Abstract
Regulatory T cells (Tregs) provide an essential tolerance mechanism to suppress the immune response. Induced Tregs hold the potential to treat autoimmune diseases in adoptive therapy and can be produced with stimulating signals to CD3 and CD28 in presence of the cytokine TGF-β and IL-2. This report examines the modulation of human Treg induction by leveraging the ability of T cells to sense the mechanical stiffness of an activating substrate. Treg induction on polyacrylamide gels (PA-gels) was sensitive to the substrate’s elastic modulus, increasing with greater material stiffness. Single-cell RNA-Seq analysis revealed that Treg induction on stiffer substrates involved greater use of oxidative phosphorylation (OXPHOS). Inhibition of ATP synthase significantly reduced the rate of Treg induction and abrogated the difference among gels while activation of AMPK (AMP-activated protein kinase) increased Treg induction on the softer sample but not on the harder sample. Treg induction is thus mechanosensitive and OXPHOS dependent, providing new strategies for improving the production of these cells for cellular immunotherapy.
Keywords: Treg induction, Metabolism, Transcriptomes, Mechanosensing
Introduction
Regulatory T cells are essential for immune homeostasis and self-tolerance. These cells prevent autoimmune diseases and chronic inflammation by selectively suppressing immune responses via inhibition of T cell proliferation, cytotoxic function, and cytokine production. Correspondingly, a reduction in activity or quantity of Tregs leads to an increased susceptibility to autoimmune diseases including multiple sclerosis, rheumatoid arthritis, and Type-I diabetes [1]. One step further, augmenting Treg function by adoptive transfer has shown preliminary success in the treatment of Type-I diabetes [2] and graft rejection [3]. To be effective, this approach requires a large number of high-quality cells, and the production of sufficient quantities of this “living drug” poses a current challenge in cellular immunotherapy.
One source of these cells is thymic (tTregs) Tregs, which develop in the thymus in vivo from immature thymocytes in response to self-antigen stimulation. Tregs can also be induced in the periphery (pTreg) from conventional T cells in response to specific microenvironments, maintaining tolerance to commensal bacteria, foods, allergens, and the fetus in a pregnant mother [4]. Tregs can also be induced in vitro (iTregs) using T cell activation signals and polarizing cytokines including IL-2 and TGF-β. IL-2 is essential for Treg survival, proliferation, and stability [5]. TGF-β1 promotes the binding of NFAT and Smad3 to the conserved non-coding sequence-1 enhancing histone acetylation and induction of FOXP3 [6], a transcription factor that is a master regulator of Treg development and function [7].
Current clinical trials focus predominantly on tTregs that have undergone polyclonal expansion [8]. However, tTregs constitute only 5–10% of CD4+ T cells, making large-scale production difficult. Reduced stability of tTregs in inflammatory environments [3] may also contribute to the limited success of these cells in adoptive transfer immunotherapy. In contrast, iTregs showed improved retention of FOXP3 and suppressive capacity under inflammatory conditions [9]. Notably, iTregs generated in vitro via the TGF-β-dependent pathway were found to be functionally equivalent to iTreg cells derived in vivo [10]. Lastly, a first-in-man iTreg clinical trial demonstrated the safety of iTreg adoptive therapy in graft-versus-host disease [11]. Thus, iTregs are an important regulatory subset and have much promise for adoptive cell transfer.
Optimization of the chemical environment for iTreg differentiation is being robustly pursued as a method to improve production. As a complementary approach, this study seeks to improve Treg induction by modulating the mechanical stiffness of substrates used to activate T cells. The mechanical presentation of ligands plays an important role in T cell function since soluble activating antibodies (anti-CD3) fail to promote proper T cell activation [12]. Subsequent studies showed that both mouse and human T cells can sense the mechanical stiffness of a surface with activating antibodies [13–15]. The induction of mouse CD4+ T cells into Foxp3+ iTregs was also demonstrated to be mechanosensitive [5]. However, mouse and human Tregs exhibit well-recognized differences in function including FOXP3 expression, stability, and suppressive capacity [16]. In addition, activation of mouse and human CD4+ T cells show contrasting responses to substrate stiffness [13, 14], complicating the interpretation of mechanosensing between species. This report consequently focuses on the induction of iTregs from primary human T cells to explore the use of mechanosensing to improve cellular immunotherapy.
Materials and Methods
Statistics.
Normality tests (D’Agostino&Pearson test) were performed before using parametric methods. Parametric data were analyzed with standard one- and two-way ANOVA with Tukey multiple comparisons. Non-parametric data were analyzed with the Mann-Whitney test or Kruskal-Wallis test depending on the number of groups. All analyses were conducted on GraphPad Prism 9 (α = 0.05). All error bars are standard deviations unless otherwise stated.
Study approval.
Primary human T cells were isolated from blood products (New York Blood Center) from healthy donors that were provided without donor-identifying information. As the origin of these cells cannot be identified, this study is exempt from DHHS regulations based on §46.104(d)(4).
Polyacrylamide gel preparation.
Polyacrylamide gels (PA-gels) were prepared following established protocols [15]. Substrate stiffness was modulated by varying the ratio of acyl monomers with bis-acrylamide crosslinkers (Table 2). A gel master mix consisted of 40% acrylamide and 2% bis-acrylamide in deionized water. This master solution was supplemented with a 1:100 ratio of 10% ammonium persulfate (APS) and a 1:1000 ratio of Tetramethylethylenediamine (TEMED) to promote polymerization, as well as streptavidin acrylamide for the subsequent capture of biotinylated antibodies.
Table 2.
Composition of PA-gels and measured Youngs’s modulus values.
| Measured E(kPa) | Acrylamide(%w/v) | Bis-acrylamide(%w/v) | Strep-Acryl concentration (%w/v) |
|---|---|---|---|
|
| |||
| 7.5 | 4 | 0.03 | 0.167 |
| 50 | 6.667 | 0.00667 | 0.333 |
| 140 | 6.667 | 0.5333 | 0.167 |
To activate T cells, the streptavidin-acrylamide-containing gels were coated with 1 μg/ml of biotinylated goat-anti-mouse for 2 hours at RT (Room Temperature), blocked with 5% BSA (Bovine Serum Albumin) for 2 hours at RT, and coated with a 50 μg/ml cocktail of 1:4 of anti-CD3 (clone OKT3) and anti-CD28 (clone 9.3) in PBS overnight at 4°C. Gel surface coating concentrations were evaluated using 10 μg/ml of OKT3 Alexa Fluor488 conjugated antibody, and confocal microscopy was used to capture the distribution and intensity of the conjugated antibody.
PA gel stiffness characterization.
The Young’s modulus (E) of prepared gels was measured by indentation [15]. Slabs of polyacrylamide gels were deformed with a flat cylindrical head and using a calibrated mass. The material’s Young’s modulus was estimated from the head diameter (D, 12mm), deflection (h), weight (m), gravitational constant (g), and Poisson ratio (v) of 0.5 assuming Hertzian contact.
Cell isolation and culture.
PBMCs (Peripheral Blood Mononuclear Cells) were isolated from Leukopaks using a Ficoll density gradient centrifugation technique. CD4+CD25− T cells were isolated from PBMC with a human CD4+CD25+ Regulatory T Cell Isolation Kit from Miltenyi using a two-step negative isolation where non-CD4 cells were depleted and then CD25+ cells were depleted. CD4+CD25− T cells were then frozen in 40% FBS (Fetal Bovine Serum) and 10% DMSO in complete cell culture media. The complete cell culture media consists of RPMI 1640 supplemented with 10 mM HEPES, 10 mM L-glutamine, 10% FBS, 0.34% β-mercaptoethanol, and 10 mM penicillin-streptomycin. Standard culture conditions were 37°C, humidified 5% CO2/balance air environment.
Treg induction.
500k cells/ml of CD4+CD25− T cells were seeded on coated PA-gels with 10 ng/ml of IL-2 and TGF-β and 1 mM of sodium pyruvate and MEM non-essential amino acids for 72 hours [5]. Dynabeads and unstimulated samples served as the positive and the negative control, respectively. After incubation, T cells were harvested and then permeabilized with a True-Nuclear™ Transcription Factor Buffer Set following the manufacturer’s protocol.
