Abstract
Asthma is a significant health-care burden that has great impact on the quality of life of patients and their families. The limited amount of previously reported data and complicated pathophysiology of asthma make it a difficult to treat and significant economic burden on public healthcare systems. Ferula asafoetida is an herbaceous, monoecious, perennial plant of the Umbelliferae family. In Asia, F. asafoetida is used to treat a range of diseases and disorders, including asthma. Several in vitro studies demonstrated the therapeutic efficacy of F. asafoetida against asthma. Nevertheless, the precise molecular mechanism is yet to be discovered. In the framework of current study, network pharmacology approach was used to identify the bioactive compounds of F. asafoetida in order to better understand its molecular mechanism for the treatment of asthma. In present work, we explored a compound-target-pathway network and discovered that assafoetidin, cynaroside, farnesiferol-B, farnesiferol-C, galbanic-acid, and luteolin significantly influenced the development of asthma by targeting MAPK3, AKT1 and TNF genes. Later, docking analysis revealed that active constituents of F. asafoetida bind stably with three target proteins and function as asthma repressor by regulating the expression of MAPK3, AKT1 and TNF genes. Thus, integration of network pharmacology with molecular docking revealed that F. asafoetida prevent asthma by modulating asthma-related signaling pathways. This study lays the basis for establishing the efficacy of multi-component, multi-target compound formulae, as well as investigating new therapeutic targets for asthma.
Keywords: Ferula asafoetida, Asthma, Network pharmacology, Molecular docking
1. Introduction
Asthma refers to a common respiratory disease with high morbidity and mortality. Asthma has affected more than 300 million individuals around the globe which makes it the most common respiratory disease (Szefler et al., 2014).Asthma is a chronic inflammatory disease of the airways that causes wheezing and other symptoms like coughing, dyspnea, and chest tightness, as well as fluctuating expiratory airway blockage. Asthma has become a noteworthy health issue (Masoli et al., 2004) due to its increasing morbidity rate throughout the past few years globally due to environmental and lifestyle changes (Selgrade et al., 2006). Currently, inhaled corticosteroids (ICS) are the effective for the long-term asthma treatment and the addition of long-acting β2-agonists further help in controlling asthma (Choby and Lee, 2015). Long-term exposure of ICS may lead to adverse effects on the patient’s health (Heffler et al., 2018). Alternative low-toxicity therapies need to be developed to mitigate later on the risks of asthma attacks. For better treatment of diseases, traditional herbal medicines are being used widely since last decade and as reported by WHO, approximately 11 % of drugs are from plant origin (Veeresham, 2012). In Pakistan respiratory diseases such as asthma is common because of severe environmental conditions and limited access of population to medical facilities. People usually rely on herbal medicine to cure respiratory diseases and it is also practiced around the globe (Alamgeer et al., 2018). Pakistan produces a variety of medicinal plants with over 6000 species in account of its diverse climate conditions (Ali and Qaiser, 1986). Multiple studies demonstrated that few distinct natural components and herbs can express anti-inflammatory properties (Plaeger, 2003). Ferula asafoetida belongs to the family Umbelliferae (Apiaceae). It is well known anti-inflammatory herbal medicine which can prohibit and cure inflammation caused by asthma (Shahrajabian et al., 2021a). Reportedly F. asafoetida has specific compounds such as α-pinene, umbelliferone, ferulic acid and luteolin showing anti-inflammatory pharmacological properties that can be utilized for the treatment of asthma (Mahendra and Bisht, 2012a), the process of anti-inflammatory activity of F. asafoetida against asthma until now is un-clarified.
Being an emerging area of pharmacology, “network pharmacology” is considered a new approach to drug designing. To date, this technique has been successful in elucidating the multi-target effects of medicinal plants for curing enormous types of diseases and disorders. The traditional medicine is one of the safest medicines demonstrated by extensive studies. Network pharmacology, a systematic biology approach has revolutionized the interaction studies between active herbal ingredients and potential targets of disease. It provides feasible and reliable ways to explore potential molecular mechanisms and key targets. The concept of network pharmacology relies on the concept of “network target, multicomponent therapeutics”, which shifts the paradigm away from the concept of one gene, one target, and one disease.
The compounds of F. asafoetida and the putative mechanism behind its anti-asthmatic activity were investigated utilizing network pharmacology integrating with molecular docking in the current study. According our understanding this is the leading study to classify the underlying mechanism of F. asafoetida for asthma treatment using bioinformatics and network pharmacology.
2. Methodology
2.1. Active compounds in F. Asafoetida
Information related to active compound of F. asafoetida was retrieved from literature and publically available databases. F. asafoetida related compounds were collected from phytochemical based databases, including traditional Chinese medicines integrated database (TCMSP) (Ru et al., 2014), KNApSAcK-3D (Nakamura et al., 2013), and Dr. Duke's Phytochemical and Ethnobotanical Database (Lans and van Asseldonk, 2020). The keyword “F. asafoetida” was used in the databases, while literature mining was conducted on PubMed, and Google Scholar. All of the predicted F. asafoetida constituents were virtually screened based on oral bioavailability (OB) and drug likeness (DL). Only those compounds were preferred for subsequent analysis which met the precise standards of DL ≥ 0.18 and OB ≥ 30 %. Regarding this, Admet SAR2.0 (Yang et al., 2019) and Molsoft were used to compute the OB and DL of all active constituents, respectively. Meanwhile, PubChem (Wang et al., 2009) and ChemSpider (Pence and Williams, 2010) were used to collect chemical information of predicted compounds such as CID number, structure, and molecular weight.
2.2. Target prediction
Prediction of target genes is an initial stage to reveal the molecular interaction of therapeutic plants for treatment of various types of diseases and disorders. Based on simplified molecular-input line-entry system (SMILES), targeted gene(s) of compounds were retrieved through Swiss Target Prediction (Gfeller et al., 2014) and STITCH (Kuhn et al., 2008) databases. In case of STITCH database, SMILES of each compound were inputted with the search restricted to “Homo sapiens”. And accumulating the targets with a combined-score ≥ 0.7 On the other hand, the SMILES numbers of compounds were then uploaded into the Swiss Target Prediction online platform. As the predicted of target genes were performed on the basis of structural similarity using a reverse pharmacophore combination method consequently, targets with probability ≥ 0.7 was selected.
