Abstract
Adipose tissue is a functional endocrine organ comprised of adipocytes and other cell types that are known to secrete a multiplicity of adipose-derived factors, including lipids and proteins. It is well established that adipose tissue and its secretome can impact systemic energy homeostasis. The endocrine and paracrine effects of adipose-derived factors have been widely studied over the last several decades. Owing to technological advances in genomics and proteomics, several additional adipose-derived protein factors have recently been identified. By learning from previous efforts, the next challenge will be to leverage these discoveries for the prevention or treatment of metabolic disorders. Here, we discuss recently discovered adipose-derived proteins secreted from white or brown adipose tissue and the opportunities and challenges of translating these biological findings into disease therapeutics.
Introduction
Adipose tissue is a mosaic organ containing mature adipocytes, immune cells, neurons, and stromal vascular cells such as progenitors, fibroblasts and epithelial cells. The adipose tissue was previously viewed as an organ mainly involved in lipid storage [1]. Studies over the last decades have broadened the functional importance of adipose tissue on whole-body energy regulation. It is now recognized that adipose tissue is a principal site for hormone production [2] and a critical regulator of body temperature by controlling non-shivering thermogenesis and providing thermal isolation [3]. Some of the endocrine and paracrine effects of the adipose tissue are mediated through the secretion of adipose-derived proteins or cytokines commonly referred to as adipokines. For example, leptin, the cytokine-like 16 kDa adipose-secreted protein discovered by positional cloning of the ob/ob gene, is one of the most well-studied adipokines [4]. The importance of leptin in regulating food intake and energy expenditure is exemplified in ob/ob mice and genetically leptin-deficient humans, resulting in severe hyperphagia and obesity. The discovery of leptin represents a translational success story contributing to important biological insights resulting in the synthetic leptin analog Metreleptin as the first FDA-approved therapy for generalized lipodystrophy [5].
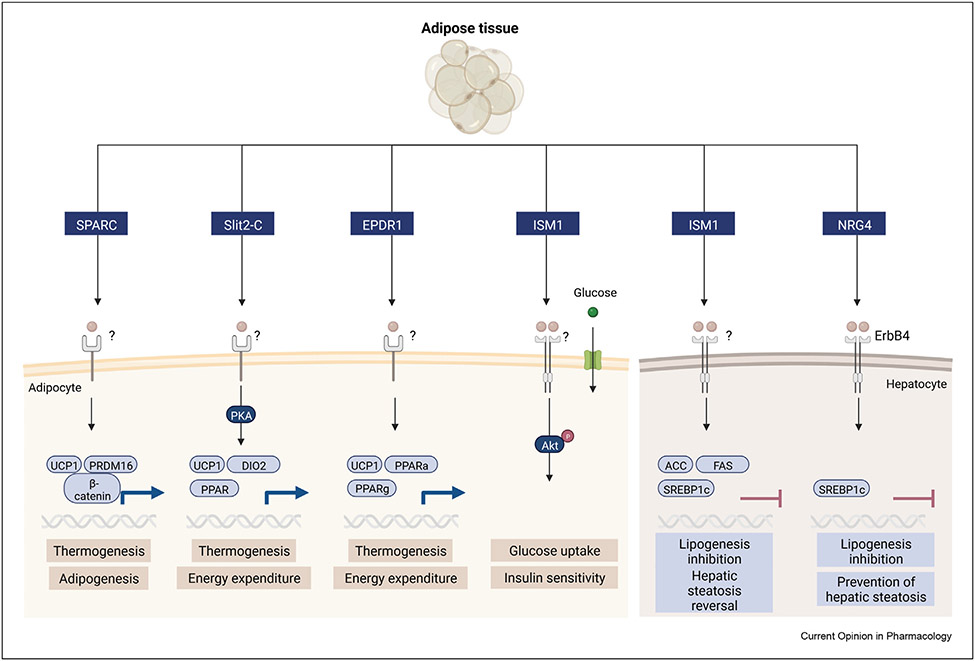
The adipose secretome is an intriguing area of intense research and many adipose-derived proteins, metabolites and lipids have been identified over the last decades. The secretome refers to the secreted components from a given cell type or tissue, including proteins, peptides, metabolites, lipids, and more recently, exosomes. The discoveries, functions and therapeutic potential of adipose factors identified during the last decades, including adiponectin, leptin, TNF-α, adipsin, chemerin, and resistin have been comprehensively reviewed elsewhere [6]. While we will not directly cover these protein hormones in this review, they serve as important benchmarks for investigating and determining the biological functions of newly discovered secreted factors. The expression and secretion of numerous secreted factors are dysregulated in obesity, metabolic syndrome, and type 2 diabetes, but have less often been comprehensively functionally characterized. Here, we focus on adipose-derived proteins for which a function in regulating peripheral or central metabolism has been identified in the past five years. These include secreted protein acidic and rich in cysteine (SPARC), Slit2-C, mammalian ependymin-related protein 1 (EPDR1), neuregulin-4 (NRG4) and Isthmin-1 (ISM1). While these factors are predominantly derived from adipocytes, they are likely also secreted from other cell types or other tissues which will not be discussed in this review. First, we cover their in vitro and in vivo phenotypes in both gain- and loss-of-function models and their mechanisms of action. Second, we highlight some key properties and lay the foundation for additional work aimed at understanding their mechanism of action and translational potential. Figure 1.
