3cr1 | Fwd – AAAAACACTGGATTTCAGGGGC Rev – CAACCAACACAGGAACAGGGAG |
Anderson et al., 2019 |
| Mertk | Fwd – CGCTCTGGAGTGGAGGCAC Rev – AAACGCAACAGGAGGTAGGAG |
Anderson et al., 2019 |
| CD68 | Fwd – GGACACTTCGGGCCATGTTT Rev – CTTACACAGTGGACTGGGGC |
Anderson et al., 2019 |
| C1qb | Fwd – ATGGATGCGTAATCACGGGG Rev – GTCTGGGTTTCAGGCAGTCAAG |
Anderson et al., 2019 |
| C3 | Fwd – TCTGACCTCTGGGGAGAAAAGC Rev – TGGGACAACCATAAACCACCATAG |
Anderson et al., 2019 |
| Cd11b | Fwd – TGTGGACTCTCATGCCTCCT Rev – TGGTCATCTCTGAAGCCGTG |
Anderson et al., 2019 |
| VNR | Fwd – CGTCCTCCAGGATGTTTCTCC Rev – GCTTTGACCTCACAGAGGC |
Anderson et al., 2019 |
| Ccr2 | Fwd – GCTGTGTTTGCCTCTCTACCAG Rev – CAAGTAGAGGCAGGATCAGGCT |
NCBI Primer-BLAST |
| Mfge8 | Fwd – GAGCAACAGTGCCAAGGAATGG Rev – ACTGTGGGCTACCTTGTAGGAC |
NCBI Primer-BLAST |
| Tyrobp | Fwd – GGACCCGGAAACAACACATTG Rev – GATCCCAGAGAGGGCTTGTT |
Anderson et al., 2019 |
| LRP | Fwd – GTGTCCAACTGCACAGCAAG Rev – GCAGACGTGAATGTCGCAAT |
Anderson et al., 2019 |
| Softwares | ||
| Imaris, version 9 | Bitplane | http://www.bitplane.com/imaris |
| FIJI | SciJava | https://fiji.sc |
| GraphPad Prism, version 9 | GraphPad Software | http://www.graphpad.com/scientific-software/prism |
| Others | ||
| BTX Harvard Apparatus Tweezertrodes | BTX | Cat#BTX450166 |
| BTX Electroporation System | BTX | Cat#ECM830 |
| Capillary Glass | Sutter Instrument | Cat#BF100-50-10 |
| Fluoview FV1200 confocal microscope | Olympus | Model FV1200 |
| STORM microscope | Bruker | Model Vutara 352 |
| Celeris ERG system | Diagnosys LLC | N/A |
| Kimble Kontes Pellet Pestle homogenizer | DWK Life Sciences | Cat#749540-0000 |
| CFX384 Touch Real-Time PCR Detection System | BioRad | N/A |
Plasmid construction
pCAG and pCAG-GFP vectors were kindly provided by Dr. Elizabeth Zuniga-Sanchez at Baylor College of Medicine. In brief, the pCAG vector was generated by cloning the promoter region of the original pCAG-IRES-GFP (Matsuda and Cepko, 2004; 2007) plasmid into the pcDNA3.1 vector (Invitrogen). The pCAG-GFP construct was generated by adding GFP to the pCAG (in pcDNA3.1) vector. Coding sequences for either SIRPα and CD47 (MG208194 and MG204706, Origene) were removed and cloned downstream of the CAG promoter in the pCAG vector. These vectors were then expressed in combination with the pCAG-GFP to allow for fluorescent visualization.
Electroporation
For SIRPα and CD47 over-expression, retinas of neonatal pups (12–24 h) were electroporated with the expression plasmids detailed above using a modified version of the protocol developed by Cepko and colleagues (Matsuda and Cepko, 2004). Briefly, sharp end glass micropipettes (Sutter Instrument) were loaded with 5–8 μl of DNA (diluted to a final concentration of 4 μg/μl) mixed with Fast Green Dye (0.2X) and were used to deliver 2–3 μl DNA into the subretinal space. Following injection, five current pulses (80V, 50ms duration, 950ms interval) were applied across the pup head using Tweezer electrodes (Harvard Apparatus).
Electroretinography
P21 SIRPαNEURON (n=5), SIRPαMICROGLIA (n=3), and respective littermate control (n=4, n=7) animals were dark-adapted overnight and then anesthetized with isoflurane (3% induction, 1.5% maintenance) carried in oxygen at a flow rate of 1 L/min using a vaporizer. Animals were placed on a heated platform, and eyes were dilated with phenylephrine hydrochloride and tropicamide. A contact lens-style electrode in contact with Gonak solution was placed on each cornea. A reference electrode was placed at the forehead, and a ground electrode was placed at the hip. Scotopic responses were elicited in the dark with flashes ranging from 0.003 cd*s/m2 to 20 cd*s/m2 using the Diagnosys Celeris ERG system. Electroretinograms were recorded from both eyes simultaneously.