Staining and flow cytometry.
FOXP3 expression was assessed with Alexa Fluor 488 anti-human FOXP3 antibody following the manufacturer’s protocol with a staining concentration of 1:20 at RT for 30 minutes. The mitochondrial potential was measured with MitoTracker Red CMXRos in a final concentration of 250 nM at 37°C for 30 minutes. CD25 expression was detected with anti-human APC REAfinity CD25 antibody (manufacturer’s protocol 1:50) at RT for 30 minutes. The live Tregs were stained with CD4 antibody in BV421 (1:20), CD25 antibody in APC (1:50), and CD127 antibody in FITC (1:20) at 4°C for 20 minutes before sorting. Data were collected using a FACSCanto II. Live sorting of (CD4+CD25+CD127−) Tregs was performed with BB Influx cell sorter. All analysis was performed on FCS Express V6 (De Novo). Dead cells were excluded only from analysis by forward/side scatter gating. Doublets were excluded as well. The rest of cells were then used for further analysis.
Treg inhibitor studies.
Blebbistatin was dissolved in DMSO, and a final concentration of 40 μM was used during seeding. A final concentration of 250 μM AICAR in deionized water was added 18 hours after cell seeding. Oligomycin was also dissolved in DMSO and added at 0 hour, 24 hours, and 48 hours after activation (2.5 ng/ml). DMSO concentration was maintained at 0.1% for all conditions and DMSO control.
Treg suppressive assay.
T cells from day 3 were continued in culture supplemented with IL-2 (no TGF-β). On day 6, live cells were collected to sort for the CD4+CD25+CD127− population with BB Influx. After overnight resting in 10 ng/ml of IL-2, Treg suppressive assay was set up according to an established protocol [17]. Tresponders (Conventional T cells (Tconvs)) were thawed on day 6 and rested overnight as well. Tregs/Tresponders ratios of 1:1, and 1:2 were utilized to assess Treg functionality. Tregs were counted and suspended in 1 million cells/ml. The initial concentration of 50k Tregs in 100 μL was seeded on a 96 well-round bottom plate, then serial dilution was performed with 50 μL of media. 25k cells of Tconvs stained with CSFE in 50 μL were added to each well. 100 μL of Dynabeads in media were then included in the culture, generating a 1:3 ratio of Tconvs to Dynabeads. The Tconvs stimulated with Dynabeads sample was used for T cell activation control, and the Tconvs sample alone was used as unstimulated control. Dynabeads were detached after 3 days of induction, then flow cytometry was performed with BD CantoII to measure CSFE fluorescence intensity. The percent suppression was analyzed with FCS express by creating a marker excluding unstimulated control and calculated based on an established protocol [18].
Single-cell RNA-Seq.
Single-cell RNA-Seq was performed on cells harvested from 7.5 kPa and 140 kPa with 2 replicates each. 4 samples were barcoded and processed together (10X Genomics), and libraries were constructed with Single-cell 3’ v2 chemistry. The completed library was sequenced on a HiSeq 2500. Data were analyzed using the Cell Ranger 3.0.2 by aligning reads to the GRCh38 genome. There are 4148 cells with 2591 median genes per cell.
Computational Analysis.
Analyses of scRNA-Seq data were performed with the Seurat package in R [19]. Cells with low-quality transcriptomes (< 500 detected genes), doublets (>8000 genes), and dead/stressed cells (10% >percent mitochondrial counts > 0.2%) were removed from the analysis. After filtering, there are 4005 cells and 33538 genes detected. Unsupervised cell clustering was conducted using the FindClusters() function from Seurat. The resolution was determined based on Silhouette scores, and a 0.1 resolution gave rise to the highest silhouette score of 0.593. Cluster annotations were determined based on Treg markers (IL2RA+(CD25+), FOXP3+, ITGAE+(CD103+)). Differential expression analysis was performed on Tregs and Tconvs respectively between hard and soft samples with FindMarkers (). All the differential expression analyses utilized the Wilcoxon rank-sum test. Results were filtered with an adjusted p-value below 0.05. Volcano plots were generated with the package, EnhancedVolcano. Over-representation analysis was carried out with enrichGO (ont = “BP”) using clusterProfiler in R [20]. Trajectory analysis was performed with SCORPIUS with annotation from Seurat [21]. Genes contributed to Treg induction were determined with the function gene_importance (). Adjusted p-values were calculated with Benjamini-Hochberg correction, and genes were filtered with an adjusted p-value less than 0.05.
Results
Induction of regulatory T cells on polyacrylamide gels is stiffness-dependent
The ability of human CD4+CD25− T cells to sense the mechanical properties of an activating substrate in Treg induction was examined using polyacrylamide gels (Fig. 1A). The Young’s modulus (E) of these substrates ranged from 7.5 kPa to 140 kPa (Fig. 1B), covering a physiological range of stiffness representing liver to bone tissue [22–24]. These gels incorporated an acryl-modified streptavidin to allow subsequent binding of a biotinylated goat anti-mouse antibody, which then captured antibodies OKT3 and 9.3 that bind to and activate CD3 and CD28, respectively. An indirect binding approach allows immobilization of OKT3 and 9.3 without biotinylation of these antibodies, which could affect protein function. The amount of primary antibody (OKT3) presented on each surface was normalized by controlling the concentration of streptavidin acrylamide as shown in Fig. S1A [13, 15].
Figure 1.
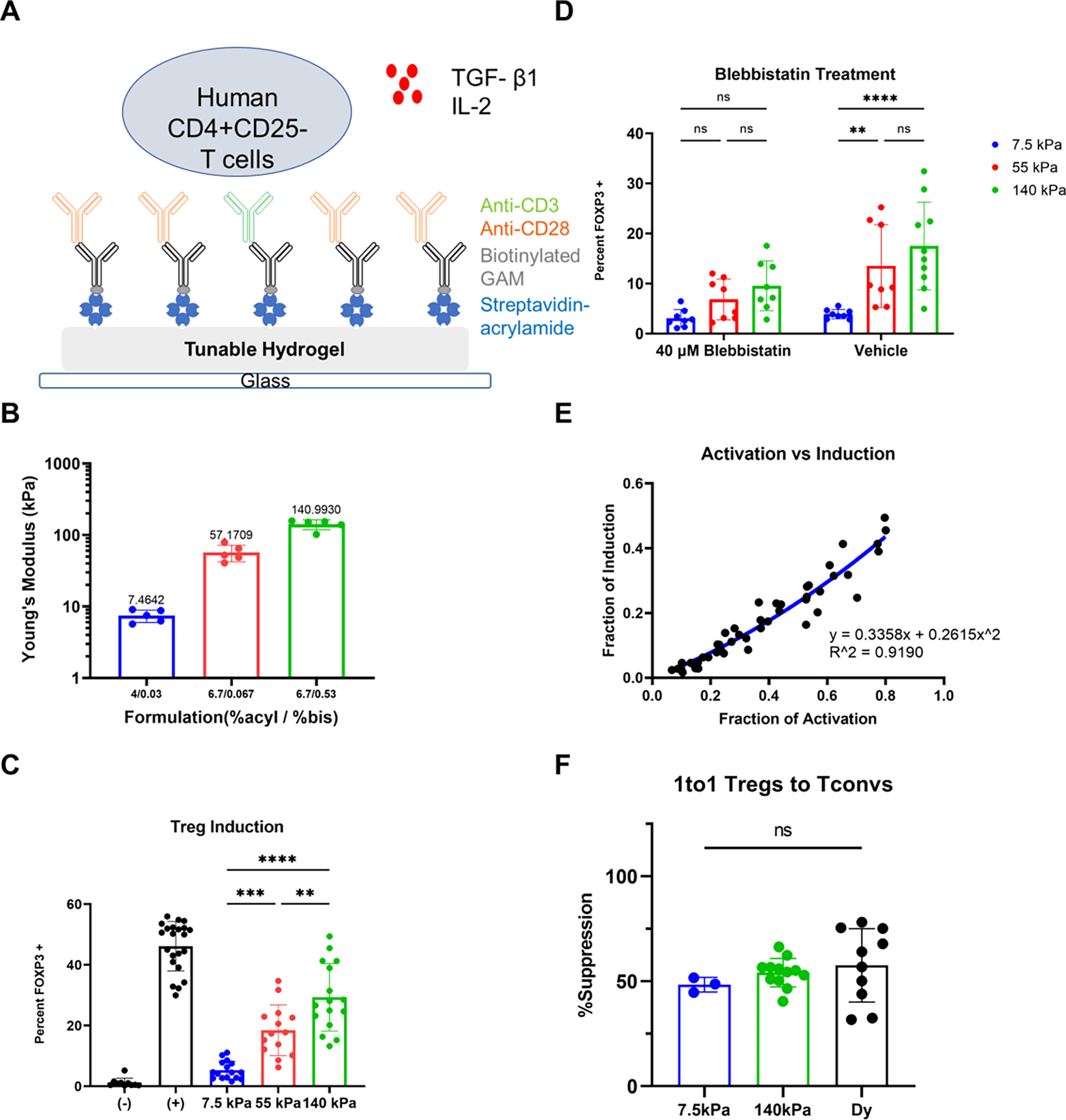