Figure 1. Adipose tissue-derived factors and their proposed roles in metabolism.
Adipose-derived protein factors are released from adipocytes or preadipocytes. Their proposed roles in metabolism include thermogenesis (SPARC, Slit2-C, EPDR1), adipogenesis (SPARC), glucose uptake (Ism1) and lipogenesis (Ism1, NRG4).
Recent adipose-secreted proteins and their functions in metabolism
SPARC
Secreted protein acidic and rich in cysteine (SPARC), also known as osteonectin or BM-40 (basement-membrane protein 40) is a widely expressed protein that was initially discovered as a blood-secreted factor involved in bone formation [7]. While the function of SPARC has been largely studied in wound healing, tissue remodeling and cancer development [8,9], SPARC also appears to play a role in glucose homeostasis and adipogenesis, although its functional role in metabolism is still under debate. The expression of SPARC in adipose tissue is increased in genetically obese mice and in high-fat diet-induced obese mice [10,11]. In humans, plasma levels are positively associated with body mass index [12]. Moreover, elevated circulating SPARC levels positively correlate with insulin resistance, dyslipidemia and inflammation in women with gestational diabetes [13]. Overall, these data suggest that SPARC might respond to nutritional changes or might increase as a result of insulin resistance.
Functionally, conflicting in vitro studies point to SPARC as a beneficial factor in the induction of thermogenesis in adipocytes in white fat depots [14]. This process, also called “browning” or “beiging” is associated with improved metabolic health. SPARC also appears to inhibit adipogenesis and induce insulin resistance, which is at odds with the process of browning [15]. Shen et al. showed that stable overexpression of SPARC in 3T3-L1 cells induced insulin resistance as demonstrated by lower insulin-stimulated glucose uptake, a lower expression of adipocyte GLUT4 and an increased expression of inflammatory cytokines [15]. In 2020, Mukherjee et al. demonstrated that SPARC stimulates Ucp1 and Prdm16 expression in 3T3-L1 cells and in HIB1B brown adipocytes [14]. Using siRNA-mediated knockdown, they also show that SPARC is required for the expression of brown fat markers in white and brown adipocytes. Necessity and sufficiency experiments suggest that SPARC regulates lipogenesis gene expression while concomitantly promoting lipolysis and fat oxidation in adipocytes. In addition, recombinant SPARC protein treatment suppresses C/Ebpα and Pparγ expression, two key transcription factors involved in adipogenesis [14]. SPARCs inhibitory effect on adipogenesis is likely occurring by direct action on preadipocytes, as recombinant SPARC treatment in cells inhibits adipogenesis, in part by enhancing the Wnt-β-catenin pathway during differentiation [16]. Although the receptor for SPARC is unknown, some data suggest that SPARC induces signal transduction through interacting with cell-surface integrins. It has also been proposed that SPARC can bind PDGF and VEGF and inhibit their binding to their cognate receptors [17] but whether SPARC regulates adipogenesis by signaling via growth factors remains unclear.
SPARC knock-out (KO) mice fed a high-fat diet have higher body weight as compared to wild-type mice [18,19]. In addition, several studies have reported that SPARC-KO mice have larger visceral and subcutaneous fat depots in comparison with wild-type mice [20,21]. Bradshaw et al. showed that deletion of SPARC in mice increases the average size of adipocytes as well as their number per fat pad and proposed that SPARC limits adipogenesis through its effect in ECM remodeling [20]. However, the control of fat deposition by SPARC protein needs further investigation in obese animal models. SPARC-KO mice are hyperglycemic, and present with lower glucose tolerance and a decreased insulin secretion, a phenotype likely secondary to the higher body weight [22]. It remains to be determined whether any of the observed phenotypes in vitro, including browning and lipogenesis, contributes to insulin resistance in SPARC-KO mice. To our knowledge, no pharmacological studies of recombinant SPARC in animal models have been performed. Collectively, these data suggest that SPARC may drive beneficial effects but further mechanistic studies, including detailing the cellular target and the direct mechanism of action using recombinant SPARC protein, are needed to determine any future therapeutic potential.
