Abstract
G-quadruplexes (G4s) are non-canonical structures formed in guanine-rich sequences through stacked guanine tetrads by Hoogsteen hydrogen bonding. Several studies have demonstrated the existence of G4s in the genome of various organisms, including humans, and have proposed that G4s play a regulatory role in various cellular functions. However, little is known regarding the dissemination of G4s in mitochondria. In this review, we report the observation that the number of potential G4-forming sequences in the mitochondrial genome increases with the evolutionary complexity of different species, suggesting that G4s play a beneficial role in higher-order organisms. Here we discuss the possible function of G4s in mitochondrial DNA and long noncoding RNA and their role in various biological processes.
Keywords: G-quadruplexes, mitochondria, evolution, long noncoding RNA
G-quadruplex and its Biological role
Nucleic acids are known to form structures other than the Watson–Crick canonical double-helical structure. A G-quadruplex (G4) is a stable secondary structure of nucleic acid that can arise from single-stranded G-rich DNA and RNA sequences (Figure 1).[1] The formation of G4 tetrameric structures was reported in 1962[2] after observation of the self-assembly of guanylic acid,[3] decades before the proposed DNA structure by Watson and Crick. G4s are formed by Hoogsteen hydrogen bonding between four guanines to form a planar G-tetrad and exhibit extremely high stability under physiological conditions in the presence of monovalent metal cations (such as K+, Na+, and Li+) and resistance to degradation by nucleases. G4s can fold into various topologies based on the orientation of the G-tract and the interconnecting loops and these polymorphic structures have an important influence on G4-related biological functions.[4] Computational tools are available to identify potential G4-forming sequences, based on algorithms;[5] and G4-specific probes, antibodies[6], and G4 sequencing[7] are developed to study G4s in cells.
Figure 1.
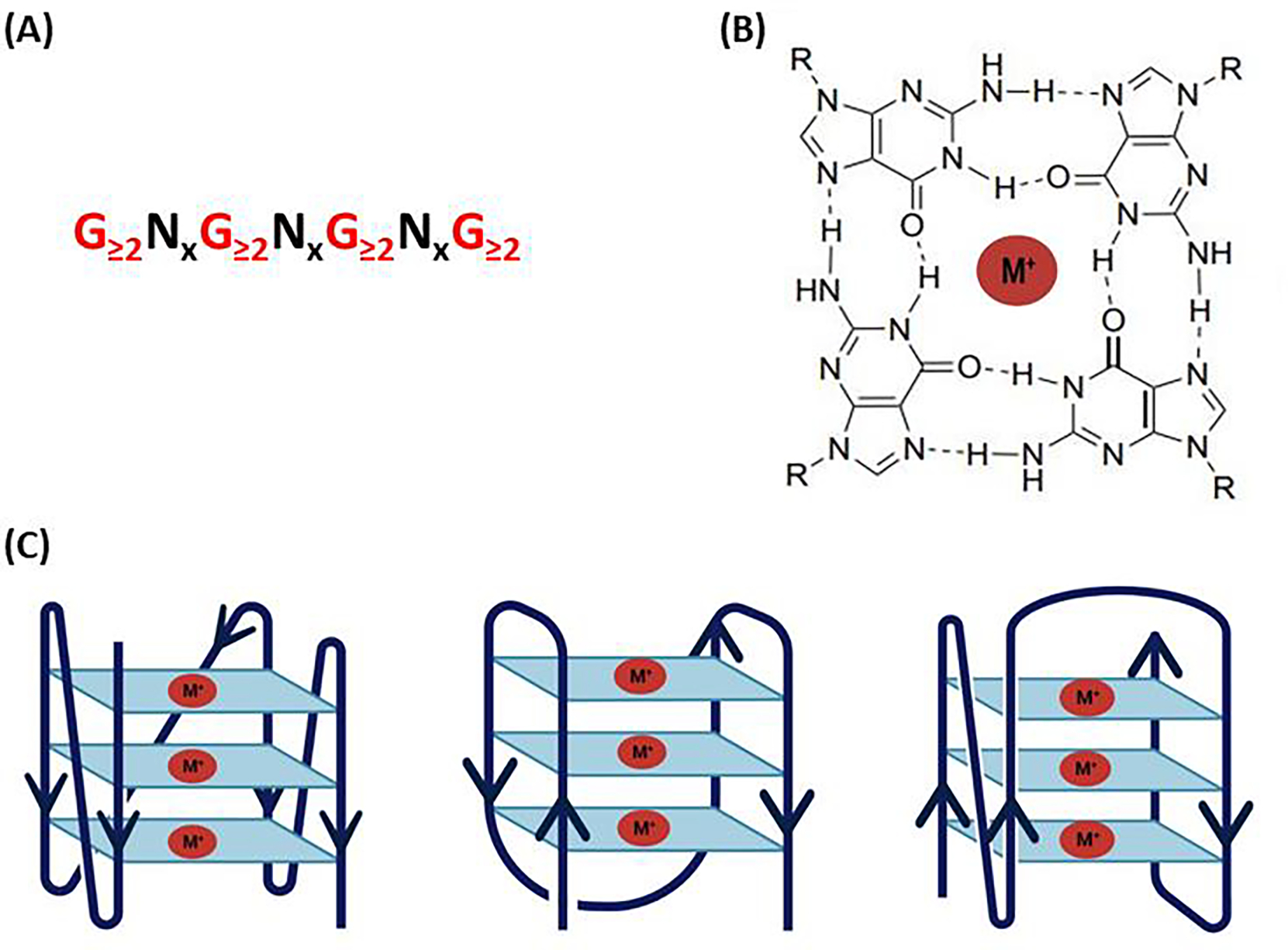
G-quadruplex structure and topologies (A) The nucleotide sequence of the G-4 motif, where N denotes the loop sequences. (B) A planar guanine tetrad formed by Hoogsteen bonds and stabilized by metal cation M+. (C) Schematic representation of some topologies of G4.
The ability of genomic DNA to form G4s was first reported for the guanine-rich sequences from the immunoglobulin switch region[8] and subsequently, the crystal structure of G4 in the human telomeric DNA sequence was elucidated.[9] The formation of G4s in the guanine-rich telomere repeat plays a role in genome stability and inhibits telomerase function. Since then, numerous studies have reported G4 formation mostly in the promoter region of various genes, such as c-MYC,[10] VEGF,[11] and HIF1α.[12] G4-forming sequences present in the promoter region were proposed to function as transcription repressors by stabilizing the G4 structures, using ligands such as TMPyP4, BRACO19, and pyridostatin (PDS).[13] Using computational approaches, several G4-forming sites were also found to be enriched in the promoter region of various oncogenes in the genome.[14,15]These findings raised the possibility of developing ligands to target G4s for various therapeutic interventions. However, G4 formation in promoters resulting in transcription suppression within the chromatin is not fully understood and a study by genome-wide G4 mapping identified that G4 ligand PDS elicited DNA damage and caused gene downregulation.[16] Also, contrary to the finding that G4s play a vital role in transcription suppression in the promoter region, other researchers have reported a role as an enhancer of various genes[17] by mechanisms such as guanine oxidation[18] and transcription factor binding.[19] Further accumulating scientific data identifies G4 has the potential to alter gene expression at many different levels and perturb the chromatin architecture and act as a regulatory element in epigenetics. (see review by Robinson and colleagues[20]) G4 further can affect the stability of the genome and manipulate the DNA damage response (DDR) as a valuable anticancer strategy. (see review by David and colleagues[21])
In addition to DNA, G4s can also form in RNA and have been implicated in a wide range of biological processes. Compared to their DNA counterpart, RNA G4s are relatively stable, unconstrained, and can readily form secondary structures. Although studies of G4s are mainly focused on DNA, many studies also report the potential of RNA G4s and their biological significance. (see review by Kwon and colleagues[22]) Computational analysis identified nearly 3,000 putative RNA G4-forming sites in the 5′-UTR of mRNA in the human genome revealing their potential in cellular function.[23] Recent studies employed a G4-specific probe to perform BioTASQ G4RP-seq and identify a large number of G4-forming RNA, demonstrating the existence of RNA G4 in in-vivo conditions.[24] The formation of G4 in the 5′-UTR of mRNA inhibited the translation of several genes, such as NRAS[23] and BCL-2,[25] and in cap-independent translation, G4 conferred IRES-mediated translation initiation in hVEGF[26] and FGF2.[27] Furthermore, RNA G4s play multiple roles in cells, such as splicing regulation, mRNA localization, and miRNA maturation; and G4-binding proteins were found to play an essential role in cellular processes. Hence, G4 formation and its functional roles have gained considerable attention, with efforts to explore its role in biological processes beyond a simple roadblock of the transcription process.
Many studies of G4s are focused solely on the nuclear genome; but little is known about G4s and their functional role in mitochondria, despite an abundance of guanine and a favorable G4-forming environment in mitochondria. A review by Falabella and colleagues [28] elaborates on the possible regulatory function of G4 in mitochondria but it majorly focused on DNA G4 and its role in transcription, translation, and genome instability. While in this review we highlight mitoG4 from an evolutionary perspective reporting an increase in potential G4-forming sequences from lower organisms to higher-order organisms, reflecting its gradual evolution. Furthermore, we discuss RNA G4s and their abundance in mitochondrial long noncoding RNA and their potential roles in translation, granules, and reactive oxygen species scavenging. Finally, we discuss the experimental tools available to study mitochondrial G4s and insight into their future perspectives and challenges.
