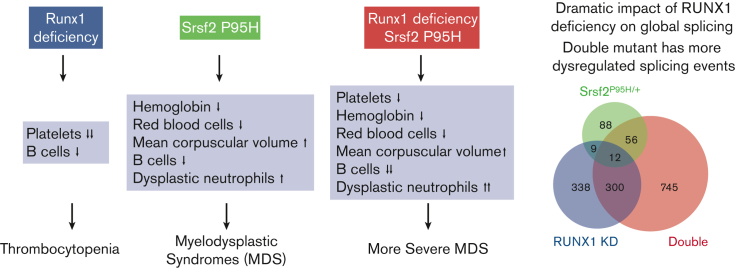
Key Points
-
•
Coexistence of the Srsf2 P95H mutation and Runx1 deficiency recapitulates the multilineage hematopoietic defects observed in MDS.
-
•
RUNX1 deficiency strikingly alters global splicing patterns and synergizes with the Srsf2 P95H mutation to affect DNA damage response genes.
Visual Abstract
Abstract
Myelodysplastic syndromes (MDSs) are a heterogeneous group of hematologic malignancies with a propensity to progress to acute myeloid leukemia. Causal mutations in multiple classes of genes have been identified in patients with MDS with some patients harboring more than 1 mutation. Interestingly, double mutations tend to occur in different classes rather than the same class of genes, as exemplified by frequent cooccurring mutations in the transcription factor RUNX1 and the splicing factor SRSF2. This prototypic double mutant provides an opportunity to understand how their divergent functions in transcription and posttranscriptional regulation may be altered to jointly promote MDS. Here, we report a mouse model in which Runx1 knockout was combined with the Srsf2 P95H mutation to cause multilineage hematopoietic defects. Besides their additive and synergistic effects, we also unexpectedly noted a degree of antagonizing activity of single mutations in specific hematopoietic progenitors. To uncover the mechanism, we further developed a cellular model using human K562 cells and performed parallel gene expression and splicing analyses in both human and murine contexts. Strikingly, although RUNX1 deficiency was responsible for altered transcription in both single and double mutants, it also induced dramatic changes in global splicing, as seen with mutant SRSF2, and only their combination induced missplicing of genes selectively enriched in the DNA damage response and cell cycle checkpoint pathways. Collectively, these data reveal the convergent impact of a prototypic MDS-associated double mutant on RNA processing and suggest that aberrant DNA damage repair and cell cycle regulation critically contribute to MDS development.
Introduction
Myelodysplastic syndromes (MDSs) are complex diseases characterized by ineffective hematopoiesis, multilineage dysplasia, peripheral cytopenia, and an elevated propensity to progress to acute myeloid leukemia (AML).1 Large-scale sequencing studies of patients with MDS have identified mutations in multiple classes of genes, including splicing factors, transcription factors, epigenetic modifiers, and cell-signaling proteins.2,3 A subset of patients with MDS carry mutations in more than 1 gene, but mutations rarely occur in multiple genes that belong to the same family. This indicates that the onset of MDS results from the alteration of multiple programs rather than the lethal inactivation of a single program. Although single gene mutations have been modeled to evaluate their contributions to disease-related phenotypes, limited information is available on how double mutants may cooperatively affect disease-relevant pathways.
RUNX1 is a master regulator of hematopoiesis and one of the most frequently mutated transcription factors in MDS, accounting for 10% to 15% of cases.2,4,5 RUNX1 mutations are prevalent in patients with high-risk MDS, leading to shorter survival.4,5 Mutations are dispersed throughout the gene and subdivided into 2 distinct functional classes: N-terminal mutations within the runt homology domain that disrupt DNA binding, and C-terminal mutations that attenuate transcriptional activity.6,7 Importantly, most RUNX1 mutations are either loss-of-function or act in a dominant-negative fashion to regulate gene expression.7
Relative to RUNX1, mutations in the splicing factors SRSF2 (also known as SC35), SF3B1, U2AF1, and ZRSR2 are exceedingly common in patients with MDS, accounting for up to 60% in certain cohorts.8 These mutations typically arise early in hematopoiesis as disease-initiating events, but rarely coexist in patients with single MDS.2,9, 10, 11, 12, 13, 14 SRSF2 mutations, which occur almost exclusively at position P95, are detected in 10% to 20% of patients with MDS and 30% to 50% of patients with chronic myelomonocytic leukemia with poor survival and prognosis.3,10,15,16 Several recent reports demonstrate that SRSF2 plays a key role in hematopoiesis and its mutations are causal to MDS.17, 18, 19, 20 Because SRSF2 is an auxiliary splicing factor that binds to exonic splicing enhancers on premessenger RNAs, mutations at P95 affect global RNA splicing by changing its binding preference, leading to missplicing of critical transcripts to induce myeloid malignancies.17,21
SRSF2 mutations are significantly associated with RUNX1 mutations in several patient cohorts,2,3,15,16,22,23 and their coexistence has been linked to inferior prognosis among patients with MDS.23,24 Considering these genes are in different families with divergent functions, namely transcription and splicing, this pair of mutations is ideal for studying how double mutants may cooperate in MDS pathogenesis.
Here, we devised a mouse model to compare the effects of Runx1 knockout, Srsf2 P95H mutation, and both genetic abnormalities in vivo. We observed more severe MDS-related phenotypes in double mutant mice compared with single mutants, including peripheral blood pancytopenia and multilineage dysplasia. To gain further disease relevance in humans and to enable molecular dissection in isogenic cellular models, we also generated human K562 cell lines that carry the same set of single or double mutations. Unexpectedly, we found that besides the altered transcription program, RUNX1 deficiency also modulated global splicing. When coupled with the SRSF2 P95H mutation, the double mutant synergistically impaired genes involved in the DNA damage response and cell cycle regulation. These findings reveal a much broader function of RUNX1 in regulating gene expression at both the transcription and posttranscriptional levels and illuminate how double mutants drive MDS development through their additive, synergistic, and antagonizing effects in different hematopoietic lineages.
Materials and methods
Please refer to the supplemental Methods for the complete description of all methods.
Mice
C57BL/6 (CD45.2) and congenic B6.SJL-Ptprca Pep3b/BoyJ (PEP3 and CD45.1) mice were obtained from Jackson Laboratory. Conditional Runx1 knockout and Mx1-Cre mice were gifts from Nancy Speck.25 Conditional Srsf2 P95H/+ mice were previously described.17 Polyinosine-polycytosine (pIpC) (Sigma) was injected intraperitoneally into mice at 12 μg/g every other day for a total of 3 injections. Genotyping polymerase chain reaction was performed using the primers previously described.17,25 Peripheral blood parameters were measured with the scil Vet abc Plus (Scil Animal Care Company, Grayslake, IL) using 50 μL of whole blood drawn from the submandibular vein. Peripheral blood smears were prepared using 2 μL of whole blood and stained with Wright-Giemsa stain (Sigma). Images were acquired on an Olympus BX51 microscope equipped with a DP71 camera and DP Controller/DP Manager software (Olympus, Tokyo, Japan). All procedures were approved by the Institutional Animal Care and Use Committee.
BMT
For competitive and noncompetitive bone marrow transplantation (BMT), total bone marrow (BM) cells were harvested from untreated donor mice. In noncompetitive settings, 2 million cells suspended in 200 μL phosphate-buffered saline were IV injected into lethally irradiated (900 rad) CD45.2 recipient mice. In competitive settings, CD45.2+ test cells were mixed 1:1 with CD45.1+ competitor cells, and 2 million cells suspended in 200 μLphosphate-buffered saline were IV injected into lethally irradiated CD45.1 recipient mice.