Slit2-C
Slit2 is an extracellular matrix protein belonging to the Slit family. Slit2 was first identified by Tessier-Lavigne’s group as a protein important for brain development and axon formation [23,24]. They found that this large protein is cleaved into N-terminal and C-terminal fragments, with the N-terminal protein controlling axon guidance. In 2016, the 50 kDa Slit2-C fragment was also found to be expressed and secreted by mature fat cells in a quantitative proteomics screen from primary cultures of inguinal mature adipocytes. In adipocytes, Slit2 expression is under the control of PRDM16, a transcriptional regulator of brown and beige fat [25]. Overexpression of the C-terminal cleavage form, but not the N-terminal fragment, is sufficient to induce PKA signaling and the expression of thermogenic genes in adipocytes and to increase whole-body energy expenditure [25]. These results suggest distinct functions of the various proteolytic fragments of the same parent protein. Slit2-C overexpression also lowers BAT and iWATweight by 20—30% and improves glucose tolerance in diet-induced obese mice, but pharmacological administration of Slit2-C was not tested in this study [25].
In 2017, Kang et al. quantified the serum level of Slit2 in humans to probe its relevance for type 2 diabetes [26]. They found that circulating Slit2 correlates negatively with HbA1c and with fasted and fed serum glucose levels, suggesting Slit2 as a biomarker of glucose homeostasis [26]. However, no significant differences in serum Slit2 levels between control and diabetic patients were identified. In 2021, Liu et al. studied the effect of recombinant Slit2 in a rat model of coronary heart disease, based on the observation that Slit2 expression is lower in plasma and heart tissues of rats with coronary heart disease [27]. Notably, daily I.V injections of recombinant Slit2 for seven days are sufficient to lower blood lipids, myocardial fibrosis while improving cardiac function with a dose-sensitive effect, ranging from 1 to 10 μg/kg Slit2. Although this study did not investigate the specific function of the C-terminal fragment of Slit2, it suggests a role for Slit2 in improving cardiovascular disease.
The proposed receptors for Slit2-C are plexinA1 and the proteoglycan glypican-1 (GPC-1), although direct ligand-receptor binding experiments are still lacking [28]. The Slit2-C/plexinA1 axis appears to be involved in axon guidance in the spinal cord during central nervous system development [29], but to what extent plexinA1 is mediating the effects on adipose browning is still unknown. Future studies on the functional receptors responsible for the metabolic effects of Slit2, the precise mechanism of action, and the enzymatic processing of Slit2-C are important to fully understand this pathway and its potential therapeutic use.
EPDR1
Mammalian ependymin-related protein 1 (EPDR1) is widely expressed across tissues [30] and was proposed as a novel brown-fat derived factor in 2019 based on its increase during brown fat differentiation [31]. EPDR1 is also present in the secretome of beige adipocytes [25] and was identified as a putative metabolic factor in a proteomics analysis comparing human brown and white adipocytes [31]. EPDR1 knockdown lowers oxygen consumption rates of cultured human primary brown adipocytes, presumably by regulating thermogenesis [31]. By performing proteomic analysis of human brown adipocytes, Deshmukh et al. showed that EPDR1 knockdown lowers mitochondrial proteins. Consistent with a role in thermogenesis, EPDR1 knockdown blunts the response to norepinephrine-induced expression of thermogenic genes, including Ucp1, Dio2, Ppara, Ppargc1a, suggesting a role for EPDR1 in response to adrenergic signaling [31].
EPDR1-KO mice demonstrate a 50% increase in body fat compared to control mice on a chow diet, supporting a role in activating thermogenesis in vivo. Furthermore, EPDR1 deficiency lowers in vivo oxygen consumption rate and physical activity without affecting food intake, strongly indicating that EPDR1 controls energy expenditure in mice [31]. These experiments suggest a potential beneficial role for EPDR1 in obesity. However, increased whole-body energy expenditure was not recapitulated by daily dosing of recombinant EPDR1 for 21 days in mice fed a high-fat diet. The reason for the lack of efficacy of the recombinant protein in mice is unclear, but the authors speculated that the room temperature housing conditions may have masked the effects of EPDR1. Based on EPDR1’s crystal structure that was solved in 2019 [32,33], EPDR1 may also have a function in lipid transport, particularly in the transport of neuronal lipids. In conclusion, EPDR1 is a new metabolic regulator important for brown fat commitment and energy homeostasis. Future studies to determine EPDR1’s mechanism of action and its direct signaling action on its receptor might guide the development of agonists with better efficacy in vivo.