Mitochondria
Mitochondria are known as the powerhouse of the cell because of their ability to generate adenosine triphosphate, which is essential for normal cellular function. They also play a vital part in various cellular functions, including calcium homeostasis, apoptosis, stem cell generation, and heme synthesis.[29] Most eukaryotic mitochondrial DNAs (mtDNAs) are double-stranded, circular molecules that are typically present at several hundred to thousands of copies per cell. mtDNA lacks histones but is usually packaged into slightly elongated DNA–protein structures known as mitochondrial nucleoids, consisting of 1–2 genome copies per nucleoid.[30] In the majority of eukaryotes, nearly 90% of mtDNA comprises coding regions, unlike nuclear DNA, and its genetic code differs slightly from that of nuclear DNA.[31] Most mammalian mtDNAs encode 37 genes, including 13 genes that form the essential subunits of the mitochondrial respiratory chain complexes, however the organism’s remaining ~77 respiratory chain subunits are encoded by the nuclear genome.[30]
Mitochondrial DNA consists of two strands that can be distinguished by their nucleotide composition and are termed heavy (H) and light (L) strands (Figure 2). Typically, mtDNA strands are separated on the basis of density using the classical biochemical technique of ultracentrifugation, producing the H strand, which has a high guanine (G) + thymidine (T) content, and the L strand, which has a low G + T content.[32] Mitochondria are proposed to have evolved from endosymbiotic bacteria, and phylogenetic analysis confirms that the lineage of mtDNA is closely related to that of alphaproteobacteria.[33,34] This has raised a number of intriguing questions for the fields of mitochondrial and evolutionary research, such as how mitochondria integrate and adapt within the host, and whether mitochondria played an important role in the transition from prokaryote to eukaryote.
Figure 2.
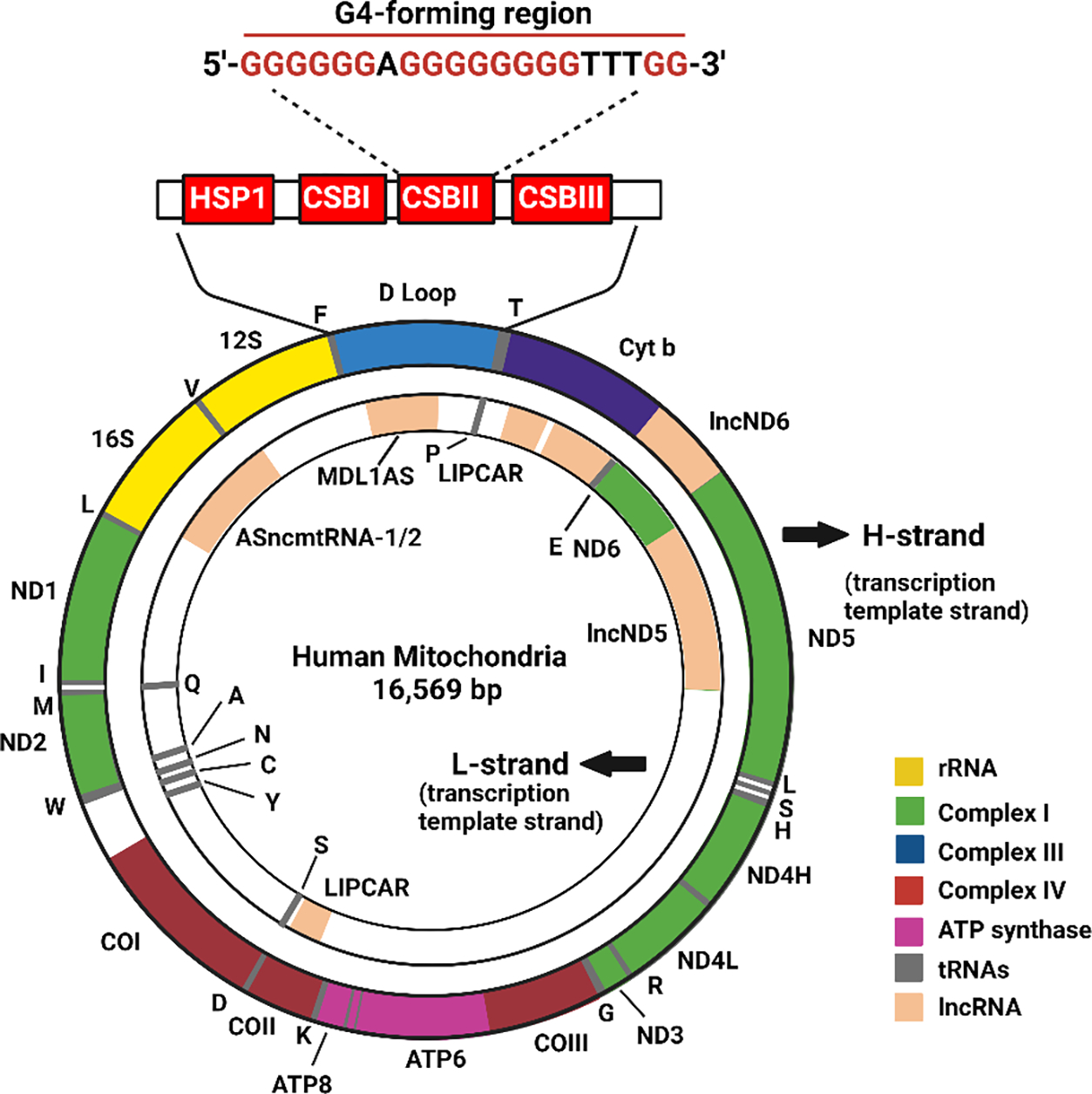
Map of human mitochondrial DNA. The highlighted region in the D loop indicates the conserved block sequence II (CSBII), G4-forming sequence, which is highly conserved in most vertebrates.
Genomic DNA G-quadruplex Evolution
Many computational analyses[5] and in-vitro[35–37] and in-vivo[38,39] techniques have been developed to identify and validate G4 sequences across the genome of various organisms, but the role of secondary DNA structures and their contribution to the evolution of each species is not well explored. However, a study employing G-quadruplex sequencing (G4-seq) mapped the DNA G4 formation in 12 species widely used as model organisms (i.e., multiple species of bacteria, plants, and eukaryotes, including human, mouse, and drosophila)[40], and found the experimentally observed G4s to be diverse in sequence composition and genomic location across different organisms. G4s were strongly depleted in the genome of bacteria and yeast, but interestingly, G4s were found to be enriched in gene promoters and transcription start sites (TSS) of mammals, which suggests a specific role of G4 in transcription. Another study by Ding and colleagues[41] that analyzed the quadruplex-forming sequences (PQSs) of microbial genomes found that enrichment of PQSs was randomly distributed in thermophilic organisms, while in the order of Deinococcales the PQS was enriched and biased in the TSS of genes. This led to the hypothesis that the different bacteria evolved G4s for different beneficial functions, such as gene regulation or thermal stability at high temperatures. Another study using comparative bioinformatics analysis of seven species of Saccharomyces revealed that G4 structures are relatively more conserved than expected by chance throughout evolution.[42] The conserved G4 motifs maintained a strong association with promoters and rDNA, but not with double-strand break sites (DSBs), supporting the theory that G4 has in vivo functions that are evolutionarily constrained. A recent study comparing the genomes of 37 evolutionary-representative species found that G4 number, length, and density generally increased with evolution.[43] This study also found G4-bearing genes particularly enriched at the TSS in the higher organisms and identified an antagonistic relationship between G4s and DNA methylation sites. It was hypothesized that the increase in G4 structures might facilitate the development of new gene regulatory mechanisms to achieve increasingly complex cellular, physiological, and behavioral activities.
Mitochondrial G-quadruplex Evolution
Mitochondrial DNA can form G4 secondary structures more easily than genomic DNA due to more favorable conditions. Most importantly, mtDNA is known to have an excessive number of guanine in the H strand compared with the L strand, violating the second parity rule, allowing more free guanine available for secondary-structure formation. This asymmetry is quantified in terms of AT skew (A − T)/(A + T) and GC skew (G − C)/(G + C), which studies suggest may be associated with replication.[44] The replication of mtDNA is highly asymmetric at the origin of replication at the H strand (OriH) in a unidirectional manner, remaining single-stranded until the synthesis of L strand replication starts 11 kb downstream.[45] Mitochondrial replication is very slow, taking about 2 hours; during this time, the single-stranded G-rich H strand has a very high opportunity to form G4 structures.[46] Also, mitochondria maintain a K+ concentration of 150–180 mM, which is very favorable for forming G4 structures.[47]
To understand the mitochondria G4 from an evolutionary perspective we studied mtDNA of 16 different biological model organisms belonging to various families and mapped the G4-forming sequences using QGRS mapper.[48] The parameters for G4-forming sequences were set to a maximum length of 30 and a minimum of two G4 groups. Most mitochondrial genomes range between 14 and 17 kb in size, although plasmodium mtDNA was a relatively small exception at 6 kb. Interestingly, we found an increase in G4-forming motifs in higher-order vertebrates compared with primitive eukaryotes, indicating a gradual evolution of G4s. Figure 3 illustrates the phylogeny tree of the mitochondrial genomes studied with their respective size, GC content, and the number of G4-forming sequences in the H and L strands. The analyses revealed that as species evolved, the G4 motif density increased, although the mitochondrial genome size remained largely the same, suggesting no association with genome size. The circos plot in Figure 4a illustrates the evolutionarily increasing GC content and high GC skew of the mtDNA, demonstrating a correlation between these characteristics and higher-order organisms. The density of G4 motifs in these species indicated an even distribution within the mitochondrial genome, but the motifs were clustered densely in the higher taxonomic species with a stepwise reduction to lower organisms. To test the influence of GC content on G4 formation we plotted the GC content of each species relative to their respective G4 motifs (Figure 4c) and found a high correlation between the PQS and GC content (R=0.8557 by Pearson, and S=0.8939 by Spearman).