Prediction of target genes related to disease was performed using GeneCards (Rebhan et al., 1998) and DisGeNET (Piñero et al., 2015) databases. Furthermore, these databases offer brief genomic data as well as functional annotations for well-studied human genes. All redundant genes were removed, and the common name of the target gene was obtained using UniProtKB function in UniProt (Bateman, 2019), with “Homo sapiens” as the organism. The final overlapping target genes between F. asafoetida and asthma-targeted genes were recognized and visualized by Venny 2.1.
2.3. Network construction
The protein–protein interactions (PPI) network of overlapped genes were obtained through STRING (von Mering et al., 2003) database, which offers experimental as well as predicted relationship information based on systematic co-expression approach, observation of shared selective signals across genomes, and automated scientific literature text mining. In framework of current study, interactions having combined score ≥ 0.4 were selected for further analysis. Cytoscape 3.2.0 (Kohl et al., 2011) was then used to generate three types of networks to further analyze the multi-scale action mechanisms of F. asafoetida compound for treatment of asthma (1) network of compound-target, (2) Target-pathway network, and (3) compound-target-pathway network. In network, nodes represented the compound, targets, and pathways while solid lines described the interaction between nodes. The degree that is a topological parameter reveals the significance of component/target/pathway in the network.
2.4. Gene ontology and KEGG pathway enrichment
Database for annotation, visualization, and integrated discovery [DAVID; (Dennis et al., 2003)] online functional annotation software was used to conduct gene ontology (GO) and KEGG pathway analysis. DAVID was used to predict the role of overlapped target genes at three contracting levels: Molecular function (MF), Biological process (BP), and Cellular component (CC). Later, on basis of P value, a bubble map was created using ggplot2 package present in R. GO terms and KEGG pathways with p value < 0.05 were examined statistically significant.
2.5. Molecular docking
A molecular docking technique was used to validate important targets. In the drug discovery toolbox, molecular docking has become a lightning rod and the most appropriate technique. It allows for the identification of interaction that keep ligands attached to their respective proteins. The crystalline structures of potential targets were collected from RCSB Protein Data Bank (PDB) (https://www.rcsb.org/) (Sussman et al., 1998). The PDB has offered as a central repository for knowledge on protein 3D structures. Following that, the site finder tool from the Molecular operating environment (MOE) (Althagafi et al., 2019) was employed to predict the binding pockets of target proteins, and PyRx (Dallakyan and Olson, 2015) software was used to execute target docking between target proteins and key active compounds. Only those docked poses were selected having lower root mean square deviation (RMSD) and binding free energy. The docking score between proteins and compounds was used as primary evaluation criteria for screening of candidate compounds and their putative target. Moreover, Discovery studio (Shaweta et al., 2021) was used to visualize the interaction present between key active compounds and predicted proteins/targets.
2.6. ADMET profiling
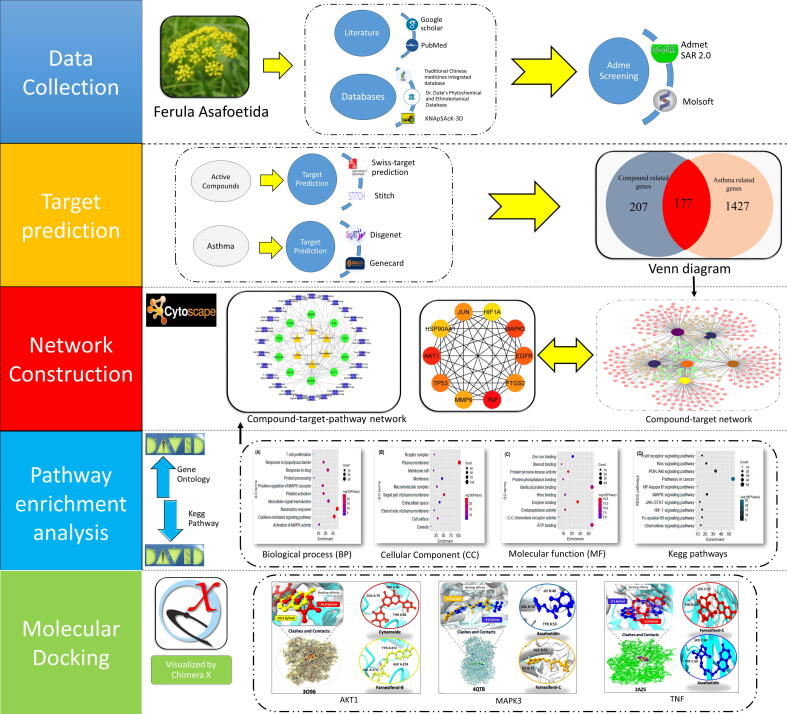
Chemical absorption, distribution, metabolism, excretion, and toxicity (ADMET), plays a key role in drug discovery and development. ADMET profiling aids in achieving the best possible balance of properties necessary for drug discovery that is both effective as well as safe. In this regard, the physiochemical characteristics of predicted compounds were identified through SWISS ADME (Daina et al., 2017), ADMETLAB 2.0 (Xiong et al., 2021) is used to predict the AMES test and PROTOX II (Banerjee et al., 2018) an online server is use to predict toxicity such as mutagenicity, carcinogenicity, and cytotoxicity. The current study’s workflow is depicted in Fig. 1.
Fig. 1.
Graphical synopsis representation of network pharmacology approach explaining the comprehensive procedure used in the identification of potential compounds and their potent targets against asthma.
3. Results
3.1. Active compounds of F. Asafoetida
Pharmacokinetic properties can be fruitful for understanding molecular mechanisms. Regarding this, DL and OB of plant related compounds were analyzed in order to better understand their action mechanism for the treatment of asthma. A total of 50 compounds of F. asafoetida were retrieved from the databases while 13 constituents were collected with the help of literature. Among these 63 compounds, a total of 6 compounds (assafoetidin, cynaroside, farnesiferol B, farnesiferol C, galbanic acid, and luteolin) were considered for further analysis as these compounds met the precise standard of OB ≥ 30 % and DL ≥ 0.18 (Table 1).