Ex vivo phagocytosis assay
We performed ex vivo phagocytosis assays as previously described (Wang et al., 2021b). In brief, freshly dissected retinas from P9 control and SIRPαNEURON; Cx3cr1GPF/+ animals were incubated in 1 mg/mL pHrodo Red-conjugated zymosan bioparticles (Thermo Fisher Scientific) resuspended in culture media of 1:1 mixture of DMEM and F-12 supplemented with B27(50X), BDNF(50X), and penicillin-streptomycin(100X) at 37 °C with gentle agitation. Retinas were subsequently washed three times with PBS and dissociated using cysteine-activated papain for 8 min at 37°C. Digestion was inactivated by the addition of medium containing ovomucoid (1.5 mg/mL), BSA(1.5 mg/mL) and DNase I (67 U/mL), followed with gentle mechanical dissociation by pipetting up and down with a P1000 tip. The sample was spun at 30 g for 20 sec, and supernatant containing cells was passed through a 40 μm strainer. This process was repeated until all cells were dissociated. Cells were then spun at 350 g at 4°C for 10 min and resuspended in 500 μL of MEM-B (no glutamine with 4% Bovine Serum Albumin media) with 0.5 μg/mL DAPI. Flow cytometry data were collected in a BD LSR II Cytometer and analyzed using FlowJo 9 to compute the percentage of GFP-positive microglia that were also positive for pHrodo Red.
Immunoblotting analysis
WT (P4, P6, P9, P14), SIRPαNEURON (P9), and SIRPαMICROGLIA (P9) retinas were dissected and snap frozen on dry ice. Frozen tissues were then transferred into a RIPA buffer containing cOmplete protease inhibitor (Roche, 1:50), phosphatase inhibitor I (Calbiochem, 1:100), and phosphatase inhibitor II (Calbiochem, 1:100). Samples were manually homogenized with a Kimble Kontes Pellet Pestle homogenizer (DWK Life Sciences). For each sample, 10 μg of protein was loaded and separated by SDS-PAGE on 10% tris-glycine gels before transferred onto nitrocellulose membranes. Blots were blocked in blocking buffer (5% BSA, 0.05% Tween 20 in TBS) for 1 h and then probed with primary antibodies overnight at 4°C in 5% BSA. Blots were subsequently washed and stained with secondary antibodies for 1 h at room temperature. FIJI was used to perform densitometry analysis of bands.
STORM imaging
Samples were prepared and imaged as described in Albrecht et al., 2021. In brief, eyes were harvested from P9 animals and fixed in 4% PFA for 45 min at room temperature. Eye cups were subsequently dissected, and the cornea and lens were removed. Following cryoprotection in 30% sucrose, eyes were embedded in OCT compound (VWR) and sectioned at 10 μm thickness. Cryosections were incubated with a 3% normal donkey serum and 0.3% Triton X-100 solution in PBS for 1 h, and then with primary antibodies overnight at 4°C. After washing, secondary antibodies were applied and incubated for 1 h at room temperature. Images were acquired on a Bruker Vutara 352 (Bruker, Billerica, MA) using a 60X water objective (UPLSAPO60XW) at an axial step size of 200 nm. 3D-structured images of OPL synapses were generated using the Ordering Points to Identify the Clustering Structure (OPTICS) algorithm. To analyze images, a general particle distance of 0.16 μm and a particle count per cluster of 25 was used for all channels on all images.
QUANTIFICATION AND STATISTICAL ANALYSIS
Histological quantification
Microglia density quantification
To quantify wildtype microglia cell density at P6, P9, and P14, three independent fields of view (635.90 μm × 635.90 μm) from one retina were imaged per animal, and three animals were imaged (n=3). The number of microglia was subsequently counted in each field, and density of microglia was calculated by dividing the total number of microglia in each field by the image area.
Microglia morphology quantification
To assess microglia morphology at P9, whole-mount retinas were stained for Iba1. For each genotype, n ≥ 3 animals were imaged. Three 635.90 μm × 635.90 μm image fields were sampled in each animal. The number of microglia process endpoints and the total branch length were quantified as previously described (Young and Morrison, 2018). In brief, each image was skeletonized after optimization and transformed into a binary image. Individual microglia endpoints and branch length were summed and divided by the total number of microglia using the Analyze Skeleton Plugin in FIJI. Microglia soma size was measured using the Free-hand selection and Measure tools in FIJI. A minimum of ten randomly selected microglia were measured for soma size in each image. Phagocytic cups were identified as cup-shaped invaginations at the tip of Iba1-positive microglial processes and were quantified using the Cell Counter tool in FIJI. The average number of phagocytic cups per cell was calculated by dividing the total number of phagocytic cups by the total number of microglia with cups in each image. The percentage of microglia with cups was calculated by dividing the number of microglia with cups by the total number of microglia in a given image.