Figure 3.
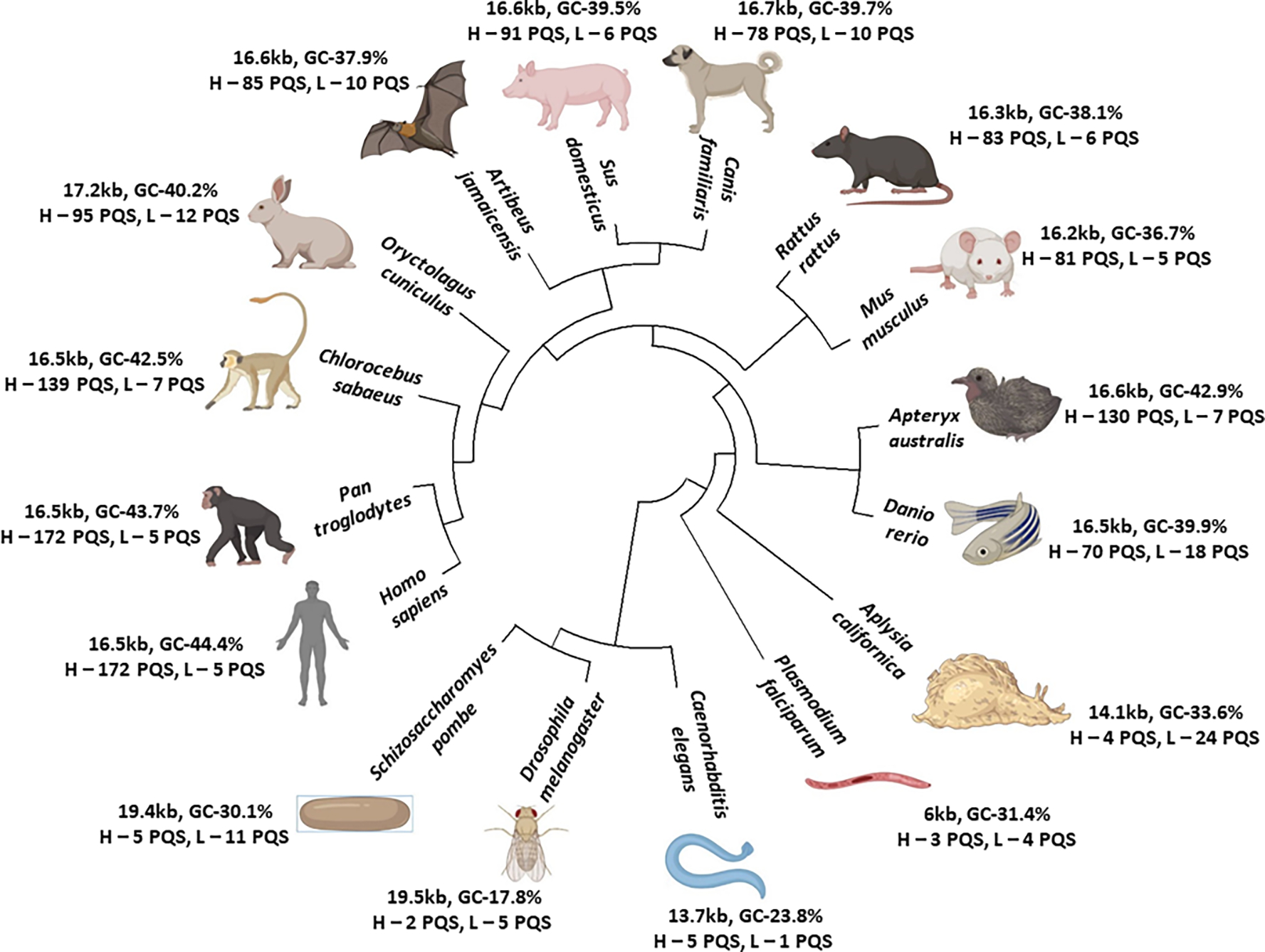
The phylogeny tree of 16 species together with their mitochondrial genome size. The GC content and the number of potential G4-forming sequences (PQS) in the heavy (H) and light (L) strands is denoted.
Figure 4.
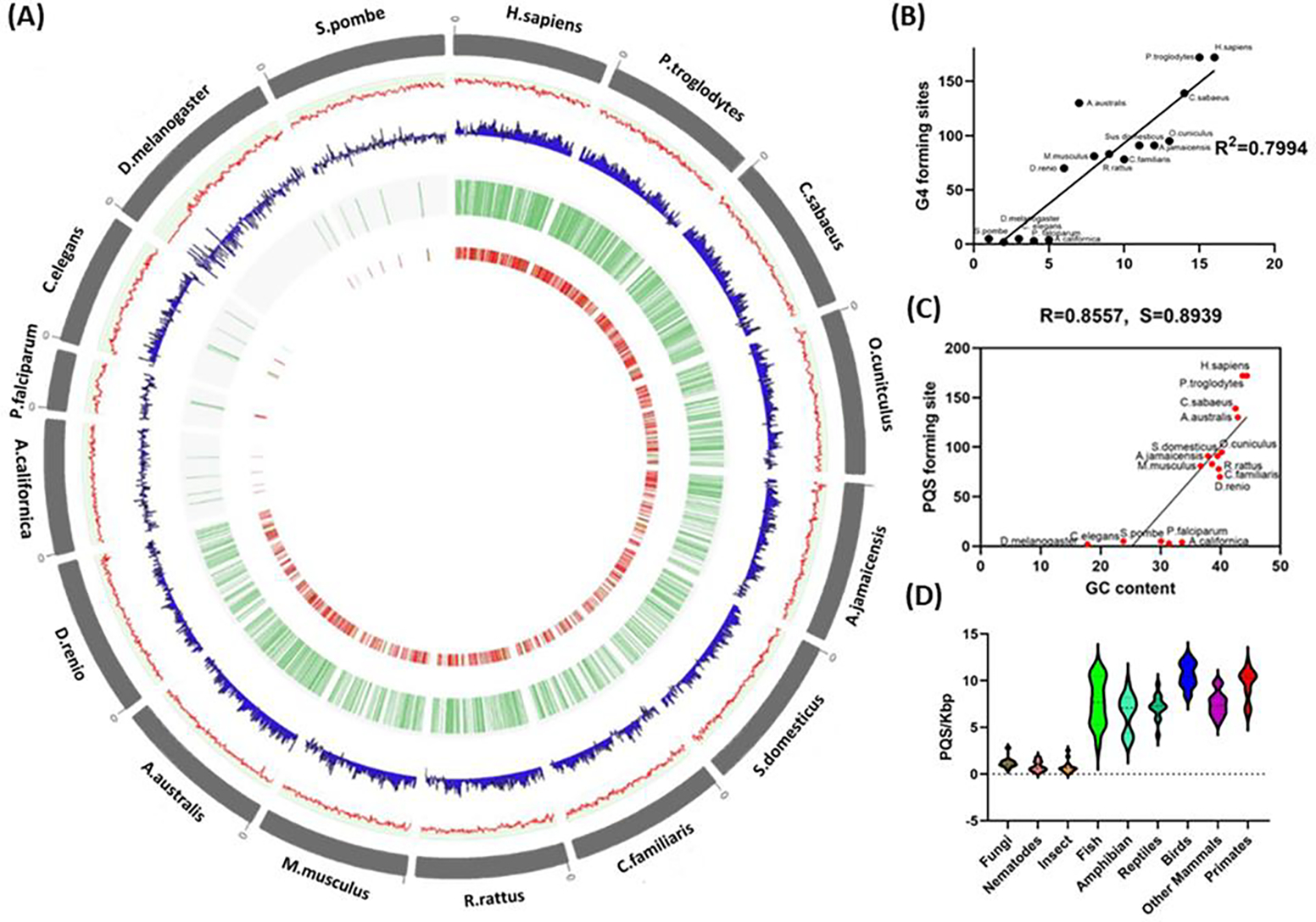
(A) Circos plot of the representative mitochondrial genomes. The outer circle represents the organism and its mtDNA in gray followed by the GC content in the form of a line plot in red, further followed by GC skew in blue. The next circle indicates the localization of the G4 plotted in the form of tiles in green, and the inner circle represents the score of the G4-forming regions plotted in the form of a heatmap. b) The GC content of the mitochondrial genomes of the 16 species. (B) Potential G4-forming motifs in the mitochondrial heavy strand of the 16 species. The x-axis represents the ranking of organisms based on their complexity by phylogeny analysis, from lower- to higher-order organisms. (C) The scatter plot indicates the PQS-forming sequence for all species on the y-axis with respect to GC content on the x-axis. The R and S values at the top denote the Pearson correlation coefficient (R) and the Spearman correlation coefficient (S). (D) The frequency of PQS-forming sites, relative to genome length expressed as PQS/kbp in the different taxonomy sub-groups.