Table 1.
Information of potential active compounds of F. asafoetida.
| Molecule Name | Molecular weight MW ≤ 500 |
Drug likeness (DL ≥ 0.18 %) |
Oral Bioavailability (OB ≥ 30 %) | Structure | PubChem ID |
|---|---|---|---|---|---|
| Assafoetidin | 382.5 | 0.67 | 0.65 | 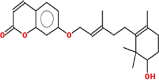 |
131,751,454 |
| Cynaroside | 448.4 | 0.60 | 0.75 | 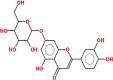 |
5,280,637 |
| Farnesiferol B | 382.5 | 0.27 | 0.54 | 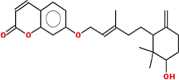 |
44,457,243 |
| Farnesiferol C | 382.5 | 0.23 | 0.67 | 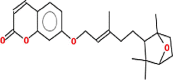 |
15,559,239 |
| Galbanic acid | 398.5 | 0.34 | 0.58 | 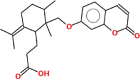 |
7,082,474 |
| Luteolin | 286.24 | 0.38 | 0.55 | 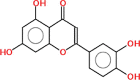 |
5,280,445 |
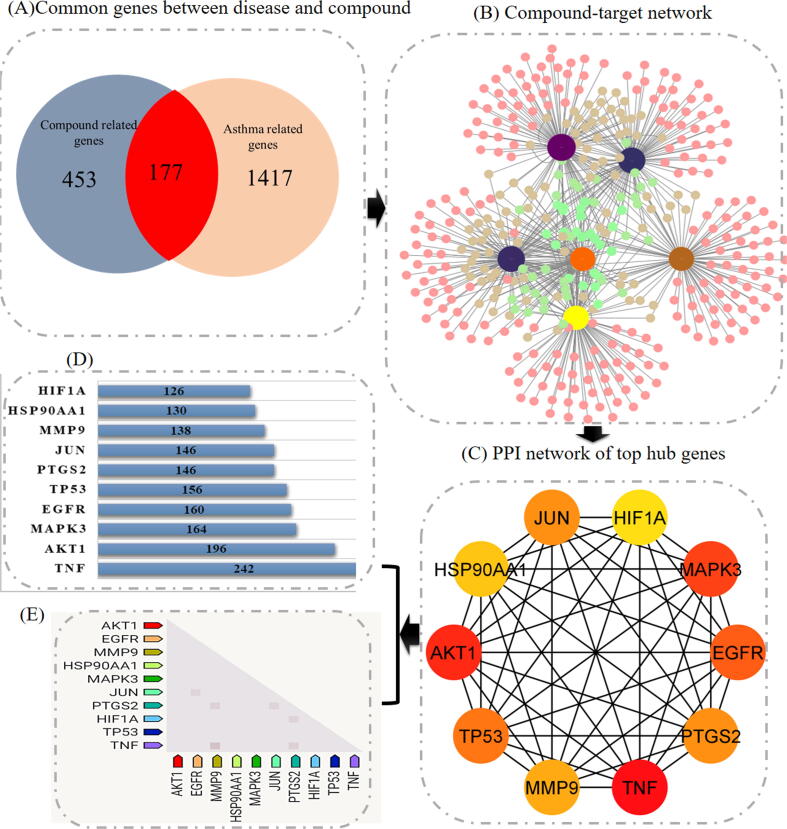
In addition, the STITCH and Swiss Target Prediction databases yielded 630 possible target genes for six active components. Following the discovery of promising targets of a compound, 1593 genes associated with asthma were collected from the GeneCards and DisGeNET databases. Later, a Venn diagram was utilized to forecast the overlapping targets of asthma and F. asafoetida-related compounds. Finally, 177 putative anti-asthmatic genes found in F. asafoetida were chosen as major targets. (Fig. 2A).
Fig. 2.
Network Pharmacology-based prediction of multi-compound, multi-target, and multi-pathway for curing Asthma (A) Venn diagram (B) Network diagram of significant compounds and their targets (C) Top 10 genes categorize by degree method (D) The bar plot of the ten genes (E) remark expression of 10 target genes in Homo sapiens.
3.2. Construction of compound-target network
F. asafoetida yielded a total of 6 putative active compounds. These 6 constituents corresponded to a number of different targets. Further, 6 plant related compounds, and the 'active compound-genes' network was built using 177 major targets and their related pathways. (Fig. 2B). This is clear evidence that multiple targets may have a synergistic impact when F. asafoetida acts as an anti-asthmatic drug. Following that, the degree of these 6 compounds in the compound-genes- pathways network were then figure out and summarized in Table 2. As highlighted in table, cynaroside and luteolin belong to class flavonoids while assafoetidin, farnesiferol B, farnesiferol C, and galbanic acid belongs to class sesquiterpene coumarins.
Table 2.
Degree of 6 significant compounds examine with the help of network analyzer in Cytoscape.
| Molecule Name | Class | Degree |
|---|---|---|
| Assafoetidin | Sesquiterpene coumarins | 110 |
| Cynaroside | Flavonoids | 117 |
| Farnesiferol B | Sesquiterpene coumarins | 97 |
| Farnesiferol C | Sesquiterpene coumarins | 99 |
| Galbanic acid | Sesquiterpene coumarins | 98 |
| Luteolin | Flavonoids | 109 |
3.3. Protein-Protein interaction (PPI) construction and analysis
177 overlapping genes were then submitted to STRING database for PPI network construction. The interplay between many targets throughout disease development is depicted in the PPI network by nodes and their related interactions. Later, PPI network of overlapped genes was analyzed using a network analyzer tool (Fig. 2C). TNF (2 4 2), AKT1 (1 9 6), MAPK3 (1 6 4), EGFR (1 6 0), TP53 (1 5 6), PTGS2 (1 4 6), JUN (1 4 6), MMP9 (1 3 8), HSP90AA1 (1 3 0), and HIF1A (1 2 6) showed a highest degree (Fig. 2D). The highest degree indicates that the targeted genes are highly connected with one another, implying that all of these genes may be potential targets. When these results were compared to those provided by functional annotation, three genes namely AKT1, MAPK3, and TNF were predicted as the principal anti-asthmatic targets of F. asafoetida and selected for molecular docking analysis.
