Abstract
The developmental process of central nervous system (CNS) myelin sheath formation is characterized by well-coordinated cellular activities ultimately ensuring rapid and synchronized neural communication. During this process, myelinating CNS cells, namely oligodendrocytes (OLGs), undergo distinct steps of differentiation, whereby the progression of earlier maturation stages of OLGs represents a critical step toward the timely establishment of myelinated axonal circuits. Given the complexity of functional integration, it is not surprising that OLG maturation is controlled by a yet fully to be defined set of both negative and positive modulators. In this context, we provide here first evidence for a role of lysophosphatidic acid (LPA) signaling via the G protein-coupled receptor LPA6 as a negative modulatory regulator of myelination-associated gene expression in OLGs. More specifically, cell surface accessibility of LPA6 was found to be restricted to the earlier maturation stages of differentiating OLGs, and OLG maturation was found to occur precociously in Lpar6 knockout mice. To further substantiate these findings, a novel small molecule ligand with selectivity for preferentially LPA6 and LPA6 agonist characteristics was functionally characterized in vitro in primary cultures of rat OLGs and in vivo in the developing zebrafish. Utilizing this approach, a negative modulatory role of LPA6 signaling in OLG maturation could be corroborated. During development, such a functional role of LPA6 signaling likely serves to ensure timely coordination of circuit formation and myelination. Under pathological conditions as seen in the major human demyelinating disease multiple sclerosis (MS), however, persistent LPA6 expression and signaling in OLGs can be seen as an inhibitor of myelin repair. Thus, it is of interest that LPA6 protein levels appear elevated in MS brain samples, thereby suggesting that LPA6 signaling may represent a potential new druggable pathway suitable to promote myelin repair in MS.
Keywords: oligodendrocyte, lipid signaling, myelination, multiple sclerosis, small molecule ligands, computational ligand-protein docking
Graphical Abstract

INTRODUCTION
Myelination in the central nervous system (CNS) of higher vertebrates has evolved to allow rapid and efficient signal propagation within the physical constraints imposed by the skull and vertebrae (Zalc 2016; Zalc et al. 2008). During development, this process is defined by a tightly regulated differentiation program during which CNS myelinating cells, namely oligodendrocytes (OLGs), undergo a stepwise progression, from lineage committed progenitor cells and through stages of maturing OLGs, to ultimately convert into myelinating and then myelin maintaining OLGs (Sock & Wegner 2021; Elbaz & Popko 2019; Emery & Lu 2015). In this scenario, each of the OLG maturation stages is characterized by a distinct gene expression profile that is determined by both intrinsic mechanisms and extrinsic factors (Adams et al. 2021; Baydyuk et al. 2020; Liu et al. 2016; Mitew et al. 2014; Wheeler & Fuss 2016). Despite the critical importance of this process for the functional acuity of neuronal signal propagation in the CNS, the regulatory molecular pathways are still not fully understood.
Notably, our previous studies identified signaling initiated by the extracellular lipid signaling molecule lysophosphatidic acid (LPA) as a critical regulator of the transcriptional program regulating OLG maturation (Yuelling et al. 2012; Wheeler et al. 2015; Wheeler & Fuss 2016). In general, LPA exerts its biological effects via activation of a family of G protein-coupled receptors with currently six members identified in mammals, namely LPA1-6 encoded by Lpar1-6 (Kano et al. 2021; Yung et al. 2014; Kihara et al. 2014; Hecht et al. 1996). With the exception of Lpar5, all of these receptors are, at least to some extent, expressed by cells of the mammalian OLG lineage, whereby at more mature stages the expression of Lpar1 and Lpar6 prevails (Marisca et al. 2020; Suckau et al. 2019; Wheeler et al. 2015; Marques et al. 2016; Zhang et al. 2014; Nogaroli et al. 2009; Cahoy et al. 2008; Yu et al. 2004; Stankoff et al. 2002; Weiner et al. 1998). Loss-of-function of Lpar1 has only in the Malága variant of Lpar1 null mice been described to affect CNS myelination by causing defects in protein trafficking without affecting the transcriptional regulation of OLG maturation (Garcia-Diaz et al. 2015; Anliker et al. 2013; Gennero et al. 2011; Estivill-Torrus et al. 2008). Thus, we focused our studies here on LPA6, the most recently identified member of the LPA receptor family (Yanagida et al. 2009; Pasternack et al. 2008; Shimomura et al. 2008), for which a role in OLG maturation and CNS myelination had, to the best of our knowledge, not yet been explored. Our findings presented here uncover LPA6 signaling as a new modulator that attenuates gene expression associated with OLG maturation, and they introduce a novel small molecule ligand with preferential selectivity for LPA6 and functional applicability in an in vitro as well as in vivo model of OLG maturation. Interestingly, our data also provide initial evidence for dysregulated LPA6 signaling in the major human demyelinating disease multiple sclerosis (MS), in which alterations in OLG gene expression profiles have been associated with pathology and disease progression (Schirmer et al. 2019; Jäkel et al. 2019; Falcão et al. 2018; Duncan et al. 2017; Kuhlmann et al. 2008; Chang et al. 2002). Thus, taken together, our studies identify LPA6 signaling as a novel mechanism modulating OLG maturation and they suggest LPA6 as a potential new druggable target for treating CNS diseases associated with myelin deficits.
MATERIALS AND METHODS
Reagents
LPA (LPA 18:1, 1-oleoyl-LPA) was purchased from Avanti Polar Lipids (Cat. No 857130, Alabaster, AL) and stored at −20 °C (10mg/ml in chloroform). (2S)-OMPT (L-sn-1-O-oleoyl-2-O-methylglyceryl-3-phosphothionate) was purchased from Echelon Biosciences (Cat. No L-9418, Salt Lake City UT) and stored at −20°C (10mM in dimethyl sulfoxide (DMSO)). GL-8-28 (Fig. 3) was synthesized by us and stored at −20°C (10mM in DMSO). Unless otherwise stated, all other reagents were purchased from Sigma-Aldrich (St. Louis, MO) or Thermo Fisher Scientific (Waltham, MA). Details to antibodies and PCR primers are shown in Tables 1 and 2.
Fig. 3.

Chemical structures of LPA (18:1; 1-oleyl-LPA), (2S)-OMPT (L-sn-1-O-oleoyl-2-O-methylglyceryl-3-phosphothionate), and GL-8-28. For the design of GL-8-28, a polar head was kept in place to ensure LPA receptor recognition, an amide bond was applied to replace the ester bond present in the natural ligand LPA to achieve higher stability in biological systems, and a long aliphatic tail was maintained.
Table 1.
Antibody RRIDs and Concentrations or Dilutions used in the assays listed.
| RRID | Assay (concentration or dilution) | |
|---|---|---|
| Primary Antibodies | ||
| A2B5 (clone 105) | CVCL_7946 | Immunopanning (15 μg/mL) Immunocytochemistry (undiluted hybridoma supernatant) |
| Anti-APC (CC-1) | AB_2057371 | Immunohistochemistry (1:100) |
| Anti-CD16/CD32 (clone 93) | AB_467133 | Flow cytometry (1:100) |
| Anti-GAPDH | AB_2107445 | Western blot (1:10,000) |
| Anti-LPA4 | AB_2340992 | Flow cytometry (1:100) Western blot (1:100) |
| Anti-LPA6 | AB_2340993 | Flow cytometry (1:100) Western blot (1:100) Immunocytochemistry (1:100) |
| Anti-MBP (SMI99P) | AB_2314772 | Immunocytochemistry (1:100) |
| O4 (clone O4) | CVCL_Z932 | Immunocytochemistry (undiluted hybridoma supernatant) |
| Anti-Olig2 | AB_10807410 | Immunohistochemistry (1:100) |
| Rabbit IgG isotype control | AB_2811130 | Flow cytometry (1:100) |
| Secondary Antibodies | ||
| Goat anti-rabbit IgG, AlexaFluor 488 conjugated | AB_143165 | Flow cytometry (1:100) Immunocytochemistry (1:500) Immunohistochemistry (1:500) |
| Goat anti-Mouse IgG2b, AlexaFluor 568 conjugated | AB_2535780 | Immunohistochemistry (1:500) |
| Goat anti-Mouse IgG2b, AlexaFuor 594 conjugated | AB_2535781 | Immunocytochemistry (1:500) |
| Goat anti-Mouse IgM, AlexaFluor 633 conjugated | AB_2535715 | Immunocytochemistry (1:500) |
| Goat anti-Mouse IgG2a, AlexaFluor 633 conjugated | AB_2535775 | Immunohistochemistry (1:500) |
| Goat anti-Mouse IgG. IRDye 680 | AB_621840 | Western blot (1:10,000) |
| Goat anti-Rabbit IgG, IRDye 800CW conjugated | AB_10796098 | Western blot (1:10,000) |
Table 2.
List of primer sequences used for RT-qPCR analysis. actb2, beta actin; cldnk, claudin k; Cnp, 2′,3′ cyclic nucleotide phosphodiesterase; ef1a, elongation factor 1-alpha; Egr1, early growth response 1; Olig2, oligodendrocyte transcription factor 2; Pgk1, phosphoglycerate kinase 1; plp1b, proteolipid protein 1b; Ppia, peptidylprolyl isomerase A (cyclophilin A); Rpl13a, ribosomal protein L13a; Ugt8, UDP glycosyltransferase 8.
| Forward primer (5’-3’) | Reverse primer (5’-3’) | |
|---|---|---|
| Mouse Genes | ||
| Olig2 | ACCGTTAACACGAGGGGCAA | TTAGGAAGCGGCGCAGTACA |
| Cnp | ATGCCCAACAGGATGTGGTG | AGGGCTTGTCCAGGTCACTT |
| Ugt8 | AGGAGCTCTGGGGAGATTGC | TTTGAATGGCCAAGCAGGTCA |
| Egr1 | CCTGACCACAGAGTCCTTTTCT | AAAGTGTTGCCACTGTTGGG |
| Mouse Reference Genes | ||
| Ppia | GGAGACGAACCTGTAGGACG | GATGCTCTTTCCTCCTGTGC |
| Pgk1 | ATGCAAAGACTGGCCAAGCTAC | AGCCACAGCCTCAGCATATTTC |
| Rpl13a | GCGCCTCAAGGTGTTGGATG | CGCCCCAGGTAAGCAAACTTTC |
| Zebrafish Genes | ||
| cldnk | TGGCATTTCGGCTCAAGCTCTGGA | GGTACAGACTGGGCAATGGACCTGA |
| plp1b | TGCCATGCCAGGGGTTGTTTGTGGA | TGGCGACCATGTAAACGAACAGGGC |
| Zebrafish Reference Genes | ||
| ef1a | GTACTACTCTTCTTGATGCCC | GTACAGTTCCAATACCTCCA |
| actb2 | CCCTGTTCCAGCCATCCTT | TTGAAAGTGGTCTCGTGGATACC |
| rpl13a | TCTGGAGGACTGTAAGAGGTATGC | AGACGCACAATCTTGAGAGCAG |
Chemical synthesis and characterization of compound GL-8-28
All reagents were purchased from Sigma-Aldrich or as otherwise stated. Melting points were obtained with a Fisher Scientific micro melting point apparatus and are uncorrected. All infrared (IR) spectra were recorded on a Nicolet Avatar 360 FT-IR Instruments. Proton (300 MHz) and Carbon-13 (75 MHz) nuclear magnetic resonance (NMR) spectra were recorded at ambient temperature with tetramethylsilane as the internal standard on either a Varian Gemini-300MHz “Tesla” spectrometer or Varian Mercury-300MHz NMR spectrometer. Gas chromatography/mass spectrometry (GC/MS) analysis was performed on a Hewlett Packard 6890 (Palo Alto, CA). Thin layer chromatography (TLC) analyses were carried out on Analtech Uniplate F254 plates. Chromatographic purification was carried out on silica gel columns (230~400 mesh, Merck). Yields were not maximized.
Compound GL-8-28 was prepared in four steps. First, 3-trimethylsilanyloxy-propylamine was prepared following a previously reported method (Mormann & Leukel 1988). The mixture of 3-amino-1-propanol (1.50 g, 20 mmol), hexamethyldisilazane (1.78 g, 11 mmol) and trimethylsilane chloride (catalytic amount) was heated at 140 °C for two hrs. After cooled down, the mixture was concentrated under vacuum to give 2.09 g light yellow oil, in 68% yield. 1H NMR (300MHz, DMSO): δ 3.70(t, J= 7.5Hz, 2 H), 2.80(t, J= 6.6 Hz, 2 H), 1.69(m, 2 H), 0.14 (s, 9 H). Second, octadec-9(Z)-enoic acid (3-trimethylsianyloxy-propyl)-amide was prepared. The mixture of octadec-9(Z)-enoic acid (3.59 g, 12.6 mmol) and thionyl chloride (3.0 g, 25.2 mmol) in chlorobenzene (40 mL) was heated to reflux overnight. After concentrated to remove solvent and the excess thionyl chloride, the residue was dissolved in dry dichloromethane (10 mL). Then the resulting solution was added into a solution of 3-trimethylsilanyloxy-propylamine (1.16 g, 6.3 mmol) in dry dichloromethane (90 mL) at 0 °C. The mixture was stirred overnight. The dichloromethane layer was washed with water (30 mL x 3), brine and dried over sodium sulfate. After filtered and concentrated, the resulting residue was purified by silica gel column, hexane and ethyl acetate (4: 1), to give 1.4 g product in 61% yield. 1H NMR (300MHz, CDCl3): δ 5.76 (br, 1 H), 5.37 (m, 2 H), 4.17 (t, J= 6.0Hz, 2 H), 3.33 (t, J= 6.6 Hz, 2 H), 2.34 (t, J= 7.2Hz, 2 H), 2.19 (m, 2 H), 2.05 (m, 2 H), 1.86 (m, 2 H), 1.65 (m, 2 H), 1.29 (m, 16 H), 0.90 (t, J= 6.6Hz, 3 H), 0.13 (s, 9 H); 13C NMR (75MHz, CDCl3) δ: 172.74, 129.59, 129.31, 61.23, 36.47, 35.71, 33.88, 31.49, 31.48, 29.35, 29.34, 29.27, 29.11, 29.10, 28.91, 28.90, 28.73, 28.72, 28.45, 26.80, 25.35, 24.55, 22.27, 13.71. Third, octadec-9(Z)-enoic acid (3-hydroxy-propyl)-amide was prepared. In an ice-water bath, the solution of octadecanoic acid (3-trimethylsilanyloxy-propyl)-amide (500 mg, 1.2 mmol) in ethanol (10 mL) was added 1N HCl (5 mL). The mixture was allowed to stir at room temperature for 5 hrs. Filtration gave 380 mg white powder in 93% yield. 1H NMR (300MHz, CDCl3): δ 6.05 (br, 1 H), 5.36 (m, 2 H), 3.63 (m, 2 H), 3.43 (q, J= 6.0 Hz, 2 H), 2.21 (m, 2 H), 2.03 (m, 4 H), 1.70 (m, 4 H), 1.30 (m, 20 H), 0.90 (t, J= 6.9Hz, 3 H); 13C NMR (75MHz, CDCl3) δ: 174.18, 129.57, 129.28, 58.68, 36.29, 35.68, 31.92, 31.47, 29.34, 29.28, 29.09, 28.89, 28.88, 28.84, 28.83, 28.71, 26.79, 26.74, 25.40, 22.26, 13.70. Last, sulfuric acid mono-(3-octadec-9(Z)-enoylamino-propyl) ester (GL-8-28) was prepared based on a reported method (Pogorevc & Faber 2002). To a suspension of sodium hydride (16.3 mg, 0.67 mmol) in dimethylformamide (DMF) (2 mL) was added octadecanoic acid (3-hydroxy-propyl)-amide (105 mg, 0.31mmol). After stirred at room temperature for 1 hour, the resulting mixture was added the solution of sulfur trioxide triethylamine complex (67 mg, 0.37 mmol) in DMF (1 mL). The reaction mixture was stirred overnight. After concentrated under vacuum to remove DMF, the residue was purified by silica gel column, dichloromethane and methanol (10:1) to give 65 mg gum, in 50% yield. 1H NMR (300MHz, CDCl3): δ 5.37 (m, 2 H), 4.06 (t, J=6.6Hz, 2 H), 3.23 (m, 2 H), 2.21 (t, J= 7.5Hz, 2 H), 2.04 (m, 2 H), 1.87 (m, 2 H), 1.62 (m, 2 H), 1.34 (m, 20 H), 0.93 (t, J= 6.6Hz, 3 H); 13C NMR (75MHz, CD3OD) δ: 174.34, 128.92, 128.81, 64.75, 46.03, 35.27, 31.14, 28.92, 28.91, 28.69, 28.53, 28.42, 28.41, 28.33, 28.26, 26.21, 25.15, 21.82, 12.57, 7.34; MS (ESI) m/z: 420.3(M+H)+.