Furthermore, an examination of PQS frequency sites relative to genome length of 140 organisms of different taxonomy (Supplementary S1) revealed that it was associated with evolutionary distance. Closely related vertebrates such as birds, reptiles, mammals, amphibians, and fish had the highest PQS frequencies, while the lower taxonomy group, such as fungi, nematodes, and insects had the lowest frequency (Figure 4d). Among the different orders, we found a strikingly high G4 frequency in the avian species. The mtDNA organization among the vertebrates is very similar, with non-coding sequences grouped together in the control region (CR), which controls mtDNA replication and transcription.[66] Interestingly, birds have an additional control region termed the pseudo control region (YCR), which is unique to avian species.[49–51] This YCR was found to have a strong association with an increased life span, despite birds having characteristics that generally have a negative effect on longevity by increasing reactive oxygen species (ROS), including higher metabolic rates, high blood glucose levels, and high body temperature.[52] One of the well-studied G4s in mtDNA lies in the CSBII (GCGGGGGAGGGGGGGTTTG) of the control region and it is a highly conserved sequence among vertebrates.[53] More surprisingly, the same G4-forming sequence was also found in the YCR region of birds, showing its important role and evolutionary conservation in mtDNA.[54] A potential role of the conserved G4 in CSBII is the formation of an R-loop Hybrid G4 three-stranded nucleic acid structure that consists of a DNA:RNA hybrid and a displaced strand of DNA[55], which plays an important role in mitochondrial replication and transcription.[56] Furthermore, the G4 stabilized R-loop is reported to increase transcription by a mechanism involving successive rounds of R-loop formation.[57] Hence, the duplication of the control region might provide an advantage for efficient replication and transcription, generating a larger number of mitochondria that can lead to more effective energy production for flight. However, the reason why the G4 is conserved in the YCR and how it contributes to the longevity of birds requires experimental validation. Recently Judy and colleagues identified co-formation of G-quadruplexes and R-loops spatially linked into unique structures called G-loops at telomere region.[58] It would be interesting to evaluate if such pronounced structures can be formed in mitochondria.
Control and Regulation of G4 in Mitochondria
G4 formation depends on the sequence composition within the G4 motif and its flanking sequences, while its stability depends on the number of G-tetrads, the loop length, and its topology. G4 formation is also highly dependent on monovalent cations, such as K+, Na+, NH4+, and Li+, and on the presence of K+ and Na+ in the cell environment.[59] Furthermore, the presence of molecular crowding and the induction of negative DNA super helicity during the transcription process can influence G4 formation.[60] In general, RNA G4s are more thermally stable than their DNA counterparts, but are limited to a parallel confirmation only, in contrast to different topologies adapted by DNA G4s.
G4s within a cell are highly regulated by G4-binding proteins (G4BPs) that can stabilize or destabilize G4 formation, influencing biological processes such as transcription, translation, and genomic stabilization.[61] Various techniques employing G4 bait and mass spectrometry[62,63] have been developed to profile G4-interacting proteins in cells. Most G4BPs belong to the helicase family such as DEAH-box helicase (DHX36),[64] RecQ class (Bloom protein (BLM),[65] and Werner’s syndrome protein (WRN).[66] Some G4BPs can also recognize specific sequences and bind to selective G4 topology. For example, POT1 is known to selectively bind to the telomeric anti-parallel G4 and promotes unfolding and refolding of G4 with the TPP1 complex.[63,64] Many G4BPs that bind to DNA G4s can also bind to RNA G4 because of their structural similarity. RNA G4BPs include DHX36, nucleolin, heterogeneous nuclear ribonucleoproteins (hnRNP), serine/arginine-rich splicing factors (SRSF), fragile X mental retardation 2 (FMR2), and ribosomal proteins.[61] However, there are still other G4BPs to be discovered and a full understanding of their effect on biological functions requires further study.
While many studies have focused on G4BPs in the nucleus and cytoplasm, there are reports of G4BPs in mitochondria that regulate mitochondrial G4s. It is reported that TWINKLE can unwind the mitochondrial G4 structures inefficiently close to the DNA deletion breakpoint, and assist mitochondrial replication machinery and prevent G4 from causing instability.[67] A knockdown of TWINKLE caused severe mtDNA depletion and affected the respiratory chain, demonstrating that TWINKLE is essential for replication.[68] Another example is Pif1, a DNA helicase that displays 5′–3′ helicase activity on both DNA/DNA and DNA/RNA hybrids. It can unwind G4 structures, and is located in both nucleus and mitochondria.[69,70] PIF1 helicase inactivation has been demonstrated to cause mitochondrial myopathy in mice, suggesting that PIF1 helicase plays a role in the regulation of G4.[71] Similar to PIF1, DNA2 Nuclease/Helicase has also been reported to unwind G4, and its mutation has been shown to impair mitochondrial function, suggesting that DNA2 also plays an important role in the regulation of G4.[72,73] While other helicases with G4 unwinding function such as RECQL4 are reported to localize in mitochondria, their role in mtDNA G4 is not yet identified.[74] The mitochondrial transcription factor A (TFAM), a high-mobility group (HMG)-box protein, is the major binding protein of human mtDNA and plays a critical role in its expression and maintenance. It is reported to bind to mtDNA G4 with an affinity similar to double-stranded DNA, suggesting functional recognition of G4 in mitochondria.[75]
The transcription of mtDNA starts in the D-loop and continues throughout the entire genome, and as a result, a large number of non-coding RNA (ncRNA) transcripts are released.[76] These ncRNA are relatively enriched in guanine, leading to a high G4-forming capability. A recent study identified that the protein G-rich RNA sequence binding factor 1 (GRSF1) was able to localize in mitochondria[77] and melt G4 structures in mtRNAs that facilitated their degradation with Suv3-PNPase complex.[78] Interestingly, this protein is present exclusively in vertebrates that have a high number of G4 motifs. We hypothesize that GRSF1 evolved to regulate and control G4 formation in mitochondria as it transitioned from G4-poor to G4-rich sequences. GRSF1 together with SUV3 helicase aids in the regulation of G4-rich lncRNA, and plays an important role in maintaining mitochondrial homeostasis.[79] The RNA-binding protein, SLIRP, was reported to be localized in mitochondria and to regulate mitochondrial protein synthesis; and it was identified as a G4-interacting protein at low nanomolar concentrations.[80,81] G4BPs influence biological processes by either resolving or binding the G4, but these effects do not always equate with the abundance of G4 motifs in the mtDNA. These reports support the likelihood of discovering novel proteins that play a role in mitochondrial maintenance, stability, and evolution.
Role of G4 in Mitochondrial Replication and Transcription
In mammals, mtDNA replicates in a distinct manner compared with nuclear DNA. Because of the circular nature of mitochondria DNA, the transcription and replication machinery may collide, which can have a detrimental effect on mtDNA gene expression.[82,83] The mitochondrial RNA polymerase (mitoRNAP) transcribes the mtDNA and generates the primers for replication, while the mitochondrial transcription elongation factor, TEFM, plays a key role in replication-transcription.[82] Transcription of human mtDNA is directed by two promoters, the light-strand promoter (LSP) and the heavy-strand promoter (HSP) located in opposing mtDNA strands, which results in polycistronic transcripts that undergo extensive processing and it is terminated at the CSBII. Transcription termination is a result of the formation of R-loops and the it is more efficient when the CSBII has more G tract in G6AG8, rather than the rare variant G5AG7, which suggests an effect of G4 on transcription.[82] The presence of G4 structures is known to cause genome instability, and plays an important role in various cancers and genetic diseases.[84,85] The distribution of G4 in the genome is not random, and it is often localized with chromosomal breakage points, further highlighting their importance in the genome.[84] Because of the presence of a higher number of potential G4 sequences in mitochondria, they may play an important role in mtDNA instability. Mitochondrial DNA deletions are prominent in cancer and genetic disorders and also play a role in aging.[86,87] The stalling of mitochondrial replication machinery by G4 sequences during DNA synthesis is a prominent source of mitochondrial genome instability.[67] We have found that G4 sequences have drastically accumulated in higher-order organisms, leading us to propose that it may possess important functional roles, despite its deleterious effect on mtDNA stability.
In recent years, it has been reported that phase separation plays a key role in many critical nuclear functions such as transcription[88], chromatin higher-order structure organization[89], membrane-less organelles[90], and histone modification.[91,92] Phase separation is driven by weak multivalent interactions between nucleic acid and nucleic acid-binding proteins that have intrinsically disordered regions (IDRs) that are sensitive to the NaCl concentration.[93] While studies of phase separation have traditionally focused on proteins, recent studies reveal that nucleic acid secondary structure can also trigger phase separation even in the absence of proteins. G4s can undergo liquid–liquid phase separation (LLPS) by connecting multiple nucleic-acid strands in an intermolecular configuration, or by π-stacking between G-tetrads of different G4s. Such interactions form droplets by combining G4 and histone H1 that mediate dynamic chromatic condemnation in the nucleus.[94] In the mitochondrial context, it was recently discovered that the TFAM undergoes phase separation with mtDNA to drive nucleoid self-assembly that regulates the mitochondrial transcription process.[95] Therefore, we propose that the guanine-dense mitochondrial DNA might also be key in promoting LLPS.
ROS, produced mostly by mitochondria, contribute significantly to mitochondrial damage, but also play a prominent role in redox signaling to other parts of the cell.[96] Among the four bases, guanine has the lowest redox potential, being oxidized by ROS generation into 8-oxo-7,8-dihydroguanine (8OG).[97] Therefore, G4s are an easy target for guanine oxidation, potentially affecting their stability. An interesting role of G4s is the epigenetic control of genes based on their 8OG modification. DNA damage inflicted on the promoter G4 region can upregulate or downregulate gene expression.[98] Furthermore, G4-rich DNA was found to accumulate in the cytoplasm and to participate in stress granule assembly upon oxidative stress, thereby also regulating gene expression.[99] Given the abundance of ROS in mitochondria, we wonder whether this can regulate mitochondrial gene expression and DNA damage responses, similar to nuclear DNA, but this question has yet to be evaluated in detail.