Effect of substrate stiffness on Treg induction. (A) Schematic of PA-gels coated with activating antibodies as Treg induction platform. (B) Mechanical testing via indentation indicates PA-gels with varying ratios of acrylamide and bis-acrylamide (n = 5 gels). (C) Treg induction on PA-gels with different stiffness (7 individual experiments and 2 replicates each with 4 donors, one-way ANOVA, ** p < 0.01, *** p < 0.001, **** p<0.0001). “+” corresponds to Dynabeads positive control, whereas “- “means unstimulated negative control without activating antibodies and cytokines. (D) Effect of blebbistatin on mechanosensing of Treg induction. 40 μM blebbistatin was introduced at the beginning of the culture (4 individual experiments and 2 replicates each, two-way ANOVA with Tukey’s multiple comparisons, ** p < 0.01, **** p<0.0001). (E) The correlation between activation and induction (data from 7.5 kPa, 55 kPa, and 140 kPa in (C), and the second-order polynomial fit was identified with the Extra sum-of-squares F test (α = 0.05). (F) Treg suppressive capacity represented as percent suppression with 1to1 ratio of Tregs to Tconvs (5 independent experiments, and the number of replicates depends on the number of cells obtained after sorting, Dy- Dynabeads, one-way ANOVA)
Treg induction was promoted by supplementing media with IL-2 and TGF-β1 (Fig. 1A) and measured as the percentage of cells that were FOXP3+ on day 3 [25]. Most prominently, Treg induction was directly correlated with substrate stiffness demonstrating that this process is mechanosensitive (Fig. 1C). Intriguingly, reducing the concentration of streptavidin in the gels (and thus the amount of OKT3 and 9.3) to 10% of the standard condition yielded a significant difference between 7.5 kPa and 55 kPa but not between 55 kPa and 140 kPa (Fig. S1C). Reducing the amount of activating antibodies leads to a more biphasic mechanosensing response, in keeping with an earlier report examining the activation and expansion of T cells [15]. Decreasing the concentration of streptavidin by an additional factor of 10 further lowered both activation and induction. To confirm that T cells were responding to mechanical aspects of the substrates, actomyosin contractility was inhibited using the myosin II inhibitor, blebbistatin. Inhibition of actomyosin contractility in this manner abrogated differences in FOXP3 expression as a function of substrate rigidity (Fig. 1D), demonstrating the significance of actomyosin contractility and mechanosensing in Treg induction.
Notably, Treg induction increased monotonically with cell activation (percentage of cells showing increased size and granularity, Fig. S2A), as shown in Fig. 1E. This relationship between induction and activation was fit significantly better by a second-order polynomial than a purely linear relationship (P= 0.0006, Extra sum-of-squares F test), indicating that the observed impact of substrate stiffness on induction is not simply explained by an increase in activation. In addition, the expression of CD25 also exhibited a mechanosensing response (Fig. S2C), and a higher percentage of the FOPX3+ population was observed in the stiffer substrate (Fig. S2D). Analysis of induction vs. the T cell activation marker CD25 showed a similar result (Fig. S2E), as well as for the data from blebbistatin treatment (Fig. S2F). Interestingly, blebbistatin treatment reduced the correlation between activation and induction, and induction reached a plateau as activation increased, thus abrogating the difference in mechanosensing response on Treg induction.
Finally, the longer-term functionality of induced Tregs was then tested through suppression assays using cells that were expanded for an additional 3 days. FOXP3 expression at this 6-day time point was higher than at 3 days and greater on 110 kPa vs. 7.5 kPa substrates (Fig. S3A). While an earlier report revealed a transient increase and mechanosensitive response of FOXP3 gene transcription in conventional T cells during activation [23], the 3-day response observed under suppressive conditions was maintained over longer time points. Live Tregs, identified as CD4+CD25+CD127− (of which typically > 60% expressed FOXP3), were tested for inhibition of Dynabeads-stimulated Tconvs. Tregs induced on PA-gels exhibited similar suppressive capacities as Tregs induced with Dynabeads (Fig. 1F) when mixed 1:1 with Tconvs. Tconvs that were stimulated in the absence of Tregs served as positive controls and exhibited greater proliferation. No difference in per-cell suppressive capacity was observed between Tregs induced using PA-gels or Dynabeads showing equal functionality across these substrates. This functionality was also observed at lower Treg: Tconv ratios for cells activated on PA-gels of different stiffness (Fig. S3B). Therefore, varying the stiffness of PA-gels can generate different numbers of Tregs with similar suppressive capacities. To identify additional mechanisms that influence Treg induction, we next conducted transcriptional analysis on Tregs induced on substrates of different stiffness.
Genes in OXPHOS are upregulated in Tregs on the stiffer gel and increased during Treg induction
Single-cell RNA-Seq (scRNA-Seq) was conducted on cells that were activated for three days using the substrates that produced the largest difference in induction (7.5 kPa and 140 kPa), denoted as soft and hard in the figures. The resultant transcriptional data was projected in a UMAP (Fig. 2A), then the clustering analysis was performed using the Seurat package, revealing 2 main populations. The top 10 markers for each cluster were plotted in Fig. S4A. Treg positive markers including FOXP3, IL2RA, and ITGAE were associated with the population on the right, while the Treg negative marker IL7A was not (Fig. 2B). Thus, the cluster on the right was denoted as Tregs, while the counterpart on the left was denoted as Tconvs. Tconvs are CD4+ T cells that are not enriched for Treg markers.
Figure 2.