Engulfment analysis
P9 retinas were harvested using the same methods described for immunohistochemistry and were stained for Iba1 and CD68 in whole-mount preparations. For each genotype, n ≥ 3 animals were imaged. For each animal, at least 15 microglia residing in the OPL were imaged on an Olympus Fluoview FV1200 confocal microscope at 20X using a step size (Z) of 0.5 μm. The images were then processed and analyzed using FIJI and IMARIS (Bitplane) as previously described (Schafer et al., 2014). Briefly, Iba1-positive microglia and CD68-positive lysosomes were 3D-reconstructed using the volume surface rendering function in IMARIS 9.2, and their respective volumes were determined. Any CD68 signal outside the Iba1-positive microglia was masked in the image using the mask function. The percent volume of CD68-positive lysosomes was determined by dividing the volume of the internal CD68 staining (μm3) by the volume of the Iba1-positive microglia (μm3). The CD68 mean fluorescence intensity was determined by dividing the total CD68 signal by the image field area after background signal was subtracted. In the over-expression experiment, engulfment of GFP-positive neural materials inside Iba1-positive microglia was 3D-reconstructed using the same method. The percent volume of GFP-positive neural material inside microglia was determined by dividing the volume of the internal GFP staining by the volume of the microglia. All analyses were performed blind to the experimental conditions.
Synapse quantification
Immunohistochemistry with the ribbon synapse marker RIBEYE was performed on P21 retina cryosections as described above. For each genotype, n ≥ 3 animals were imaged and three independent fields of view in the OPL were captured per animal (60X objective, 2X zoom) using a 20 μm Z-stack comprised of a 0.5 μm step size. Images were subsequently quantified for the number of RIBEYE-positive ribbon synapses in every fifth Z-plane using the Cell Counter tool in FIJI. Synapse numbers were then averaged per animal. RIBEYE mean fluorescence intensity was determined by dividing the total RIBEYE signal by the OPL area after background signal was subtracted using the Freehand and Measure tools in FIJI. All analyses were performed blind to the genotype.
Colocalization quantification
To quantify the degree to which SIRPα or CD47 co-localized with either presynaptic markers (mCAR, PSD95, Vglut1) or postsynaptic markers (Calbindin, SCGN), we calculated the Manders’ Colocalization Coefficients (MCC) for each combination of markers using the FIJI plugin JACoP (Just Another Co-localization Plug-in) (Dunn et al., 2011). n ≥ 3 animals were imaged, and at least two independent fields of view in the OPL were captured per animal using an Olympus Fluoview FV1200 confocal microscope.
Statistical analysis
Statistical analyses of the mean fluorescence intensity, the number of RIBEYE synapses, the number of process endpoints per microglia, the summed process length of microglia, microglia soma size, the percent CD68 and engulfment volume, the percentage of microglia with phagocytic cups, the number of phagocytic cups per microglia, the percent colocalization, and scotopic responses were performed using either unpaired Student’s t-test, one-way ANOVA followed by Bonferroni correction, or two-way ANOVA followed by Bonferroni correction in Prism GraphPad 8.0. P values < 0.05 were considered statistically significant.
Supplementary Material
Highlights.
Neuronal SIRPα production spatiotemporally aligns with peak microglial phagocytosis.
Neuronal, but not microglial, SIRPα permits microglial phagocytosis and synapse refinement.
Prolonging neuronal SIRPα expression is sufficient to extend microglia phagocytosis.
Neuronal SIRPα alters microglial phagocytosis by limiting neuronal CD47 accessibility.