Results
Srsf2 mutation and Runx1 deficiency cooperatively induce pervasive MDS-like phenotypes
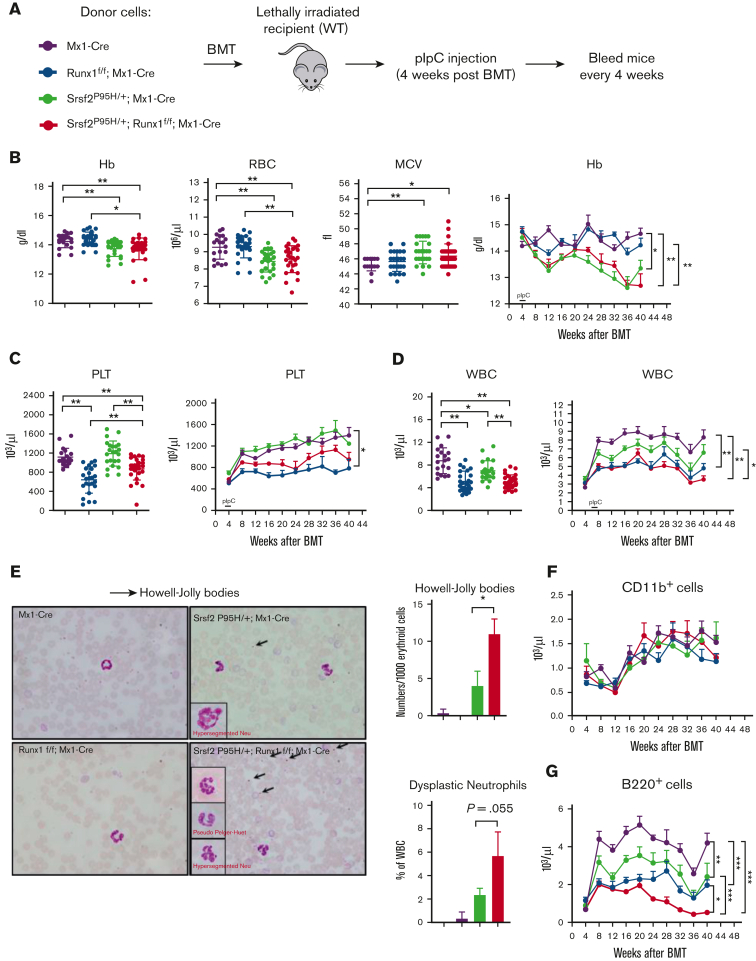
To investigate whether the coexistence of Srsf2 and Runx1 mutations may cooperatively impair hematopoiesis in vivo, we established a mouse model (Srsf2 P95H/+:Runx1 f/f:Mx1-Cre) by crossing Srsf2-P95H conditional knockin (P95H/+) mice17 with Runx1 conditional knockout mice (Runx1 f/f),25 both carrying a pIpC-inducible Mx1-Cre transgene. We chose Runx1 knockout to model dominant-negative RUNX1 mutations that are often seen in patients with MDS.7 The Srsf2 P95H allele frequency (50%) and Runx1 knockout (100%) were confirmed by RNA sequencing (RNA-seq) (supplemental Figure 1A-B). To determine the phenotypic effects of single and double mutations in a hematopoietic cell intrinsic manner, we transplanted mouse BM mononuclear cells collected from Mx1-Cre control, Runx1 f/f:Mx1-Cre, Srsf2 P95H/+:Mx1-Cre, and Srsf2 P95H/+:Runx1 f/f:Mx1-Cre conditional mutant mice into lethally irradiated recipients and induced Cre expression by pIpC injection 4 weeks after transplantation (Figure 1A).
Figure 1.
Coexistence of the Srsf2 P95H mutation and Runx1 deficiency leads to MDS phenotypes in vivo. (A) Schematic diagram of the noncompetitive BMT experiments. (B) Total amount of hemoglobin (Hb), number of red blood cells (RBCs), and mean corpuscular volume (MCV) in the peripheral blood of recipient mice at 16 weeks after BMT (wild-type [WT], n = 19; Srsf2P95H/+, n = 23; Runx1 knockout, n = 24; double mutant, n = 27); time course analysis of Hb. (C) Total number of platelets (PLTs) in the peripheral blood of recipient mice at 16 weeks after BMT (WT, n = 19; Srsf2P95H/+, n = 23; Runx1 knockout, n = 24; double mutant, n = 27); time course analysis of platelet. (D) Total number of white blood cells (WBCs) in the peripheral blood of recipient mice at 16 weeks after BMT (WT, n = 19; Srsf2P95H/+, n = 23; Runx1 knockout, n = 24; double mutant, n = 27); time course analysis of WBC. (E) Peripheral blood smears of recipient mice. Dysplastic cells are indicated and include hyper/hyposegmented neutrophils (insets) and Howell-Jolly bodies (nuclear remnants) in RBCs (arrows). Smears are representative of 3 mice per genotype. The frequencies of dysplastic erythroid cells and neutrophils in each genotype are indicated in the graphs to the right. Absolute numbers of myeloid cells (Cd11b+) (F) and B cells (B220+) (G) in the peripheral blood of mice at the indicated times after BMT. Data are mean ± standard error of the mean (SEM). Significance was determined by 1-way analysis of variance (ANOVA) with Tukey post hoc test. ∗P < .05, ∗∗P < .01.
Because MDS is diagnosed by the presence of peripheral blood cytopenias and dysplastic cell morphology of various lineages,1 we first examined the peripheral blood of transplanted mice. We found that double mutant mice exhibited anemia as indicated by reduced Hb and RBCs accompanied with increased MCV, a phenotype driven by the Srsf2 P95H mutation (Figure 1B). Double mutant mice also displayed thrombocytopenia (Figure 1C), a phenotype that was predominately driven by Runx1 deficiency. These observations illustrate the additive effects of single mutants on MDS-related phenotypes by separately affecting different hematopoietic cell populations. By contrast, both mutations negatively affected the WBC population and additively contributed to more severe leukopenia in double mutant mice (Figure 1D). Altogether, double mutant mice exhibited more pervasive peripheral blood cytopenia than single mutant mice.
Next, we examined cell morphology in the peripheral blood of transplanted mice. Srsf2 P95H single mutant mice had a pronounced increase in hyposegmented and hypersegmented neutrophils and RBCs with Howell-Jolly bodies, in agreement with previous reports that these single mutant mice have MDS (Figure 1E).17 Double mutant mice had significantly more dysplastic RBCs and a trend toward more dysplastic neutrophils, indicative of a more severe MDS phenotype.
To further characterize the WBC defects, we measured the absolute numbers of lymphoid and myeloid cells. There was no significant difference in myeloid cells (CD11b+) among the 4 genotypes, but all 3 mutant mice groups had fewer B cells (B220+) than WT mice, and double mutant mice had significantly fewer B cells than either single mutant (Figure 1F-G). In agreement with the reduction in lymphoid cells in the peripheral blood, we also observed a significantly reduced proportion of B cells among WBCs in the BM and spleen (supplemental Figure 1C-D). Finally, we observed splenomegaly in double mutant mice, a phenotype driven by Runx1 deficiency (supplemental Figure 1E).
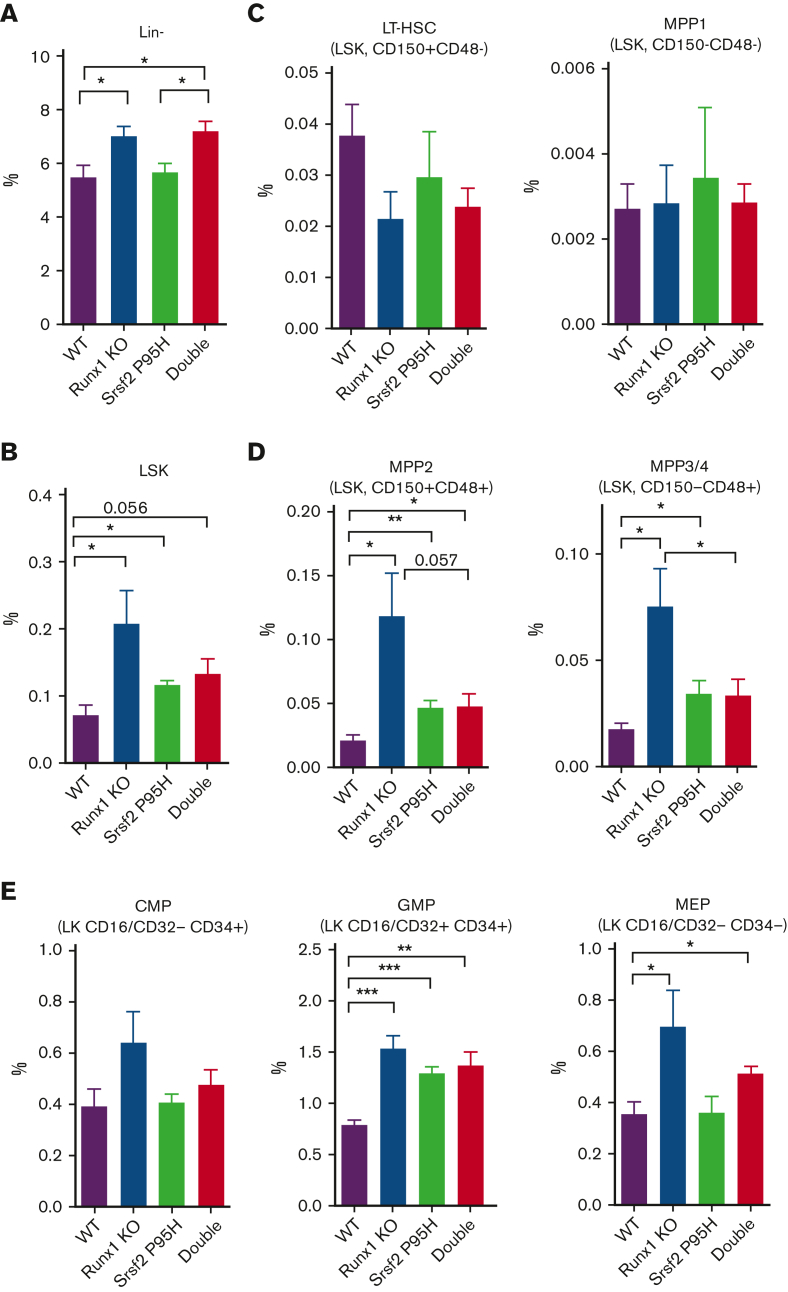
Defective HSPCs in single and double mutant mice
To delineate the cellular source of defective hematopoiesis, we next examined the hematopoietic stem and progenitor cell (HSPC) compartments in single and double mutant mice. In patients with MDS, there is an expansion of hematopoietic stem cells (HSCs) and common myeloid progenitors (CMPs) or granulocyte-monocyte progenitors (GMPs), indicative of a preleukemic stage.26 Indeed, we detected the expansion of lineage− (Lin−) cells and Lin−Sca-1+c-Kit+ (LSK) cells in the BM (Figure 2A-B) and spleen (supplemental Figure 2A-B) of single Runx1 deficient mice, as previously reported,25,27 and double mutant mice. The Srsf2 P95H mutant showed some modest effects on LSK cell expansion, but its combination with Runx1 deficiency seemed to mitigate, not exacerbate, the phenotype (Figure 2B; supplemental Figure 2B).