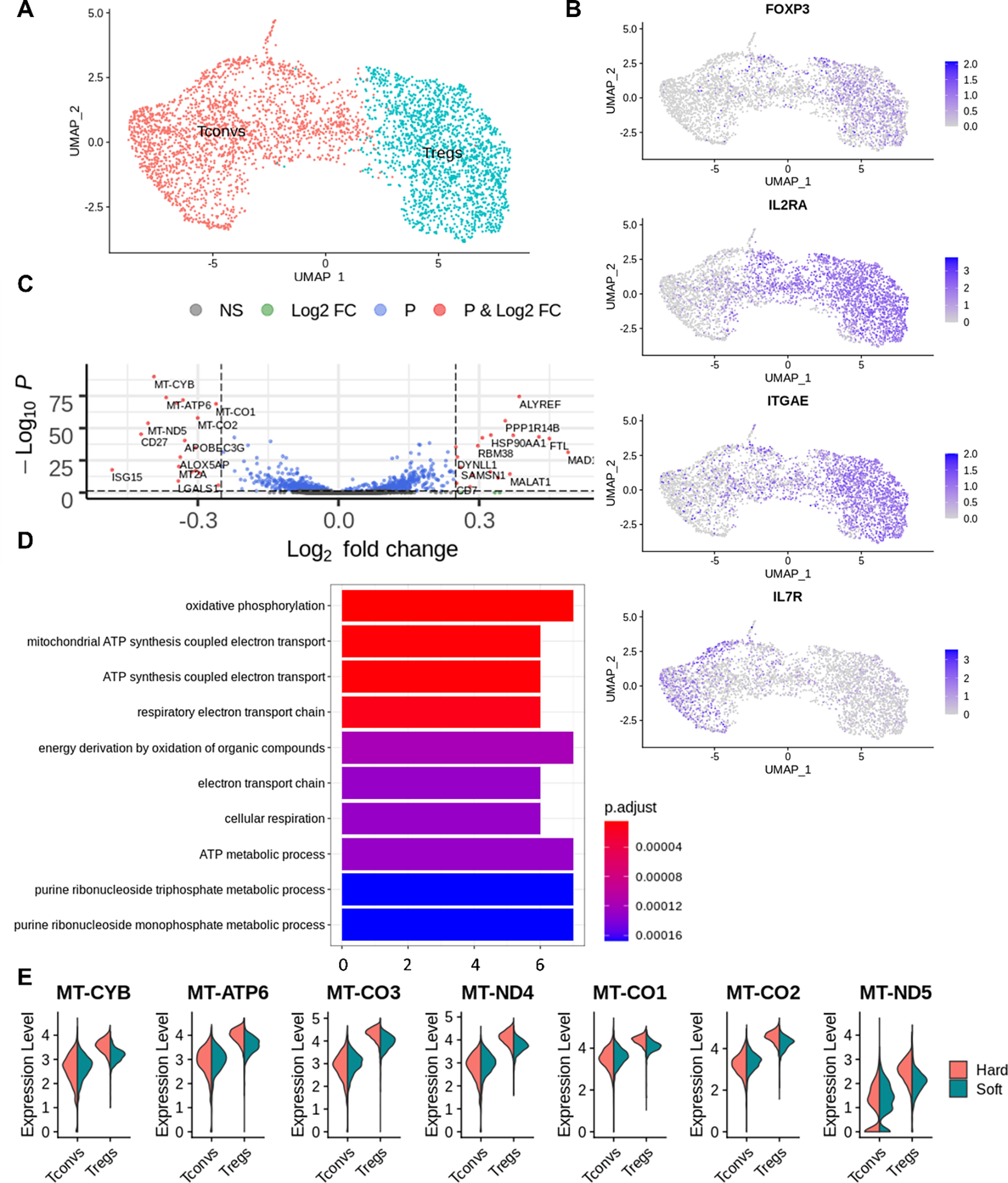
Clustering and differential expression analysis of Tregs with Seurat. (A) Cells were clustered into 2 populations in Seurat. (B) FOXP3, IL2RA, ITGAE, IL7R expression on both hard and soft samples’ UMAP projection. (C) Volcano plot of differentially expressed genes from Tregs comparing the stiffer sample to the softer sample. (D) Over-representation analysis of differential expressed genes from Tregs when comparing the soft and hard samples. (E) Violin plot of differentially expressed genes from (C) involved in oxidative phosphorylation.
Differential expression analysis was then conducted to compare Tregs from the hard and the soft substrates, identifying 37 differentially expressed genes (Fig. 2C). Seven (upregulated on the hard substrate) out of these genes were mitochondrial genes that encode for proteins involved in oxidative phosphorylation (Fig. 2E). In addition, over-representation analysis revealed that oxidative phosphorylation pathway was upregulated on Treg population induced on the harder substrate relative to Tregs induced on the softer substrate (Fig. 2D). The same analysis was carried out to compare Tconvs between the hard and the soft substrates, yielding 143 differentially expressed genes (Fig. S4D). Furthermore, the expression of mitochondrial genes (including OXPHOS genes) was higher in the Treg population relative to the Tconv population. Thus, OXPHOS could play a role in the mechanosensing of Treg induction, as it is upregulated in the process of Treg induction, and differentially expressed on Tregs between substrates.
T cell metabolic remodeling occurs before the completion of Treg induction
While scRNA-Seq analysis identified two populations, Tconvs and Tregs were not fully separated on the projected UMAP, suggesting a progression from conventional T cells to regulatory T cells. To characterize this progression in detail we applied SCORPIUS, a trajectory inference algorithm [21]. This analysis identified a linear trajectory from Tconvs to Tregs (Fig. 3A) and 347 genes that changed along this transition (genes of importance). These genes were then grouped into 6 different modules by the algorithm (Fig. 3C). Genes in module 1 are upregulated in Tconvs and enriched for translational and ribosomal activity. The rest of the modules are all upregulated in Tregs. Module 2, which shows the greatest increase over the trajectory, contains gene-linked ATP metabolic processes, glycolysis, and oxidative phosphorylation. Modules 3 and 4 are associated with mitochondrial gene expression and translation. Modules 5 and 6 are involved in cell cycle. IL2RA and ITGAE are from modules 3 and 4, respectively. The expressions of Treg markers are visualized in the embedding of SCORPIUS. FOXP3, IL2RA, and ITGAE were expressed on the Treg population (Fig. 3B). Finally, plotting the distribution of cells along pseudotime reveals two distinct peaks for the hard sample, as opposed to flatter peaks with more populated cells in the intermediate state for the soft sample (Fig. 3D).
Figure 3.
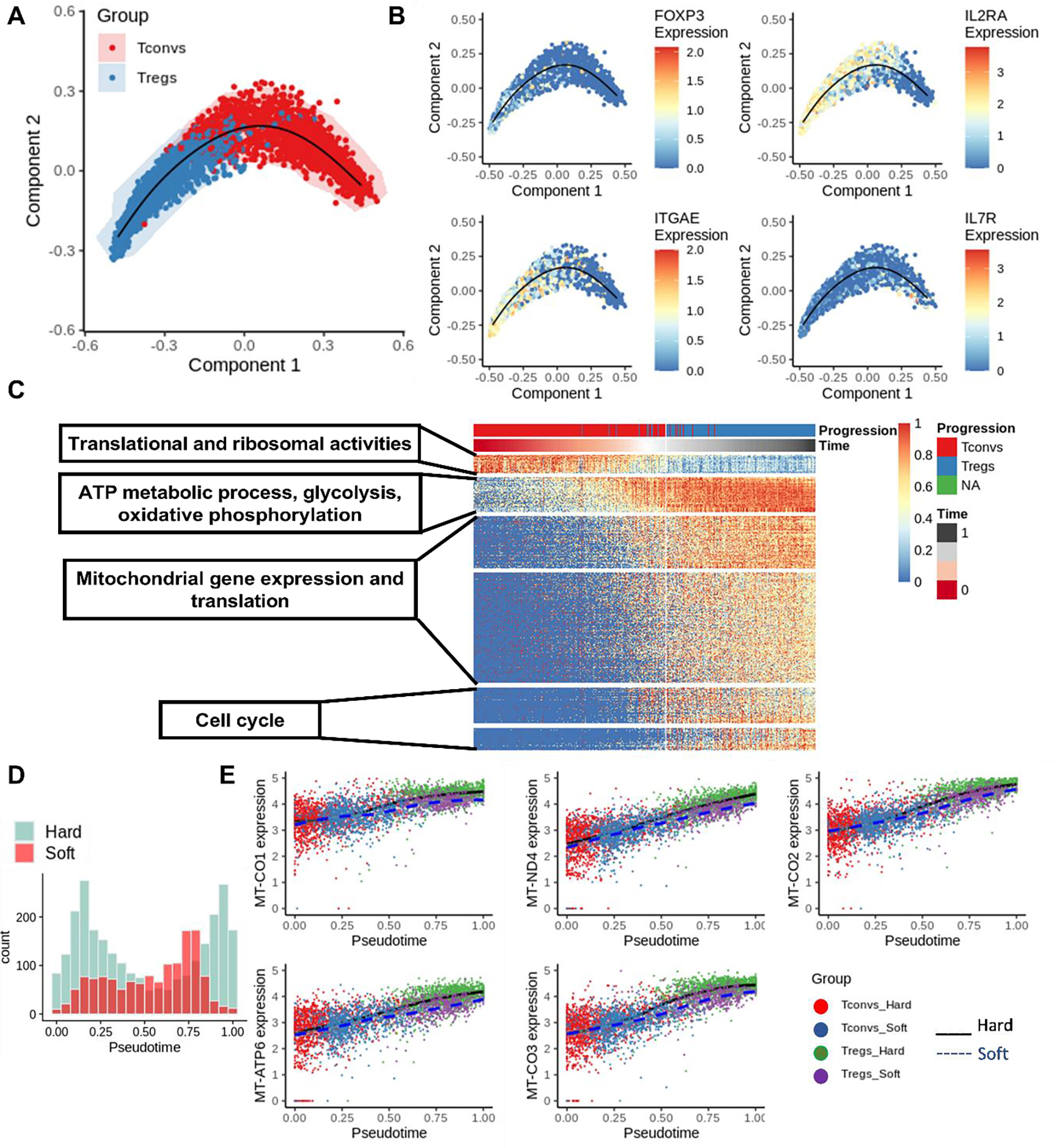
Trajectory analysis of Treg induction with SCORPIUS. (A) Trajectory analysis was performed with SCORPIUS with T cells from both hard and soft samples combined. (B) Treg signatures were plotted on the embedding from SCORPIUS. (C) Important genes that contribute to Treg induction were organized into different modules (p.adjust <0.05). (D) Histogram of cells along the trajectory on soft and hard samples, respectively. (E) The expression of mitochondrial genes was plotted along with the pseudotime projection.