Acknowledgments
We thank Dr. Beth Stevens for the SIRPαF/F mice, Dr. Elizabeth Zuniga-Sanchez for plasmids, and members of our laboratory for scientific discussions and advice. This work was supported by the National Institutes of Health (NIH, R01EY030458, DP2EY02798, and R21AG074163 to M.A.S. and R01MH113743 and R01NS117533 to D.P.S), the Ted Nash Long Life Foundation, the Mallinckrodt Foundation, and the Bright Focus Foundation. C.A.B. was supported by the National Eye Institute under award number T32EY007001. N.E.A. was supported by the National Institute of General Medical Sciences under award number T32GM088129. This project was supported by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the CPRIT Core Facility Support Award (CPRIT-RP180672), the NIH (CA125123 and RR024574), and the assistance of Joel M. Sederstrom. The availability of the Knockout Mouse Project lines was supported by KOMP2 awards UM1HG006348, U42OD11174, and U54HG006348.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Inclusion and Diversity
We worked to ensure sex balance in the selection of non-human subject. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper received support from a program designed to increase minority representation in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams S, van der Laan LJ, Vernon-Wilson E, Renardel de Lavalette C, Dopp EA, Dijkstra CD, Simmons DL, and van den Berg TK (1998). Signal-regulatory protein is selectively expressed by myeloid and neuronal cells. J Immunol 161, 1853–1859. [PubMed] [Google Scholar]
- Albrecht NE, Jiang D, Akhanov V, Hobson R, Speer CM, Robichaux MA, and Samuel MA (2022). Rapid 3D-STORM imaging of diverse molecular targets in tissue. Cell Rep Methods 2, 100253. 10.1016/j.crmeth.2022.100253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann C, and Schmidt MHH (2018). The Role of Microglia in Diabetic Retinopathy: Inflammation, Microvasculature Defects and Neurodegeneration. Int J Mol Sci 19. 10.3390/ijms19010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SR, Roberts JM, Zhang J, Steele MR, Romero CO, Bosco A, and Vetter ML (2019). Developmental Apoptosis Promotes a Disease-Related Gene Signature and Independence from CSF1R Signaling in Retinal Microglia. Cell Rep 27, 2002–2013 e2005. 10.1016/j.celrep.2019.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azcutia V, Bassil R, Herter JM, Engelbertsen D, Newton G, Autio A, Mayadas T, Lichtman AH, Khoury SJ, Parkos CA, et al. (2017). Defects in CD4+ T cell LFA-1 integrin-dependent adhesion and proliferation protect Cd47−/− mice from EAE. J Leukoc Biol 101, 493–505. 10.1189/jlb.3A1215-546RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barclay AN, and Brown MH (2006). The SIRP family of receptors and immune regulation. Nat Rev Immunol 6, 457–464. 10.1038/nri1859. [DOI] [PubMed] [Google Scholar]
- Bessis A, Bechade C, Bernard D, and Roumier A (2007). Microglial control of neuronal death and synaptic properties. Glia 55, 233–238. 10.1002/glia.20459. [DOI] [PubMed] [Google Scholar]
- Brooke G, Holbrook JD, Brown MH, and Barclay AN (2004). Human lymphocytes interact directly with CD47 through a novel member of the signal regulatory protein (SIRP) family. J Immunol 173, 2562–2570. 10.4049/jimmunol.173.4.2562. [DOI] [PubMed] [Google Scholar]
- Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT, Jung S, and Waisman A (2005). A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods 2, 419–426. 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- Burger CA, Jiang D, Li F, and Samuel MA (2020). C1q Regulates Horizontal Cell Neurite Confinement in the Outer Retina. Front Neural Circuits 14, 583391. 10.3389/fncir.2020.583391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao MP, Majeti R, and Weissman IL (2011). Programmed cell removal: a new obstacle in the road to developing cancer. Nat Rev Cancer 12, 58–67. 10.1038/nrc3171. [DOI] [PubMed] [Google Scholar]
- Chao MP, Weissman IL, and Majeti R (2012). The CD47-SIRPalpha pathway in cancer immune evasion and potential therapeutic implications. Curr Opin Immunol 24, 225–232. 10.1016/j.coi.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TT, Brown EJ, Huang EJ, and Seaman WE (2004). Expression and activation of signal regulatory protein alpha on astrocytomas. Cancer Res 64, 117–127. 10.1158/0008-5472.can-3455-2. [DOI] [PubMed] [Google Scholar]
- Chuang W, and Lagenaur CF (1990). Central nervous system antigen P84 can serve as a substrate for neurite outgrowth. Dev Biol 137, 219–232. 10.1016/0012-1606(90)90249-i. [DOI] [PubMed] [Google Scholar]