3.4. Gene ontology and KEGG pathway enrichment
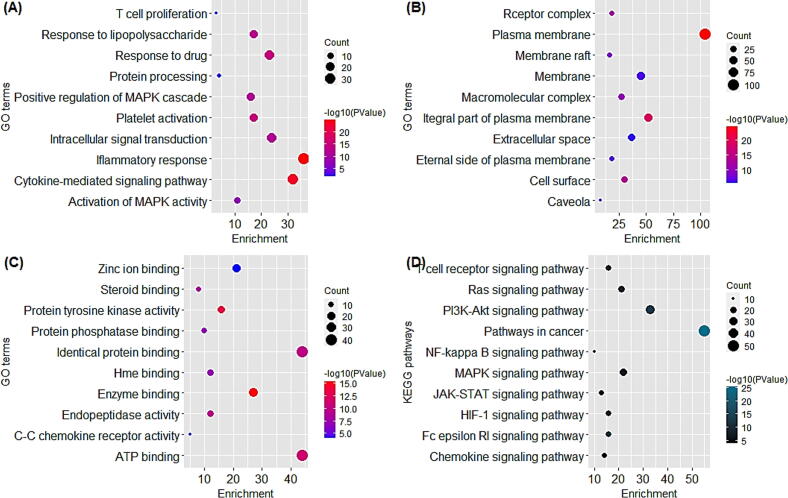
The pathway and enrichment result displayed biological function of 177 common targets. According to GO enrichment results, the pathway and enrichment analysis revealed the biological function of 177 common targets; the target genes were mainly involved in T cell proliferation, inflammatory responses, and activation of MAPK activity, C—C chemokine receptor activity, and response to drug and so on. While KEGG pathway results disclose that the genes are significantly enriched in PI3K-Akt signaling pathway, T cell receptor signaling pathway, Ras signaling pathway, MAPK signaling pathway, chemokine signaling pathway, NF-kappa B signaling pathway, pathways in cancer, and Fc epsilon RI signaling pathway (Fig. 3). At last, KEGG pathway results disclose that AKT1, MAPK3, and TNF were significantly enriched genes (Fig. 4).
Fig. 3.
Representation of functional annotation and enriched pathways in form of Bubble Plot (A) GO in terms of Biological processes (B) GO in terms of Cellular Components (C) GO in terms of Molecular function (D) KEGG pathway analysis.
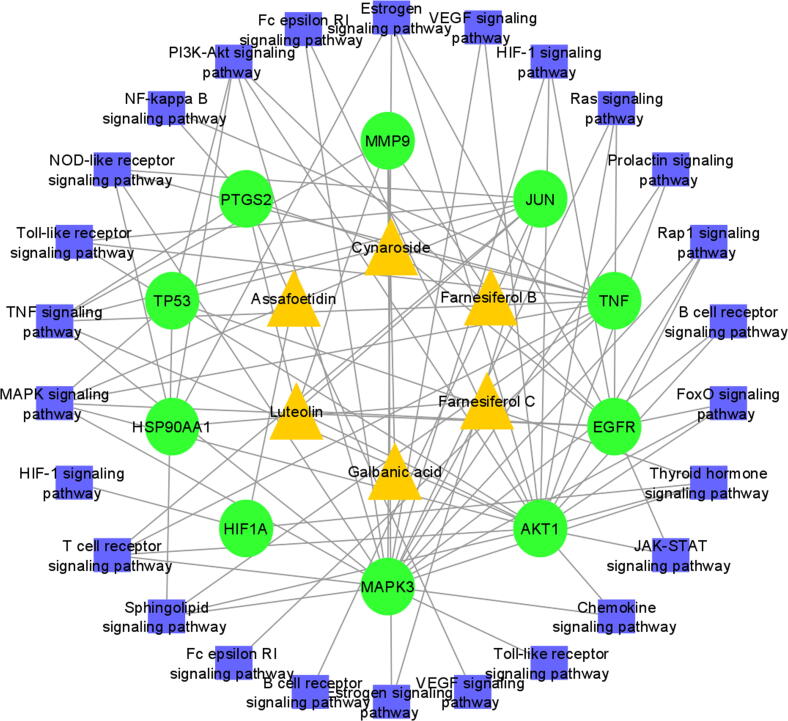
Fig. 4.
Pathways induced by F. asafoetida. The green nodes show the hub genes, the yellow nodes represent active compounds and the blue nodes are the pathways associated with the core targets.
3.5. Molecular docking analysis
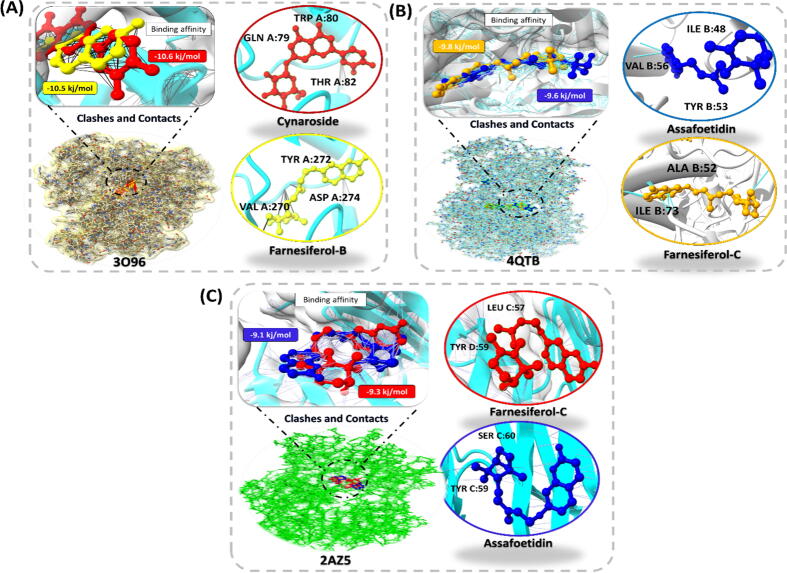
PyRx was utilized to perform molecular docking among the key active compounds present in F. asafoetida (Assafoetidin, Cynaroside, Farnesiferol-B Farnesiferol-C, Galbanic acid and Luteolin) and potential targets namely TNF, MAPK3, AKT1. Docking analysis was performed in order to screen key active targets to minimize risk of asthma. Binding affinity along with the RMSD value of compounds was shown in Table 3. The result indicates that AKT1 (3O96) had the maximum binding affinity as well as RMSD value with Cynaroside and Farnesiferol-B, TNF (2AZ5) firmly bind with the Assafoetidin and Farnesiferol-C, and MAPK3(4QTB) stably bind with Assafoetidin and Farnesiferol-C. It is noteworthy, that active compounds of F. asafoetida bind stably with three potent targets in order to control the asthma. All significant drug candidates showed van der wall interaction, carbon hydrogen bond, pi-anion, pi-pi stacked and conventional hydrogen bond present in the form of 2D diagram mentioned in (Supplementary files S1-S3). Overall this result provides us a clear evidence that these 6 bio-active compounds of F. asafoetida plays a substantial role for the treatment of asthma. Moreover, (Fig. 5) represent the strongest binding affinity between target and compound.