To further understand the mechanosensing of Treg induction, we compared differentially expressed genes (Fig. 2C) and genes that contributed to Treg induction (Fig. 3C). Ten genes were identified, and 5 out of these 10 genes (MTND4, MTATP6, MTCO3, MTCO2, MTCO1) were involved in the pathway of OXPHOS (in Module 2). Expression of these 5 genes along the Treg induction trajectory is visualized in a single cell level, comparing hard and soft samples, in Fig. 3E. Differences in mitochondrial gene expression start at approximately 0.3 pseudotime, whereas the cutoff between Tregs and Tconvs is approximately 0.5 pseudotime. This potentially indicates that stiffness-mediated T cell metabolic remodeling occurs before the completion of Treg induction. The modulation of metabolic activity can potentially lead to a different level of induction.
The impact of OXPHOS on Treg induction is stiffness-dependent
Our scRNA-Seq data support the current understanding that oxidative phosphorylation is important for the mechanosensing of Treg induction. To confirm this finding, the final stages of OXPHOS were inhibited using oligomycin. The introduction of oligomycin at the early time point significantly reduced the percent of FOXP3 expressing cells and eliminated the difference in Treg induction among gels. Interestingly, introducing this compound at a later time point did not affect the rate of Treg induction (Fig. 4A). These results suggest that metabolic regulation occurs at an early stage, which agrees with findings from our previous trajectory analysis.
Figure 4.
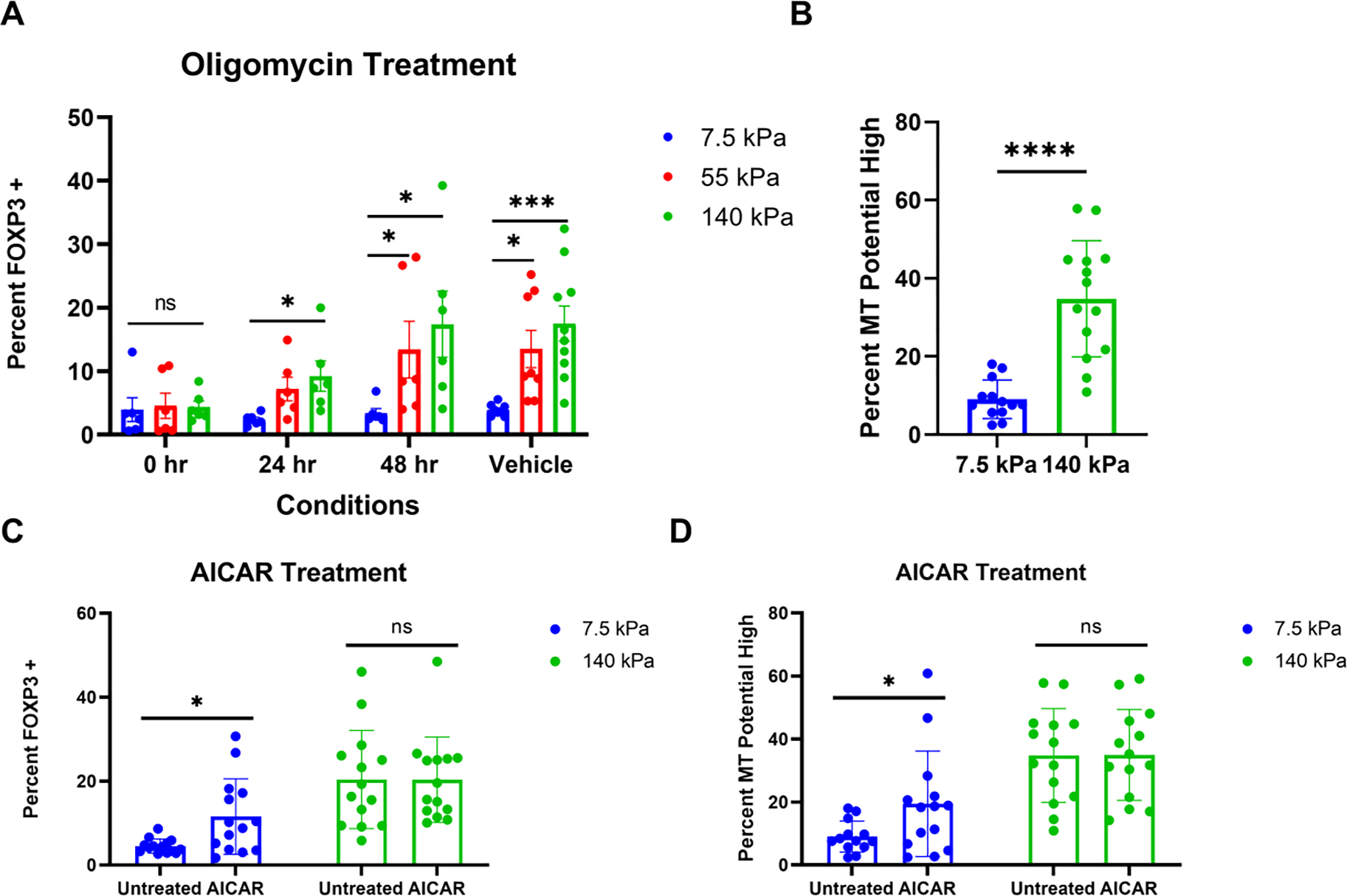
Stiffness-dependent oxidative phosphorylation modulates Treg induction. (A) Effect of OXPHOS inhibition on Treg induction. Oligomycin was added to the culture media at 0 hour, 24 hours, and 48 hours after seeding. For the Vehicle control, DMSO was added in the culture at the 0 hour time point (3 individual experiments and 2 replicates each, non-parametric, Kruskal-Wallis test with Dunn’s multiple comparisons was used for comparison between gels for each time point/vehicle, * p < 0.05, *** p < 0.001). (B) Mitochondrial potential on PA-gels. 72 hours after induction, cells were treated with MitoTracker Red, and mitochondria potential high is measured relative to the stained unstimulated control (7 individual experiments and 2 replicates each with 4 donors, parametric, unpaired t-test, **** p < 0.0001). (C, D) Effect of OXPHOS on mechanosensing of Treg induction. 250 μM of AICAR were introduced in the culture at 18 hours of the induction (7 individual experiments and 2 replicates each with 4 donors, one outlier found with the ROUT method in D was excluded, non-parametric, Mann-Whitney test was used to compare untreated to AICAR for each substrate, * p < 0.05).
As a complementary approach to understanding OXPHOS in mechanosensitive Treg induction, AICAR (5-Aminoimidazole-4-carboxamide ribonucleotide) was used to upregulate AMP-activated protein kinase by mimicking AMP thus promoting fatty acid uptake and increasing OXPHOS [26, 27]. Treatment with AICAR significantly increased Treg induction on the softer sample but not on the harder counterpart. In addition, the rate of Treg induction on AICAR-treated soft samples is still lower than on untreated stiffer samples (Fig. 4C). This means mechanical cues could be more effective in regulating metabolic activity than chemical modulators could. The impact of AICAR is metabolic-dependent since a higher percentage of cells with upregulated mitochondrial potential was only observed on the softer substrate (Fig. 4D). Mitochondria potential across the inner membrane is necessary for oxidative phosphorylation [28]. Cells on the harder sample also exhibited elevated mitochondrial potential relative to the softer substrate (Fig. 4B). Thus, the softer substrate may not properly regulate the metabolic remodeling that is essential for Treg induction. In addition, the modulation of metabolic activity can further control Treg induction.