- Comu S, Weng W, Olinsky S, Ishwad P, Mi Z, Hempel J, Watkins S, Lagenaur CF, and Narayanan V (1997). The murine P84 neural adhesion molecule is SHPS-1, a member of the phosphatase-binding protein family. J Neurosci 17, 8702–8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datwani A, McConnell MJ, Kanold PO, Micheva KD, Busse B, Shamloo M, Smith SJ, and Shatz CJ (2009). Classical MHCI molecules regulate retinogeniculate refinement and limit ocular dominance plasticity. Neuron 64, 463–470. 10.1016/j.neuron.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Wang J, Huang M, Chen Z, Liu J, Zhang Q, Zhang C, Xiang Y, Zen K, and Li L (2021). Loss of microglial SIRPalpha promotes synaptic pruning in preclinical models of neurodegeneration. Nat Commun 12, 2030. 10.1038/s41467-021-22301-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucey MA, Scarpellino L, Zimmer J, Guillaume P, Luescher IF, Bron C, and Held W (2004). Cis association of Ly49A with MHC class I restricts natural killer cell inhibition. Nat Immunol 5, 328–336. 10.1038/ni1043. [DOI] [PubMed] [Google Scholar]
- Dunn KW, Kamocka MM, and McDonald JH (2011). A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol 300, C723–742. 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes ML, and McAllister AK (2015). Immune mediators in the brain and peripheral tissues in autism spectrum disorder. Nat Rev Neurosci 16, 469–486. 10.1038/nrn3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipello F, Morini R, Corradini I, Zerbi V, Canzi A, Michalski B, Erreni M, Markicevic M, Starvaggi-Cucuzza C, Otero K, et al. (2018). The Microglial Innate Immune Receptor TREM2 Is Required for Synapse Elimination and Normal Brain Connectivity. Immunity 48, 979–991 e978. 10.1016/j.immuni.2018.04.016. [DOI] [PubMed] [Google Scholar]
- Ford AL, Goodsall AL, Hickey WF, and Sedgwick JD (1995). Normal adult ramified microglia separated from other central nervous system macrophages by flow cytometric sorting. Phenotypic differences defined and direct ex vivo antigen presentation to myelin basic protein-reactive CD4+ T cells compared. J Immunol 154, 4309–4321. [PubMed] [Google Scholar]
- Fu R, Shen Q, Xu P, Luo JJ, and Tang Y (2014). Phagocytosis of microglia in the central nervous system diseases. Mol Neurobiol 49, 1422–1434. 10.1007/s12035-013-8620-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y, Lagutin O, Hogan BL, and Oliver GC (2000). Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis 26, 130–132. [PubMed] [Google Scholar]
- Gardai SJ, McPhillips KA, Frasch SC, Janssen WJ, Starefeldt A, Murphy-Ullrich JE, Bratton DL, Oldenborg PA, Michalak M, and Henson PM (2005). Cell-surface calreticulin initiates clearance of viable or apoptotic cells through trans-activation of LRP on the phagocyte. Cell 123, 321–334. 10.1016/j.cell.2005.08.032. [DOI] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, and Rodewald HR (2015). Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551. 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunner G, Cheadle L, Johnson KM, Ayata P, Badimon A, Mondo E, Nagy MA, Liu L, Bemiller SM, Kim KW, et al. (2019). Sensory lesioning induces microglial synapse elimination via ADAM10 and fractalkine signaling. Nat Neurosci 22, 1075–1088. 10.1038/s41593-019-0419-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han MH, Lundgren DH, Jaiswal S, Chao M, Graham KL, Garris CS, Axtell RC, Ho PP, Lock CB, Woodard JI, et al. (2012). Janus-like opposing roles of CD47 in autoimmune brain inflammation in humans and mice. J Exp Med 209, 1325–1334. 10.1084/jem.20101974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes BH, Tsai RK, Dooling LJ, Kadu S, Lee JY, Pantano D, Rodriguez PL, Subramanian S, Shin JW, and Discher DE (2020). Macrophages show higher levels of engulfment after disruption of cis interactions between CD47 and the checkpoint receptor SIRPalpha. J Cell Sci 133. 10.1242/jcs.237800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, Merry KM, Shi Q, Rosenthal A, Barres BA, et al. (2016). Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science 352, 712–716. 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks BM, and Chen C (2007). Critical periods in the visual system: changing views for a model of experience-dependent plasticity. Neuron 56, 312–326. 10.1016/j.neuron.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, and Shatz CJ (2000). Functional requirement for class I MHC in CNS development and plasticity. Science 290, 2155–2159. 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume DA, Perry VH, and Gordon S (1983). Immunohistochemical localization of a macrophage-specific antigen in developing mouse retina: phagocytosis of dying neurons and differentiation of microglial cells to form a regular array in the plexiform layers. J Cell Biol 97, 253–257. 10.1083/jcb.97.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa-Sekigami T, Kaneko Y, Okazawa H, Tomizawa T, Okajo J, Saito Y, Okuzawa C, Sugawara-Yokoo M, Nishiyama U, Ohnishi H, et al. (2006). SHPS-1 promotes the survival of circulating erythrocytes through inhibition of phagocytosis by splenic macrophages. Blood 107, 341–348. 10.1182/blood-2005-05-1896. [DOI] [PubMed] [Google Scholar]
- Jiang D, Burger CA, Casasent AK, Albrecht NE, Li F, and Samuel MA (2020). Spatiotemporal gene expression patterns reveal molecular relatedness between retinal laminae. J Comp Neurol 528, 729–755. 10.1002/cne.24784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Lagenaur CF, and Narayanan V (1999). Integrin-associated protein is a ligand for the P84 neural adhesion molecule. J Biol Chem 274, 559–562. 10.1074/jbc.274.2.559. [DOI] [PubMed] [Google Scholar]