Table 3.
Binding energy and interactions of active compounds and their associated three target proteins.
| Target proteins (PDB ID) | Compounds | Binding Affinity | RMSD | Interacting residues |
|---|---|---|---|---|
| 2AZ5 | Assafoetidin | −9.1 | 3.4 | LEU C:57, TYR C:59, TYR D:59, SER C:60, LEU D:120, GLY D:121 |
| Farnesiferol-C | −9.2 | 2.9 | LEU C:57, LEU D:57, TYR C:59, TYR D:59, TYR D:119, TYR D:151 | |
| 3O96 | Cynaroside | −10.6 | 1.4 | GLN A:79, TRP A:80, THR A:82, SER A:205, LEU A:264, LYS A:268, VAL A:270, ASP A:292 |
| Farnesiferol-B | −10.5 | 2.8 | TRP A:80, ILE A:84, SER A:205, LYS A:268, VAL A:270, TYR A:272, ASP A:274 | |
| 4QTB | Assafoetidin | −9.6 | 1.59 | ILE B:48, TYR B:53, VAL B:56, ILE B:73, ARG B:84, GLU B:88, LEU B:173, CYS B:183 |
| Farnesiferol-C | −9.8 | 1.67 | ALA B:52, TYR B:53, VAL B:56 LYS B:71, ILE B:73, ARG B: 84, LEU B:173, CYS B:183 |
Fig. 5.
Representation of docked complexes of three target genes namely AKT1 (3O96), MAPK3 (4QTB) and TNF (2AZ5) having strong binding affinity with their compounds.
3.6. ADMET profiling
Drug development require assessment of absorption, distribution metabolism and excretion (ADME), A vast number of molecular structures are assessed using a variety of criteria in order to guide the selecting of which chemicals to generate, test, and promote, with the ultimate goal of determining which have the best possibility of becoming useful medicines for patients. Moreover, ADME analysis is a demanding process in drug discovery. This is attained through SWISSADME database and results indicate all the compounds have satisfactory pharmacokinetic properties (Table 4). The results of ADMET profiling of selected drug candidate reveal that there is no or less side effect of pharmacokinetic properties of the key active compounds. The ADME properties of the key active compounds for distinct models, such as BBB permeant P-gp substrate, CYP1A2 inhibitor, CYP2C9 inhibitor showed positive result in some potential compounds that indicate the compound capability to act as a drug candidate. Moreover, the key active compounds show the non-toxic behavior, suggested that these compounds can act a good drug candidate in pre-clinical trials.
Table 4.
ADMET profiling of active compounds.
| Compounds | Assafoetidin | Cynaroside | Farnesiferol-B | Farnesiferol-C |
|---|---|---|---|---|
| GI absorption | High | Low | High | High |
| BBB permeant | Yes | No | Yes | Yes |
| P-gp substrate | No | Yes | No | Yes |
| CYP1A2 inhibitor | No | No | No | No |
| CYP2C19 inhibitor | Yes | No | Yes | Yes |
| CYP2C9 inhibitor | Yes | No | Yes | Yes |
| CYP2D6 inhibitor | Yes | No | Yes | Yes |
| CYP3A4 inhibitor | Yes | No | Yes | Yes |
| Toxicity | ||||
| Acute toxicity | Negative | Negative | Negative | Negative |
| Mutagenicity | In-active | In-active | In-active | In-active |
| Carcinogenicity | In-active | In-active | In-active | In-active |
4. Discussion
Asthma is dreadful heterogeneous inflammatory disease of the human respiratory tract with increase morbidity rate throughout the world. The rate of asthmatic patient rise approximately 2.5 million each year (D’Amato et al., 2016). Asthma cannot be effectively treated in many cases, despite the enormous availability of drugs and therapeutic approaches. Moreover, 50 % of asthmatic patients have symptoms on regular basis; nearly all patients have difficulty in breathing and coughing (Barnes et al., 2005). It is reported that, an oral anti-asthmatic medicine has a major side effect on a patient health (Cooper et al., 2015). Moreover, the advancement of novel drug is often restricted due to the lack of absorption, distribution, metabolism, and excretion (ADME) properties, and these highly-priced natures imposes an additional trouble in the process of drug development (M. Honorio et al., 2013). Therefore, ADME based screening has acquired more attention from scientists studying the drug discovery process. For this purpose, the use of network pharmacology can aid in the systematic and holistic evaluation by spotting key bioactive compounds and the potent targets from a large amount of data. Based on systems biology, high throughput screening and network pharmacology can help to discover promising candidate genes to control asthma. In order to analyze the interaction between body and drug based upon system biology, high throughput screening and network pharmacology to discover promising candidate genes to control asthma.
Medicinal plants are used as a source of drugs which undergoes significant resurgence now a day because of their less toxic effects and reduced side effects as compared to several synthetic drugs (Aslam and Ahmad, 2016). According to the W.H.O 80 % of the population depend upon herbal medicines because of their remarkable applications in our lifestyle (Cragg and Newman, 2001).The identification of potent active compounds that prevent the ailments and diseases will be regarded as of great concern in current era.