Discussion
Engineering of materials used in production of T cells has great potential for improving cellular immunotherapy. This report leverages the ability of T cells to respond to the mechanical rigidity of a substrate toward improving Treg induction. Building on previous reports of stiffness-dependent T cell activation and expansion [13–15, 23, 29], this report demonstrates that Treg induction is sensitive to the stiffness of polyacrylamide hydrogels providing a simple material parameter that can be adjusted to improve cell production. Moreover, the effect of stiffness on induction is not simply proportional to changes in activation (Fig. 1E), indicating that additional processes impact Treg generation. This is further supported by single-cell transcriptomic analysis which shows that activation using a stiffer substrate promotes higher use of OXPHOS and stronger differentiation during induction (Fig. 2E, Fig. 3E).
These stiffness-induced changes are part of a larger metabolic program that is central to the T cell response. Resting T cells are normally associated with low metabolic function. Upon activation, T cells upregulate metabolic pathways to accommodate the increase in energy use and biosynthesis [30]. In response to mechanical forces, cells increase ATP production to provide energy to reinforce the actin cytoskeleton so that cells can resist physiological forces. Mitochondria were also recruited to immune synapse during T cell activation to generate local ATP to meet the energy demand [31, 32]. While mitochondrial oxidative metabolism and OXPHOS are more efficient in producing energy, glycolysis more quickly responds to meet energy demand. Glycolysis consequently tends to support the function of pro-inflammatory cells such as effector T cells and M1 macrophages, while OXPHOS and fatty acid oxidation tend to be used by anti-inflammatory cells including M2 macrophages and Tregs [30, 33, 34]. In addition, it has been reported that stiffer substrates induced a higher rate of oxidative phosphorylation [32]. Thus, promoting OXPHOS on stiffer surfaces may encourage Treg induction.
The detailed mechanisms by which substrate stiffness modulates metabolism remain unclear. Key components of metabolic regulation in T cells include mTOR and AMPK. mTOR regulates cell proliferation, metabolism, and differentiation. AMPK is a fuel sensor that promotes the production of ATP. mTOR is regulated by the PI3K Akt pathway that is TCR signaling dependent. Activated AMPK has been shown to negatively regulate the mTOR pathway and promote mitochondrial biogenesis and OXPHOS capacity [27, 35–37]. The inhibition of mTOR in Tregs diminishes glucose uptake, promoting a stable Treg phenotype [30]. The results presented here suggest that mechanosensing in Treg induction is OXPHOS-dependent as the inhibition of ATP synthase reduced Treg induction, and AMPK activation increased the rate of Treg induction and mitochondrial potential on the soft substrate only.
We are not alone in discovering the link between mechanosensing and metabolic activity. Saitakis et al. also found that metabolism was associated with TCR-mediated stiffness-sensing response by studying T cell activation on polyacrylamide gels (0.5 kPa – 100 kPa) [23]. Our study focuses on the mechanosensing of Treg differentiation and demonstrated that the modulation of metabolism can also regulate T cell differentiation. Our single-cell RNA Seq data reveals that signal transduction by the p53 class mediator pathway only contributes to the trajectory of Treg induction on the soft sample, but not the hard sample (Fig. S5 E, G). Substrate stiffness was reported to influence p53 activities and ROCK2 expression that induces activation of myosin II [38], and p53 was found to regulate both metabolic gene and cell cycle genes. Thus, p53 could be another regulator for mechanosensing of Treg induction, however, in vitro studies need to be conducted to confirm this hypothesis and elucidate its detailed mechanism.
On the other hand, TCR-independent regulations on metabolism via mechanical cues were also reported by several studies. Meng et al. identified Yes-associated protein (YAP) as the mechanosensor that senses mechanical changes and controls metabolic reprogramming in mice T cells [39]. YAP reads a wide range of mechanical cues and translates them into cell-specific downstream signaling, and YAP activation is associated with actin contractility and increased tension of the actin cytoskeleton [40]. YAP-1 was also reported to promote Treg differentiation by upregulating TGFBR2, but the expression of YAP-1 was not detected in our transcriptomic data and TGFBR2 (upregulated in Tregs) did not show a mechanosensitive response (Fig. S6). Park et al. also demonstrated that the softer substrate induced downregulation of glycolysis via proteasomal degradation of PFK, which is linked to stress fiber via TRIM21 on epithelial cells [41]. Actomyosin stress fiber is a key component for cells to probe the mechanics of the extracellular environment through integrin-mediated focal adhesions [42, 43]. However, TRIM21, PFKP, PFKL, and PFKM (PFK genes) expression levels were very similar between the hard and soft substrates (Fig. S6). Thus, TCR-dependent and TCR-independent mechanosensing pathways could have different regulatory mechanisms. This interaction between mechanotransduction and metabolism is a rapidly emerging area of interest with implications across a wide range of diseases.
Mechanosensing affects how T cells manage their energy demand for proliferation and differentiation, which can also be leveraged to promote the production of Treg for adoptive therapy. The goal of this project is not to optimize the Treg induction protocol for clinical production, but to understand the mechanism of mechanosensing in Treg induction. To improve Treg production, it is necessary to optimize both chemical modulators and mechanical cues, including the concentration of polarizing molecules (TGF-β, IL-2, all-trans retinoic acid, rapamycin, and butyrate), metabolic modulators, activating substrates, the duration of TCR stimulation, and the concentration of activating antibodies. Thus, one approach can start with optimizing material stiffness for Treg induction combined with promoting OXPHOS through the use of pharmacological molecules. However, iTregs were reported to be not as stable as expanded tTregs [44], and iTreg suppressive function could be compromised in vivo [45]. The stability and functionality of iTregs should also be assessed by measuring Treg epigenetic profiles and suppressive capacities. Insights into how to generate a high quantity of functional and stable Tregs will be a huge step forward for generating easily accessible Treg therapeutic products.
Supplementary Material
Figure S1. (A) Streptavidin-acrylamide concentrations were varied to obtain a similar coating of antibodies on PA-gels, confirmed by fluorescence intensities on the surface of gels (n = 4 gels for each formulation, Kruskal-Wallis test with Dunn’s multiple comparisons). (B) Representative gating strategies for FOXP3 expression from Tregs induced on PA-gels. (C) Treg induction with various concentrations of streptavidin-acrylamide (4 independent experiments and 2 replicates each for 0.1X and 0.01X, two-way ANOVA with Tukey’s multiple comparisons * p < 0.05, *** p < 0.001, **** p < 0.0001). (D) A representative gating strategy for percent mitochondrial high cells for Fig. 4.
Figure S2. (A) Gating of activated T cells on the forward scattering and side scattering of flow cytometry data (B) Representative flow gating strategy for CD25+ population (APC-A, CD25). (C) Percent CD25+ on both 7.5 kPa and 140 kPa (3 independent experiments and 2 replicates each, Mann-Whitney test, ** p < 0.01). (D) Percent of CD25+ population in FOXP3+ population (3 independent experiments and 2 replicates each, One-way ANOVA, *** p < 0.001). (E) Fraction of CD25+ versus fraction of induction, measured as fraction of cells that are FOXP3+, (Extra sum-of-squares F test (α = 0.05), 3 independent experiments and 2 replicates each). (F) Activation vs induction of all the data from blebbistatin treatment and the second-order polynomial fit was identified with the Extra sum-of-squares F test (α = 0.05).
Figure S3. (A) Percent FOXP3+ on day 6 (7.5 kPa, and 140 kPa, Mann-Whitney test was used to compare gels on day 3 and day 6 individually, **** p < 0.0001). (B) Treg suppressive capacities with 1to2 ratio of Tregs to Tconvs (n = 5 independent experiments, and the number of replicates depends on the number of cells obtained after sorting, unpaired t-test). (C) Representative gating strategies for calculation of percent suppression with a 1to1 ratio of Tregs to Tconvs samples. Tconvs only is the positive control without Tregs with Dynabeads. (D) Representative CSFE(FITC-A) vs FSC-A density plots for 1to1 ratio of Tregs to Tconvs samples.