Experimental Animals
Lpar6 KO mice (Accession No. CDB0977K: http://www.clst.riken.jp/arg/mutant%20mice%20list.html)) were generated as previously described (Hata et al. 2016); these mice were housed at Akita University (Japan) under temperature-controlled conditions with ad libitum access to food and water in a 12 hr dark/light cycle, and kept on a C57BL/6N background. In total, 17 mice (males and females) were used in these studies. No animals were excluded from the analyses described here, and no exclusion criteria were pre-determined. Zebrafish embryos were obtained through natural matings, raised at 28.5°C, and staged according to morphological criteria and hrs postfertilization (hpf) (Kimmel et al. 1995). Wild-type zebrafish were of the AB or NHGRI-1 strains. Sprague Dawley female rats with early postnatal litters were obtained from Envigo/Harlan Laboratories (Indianapolis, IN). All animal studies were approved by the Institutional Animal Care and Use Committees at Virginia Commonwealth University (IACUC #AM10229 and #AM10189) and Akita University. Unless otherwise stated, n = number of animals used when multiple animals were compared. This study was not preregistered, and no blinding or randomization was performed.
Human Tissue Samples
Human white matter tissue samples from non-MS and MS donors were obtained from the Netherlands Brain Bank, Netherlands Institute for Neuroscience, Amsterdam (open access: www.brainbank.nl). All material has been collected from donors for or from whom a written informed consent for a brain autopsy and the use of the material and clinical information for research purposes had been obtained by the Netherlands Brain Bank. Based on medical history and autopsy evaluation, none of the MS individuals included in this study presented any neurodegenerative disorders other than MS. Brains were classified as MS or non-MS based on medical history and post-mortem pathological analyses. Details to the demographics of the donor patients are shown in Table 3.
Table 3.
Demographics of non-multiple sclerosis and multiple sclerosis patients. MS: Multiple Sclerosis, SPMS: Secondary Progressive Multiple Sclerosis, PPMS: Primary Progressive Multiple Sclerosis, PMI (hrs): Postmortem Interval in hours.
| MS type | Age (yrs)/Sex | PMI (hrs) | |
|---|---|---|---|
| Control 1 | Non-MS | 90/F | 7.25 |
| Control 2 | Non-MS | 84/M | - |
| Control 3 | Non-MS | 51/M | 7.5 |
| Control 4 | Non-MS | 87/F | 8 |
| Control 5 | Non-MS | 86/F | 13.3 |
| MS 1 | SPMS | 49/M | 8 |
| MS 2 | MS | 44/M | 10 |
| MS 3 | MS | 57/F | 8.75 |
| MS 4 | SPMS | 75/M | 7.5 |
| MS 5 | SPMS | 59/F | 4.75 |
| MS 6 | MS | 68/F | 8.25 |
| MS 7 | SPMS | 45/M | 7.75 |
| MS 8 | PPMS | 57/F | 5.75 |
| MS 9 | PPMS | 74/F | 5.5 |
| MS 10 | SPMS | 48/F | 11.75 |
| MS 11 | MS | 60/F | 10.5 |
| MS 12 | MS | 56/F | 8.25 |
| MS 13 | SPMS | 78/M | 8.75 |
| MS 14 | MS | 53/M | 10 |
Cell Culture
Primary OLG progenitors were isolated from postnatal day 2 (P2) rat brains by A2B5 immunopanning as described previously (Barres et al. 1992; Martinez-Lozada et al. 2014; Wheeler et al. 2015). For each individual experiment, brains from all animals (8 to 12; male and female pups) of an entire litter were pooled. For immunopanning, supernatants of A2B5 hybridoma cells (ATCC, Manassas, VA) cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Cat. No. 11995040, Gibco/Thermo Fisher Scientific, Waltham, MA) containing 10% fetal bovine serum (FBS; Cat. No. SH30071.03, HyClone, Cytvia, Marlborough, MAre) and 1% antibiotic-antimycotic (Cat. No. 15240062, Gibco/Thermo Fisher Scientific, Waltham, MA) were used directly or A2B5 antibodies were purified by ammonium sulfate precipitation (Andrew et al. 2009) and used at a concentration of 15 μg/mL as determined by ELISA (Mouse IgM ELISA Kit, Cat. No. ab133047, Abcam, Cambridge, MA). Isolated OLG progenitors were plated onto fibronectin (10 μg/ml; Cat. No 3416355MG or 341668500UG, MilliporeSigma, Burlington, MA)-coated tissue culture dishes, glass coverslips, or 96-well plates and cultured at 37°C and 5% CO2 in serum-free proliferation medium [DMEM containing N2 supplement (Cat. No. 17502001, Thermo Fisher Scientific, Waltham, MA), 5 ng/mL human platelet-derived growth factor (PDGF, Cat No. 300-176P, Gemini Bio-Products, West Sacramento, CA) and 5 ng/mL basic fibroblast growth factor (bFGF, Cat. No. 300-113P, Gemini Bio-Products, West Sacramento, CA)] for 24 hrs, after which cells were allowed to differentiate in serum-free differentiation medium [DMEM containing N2 supplement (Thermo Fisher Scientific, Waltham, MA) and 40 ng/mL tri-iodo-thyronine (T3; Cat. No. T2877, Sigma, St Louis, MO)] over the time periods indicated. Differentiating OLGs were collected for RNA isolation, and supernatants [phenol red-free DMEM, Cat. No. 31053036, Gibco/Thermo Fisher Scientific, Waltham, MA with added L-glutamine (Cat. No 25030, Gibco/Thermo Fisher Scientific, Waltham, MA) and sodium pyruvate (Cat. No. 11360070 Gibco/Thermo Fisher Scientific, Waltham, MA)] were collected for determining cell viability using the Thermo Scientific Pierce LDH Cytotoxicity Assay Kit (Cat. No. 88954, Thermo Fisher Scientific, Waltham, MA) and a PHERAstar plate reader (BMG LABTECH Inc, Cary, NC). For all primary OLG cell culture studies, at least three independent experiments were performed, whereby an independent experiment refers to an experiment in which cells were isolated from a separate P2 rat litter at an independent time point (day) and treated separately from all other independent experiments.
For the determination of LPA receptor selectivity, the PRESTO-Tango assay was used (Kroeze et al. 2015). HTLA cells, (an HEK293 cell line stably expressing a tTA-dependent luciferase reporter and a β-arrestin2-TEV fusion gene) were kindly provided by Dr. Wesley Kroeze and maintained in DMEM supplemented with 10% FBS, 2 μg/ml puromycin (Cat. No. A1113803, Gibco/Thermo Fisher Scientific, Waltham, MA) and 100 μg/ml hygromycin B (Cat. No. 10687010, Thermo Fisher Scientific, Waltham, MA) in a humidified atmosphere at 37°C in 5% CO2. This cell lines is not listed as commonly misidentified cell line by the International Cell Line Authentication Committee (ICLAC). TANGO-ized plasmid constructs for LPAR1, 2 and LPAR4-6 (Kroeze et al. 2015) were gifts from Dr. Bryan Roth and obtained through Addgene (LPAR1-Tango, RRID: Addgene_66418; LPAR2-Tango, RRID: Addgene_66419; LPAR4-Tango, RRID: Addgene_66420; LPAR5-Tango, RRID: Addgene_66421; LPAR6-Tango, RRID: Addgene_66422). The LPAR3-TANGO plasmid construct was designed analogous to all other TANGO-ized G protein-coupled receptor plasmid constructs and synthesized through Thermofisher Scientific’s Synthetic Biology and Genome Engineering service. Plasmid constructs were confirmed by sequencing and purified using an EndoFree plasmid maxi prep kit (Cat. No. 12362, Quiagen Inc, Valencia, CA). For transfection, HTLA cells were plated at a density of 1x106 cells per well into 12-well cell culture plates and transfected the following day (day 2) with 4 μg (LPA1-5) or 8 μg (LPA6) of ‘TANGO-ized’ LPA receptor plasmid construct per well using Lipofectamine 2000 (Cat. No. 11668019, Thermo Fisher Scientific, Waltham, MA) in serum-free Opti-MEM medium (Cat. No. 31985070, Gibco/Thermo Fisher Scientific, Waltham, MA) without antibiotics. On day 3, cells were re-plated at 1x105 cells per well into poly-L-lysine coated 96-well white, clear flat bottom microplates (Cat. No. CLS3610, Corning Life Sciences, Tewksbury, MA) in Opti-MEM medium containing 10% charcoal-stripped FBS (Cat. No. 12676029, Gibco/ThermoFisher Scientific, Waltham, MA). On day 4, 5x drug stimulation solutions were prepared in filter-sterilized assay buffer, which consisted of 20 mM HEPES in Hank’s Balanced Salt Solution (HBSS, Cat. No. 14170112, Gibco/Thermo Fisher Scientific, Waltham, MA), pH 7.4, and 20 μL were added to each well containing 80 μL of medium. Constitutive activity controls received only the assay buffer. On day 5 (after about 18 hrs of incubation), luminescence, as a measure for receptor activation, was determined using the Bright-Glo Luciferase Assay System (Cat. No. E2610, Promega, Madison, WI) and a PHERAstar plate reader (BMG LABTECH Inc, Cary, NC).
Treatment with Pharmacological Compounds
Zebrafish embryos (at 24 hpf) and primary cultures of differentiating OLGs (at 24 hrs, i.e. at the time of switch to differentiation medium) were treated with LPA, (2S)-OMPT or GL-8-28 at the concentrations indicated. LPA was dissolved in DMEM containing 0.1% fatty acid-free bovine serum albumin (BSA; Cat. No. A8806, Sigma, St. Louis MO), while (2S)-OMPT and GL-8-28 were dissolved in dimethyl sulfoxide (DMSO), resulting in a final experimental concentration of 0.1% DMSO. Vehicle treatments were used as controls.
Immunocytochemistry, Immunohistochemistry, RNAscope, and Confocal Microscopy
Differentiating OLGs, plated on fibronectin-coated coverslips, were immunostained live (O4 or A2B5 hybridoma supernatants, anti-LPA6 antibodies) or after fixation in 4% paraformaldehyde/PBS (anti-MBP, anti-LPA6 antibodies). In combined double staining procedures, live staining was performed first, followed by fixation. Fixed cells were permeabilized using 0.5% Triton X-100/PBS, and nonspecific binding sites were blocked by incubation in 10% FCS/DMEM (live cells) or 2% BSA/PBS (fixed cells) for 30 min at room temperature. Primary antibodies were diluted in 10% FCS/DMEM (live cells) or 1% BSA/PBS (fixed cells), and cells were incubated for 1 hr at room temperature followed by incubation with secondary antibodies diluted in 1% BSA/PBS for 30 min or 1 hr at room temperature. Nuclei were counterstained using Hoechst 33342 (Cat. No. 14533; MilliporeSigma, Burlington MA), and sections were mounted using Vectashield (Cat. No. H-1000-10, Vector Laboratories, Burlingame, CA).