Mitochondrial RNA G4
The human mitochondrial genome is densely packed with genetic information that encodes 2 rRNAs, 22 tRNAs, and 13 mRNAs for the oxidative phosphorylation system. The entire mitochondrial genome is transcribed from both H and L strands as long polycistronic transcripts that undergo multiple processing before becoming functional. While most mRNAs are transcribed from the template of the G-rich H strand under the control of the HSP, the complementary L strand serves as the template for ND6 mitochondrial mRNA from the LSP. The transcribed polycistronic RNA undergoes processing at ribonucleoprotein structures called mitochondrial RNA granules (MRGs) before protein synthesis.[100,101] Several proteins associated with the processing and maturation of primary transcripts have been identified in MRGs, suggesting their role in the regulation of mitochondrial translation.[102]
Genomes generate a large number of long non-coding RNAs (lncRNAs), sequences longer than 200 nucleotides that do not translate into protein. Via their interaction with DNA, RNA, and proteins, lncRNAs are linked to gene regulation, epigenetic control of chromatin structure, and membrane-less bodies, respectively.[103] The importance of lncRNA in evolution is reflected by its increasing presence in higher-order organisms, contributing to development and differentiation processes.[104] The formation of G4s is reported in lncRNAs such as TERRA[105] and MALAT[106], with interest growing in identifying their functional role.
In mitochondria, while the L strand was known to be a template for ND6 and several tRNAs, it was reported also to generate several lncRNAs, which contain a high number of potential G4-forming motifs.[76,107] The regions of the mitochondrial genome complementary to the genes that encode ND5, ND6, and Cytb mRNAs were found to have high levels of lncRNAs.[76] Using QGRS mapper, we identified 21 G4-forming motifs in guanine-rich lncND5, and 17 in lncCYTB. In a different study, the mitochondrial lncRNA, LIPCAR, was found to be a biomarker associated with chronic heart failure, although the actual mechanism remains elusive. Another heart failure study using deep sequencing of RNA revealed an abundance of mRNA (37%) and lncRNA (71%) of mitochondrial origin.[108] Given its abundance and G4 enrichment, it is possible that lncRNA generated from mitochondria may have a biological significance that has yet to be identified.
Newly transcribed RNA and RNA processing protein form MRGs in mitochondria, although the exact mechanism of their formation is not fully understood.[109] The presence of G4 structures forms droplets by LLPS without the presence of protein in short-hair root RNA[110] and ALS/FTD-associated C9ORF72 repeat RNA.[111] Considering that this lncRNA with the presence of G4 may form droplets, it may constitute mitochondrial granules, but whether the G4-enriched lncRNA alone or other molecular determinants such as protein promotes the formation of granules has yet to be elucidated. Apart from producing ATP for cells, mitochondria also generate heme, an essential cofactor for many enzymes, by inserting ferrous iron into protoporphyrin IX using ferrochelatase. Free heme, however, is toxic even at very low levels, catalyzing the formation of ROS and inducing oxidative stress. This risk is mitigated by G4 because it can sequester heme in live cells, thus preventing oxidative DNA damage.[112,113] Earlier studies show that G4 in complex with porphyrins such as heme have peroxidase and peroxygenase enzyme-like activity capable of reducing hydrogen peroxidase and other hydroperoxidases.[114] Furthermore, the DNA and RNA sequences that form parallel-stranded G4 were found to be optimal for heme binding and peroxidase activity.[115] It has been reported that in Alzheimer’s disease, amyloid-β-peptide sequesters heme, leading to heme deficiency and strong peroxidase activity at the intracellular level.[116,117] Given this evidence, we hypothesize that mitochondrial DNA and lncRNA can sequester free heme and protect cells from ROS arising from G4/heme complexes.
Targeting MitoG4s
In living cells, the presence of G4s in genomic DNA has been demonstrated with various imaging and sequencing techniques. G4 ligands cannot easily target mtDNA due to the highly dense, impermeable mitochondrial inner membrane.[118] The first direct evidence of mitochondrial G4 was reported by developing a fluorescent G4 ligand, 3,6-bis(1-methyl-4-vinylpyridinium) carbazole diiodide (BMVC), which was found to localize within mitochondria.[119] It was found that a sufficient quantity of BMVC suppressed mtDNA gene expression, eventually inducing cell death. Using live-cell imaging, another G4-ligand, RHPS4, was found to be localized primarily in mitochondria, even at low doses. Treatment with RHPS4 was also found to induce acute inhibition of mitochondrial transcript elongation, leading to respiratory complex depletion.[120] Further, mitochondria-specific probes targeting G-quadruplexes for live-cell imaging were developed recently.[121] The probe showed high selectivity almost 1000-fold higher than that of mitochondrial double-stranded and sensitivity towards mitoG4 making it a suitable candidate for monitoring the dynamic process of mitochondria. More recently mtDNA G4 targeting probe integrating active photosensitizer and mitochondrial targeting functional group was developed for mtDNA G4 sensing. [122] Further Hu developed near-infrared fluorescent ligand for tracking mitochondrial DNA G4.[123] Another study developed a set of chemical probes to thoroughly investigate mtDNA G4s: MitoISCH, a mtDNA G4-specific fluorescent probe; and MitoPDS, a mitochondrial DNA-targeted, G4-stabilizing agent.[124] MitoPDS caused glycolysis-related gene activation in cancer cells, revealing a connection of mtDNA G4s to glycolysis. Many types of cancer display increased glycolysis even in the presence of oxygen and competent mitochondrial function,[125] and mtDNA instability contributes to the enhancement of glycolysis in cancer cells.[126] Given the increase in mtDNA instability caused by G4 stabilization, it is hoped that unraveling the associated mechanism will provide insights that will facilitate the development of cancer treatments.
Concluding Remarks and Future Perspectives
Evolutionary erosion of GC-rich isochores in the nuclear genome is a common trend in most organisms, but mitochondria were found to transform from GC poor to GC rich as evolution progressed. The enrichment of G4 in birds and primates is particularly interesting, and suggests a need for further study of how these adaptions facilitate advantages in higher organisms (related to aging, meeting the needs of complex cellular mechanisms, and physiological activities, etc.). While the role of mtDNA G4 has been explored, we believe it is worth investigating G-rich lncRNA produced by mtDNA, and its biological role inside mitochondria and the cytoplasm. Current knowledge of mitochondrial G4s and their functional relevance is limited because of a lack of tools, unlike nuclear G4s for which many probes, ligands, antibodies, and sequencing techniques have been developed. We believe that the potential role of G4 structures in mitochondria can be unraveled by developing probes capable of infiltrating these respiratory organelles. With many questions still unanswered (see outstanding questions), we believe that the field of mitochondrial G4 research will continue to grow and provide a better understanding of secondary structures and their role in the evolution of higher organisms and their cellular processes.
Outstanding question.
G4 and mitochondria evolution: Why does guanine increase in heavy strands despite its deleterious effects? How does mtDNA G4 contribute to the evolution of higher organisms? Do they evolve to meet the higher organism physiological demand? Does mtDNA G4 plays role in an organism’s lifespan?
Does mtG4 evolve in birds to meet its energy needs? How does the pseudo-control region (YCR) in birds contribute to a bird’s lifespan? If so why the conserved G4 in the control region is also present in YCR? Does it contribute to enhanced mitochondrial replication and transcription?
What is the functional role of G-rich lncRNA produced by mitochondria? Does it play an important role in mitochondrial function? Does the lncRNA from mitochondria shuttle to the cytoplasm or nucleus as a molecular signal? Why do the levels of lncRNA from mitochondria vary in cardiac diseases?
lncRNA G4s and granules: Does G4 in the mitochondrial lncRNA contribute to RNA granules? If so are there any other molecular determinants govern its formation? Does it play important role in Heme scavenging?
Interplay between mtDNA G4s and Glycolysis: How does the G4 in mtDNA contribute to enhanced glycolysis in cancer cells? How does it activate glycolysis-related genes? Does mtDNA G4 control mitochondrial oxidative phosphorylation? Do the mtDNA G4s play a key role in the Warburg effect?
Supplementary Material
Highlights.
G4s are nucleic acid secondary structures consisting of stacked planar G-tetrads. Their ability to influence biological processes such as replication, transcription, and genome instability has been observed in mitochondria.
Mitochondrial G4 forming sequence has increased in the higher organism with evolution showing its key role in mitochondrial regulation.
We discuss the possible function of DNA G4 in mitochondria in various functions such as transcription, genome stability, and their regulation.
We report the potential G4 forming sequences in the long non-coding RNA produced in mitochondria and their possible role in RNA granules, Heme scavenging.
Acknowledgments
We express sincere thanks for a Grant-in-Aid Priority Research (16H06356 to H. S.) and 20H05936 from JSPS. This work was also supported by AMED [JP20am0101101 to H. S. (Platform Project for Supporting Drug Discovery and Life Science Research (BINDS) and NIH award R01CA236350 to H. S. The authors sincerely acknowledge Jeffrey Friedl for help in proofreading the manuscript. We also acknowledge Shinjiro Suzuki, Manendra Lankadasari, Vipin Mohan Dan, and Vinay Rajput for their input. Figures 2, 5, 6 are created using Biorender.
Figure 5. Role of G4 in mtDNA.