Asafoetida is an oleo-gum resin obtained from F. asafoetida plant (Umbelliferae family) abundantly present in central Asia. The main chemical constituents of F. asafoetida include resin, flavonoid, essential oil, terpenoids, coumarins and sesquiterpene. This plant has been expressed to hold anti-tumor, anti-bacterial, and anti-inflammatory effects (Mahendra and Bisht, 2012b). Clinically, F. asafoetida is used to treat asthma, menstrual pain, and reduce headaches (Shahrajabian et al., 2021b). Traditional uses of F. asafoetida include the treatment of strep throat, asthma, ulcer, seizure disorders, stomach pain, diarrhea, pneumonia, parasitic infections, weak digestion, and illness (Amalraj and Gopi, 2017).This research has been designed to screen potent bioactive components of F. asafoetida and their potential targets to reveal the mechanisms for the cure of asthma. Notably, the result of screening represented that flavonoids and sesquiterpene coumarins were the major bioactive compounds of F. asafoetida which plays a substantial role in the asthma treatment by affecting TNF, AKT1 and MAPK3 genes. The construction of compound-target pathway network discovered that Galbanic acid, Cynaroside, Farnesiferol-B, Farnesiferol-C, Luteolin and Assafoetidin had a firm interdependency in a network, associated that it has anti-asthmatic effects. Furthermore, the results of molecular docking also approved that there exists a strong binding energy present between the active compounds and the potential targets. Finally, the ADME profiling also strengthen our finding that the resulted compounds can act as a drug candidate in pre-clinical phase.
GO functional annotation was carried out to gain more knowledge about the biological processes from CC, MF, and BP aspect. CC, MF, and BP exhibit the sequence of occurrence obtained by the orderly composite of cellular localization, molecular functions of proteins targets and their biological process, respectively. The gene enrichment analysis revealed that main pathways of F. asafoetida for the treatment of asthma includes Fc epsilon RI signaling pathway, PI3K-AKT signaling pathway, T cell receptor signaling pathway, HIF-1 signaling pathway, Ras signaling pathway, MAPK signaling pathway, VEGF signaling pathway, Chemokine signaling pathway and Pathway in cancer. Intriguingly, activation of the MAPK signaling system regulates airway inflammation in asthma patients (Alam and Gorska, 2011). Previous studies reported that, excitation of the PI3K-Akt signaling pathway can significantly raise respiratory epithelial–mesenchymal evolution which may facilitate in treatment of asthma (Yoo et al., 2017). The VEGF signaling pathway plays a potent role by stimulating the proliferation of cells and response to airway inflammation (Samitas et al., 2016). Hypoxia-inducible factor (HIF)-1 is a chief activator of inflammatory responses that is elevated in asthma alveolar macrophages (Dewitz et al., 2017). It is noteworthy that TNF released in large amounts in chronic bronchitis airways, which have a potent role in the development of bronchial hyper responsiveness (Babu et al., 2004). In summary F. asafoetida for the treatment of asthma through P13-AKT, Neuroactive ligand-receptor interaction, VEGF, MAPK, Toll like receptor, FoxO, Chemokine and other signaling pathways, that is compatible with the current results of this study.
In docking analysis, the binding affinity with the top docking score was preferred to analyze the interaction occurs joining active compounds and the potential targets. The docking results shows that each of the 6 active compounds have best binding affinity to all three targets namely MAPK3, TNF and AKT1 that play a key role in the multiple pathways to treat asthma. These results indicate that key active compounds present in F. asafoetida could mitigate airway inflammation through binding with MAPK3, TNF and AKT1. Finally, the corresponding ADME properties of 6 active compounds for distinct models such as GI absorption, BBB permeant and P-gp substrate, showed positive results that greatly holdup active compounds acceptability as drug candidate in pre-clinical and clinical studies.
Network pharmacology is an emerging technique based on systems pharmacology that examines the consequences of drugs on the interactome that involves the molecular interactions. It is a powerful tool to examine and analyze vast accumulated data that is generated as a consequence of the improvement in omics technologies. It differs from previous drug discovery processes which are based upon one disease one target, on gene approaches that are not sufficiently predictive as evident from high clinical attrition rate that usually ends up with the financial loss to the companies. Moreover, one target; one drug paradigm is an another issue where one drug acting on the single target and this paradigm also fail because we selectively design drug that interact with multiple targets rather than one identified diseased target within the biological system. So the network pharmacology-one of the holistic insights in the process of drug discovery aids in the development of useful network models and the prediction of drug targets in accordance with freely available database and may helpful in designing effective multi-target therapies. Moreover, it also provides the formation of network model of drug-target-disease by utilizing computational bioinformatics tools and high throughput screening. As a result network pharmacology holds promising approach for the recognition of novel targets and identification of drug with the assistance of medicinal plants that decrease the side effect rates and also reduce the cost rate of drug development (Zhang et al., 2019).
However, this study has some limitations. For example, the gathering of active ingredients and therapeutic targets is insufficient, and network pharmacology outcomes have not been checked in animal model or clinical trials. New detection technologies, such as mass spectrometry, two-dimensional liquid chromatography, liquid chromatography, can improve follow-up research in near future. Furthermore, animal research and clinical trials should be used to validate the major signaling pathways of F. asafoetida in the treatment of asthma, in order to give scientific validity and thoughts for better asthmatic therapies by utilizing integrated western and traditional herb medicine.
5. Conclusion
Network pharmacology was applied in the present study for identification of mechanism of the plant F. asafoetida for the treatment of asthma. Traditional herbal medicine aims to improve the patient whole-body balance by using herbal formula having suppressed side effects. Our findings suggested that these compounds (Cynaroside, Farnesiferol-B, Farnesiferol-C and Assafoetidin) can play a significant role to elucidate the mechanism of F. asafoetida for the treatment of asthma. Thus, network pharmacology has vast number of applications and serves as a promising approach for the future drug discovery process to diminish the incidence of asthma.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Muhammad Qasim, Email: qasimawan@gcuf.edu.pk.