- Jordao MJC, Sankowski R, Brendecke SM, Sagar, Locatelli G, Tai YH, Tay TL, Schramm E, Armbruster S, Hagemeyer N, et al. (2019). Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363. 10.1126/science.aat7554. [DOI] [PubMed] [Google Scholar]
- Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, and Littman DR (2000). Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 20, 4106–4114. 10.1128/MCB.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonenkov A, Chen Z, Sures I, Wang H, Schilling J, and Ullrich A (1997). A family of proteins that inhibit signalling through tyrosine kinase receptors. Nature 386, 181–186. 10.1038/386181a0. [DOI] [PubMed] [Google Scholar]
- Kim IJ, Zhang Y, Meister M, and Sanes JR (2010). Laminar restriction of retinal ganglion cell dendrites and axons: subtype-specific developmental patterns revealed with transgenic markers. J Neurosci 30, 1452–1462. 10.1523/JNEUROSCI.4779-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinuthia UM, Wolf A, and Langmann T (2020). Microglia and Inflammatory Responses in Diabetic Retinopathy. Front Immunol 11, 564077. 10.3389/fimmu.2020.564077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima Y, Volkmer JP, McKenna K, Civelek M, Lusis AJ, Miller CL, Direnzo D, Nanda V, Ye J, Connolly AJ, et al. (2016). CD47-blocking antibodies restore phagocytosis and prevent atherosclerosis. Nature 536, 86–90. 10.1038/nature18935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning N, Bo L, Hoek RM, and Huitinga I (2007). Downregulation of macrophage inhibitory molecules in multiple sclerosis lesions. Ann Neurol 62, 504–514. 10.1002/ana.21220. [DOI] [PubMed] [Google Scholar]
- Lall D, Lorenzini I, Mota TA, Bell S, Mahan TE, Ulrich JD, Davtyan H, Rexach JE, Muhammad A, Shelest O, et al. (2021). C9orf72 deficiency promotes microglial-mediated synaptic loss in aging and amyloid accumulation. Neuron 109, 2275–2291 e2278. 10.1016/j.neuron.2021.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman EK, Wilton DK, Litvina EY, Welsh CA, Chang ST, Frouin A, Walker AJ, Heller MD, Umemori H, Chen C, and Stevens B (2018). CD47 Protects Synapses from Excess Microglia-Mediated Pruning during Development. Neuron 100, 120–134 e126. 10.1016/j.neuron.2018.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Jiang D, and Samuel MA (2019). Microglia in the developing retina. Neural Dev 14, 12. 10.1186/s13064-019-0137-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, Huang HY, Shang Y, Oldham MC, Martens LH, Gao F, et al. (2016). Progranulin Deficiency Promotes Circuit-Specific Synaptic Pruning by Microglia via Complement Activation. Cell 165, 921–935. 10.1016/j.cell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K, Kobayashi Y, Udagawa N, Uehara S, Ishihara A, Mizoguchi T, Kikuchi Y, Takada I, Kato S, Kani S, et al. (2012). Wnt5a-Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nat Med 18, 405–412. 10.1038/nm.2653. [DOI] [PubMed] [Google Scholar]
- Majeti R, Chao MP, Alizadeh AA, Pang WW, Jaiswal S, Gibbs KD Jr., van Rooijen N, and Weissman IL (2009). CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell 138, 286–299. 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mass E, Ballesteros I, Farlik M, Halbritter F, Gunther P, Crozet L, Jacome-Galarza CE, Handler K, Klughammer J, Kobayashi Y, et al. (2016). Specification of tissue-resident macrophages during organogenesis. Science 353. 10.1126/science.aaf4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matozaki T, Murata Y, Okazawa H, and Ohnishi H (2009). Functions and molecular mechanisms of the CD47-SIRPalpha signalling pathway. Trends Cell Biol 19, 72–80. 10.1016/j.tcb.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Matsuda T, and Cepko CL (2004). Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci U S A 101, 16–22. 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, and Cepko CL (2007). Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A 104, 1027–1032. 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi ZP, Jiang P, Weng WL, Lindberg FP, Narayanan V, and Lagenaur CF (2000). Expression of a synapse-associated membrane protein, P84/SHPS-1, and its ligand, IAP/CD47, in mouse retina. J Comp Neurol 416, 335–344. [PubMed] [Google Scholar]