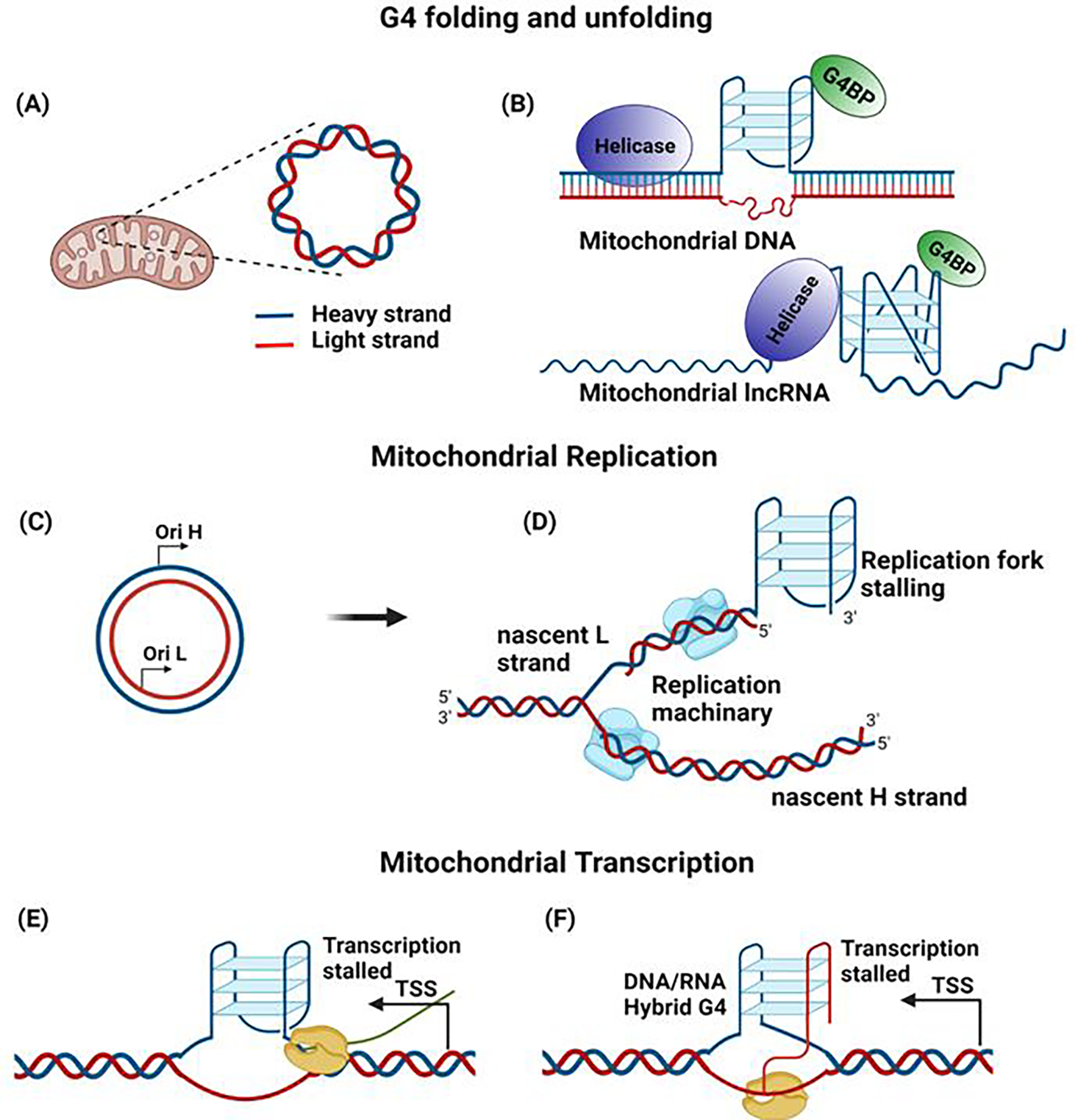
(A) Mitochondrial DNA is represented as an H strand and an L strand, based on guanine content. (B) The presence of proteins that can bind and stabilize G4 (G4BP-G4 binding protein), as well as helicases that can unwind G4, can influence the G4 in mitochondria. (C) The mitochondrial replication origin in the H and L strand (OH & OL) initiates DNA replication in the respective strands. (D) The formation of G4 can influence mitochondrial DNA replication by stalling the process. (E) The presence of G4 in the transcription start site (TSS) can block the progression of mtRNA polymerase, resulting in altered transcription. (F) The formation of hybrid DNA:RNA G4 in the CSBII region terminates the transcription.
Figure 6 – Role of G4 in RNA.
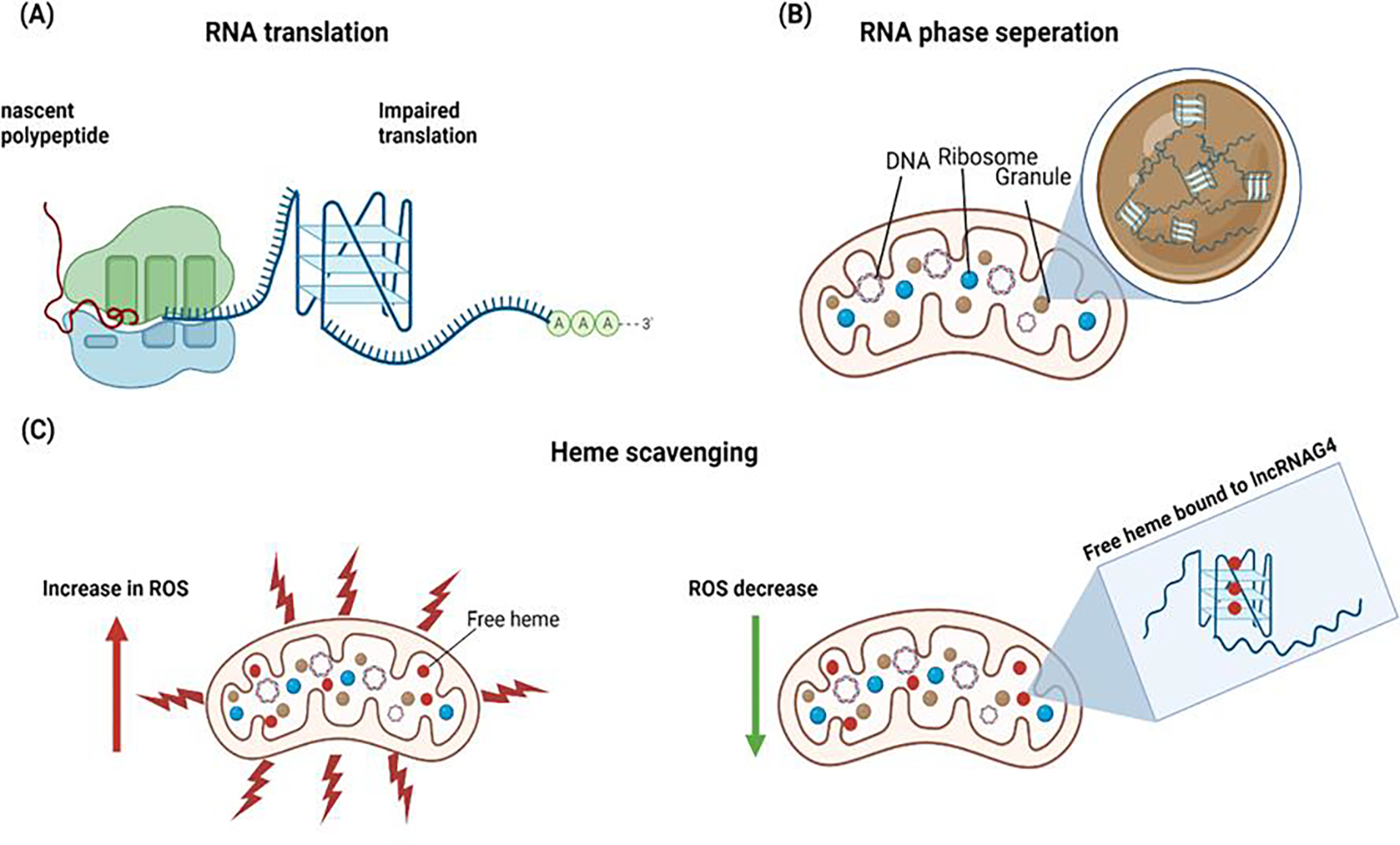
(A) Formation of RNA G-quadruplex can impair the translation of mRNA rich in guanine. (B) The lncRNA with G-quadruplex structures can trigger phase separation and contribute to RNA granules. The lncRNA can scavenge the free heme in the mitochondria and reduce reactive oxygen species (ROS), thereby reducing oxidative stress.
Glossary
- Hoogsteen hydrogen bonding
It is a variation of base-pairing in nucleic acids where two nucleobases, one on each strand can be held together by hydrogen bonds in the major groove.
- G-quadruplex
G-quadruplex secondary structures (G4) are formed in nucleic acids sequences that are rich in guanine where four guanine bases can associate through Hoogsteen hydrogen bonding to form a square planar structure called a guanine tetrad.
- Mitochondrial heavy and light strand
The mtDNA consists of “light” and “heavy” strands, with the heavy strand containing a higher proportion of guanine and adenine nucleotides and the light strand containing a higher proposition of cytosine and thymine respectively.
- GC skew
GC skew denotes the relative excess of G nucleotides over C nucleotides on the one strand compared to other and it is calculated by GC skew = (G − C)/(G + C).
- Circos plot
Circos visualizes data in a circular layout ideal for exploring relationships between objects or positions. It greatly enhances the visualization of scientific results especially in the genomics field.