Figure 2.
Characterization of HSPCs in the BM of single and double mutant mice. Percentages of HSPCs from mouse BM analyzed by flow cytometry 20 weeks after BMT (WT, n = 5; Srsf2P95H/+, n = 5; Runx1 knockout, n = 5; double mutant, n = 6). (A) Lin− cells. (B) Lin−Sca-1+c-Kit+ (LSK) cells. (C) Long-term HSCs (LT-HSCs; LSK CD150+CD48−) and multipotent progenitor 1 cells (MPP1; LSK CD150−CD48−). (D) MPP2 (LSK CD150+CD48+) and MPP3/4 (LSK CD150−CD48+). (E) CMPs (Lin−Sca-1−c-Kit+CD16/CD32−CD34+), GMPs (Lin−Sca-1−c-Kit+CD16/CD32+CD34+), and megakaryocyte-erythrocyte progenitors (MEPs; Lin−Sca-1−c-Kit+CD16/CD32−CD34−). Data are mean ± SEM. Significance was determined by 1-way ANOVA with Tukey post hoc test. ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. KO, knockout.
On closer examination of LT-HSCs and various populations of MPPs, we found that Runx1 deficiency selectively expanded the CD150−CD48+ MPP2 and CD150+CD48+ MPP3/4 cell populations (Figure 2C-D; supplemental Figure 2C-D). Interestingly, the Srsf2 P95H mutation modestly contributed to MPP2 and MPP3/4 cell expansion, but its combination with Runx1 deficiency suppressed this phenotype in the BM (Figure 2D), although this suppression effect was less obvious in the spleen (supplemental Figure 2D).
On further differentiation, the impacts of Runx1 deficiency on both single and double mutant mice were transmitted to MPP-derived CMPs and then to CMP-derived MEPs and GMPs in both the BM (Figure 2E) and spleen (supplemental Figure 2E). However, at these stages, the suppressive effect of the Srsf2 P95H mutation became obscured, and further differentiation of MEP cells segregated the impact of the Srsf2 P95H mutation on erythrocytes and that of Runx1 deficiency on megakaryocytes (Figure 1B-C). These findings revealed that in addition to the independent and/or cooperative impacts of single mutations, the combination of Srsf2 P95H mutation with Runx1 deficiency also causes hematopoietic abnormalities by means of their antagonizing effects in specific hematopoietic progenitors.
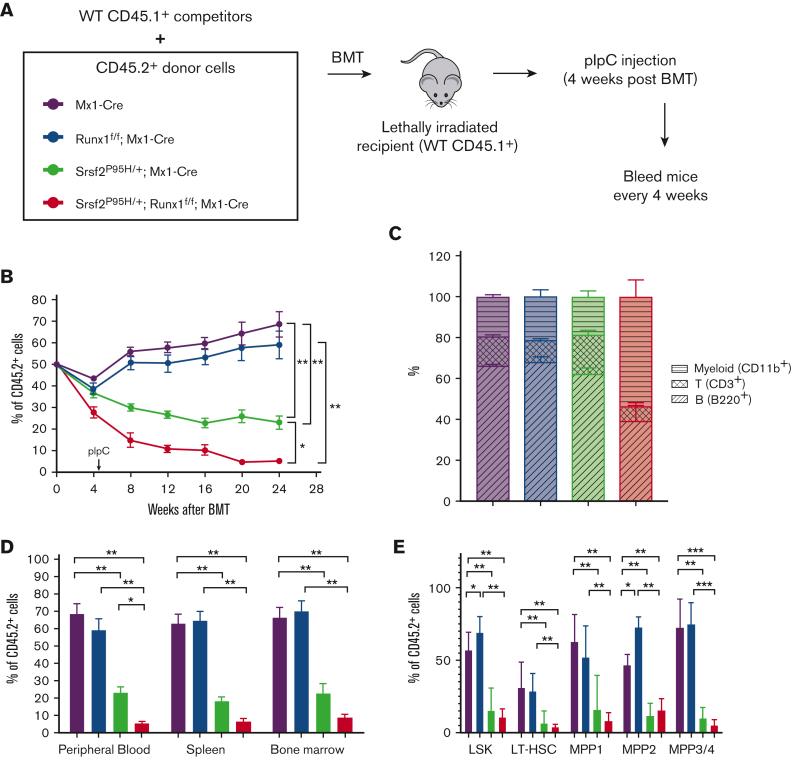
Competitive disadvantage of HSPCs in mutant mice
In addition to changes in HSPC number in patients with MDS, HSPC function is compromised, resulting in defective differentiation and the observed peripheral cytopenias and dysplasia.17,26,28 Therefore, we performed competitive BMT using CD45 congenic mice to analyze HSC fitness in vivo. We mixed CD45.1 competitor BM of WT mice with CD45.2 BM of individual mutant mice in a 1:1 ratio, transplanted these cells into lethally irradiated recipients, and injected the mice with pIpC 4 weeks after transplantation (Figure 3A). Then, we evaluated the chimerism by determining the percentage of CD45.1 and CD45.2 populations in the peripheral blood over time. Although Runx1 deficiency had a minor impact, the Srsf2 P95H mutation conferred a significant competitive disadvantage in this repopulation assay, as previously reported,17 and the double mutant further diminished the competitiveness throughout the experiment (Figure 3B). Interestingly, we observed fewer CD3+ T cells among the double mutant CD45.2 cells at 12 weeks after transplantation (supplemental Figure 3A) and a substantially reduced proportion of B220+ and CD3+ lymphoid cells at 24 weeks after transplantation (Figure 3C), analogous to our observation in the noncompetitive BMT experiments (Figure 1F-G).
Figure 3.
Runx1 loss exacerbates the competitive disadvantage of Srsf2 P95H HSPCs. (A) Schematic diagram of the competitive BMT experiments. (B) Percentage of CD45.2+ cells, as measured by flow cytometry, in the peripheral blood of mice at the indicated time following competitive BMT (WT, n = 6; Srsf2P95H/+, n = 8; Runx1 knockout, n = 7; double mutant, n = 7). (C) Percentages of T cells (CD3+), myeloid cells (CD11b+), and B cells (B220+) in CD45.2+ peripheral blood cells of mice 24 weeks after transplantation (WT, n = 6; Srsf2P95H/+, n = 8; Runx1 knockout, n = 7; double mutant, n = 7). (D) CD45.2 chimerism of all cells in the peripheral blood, spleen, and BM of mice at 24 weeks after transplantation (WT, n = 6; Srsf2P95H/+, n = 8; Runx1 knockout, n = 7; double mutant, n = 7). (E) CD45.2 chimerism of each stem and progenitor cell compartment in the BM of mice 24 weeks after transplantation (WT, n = 6; Srsf2P95H/+, n = 8; Runx1 knockout, n = 7; double mutant, n = 7). The surface markers indicative of each progenitor population are listed in Figure 2A-D. Data are mean ± SEM. Significance was determined by 1-way ANOVA with Tukey post hoc test. ∗P < .05, ∗∗P < .01.