NRG4
Neuregulins (NRGs) have emerged as interesting and potent candidates for improving vital aspects of metabolic health [34]. NRG is a subfamily of EGF-ligands that contains four members (NRG1 to NRG4) that binds to the ErbB tyrosine kinase receptor family. Among them, NRG4 was identified as a brown-fat secreted factor regulating energy metabolism [35,36]. NRG4 is also expressed in white adipose tissue and to a lesser extent in the liver. NRG4 expression is upregulated in white fat upon cold exposure and during brown adipocyte differentiation [35,36]. Recently, Comas et al. demonstrated that NRG4 expression correlates with the expression of thermogenic genes and identified NRG4 as a marker of beige-adipocytes in humans [37]. Furthermore, adipose tissue expression of NRG4 is decreased in obese mice and humans and serum NRG4 levels are inversely associated with metabolic syndrome, cardiovascular diseases and non-alcoholic fatty liver disease (NAFLD) in obese humans [38-41]. These data indicate that impaired NRG4 secretion may promote metabolic dysfunction. Wang et al. demonstrated that deletion of NRG4 induces glucose intolerance and insulin resistance and exacerbates hepatic steatosis in diet-induced obese mice [35]. Conversely, transgenic overexpression of NRG4 in mice fed a high-fat diet decreases fat mass, improves glucose tolerance, increases energy expenditure and alleviates diet-induced fatty liver [35,42].
Mechanistically, NRG4 binds to the tyrosine kinase receptors ErbB3 and ErbB4 and lowers de novo lipogenesis in hepatocytes by reducing the expression of the lipogenic genes downstream of srebp1c. NRG4 also protects from hepatic steatosis by accelerating fatty acid oxidation [42]. Taken together, these data strongly indicate that NRG4 regulates energy, glucose and lipid metabolism and have beneficial effects by improving NAFLD characteristics. While NRG4 was not tested as a pharmacological agent in these studies, the close family member NRG1 acting on the same receptors, has undergone extensive protein engineering to optimize its efficacy in reducing metabolic syndrome. Treatment with an NRG1 protein fused to an Fc domain to extend half-life lowers blood glucose, improves insulin sensitivity, and suppresses food intake in high-fat diet-fed mice [43]. Importantly, given the role of EGFR family receptors in cancer progression, it is plausible that pharmacological activation of the NRG4-Erbb4 signaling could be associated with adverse effects. However, little is known about ErbB4 signaling in cancer, but the data so far points towards a suppressive rather than protumorigenic effect of ErbB4, at least in the development of hepatocellular carcinoma [44]. The potential impact of NRG4/Erbb4 in tumorigenesis requires further study to assess the safety of neuregulin-based therapies.
Isthmin-1
Isthmin (ISM1) was originally identified as a gene expressed in the Xenopus midbrain-hindbrain organizer called Isthmus [45]. ISM1 is a 50 kDa secreted protein with a central thrombospondin domain and an adhesion-associated domain in MUC4 and other proteins (AMOP) domain. ISM1 has been shown to play a role in craniofacial development in humans and ISM1 knockdown in Xenopus laevis embryos results in craniofacial dysmorphologies [46]. Surprisingly, whole body Ism1 deletion in mice and Ism1 knockdown in zebrafish are not associated with craniofacial defects [47,48], pointing to potentially significant species differences in expression or function. Mouse Ism1 is expressed in adipocytes, immune cells and fibroblasts, while human ISM1 expression is highest in the thyroid gland but is also expressed in adipose tissue. Recently, Ism1 was identified by our lab as a functional regulator of metabolism [47]. In mice, Ism1 is secreted from mature adipocytes and has a dual role in increasing adipose glucose uptake while suppressing hepatic lipid synthesis [47]. Whole-body Ism1-KO mice are glucose intolerant and show higher insulin resistance. Likewise, Ism1 knockdown in adipocytes blunts the insulin-stimulated glucose uptake and insulin-stimulated signaling [47]. Ism1 binds to an unknown receptor and activates the PI3K-AKT pathway independently of the insulin receptor, or IGF1R to promote glucose uptake. While Ism1 and insulin share some common downstream signaling targets and glucoregulatory functions, there is also an important divergence in signaling pathways downstream of AKT, leading to Ism1-mediated reductions in insulin-stimulated de novo lipogenesis. The mechanism appears to occur by decreasing the expression of insulin-induced lipogenic genes such as Srebp1c, Fas and Acc, while also turning on anabolic pathways associated with protein synthesis [47]. Furthermore, therapeutic dosing of recombinant Ism1 improves glucose tolerance in diet-induced obese mice and reverses the lipid accumulation seen in established hepatic steatosis in a diet-induced fatty liver mouse model [47]. The precise mechanism behind its ability to regulate lipid metabolism, and whether the effect of Ism1 in the liver is dependent on endocrine signaling from the adipose tissue remains to be determined.