Figure S4. (A) Top 10 markers of Treg and Tconv populations. (B, C) Over-representation analysis of markers from Treg and Tconvs, respectively. (D) Volcano plot of differentially expressed genes from Tconvs comparing the stiffer sample to the softer sample. (E) Over-representation analysis of differential expressed genes from Tconvs when comparing the stiffer sample to the softer sample.
Figure S5. Trajectory analyses on either the hard or the soft samples. (A, B) Trajectory analyses were performed with SCORPIUS with T cells from either the hard (A) or the soft (B) sample separately. (C) Histogram of cells along the trajectory on either the hard or the soft sample. (D, F) Important genes that contribute to Treg induction were organized into modules for either the hard (D) or the soft (F) sample. (E, G) Over-representation analysis of genes uniquely identified in either the hard (E) or the soft (G) sample.
Figure S6. (A) Violin plots of SMAD2, YAP1, TAZ, SMAD4, TGFBR2, TRIM21, PFKP, PFKM, and PFKL.
Table 1.
Reagents and resources.
| REAGENT | SOURCE | IDENTIFIER | City | Country |
|---|---|---|---|---|
|
| ||||
| Biotinylated Goat-Anti-Mouse | Biolegend | 405303 | San Diego | CA |
| Mouse-Anti-Human CD3 (Clone, OKT3) | Bioxcell | Be0001-2 | Lebanon | NH |
| Mouse-Anti-Human CD28 (Clone, 9.3) | Bioxcell | Be0248 | Lebanon | NH |
| Leukopak | New York Blood Center | New York | NY | |
| Acrylamide (40%) | Sigma-Aldrich | 01697 | St. Louis | MO |
| Bis-Acrylamide (2%) | Fisher Scientific | BP1404-250 | Hampton | NH |
| Ammonium Persulfate (APS) | Sigma-Aldrich | A3678 | St. Louis | MO |
| Tetramethylethylenediamine (TEMED) | Sigma-Aldrich | T7024 | St. Louis | MO |
| Streptavidin-Acrylamide | ThermoFisher | S21379 | Waltham | MA |
| Blebbistatin | STEMCELL | 72402 | Vancouver | Canada |
| AICAR | Cell Signaling Technology | 9944 | Danvers | MA |
| Oligomycin | MilliporeSigma | O4876 | Burlington | MA |
| Alexa Fluor 488 anti-human FOXP3 Antibody (Clone, 206D) | BioLegend | 320111 | San Diego | CA |
| CD25 Antibody, anti-human, REAfinity APC (Clone, REA570) | Miltenyi Biotec | 130-113-284 | Bergisch Gladbach | Germany |
| CD3 Mouse anti-Human, Alexa Fluor 488, Clone OKT3 | eBioscience | 50-112-9566 | Waltham | MA |
| MitoTracker Red CMXRos | ThermoFisher | M7512 | Waltham | MA |
| RPMI 1640 | ThermoFisher | 21870092 | Waltham | MA |
| HEPES Buffer | ThermoFisher | 15630 | Waltham | MA |
| L-Glutamine | ThermoFisher | 25030 | Waltham | MA |
| Fetal Bovine Serum (FBS) | ThermoFisher | Gibco-Performance 26140-079 | Waltham | MA |
| β -Mercaptoethanol | Sigma-Aldrich | M3148 | St. Louis | MO |
| Penicillin-Streptomycin | ThermoFisher | 15140122 | Waltham | MA |
| Ficoll-Paque Plus | Cytiva | 17144002 | Marlborough | MA |
| CD4+CD25+Regulatory T Cell Isolation Kit | Miltenyi Biotec | 130-091-301 | Bergisch Gladbach | Germany |
| IL-2 | PeproTech | 200-02 | East Windsor | NJ |
| TGF- β1 | PeproTech | 100-21C | East Windsor | NJ |
| Sodium Pyruvate (100 Mm) | ThermoFisher | 11360070 | Waltham | MA |
| MEM Non-Essential Amino Acids Solution (100X) | ThermoFisher | 11140050 | Waltham | MA |
| True-Nuclear™ Transcription Factor Buffer Set | BioLegend | 424401 | San Diego | CA |
| Dynabeads | ThermoFisher | 11132D | Waltham | MA |
| CSFE | ThermoFisher | C34554 | Waltham | MA |
| FITC anti-human CD127 antibody (Clone, A019D5) | Biolegend | 351312 | San Diego | CA |
| BV421 anti-human CD4 antibody (Clone, OKT4) | Biolegend | 317434 | San Diego | CA |
Acknowledgments
This work was supported in part by the National Institutes of Health (U24AI118669 and R01AI110593 to L.C.K.). This material is based upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. (NSF DGE- 2036197 to L. S.). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. These studies used the resources of the Herbert Irving Comprehensive Cancer Center Flow Cytometry Shared Resources funded in part through Center Grant P30CA013696. This research was funded in part through the NIH/NCI Cancer Center Support Grant P30CA013696 and used the Genomics and High Throughput Screening Shared Resource. This publication was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant Number UL1TR001873. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We would like to express our great appreciation to Professor Elham Azizi for her valuable and constructive suggestions for the single-cell RNA Seq analysis. Lastly, we appreciate Dr. Dennis J. Yuan’s fruitful discussion throughout this project and insightful input toward this work.
Footnotes
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data availability
The data that support the findings of this report are available from the authors upon reasonable request.
References
- 1.Cools N, et al. , Regulatory T cells and human disease. Clin Dev Immunol, 2007. 2007: p. 89195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bluestone JA, et al. , Type 1 diabetes immunotherapy using polyclonal regulatory T cells. Sci Transl Med, 2015. 7(315): p. 315ra189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lan Q, et al. , Induced Foxp3(+) regulatory T cells: a potential new weapon to treat autoimmune and inflammatory diseases? J Mol Cell Biol, 2012. 4(1): p. 22–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanamori M, et al. , Induced Regulatory T Cells: Their Development, Stability, and Applications. Trends Immunol, 2016. 37(11): p. 803–811. [DOI] [PubMed] [Google Scholar]
- 5.Nataraj NM, et al. , Ex vivo induction of regulatory T cells from conventional CD4(+) T cells is sensitive to substrate rigidity. J Biomed Mater Res A, 2018. 106(12): p. 3001–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tone Y, et al. , Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol, 2008. 9(2): p. 194–202. [DOI] [PubMed] [Google Scholar]
- 7.Vent-Schmidt J, et al. , The role of FOXP3 in regulating immune responses. Int Rev Immunol, 2014. 33(2): p. 110–28. [DOI] [PubMed] [Google Scholar]
- 8.Romano M, et al. , Past, Present, and Future of Regulatory T Cell Therapy in Transplantation and Autoimmunity. Front Immunol, 2019. 10: p. 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong N, et al. , Antigen-specific transforming growth factor beta-induced Treg cells, but not natural Treg cells, ameliorate autoimmune arthritis in mice by shifting the Th17/Treg cell balance from Th17 predominance to Treg cell predominance. Arthritis Rheum, 2012. 64(8): p. 2548–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haribhai D, et al. , A requisite role for induced regulatory T cells in tolerance based on expanding antigen receptor diversity. Immunity, 2011. 35(1): p. 109–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacMillan ML, et al. , First-in-human phase 1 trial of induced regulatory T cells for graft-versus-host disease prophylaxis in HLA-matched siblings. Blood Adv, 2021. 5(5): p. 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mescher MF, Surface contact requirements for activation of cytotoxic T lymphocytes. J Immunol, 1992. 149(7): p. 2402–5. [PubMed] [Google Scholar]
- 13.Judokusumo E, et al. , Mechanosensing in T lymphocyte activation. Biophys J, 2012. 102(2): p. L5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Connor RS, et al. , Substrate rigidity regulates human T cell activation and proliferation. J Immunol, 2012. 189(3): p. 1330–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan DJ, Shi LT, and Kam LC, Biphasic response of T cell activation to substrate stiffness. Biomaterials, 2021. 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, et al. , Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur J Immunol, 2007. 37(1): p. 129–38. [DOI] [PubMed] [Google Scholar]
- 17.Collison LW and Vignali DA, In vitro Treg suppression assays. Methods Mol Biol, 2011. 707: p. 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMurchy AN and Levings MK, Suppression assays with human T regulatory cells: a technical guide. Eur J Immunol, 2012. 42(1): p. 27–34. [DOI] [PubMed] [Google Scholar]