Next, we analyzed the chimerism of total cells in the spleen and BM 24 weeks after transplantation. In agreement with the trend in peripheral blood, the Srsf2 P95H mutation conferred a competitive disadvantage that was further exacerbated by Runx1 loss (Figure 3D). We also examined the competitive disadvantage in individual HSPC populations. We observed a similar expansion of the MPP2 population in Runx1-deficient LSK cells (Figure 3E), as seen in the noncompetitive environment (Figure 2D). However, unlike the minor impact of the Srsf2 P95H mutant in the noncompetitive environment, we saw a major loss of competitiveness across all progenitor subpopulations in the competitive environment (Figure 3E). Double mutant cells exhibited further loss of chimerism, though it was not significantly lower than single Srsf2 P95H mutant cells (Figure 3E). Altogether, these data demonstrated that the Srsf2 P95H mutation is largely responsible for the competitive disadvantage of hematopoietic progenitors, which is further diminished by Runx1 loss. This reduction in HSC fitness contributes to the peripheral blood pancytopenia of double mutant mice, indicative of a worsened MDS phenotype.
Global gene expression dysregulated by single and double mutations
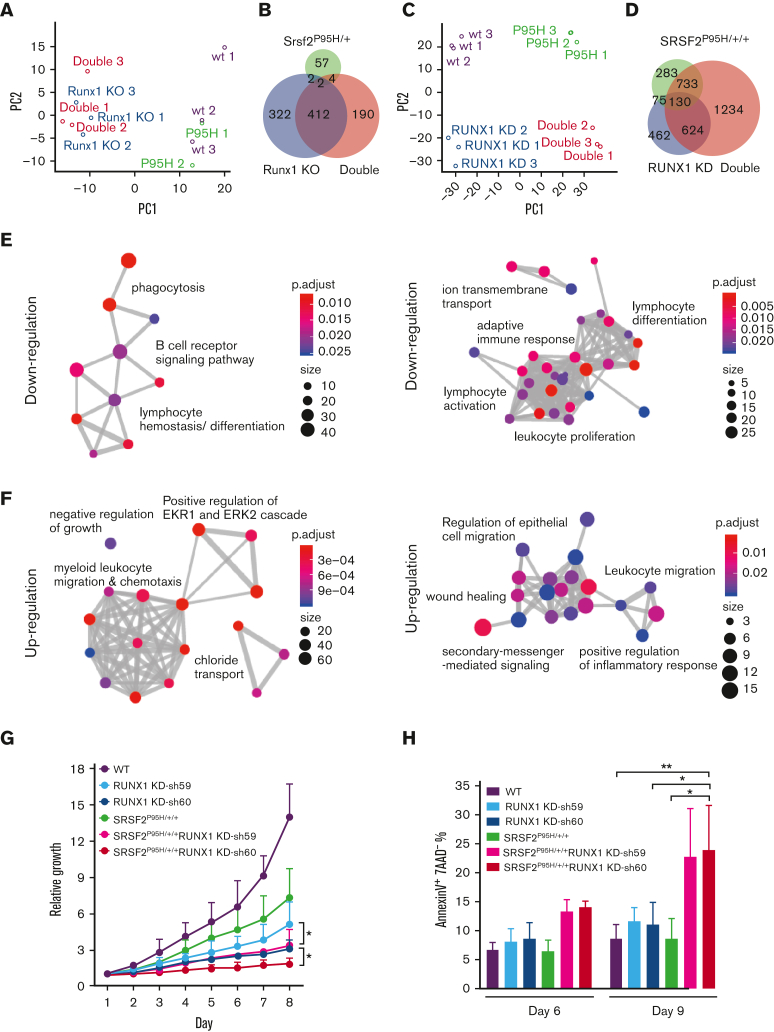
We next explored the mechanistic basis of the magnified MDS phenotype by the coexistence of SRSF2 and RUNX1 mutations. To compare altered gene expression in vivo, we performed RNA-seq on sorted Lin− c-Kit+ (LK) hematopoietic progenitors from the BM of mice from each genotype 4 weeks following pIpC injection (supplemental Figure 4A). To overcome the heterogeneity of isolated hematopoietic cells from mice and to link potential molecular abnormalities in the context of human cells, we also obtained K562 cells previously engineered by CRISPR knockin to harbor the SRSF2 P95H mutation in 1 of 3 SRSF2 alleles29 (supplemental Figure 4B) and performed short hairpin RNA–mediated RUNX1 knockdown in both parental and SRSF2 P95H knockin cells (supplemental Figure 4C). This established a set of isogenic K562 cell lines: WT, SRSF2 P95H knockin (SRSF2P95H/+/+), RUNX1 knockdown (RUNX1 knockdown), and SRSF2 P95H knockin with RUNX1 knockdown (double).
Considering RUNX1 as a transcription factor30 and a previously uncovered function of SRSF2 in transcription,31,32 we hypothesized that the combination of these mutations might promote more severe MDS phenotypes by jointly altering global gene expression. In LK cells, unsupervised principal component analysis of the RNA-seq data showed that the Srsf2 P95H mutant cells were more like WT cells, whereas Runx1 deficient cells were like the double mutant cells (Figure 4A). This was reflected by minor gene expression alterations in Srsf2 P95H mutant mice but a drastically altered gene expression program in Runx1 deficient mice (supplemental Figure 5A; Figure 4B). In contrast, RNA-seq performed on single and double mutant K562 cells upon RUNX1 knockdown (supplemental Figure 5B) showed that SRSF2P95H/+/+ and RUNX1 knockdown cells were both well segregated from WT and double mutant cells (Figure 4C), supported by dramatically induced gene expression changes in these isogenic K562 cell lines (supplemental Figure 5C). The double mutant recapitulated much of the dysregulated gene expression observed in single mutant cells and displayed a clear synergy as there were numerous additionally altered gene expression events (Figure 4D). The distinctions between LK and K562 cells, especially regarding the contribution of the SRSF2 P95H mutation, might reflect differences between mice and humans, their different stages in hematopoiesis, and/or the effect of other mutations in K562 cells.
Figure 4.
Global gene expression is dysregulated by single and double mutants. (A) Unsupervised principal component analysis of differentially expressed genes in murine LK cells of each genotype (WT, n = 3; Srsf2P95H/+, n = 2; Runx1 knockout, n = 3; double mutant, n = 3). (B) Venn diagram showing the overlap between differentially expressed genes of each genotype compared with WT murine LK cells. (C) Unsupervised principal component analysis of differentially expressed genes in K562 cells of each genotype (n = 3 for each genotype). (D) Venn diagram showing the overlap between differentially expressed genes of each genotype compared with WT K562 cells. (E) GO enrichment analysis of the downregulated genes in double mutant K562 cells compared with WT (left) and in double mutant murine LK cells compared with WT (right). (F) GO enrichment analysis of the upregulated genes in double mutant K562 cells compared with WT (left) and in double mutant murine LK cells compared with WT (right). ClusterProfiler33 was used to refine overlapping GOs by calculating enriched functional categories of individual gene clusters for panels E and F. The colors of the circles indicate the P values and the sizes of the circles indicate the number of genes in each GO term. (G) Growth curves of single/double mutant K562 cells. Cells were seeded in triplicate on day 1 and counted daily using trypan blue exclusion. Data are mean ± standard deviation of 3 independent experiments. ∗P < .05. (H) Percentages of early apoptotic cells (annexinV+ 7AAD−) in single/double mutant K562 cells were analyzed by flow cytometry on day 6 and 9 after short hairpin RNA transduction and puromycin selection. Cells were seeded in duplicate in 3 independent experiments. ∗P < .05, ∗∗P < .01. GO, gene ontology; KD, knockdown.
To gain insights into the combined contribution of the Srsf2 P95H mutation and Runx1 deficiency in MDS pathogenesis, we examined the functional classes of genes altered by the double mutant in both murine and human contexts. GO cluster analysis of downregulated genes in double mutant cells of both murine and human origins revealed enrichment of lymphocyte differentiation (Figure 4E), which likely contributed to the B-cell reduction observed in double mutant mice. GO analysis of upregulated genes in double mutant cells uncovered enrichment of gene expression signatures associated with negative regulation of cell growth and cellular apoptosis (ERK1/ERK2 regulation) (Figure 4F).34,35 Therefore, we tested whether the coexistence of the SRSF2 P95H mutation and RUNX1 knockdown affected cellular fitness. Indeed, double mutant K562 cells exhibited the most severe growth defect (Figure 4G) and significantly more apoptotic cells than WT or single mutant cells (Figure 4H). These functional consequences likely provide the selection pressure on various differentiated cell types.