The fact that ISM1 activates the PI3K-AKT pathway raises the potential concern that ISM1 may drive carcinogenesis. Previous studies suggest that ISM1 instead decreases tumor growth by inducing apoptosis and inhibiting angiogenesis in mice [49,50]. However, those studies made use of recombinant protein produced in bacteria that lack glycosylation machinery. It is not known whether mammalian recombinant ISM1 inhibits angiogenesis. Nevertheless, the potential role of ISM1 in promoting tumorigenesis needs to be further explored, notably after long-term therapeutic dosing in rodent and non-rodent models to better evaluate the safety of ISM1 therapeutics. In conclusion, ISM1 regulates glucose and lipid metabolism and might represent a new therapeutic target for metabolic disorders. Further work to better understand the receptor and its endocrine functions in energy metabolism will facilitate translational studies.
Therapeutic opportunities and challenges
Over the past decade, many adipokines with promising and interesting abilities to regulate metabolism have been identified, and as such, represent potential new therapeutic opportunities for metabolic diseases. To date, > 200 therapeutics peptides and proteins and their drug analogs, including a few number of adipokines as protein therapeutics, have been FDA approved [51]. Understanding the biology, including receptor identification and studies of the receptor-ligand binding affinity properties are essential for the development of proteins as therapeutic agents [52]. Identifying the receptors and their tissue expression will also be important to evaluate potential adverse on-target effects on other organs. While some of the recent adipokines described here show therapeutic potential in preliminary work, most insights have been accomplished through loss- and gain-of-function models in vitro and in vivo, which do not always mirror the agonist action at pharmacological doses. Although the metabolic functions of these secreted factors have been validated in vivo using recombinant protein treatments (EPDR1 and ISM1) and/or whole-body knock-out mice (SPARC, EPDR1, NRG4 and ISM1), adipose tissue-specific loss- and gain-of-function studies are still required to fully understand their mechanism of action. Additionally, further studies on the contribution of each relative fat depot in the secretion of the protein factors described in this review are required to fully understand their biological relevance and therapeutic potential. For some ligands, the receptors are unknown, and for future therapeutic use, identification of the receptor and downstream signaling pathways will be essential to establish the tissue-specific contributions, to enable regulation of their bioactivities, and to develop specific bioassays. Among the proteins described in this review, direct downstream signaling pathways upon recombinant protein treatment have been performed for SPARC, ISM1 and NRG4. NRG4 is the only ligand for which a receptor responsible for the metabolic effects has been established.
Furthermore, rigorous protein quality control, pharmacokinetic and efficacy studies are necessary to facilitate translation. A short half-life will potentially limit the therapeutic efficacy and could require repeated dosing. As an example, the half-life of unmodified Ism1 is 70 min, likely requiring daily dosing for sufficient efficacy. PEGylation and Fc fusion strategies are frequently used to increase the circulating half-life of a drug. Likewise, glycosylation alterations can be used to affect the ligand binding to its receptor and overall bioactivity. Except for NRG1-Fc fusion proteins, modifications of the adipose-derived factors described here have not been performed. Moreover, approaches based on library screens and random protein mutagenesis have been used to generate receptor super-agonists or antagonists, as exemplified with CLCF1 (cardiotrophin-like cytokine factor 1), a cytokine from the IL-6 family [53]. Such approaches would generate unique and more potent receptor signaling agonists at lower concentrations.
Finally, in vivo studies in rodent and non-rodent models are essential to evaluate and modulate protein safety including on and off-target toxicity, immunogenicity [54], and mitogenic capacity. For example, Aldafermin, an engineered non-tumorigenic analog of the human hormone fibroblast growth factor 19 (FGF19) differs from the endogenous human FGF19 by having 3 amino acid substitutions (A30S, G31S, and H33L) and a 5-amino acid deletion in the N-terminal region. These modifications render the protein inactive in promoting mitogenesis while still maintaining its metabolic benefits [55,56]. Furthermore, immunogenicity, the unwanted immune response of the body against the therapeutic protein, needs to be considered in these studies, which can result in allergic reactions or loss of activity of the drug [57].