- Miksa M, Komura H, Wu R, Shah KG, and Wang P (2009). A novel method to determine the engulfment of apoptotic cells by macrophages using pHrodo succinimidyl ester. J Immunol Methods 342, 71–77. 10.1016/j.jim.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagappan-Chettiar S, Johnson-Venkatesh EM, and Umemori H (2018). Tyrosine phosphorylation of the transmembrane protein SIRPalpha: Sensing synaptic activity and regulating ectodomain cleavage for synapse maturation. J Biol Chem 293, 12026–12042. 10.1074/jbc.RA117.001488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Koren EG, Yu C, Klingeborn M, Wong AYW, Prigge CL, Mathew R, Kalnitsky J, Msallam RA, Silvin A, Kay JN, et al. (2019). Microglial Function Is Distinct in Different Anatomical Locations during Retinal Homeostasis and Degeneration. Immunity 50, 723–737 e727. 10.1016/j.immuni.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenborg PA, Gresham HD, and Lindberg FP (2001). CD47-signal regulatory protein alpha (SIRPalpha) regulates Fcgamma and complement receptor-mediated phagocytosis. J Exp Med 193, 855–862. 10.1084/jem.193.7.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, and Lindberg FP (2000). Role of CD47 as a marker of self on red blood cells. Science 288, 2051–2054. 10.1126/science.288.5473.2051. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456–1458. 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Perry VH, Nicoll JA, and Holmes C (2010). Microglia in neurodegenerative disease. Nat Rev Neurol 6, 193–201. 10.1038/nrneurol.2010.17. [DOI] [PubMed] [Google Scholar]
- Punal VM, Paisley CE, Brecha FS, Lee MA, Perelli RM, Wang J, O’Koren EG, Ackley CR, Saban DR, Reese BE, and Kay JN (2019). Large-scale death of retinal astrocytes during normal development is non-apoptotic and implemented by microglia. PLoS Biol 17, e3000492. 10.1371/journal.pbio.3000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin JM, Yan HX, Liu SQ, Wan XW, Zeng JZ, Cao HF, Qiu XH, Wu MC, and Wang HY (2006). Negatively regulating mechanism of Sirpalpha1 in hepatocellular carcinoma: an experimental study. Hepatobiliary Pancreat Dis Int 5, 246–251. [PubMed] [Google Scholar]
- Rashid K, Akhtar-Schaefer I, and Langmann T (2019). Microglia in Retinal Degeneration. Front Immunol 10, 1975. 10.3389/fimmu.2019.01975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JT, Morganti JM, Bachstetter AD, Hudson CE, Peters MM, Grimmig BA, Weeber EJ, Bickford PC, and Gemma C (2011). CX3CR1 deficiency leads to impairment of hippocampal cognitive function and synaptic plasticity. J Neurosci 31, 16241–16250. 10.1523/JNEUROSCI.3667-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumier A, Bechade C, Poncer JC, Smalla KH, Tomasello E, Vivier E, Gundelfinger ED, Triller A, and Bessis A (2004). Impaired synaptic function in the microglial KARAP/DAP12-deficient mouse. J Neurosci 24, 11421–11428. 10.1523/JNEUROSCI.2251-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MW, and Stevens B (2017). Microglia emerge as central players in brain disease. Nat Med 23, 1018–1027. 10.1038/nm.4397. [DOI] [PubMed] [Google Scholar]
- Samuel MA, Zhang Y, Meister M, and Sanes JR (2011). Age-related alterations in neurons of the mouse retina. J Neurosci 31, 16033–16044. 10.1523/JNEUROSCI.3580-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S, Zuniga-Sanchez E, Kurmangaliyev YZ, Cousins H, Patel M, Hernandez J, Zhang KX, Samuel MA, Morey M, Sanes JR, and Zipursky SL (2018). Role for Wnt Signaling in Retinal Neuropil Development: Analysis via RNA-Seq and In Vivo Somatic CRISPR Mutagenesis. Neuron 98, 109–126 e108. 10.1016/j.neuron.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Heller CT, and Stevens B (2014). An engulfment assay: a protocol to assess interactions between CNS phagocytes and neurons. J Vis Exp. 10.3791/51482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, and Stevens B (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705. 10.1016/j.neuron.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellgren CM, Gracias J, Watmuff B, Biag JD, Thanos JM, Whittredge PB, Fu T, Worringer K, Brown HE, Wang J, et al. (2019). Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nat Neurosci 22, 374–385. 10.1038/s41593-018-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra A, Abiega O, Shahraz A, and Neumann H (2013). Janus-faced microglia: beneficial and detrimental consequences of microglial phagocytosis. Front Cell Neurosci 7, 6. 10.3389/fncel.2013.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman SM, and Wong WT (2018). Microglia in the Retina: Roles in Development, Maturity, and Disease. Annu Rev Vis Sci 4, 45–77. 10.1146/annurev-vision-091517-034425. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, et al. (2011). A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474, 337–342. 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, et al. (2007). The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178. 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- Swanson JA (2008). Shaping cups into phagosomes and macropinosomes. Nat Rev Mol Cell Biol 9, 639–649. 10.1038/nrm2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S (2018). Molecular functions of SIRPalpha and its role in cancer. Biomed Rep 9, 3–7. 10.3892/br.2018.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth AB, Terauchi A, Zhang LY, Johnson-Venkatesh EM, Larsen DJ, Sutton MA, and Umemori H (2013). Synapse maturation by activity-dependent ectodomain shedding of SIRPalpha. Nat Neurosci 16, 1417–1425. 10.1038/nn.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay ME, Lowery RL, and Majewska AK (2010). Microglial interactions with synapses are modulated by visual experience. PLoS Biol 8, e1000527. 10.1371/journal.pbio.1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemori H, and Sanes JR (2008). Signal regulatory proteins (SIRPS) are secreted presynaptic organizing molecules. J Biol Chem 283, 34053–34061. 10.1074/jbc.M805729200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umpierre AD, and Wu LJ (2021). How microglia sense and regulate neuronal activity. Glia 69, 1637–1653. 10.1002/glia.23961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beek EM, Cochrane F, Barclay AN, and van den Berg TK (2005). Signal regulatory proteins in the immune system. J Immunol 175, 7781–7787. 10.4049/jimmunol.175.12.7781. [DOI] [PubMed] [Google Scholar]