Dramatic impact of RUNX1 deficiency on global splicing
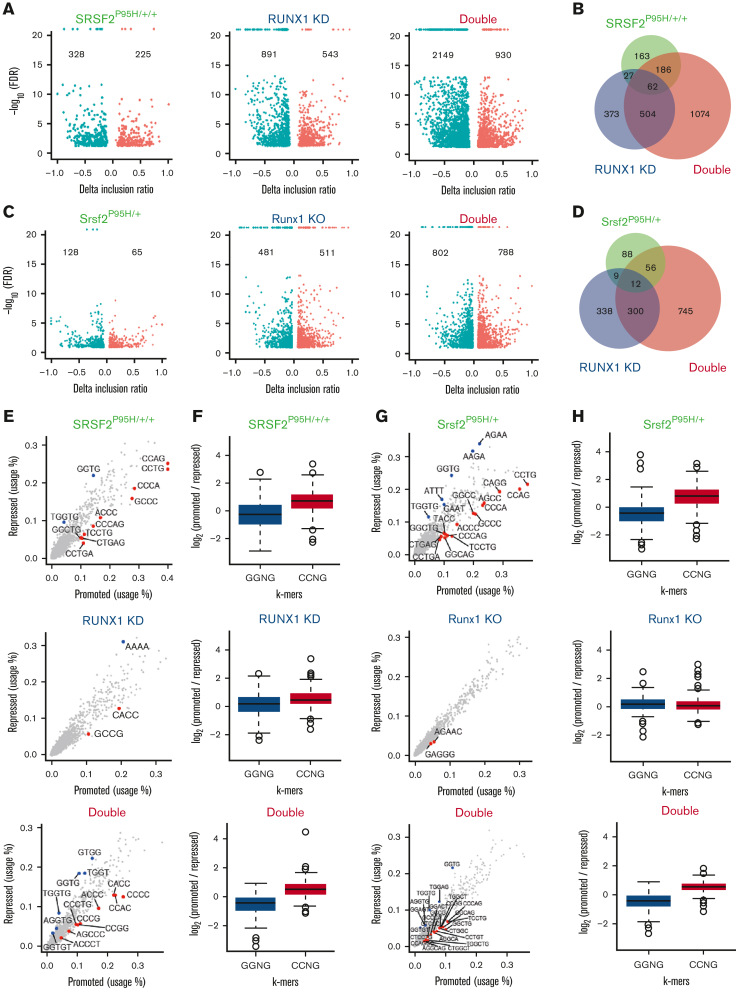
Because transcription factors may also directly or indirectly modulate splice site selections and SRSF2 is a well-established splicing regulator, we next analyzed the impact of these mutations on RNA splicing by using the RNA-seq data generated from murine LK cells and human K562 isogenic cell lines. Interestingly, principal componenet analyses of alternative splicing patterns in both the human and murine systems revealed that cells harboring the SRSF2 P95H mutation were similar to WT cells, whereas RUNX1-deficient cells more closely resembled double mutant cells (supplemental Figure 6A-B). This resulted from the unexpected impact of Runx1 deficiency on splicing, which was more pronounced than that induced by the SRSF2 P95H mutation (Figure 5A,C). Double mutant cells exhibited substantially more dysregulated splicing events than single mutant cells in both K562 cells and LK progenitors (Figure 5B,D). In all 3 mutant genotypes of both human and murine cells, skipped exon (SE) events were the most common splicing abnormalities detected (supplemental Figure 6C-D).
Figure 5.
RUNX1 deficiency dramatically affects global RNA spicing. (A) Volcano plots of significantly altered splice events in SRSF2P95H/+/+, RUNX1 knockdown, and double mutant K562 cells compared with WT cells. (B) Venn diagram showing the overlap between splicing events that were significantly dysregulated in single/double mutant K562 cells relative to WT cells. (C) Volcano plots of significantly altered splice events in Srsf2P95H/+, Runx1 knockout, and double mutant murine LK cells compared with WT LK cells. (D) Venn diagram showing the overlap between splicing events that were significantly dysregulated in single/double mutant LK cells relative to WT LK cells. (E) Scatter plots showing the use of 4, 5, or 6-mer nucleotide sequences on promoted vs repressed exons in single/double mutant cells relative to WT K562 cells. (F) Bar plots quantifying the enrichment of CCNG/GGNG (N = any nucleotide) exonic splicing enhancer motifs adjacent to differentially spliced cassette exons that were promoted vs repressed in single/double mutant K562 cells relative to WT K562 cells (SRSF2P95H/+/+: P < 2.2e-16; RUNX1 knockdown: P = 1.18e-6; double mutant: P < 2.2e-16). (G) Scatter plots showing the use of 4, 5, or 6-mer nucleotide sequences on promoted vs repressed exons in single/double mutant murine LK cells relative to WT LK cells. (H) Bar plots quantifying the enrichment of CCNG/GGNG exonic splicing enhancer motifs adjacent to differentially spliced cassette exons that were promoted vs repressed in single/double mutant murine LK cells relative to WT cells (Srsf2P95H/+: P < 2.2e-16; Runx1 knockout: P = .1443; Double mutant: P < 2.2e-16). FDR, false discovery rate.
To understand how RUNX1 is involved in splicing regulation, we analyzed sequence motifs that were significantly enriched in enhanced or repressed exons in all 3 mutational contexts. As shown previously, the repressed and enhanced exons in single SRSF2 P95H mutant cells were enriched for GGNG and CCNG motifs, respectively,17 in both human K562 cells (Figure 5E-F) and murine LK cells (Figure 5G-H). RUNX1-deficient cells did not exhibit this motif bias, and double mutant cells resembled the single SRSF2 P95H mutant cells. These data suggest that RUNX1 may affect splicing by altering the expression of diverse classes of RNA-binding proteins (RBPs). Supporting this hypothesis, we detected altered expression of 28 RBPs in both RUNX1 knockdown and double mutant K562 cells (supplemental Figure 6E), 10 of which have documented roles in splicing regulation (supplemental Figure 6F).
Synergistic impact of RUNX1 deficiency and SRSF2 mutation on the DNA damage response and cell cycle checkpoint
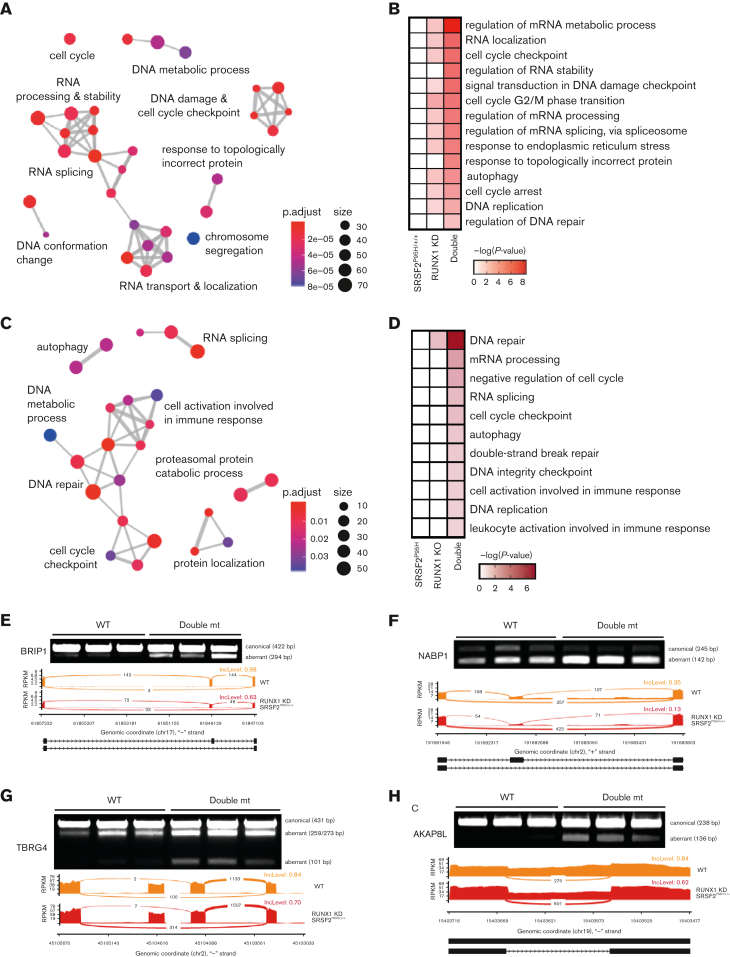
Given the synergy between Runx1 deficiency and Srsf2 P95H mutation in LK progenitors that was more pronounced at the level of splicing than transcription, we hypothesized that the exacerbated splicing dysregulation in double mutant cells might contribute to more severe MDS phenotypes. We first performed GO enrichment analysis of the genes with altered splicing (Figure 6A). Interestingly, among genes with dysregulated splicing in SRSF2P95H/+/+ K562 cells, we found no significant GO enrichment, whereas in RUNX1 knockdown cells, we identified the enrichment of GO clusters related to RNA splicing/processing/location, DNA replication, and cell cycle checkpoint regulation (Figure 6B). Importantly, we found even stronger enrichment of those GO clusters and additional ones, such as a cluster associated with DNA damage, in double mutant K562 cells (Figure 6B). These affected pathways were more pronounced in murine LK cells, where only the double mutant cells demonstrated significantly enriched GO clusters (Figure 6C-D). Considering the known roles of DNA damage and aberrant cell cycle regulation in MDS,36, 37, 38, 39, 40 these enriched pathways support a link between exacerbated splicing defects in double mutant cells and worsened MDS phenotypes. For example, among synergistically altered splicing events in LK progenitors, we identified increased exon skipping in Fanca and Atm genes involved in DNA damage response (supplemental Figure 7A) and Fyn and Plcb2 genes important for B-cell development (supplemental Figure 7B), which may contribute to the B-cell defects observed in double mutant mice.