In conclusion, there have been tremendous efforts in the past decades to identify new adipose-derived secreted factors that regulate peripheral tissue and systemic metabolism. This emerging picture supports the diversity in the regulation of whole-body metabolism, while also providing a framework for future discoveries. Although further studies detailing the mechanisms and structural properties are required, the recently identified factors discussed in this review demonstrate promising and interesting biological effects in vitro and in vivo. They represent possible future targets to understand the biology, and possibly treat or prevent metabolic diseases.
Acknowledgements
K.J.S. was supported by NIH grants DK125260, DK111916, the Stanford Diabetes Research Center P30DK116074, the Jacob Churg Foundation, the McCormick and Gabilan Award, the Weintz Family COVID-19 research fund, American Heart Association (AHA), the Stanford School of Medicine, and the Stanford Cardiovascular Institute (CVI). M.Z. was supported by the American Heart Association (AHA) postdoctoral fellowship (905674). L. C. was supported by Stanford School of Medicine Dean’s Postdoctoral Fellowship. N.B.D.S. was supported by the Carlsberg Foundation Internationalization Fellowship. The figure was created with Biorender.com.
Abbreviations
- ACC
acetyl-CoA carboxylase
- C/EBPα
CCAAT/enhancer-binding protein
- c-FLIPL
cellular caspase 8 (FLICE)-like inhibitory protein
- DIO2
iodothyronine Deiodinase 2
- ECM
extracellular matrix
- EGFR
epidermal growth factor receptor
- FAS
fatty acid synthase
- FGF
fibroblast growth factor
- GLUT4
glucose transporter type 4
- GSK-3β
glycogen synthase kinase-3beta
- HbA1c
hemoglobin A1c
- IGF1R
insulin-like growth factor receptor 1
- PDGF
Platelet-derived growth factor
- PKA
protein kinase A
- PPARα
Peroxisome proliferator-activated receptor alpha
- PPARγ
Peroxisome proliferator-activated receptor gamma
- PPARGC1A
PPARγ Coactivator 1 Alpha
- PRDM16
PR domain-containing 16
- SREBP1
sterol regulatory element-binding protein-1
- TGF-β
transforming growth factor-beta
- TNF-α
tumor-necrosis factor-alpha
- UCP1
uncoupling protein 1
- VEGF
vascular endothelial growth factor
Footnotes
Conflict of interest statement
K.J.S. has a patent for “Methods for identification, assessment, prevention, and treatment of metabolic disorders using Slit2.” US Patent. 15/741, 326. WO/2017/011763. The other authors have no conflicts of interests.
CRediT authorship contribution statement
Writing - original draft; Voilquin, L., Svensson K.J.: Writing - review & editing, Voilquin, L., Zhao. M, Danneskiold-Samsøe, N.B., Allen, H. Funding acquisition; Svensson, K.J.
References
- 1.Spiegelman BM, Flier JS: Obesity and the regulation of energy balance. Cell 2001, 104:531–543. [DOI] [PubMed] [Google Scholar]
- 2.Kershaw EE, Flier JS: Adipose tissue as an endocrine organ. J Clin Endocrinol Metab 2004, 89:2548–2556. [DOI] [PubMed] [Google Scholar]
- 3.Wu J, Cohen P, Spiegelman BM: Adaptive thermogenesis in adipocytes: is beige the new brown? Genes Dev 2013, 27:234–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y, et al. : Positional cloning of the mouse obese gene and its human homologue. Nature 1994, 372:425–432. [DOI] [PubMed] [Google Scholar]
- 5.Chou K, Perry CM, Metreleptin: First global approval. Drugs 2013, 73:989–997. [DOI] [PubMed] [Google Scholar]
- 6.Recinella L, et al. : Adipokines: new potential therapeutic target for obesity and metabolic, rheumatic, and cardiovascular diseases. Front Physiol 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Termine JD, et al. : Osteonectin, a bone-specific protein linking mineral to collagen. Cell 1981, 26:99–105. [DOI] [PubMed] [Google Scholar]
- 8.Chlenski A, Cohn SL: Modulation of matrix remodeling by SPARC in neoplastic progression. Semin Cell Dev Biol 2010, 21:55–65. [DOI] [PubMed] [Google Scholar]
- 9.Nagaraju GPC, Sharma D: Anti-cancer role of SPARC, an inhibitor of adipogenesis. Cancer Treat Rev 2011, 37:559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tartare-Deckert S, Chavey C, Monthouel MN, Gautier N, Van Obberghen E: The matricellular protein SPARC/osteonectin as a newly identified factor up-regulated in obesity. J Biol Chem 2001, 276:22231–22237. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi M, et al. : The expression of SPARC in adipose tissue and its increased plasma concentration in patients with coronary artery disease. Obes Res 2001, 9:388–393. [DOI] [PubMed] [Google Scholar]