Muhammad Abdullah, Email: mabdulla23@gcuf.edu.pk.
Usman Ali Ashfaq, Email: ashfaqua@gcuf.edu.pk.
Fatima Noor, Email: fatimanooor@gcuf.edu.pk.
Nazia Nahid, Email: nazianahid@gcuf.edu.pk.
Ahmad Alzamami, Email: aalzamami@su.edu.sa.
Norah A Alturki, Email: noalturki@ksu.edu.sa.
Mohsin Khurshid, Email: mohsinkhurshid@gcuf.edu.pk.
References
- Alam R., Gorska M.M. MAPK Signaling and ERK1/2 bistability in Asthma. Clin Exp Allergy. 2011;41:149. doi: 10.1111/J.1365-2222.2010.03658.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamgeer, Younis W., Asif H., Sharif A., Riaz H., Bukhari I.A., Assiri A.M. Traditional medicinal plants used for respiratory disorders in Pakistan: A review of the ethno-medicinal and pharmacological evidence Milen Georgiev, Ruibing Wang. Chinese Medicine (United Kingdom) 2018;13:1–29. doi: 10.1186/S13020-018-0204-Y/TABLES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S.I., Qaiser M. A phytogeographical analysis of the phanerogams of Pakistan and Kashmir. Proc., Sect. B Biol. sci. 1986;89:89–101. [Google Scholar]
- Althagafi, I., El-Metwaly, N., Farghaly, T.A., 2019. New Series of Thiazole Derivatives: Synthesis, Structural Elucidation, Antimicrobial Activity, Molecular Modeling and MOE Docking. Molecules Vol. 24, Page 1741 24, 1741. https://doi.org/10.3390/MOLECULES24091741 [DOI] [PMC free article] [PubMed]
- Amalraj A., Gopi S. Biological activities and medicinal properties of Asafoetida: A review. J. Tradit. Complement. Med. 2017;7:347–359. doi: 10.1016/J.JTCME.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam M.S., Ahmad M.S. Worldwide Importance of Medicinal Plants: Current and Historical Perspectives. Recent Adv Biol Med. 2016;2:909. doi: 10.18639/RABM.2016.02.338811. [DOI] [Google Scholar]
- Babu K.S., Davies D.E., Holgate S.T. Role of tumor necrosis factor alpha in asthma. Immunol. Allergy Clin. North Am. 2004;24:583–597. doi: 10.1016/J.IAC.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Banerjee P., Eckert A.O., Schrey A.K., Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018;46:W257–W263. doi: 10.1093/NAR/GKY318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes P., Virchow J.C., Sanchis J., Welte T., Pedersen S. Asthma management: important issues. Eur. Respir. Rev. 2005;14:147–151. doi: 10.1183/09059180.05.00009704. [DOI] [Google Scholar]
- Bateman A. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/NAR/GKY1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choby G.W., Lee S. Pharmacotherapy for the treatment of asthma: current treatment options and future directions. Int Forum Allergy Rhinol. 2015;5:S35–S40. doi: 10.1002/ALR.21592. [DOI] [PubMed] [Google Scholar]
- Cooper V., Metcalf L., Versnel J., Upton J., Walker S., Horne R. Patient-reported side effects, concerns and adherence to corticosteroid treatment for asthma, and comparison with physician estimates of side-effect prevalence: a UK-wide, cross-sectional study. NPJ Prim Care Respir Med. 2015;25:15026. doi: 10.1038/NPJPCRM.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg G.M., Newman D.J. Natural product drug discovery in the next millennium. Pharm. Biol. 2001;39(Suppl 1):8–17. doi: 10.1076/PHBI.39.S1.8.0009. [DOI] [PubMed] [Google Scholar]
- D’Amato G., Vitale C., Molino A., Stanziola A., Sanduzzi A., Vatrella A., Mormile M., Lanza M., Calabrese G., Antonicelli L., D’Amato M. Asthma-related deaths. Multidiscip. Respir. Med. 2016;11:1–5. doi: 10.1186/S40248-016-0073-0/TABLES/1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7(1) doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallakyan S., Olson A.J. Small-Molecule Library Screening by Docking with PyRx. Methods Mol. Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- Dennis G., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:1–11. doi: 10.1186/GB-2003-4-9-R60/TABLES/3. [DOI] [PubMed] [Google Scholar]
- Dewitz C., McEachern E., Shin S., Akong K., Nagle D.G., Broide D.H., Akuthota P., Crotty Alexander L.E. Hypoxia-inducible factor-1α inhibition modulates airway hyperresponsiveness and nitric oxide levels in a BALB/c mouse model of asthma. Clin. Immunol. 2017;176:94–99. doi: 10.1016/J.CLIM.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Gfeller D., Grosdidier A., Wirth M., Daina A., Michielin O., Zoete V. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res. 2014;42:W32–W38. doi: 10.1093/NAR/GKU293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffler E., Madeira L.N.G., Ferrando M., Puggioni F., Racca F., Malvezzi L., Passalacqua G., Canonica G.W. Inhaled Corticosteroids Safety and Adverse Effects in Patients with Asthma. J. Allergy Clin. Immunol. Pract. 2018;6:776–781. doi: 10.1016/J.JAIP.2018.01.025. [DOI] [PubMed] [Google Scholar]
- M. Honorio, K., L. Moda, T., D. Andricopulo, A., 2013. Pharmacokinetic properties and in silico ADME modeling in drug discovery. Med Chem 9, 163–176. https://doi.org/10.2174/1573406411309020002. [DOI] [PubMed]
- Kohl M., Wiese S., Warscheid B. Cytoscape: Software for Visualization and Analysis of Biological Networks. Methods Mol. Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- Kuhn M., von Mering C., Campillos M., Jensen L.J., Bork P. STITCH: interaction networks of chemicals and proteins. Nucleic Acids Res. 2008;36:D684–D688. doi: 10.1093/NAR/GKM795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans, C., van Asseldonk, T., 2020. Dr. Duke’s Phytochemical and Ethnobotanical Databases, a Cornerstone in the Validation of Ethnoveterinary Medicinal Plants, as Demonstrated by Data on Pets in British Columbia 219–246. https://doi.org/10.1007/978-3-030-44930-8_10.