Tissue sections for immunohistochemistry and RNAscope were prepared in principle as described previously (Dupree et al. 1999; Benusa et al. 2017). Briefly, mice were anesthetized by intraperitoneal injection of 0.8ml/20 g (of mouse body weight) 2-2-2 tribromoethanol (20g/ml; Cat. No. T48402, Sigma-Aldrich, St. Louis, MO) and then transcardially perfused with 4% paraformaldehyde in 0.1 M Millonig’s phosphate buffer (Karlsson & Schultz 1965), brains were removed, post-fixed for 24 hrs in perfusion fixative, cryoprotected by immersion in 30% sucrose in PBS for 48 hrs, and then embedded and frozen in Tissue-Tek O.C.T. compound (Cat. No. 4583, Sakura Finetek USA, Torrance, CA). For immunohistochemistry, serial coronal sections (40-μm) were prepared using a Leica CM 1850 cryostat (Leica Biosystems, Buffalo Grove, IL) and stored at −80°C. Tissue sections were permeabilized for 10 minutes in ice-cold acetone and blocked for 1 hr at room temperature using PBS containing 1% Triton X-100 and 5% cold fish gelatin (Cat. No. 50-259-35, Electron Microscopy Science, Hatfield, PA). Primary antibodies were diluted in blocking solution, and sections were incubated for 48 hrs at 4°C followed by incubation with secondary antibodies for 90 min at room temperature. Nuclei were counterstained using Hoechst 33342 (Cat. No. 14533; MilliporeSigma, Burlington MA), and sections were mounted using Vectashield (Cat. No. H-1000-10, Vector Laboratories, Burlingame, CA). For RNAscope, coronal sections (15-μm) were prepared using a Leica CM 1850 cryostat (Leica Biosystems, Buffalo Grove, IL), sections were mounted on Leica Bond Plus slides (Cat. No. S21.2113.A, Leica Biosystems, Deer Park, IL) and post-fixed as follows: 60°C for 30 min, 4% paraformaldehyde in PBS for 15 min at 4°C, 50% ethanol for 5 min at room temperature, 70% ethanol for 5 min at room temperature, 100% ethanol for 5 min at room temperature (twice), air dried for 5 min, and stored at −80°C. RNAScope was performed using a Leica Biosystems Bond RX automated immunohistochemistry/in situ hybridization staining system (Leica Biosystems, Deer Park, IL) located within VCU’s Tissue and Data Acquisition and Analysis Core and the following RNAscope 2.5 LS Probe probes (all from Advanced Cell Diagnostics, Inc., Newark, CA): Mm-Lpar6 (Cat. No. 318358), Mm-Olig2-C2 (Cat. No. 447098-C2), Mm-Plp1-C4 (Cat. No. 428188-C4); for fluorescent detection, RNAscope LS Fluorescent Reagent and RNAscope LS 4-Plex Ancilliary kits (Cat Nos. 322800 and 322830, Advanced Cell Diagnostics, Inc., Newark, CA) were used in combination with the following Opal fluorophores (Akoya Biosciences, Marlborough, MA): Opal 690 (Cat. No. PN FP1497001KT), Opal 620 (Cat. No. PN FP1495001KT), Opal 520 (Cat. No. PN FP1487001KT).
All confocal images were collected using a Zeiss LSM 710 or LSM 880 confocal laser scanning microscope (Carl Zeiss Microscopy, LLC, Thornwood, NY) located within VCU’s Microcopy Shared Resource. For immunostained cells, confocal z-stacks, each spanning the optical distance of the entire cell, were collected at 0.48 μm intervals using a 63x oil-immersion objective with a numerical aperture of 1.4 and the following settings: a pinhole size of one Airy unit, a dimension of 1912x1912 pixels and 4 times line averaging. For immunostained sections, confocal z-stacks, each spanning an optical distance of 15μm, were collected at 1 μm intervals using a 40x oil-immersion objective with a numerical aperture of 1.3 and the following settings: a pinhole size of one Airy unit, a dimension of 1248x1248 pixels and 4 times line averaging. For sections labeled by RNAscope, confocal z-stacks, each spanning an optical distance of 15μm, were collected at 0.5 μm intervals using a 63x oil-immersion objective with a numerical aperture of 1.4 and the following settings: a pinhole size of one Airy unit, a dimension of 1200x1200 pixels and 4 times line averaging. For image acquisition and the generation of maximum intensity projections ZEN imaging software (Carl Zeiss Microscopy, LLC, Thornwood, NY; RRID:SCR_013672) was used.
Western Blot Analysis
For Western blot analysis, human tissue samples were homogenized in lysis buffer (10 μL per mg tissue; phosphate-buffered saline (PBS), 2 mM EDTA) containing 1x Halt protease and phosphatase inhibitor cocktail (Cat. No. 78430, Thermo Fisher Scientific, Waltham, MA). After centrifugation at 15,000 rpm for 15 min at 4°C, protein concentrations of the supernatants were determined using the BCA protein assay kit (Cat. No. 23225, Pierce/Thermo Fisher Scientific, Waltham, MA). Samples were heat-denatured (90°C for 5 min) in Laemmli sample buffer (Cat. No. S3401, MilliporeSigma, Burlington, MA), and equal amounts of denatured protein (60 μg) were separated by electrophoresis through 4-20% gradient sodium dodecyl sulfate (SDS)-polyacrylamide gels (Cat. No. 4561094, Bio-Rad Laboratories, Hercules, CA). Separated proteins were subsequently electroblotted onto Immobilon-P polyvinylidene difluoride (PVDF) membranes (Cat. No. IPVH00010, MilliporeSigma, Burlington, MA). Total protein levels were determined using Revert 700 Total Protein Stain (Cat. No. 926-11011, LI-COR Biosciences, Lincoln, NE) and membranes were subsequently incubated in blocking buffer (Cat. No. 927-70001, LI-COR Biosciences, Lincoln, NE) for 1 hr at room temperature prior to incubation with primary antibodies diluted in blocking buffer (48 hrs at 4°C). Bound primary antibodies were detected using IRDye 680RD- or 800CW-conjugated secondary antibodies (LI-COR Biosciences, Lincoln, NE) diluted in blocking buffer (3 hrs at room temperature). For quantification, membranes were imaged using an Odyssey infrared imaging system and analyzed using Image Studio and Empiria Studio software packages (LI-COR Biosciences, Lincoln, NE).
Flow Cytometry
Differentiating OLGs were collected after 48 hrs culture in serum-free differentiation medium using Accutase cell detachment solution (Cat. No. A6964, MilliporeSigma, Burlington, MA). Cells were counted, and 106 cells per sample were immunostained for analysis. Cells-only controls were placed immediately into flow cytometry buffer [0.5% bovine serum albumin (BSA) and 2mM EDTA in phosphate-buffered saline (PBS)]. All immunostaining procedures were performed on ice in PBS containing 0.5% BSA and 2mM EDTA. Fc receptors were blocked by incubation with anti-mouse CD16/32 for 15 minutes, and cells were incubated for 30 min first with anti-LPA receptor antibodies or their respective isotype controls and then with AlexaFluor 488-conjugated secondary antibodies. Cells were washed twice and resuspended in flow cytometry buffer. Immediately prior to flow analysis, 7-AAD viability stain (Cat. No. A1310, Invitrogen/eBioscience/Thermo Fisher Scientific, Waltham, MA) was added, and samples were run on a LSRFortessa-X20 flow cytometer (BD Biosciences, San Jose, CA) at room temperature using a 488 nm laser (>20mW power, Coherent solid-state) and 530/30 nm and 647/20 nm bandpass filters. Settings were carefully determined empirically and exactly reproduced in each experiment. Gates were demarcated to count 488-positive (7-AAD-negative) cells up to 10,000 events. FACSDIVA software (BD Biosciences, San Jose, CA) was used for acquisition, and data were analyzed using FCS Express Flow Cytometry software (DeNovo Software, Pasadena, CA).
RNA Isolation and RT-qPCR
RNA was purified using RNeasy Mini (cultured OLGs) or Midi (zebrafish embryos) Kits (Cat Nos. 74104 and 75144, Qiagen LLC, Germantown, MD). For the isolation of RNA from cultured rodent OLGs, cells were plated on fibronectin-coated 12 well plates (1x106 cells per well) and collected in RTL lysis buffer (Qiagen LLC, Germantown, MD) containing 1% 2-mercaptoethanol. For RNA isolation from zebrafish embryos at 48 hpf, embryos were anesthetized using 0.015% tricaine methanesulfonate (Syncaine, Syndel/Western Chemical, Ferndale, WA), chorions were removed using 2mg/mL Pronase (Cat. No. 11459643001, Sigma, St Louis, MO), and pools of about 20 embryos were homogenized in RTL lysis buffer (included in RNeasy kits, Qiagen LLC, Germantown, MD) containing 1% 2-mercaptoethanol. All RNA samples were treated with DNase (DNA-Free Kit; Cat. No. AM1906, Applied Biosystems/Thermo Fisher Scientific, Waltham, MA). RNA concentrations and purity were determined using a 2100 Bioanalyzer and RNA 6000 Pico kits (Cat. No. 5067-1513, Agilent, Santa Clara, CA); samples with an RNA integrity number above 7 were used for further analysis. Oligo-dT- and random hexamer-primed cDNAs were synthesized from 50 ng to 1 μg of RNA using Omniscript or Sensiscript Reverse Transcription Kits (Cat. Nos. 205113 and 205213, Qiagen LLC, Germantown, MD) according to the guidelines of the manufacturer. RNA samples were normalized to the same approximate concentration, and the same amount of RNA was used for all conditions of an individual independent RT-qPCR experiment.
For all RT-qPCR experiments the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines were followed (Bustin et al. 2009). Briefly, RT-qPCR primers (Table 3) were designed and in silico tested for specificity using the National Center for Biotechnology Information’s basic local alignment search tool (Primer-BLAST) (Ye et al. 2012). All primers were designed to amplify all known splice variants, and for all primer pairs melting curves were used to ensure specificity. cDNA reactions without reverse transcriptase were performed for all samples to ensure no-reverse-transcriptase quantitation cycle (Cq) numbers of at least five cycles below the lowest Cq for any of the experimental samples. RT-qPCR reactions with three technical replicates per sample were performed on a CFX96 real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA) using the iTaq Universal SYBR Green Supermix (Cat. No. 1725121, Bio-Rad Laboratories, Hercules, CA). PCR conditions were as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 15 s, 58°C for 30 s, and 95°C for 10 s. Expression levels were determined using the ΔΔCT method relative to the geometric mean of the three reference genes (Livak & Schmittgen 2001).
Homology Modeling and Molecular Docking
The published zebrafish LPA6 crystal structure was utilized as the template protein to build the 3D conformations of human LPA6 (Taniguchi et al. 2017). The amino acid sequence for human LPA6 was downloaded from UniProtKB (primary accession number P43657). The sequence alignment between target (human LPA6) and template (zebrafish LPA6) sequences were performed by the program ClustalX 2.1 (Larkin et al. 2007) with default parameters. Based on the sequence alignments, 100 3D models of LPA6 were built by MODELLER 9.19. The 3D models with the highest DOPE assessment score were selected as the optimal homology model for human LPA6 (Shen & Sali 2006).
GL-8-28 was sketched in Sybyl-X 2.0 and further energy minimized to a gradient of 0.05 with Gasteiger-Hückel charges assigned under the Tripos force field (TAFF) (Ballesteros & Weinstein 1995). The molecular docking study was performed by GOLD 2020 (Cambridge Crystallographic Data Centre, CCDC, Cambridge, UK) to obtain the ligand-receptor complex. The putative docking sites of the human LPA6 model were defined by 10 Å around the α-carbon atom of V5.39 of LPA6, which formed directly hydrophobic interaction with the LPA species in the crystal structure of the zebrafish LPA6 (Taniguchi et al. 2017). Except for the above parameters, molecular docking studies were conducted with standard default settings. Fifty docking solutions were generated in the respective optimal homology model of LPA6. The docking solutions of GL-8-28 with the highest CHEM-PLP score within the fifty docking solutions were chosen as the optimal docking poses in the homology model of human LPA6.
Statistical Analysis
GraphPad Prism (GraphPad Software Inc., San Diego, CA; RRID:SCR_002798) was used for all statistical analyses. Prior sample calculations were not performed; sample size was estimated based on previous studies of a similar nature (Dennis et al. 2008; Lafrenaye & Fuss 2011; Martinez-Lozada et al. 2014; Thomason et al. 2022; Waggener et al. 2013; Wheeler et al. 2015). Data were assessed for normality using the Shapiro-Wilk normality test prior to analysis. Data compared with a set control value lacking variability were analyzed using the one-sample t test (Dalgaard 2008; Skokal & Rohlf 1995) and presented in graphs showing individual data points plus means with SEM. For comparing multiple groups one-way ANOVA or in case of multiple data points per animal nested one-way ANOVA was used. PRESTO-Tango assays were analyzed using dose-response curves, and data are presented as means ± SEMs for each agonist concentration plus dose-response curves. p≥0.05 was used as threshold for significant for all statistical tests used. EC50 values were calculated from the dose-response curves, and the percent maximum activity was calculated using the following formula: % maximum activity = (maximum response of GL-8-28 − constitutive activity)/(maximum response of full agonist − constitutive activity) x 100.
RESULTS
LPA6 is expressed by OLG lineage cells and uniquely localized at the surface of earlier maturation stages
In order to gain a deeper understanding of the contribution of individual LPA receptors to the regulation of OLG maturation, we focused our studies here on the LPA receptor LPA6, which has previously been shown to be expressed by OLG lineage cells (Suckau et al. 2019; Wheeler et al. 2015). To better define the expression and surface localization of LPA6, enriched cultures of rat brain-derived differentiating OLGs were immunolabeled using anti- LPA6 antibodies recognizing the second extracellular loop region of LPA6 in combination with the following antibodies marking selective OLG maturation stages: A2B5 (OLG progenitor cells; (Duchala et al. 1995), O4 (maturing OLGs(Duchala et al. 1995), anti-MBP (mature OLGs(Dubois-Dalcq et al. 1986). As shown in Fig. 1A, upon fixation and permeabilization of cultured cells, LPA6 protein could be detected at all stages of the OLG lineage. Similarly, in the 3-week-old mouse corpus callosum, Lpar6 mRNA was detected in all cells expressing Olig2, a marker for all stages of the OLG lineage in rodents (Zhou et al. 2000; Lu et al. 2000; Wegner 2001), including later stages of maturing OLGs as identified by the presence of Plp1 mRNA (Dubois-Dalcq et al. 1986; Duchala et al. 1995) (Fig. 1C). These findings are consistent with previously published mRNA profiling data (Suckau et al. 2019; Marques et al. 2016; Zhang et al. 2014). Interestingly, however, when using a live staining protocol, surface localization of LPA6 was found to be restricted to earlier maturation stages of differentiating OLGs (Fig. 1B). Taken together, these data indicate that while LPA6 is expressed throughout the OLG lineage, its functional roles are likely restricted to earlier maturation stages of differentiating OLGs.
Fig. 1.

LPA6 is expressed throughout the OLG lineage but surface localization is restricted to earlier maturation stages of differentiating OLGs. A. and B. Representative confocal images of cultured rat brain-derived OLG lineage cells immunolabeled with antibodies detecting an extracellular surface epitope of LPA6 in combination with the following antibodies marking selective maturation stages: A2B5 (OLG progenitor cells) (Duchala et al. 1995), O4 (maturing OLGs) (Duchala et al. 1995), anti-MBP (mature OLGs) (Dubois-Dalcq et al. 1986). Cells were labeled after fixation and permeabilization (A) or live (B). Scale bar: 10 μm. C. Representative confocal images of the 3-week-old mouse corpus callosum, triple labeled using RNAScope for mRNAs encoding Lpar6, Olig2 to mark all OLG lineage cells (Zhou et al. 2000; Lu et al. 2000; Wegner 2001) and Plp1 to mark later OLG maturation stages (Dubois-Dalcq et al. 1986; Duchala et al. 1995). Stars indicate nuclei of Olig2-positive cells. Scale bar: 20 μm.