- 12.Kos K, et al. : Regulation of the fibrosis and angiogenesis promoter SPARC/osteonectin in human adipose tissue by weight change, leptin, insulin, and glucose. Diabetes 2009, 10.2337/db09-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.X L, et al. : Elevated plasma SPARC levels are associated with insulin resistance, dyslipidemia, and inflammation in gestational diabetes mellitus. PLoS One 2013, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukherjee S, Choi MJ, Kim SW, Yun JW: Secreted protein acidic and rich in cysteine (SPARC) regulates thermogenesis in white and brown adipocytes. Mol Cell Endocrinol 2020, 506, 110757. [DOI] [PubMed] [Google Scholar]
- 15.Shen Y, et al. : SPARC is over-expressed in adipose tissues of diet-induced obese rats and causes insulin resistance in 3T3-L1 adipocytes. Acta Histochem 2014, 116:158–166. [DOI] [PubMed] [Google Scholar]
- 16.Nie J, Helene Sage E: SPARC inhibits adipogenesis by its enhancement of β-catenin signaling. J Biol Chem 2009, 284:1279–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ew R, Tf L, Ml I-A, R R, Eh S: The extracellular glycoprotein SPARC interacts with platelet-derived growth factor (PDGF)-AB and -BB and inhibits the binding of PDGF to its receptors. Proc Natl Acad Sci U S A 1992, 89:1281–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nie J, Sage EH: SPARC functions as an inhibitor of adipogenesis. J Cell Commun Signal 2009, 33:247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nie J, Bradshaw AD, Delany AM, Sage EH: Inactivation of SPARC enhances high-fat diet-induced obesity in mice. Connect Tissue Res 2011, 52:99–108, 10.3109/03008207.2010.483747. [DOI] [PubMed] [Google Scholar]
- 20.Bradshaw AD, Graves DC, Motamed K, Sage EH: SPARC-null mice exhibit increased adiposity without significant differences in overall body weight. Proc Natl Acad Sci U S A 2003, 100:6045–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazzolini G, et al. : SPARC expression is associated with hepatic injury in rodents and humans with non-alcoholic fatty liver disease. Sci Rep 2018, 8, 10.1038/s41598-017-18981-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atorrasagasti C, et al. : SPARC is required for the maintenance of glucose homeostasis and insulin secretion in mice. Clin Sci 2019, 133:351–365. [DOI] [PubMed] [Google Scholar]
- 23.Kuan HW, et al. : Biochemical purification of a mammalian slit protein as a positive regulator of sensory axon elongation and branching. Cell 1999, 96:771–784. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen Ba-Charvet KT, et al. : Diversity and specificity of actions of Slit2 proteolytic fragments in axon guidance. J Neurosci 2001, 21:4281–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svensson KJ, et al. : A secreted slit2 fragment regulates adipose tissue thermogenesis and metabolic function. Cell Metabol 2016, 23:454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang YE, Choung S, Lee JH, Kim HJ, Ku BJ: The role of circulating slit2, the one of the newly batokines, in human diabetes mellitus. Endocrinol Metabol 2017, 32:383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J-W, Liu H-T, Chen L: The therapeutic role of Slit2 in anti-fibrosis, anti-inflammation and anti-oxidative stress in rats with coronary heart disease. Cardiovasc Toxicol 2021, 1:1–11. [DOI] [PubMed] [Google Scholar]
- 28.Ronca F, Andersen JS, Paech V, Margolis RU: Characterization of slit protein interactions with glypican-1. J Biol Chem 2001, 276:29141–29147. [DOI] [PubMed] [Google Scholar]
- 29.Delloye-Bourgeois C, et al. : PlexinA1 is a new Slit receptor and mediates axon guidance function of Slit C-terminal fragments. Nat Neurosci 2015, 18:36–45. [DOI] [PubMed] [Google Scholar]
- 30.Chen R, Zhang Y: EPDR1 correlates with immune cell infiltration in hepatocellular carcinoma and can be used as a prognostic biomarker. J Cell Mol Med 2020, 24:12107–12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deshmukh AS, et al. : Proteomics-based comparative mapping of the secretomes of human Brown and white adipocytes reveals EPDR1 as a novel batokine. Cell Metabol 2019, 30:963–975. e7. [DOI] [PubMed] [Google Scholar]