- R-loop
During transcription, the nascent RNA strand can base pair with its template DNA, displacing the non-template strand as ssDNA and forming a structure called an R-loop.
Footnotes
Declaration of interests
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bochman ML et al. (2012) DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gellert M et al. (1962) Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U. S. A. 48, 2013–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bang I (1910) Untersuchungen über die Guanylsäure. Biochemische Zeitschrift 26, 293–311 [Google Scholar]
- 4.Burge S et al. (2006) Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 34, 5402–5415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lombardi EP and Londoño-Vallejo A (2020) A guide to computational methods for G-quadruplex prediction. Nucleic Acids Res. 48, 1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monchaud D (2020) Quadruplex detection in human cells. In Annual Reports in Medicinal Chemistry 54 (Neidle S, ed), pp. 133–160, Elsevier [Google Scholar]
- 7.Kwok CK and Merrick CJ (2017) G-quadruplexes: Prediction, characterization, and biological application. Trends Biotechnol. 35, 997–1013 [DOI] [PubMed] [Google Scholar]
- 8.Sen D and Gilbert W (1988) Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334, 364–366 [DOI] [PubMed] [Google Scholar]
- 9.Parkinson GN et al. (2002) Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 417, 876–880 [DOI] [PubMed] [Google Scholar]
- 10.Siddiqui-Jain A et al. (2002) Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U. S. A. 99, 11593–11598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dexheimer TS et al. (2006) Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 128, 5404–5415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Armond R et al. (2005) Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry 44, 16341–16350 [DOI] [PubMed] [Google Scholar]
- 13.Neidle S (2016) Quadruplex nucleic acids as novel therapeutic targets. J. Med. Chem. 59, 5987–6011 [DOI] [PubMed] [Google Scholar]
- 14.Huppert JL and Balasubramanian S (2007) G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 35, 2105–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halder K et al. (2009) Genome-wide analysis predicts DNA structural motifs as nucleosome exclusion signals. Mol. Biosyst. 5, 1703–1712 [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez R et al. (2012) Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 8, 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lago S et al. (2021) Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat. Commun. 12, 3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleming AM and Burrows CJ (2020) Interplay of guanine oxidation and G-quadruplex folding in gene promoters. J. Am. Chem. Soc. 142, 1115–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spiegel J et al. (2021) G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol. 22, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson J et al. (2021) DNA G-quadruplex structures: more than simple roadblocks to transcription? Nucleic Acids Res. 49, 8419–8431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zell J et al. (2021) DNA folds threaten genetic stability and can be leveraged for chemotherapy. RSC Chem Biol 2, 47–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyu K et al. (2021) RNA G-quadruplexes (rG4s): genomics and biological functions. Nucleic Acids Res. 49, 5426–5450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumari S et al. (2007) An RNA G-quadruplex in the 5’ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 3, 218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang SY et al. (2022) Global mapping of RNA G-quadruplexes (G4-RNAs) using G4RP-seq. Nat. Protoc. 17, 870–889 [DOI] [PubMed] [Google Scholar]
- 25.Shahid R et al. (2010) The BCL-2 5’ untranslated region contains an RNA G-quadruplex-forming motif that modulates protein expression. Biochemistry 49, 8300–8306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris MJ et al. (2010) An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Am. Chem. Soc. 132, 17831–17839 [DOI] [PubMed] [Google Scholar]
- 27.Bonnal S et al. (2003) A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 278, 39330–39336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Falabella M et al. (2019) Potential Roles for G-Quadruplexes in Mitochondria. Curr. Med. Chem. 26, 2918–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malinee M and Sugiyama H (2021) Impact of Reactive Oxygen Species and G-Quadruplexes in Telomeres and Mitochondria. In Creative Complex Systems (Nishimura K et al. , eds), pp. 249–274, Springer; Singapore [Google Scholar]
- 30.Filograna R et al. (2021) Mitochondrial DNA copy number in human disease: the more the better? FEBS Lett. 595, 976–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chinnery PF and Hudson G (2013) Mitochondrial genetics. Br. Med. Bull. 106, 135–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taanman JW (1999) The mitochondrial genome: structure, transcription, translation and replication. Biochim. Biophys. Acta 1410, 103–123 [DOI] [PubMed] [Google Scholar]
- 33.Gray MW (2012) Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 4, a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roger AJ et al. (2017) The Origin and Diversification of Mitochondria. Curr. Biol. 27, R1177–R1192 [DOI] [PubMed] [Google Scholar]
- 35.Feng Y et al. (2022) Epigenomic features of DNA G-quadruplexes and their roles in regulating rice gene transcription. Plant Physiol. 188, 1632–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwok CK et al. (2016) rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat. Methods 13, 841–844 [DOI] [PubMed] [Google Scholar]
- 37.Dumetz F et al. (2021) G-quadruplex RNA motifs influence gene expression in the malaria parasite Plasmodium falciparum. Nucleic Acids Res. 49, 12486–12501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X et al. (2020) RNA G-quadruplex structures exist and function in vivo in plants. Genome Biol. 21, 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hänsel-Hertsch R et al. (2016) G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 48, 1267–1272 [DOI] [PubMed] [Google Scholar]
- 40.Marsico G et al. (2019) Whole genome experimental maps of DNA G-quadruplexes in multiple species. Nucleic Acids Res. 47, 3862–3874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding Y et al. (2018) Case studies on potential G-quadruplex-forming sequences from the bacterial orders Deinococcales and Thermales derived from a survey of published genomes. Sci. Rep. 8, 15679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capra JA et al. (2010) G-quadruplex DNA sequences are evolutionarily conserved and associated with distinct genomic features in Saccharomyces cerevisiae. PLoS Comput. Biol. 6, e1000861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu F et al. (2021) Genome-wide analysis of DNA G-quadruplex motifs across 37 species provides insights into G4 evolution. Commun Biol 4, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hubert B (2022) SkewDB, a comprehensive database of GC and 10 other skews for over 30,000 chromosomes and plasmids. Sci. Data 9, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka M and Ozawa T (1994) Strand asymmetry in human mitochondrial DNA mutations. Genomics 22, 327–335 [DOI] [PubMed] [Google Scholar]
- 46.Clayton DA (1982) Replication of animal mitochondrial DNA. Cell 28, 693–705 [DOI] [PubMed] [Google Scholar]
- 47.Garlid KD (1996) Cation transport in mitochondria--the potassium cycle. Biochim. Biophys. Acta 1275, 123–126 [DOI] [PubMed] [Google Scholar]
- 48.Kikin O et al. (2006) QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 34, W676–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haring E et al. (2009) Evolution of a pseudo-control region in the mitochondrial genome of Palearctic buzzards (genus Buteo). J. Zoolog. Syst. Evol. Res. 37, 185–194 [Google Scholar]
- 50.Väli U (2002) Mitochondrial pseudo-control region in old world eagles (genus Aquila). Mol. Ecol. 11, 2189–2194 [DOI] [PubMed] [Google Scholar]
- 51.Schirtzinger EE et al. (2012) Multiple independent origins of mitochondrial control region duplications in the order Psittaciformes. Mol. Phylogenet. Evol. 64, 342–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Skujina I et al. (2016) Duplication of the mitochondrial control region is associated with increased longevity in birds. Aging (Albany NY) 8, 1781–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pham XH et al. (2006) Conserved sequence box II directs transcription termination and primer formation in mitochondria. J. Biol. Chem. 281, 24647–24652 [DOI] [PubMed] [Google Scholar]
- 54.Bohálová N et al. (2022) Conservation and over-representation of G-quadruplex sequences in regulatory regions of mitochondrial DNA across distinct taxonomic sub-groups. Biochimie 194, 28–34 [DOI] [PubMed] [Google Scholar]
- 55.Crossley MP et al. (2019) R-loops as cellular regulators and genomic threats. Mol. Cell 73, 398–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wanrooij PH et al. (2012) A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop. Nucleic Acids Res. 40, 10334–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee C-Y et al. (2020) R-loop induced G-quadruplex in non-template promotes transcription by successive R-loop formation. Nat. Commun. 11, 3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang SY et al. (2021) G-quadruplexes mark alternative lengthening of telomeres. NAR Cancer 3, zcab031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saintomé C et al. (2016) The exception that confirms the rule: a higher-order telomeric G-quadruplex structure more stable in sodium than in potassium. Nucleic Acids Res. 44, 2926–2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shrestha P et al. (2017) Confined space facilitates G-quadruplex formation. Nat. Nanotechnol. 12, 582–588 [DOI] [PubMed] [Google Scholar]
- 61.Meier-Stephenson V (2022) G4-quadruplex-binding proteins: review and insights into selectivity. Biophys. Rev. DOI: 10.1007/s12551-022-00952-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Makowski MM et al. (2018) Global profiling of protein–DNA and protein–nucleosome binding affinities using quantitative mass spectrometry. Nat. Commun. 9, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X et al. (2021) Chemical profiling of DNA G-quadruplex-interacting proteins in live cells. Nat. Chem. 13, 626–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen MC et al. (2018) Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature 558, 465–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mohaghegh P et al. (2001) The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 29, 2843–2849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheok CF et al. (2005) Roles of the Bloom’s syndrome helicase in the maintenance of genome stability. Biochem. Soc. Trans. 33, 1456–1459 [DOI] [PubMed] [Google Scholar]