During development, LPA6 functions as a negative modulator of OLG maturation
After having established that LPA6 is expressed by OLGs, we next assessed potential functional effects of a loss of LPA6 on OLG maturation via the use of Wt and Lpar6 KO mice (Hata et al. 2016). More specifically, OLG maturation was analyzed in the corpus callosum and cortex at 2 and 3 weeks of age, a developmental time point that coincides with a rapid phase of myelination (Bergles & Richardson 2015; Sturrock 1980) associated with active proliferation of OLG lineage cells (Spitzer et al. 2019; Young et al. 2013; Sturrock 1983). As shown in Fig. 2B, based on immunostaining with anti-Olig2 antibodies, known to mark all OLG lineage cells in rodents (Wegner 2001; Zhou et al. 2000; Lu et al. 2000), no changes in the total number of OLG lineage cells were noted. Co-immunostaining with CC1 antibodies, labeling maturing OLGs (Bin et al. 2016; Fuss et al. 2000), on the other hand, revealed an increased percentage of maturing OLG lineage cells at 3 but not 2 weeks of age (Fig. 2C, D). Thus, there is a precocious appearance of maturing OLGs during the rapid phase of myelination in the corpus callosum and cortex of Lpar6 KO mice. This alteration appears largely due to an acceleration of OLG maturation rather than an overall increase in the number of OLG lineage cells, and it provides first evidence for a role of LPA6 in modulating the timing of OLG maturation in a negative regulatory, or restraining, fashion.
Fig. 2.
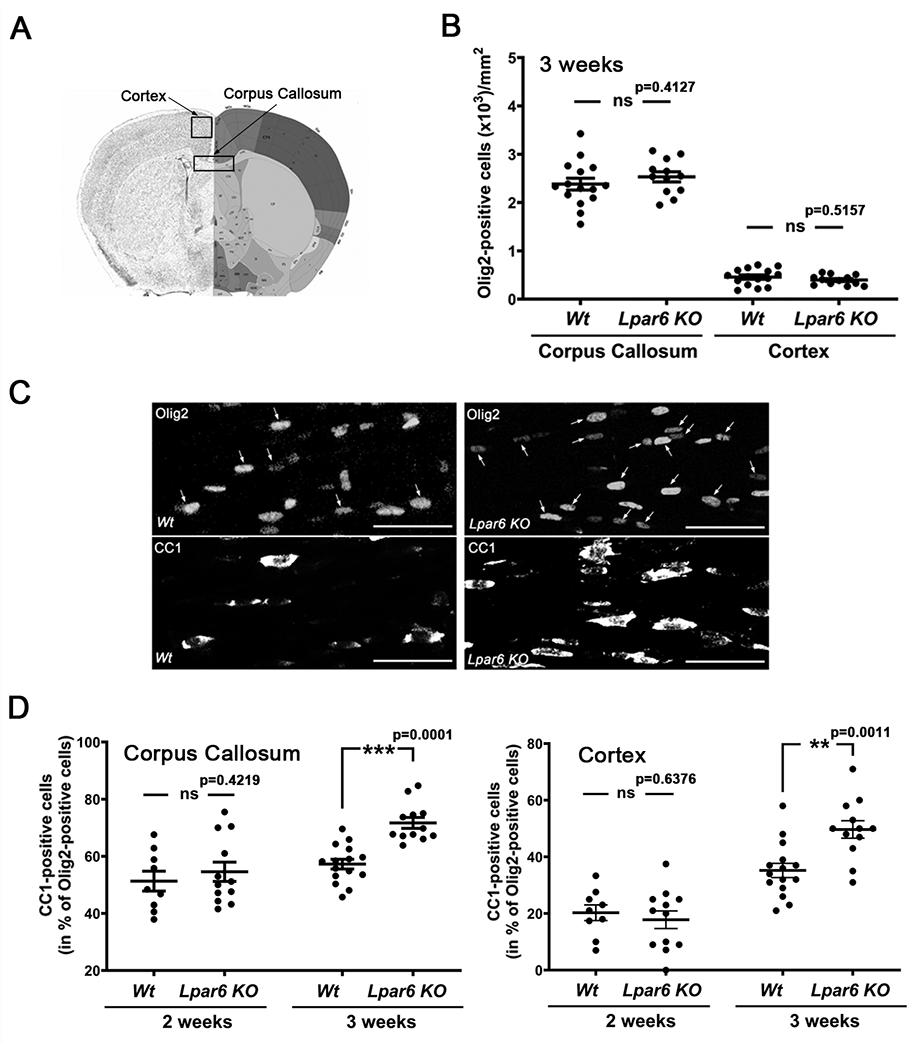
Developmental maturation of OLGs is accelerated in Lpar6 KO mice. A. Coronal brain section marking brain regions (Image credit: Allen Mouse Brain Atlas, Allen Institute). B. Graph depicting the number of Olig2-positive OLG lineage cells per mm2 in sections of 15 μm depth from 3-week-old Wt and Lpar6 KO mice as determined by immunostaining and subsequent image analysis. C. Representative confocal images of the corpus callosum in 3-week-old Wt and Lpar6 KO mice double-immunostained with an anti-Olig2 antibody to mark all OLG lineage cells (Zhou et al. 2000; Lu et al. 2000; Wegner 2001) and with the CC1 antibody to identify mature OLGs (Bin et al. 2016; Fuss et al. 2000); arrows mark CC1-Olig2 double-positive cells. Scale bar: 50 μm. D. Graphs depicting the percentage of mature CC1-Olig2 double-positive OLGs in the midcaudal corpus callosum (left) and motor cortex (right) of Wt and Lpar6 KO mice at 2 weeks and 3 weeks of age as determined by immunostaining and subsequent image analysis. D. Dots in all graphs represent 3 fields of view (40x objective) from non-adjacent sections for each individual animal. ***p ≤ 0.001, **p ≤ 0.01 ns p>0.05, 3 fields of view from 5 (3-week-old Wt mice) or 4 (2-week-old Wt and Lpar6 KO mice, 3 week Lpar6 KO mice) animals, nested two-tailed t-Test, t=0.8708 and df=7 (B, corpus callosum), t=0.6845 and df=7 (B, cortex), t=0.8294 and df=13 (D, two weeks), t=4.517 and df=25 (D, three weeks), t=0.5011 and df=5 (E, two weeks), t=3.680 and df=25 (three weeks).
The small molecule GL-8-28 represents a dual LPA4/6 ligand with preferential agonist activity at LPA6
While the data thus far support a negative modulatory role of LPA6 in the timing of OLG maturation, they do not directly address LPA6 signaling in OLGs and the potential of pharmacological accessibility for LPA6 initiated signaling. Hence, we turned our attention to a library of small molecules that had been designed and prepared by us as potential LPA receptor ligands. We first used the PRESTO-Tango GPCR assay system (Kroeze et al. 2015) to examine agonist activity at the six known LPA receptors (LPA1-6). For LPA1-5, the well-established endogenous agonist LPA (18:1) served as positive control, while, due to the much lower affinity of LPA (18:1) for LPA6, the LPA phosphorothioate analog (2S)-OMPT was used for assaying LPA6 ligands (Jiang et al. 2013; Yanagida et al. 2009; Inoue et al. 2011; Inoue et al. 2012). As shown in Fig. 4, one of the compounds from our library, designated here as GL-8-28 (Fig. 3), was found to partially activate both LPA4 and LPA6. Notably, based on estimated EC50 calculations, the potency of GL-8-28 appears to be approximately five times higher for LPA6 compared to LPA4. Neither of the remaining known LPA receptors were activated by GL-8-28 above a level of 20% of control (Fig. 4B), thus defining GL-8-28 as a dual LPA4/6 partial agonist with reasonable selectivity to LPA6 over LPA4 as well as other LPA receptors. This observation was further corroborated by molecular modeling and ligand-receptor docking using the published zebrafish LPA6 crystal structure (Taniguchi et al. 2017) as a template to build the 3D conformations of human LPA6 bound to the ligand GL-8-28. As shown in Fig. 5, the polar head group of GL-8-28 interacts with the conserved positively charged residues K1.31, R2.60, R6.62, and R7.32, previously shown to be particularly important for ligand recognition and receptor activation (Taniguchi et al. 2017). Agonist binding to the LPA6 receptor has been proposed to induce a conformational inward shift of the transmembrane helices 6 (TM6) and 7 (TM7) resulting in direct interaction with all of the above conserved positively charged residues with the ligand headgroup; this shift is not depicted in Fig. 5. The long aliphatic chain of GL-8-28 is positioned within a hydrophobic environment formed at the TM4-TM5 interfaces; this hydrophobic cleft has been implicated in binding of the acyl chain of LPA (Taniguchi et al. 2017). Thus, GL-8-28 displays ligand-receptor interactions that involve binding pocket residues predicted to be crucial for binding of the endogenous agonist LPA and subsequent receptor activation.
Fig. 4.

GL-8-28 functions as an LPA4/6 dual ligand with preferential LPA6 agonist activity. A. Graphs depicting dose response curves as determined by the PRESTO-Tango GPCR assay system (Kroeze et al. 2015). Note, that LPA (18:1) was used as previously established agonist for LPA1-5, while (2S)-OMPT was used for LPA6 (Jiang et al. 2013). Means ± SEM are shown for at least five independent experiments performed in triplicates. Significant agonist activity for GL-8-28 is defined as above 20% of control (see B). Note that the estimated potency for GL-8-28 is much higher for LPA6 compared to LPA4. B. Graph depicting the percentage of GL-8-28 activity compared to control. Data points represent independent experiments (n=5 (LPA5), 6 (LPA2), 7 (LPA3), 9 (LPA1,4), 10 (LPA6)) performed in triplicates. *p≤0.05, **p≤0.01, one-sample two tailed t-test, theoretical mean set to 20% for LPA1,2,4 and 6; to 2% for LPA3, t=0.3839 and df=8 (LPA1), t=0.1326 and df=5 (LPA2), t=0.2239 and df=6 (LPA3), t=3.515 and df=8 (LPA4), t=0.5633 and df=4 (LPA5), t=3.230 and df=9 (LPA6).
Fig. 5.

GL-8-28 displays ligand-receptor interactions that involve binding pocket residues predicted to be crucial for binding of the endogenous agonist LPA and receptor activation. The binding pocket of LPA6 is shown as cartoon model (lavender) and the ligand GL-8-28 is depicted as stick model (green carbon atoms). Key amino acid residues binding with GL-8-28 are shown as sphere model. Arrowheads (gray) point to conserved positively charged residues implicated in head group binding and receptor activation (Taniguchi et al. 2017)}; positively charged residues (pink), hydrophobic residues (orange), oxygen atoms (red), nitrogen atoms (blue), sulfonate atoms (yellow).
Pharmacological activation of LPA6 signaling attenuates OLG maturation
In order to directly assess the effect of LPA6 activation on OLG maturation, we treated primary cultures of differentiating rat brain-derived OLGs with the LPA6 preferential agonist GL-8-28. In agreement with our live staining experiments (see Fig. 1) surface localization was detected by flow cytometry on a subpopulation of OLG lineage cells (approximately 43%; see Fig. 6 A’). In contrast, only very few cells (approximately 6%) displayed detectable levels of surface localization for LPA4 (Fig. 6 A’). Thus, any effects exerted by GL-8-28 are mediated primarily by its LPA6 agonist activity and are directed at earlier maturation stages of differentiating OLGs (see Fig. 1). As shown in Fig. 6D, in the presence of the highest concentration tested, i.e. 10 μM, GL-8-28 treatment led to significantly reduced mRNA levels for OLG expressed genes associated with OLG maturation and CNS myelination, namely 2,′3′-cyclic nucleotide 3′-phosphodiesterase (Cnp) (Kurihara et al. 1992; Gravel et al. 1998) and UDP glycosyltransferase 8 (Ugt8) (Gard & Pfeiffer 1990; Bosio et al. 1996). This outcome was not found to be associated with a cytotoxic effect (Fig. 6B) or a change in the expression of Olig2 (Fig. 6C), a transcription factor expressed throughout the OLG lineage in rodents (Zhou et al. 2000; Lu et al. 2000; Wegner 2001). Additionally, at the 10 μM concentration, GL-8-28 significantly increased the expression of the transcriptional repressor Egr1 (Swiss et al. 2011; Sock et al. 1997), a known inhibitor of the gene expression profile associated with OLG maturation and CNS myelination. In this context, it is important to note that downregulated Egr1 expression concomitant with OLG differentiation was previously shown to occur upon thyroid hormone application (Swiss et al., 2011), thus demonstrating that in differentiating OLGs upregulation of Egr1 is not generally associated with a stimulus evoked response as for example seen in the context of neuronal activity and plasticity (Duclot & Kabbaj 2017).
Fig. 6.

The preferential LPA6 agonist GL-8-28 attenuates maturation in primary cultures of rat OLGs. A. Scheme depicting the experimental paradigm; the inset in A’ represents the percent of LPA4 (left) and LPA6 (right) positive OLGs at 48 h, as determined by flow cytometry. B-E. Graphs depicting the extracellular LDH levels (B) and mRNA levels as determined by RT-qPCR analysis (C-E). Levels for control (vehicle-treated) cells were set to 100% (horizontal dotted line) and relative levels were calculated accordingly. Individual data points represent independent experiments, means ± SEM are shown as horizontal lines with error bars. *p≤0.05 and ****p≤0.0001, one sample two-tailed t-test, B: n=6, t=1.897 and df=5; C: 1 nM: n=4, p=0.7220, t=0.3909, df=3, 10nM: n=6, p=0.0523, t=2.534, df=5, 100nM: n=7, p=0.6686, t=0.4498, df=6, 1μM: n=5, p=0.0508, t=2.760, df=4, 10μM: n=5, p=0.2441, t=1.365, df=4; D: 1 nM: n=4, p=0.0542, t=3,079, df=3 (CNP); n=5, p=0.2653, t=1.294, df=4 (Ugt8) 10nM: n=7, p=0.0606, t=2.306, df=6 (CNP); n=6, p=0.1412, t=1.746, df=5 (Ugt8), 100nM: n=7, p=0.5995, t=0.5543, df=6 (CNP); n=6, p=0.2538, t=1.289, df=5 (Ugt9), 1μM:n=5, p=0.6630, t=0.4697, df=4 (CNP); n=5, p=0.4583, t=0.8188, df=4 (Ugt8), 10μM:n=5, p˂0.0001, 4t=21.56, df=4 (CNP), n=5, p˂0.0001, t=18.93, df=4, E: 1 nM: n=5, p=0.5163, t=0.7111, df=4, 10nM: n=7, p=0.4297, t=0.8465, df=6, 100nM: n=7, p=0.8491, t=0.1986, df=6, 1μM: n=5, p=0.4981, t=0.7442, df=4, 10μM: n=5, p=0.0300, t=3.299, df=4.