- 19.Stuart T, et al. , Comprehensive Integration of Single-Cell Data. Cell, 2019. 177(7): p. 1888–1902 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu G, et al. , clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS, 2012. 16(5): p. 284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cannoodt R, et al. , SCORPIUS improves trajectory inference and identifies novel modules in dendritic cell development. bioRxiv, 2016: p. 079509. [Google Scholar]
- 22.Huang G, et al. , Engineering three-dimensional cell mechanical microenvironment with hydrogels. Biofabrication, 2012. 4(4): p. 042001. [DOI] [PubMed] [Google Scholar]
- 23.Saitakis M, et al. , Different TCR-induced T lymphocyte responses are potentiated by stiffness with variable sensitivity. Elife, 2017. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells RG, Tissue mechanics and fibrosis. Biochim Biophys Acta, 2013. 1832(7): p. 884–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt A, et al. , Time-resolved transcriptome and proteome landscape of human regulatory T cell (Treg) differentiation reveals novel regulators of FOXP3. BMC Biol, 2018. 16(1): p. 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gualdoni GA, et al. , The AMP analog AICAR modulates the Treg/Th17 axis through enhancement of fatty acid oxidation. FASEB J, 2016. 30(11): p. 3800–3809. [DOI] [PubMed] [Google Scholar]
- 27.Brandauer J, et al. , AMP-activated protein kinase controls exercise training- and AICAR-induced increases in SIRT3 and MnSOD. Front Physiol, 2015. 6: p. 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hroudova J and Fisar Z, Control mechanisms in mitochondrial oxidative phosphorylation. Neural Regen Res, 2013. 8(4): p. 363–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dang A, et al. , Enhanced activation and expansion of T cells using mechanically soft elastomer fibers. Adv Biosyst, 2018. 2(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kempkes RWM, et al. , Metabolic Pathways Involved in Regulatory T Cell Functionality. Front Immunol, 2019. 10: p. 2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quintana A, et al. , T cell activation requires mitochondrial translocation to the immunological synapse. Proc Natl Acad Sci U S A, 2007. 104(36): p. 14418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papalazarou V, et al. , The creatine-phosphagen system is mechanoresponsive in pancreatic adenocarcinoma and fuels invasion and metastasis. Nat Metab, 2020. 2(1): p. 62–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerriets VA, et al. , Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest, 2015. 125(1): p. 194–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie D, et al. , Foxp3 drives oxidative phosphorylation and protection from lipotoxicity. JCI Insight, 2017. 2(3): p. e89160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agarwal S, et al. , AMP-activated Protein Kinase (AMPK) Control of mTORC1 Is p53- and TSC2-independent in Pemetrexed-treated Carcinoma Cells. J Biol Chem, 2015. 290(46): p. 27473–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaube B, et al. , AMPK maintains energy homeostasis and survival in cancer cells via regulating p38/PGC-1alpha-mediated mitochondrial biogenesis. Cell Death Discov, 2015. 1: p. 15063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winder WW, et al. , Activation of AMP-activated protein kinase increases mitochondrial enzymes in skeletal muscle. J Appl Physiol (1985), 2000. 88(6): p. 2219–26. [DOI] [PubMed] [Google Scholar]
- 38.Ebata T, et al. , Substrate Stiffness Influences Doxorubicin-Induced p53 Activation via ROCK2 Expression. Biomed Res Int, 2017. 2017: p. 5158961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meng KP, et al. , Mechanosensing through YAP controls T cell activation and metabolism. J Exp Med, 2020. 217(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panciera T, et al. , Mechanobiology of YAP and TAZ in physiology and disease. Nat Rev Mol Cell Biol, 2017. 18(12): p. 758–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park JS, et al. , Mechanical regulation of glycolysis via cytoskeleton architecture. Nature, 2020. 578(7796): p. 621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanchanawong P, et al. , Nanoscale architecture of integrin-based cell adhesions. Nature, 2010. 468(7323): p. 580–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Case LB and Waterman CM, Integration of actin dynamics and cell adhesion by a three-dimensional, mechanosensitive molecular clutch. Nat Cell Biol, 2015. 17(8): p. 955–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossetti M, et al. , Ex vivo-expanded but not in vitro-induced human regulatory T cells are candidates for cell therapy in autoimmune diseases thanks to stable demethylation of the FOXP3 regulatory T cell-specific demethylated region. J Immunol, 2015. 194(1): p. 113–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt A, et al. , Comparative Analysis of Protocols to Induce Human CD4+Foxp3+ Regulatory T Cells by Combinations of IL-2, TGF-beta, Retinoic Acid, Rapamycin and Butyrate. PLoS One, 2016. 11(2): p. e0148474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. (A) Streptavidin-acrylamide concentrations were varied to obtain a similar coating of antibodies on PA-gels, confirmed by fluorescence intensities on the surface of gels (n = 4 gels for each formulation, Kruskal-Wallis test with Dunn’s multiple comparisons). (B) Representative gating strategies for FOXP3 expression from Tregs induced on PA-gels. (C) Treg induction with various concentrations of streptavidin-acrylamide (4 independent experiments and 2 replicates each for 0.1X and 0.01X, two-way ANOVA with Tukey’s multiple comparisons * p < 0.05, *** p < 0.001, **** p < 0.0001). (D) A representative gating strategy for percent mitochondrial high cells for Fig. 4.
Figure S2. (A) Gating of activated T cells on the forward scattering and side scattering of flow cytometry data (B) Representative flow gating strategy for CD25+ population (APC-A, CD25). (C) Percent CD25+ on both 7.5 kPa and 140 kPa (3 independent experiments and 2 replicates each, Mann-Whitney test, ** p < 0.01). (D) Percent of CD25+ population in FOXP3+ population (3 independent experiments and 2 replicates each, One-way ANOVA, *** p < 0.001). (E) Fraction of CD25+ versus fraction of induction, measured as fraction of cells that are FOXP3+, (Extra sum-of-squares F test (α = 0.05), 3 independent experiments and 2 replicates each). (F) Activation vs induction of all the data from blebbistatin treatment and the second-order polynomial fit was identified with the Extra sum-of-squares F test (α = 0.05).
Figure S3. (A) Percent FOXP3+ on day 6 (7.5 kPa, and 140 kPa, Mann-Whitney test was used to compare gels on day 3 and day 6 individually, **** p < 0.0001). (B) Treg suppressive capacities with 1to2 ratio of Tregs to Tconvs (n = 5 independent experiments, and the number of replicates depends on the number of cells obtained after sorting, unpaired t-test). (C) Representative gating strategies for calculation of percent suppression with a 1to1 ratio of Tregs to Tconvs samples. Tconvs only is the positive control without Tregs with Dynabeads. (D) Representative CSFE(FITC-A) vs FSC-A density plots for 1to1 ratio of Tregs to Tconvs samples.
Figure S4. (A) Top 10 markers of Treg and Tconv populations. (B, C) Over-representation analysis of markers from Treg and Tconvs, respectively. (D) Volcano plot of differentially expressed genes from Tconvs comparing the stiffer sample to the softer sample. (E) Over-representation analysis of differential expressed genes from Tconvs when comparing the stiffer sample to the softer sample.
Figure S5. Trajectory analyses on either the hard or the soft samples. (A, B) Trajectory analyses were performed with SCORPIUS with T cells from either the hard (A) or the soft (B) sample separately. (C) Histogram of cells along the trajectory on either the hard or the soft sample. (D, F) Important genes that contribute to Treg induction were organized into modules for either the hard (D) or the soft (F) sample. (E, G) Over-representation analysis of genes uniquely identified in either the hard (E) or the soft (G) sample.
Figure S6. (A) Violin plots of SMAD2, YAP1, TAZ, SMAD4, TGFBR2, TRIM21, PFKP, PFKM, and PFKL.
Data Availability Statement
The data that support the findings of this report are available from the authors upon reasonable request.