- Mahendra P., Bisht S. Ferula asafoetida: Traditional uses and pharmacological activity. Pharmacogn. Rev. 2012;6:141. doi: 10.4103/0973-7847.99948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoli M., Fabian D., Holt S., Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004;59(5):469–478. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Shimura N., Otabe Y., Hirai-Morita A., Nakamura Y., Ono N., Ul-Amin M.A., Kanaya S. KNApSAcK-3D: A Three-Dimensional Structure Database of Plant Metabolites. Plant Cell Physiol. 2013;54:e4–e. doi: 10.1093/PCP/PCS186. [DOI] [PubMed] [Google Scholar]
- Pence H.E., Williams A. ChemSpider: An Online Chemical Information Resource. J. Chem. Educ. 2010;87:1123–1124. doi: 10.1021/ED100697W. [DOI] [Google Scholar]
- Piñero J., Queralt-Rosinach N., Bravo À., Deu-Pons J., Bauer-Mehren A., Baron M., Sanz F., Furlong L.I. DisGeNET: a discovery platform for the dynamical exploration of human diseases and their genes. Database. 2015;2015:1–17. doi: 10.1093/DATABASE/BAV028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaeger S.F. Clinical Immunology and Traditional Herbal Medicines. Clin. Vaccine Immunol. 2003;10:337–338. doi: 10.1128/CDLI.10.3.337-338.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebhan M., Chalifa-Caspi V., Prilusky J., Lancet D. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics. 1998;14:656–664. doi: 10.1093/BIOINFORMATICS/14.8.656. [DOI] [PubMed] [Google Scholar]
- Ru J., Li P., Wang J., Zhou W., Li B., Huang C., Li P., Guo Z., Tao W., Yang Y., Xu X., Li Y., Wang Y., Yang L. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:1–6. doi: 10.1186/1758-2946-6-13/FIGURES/2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samitas K., Poulos N., Semitekolou M., Morianos I., Tousa S., Economidou E., Robinson D.S., Kariyawasam H.H., Zervas E., Corrigan C.J., Ying S., Xanthou G., Gaga M. Activin-A is overexpressed in severe asthma and is implicated in angiogenic processes. Eur. Respir. J. 2016;47:769–782. doi: 10.1183/13993003.00437-2015. [DOI] [PubMed] [Google Scholar]
- Selgrade M.K., Lemanske R.F., Gilmour M.I., Neas L.M., Ward M.D.W., Henneberger P.K., Weissman D.N., Hoppin J.A., Dietert R.R., Sly P.D., Geller A.M., Enright P.L., Backus G.S., Bromberg P.A., Germolec D.R., Yeatts K.B. Induction of Asthma and the Environment: What We Know and Need to Know. Environ. Health Perspect. 2006;114(4):615–619. doi: 10.1289/ehp.8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrajabian M.H., Sun W., Soleymani A., Khoshkaram M., Cheng Q. Asafoetida, God’s Food, a Natural Medicine. Pharmacognosy Communications. 2021;11:36–39. doi: 10.5530/PC.2021.1.8. [DOI] [Google Scholar]
- Shaweta, S., Akhil, S., Utsav, G., 2021. Molecular Docking studies on the Anti-fungal activity of Allium sativum (Garlic) against Mucormycosis (black fungus) by BIOVIA discovery studio visualizer 21.1.0.0. Annals of Antivirals and Antiretrovirals 028–032. https://doi.org/10.17352/AAA.000013.
- Sussman, J.L., Lin, D., Jiang, J., Manning, N.O., Prilusky, J., Ritter, O., Abola, E.E., 1998. Protein Data Bank (PDB): Database of Three-Dimensional Structural Information of Biological Macromolecules. urn:issn:0907-4449 54, 1078–1084. https://doi.org/10.1107/S0907444998009378. [DOI] [PubMed]
- Szefler S.J., Chmiel J.F., Fitzpatrick A.M., Giacoia G., Green T.P., Jackson D.J., Nielsen H.C., Phipatanakul W., Raissy H.H. Asthma Across the Ages: Knowledge Gaps in Childhood Asthma Prepared for the 2014 theme issue in the Journal of Allergy and Clinical Immunology. J. Allergy Clin. Immunol. 2014;133(1):3–13. doi: 10.1016/j.jaci.2013.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeresham C. Natural products derived from plants as a source of drugs. J. Adv. Pharm. Technol. Res. 2012;3:200. doi: 10.4103/2231-4040.104709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C., Huynen M., Jaeggi D., Schmidt S., Bork P., Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/NAR/GKG034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xiao J., Suzek T.O., Zhang J., Wang J., Bryant S.H. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/NAR/GKP456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong G., Wu Z., Yi J., Fu L., Yang Z., Hsieh C., Yin M., Zeng X., Wu C., Lu A., Chen X., Hou T., Cao D. ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 2021;49:W5–W14. doi: 10.1093/NAR/GKAB255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Lou C., Sun L., Li J., Cai Y., Wang Z., Li W., Liu G., Tang Y. admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics. 2019;35:1067–1069. doi: 10.1093/BIOINFORMATICS/BTY707. [DOI] [PubMed] [Google Scholar]
- Yoo E.J., Ojiaku C.A., Sunder K., Panettieri R.A. Phosphoinositide 3-Kinase in Asthma: Novel roles and therapeutic approaches. Am. J. Respir. Cell Mol. Biol. 2017;56:700–707. doi: 10.1165/RCMB.2016-0308TR/SUPPL_FILE/DISCLOSURES.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Zhu X., Bai H., Ning K. Network pharmacology databases for traditional Chinese medicine: Review and assessment. Front. Pharmacol. 2019;10:123. doi: 10.3389/FPHAR.2019.00123/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
Further Reading
- Asthma - Symptoms and causes - Mayo Clinic [WWW Document], n.d. URL https://www.mayoclinic.org/diseases-conditions/asthma/symptoms-causes/syc-20369653 (accessed 12.22.21).
- Res, I.H.-P.J.M., 2004, medicine, n.d. Safety of medicinal plants. Citeseer.