Figure 6.
Mutant SRSF2 and RUNX1 deficiency synergistically affect the DNA damage response and cell cycle. (A) GO enrichment cluster analysis of the dysregulated splicing events in double mutant (mt) cells compared with WT K562 cells. (B) Heatmap of the most significantly enriched GO categories of misspliced transcripts in each single/double mutant K562 genotype relative to WT K562 cells. (C) GO enrichment cluster analysis of the dysregulated splicing events in double mutant murine LK cells compared with WT cells. ClusterProfiler33 was used to refine overlapping GOs by calculating enriched functional categories of individual gene clusters in panels A and C. The colors of the circles indicate the P values and the sizes of the circles indicate the number of genes in each GO term. (D) Heatmap of the most significantly enriched GO categories of misspliced transcripts in single and double mutant LK cells relative to WT cells. (E-H) Reverse transcription polymerase chain reaction validation and sashimi plots depicting the abnormal splicing of genes related to the DNA damage checkpoint (BRIP1), DNA repair (NABP1), and cell cycle (TBRG4 and AKAP8L). bp, base pair; mRNA, messenger RNA; RPKM, reads per kilo base per million mapped reads.
To link specific splicing alterations to aberrant DNA damage responses and cell cycle regulation, we validated SE events of genes in these critical MDS-related pathways. In all the selected genes, exon skipping would induce a frameshift that produces a truncated protein or reduces expression of the corresponding protein, impeding DNA damage repair or dysregulating cell cycle progression. By reverse transcription polymerase chain reaction in K562 cells, we confirmed double mutant–induced exon skipping in the Fanconi anemia J gene, BRIP1 (also known as FANCJ, BACH1) (Figure 6E), which encodes a DEAH helicase that interacts with BRCA1 and has BRCA1-dependent DNA repair and checkpoint functions.41,42 Exclusion of the cassette exon in BRIP1 transcripts results in a truncated protein, which is in line with the causal role of inactivating truncation and point mutations of BRIP1 in Fanconi anemia.43, 44, 45, 46, 47 We similarly confirmed double mutant–induced exon skipping in NABP1 (also known as SOSS-B2), a component of the SOSS complex that senses single-stranded DNA and promotes DNA repair (Figure 6F),48,49 and in FANCD2/FANCI-associated nuclease 1 (FAN1), which plays a critical role in DNA interstrand crosslink repair (supplemental Figure 7C).50, 51, 52 We also validated SEs in genes involved in cell cycle progression, including TBRG453 and AKAP8L54 (Figure 6G-H).
In addition to missplicing of genes involved in DNA damage and cell cycle regulation, there were several other interesting genes with altered splicing that may have implications on MDS severity. For example, we detected 2 exon skipping events in PFKM (supplemental Figure 7D), which encodes Phosphofructokinase Muscle, a key regulatory enzyme of glycolysis, with deficient activity linked to hemolytic anemia.55,56 Lastly, we observed significant enrichment of misspliced genes involved in RNA processing and metabolism, such as EXOSC9 (supplemental Figure 7E),57, 58, 59 illuminating the possibility that aberrant splicing of RNA processing regulators may further amplify splicing defects in double mutant cells, contributing to worsened MDS phenotypes.
Discussion
Here, we generated a mouse model to study the phenotypic and mechanistic impacts of the SRSF2 P95H mutation in combination with RUNX1 deficiency, mutations significantly cooccurring in patients with MDS.2,3,15,16,22,23 We focused on homozygous Runx1 knockout to model dominant-negative RUNX1 mutations. Future studies will be required to test the impact of heterozygous Runx1 knockout, modeling loss-of-function mutations, which may produce distinct phenotypes from those observed here. Using our double mutant mouse model, we demonstrated that the combination of splicing and transcription perturbation promotes more severe MDS phenotypes than either single mutation. In addition, we uncovered a surprising role for the transcription factor RUNX1 in splicing and underscored the importance of the DNA damage response in MDS pathogenesis.
In our mouse model, double mutant mice had more pervasive MDS-related phenotypes than single mutant mice because more cell types were affected by the coexisting mutations. For instance, double mutant mice exhibited aberrant hematopoietic phenotypes related to either Srsf2 P95H (leukopenia, anemia, and dysplastic neutrophils) or Runx1 deficiency (leukopenia, thrombocytopenia, myeloid cell expansion, and MPP cell expansion). Because the mutations affect mostly nonoverlapping hematopoietic cell populations, the additive effect of these coexisting mutations leads to multilineage defects. Besides such additive effects, we also observed synergistic effects of these mutations in the same cell populations, resulting in more prevalent dysplastic cellular morphology, worsened B-cell deficiency, and exacerbated competitive deficiency. Interestingly, a reduction in B cells has also been observed in patients with MDS60 and in both Srsf2 P95H17,20 and Runx1 knockout25 mouse models. Future studies are warranted to address the mechanism and impact of this unexplored phenotype.
Despite the more severe hematopoietic defects in double mutant mice, we did not observe transformation to AML. This reflects different phenotypic severities induced by different combinations of mutations, as highlighted in a previous study where the coexistence of Srsf2 and Idh2 mutations led to leukemia development, whereas the coexistence of Srsf2 and Tet2 mutations did not.61 The disease severity could be further enhanced by exposing the mice to a mutagen, as was shown in a mouse model of Runx1 deficiency in combination with the mutant splicing factor U2af1.62 In this previous study, double mutant mice did not develop AML until they were exposed to the mutagen N-ethyl-N-nitrosourea, which triggered additional mutations in Tet2, Idh1, and Gata2.62 Consequently, we speculate that certain tertiary mutations (TET2, ASXL1, and STAG2) identified in patients with MDS with double SRSF2 and RUNX1 deficiencies may contribute to leukemic transformation.2,24
Intriguingly, in both of our human and mouse isogenic models, RUNX1 deficiency resulted in profound splicing alterations and double mutant cells had the most severe splicing dysregulation. This is reminiscent of a previous study that showed patient cells harboring mutations in both SRSF2 and the epigenetic regulator IDH2 had greater splicing alterations than either single mutation alone.61 Here, we provide evidence that RUNX1 deficiency may modulate splicing by altering the expression of RBPs, likely owing to the role of RUNX1 in transcription control63 and/or by causing missplicing of splicing regulators. However, RUNX1 may also directly contribute to splicing regulation by potential RNA binding of its runt homology domain as observed in vitro64,65 or through its interaction with splicing factors during cotranscriptional splicing as observed with other transcription factors.66,67 The involvement of diverse mechanisms may explain the lack of enriched motifs associated with altered splicing events in RUNX1-deficient cells.
Importantly, among misspliced transcripts in human and mouse double mutant cells, we observed significant enrichment of genes involved in cell cycle regulation and the DNA damage response, characteristic of MDS.36,37 Our data agree with a recent report that also identified overrepresentation of misspliced cell cycle and DNA repair transcripts in human and murine SRSF2P95H/+ samples.68 These pathways were also among the top synthetic lethal interactions with SRSF2P95H/+ in a CRISPR dropout screen.68 Missplicing of target genes involved in the DNA damage response may further synergize with genomic instability and DNA damage by means of accumulated R-loops39,69 and in combination with RUNX1 deficiency–induced modulation of p53 and FANCD2 activities.70,71 Together, these findings support the therapeutic potential of targeting the cell cycle and DNA damage in patients harboring SRSF2 or RUNX1 mutations.