- 67.Bharti SK et al. (2014) DNA sequences proximal to human mitochondrial DNA deletion breakpoints prevalent in human disease form G-quadruplexes, a class of DNA structures inefficiently unwound by the mitochondrial replicative Twinkle helicase. J. Biol. Chem. 289, 29975–29993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Milenkovic D et al. (2013) TWINKLE is an essential mitochondrial helicase required for synthesis of nascent D-loop strands and complete mtDNA replication. Hum. Mol. Genet. 22, 1983–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boulé J-B and Zakian VA (2006) Roles of Pif1-like helicases in the maintenance of genomic stability. Nucleic Acids Res. 34, 4147–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Futami K et al. (2007) Mitochondrial and nuclear localization of human Pif1 helicase. Biol. Pharm. Bull. 30, 1685–1692 [DOI] [PubMed] [Google Scholar]
- 71.Bannwarth S et al. (2016) Inactivation of Pif1 helicase causes a mitochondrial myopathy in mice. Mitochondrion 30, 126–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duxin JP et al. (2009) Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Mol. Cell. Biol. 29, 4274–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ronchi D et al. (2013) Mutations in DNA2 link progressive myopathy to mitochondrial DNA instability. Am. J. Hum. Genet. 92, 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De S et al. (2012) RECQL4 is essential for the transport of p53 to mitochondria in normal human cells in the absence of exogenous stress. J. Cell Sci. 125, 2509–2522 [DOI] [PubMed] [Google Scholar]
- 75.Lyonnais S et al. (2017) The human mitochondrial transcription factor A is a versatile G-quadruplex binding protein. Sci. Rep. 7, 43992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rackham O et al. (2011) Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 17, 2085–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Antonicka H et al. (2013) The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 17, 386–398 [DOI] [PubMed] [Google Scholar]
- 78.Pietras Z et al. (2018) Dedicated surveillance mechanism controls G-quadruplex forming non-coding RNAs in human mitochondria. Nat. Commun. 9, 2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Khidr L et al. (2008) Role of SUV3 helicase in maintaining mitochondrial homeostasis in human cells. J. Biol. Chem. 283, 27064–27073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Williams P et al. (2017) Identification of SLIRP as a G quadruplex-binding protein. J. Am. Chem. Soc. 139, 12426–12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chujo T et al. (2012) LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 40, 8033–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Agaronyan K et al. (2015) Mitochondrial biology. Replication-transcription switch in human mitochondria. Science 347, 548–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pomerantz RT and O’Donnell M (2010) What happens when replication and transcription complexes collide? Cell Cycle 9, 2537–2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao J et al. (2010) Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. 67, 43–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Y et al. (2019) G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat. Commun. 10, 943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bratic A and Larsson N-G (2013) The role of mitochondria in aging. J. Clin. Invest. 123, 951–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wallace DC (2012) Mitochondria and cancer. Nat. Rev. Cancer 12, 685–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hnisz D et al. (2017) A phase separation model for transcriptional control. Cell 169, 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gibson BA et al. (2019) Organization of chromatin by intrinsic and regulated phase separation. Cell 179, 470–484.e21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Uversky VN (2017) Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr. Opin. Struct. Biol. 44, 18–30 [DOI] [PubMed] [Google Scholar]
- 91.Sanulli S et al. (2019) HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 575, 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang L et al. (2019) Histone modifications regulate chromatin compartmentalization by contributing to a phase separation mechanism. Mol. Cell 76, 646–659.e6 [DOI] [PubMed] [Google Scholar]
- 93.Kamimura YR and Kanai M (2021) Chemical insights into liquid-liquid phase separation in molecular biology. Bull. Chem. Soc. Jpn. 94, 1045–1058 [Google Scholar]
- 94.Mimura M et al. (2021) Quadruplex folding promotes the condensation of linker histones and DNAs via liquid-liquid phase separation. J. Am. Chem. Soc. 143, 9849–9857 [DOI] [PubMed] [Google Scholar]
- 95.Long Q et al. (2021) Phase separation drives the self-assembly of mitochondrial nucleoids for transcriptional modulation. Nat. Struct. Mol. Biol. 28, 900–908 [DOI] [PubMed] [Google Scholar]
- 96.Sies H et al. (2022) Defining roles of specific reactive oxygen species (ROS) in cell biology and physiology. Nat. Rev. Mol. Cell Biol. 23, 499–515 [DOI] [PubMed] [Google Scholar]
- 97.Saito I et al. (1998) Mapping of the hot spots for DNA damage by one-electron oxidation: Efficacy of GG doublets and GGG triplets as a trap in long-range hole migration. J. Am. Chem. Soc. 120, 12686–12687 [Google Scholar]
- 98.Fleming AM and Burrows CJ (2020) On the irrelevancy of hydroxyl radical to DNA damage from oxidative stress and implications for epigenetics. Chem. Soc. Rev. 49, 6524–6528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Byrd AK et al. (2016) Evidence that G-quadruplex DNA accumulates in the cytoplasm and participates in stress granule assembly in response to oxidative stress. J. Biol. Chem. 291, 18041–18057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jourdain AA et al. (2016) Mitochondrial RNA granules: Compartmentalizing mitochondrial gene expression. J. Cell Biol. 212, 611–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Antonicka H and Shoubridge EA (2015) Mitochondrial RNA granules are centers for posttranscriptional RNA processing and ribosome biogenesis. Cell Rep. 10, 920–932 [DOI] [PubMed] [Google Scholar]
- 102.Tu Y-T and Barrientos A (2015) The human mitochondrial DEAD-box protein DDX28 resides in RNA granules and functions in mitoribosome assembly. Cell Rep. 10, 854–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Statello L et al. (2021) Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fatica A and Bozzoni I (2014) Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21 [DOI] [PubMed] [Google Scholar]
- 105.Mei Y et al. (2021) TERRA G-quadruplex RNA interaction with TRF2 GAR domain is required for telomere integrity. Sci. Rep. 11, 3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mou X et al. (2022) Identification and targeting of G-quadruplex structures in MALAT1 long non-coding RNA. Nucleic Acids Res. 50, 397–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kumarswamy R et al. (2014) Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ. Res. 114, 1569–1575 [DOI] [PubMed] [Google Scholar]
- 108.Yang K-C et al. (2014) Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 129, 1009–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rey T et al. (2020) Mitochondrial RNA granules are fluid condensates positioned by membrane dynamics. Nat. Cell Biol. 22, 1180–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang Y et al. (2019) G-quadruplex structures trigger RNA phase separation. Nucleic Acids Res. 47, 11746–11754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fay MM et al. (2017) ALS/FTD-associated C9ORF72 repeat RNA promotes phase transitions in vitro and in cells. Cell Rep. 21, 3573–3584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gray LT et al. (2019) G-quadruplexes sequester free heme in living cells. Cell Chem. Biol. 26, 1681–1691.e5 [DOI] [PubMed] [Google Scholar]
- 113.Kawai K et al. (2022) Molecular imaging of labile heme in living cells using a small molecule fluorescent probe. J. Am. Chem. Soc. 144, 3793–3803 [DOI] [PubMed] [Google Scholar]
- 114.Sen D and Poon LCH (2011) RNA and DNA complexes with hemin [Fe(III) heme] are efficient peroxidases and peroxygenases: how do they do it and what does it mean? Crit. Rev. Biochem. Mol. Biol. 46, 478–492 [DOI] [PubMed] [Google Scholar]
- 115.Cheng X et al. (2009) General peroxidase activity of G-quadruplex-hemin complexes and its application in ligand screening. Biochemistry 48, 7817–7823 [DOI] [PubMed] [Google Scholar]
- 116.Atamna H et al. (2009) Human and rodent amyloid-beta peptides differentially bind heme: relevance to the human susceptibility to Alzheimer’s disease. Arch. Biochem. Biophys. 487, 59–65 [DOI] [PubMed] [Google Scholar]
- 117.Atamna H and Boyle K (2006) Amyloid-beta peptide binds with heme to form a peroxidase: relationship to the cytopathologies of Alzheimer’s disease. Proc. Natl. Acad. Sci. U. S. A. 103, 3381–3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hidaka T et al. (2017) Creation of a synthetic ligand for mitochondrial DNA sequence recognition and promoter-specific transcription suppression. J. Am. Chem. Soc. 139, 8444–8447 [DOI] [PubMed] [Google Scholar]
- 119.Huang W-C et al. (2015) Direct evidence of mitochondrial G-quadruplex DNA by using fluorescent anti-cancer agents. Nucleic Acids Res. 43, 10102–10113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Falabella M et al. (2019) G-quadruplex dynamics contribute to regulation of mitochondrial gene expression. Sci. Rep. 9, 5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.She M-T et al. (2022) Design mitochondria-specific fluorescent turn-on probes targeting G-quadruplexes for live cell imaging and mitophagy monitoring study. Chem. Eng. J. 446, 136947 [Google Scholar]
- 122.Wang Y et al. (2022) A smart small molecule as specific fluorescent probe for sensitive recognition of mitochondrial DNA G-Quadruplexes. Chem. Eng. J. 441, 135977 [Google Scholar]
- 123.Hu M-H (2021) Molecular engineering of a near-infrared fluorescent ligand for tracking mitochondrial DNA G-quadruplexes. Anal. Chim. Acta 1169, 338600. [DOI] [PubMed] [Google Scholar]
- 124.Chen X-C et al. (2021) Monitoring and modulating mtDNA G-quadruplex dynamics reveal its close relationship to cell glycolysis. J. Am. Chem. Soc. 143, 20779–20791 [DOI] [PubMed] [Google Scholar]
- 125.Vander Heiden MG et al. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Crooks DR et al. (2021) Mitochondrial DNA alterations underlie an irreversible shift to aerobic glycolysis in fumarate hydratase-deficient renal cancer. Sci. Signal. 14, eabc4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