In an attempt to extend these findings into an in vivo system and to gain initial insight into the in vivo use of GL-8-28, we took advantage of the following characteristics of the developing zebrafish. First, in this model, pharmacological agents can be taken up from the surrounding water source via passage through the skin (Rombough 2002). Second, a lack of a functional blood-brain-barrier (BBB) prior to 3 days of age (Li et al. 2017; Fleming et al. 2013) allows diffusion into the developing CNS prior to completion of myelination (Buckley et al. 2008; Jung et al. 2010; Yoshida & Macklin 2005; Brosamle & Halpern 2002), for which key regulatory mechanisms are highly conserved during the development of mammals and zebrafish (Ackerman & Monk 2016; Lyons & Talbot 2014; Preston & Macklin 2015). In this context, the expression of proteolipid protein, present in both maturing rodent (Plp1) and zebrafish (plp1a/plp1b) OLGs (Siems et al. 2021), as well as myelin protein zero (mpz) and claudin K (cldnk), found more specifically in maturing zebrafish OLGs, has been reported to significantly increase between 24 and 48 hrs post fertilization (hpf) in the CNS of the developing zebrafish embryo (Preston et al. 2019; Wheeler et al. 2015; Munzel et al. 2012; Takada & Appel 2010; Schweitzer et al. 2006; Yoshida & Macklin 2005; Brosamle & Halpern 2002). In the presence of GL-8-28, mRNA levels for all these three zebrafish genes known to be associated with OLG maturation were significantly reduced (Fig. 7). At the same time, gross anatomical features remained unchanged up to a concentration of 1 μM (Fig. 7B). Importantly, based on recently published gene expression profiling data (Marisca et al. 2020), LPA receptor expression in zebrafish OLGs appears to be limited to differentiating rather than progenitor stages of the lineage and restricted to the receptors lpar1 and lpar6a. Furthermore, equivalents of several residues found to be important for ligand binding to human LPA6 were also found to affect binding in models based on the crystallized structure of the zebrafish receptor, thus providing confidence of equivalent binding characteristics in all species analyzed here (Taniguchi et al. 2017; Raza et al. 2014). Thus, the OLG maturation attenuating effects of GL-8-28 in the developing zebrafish are likely to be mediated, at least in part, via activation of LPA6 in differentiating OLGs. However, these effects of GL-8-28 were observed at much lower concentrations as seen in the rat primary cultures, possibly due to receptor desensitization in the culture system and/or cumulative effects in the in vivo system. Nevertheless, the above data demonstrate an important functional role of LPA6 signaling in regulating the timely appearance of mature OLGs in both rodent cultures of differentiating OLG and the developing zebrafish.
Fig. 7.

The preferential LPA6 agonist GL-8-28 attenuates OLG maturation in the developing zebrafish. A. Scheme depicting the experimental paradigm. B. Representative images of control and Gl-8-28 (1μM)-treated zebrafish embryos at 48 hpf (hours post fertilization). C-E. Graphs depicting mRNA levels as determined by RT-qPCR analysis. Levels for control (vehicle-treated) zebrafish embryos were set to 100% (horizontal dotted line) and relative levels were calculated accordingly. Individual data points represent independent experiments, means ± SEM are shown as horizontal lines with error bars. plp1b: proteolipid protein, mpz: myelin protein zero, cldnk: claudin K. *p≤0.05 and **p≤0.001, one sample two-tailed t-test, C: 100pM: n=4, p=0.0084, t=6.226, df=3, 1nM: n=4, p=0.0033, t=8.621, df=3, 10nM: n=4, p=0.0087, t=6.142, df=3, 100nM: n=4, p=0.0432, t=3.377, df=3, 1μM: n=4, p=0.4916, t=0.7814, df=3 D: n=4, p=0.0004, t=17.23, df=3, 1nM: n=4, t=9.185, df=3, 10nM: n=4, p=0.0085, t=6.198, df=3, 100nM: n=4, p=0.0067, t=6.738, df=3, 1μM: n=4, p=0.9009, t=0.1353, df=3, E: 100pM: n=5, p=0.0051, t=5.573, df=4, 1nM: n=5, p=0.0109, t=4.491, df=4, 10nM: n=5, p=0.0131, t=4.258, df=4, 100nM: n=5, p=0.1138, t=0.1138, t=2.017, df=4, 1μM: n=5,p=0.1684, t=1.679, df=4.
LPA6 protein levels appear elevated in MS white matter lesions
In the context of development, the maturation attenuating functional role of LPA6 likely serves to ensure timely coordination of circuit formation and myelination (Fletcher et al. 2021). Under pathological conditions, however, persistent LPA6 expression and signaling could be seen as an inhibitor of efficient OLG maturation and myelin repair. In this regard, it is of particular interest that our analysis of human MS lesion samples (Fig. 8) revealed, on average, elevated LPA6 protein levels. Interestingly, the variance seen within the MS sample pool was significantly broader than the one for the control sample pool, an observation that is consistent with the increasingly recognized heterogeneity in MS pathology between patients (Smets et al. 2021; Patrikios et al. 2006; Lucchinetti et al. 2000). Curiously, in addition to a somewhat diffuse band with the expected apparent molecular weight of approximately 50 kDa, a sharp lower molecular weight band of approximately 40 kDa was observed in some of the MS samples, possibly reflecting differences in N-glycosylation (Suckau et al. 2019). Taken together, these data suggest that LPA6 protein levels may be increased in at lest some types of MS white matter lesions, an observation that is consistent with recent transcriptome profiling data (Elkjaer et al. 2019; Falcão et al. 2018).
Fig. 8.

LPA6 protein levels are increased in multiple sclerosis (MS) white matter lesions. A. Representative Western blot depicting LPA6 and total protein between molecular weight markers 70 kDa (top) and 38 kDa (bottom) in control (CTRL) white matter and multiple sclerosis (MS) white matter lesions; total protein per lane was used for normalization. B. Graph depicting the percentage of LPA6 protein levels in control (CTRL) white matter and MS white matter lesions (MS) (left y-axis) and the mean difference (right y-axis) as determined by Western blot analysis. Tissues were obtained from the Netherlands Brain Bank (for details see Table 2). The mean control value, normalized to total protein, was set to 100% and all other values were adjusted accordingly (average values from two independent Western blots are shown). ***p≤0.001 Welch’s two-tailed t-test (Variancep≤0.05), Welch-corrected t=3.997, df=17.00. CI, confidence interval.
DISCUSSION
In this study we identified signaling via the LPA receptor LPA6 as a novel modulator of the transcriptional regulation of OLG maturation. More specifically, our data revealed a precocious developmental appearance of OLGs expressing a marker for more mature stages of the lineage in Lpar6 KO mice. Thus, initial evidence was obtained that activation of LPA6 signaling in OLGs negatively modulates the transcriptional program associated with OLG maturation. This interpretation could be substantiated via the use of a novel LPA6 selective ligand with agonist activity, here referred to as GL-8-28; both in vitro in cultures of differentiating OLGs and in vivo in the developing zebrafish, treatment with GL-8-28 was found to impede the expression of genes known to be associated with OLG maturation. During development, such a functional role for LPA6 signaling is likely transient and serves to ensure timely coordination of circuit formation and myelination (Fletcher et al. 2021). This point of view may be supported by the apparent lack of an overt myelination phenotype in Lpar6 KO mice (Hata et al. 2016). However, future studies are needed to further substantiate such a proposed developmentally transient functional role of LPA6 signaling in maturing OLGs. Under pathological conditions accompanying white matter lesions in MS, LPA6 signaling appears to be persistent, at least in some patients. Hence, our studies reveal a novel molecular mechanism modulating OLG maturation, and they uncover a novel and conceivably druggable signaling pathway with the potential for future developments toward innovative therapeutic interventions stimulating myelin repair.
LPA signaling plays diverse roles in the regulation of OLG maturation and CNS myelination
A key step in the biosynthetic pathway(s) generating extracellular LPA is represented by the conversion from lysophosphatidylcholine (LPC) via the enzymatic activity of secreted autotaxin, also known as extracellular nucleotide pyrophosphatase-phosphodiesterase 2 (ENPP2) or lysophospholipase D (lysoPLD) (Aoki et al. 2008; Tanaka et al. 2006; van Meeteren et al. 2006; Gijsbers et al. 2003; Tokumura et al. 2002; Umezu-Goto et al. 2002). In our previous studies, we had identified OLG expressed autotaxin as a positive regulator of OLG maturation (Wheeler et al. 2015; Yuelling et al. 2012; Dennis et al. 2012; Nogaroli et al. 2009; Dennis et al. 2008; Fox et al. 2004; Fox et al. 2003). Importantly, autotaxin was found to stimulate the transcriptional expression profile leading to OLG differentiation via its enzymatic activity generating LPA (Wheeler et al. 2016; Wheeler et al. 2015; Yuelling et al. 2012). In addition, autotaxin-dependent activation of specifically LPA6 has recently been proposed to exert functional effects on cells other than OLGs (Matas-Rico et al. 2021; Okasato et al. 2021; Masago et al. 2018). Thus, a maturation attenuating role of LPA6 signaling on OLGs, as described here, may appear puzzling. However, despite the simple basic structure of LPA, i.e. a glycerol backbone connected to a phosphate head group at the sn-3 position and a fatty acid chain linked to the sn-1 or sn-2 position, variations in fatty acid side chain length, saturation and backbone position generate diversity that impacts receptor selectivity and downstream signaling outcomes (Aikawa et al. 2015; Hayashi et al. 2001; Okudaira et al. 2014; Yung et al. 2014; Okudaira et al. 2010; Yoshida et al. 2003; Baker et al. 2001; Bandoh et al. 2000). Furthermore, structure-function analyses suggest a highly localized delivery of LPA to the respective receptor to be activated, thereby refining the selectivity for LPA receptor subtype activation (Taniguchi et al. 2017; Hausmann et al. 2013; Houben et al. 2013; Fulkerson et al. 2011; Moolenaar & Perrakis 2011; Hausmann et al. 2011; Nishimasu et al. 2011; Kanda et al. 2008). Regarding LPA6, preference for 2-acyl rather than 1-acyl LPA species confer somewhat unique characteristics within the LPA receptor family (Aikawa et al. 2015; Nishimasu et al. 2011; Inoue et al. 2011; Inoue et al. 2012; Yanagida et al. 2009; Morishige et al. 2007; Tokumura et al. 2002). Thus, fine-tuned regulation of the LPA receptor profile present on OLG surfaces in combination with the availability and local delivery of specific LPA species may determine the balance between LPA-mediated maturation promoting and attenuating outcomes. Interestingly, a counter-regulatory function of LPA6, compared to one or more of the remaining known LPA receptors, has also been described for cell types other than OLGs (Matas-Rico et al. 2021; Takahashi et al. 2017; Ishii et al. 2015).
The small molecule GL-8-28 represents a novel chemical compound with preferential LPA6 agonist activity
Small molecule modulators of the LPA family of G protein-coupled receptors are emerging as promising drugs for the treatment of a variety of diseases including those affecting the nervous system (Liu et al. 2021; Geraldo et al. 2021; Yanagida & Valentine 2020; Herr et al. 2020; Stoddard & Chun 2015; Kihara et al. 2015; Yung et al. 2015). Importantly, recent progress has been made in developing ligands with high selectivity for LPA receptor subtypes, in particular LPA1-3 (Liu et al. 2021). In addition, despite the high sequence homology between LPA4 and LPA6 (Yanagida & Ishii 2011; Pasternack et al. 2008; Noguchi et al. 2003), unique ligand preferences have been described (Kano et al. 2019; Yanagida et al. 2013), thereby supporting a long-term prospect of highly selective LPA6 ligands. Intriguingly, a novel compound with selective LPA6 antagonist activity has recently been reported (Gnocchi et al. 2020).
The preferential LPA6 agonist GL-8-28 attenuates OLG maturation in both the developing zebrafish and primary cultures of differentiating OLGs.
In both the in vitro cell culture and the in vivo zebrafish model, GL-8-28 was found to attenuate the gene expression profile associated with OLG maturation. However, much higher ligand concentrations were needed in the in vitro studies. In this context, it has been well-established that LPA receptors, similar to other G protein-coupled receptors, can be subject to receptor desensitization (Solís et al. 2021; Alcántara-Hernández et al. 2015; Avendaño-Vázquez et al. 2005; Zhao et al. 2021; Urs et al. 2005). Interestingly, next to homologous desensitization by the agonist there is evidence for agonist-independent heterologous desensitization (Alcántara-Hernández et al. 2015; Castillo-Badillo et al. 2015). Thus, it is conceivable that the culture conditions used in the in vitro experiments regulate extracellular LPA levels differently than seen in vivo and/or more significantly contribute to heterologous LPA6 desensitization, thereby requiring higher GL-8-28 concentrations to trigger measurable functional outcomes. In addition, in the in vivo studies using the developing zebrafish, functional consequences of GL-8-28 on cell types other than OLGs could contribute to the observed attenuation of OLG maturation. In particular, LPA6 signaling has been implicated in the regulation of vascular development (Okasato et al. 2021; Yasuda et al. 2019; Kano et al. 2019), and OLG-vascular crosstalk has been associated with a modulation of developmental OLG differentiation (Tsai et al. 2016; Yuen et al. 2014; De La Fuente et al. 2017). Further studies will be necessary to dissect the contributions of receptor desensitization and LPA6-mediated non-OLG target effects to the differences in GL-8-28 sensitivity observed in the in vitro versus in vivo systems used here. Nevertheless, both the in vivo and in vitro data provide compelling support for a negative modulatory role of LPA6 signaling on the expression of genes associated with OLG maturation.