- 32.Wei Y, et al. : Crystal structures of human lysosomal EPDR1 reveal homology with the superfamily of bacterial lipoprotein transporters. Commun Biol 2019, 21:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park JK, et al. : Structures of three ependymin-related proteins suggest their function as a hydrophobic molecule binder. IUCrJ 2019:729–739. urn:issn:2052–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao M, Jung Y, Jiang Z, Svensson KJ: Regulation of energy metabolism by receptor tyrosine kinase ligands. Front Physiol 2020:354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang GX, et al. : The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat Med 2014, 20:1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosell M, et al. : Brown and white adipose tissues: intrinsic differences in gene expression and response to cold exposure in mice. Am J Physiol Endocrinol Metab 2014, 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comas F, et al. : Neuregulin 4 is a novel marker of beige adipocyte precursor cells in human adipose tissue. Front Physiol 2019:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai C, et al. : Association of circulating neuregulin 4 with metabolic syndrome in obese adults: a cross-sectional study. BMC Med 2016, 141:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang J, et al. : Circulating neuregulin 4 levels are inversely associated with subclinical cardiovascular disease in obese adults. Sci Rep 2016, 61:1–8. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang R, et al. : Decreased serum neuregulin 4 levels associated with non-alcoholic fatty liver disease in children with obesity. Clin Obes 2019, 9, e12289. [DOI] [PubMed] [Google Scholar]
- 41.Dai YN, et al. : A case-control study: Association between serum neuregulin 4 level and non-alcoholic fatty liver disease. Metabolism 2015, 64:1667–1673. [DOI] [PubMed] [Google Scholar]
- 42.Chen Z, et al. : Nrg4 promotes fuel oxidation and a healthy adipokine profile to ameliorate diet-induced metabolic disorders. Mol Metabol 2017, 6:863–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang P, et al. : NRG1-Fc improves metabolic health via dual hepatic and central action. JCI Insight 2018, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, et al. : ERBB4 acts as a suppressor in the development of hepatocellular carcinoma. Carcinogenesis 2017, 38:465–473. [DOI] [PubMed] [Google Scholar]
- 45.Pera EM, et al. : Isthmin is a novel secreted protein expressed as part of the Fgf-8 synexpression group in the Xenopus midbrain-hindbrain organizer. Mech Dev 2002, 116:169–172. [DOI] [PubMed] [Google Scholar]
- 46.Lansdon LA, et al. : Identification of isthmin 1 as a novel clefting and craniofacial patterning gene in humans. Genetics 2018, 208:283–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang Z, et al. : Isthmin-1 is an adipokine that promotes glucose uptake and improves glucose tolerance and hepatic steatosis. Cell Metabol 2021, 33:1836–1852. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berrun A, Harris E, Stachura DL: Isthmin 1 (ism1) is required for normal hematopoiesis in developing zebrafish. PLoS One 2018, 13, e0196872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.W X, et al. : Isthmin is a novel secreted angiogenesis inhibitor that inhibits tumour growth in mice. J Cell Mol Med 2011, 15:359–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen M, et al. : Isthmin targets cell-surface GRP78 and triggers apoptosis via induction of mitochondrial dysfunction. Cell Death Differ 2014, 215:797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Usmani SS, et al. : THPdb: database of FDA-approved peptide and protein therapeutics. PLoS One 2017, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones DS, Silverman AP, Cochran JR: Developing therapeutic proteins by engineering ligand–receptor interactions. Trends Biotechnol 2008, 26:498–505. [DOI] [PubMed] [Google Scholar]
- 53.Kim JW, et al. : Engineering a potent receptor superagonist or antagonist from a novel IL-6 family cytokine ligand. Proc Natl Acad Sci USA 2020, 117:14110–14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schellekens H: How to predict and prevent the immunogenicity of therapeutic proteins. Biotechnol Annu Rev 2008, 14:191–202. [DOI] [PubMed] [Google Scholar]
- 55.DePaoli AM, et al. : FGF19 analog as a surgical factor mimetic that contributes to metabolic effects beyond glucose homeostasis. Diabetes 2019, 68:1315–1328. [DOI] [PubMed] [Google Scholar]
- 56.Harrison SA, et al. : CLINICAL-LIVER efficacy and safety of Aldafermin, an engineered FGF19 analog, in a randomized, double-blind, placebo-controlled trial of patients with nonalcoholic steatohepatitis. Gastroenterology 2021, 160:219–231. e1. [DOI] [PubMed] [Google Scholar]
- 57.Tourdot S, Hickling TP: Nonclinical immunogenicity risk assessment of therapeutic proteins. Bioanalysis 2019, 11:1631–1643, 10.4155/bio-2018-0246. [DOI] [PubMed] [Google Scholar]