- Vasek MJ, Garber C, Dorsey D, Durrant DM, Bollman B, Soung A, Yu J, Perez-Torres C, Frouin A, Wilton DK, et al. (2016). A complement-microglial axis drives synapse loss during virus-induced memory impairment. Nature 534, 538–543. 10.1038/nature18283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Newton G, Wu L, Lin LL, Miracco AS, Natesan S, and Luscinskas FW (2021a). CD47 antibody blockade suppresses microglia-dependent phagocytosis and monocyte transition to macrophages, impairing recovery in EAE. JCI Insight 6. 10.1172/jci.insight.148719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SK, Xue Y, and Cepko CL (2021b). Augmentation of CD47/SIRPalpha signaling protects cones in genetic models of retinal degeneration. JCI Insight 6. 10.1172/jci.insight.150796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao L, Zhang J, Fariss RN, Ma W, Kretschmer F, Wang M, Qian HH, Badea TC, Diamond JS, et al. (2016). Requirement for Microglia for the Maintenance of Synaptic Function and Integrity in the Mature Retina. J Neurosci 36, 2827–2842. 10.1523/JNEUROSCI.3575-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf K, Jahchan NS, Schnorr PJ, Cristea S, Ring AM, Maute RL, Volkmer AK, Volkmer JP, Liu J, Lim JS, et al. (2016). CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer. J Clin Invest 126, 2610–2620. 10.1172/JCI81603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werneburg S, Feinberg PA, Johnson KM, and Schafer DP (2017). A microglia-cytokine axis to modulate synaptic connectivity and function. Curr Opin Neurobiol 47, 138–145. 10.1016/j.conb.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werneburg S, Jung J, Kunjamma RB, Ha SK, Luciano NJ, Willis CM, Gao G, Biscola NP, Havton LA, Crocker SJ, et al. (2020). Targeted Complement Inhibition at Synapses Prevents Microglial Synaptic Engulfment and Synapse Loss in Demyelinating Disease. Immunity 52, 167–182 e167. 10.1016/j.immuni.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham SB, Volkmer JP, Gentles AJ, Sahoo D, Dalerba P, Mitra SS, Wang J, Contreras-Trujillo H, Martin R, Cohen JD, et al. (2012). The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A 109, 6662–6667. 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton DK, Dissing-Olesen L, and Stevens B (2019). Neuron-Glia Signaling in Synapse Elimination. Annu Rev Neurosci 42, 107–127. 10.1146/annurev-neuro-070918-050306. [DOI] [PubMed] [Google Scholar]
- Wong RO, and Ghosh A (2002). Activity-dependent regulation of dendritic growth and patterning. Nat Rev Neurosci 3, 803–812. 10.1038/nrn941. [DOI] [PubMed] [Google Scholar]
- Wu CJ, Chen Z, Ullrich A, Greene MI, and O’Rourke DM (2000). Inhibition of EGFR-mediated phosphoinositide-3-OH kinase (PI3-K) signaling and glioblastoma phenotype by signal-regulatory proteins (SIRPs). Oncogene 19, 3999–4010. 10.1038/sj.onc.1203748. [DOI] [PubMed] [Google Scholar]
- Wu Y, Dissing-Olesen L, MacVicar BA, and Stevens B (2015). Microglia: Dynamic Mediators of Synapse Development and Plasticity. Trends Immunol 36, 605–613. 10.1016/j.it.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan HX, Wang HY, Zhang R, Chen L, Li BA, Liu SQ, Cao HF, Qiu XH, Shan YF, Yan ZH, et al. (2004). Negative regulation of hepatocellular carcinoma cell growth by signal regulatory protein alpha1. Hepatology 40, 618–628. 10.1002/hep.20360. [DOI] [PubMed] [Google Scholar]
- Yao C, Li G, Cai M, Qian Y, Wang L, Xiao L, Thaiss F, and Shi B (2017). Prostate cancer downregulated SIRP-alpha modulates apoptosis and proliferation through p38-MAPK/NF-kappaB/COX-2 signaling. Oncol Lett 13, 4995–5001. 10.3892/ol.2017.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91. 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young K, and Morrison H (2018). Quantifying Microglia Morphology from Photomicrographs of Immunohistochemistry Prepared Tissue Using ImageJ. J Vis Exp. 10.3791/57648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Wang J, Kong X, Li E, Liu Y, Du X, Kang Z, Tang Y, Kuang Y, Yang Z, et al. (2016). CD47 Promotes Tumor Invasion and Metastasis in Non-small Cell Lung Cancer. Sci Rep 6, 29719. 10.1038/srep29719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XF, Alam MM, Liao Y, Huang T, Mathur R, Zhu X, and Huang Y (2019). Targeting Microglia Using Cx3cr1-Cre Lines: Revisiting the Specificity. eNeuro 6. 10.1523/ENEURO.0114-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