LPA6 signaling emerges as a novel and conceivably druggable signaling pathway with the potential toward promoting myelin repair
Changes in transcriptional regulation are thought to be linked to modifications in OLG heterogeneity (Jäkel et al. 2019; Falcão et al. 2018) and inefficiencies in OLG maturation and myelin repair (Duncan et al. 2017; Kuhlmann et al. 2008; Chang et al. 2002). As cause for such dysregulated gene expression in OLGs, recent evidence points to the actions of extrinsic factors rather than intrinsic OLG defects (Saraswat et al. 2021; Golan et al. 2021; Mozafari et al. 2020; Starost et al. 2020; Kirby et al. 2019). Thus, targeting druggable signaling pathways with the capacity to modulate gene expression profiles associated with OLG maturation has emerged as a promising approach toward promoting repair of the myelin sheath in MS (Rajendran et al. 2021; Angelini et al. 2021; Jeffries et al. 2021; Jeffries et al. 2016; Mausner-Fainberg et al. 2021; Roggeri et al. 2020; Lecca et al. 2020; Wang et al. 2020; Skinner & Lane 2020; Thümmler et al. 2019; Göttle et al. 2019; Welliver et al. 2018; Petersen et al. 2021; Green et al. 2017; Gaesser & Fyffe-Maricich 2016; Mierzwa et al. 2013). Despite an increasing number of potential pathways with promyelinating characteristics, however, clinical translation has, up until now, been below expectation (Lubetzki et al. 2020; Abu-Rub & Miller 2018). In light of the evidence for increased levels of LPA6 in MS white matter lesions, targeting LPA6 signaling may, therefore, represent a promising novel approach for stimulating OLG maturation in the context of myelin repair.
Acknowledgments
This work was supported by the following grants: NIH R01NS045883 (B.F.), NIH R21NS123317 (B.F.), NMSS RG-1506-04546. Services related to microscopy, flow cytometry, and RNAScope were supported, in part, by funding from NIH-NCI Cancer Center Support Grant P30CA016059. The authors thank Dr. Wesley K. Kroeze and Dr. Bryan L. Roth (University of North Carolina at Chapel Hill) for providing the HTLA cell line and for advice in setting up the PRESTO-Tango assay.
Abbreviations:
- actb2
beta actin
- BSA
bovine serum albumin
- cldnk
claudin k
- Cnp
2′,3′ cyclic nucleotide phosphodiesterase
- CNS
central nervous system
- DMEM
Dulbecco’s modified Eagle’s medium
- DMSO
dimethyl sulfoxide
- DMF
dimethylformamide
- EAE
experimental autoimmune encephalomyelitis
- EDTA
ethylenediaminetetraacetic acid
- ef1a
elongation factor 1-alpha
- Egr1
early growth response 1
- ENPP2
ectonucleotide pyrophosphatase/phosphodiesterase 2
- FBS
fetal bovine serum
- bFGF
basic fibroblast growth factor
- GC/MS
gas chromatography/mass spectrometry
- HBSS
Hank’s balanced salt solution
- hpf
hours postfertilization
- hrs
hours
- IR
infrared
- LPA
lysophosphatidic acid
- LPC
lysophosphatidylcholine
- lysoPLD
lysophospholipase D
- MHz
megahertz
- MS
multiple sclerosis
- NMR
nuclear magnetic resonance
- OLG
oligodendrocyte
- Olig2
oligodendrocyte transcription factor 2
- (2S)-OMPT
L-sn-1-O-oleoyl-2-O-methylglyceryl-3-phosphothionate
- PA
phosphatidic acid
- PBS
phosphate-buffered saline
- PDGF
platelet-derived growth factor
- Pgk1
phosphoglycerate kinase 1
- plp1b
proteolipid protein 1b
- Ppia
peptidylprolyl isomerase A (cyclophilin A)
- Rpl13a
ribosomal protein L13a
- RT-qPCR
quantitative reverse transcription polymerase chain reaction
- SDS
sodium dodecyl sulfate (SDS)
- T3
tri-iodo-thyronine
- TAZ
transcriptional coactivator with PDZ-binding motif
- TLC
thin layer chromatography
- Ugt8
UDP glycosyltransferase 8
- YAP
Yes-associated protein
Footnotes
CONFLICT OF INTEREST DISCLOSURE
BF is a consultant for Gryphon Bio, Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Abu-Rub M and Miller RH (2018) Emerging Cellular and Molecular Strategies for Enhancing Central Nervous System (CNS) Remyelination. Brain Sci 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman SD and Monk KR (2016) The scales and tales of myelination: using zebrafish and mouse to study myelinating glia. Brain research 1641, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams KL, Dahl KD, Gallo V and Macklin WB (2021) Intrinsic and extrinsic regulators of oligodendrocyte progenitor proliferation and differentiation. Seminars in cell & developmental biology 116, 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aikawa S, Hashimoto T, Kano K and Aoki J (2015) Lysophosphatidic acid as a lipid mediator with multiple biological actions. Journal of biochemistry 157, 81–89. [DOI] [PubMed] [Google Scholar]
- Alcántara-Hernández R, Hernández-Méndez A, Campos-Martínez GA, Meizoso-Huesca A and García-Sáinz JA (2015) Phosphorylation and Internalization of Lysophosphatidic Acid Receptors LPA1, LPA2, and LPA3. PloS one 10, e0140583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew SM, Titus JA, Amin A and Coico R (2009) Isolation of murine and human immunoglobulin m and murine immunoglobulin D. Current protocols in immunology Chapter 2, Unit 2 9. [DOI] [PubMed] [Google Scholar]
- Angelini J, Marangon D, Raffaele S, Lecca D and Abbracchio MP (2021) The Distribution of GPR17-Expressing Cells Correlates with White Matter Inflammation Status in Brain Tissues of Multiple Sclerosis Patients. International journal of molecular sciences 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anliker B, Choi JW, Lin ME, Gardell SE, Rivera RR, Kennedy G and Chun J (2013) Lysophosphatidic acid (LPA) and its receptor, LPA1 , influence embryonic schwann cell migration, myelination, and cell-to-axon segregation. Glia 61, 2009–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki J, Inoue A and Okudaira S (2008) Two pathways for lysophosphatidic acid production. Biochimica et biophysica acta 1781, 513–518. [DOI] [PubMed] [Google Scholar]
- Avendaño-Vázquez SE, García-Caballero A and García-Sáinz JA (2005) Phosphorylation and desensitization of the lysophosphatidic acid receptor LPA1. The Biochemical journal 385, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DL, Desiderio DM, Miller DD, Tolley B and Tigyi GJ (2001) Direct quantitative analysis of lysophosphatidic acid molecular species by stable isotope dilution electrospray ionization liquid chromatography-mass spectrometry. Analytical biochemistry 292, 287–295. [DOI] [PubMed] [Google Scholar]
- Ballesteros JA and Weinstein H (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods in neurosciences 25, 366–428. [Google Scholar]
- Bandoh K, Aoki J, Taira A, Tsujimoto M, Arai H and Inoue K (2000) Lysophosphatidic acid (LPA) receptors of the EDG family are differentially activated by LPA species. Structure-activity relationship of cloned LPA receptors. FEBS letters 478, 159–165. [DOI] [PubMed] [Google Scholar]
- Barres BA, Hart IK, Coles HS, Burne JF, Voyvodic JT, Richardson WD and Raff MC (1992) Cell death and control of cell survival in the oligodendrocyte lineage. Cell 70, 31–46. [DOI] [PubMed] [Google Scholar]
- Baydyuk M, Morrison VE, Gross PS and Huang JK (2020) Extrinsic Factors Driving Oligodendrocyte Lineage Cell Progression in CNS Development and Injury. Neurochemical research 45, 630–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benusa SD, George NM, Sword BA, DeVries GH and Dupree JL (2017) Acute neuroinflammation induces AIS structural plasticity in a NOX2-dependent manner. Journal of neuroinflammation 14, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergles DE and Richardson WD (2015) Oligodendrocyte Development and Plasticity. Cold Spring Harbor perspectives in biology 8, a020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin JM, Harris SN and Kennedy TE (2016) The oligodendrocyte-specific antibody ‘CC1’ binds Quaking 7. Journal of neurochemistry 139, 181–186. [DOI] [PubMed] [Google Scholar]
- Bosio A, Binczek E and Stoffel W (1996) Molecular cloning and characterization of the mouse CGT gene encoding UDP-galactose ceramide-galactosyltransferase (cerebroside synthetase). Genomics 35, 223–226. [DOI] [PubMed] [Google Scholar]
- Brosamle C and Halpern ME (2002) Characterization of myelination in the developing zebrafish. Glia 39, 47–57. [DOI] [PubMed] [Google Scholar]
- Buckley CE, Goldsmith P and Franklin RJ (2008) Zebrafish myelination: a transparent model for remyelination? Dis Model Mech 1, 221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A et al. (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. The Journal of neuroscience : the official journal of the Society for Neuroscience 28, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Badillo JA, Sánchez-Reyes OB, Alfonzo-Méndez MA, Romero-Ávila MT, Reyes-Cruz G and García-Sáinz JA (2015) α1B-adrenergic receptors differentially associate with Rab proteins during homologous and heterologous desensitization. PloS one 10, e0121165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A, Tourtellotte WW, Rudick R and Trapp BD (2002) Premyelinating oligodendrocytes in chronic lesions of multiple sclerosis. N Engl J Med 346, 165–173. [DOI] [PubMed] [Google Scholar]
- Dalgaard P (2008) Introductory Statistics with R. Springer, New York. [Google Scholar]
- De La Fuente AG, Lange S, Silva ME et al. (2017) Pericytes Stimulate Oligodendrocyte Progenitor Cell Differentiation during CNS Remyelination. Cell reports 20, 1755–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis J, Morgan MK, Graf MR and Fuss B (2012) P2Y(12) receptor expression is a critical determinant of functional responsiveness to ATX’s MORFO domain. Purinergic Signal 8, 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis J, White MA, Forrest AD, Yuelling LM, Nogaroli L, Afshari FS, Fox MA and Fuss B (2008) Phosphodiesterase-Ialpha/autotaxin’s MORFO domain regulates oligodendroglial process network formation and focal adhesion organization. Molecular and cellular neurosciences 37, 412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois-Dalcq M, Behar T, Hudson L and Lazzarini RA (1986) Emergence of three myelin proteins in oligodendrocytes cultured without neurons. The Journal of cell biology 102, 384–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchala CS, Asotra K and Macklin WB (1995) Expression of cell surface markers and myelin proteins in cultured oligodendrocytes from neonatal brain of rat and mouse: a comparative study. Developmental neuroscience 17, 70–80. [DOI] [PubMed] [Google Scholar]
- Duclot F and Kabbaj M (2017) The Role of Early Growth Response 1 (EGR1) in Brain Plasticity and Neuropsychiatric Disorders. Frontiers in behavioral neuroscience 11, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan GJ, Plemel JR, Assinck P et al. (2017) Myelin regulatory factor drives remyelination in multiple sclerosis. Acta neuropathologica 134, 403–422. [DOI] [PubMed] [Google Scholar]
- Dupree JL, Girault JA and Popko B (1999) Axo-glial interactions regulate the localization of axonal paranodal proteins. The Journal of cell biology 147, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaz B and Popko B (2019) Molecular Control of Oligodendrocyte Development. Trends in neurosciences 42, 263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkjaer ML, Frisch T, Reynolds R, Kacprowski T, Burton M, Kruse TA, Thomassen M, Baumbach J and Illes Z (2019) Molecular signature of different lesion types in the brain white matter of patients with progressive multiple sclerosis. Acta neuropathologica communications 7, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B and Lu QR (2015) Transcriptional and Epigenetic Regulation of Oligodendrocyte Development and Myelination in the Central Nervous System. Cold Spring Harbor perspectives in biology 7, a020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estivill-Torrus G, Llebrez-Zayas P, Matas-Rico E et al. (2008) Absence of LPA1 signaling results in defective cortical development. Cerebral cortex (New York, N.Y.: 1991) 18, 938–950. [DOI] [PubMed] [Google Scholar]
- Falcão AM, van Bruggen D, Marques S et al. (2018) Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nature medicine 24, 1837–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A, Diekmann H and Goldsmith P (2013) Functional characterisation of the maturation of the blood-brain barrier in larval zebrafish. PloS one 8, e77548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JL, Makowiecki K, Cullen CL and Young KM (2021) Oligodendrogenesis and myelination regulate cortical development, plasticity and circuit function. Seminars in cell & developmental biology. [DOI] [PubMed] [Google Scholar]
- Fox MA, Alexander JK, Afshari FS, Colello RJ and Fuss B (2004) Phosphodiesterase-I alpha/autotaxin controls cytoskeletal organization and FAK phosphorylation during myelination. Molecular and cellular neurosciences 27, 140–150. [DOI] [PubMed] [Google Scholar]
- Fox MA, Colello RJ, Macklin WB and Fuss B (2003) Phosphodiesterase-Ialpha/autotaxin: a counteradhesive protein expressed by oligodendrocytes during onset of myelination. Molecular and cellular neurosciences 23, 507–519. [DOI] [PubMed] [Google Scholar]
- Fulkerson Z, Wu T, Sunkara M, Kooi CV, Morris AJ and Smyth SS (2011) Binding of autotaxin to integrins localizes lysophosphatidic acid production to platelets and mammalian cells. The Journal of biological chemistry 286, 34654–34663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss B, Mallon B, Phan T, Ohlemeyer C, Kirchhoff F, Nishiyama A and Macklin WB (2000) Purification and analysis of in vivo-differentiated oligodendrocytes expressing the green fluorescent protein. Developmental biology 218, 259–274. [DOI] [PubMed] [Google Scholar]
- Gaesser JM and Fyffe-Maricich SL (2016) Intracellular signaling pathway regulation of myelination and remyelination in the CNS. Experimental neurology 283, 501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Diaz B, Riquelme R, Varela-Nieto I et al. (2015) Loss of lysophosphatidic acid receptor LPA1 alters oligodendrocyte differentiation and myelination in the mouse cerebral cortex. Brain Struct Funct 220, 3701–3720. [DOI] [PubMed] [Google Scholar]
- Gard AL and Pfeiffer SE (1990) Two proliferative stages of the oligodendrocyte lineage (A2B5+O4− and O4+GalC−) under different mitogenic control. Neuron 5, 615–625. [DOI] [PubMed] [Google Scholar]
- Gennero I, Laurencin-Dalicieux S, Conte-Auriol F et al. (2011) Absence of the lysophosphatidic acid receptor LPA1 results in abnormal bone development and decreased bone mass. Bone 49, 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo LHM, Spohr T, Amaral RFD, Fonseca A, Garcia C, Mendes FA, Freitas C, dosSantos MF and Lima FRS (2021) Role of lysophosphatidic acid and its receptors in health and disease: novel therapeutic strategies. Signal transduction and targeted therapy 6, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijsbers R, Aoki J, Arai H and Bollen M (2003) The hydrolysis of lysophospholipids and nucleotides by autotaxin (NPP2) involves a single catalytic site. FEBS letters 538, 60–64. [DOI] [PubMed] [Google Scholar]
- Gnocchi D, Kapoor S, Nitti P, Cavalluzzi MM, Lentini G, Denora N, Sabbà C and Mazzocca A (2020) Novel lysophosphatidic acid receptor 6 antagonists inhibit hepatocellular carcinoma growth through affecting mitochondrial function. Journal of molecular medicine (Berlin, Germany) 98, 179–191. [DOI] [PubMed] [Google Scholar]