In summary, we have shown that Runx1 deficiency and mutant Srsf2 collaborate to impair multilineage hematopoiesis and exacerbate MDS disease phenotypes in vivo. At the genome-wide level, loss of RUNX1 dysregulates splicing outcomes and cooperates with mutant SRSF2 to further perturb the expression and splicing of key regulators involved in cell cycle control and the DNA damage response. These results illustrate how mutations in a transcription factor and splicing factor can cooperatively promote pathogenesis and support further studies to explore the therapeutic potential of targeting the DNA damage response or aberrant splicing in patients with MDS.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Acknowledgments
The authors are grateful to members of the D.-E.Z. and X.-D.F. laboratories for technical help, reagent sharing, and discussion; and to Nancy A. Speck for generously providing Runx1 conditional knockout mice. The authors also thank Kristen Jepsen from the UC San Diego Institute for Genomic Medicine Genomics Center for the help and technical support in RNA sequencing, and Jesus Olvera and Cody Fine from the UC San Diego Human Embryonic Stem Cell Core Facility for their help and technical advice during cell sorting and fluorescence-activated cell sorting.
This work was supported by grant from the National Institutes of Health (grant NIHR01DK098808) (X.-D.F. and D.-E. Z.).
Authorship
Contribution: Y.-J.H. and D.-E.Z. designed the experiments; Y.-J.H., M.Y., A.G.D., and S.M. performed the experiments; J.-Y.C. and Y.H. performed the RNA data analysis; O.A.-W. provided the SRSF2 P95H knockin mice; S.K. and R.B. provided the CRISPR-edited K562 cells; Y.-J.H., A.G.D., J.-Y.C., M.Y., L.C., X.-D.F., and D.-E.Z. participated in critical discussion; Y.-J.H., A.G.D., and X.-D.F. wrote the manuscript; and D.-E.Z. supervised data analysis and manuscript preparation.
Footnotes
The sequencing data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE200446).
Data are available on request from the corresponding author, Dong-Er Zhang (d7zhang@ucsd.edu).
The full-text version of this article contains a data supplement.
The current affiliation for J.-Y.C. is State Key Laboratory of Pharmaceutical Biotechnology, School of Life Sciences, Chemistry and Biomedicine Innovation Center, Nanjing University, Nanjing, China.
The current affiliation for L.C. is Hubei Key Laboratory of Cell Homeostasis, RNA Institute, College of Life Sciences, Wuhan University, Wuhan, China.
Contributor Information
Xiang-Dong Fu, Email: xdfu@ucsd.edu.
Dong-Er Zhang, Email: d7zhang@ucsd.edu.
Supplementary Material
References
- 1.Gangat N, Patnaik MM, Tefferi A. Myelodysplastic syndromes: contemporary review and how we treat. Am J Hematol. 2016;91(1):76–89. doi: 10.1002/ajh.24253. [DOI] [PubMed] [Google Scholar]
- 2.Haferlach T, Nagata Y, Grossmann V, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28(2):241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, Gerstung M, Malcovati L, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen CY, Lin LI, Tang JL, et al. RUNX1 gene mutation in primary myelodysplastic syndrome--the mutation can be detected early at diagnosis or acquired during disease progression and is associated with poor outcome. Br J Haematol. 2007;139(3):405–414. doi: 10.1111/j.1365-2141.2007.06811.x. [DOI] [PubMed] [Google Scholar]
- 5.Tsai SC, Shih LY, Liang ST, et al. Biological activities of RUNX1 mutants predict secondary acute leukemia transformation from chronic myelomonocytic leukemia and myelodysplastic syndromes. Clin Cancer Res. 2015;21(15):3541–3551. doi: 10.1158/1078-0432.CCR-14-2203. [DOI] [PubMed] [Google Scholar]
- 6.Ito Y, Bae SC, Chuang LS. The RUNX family: developmental regulators in cancer. Nat Rev Cancer. 2015;15(2):81–95. doi: 10.1038/nrc3877. [DOI] [PubMed] [Google Scholar]
- 7.Sood R, Kamikubo Y, Liu P. Role of RUNX1 in hematological malignancies. Blood. 2017;129(15):2070–2082. doi: 10.1182/blood-2016-10-687830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen S, Benbarche S, Abdel-Wahab O. Splicing factor mutations in hematologic malignancies. Blood. 2021;138(8):599–612. doi: 10.1182/blood.2019004260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 11.Papaemmanuil E, Cazzola M, Boultwood J, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365(15):1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walter MJ, Shen D, Ding L, et al. Clonal architecture of secondary acute myeloid leukemia. N Engl J Med. 2012;366(12):1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sperling AS, Gibson CJ, Ebert BL. The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat Rev Cancer. 2017;17(1):5–19. doi: 10.1038/nrc.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pellagatti A, Roy S, Di Genua C, et al. Targeted resequencing analysis of 31 genes commonly mutated in myeloid disorders in serial samples from myelodysplastic syndrome patients showing disease progression. Leukemia. 2016;30(1):247–250. doi: 10.1038/leu.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thol F, Kade S, Schlarmann C, et al. Frequency and prognostic impact of mutations in SRSF2, U2AF1, and ZRSR2 in patients with myelodysplastic syndromes. Blood. 2012;119(15):3578–3584. doi: 10.1182/blood-2011-12-399337. [DOI] [PubMed] [Google Scholar]
- 16.Wu SJ, Kuo YY, Hou HA, et al. The clinical implication of SRSF2 mutation in patients with myelodysplastic syndrome and its stability during disease evolution. Blood. 2012;120(15):3106–3111. doi: 10.1182/blood-2012-02-412296. [DOI] [PubMed] [Google Scholar]
- 17.Kim E, Ilagan JO, Liang Y, et al. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell. 2015;27(5):617–630. doi: 10.1016/j.ccell.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komeno Y, Huang YJ, Qiu J, et al. SRSF2 is essential for hematopoiesis, and its myelodysplastic syndrome-related mutations dysregulate alternative pre-mRNA splicing. Mol Cell Biol. 2015;35(17):3071–3082. doi: 10.1128/MCB.00202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smeets MF, Tan SY, Xu JJ, et al. Srsf2(P95H) initiates myeloid bias and myelodysplastic/myeloproliferative syndrome from hemopoietic stem cells. Blood. 2018;132(6):608–621. doi: 10.1182/blood-2018-04-845602. [DOI] [PubMed] [Google Scholar]
- 20.Kon A, Yamazaki S, Nannya Y, et al. Physiological Srsf2 P95H expression causes impaired hematopoietic stem cell functions and aberrant RNA splicing in mice. Blood. 2018;131(6):621–635. doi: 10.1182/blood-2017-01-762393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J, Lieu YK, Ali AM, et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci U S A. 2015;112(34):E4726–E4734. doi: 10.1073/pnas.1514105112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Damm F, Kosmider O, Gelsi-Boyer V, et al. Mutations affecting mRNA splicing define distinct clinical phenotypes and correlate with patient outcome in myelodysplastic syndromes. Blood. 2012;119(14):3211–3218. doi: 10.1182/blood-2011-12-400994. [DOI] [PubMed] [Google Scholar]
- 23.Gaidzik VI, Teleanu V, Papaemmanuil E, et al. RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia. 2016;30(11):2160–2168. doi: 10.1038/leu.2016.126. [DOI] [PubMed] [Google Scholar]
- 24.Ochi Y, Kon A, Sakata T, et al. Combined cohesin-RUNX1 deficiency synergistically perturbs chromatin looping and causes myelodysplastic syndromes. Cancer Discov. 2020;10(6):836–853. doi: 10.1158/2159-8290.CD-19-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Growney JD, Shigematsu H, Li Z, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106(2):494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shastri A, Will B, Steidl U, Verma A. Stem and progenitor cell alterations in myelodysplastic syndromes. Blood. 2017;129(12):1586–1594. doi: 10.1182/blood-2016-10-696062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ichikawa M, Asai T, Saito T, et al. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med. 2004;10(3):299–304. doi: 10.1038/nm997. [DOI] [PubMed] [Google Scholar]
- 28.Pang WW, Pluvinage JV, Price EA, et al. Hematopoietic stem cell and progenitor cell mechanisms in myelodysplastic syndromes. Proc Natl Acad Sci U S A. 2013;110(8):3011–3016. doi: 10.1073/pnas.1222861110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollyea DA, Harris C, Rabe JL, et al. Myelodysplastic syndrome-associated spliceosome gene mutations enhance innate immune signaling. Haematologica. 2019;104(9):e388–e392. doi: 10.3324/haematol.2018.214155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Wijnen AJ, Stein GS, Gergen JP, et al. Nomenclature for Runt-related (RUNX) proteins. Oncogene. 2004;23(24):4209–4210. doi: 10.1038/sj.onc.1207758. [DOI] [PubMed] [Google Scholar]
- 31.Mo S, Ji X, Fu XD. Unique role of SRSF2 in transcription activation and diverse functions of the SR and hnRNP proteins in gene expression regulation. Transcription. 2013;4(5):251–259. doi: 10.4161/trns.26932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji X, Zhou Y, Pandit S, et al. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153(4):855–868. doi: 10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu GC, Wang LG, Han YY, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, Martindale JL, Holbrook NJ. Requirement for ERK activation in cisplatin-induced apoptosis. J Biol Chem. 2000;275(50):39435–39443. doi: 10.1074/jbc.M004583200. [DOI] [PubMed] [Google Scholar]
- 35.Cagnol S, Chambard JC. ERK and cell death: mechanisms of ERK-induced cell death--apoptosis, autophagy and senescence. FEBS J. 2010;277(1):2–21. doi: 10.1111/j.1742-4658.2009.07366.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhou T, Hasty P, Walter CA, Bishop AJ, Scott LM, Rebel VI. Myelodysplastic syndrome: an inability to appropriately respond to damaged DNA? Exp Hematol. 2013;41(8):665–674. doi: 10.1016/j.exphem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popp HD, Naumann N, Brendel S, et al. Increase of DNA damage and alteration of the DNA damage response in myelodysplastic syndromes and acute myeloid leukemias. Leuk Res. 2017;57:112–118. doi: 10.1016/j.leukres.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 38.Ogawa S. Genetics of MDS. Blood. 2019;133(10):1049–1059. doi: 10.1182/blood-2018-10-844621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh S, Ahmed D, Dolatshad H, et al. SF3B1 mutations induce R-loop accumulation and DNA damage in MDS and leukemia cells with therapeutic implications. Leukemia. 2020;34:2525–2530. doi: 10.1038/s41375-020-0753-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen HD, Leong WY, Li W, et al. Spliceosome mutations induce R loop-associated sensitivity to ATR inhibition in myelodysplastic syndromes. Cancer Res. 2018;78(18):5363–5374. doi: 10.1158/0008-5472.CAN-17-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cantor SB, Bell DW, Ganesan S, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105(1):149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 42.Cantor S, Drapkin R, Zhang F, et al. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc Natl Acad Sci U S A. 2004;101(8):2357–2362. doi: 10.1073/pnas.0308717101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seal S, Thompson D, Renwick A, et al. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38(11):1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 44.Levitus M, Waisfisz Q, Godthelp BC, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group. J Nat Genet. 2005;37(9):934–935. doi: 10.1038/ng1625. [DOI] [PubMed] [Google Scholar]
- 45.Levran O, Attwooll C, Henry RT, et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet. 2005;37(9):931–933. doi: 10.1038/ng1624. [DOI] [PubMed] [Google Scholar]
- 46.Wu Y, Sommers JA, Suhasini AN, et al. Fanconi anemia group J mutation abolishes its DNA repair function by uncoupling DNA translocation from helicase activity or disruption of protein-DNA complexes. Blood. 2010;116(19):3780–3791. doi: 10.1182/blood-2009-11-256016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo M, Vidhyasagar V, Ding H, Wu Y. Insight into the roles of helicase motif Ia by characterizing Fanconi anemia group J protein (FANCJ) patient mutations. J Biol Chem. 2014;289(15):10551–10565. doi: 10.1074/jbc.M113.538892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J, Gong Z, Ghosal G, Chen J. SOSS complexes participate in the maintenance of genomic stability. Mol Cell. 2009;35(3):384–393. doi: 10.1016/j.molcel.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, Bolderson E, Kumar R, et al. HSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J Biol Chem. 2009;284(35):23525–23531. doi: 10.1074/jbc.C109.039586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trujillo JP, Mina LB, Pujol R, et al. On the role of FAN1 in Fanconi anemia. Blood. 2012;120(1):86–89. doi: 10.1182/blood-2012-04-420604. [DOI] [PubMed] [Google Scholar]
- 51.Smogorzewska A, Desetty R, Saito TT, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39(1):36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoshikiyo K, Kratz K, Hirota K, et al. KIAA1018/FAN1 nuclease protects cells against genomic instability induced by interstrand cross-linking agents. Proc Natl Acad Sci U S A. 2010;107(50):21553–21557. doi: 10.1073/pnas.1011081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Edwards MC, Liegeois N, Horecka J, et al. Human CPR (cell cycle progression restoration) genes impart a Far- phenotype on yeast cells. Genetics. 1997;147(3):1063–1076. doi: 10.1093/genetics/147.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Kao GD, Garcia BA, et al. A novel histone deacetylase pathway regulates mitosis by modulating Aurora B kinase activity. Genes Dev. 2006;20(18):2566–2579. doi: 10.1101/gad.1455006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Wijk R, van Solinge WW. The energy-less red blood cell is lost: erythrocyte enzyme abnormalities of glycolysis. Blood. 2005;106(13):4034–4042. doi: 10.1182/blood-2005-04-1622. [DOI] [PubMed] [Google Scholar]
- 56.Garcia M, Pujol A, Ruzo A, et al. Phosphofructo-1-kinase deficiency leads to a severe cardiac and hematological disorder in addition to skeletal muscle glycogenosis. PLoS Genet. 2009;5(8) doi: 10.1371/journal.pgen.1000615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mukherjee D, Gao M, O'Connor JP, et al. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21(1-2):165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.West S, Gromak N, Norbury CJ, Proudfoot NJ. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol Cell. 2006;21(3):437–443. doi: 10.1016/j.molcel.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 59.van Dijk EL, Schilders G, Pruijn GJ. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA. 2007;13(7):1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sternberg A, Killick S, Littlewood T, et al. Evidence for reduced B-cell progenitors in early (low-risk) myelodysplastic syndrome. Blood. 2005;106(9):2982–2991. doi: 10.1182/blood-2005-04-1543. [DOI] [PubMed] [Google Scholar]
- 61.Yoshimi A, Lin KT, Wiseman DH, et al. Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis. Nature. 2019;574(7777):273–277. doi: 10.1038/s41586-019-1618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fei DL, Zhen T, Durham B, et al. Impaired hematopoiesis and leukemia development in mice with a conditional knock-in allele of a mutant splicing factor gene U2af1. Proc Natl Acad Sci U S A. 2018;115(44):E10437–E10446. doi: 10.1073/pnas.1812669115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grinev VV, Barneh F, Ilyushonak IM, et al. RUNX1/RUNX1T1 mediates alternative splicing and reorganises the transcriptional landscape in leukemia. Nat Commun. 2021;12(1):520. doi: 10.1038/s41467-020-20848-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barton JL, Bunka DH, Knowling SE, et al. Characterization of RNA aptamers that disrupt the RUNX1-CBFbeta/DNA complex. Nucleic Acids Res. 2009;37(20):6818–6830. doi: 10.1093/nar/gkp728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fukunaga J, Nomura Y, Tanaka Y, et al. The Runt domain of AML1 (RUNX1) binds a sequence-conserved RNA motif that mimics a DNA element. RNA. 2013;19(7):927–936. doi: 10.1261/rna.037879.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10(10):1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15(10):1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu JJ, Chalk AM, Nikolic I, Simpson K, Smeets MF, Walkley C. Genome-wide screening identifies cell cycle control as a synthetic lethal pathway with SRSF2P95H mutation. Blood Adv. 2022;6(7):2092–2106. doi: 10.1182/bloodadvances.2021004571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen L, Chen JY, Huang YJ, et al. The augmented R-loop is a unifying mechanism for myelodysplastic syndromes induced by high-risk splicing factor mutations. Mol Cell. 2018;69(3):412–425.e416. doi: 10.1016/j.molcel.2017.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bellissimo DC, Speck NA. RUNX1 mutations in inherited and sporadic leukemia. Front Cell Dev Biol. 2017;5:111. doi: 10.3389/fcell.2017.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Samarakkody AS, Shin NY, Cantor AB. Role of RUNX family transcription factors in DNA damage response. Mol Cells. 2020;43(2):99–106. doi: 10.14348/molcells.2019.0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.