- Golan M, Krivitsky A, Mausner-Fainberg K, Benhamou M, Vigiser I, Regev K, Kolb H and Karni A (2021) Increased Expression of Ephrins on Immune Cells of Patients with Relapsing Remitting Multiple Sclerosis Affects Oligodendrocyte Differentiation. International journal of molecular sciences 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttle P, Förster M, Weyers V, Küry P, Rejdak K, Hartung HP and Kremer D (2019) An unmet clinical need: roads to remyelination in MS. Neurological research and practice 1, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel M, Di Polo A, Valera PB and Braun PE (1998) Four-kilobase sequence of the mouse CNP gene directs spatial and temporal expression of lacZ in transgenic mice. Journal of neuroscience research 53, 393–404. [DOI] [PubMed] [Google Scholar]
- Green AJ, Gelfand JM, Cree BA et al. (2017) Clemastine fumarate as a remyelinating therapy for multiple sclerosis (ReBUILD): a randomised, controlled, double-blind, crossover trial. Lancet (London, England) 390, 2481–2489. [DOI] [PubMed] [Google Scholar]
- Hata E, Sasaki N, Takeda A et al. (2016) Lysophosphatidic acid receptors LPA4 and LPA6 differentially promote lymphocyte transmigration across high endothelial venules in lymph nodes. International immunology 28, 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann J, Kamtekar S, Christodoulou E et al. (2011) Structural basis of substrate discrimination and integrin binding by autotaxin. Nature structural & molecular biology 18, 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausmann J, Perrakis A and Moolenaar WH (2013) Structure-function relationships of autotaxin, a secreted lysophospholipase D. Advances in biological regulation 53, 112–117. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Takahashi M, Nishida W, Yoshida K, Ohkawa Y, Kitabatake A, Aoki J, Arai H and Sobue K (2001) Phenotypic modulation of vascular smooth muscle cells induced by unsaturated lysophosphatidic acids. Circulation research 89, 251–258. [DOI] [PubMed] [Google Scholar]
- Hecht JH, Weiner JA, Post SR and Chun J (1996) Ventricular zone gene-1 (vzg-1) encodes a lysophosphatidic acid receptor expressed in neurogenic regions of the developing cerebral cortex. The Journal of cell biology 135, 1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr DR, Chew WS, Satish RL and Ong WY (2020) Pleotropic Roles of Autotaxin in the Nervous System Present Opportunities for the Development of Novel Therapeutics for Neurological Diseases. Molecular neurobiology 57, 372–392. [DOI] [PubMed] [Google Scholar]
- Houben AJ, van Wijk XM, van Meeteren LA et al. (2013) The polybasic insertion in autotaxin alpha confers specific binding to heparin and cell surface heparan sulfate proteoglycans. The Journal of biological chemistry 288, 510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Arima N, Ishiguro J, Prestwich GD, Arai H and Aoki J (2011) LPA-producing enzyme PA-PLA(1)alpha regulates hair follicle development by modulating EGFR signalling. The EMBO journal 30, 4248–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Ishiguro J, Kitamura H et al. (2012) TGFα shedding assay: an accurate and versatile method for detecting GPCR activation. Nature methods 9, 1021–1029. [DOI] [PubMed] [Google Scholar]
- Ishii S, Hirane M, Fukushima K, Tomimatsu A, Fukushima N and Tsujiuchi T (2015) Diverse effects of LPA4, LPA5 and LPA6 on the activation of tumor progression in pancreatic cancer cells. Biochemical and biophysical research communications 461, 59–64. [DOI] [PubMed] [Google Scholar]
- Jäkel S, Agirre E, Mendanha Falcão A et al. (2019) Altered human oligodendrocyte heterogeneity in multiple sclerosis. Nature 566, 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries MA, McLane LE, Khandker L, Mather ML, Evangelou AV, Kantak D, Bourne JN, Macklin WB and Wood TL (2021) mTOR Signaling Regulates Metabolic Function in Oligodendrocyte Precursor Cells and Promotes Efficient Brain Remyelination in the Cuprizone Model. The Journal of neuroscience : the official journal of the Society for Neuroscience 41, 8321–8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries MA, Urbanek K, Torres L, Wendell SG, Rubio ME and Fyffe-Maricich SL (2016) ERK1/2 Activation in Preexisting Oligodendrocytes of Adult Mice Drives New Myelin Synthesis and Enhanced CNS Function. The Journal of neuroscience : the official journal of the Society for Neuroscience 36, 9186–9200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G, Inoue A, Aoki J and Prestwich GD (2013) Phosphorothioate analogs of sn-2 radyl lysophosphatidic acid (LPA): metabolically stabilized LPA receptor agonists. Bioorganic & medicinal chemistry letters 23, 1865–1869. [DOI] [PubMed] [Google Scholar]
- Jung SH, Kim S, Chung AY, Kim HT, So JH, Ryu J, Park HC and Kim CH (2010) Visualization of myelination in GFP-transgenic zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists 239, 592–597. [DOI] [PubMed] [Google Scholar]
- Kanda H, Newton R, Klein R, Morita Y, Gunn MD and Rosen SD (2008) Autotaxin, an ectoenzyme that produces lysophosphatidic acid, promotes the entry of lymphocytes into secondary lymphoid organs. Nature immunology 9, 415–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano K, Aoki J and Hla T (2021) Lysophospholipid Mediators in Health and Disease. Annual review of pathology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano K, Matsumoto H, Inoue A, Yukiura H, Kanai M, Chun J, Ishii S, Shimizu T and Aoki J (2019) Molecular mechanism of lysophosphatidic acid-induced hypertensive response. Scientific reports 9, 2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson U and Schultz RL (1965) Fixation of the Central Nervous System from Electron Microscopy by Aldehyde Perfusion. I. Preservation with Aldehyde Perfusates Versus Direct Perfusion with Osmium Tetroxide with Special Reference to Membranes and the Extracellular Space. Journal of ultrastructure research 12, 160–186. [DOI] [PubMed] [Google Scholar]
- Kihara Y, Maceyka M, Spiegel S and Chun J (2014) Lysophospholipid receptor nomenclature review: IUPHAR Review 8. British journal of pharmacology 171, 3575–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara Y, Mizuno H and Chun J (2015) Lysophospholipid receptors in drug discovery. Experimental cell research 333, 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B and Schilling TF (1995) Stages of embryonic development of the zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists 203, 253–310. [DOI] [PubMed] [Google Scholar]
- Kirby L, Jin J, Cardona JG et al. (2019) Oligodendrocyte precursor cells present antigen and are cytotoxic targets in inflammatory demyelination. Nature communications 10, 3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeze WK, Sassano MF, Huang XP, Lansu K, McCorvy JD, Giguere PM, Sciaky N and Roth BL (2015) PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nature structural & molecular biology 22, 362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlmann T, Miron V, Cui Q, Wegner C, Antel J and Bruck W (2008) Differentiation block of oligodendroglial progenitor cells as a cause for remyelination failure in chronic multiple sclerosis. Brain : a journal of neurology 131, 1749–1758. [DOI] [PubMed] [Google Scholar]
- Kurihara T, Monoh K, Takahashi Y, Goto K and Kondo H (1992) 2’,3’-Cyclic-nucleotide 3’-phosphodiesterase. Complementary DNA and gene cloning for mouse enzyme and in situ hybridization of the messenger RNA in mouse brain. Advances in second messenger and phosphoprotein research 25, 101–110. [PubMed] [Google Scholar]
- Lafrenaye AD and Fuss B (2011) Focal adhesion kinase can play unique and opposing roles in regulating the morphology of differentiating oligodendrocytes. Journal of neurochemistry 115, 269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics (Oxford, England) 23, 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lecca D, Raffaele S, Abbracchio MP and Fumagalli M (2020) Regulation and signaling of the GPR17 receptor in oligodendroglial cells. Glia 68, 1957–1967. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen T, Miao X, Yi X, Wang X, Zhao H, Lee SM and Zheng Y (2017) Zebrafish: A promising in vivo model for assessing the delivery of natural products, fluorescence dyes and drugs across the blood-brain barrier. Pharmacological research 125, 246–257. [DOI] [PubMed] [Google Scholar]
- Liu J, Moyon S, Hernandez M and Casaccia P (2016) Epigenetic control of oligodendrocyte development: adding new players to old keepers. Current opinion in neurobiology 39, 133–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Hopkins AM and Hou J (2021) The development of modulators for lysophosphatidic acid receptors: A comprehensive review. Bioorganic chemistry 117, 105386. [DOI] [PubMed] [Google Scholar]
- Livak KJ and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif.) 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD and Rowitch DH (2000) Sonic hedgehog--regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron 25, 317–329. [DOI] [PubMed] [Google Scholar]
- Lubetzki C, Zalc B, Williams A, Stadelmann C and Stankoff B (2020) Remyelination in multiple sclerosis: from basic science to clinical translation. The Lancet. Neurology 19, 678–688. [DOI] [PubMed] [Google Scholar]
- Lucchinetti C, Bruck W, Parisi J, Scheithauer B, Rodriguez M and Lassmann H (2000) Heterogeneity of multiple sclerosis lesions: implications for the pathogenesis of demyelination. Annals of neurology 47, 707–717. [DOI] [PubMed] [Google Scholar]
- Lyons DA and Talbot WS (2014) Glial cell development and function in zebrafish. Cold Spring Harbor perspectives in biology 7, a020586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marisca R, Hoche T, Agirre E, Hoodless LJ, Barkey W, Auer F, Castelo-Branco G and Czopka T (2020) Functionally distinct subgroups of oligodendrocyte precursor cells integrate neural activity and execute myelin formation. Nature neuroscience 23, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques S, Zeisel A, Codeluppi S et al. (2016) Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science (New York, N.Y.) 352, 1326–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Lozada Z, Waggener CT, Kim K, Zou S, Knapp PE, Hayashi Y, Ortega A and Fuss B (2014) Activation of sodium-dependent glutamate transporters regulates the morphological aspects of oligodendrocyte maturation via signaling through calcium/calmodulin-dependent kinase IIbeta’s actin-binding/-stabilizing domain. Glia 62, 1543–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masago K, Kihara Y, Yanagida K, Hamano F, Nakagawa S, Niwa M and Shimizu T (2018) Lysophosphatidic acid receptor, LPA6, regulates endothelial blood-brain barrier function: Implication for hepatic encephalopathy. Biochemical and biophysical research communications 501, 1048–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matas-Rico E, Frijlink E, van der Haar Àvila I et al. (2021) Autotaxin impedes anti-tumor immunity by suppressing chemotaxis and tumor infiltration of CD8(+) T cells. Cell reports 37, 110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mausner-Fainberg K, Benhamou M, Golan M, Kimelman NB, Danon U, Marom E and Karni A (2021) Specific Blockade of Bone Morphogenetic Protein-2/4 Induces Oligodendrogenesis and Remyelination in Demyelinating Disorders. Neurotherapeutics : the journal of the American Society for Experimental NeuroTherapeutics 18, 1798–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa AJ, Zhou YX, Hibbits N, Vana AC and Armstrong RC (2013) FGF2 and FGFR1 signaling regulate functional recovery following cuprizone demyelination. Neuroscience letters 548, 280–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitew S, Hay CM, Peckham H, Xiao J, Koenning M and Emery B (2014) Mechanisms regulating the development of oligodendrocytes and central nervous system myelin. Neuroscience 276, 29–47. [DOI] [PubMed] [Google Scholar]
- Moolenaar WH and Perrakis A (2011) Insights into autotaxin: how to produce and present a lipid mediator. Nature reviews. Molecular cell biology 12, 674–679. [DOI] [PubMed] [Google Scholar]
- Morishige J, Touchika K, Tanaka T, Satouchi K, Fukuzawa K and Tokumura A (2007) Production of bioactive lysophosphatidic acid by lysophospholipase D in hen egg white. Biochimica et biophysica acta 1771, 491–499. [DOI] [PubMed] [Google Scholar]
- Mormann W and Leukel G (1988) A simple and versatile synthesis of trimethylsiloxy-substituted isocyanates Synthesis 12, 990–992. [Google Scholar]
- Mozafari S, Starost L, Manot-Saillet B et al. (2020) Multiple sclerosis iPS-derived oligodendroglia conserve their properties to functionally interact with axons and glia in vivo. Science advances 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzel EJ, Schaefer K, Obirei B et al. (2012) Claudin k is specifically expressed in cells that form myelin during development of the nervous system and regeneration of the optic nerve in adult zebrafish. Glia 60, 253–270. [DOI] [PubMed] [Google Scholar]
- Nishimasu H, Okudaira S, Hama K et al. (2011) Crystal structure of autotaxin and insight into GPCR activation by lipid mediators. Nature structural & molecular biology 18, 205–212. [DOI] [PubMed] [Google Scholar]
- Nogaroli L, Yuelling LM, Dennis J, Gorse K, Payne SG and Fuss B (2009) Lysophosphatidic acid can support the formation of membranous structures and an increase in MBP mRNA levels in differentiating oligodendrocytes. Neurochemical research 34, 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi K, Ishii S and Shimizu T (2003) Identification of p2y9/GPR23 as a novel G protein-coupled receptor for lysophosphatidic acid, structurally distant from the Edg family. The Journal of biological chemistry 278, 25600–25606. [DOI] [PubMed] [Google Scholar]
- Okasato R, Kano K, Kise R, Inoue A, Fukuhara S and Aoki J (2021) An ATX-LPA(6)-Gα(13)-ROCK axis shapes and maintains caudal vein plexus in zebrafish. iScience 24, 103254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okudaira M, Inoue A, Shuto A, Nakanaga K, Kano K, Makide K, Saigusa D, Tomioka Y and Aoki J (2014) Separation and quantification of 2-acyl-1-lysophospholipids and 1-acyl-2-lysophospholipids in biological samples by LC-MS/MS. Journal of lipid research 55, 2178–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okudaira S, Yukiura H and Aoki J (2010) Biological roles of lysophosphatidic acid signaling through its production by autotaxin. Biochimie 92, 698–706. [DOI] [PubMed] [Google Scholar]
- Pasternack SM, von Kugelgen I, Al Aboud K et al. (2008) G protein-coupled receptor P2Y5 and its ligand LPA are involved in maintenance of human hair growth. Nature genetics 40, 329–334. [DOI] [PubMed] [Google Scholar]
- Patrikios P, Stadelmann C, Kutzelnigg A et al. (2006) Remyelination is extensive in a subset of multiple sclerosis patients. Brain : a journal of neurology 129, 3165–3172. [DOI] [PubMed] [Google Scholar]
- Petersen MA, Tognatta R, Meyer-Franke A et al. (2021) BMP receptor blockade overcomes extrinsic inhibition of remyelination and restores neurovascular homeostasis. Brain : a journal of neurology 144, 2291–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorevc M and Faber K (2002) Enantioselective stereoinversion of sec-alkyl sulfates by an alkylsulfatase from Rhodococcus ruber DSM 44541. Tetrahedron: Asymmetry 13, 1435–1441. [DOI] [PubMed] [Google Scholar]
- Preston MA, Finseth LT, Bourne JN and Macklin WB (2019) A novel myelin protein zero transgenic zebrafish designed for rapid readout of in vivo myelination. Glia 67, 650–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston MA and Macklin WB (2015) Zebrafish as a model to investigate CNS myelination. Glia 63, 177–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajendran R, Böttiger G, Stadelmann C, Karnati S and Berghoff M (2021) FGF/FGFR Pathways in Multiple Sclerosis and in Its Disease Models. Cells 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza SI, Muhammad D, Jan A, Ali RH, Hassan M, Ahmad W and Rashid S (2014) In silico analysis of missense mutations in LPAR6 reveals abnormal phospholipid signaling pathway leading to hypotrichosis. PloS one 9, e104756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggeri A, Schepers M, Tiane A, Rombaut B, van Veggel L, Hellings N, Prickaerts J, Pittaluga A and Vanmierlo T (2020) Sphingosine-1-Phosphate Receptor Modulators and Oligodendroglial Cells: Beyond Immunomodulation. International journal of molecular sciences 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombough P (2002) Gills are needed for ionoregulation before they are needed for O(2) uptake in developing zebrafish, Danio rerio. J Exp Biol 205, 1787–1794. [DOI] [PubMed] [Google Scholar]
- Saraswat D, Shayya HJ, Polanco JJ et al. (2021) Overcoming the inhibitory microenvironment surrounding oligodendrocyte progenitor cells following experimental demyelination. Nature communications 12, 1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer L, Velmeshev D, Holmqvist S et al. (2019) Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature 573, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer J, Becker T, Schachner M, Nave KA and Werner H (2006) Evolution of myelin proteolipid proteins: gene duplication in teleosts and expression pattern divergence. Molecular and cellular neurosciences 31, 161–177. [DOI] [PubMed] [Google Scholar]
- Shen MY and Sali A (2006) Statistical potential for assessment and prediction of protein structures. Protein science : a publication of the Protein Society 15, 2507–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura Y, Wajid M, Ishii Y, Shapiro L, Petukhova L, Gordon D and Christiano AM (2008) Disruption of P2RY5, an orphan G protein-coupled receptor, underlies autosomal recessive woolly hair. Nature genetics 40, 335–339. [DOI] [PubMed] [Google Scholar]
- Siems SB, Jahn O, Hoodless LJ, Jung RB, Hesse D, Möbius W, Czopka T and Werner HB (2021) Proteome Profile of Myelin in the Zebrafish Brain. Frontiers in cell and developmental biology 9, 640169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner DD and Lane TE (2020) CXCR2 Signaling and Remyelination in Preclinical Models of Demyelination. DNA and cell biology 39, 3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skokal RR and Rohlf FJ (1995) Biometry: the principle and practice in biological research. W. H. Freeman and Company, New York. [Google Scholar]
- Smets I, Goris A, Vandebergh M, Demeestere J, Sunaert S, Dupont P and Dubois B (2021) Quantitative MRI phenotypes capture biological heterogeneity in multiple sclerosis patients. Scientific reports 11, 1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sock E, Leger H, Kuhlbrodt K, Schreiber J, Enderich J, Richter-Landsberg C and Wegner M (1997) Expression of Krox proteins during differentiation of the O-2A progenitor cell line CG-4. Journal of neurochemistry 68, 1911–1919. [DOI] [PubMed] [Google Scholar]
- Sock E and Wegner M (2021) Using the lineage determinants Olig2 and Sox10 to explore transcriptional regulation of oligodendrocyte development. Developmental neurobiology 81, 892–901. [DOI] [PubMed] [Google Scholar]
- Solís KH, Romero-Ávila MT, Guzmán-Silva A and García-Sáinz JA (2021) The LPA(3) Receptor: Regulation and Activation of Signaling Pathways. International journal of molecular sciences 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer SO, Sitnikov S, Kamen Y, Evans KA, Kronenberg-Versteeg D, Dietmann S, de Faria O Jr., Agathou S and Karadottir RT (2019) Oligodendrocyte Progenitor Cells Become Regionally Diverse and Heterogeneous with Age. Neuron 101, 459–471 e455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankoff B, Barron S, Allard J et al. (2002) Oligodendroglial expression of Edg-2 receptor: developmental analysis and pharmacological responses to lysophosphatidic acid. Molecular and cellular neurosciences 20, 415–428. [DOI] [PubMed] [Google Scholar]
- Starost L, Lindner M, Herold M et al. (2020) Extrinsic immune cell-derived, but not intrinsic oligodendroglial factors contribute to oligodendroglial differentiation block in multiple sclerosis. Acta neuropathologica 140, 715–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard NC and Chun J (2015) Promising pharmacological directions in the world of lysophosphatidic Acid signaling. Biomol Ther (Seoul) 23, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturrock RR (1980) Myelination of the mouse corpus callosum. Neuropathology and applied neurobiology 6, 415–420. [DOI] [PubMed] [Google Scholar]
- Sturrock RR (1983) Identification of mitotic oligodendrocytes in semithin sections of the developing mouse corpus callosum and hippocampal commissure. Journal of anatomy 137 (Pt 1), 47–55. [PMC free article] [PubMed] [Google Scholar]
- Suckau O, Gross I, Schrotter S et al. (2019) LPA1 , LPA2 , LPA4 , and LPA6 receptor expression during mouse brain development. Developmental dynamics : an official publication of the American Association of Anatomists 248, 375–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiss VA, Nguyen T, Dugas J, Ibrahim A, Barres B, Androulakis IP and Casaccia P (2011) Identification of a gene regulatory network necessary for the initiation of oligodendrocyte differentiation. PloS one 6, e18088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada N and Appel B (2010) Identification of genes expressed by zebrafish oligodendrocytes using a differential microarray screen. Developmental dynamics : an official publication of the American Association of Anatomists 239, 2041–2047. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Fukushima K, Onishi Y, Inui K, Node Y, Fukushima N, Honoki K and Tsujiuchi T (2017) Lysophosphatidic acid (LPA) signaling via LPA4 and LPA6 negatively regulates cell motile activities of colon cancer cells. Biochemical and biophysical research communications 483, 652–657. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Okudaira S, Kishi Y et al. (2006) Autotaxin stabilizes blood vessels and is required for embryonic vasculature by producing lysophosphatidic acid. The Journal of biological chemistry 281, 25822–25830. [DOI] [PubMed] [Google Scholar]
- Taniguchi R, Inoue A, Sayama M et al. (2017) Structural insights into ligand recognition by the lysophosphatidic acid receptor LPA(6). Nature 548, 356–360. [DOI] [PubMed] [Google Scholar]
- Thomason EJ, Suárez-Pozos E, Afshari FS, Rosenberg PA, Dupree JL and Fuss B (2022) Deletion of the Sodium-Dependent Glutamate Transporter GLT-1 in Maturing Oligodendrocytes Attenuates Myelination of Callosal Axons During a Postnatal Phase of Central Nervous System Development. Frontiers in cellular neuroscience 16, 905299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thümmler K, Rom E, Zeis T et al. (2019) Polarizing receptor activation dissociates fibroblast growth factor 2 mediated inhibition of myelination from its neuroprotective potential. Acta neuropathologica communications 7, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumura A, Majima E, Kariya Y, Tominaga K, Kogure K, Yasuda K and Fukuzawa K (2002) Identification of human plasma lysophospholipase D, a lysophosphatidic acid-producing enzyme, as autotaxin, a multifunctional phosphodiesterase. The Journal of biological chemistry 277, 39436–39442. [DOI] [PubMed] [Google Scholar]
- Tsai HH, Niu J, Munji R et al. (2016) Oligodendrocyte precursors migrate along vasculature in the developing nervous system. Science (New York, N.Y.) 351, 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu-Goto M, Kishi Y, Taira A et al. (2002) Autotaxin has lysophospholipase D activity leading to tumor cell growth and motility by lysophosphatidic acid production. The Journal of cell biology 158, 227–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urs NM, Jones KT, Salo PD, Severin JE, Trejo J and Radhakrishna H (2005) A requirement for membrane cholesterol in the beta-arrestin- and clathrin-dependent endocytosis of LPA1 lysophosphatidic acid receptors. Journal of cell science 118, 5291–5304. [DOI] [PubMed] [Google Scholar]
- van Meeteren LA, Ruurs P, Stortelers C et al. (2006) Autotaxin, a secreted lysophospholipase D, is essential for blood vessel formation during development. Molecular and cellular biology 26, 5015–5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggener CT, Dupree JL, Elgersma Y and Fuss B (2013) CaMKIIbeta regulates oligodendrocyte maturation and CNS myelination. The Journal of neuroscience : the official journal of the Society for Neuroscience 33, 10453–10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, He X, Meng H, Li Y, Dmitriev P, Tian F, Page JC, Lu QR and He Z (2020) Robust Myelination of Regenerated Axons Induced by Combined Manipulations of GPR17 and Microglia. Neuron 108, 876–886.e874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner M (2001) Expression of transcription factors during oligodendroglial development. Microscopy research and technique 52, 746–752. [DOI] [PubMed] [Google Scholar]
- Weiner JA, Hecht JH and Chun J (1998) Lysophosphatidic acid receptor gene vzg-1/lpA1/edg-2 is expressed by mature oligodendrocytes during myelination in the postnatal murine brain. The Journal of comparative neurology 398, 587–598. [PubMed] [Google Scholar]
- Welliver RR, Polanco JJ, Seidman RA et al. (2018) Muscarinic Receptor M(3)R Signaling Prevents Efficient Remyelination by Human and Mouse Oligodendrocyte Progenitor Cells. The Journal of neuroscience : the official journal of the Society for Neuroscience 38, 6921–6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler NA and Fuss B (2016) Extracellular cues influencing oligodendrocyte differentiation and (re)myelination. Experimental neurology 283, 512–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler NA, Fuss B, Knapp PE and Zou S (2016) HIV-1 Tat Inhibits Autotaxin Lysophospholipase D Activity and Modulates Oligodendrocyte Differentiation. ASN neuro 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler NA, Lister JA and Fuss B (2015) The Autotaxin-Lysophosphatidic Acid Axis Modulates Histone Acetylation and Gene Expression during Oligodendrocyte Differentiation. The Journal of neuroscience : the official journal of the Society for Neuroscience 35, 11399–11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida K and Ishii S (2011) Non-Edg family LPA receptors: the cutting edge of LPA research. Journal of biochemistry 150, 223–232. [DOI] [PubMed] [Google Scholar]
- Yanagida K, Kurikawa Y, Shimizu T and Ishii S (2013) Current progress in non-Edg family LPA receptor research. Biochimica et biophysica acta 1831, 33–41. [DOI] [PubMed] [Google Scholar]
- Yanagida K, Masago K, Nakanishi H, Kihara Y, Hamano F, Tajima Y, Taguchi R, Shimizu T and Ishii S (2009) Identification and characterization of a novel lysophosphatidic acid receptor, p2y5/LPA6. The Journal of biological chemistry 284, 17731–17741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagida K and Valentine WJ (2020) Druggable Lysophospholipid Signaling Pathways. Advances in experimental medicine and biology 1274, 137–176. [DOI] [PubMed] [Google Scholar]
- Yasuda D, Kobayashi D, Akahoshi N, Ohto-Nakanishi T, Yoshioka K, Takuwa Y, Mizuno S, Takahashi S and Ishii S (2019) Lysophosphatidic acid-induced YAP/TAZ activation promotes developmental angiogenesis by repressing Notch ligand Dll4. The Journal of clinical investigation 129, 4332–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S and Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Nishida W, Hayashi K, Ohkawa Y, Ogawa A, Aoki J, Arai H and Sobue K (2003) Vascular remodeling induced by naturally occurring unsaturated lysophosphatidic acid in vivo. Circulation 108, 1746–1752. [DOI] [PubMed] [Google Scholar]
- Yoshida M and Macklin WB (2005) Oligodendrocyte development and myelination in GFP-transgenic zebrafish. Journal of neuroscience research 81, 1–8. [DOI] [PubMed] [Google Scholar]
- Young KM, Psachoulia K, Tripathi RB, Dunn SJ, Cossell L, Attwell D, Tohyama K and Richardson WD (2013) Oligodendrocyte dynamics in the healthy adult CNS: evidence for myelin remodeling. Neuron 77, 873–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Lariosa-Willingham KD, Lin FF, Webb M and Rao TS (2004) Characterization of lysophosphatidic acid and sphingosine-1-phosphate-mediated signal transduction in rat cortical oligodendrocytes. Glia 45, 17–27. [DOI] [PubMed] [Google Scholar]
- Yuelling LW, Waggener CT, Afshari FS, Lister JA and Fuss B (2012) Autotaxin/ENPP2 regulates oligodendrocyte differentiation in vivo in the developing zebrafish hindbrain. Glia 60, 1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen TJ, Silbereis JC, Griveau A, Chang SM, Daneman R, Fancy SPJ, Zahed H, Maltepe E and Rowitch DH (2014) Oligodendrocyte-encoded HIF function couples postnatal myelination and white matter angiogenesis. Cell 158, 383–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung YC, Stoddard NC and Chun J (2014) LPA receptor signaling: pharmacology, physiology, and pathophysiology. Journal of lipid research 55, 1192–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung YC, Stoddard NC, Mirendil H and Chun J (2015) Lysophosphatidic Acid signaling in the nervous system. Neuron 85, 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalc B (2016) The acquisition of myelin: An evolutionary perspective. Brain research 1641, 4–10. [DOI] [PubMed] [Google Scholar]
- Zalc B, Goujet D and Colman D (2008) The origin of the myelination program in vertebrates. Current biology : CB 18, R511–512. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA et al. (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience 34, 11929–11947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Stephens T and Zhao Y (2021) Molecular Regulation of Lysophosphatidic Acid Receptor 1 Maturation and Desensitization. Cell biochemistry and biophysics 79, 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Wang S and Anderson DJ (2000) Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron 25, 331–343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


