Abstract
Background
Anti-PD-1(L1) therapies are less efficacious in patients with EGFR-mutated non-small-cell lung cancer. However, the underlying mechanism is poorly understood.
Methods
The characteristics of T cells in EGFR-mutated and wild-type tumors were analyzed based on The Cancer Genome Atlas database and clinical samples. Plasma levels of 8 T-cell-related cytokines were evaluated and its association with immunotherapy efficacy were explored. Association between EGFR signaling pathway and IL-10 was examined through tumor cell lines and clinical tumor samples. In vitro restimulation model of human CD8+T cells isolated from peripheral blood was used to analyze the impact of IL-10 on T cells. Doxycycline-inducible transgenic EGFRL858R mouse models were used to investigate the efficacy of combining recombinant mouse IL-10 protein and PD-1 blockade and its underlying mechanism in vivo.
Results
EGFR-mutated tumors showed a lack of CD8+T cell infiltration and impaired CD8+T cell cytotoxic function. The incompetent CD8+T cells in EGFR-mutated tumors were characterized as absence of CD39 expression, which defined hallmarks of cytotoxic and exhausted features and could not be reinvigorated by anti-PD-1(L1) treatment. Instead, CD39 expression defined functional states of CD8+T cells and was associated with the therapeutic response of anti-PD-1(L1) therapies. Mechanically, IL-10 upregulated CD39 expression and was limited in EGFR-mutated tumors. IL-10 induced hallmarks of CD8+T cells immunity in CD39-dependent manner. Using autochthonous EGFR L858R-driven lung cancer mouse models, combining recombinant mouse IL-10 protein and PD-1 blockade optimized antitumor effects in EGFR-mutated lung tumors.
Conclusions
Our study suggested that owing to low level of IL-10 to induce the expression of CD39 on CD8+T cells, fewer phenotypically cytotoxic and exhausted CD39+CD8+T cells in EGFR-mutated tumors could be potentially reinvigorated by anti-PD-1(L1) treatment. Hence, IL-10 could potentially serve as a cytokine-based strategy to enhance efficacy of anti-PD-1(L1) treatment in EGFR-mutated tumors.
Keywords: Lung Neoplasms, Tumor Microenvironment, Translational Medical Research
WHAT IS ALREADY KNOWN ON THIS TOPIC
EGFR-mutated non-small-cell lung cancer (NSCLC) patients responded poorly to PD-1/PD-L1 inhibitors, however, the determinants of failure remained poorly understood.
WHAT THIS STUDY ADDS
Abundant bystander CD39-CD8+T cells without cytotoxic activities were present in EGFR-mutated tumors.
EGFR-mutated tumors were lack of IL-10 to induce the expression of CD39 on CD8+T cell with cytotoxic and exhausted features that could be reinvigorated on immune checkpoint inhibitor treatment.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Using IL-10 to expand the population of CD39+CD8+T cells could be served as a strategy to potentiate antitumor effect of PD-1 blockade in EGFR-mutated NSCLC.
Introduction
Immune-checkpoint inhibitors (ICIs), especially targeting programmed cell death protein-1 (PD-1) and programmed death ligand-1 (PD-L1), have emerged as a core pillar of cancer therapy. Several PD-1/PD-L1 inhibitors have been approved as standard first-line or second-line treatment strategy in non-small-cell lung cancer (NSCLC) patients,1–3 especially for patients with PD-L1 tumor proportion score (TPS) of 50% or greater, the 5-year OS rate reached 31.9%. 4 However, patients with EGFR mutation, which is the most common oncogenic driver mutation with prevalence of approximately 50% of Asian NSCLC cases,5 were excluded in most ICI trials. Additionally, subgroup analyses of clinical trials, small phase I or II non-controlled trials or retrospective analyses from real-world data demonstrated that EGFR-mutated patients responded poorly to ICI treatment with no improvement in OS.1 3 6–10 Therefore, elucidating mechanisms of low response rate to PD-1/PD-L1 blockade will identify potential biomarkers to predict immunoresponse and explore promising therapeutics to improve OS in EGFR-mutated patients.
Despite the researchers are attempting to reveal the possible underlying mechanisms for the poor efficacy of anti-PD-1/PD-L1 treatment in EGFR-mutated NSCLC, the determinants of failure remained poorly understood.11 For instance, PD-L1 expression, as a widely adopted biomarker to stratify patients for PD-1/PD-L1 blockade,12 was found to be upregulated on activating of EGFR signaling pathway.13 However, results from ATLANTIC showed that even in patients with PD-L1 TPS ≥25%, the objective response rate (ORR) was only 12.2% in patients treated with durvalumab.14 Checkmate 026 firstly proposed that high tumor mutation burden (TMB) was associated with improved efficacy of nivolumab in NSCLC patients.15 Although previous studies demonstrated that EGFR-mutated tumors had lower somatic mutations and lower number of neoantigens compared with EGFR wild-type tumors,16 Vokes et al pointed out that TMB was not linked with response to ICI if driver mutations were present.17 Taken together, these lines of evidence address the complexity of tumor microenvironment (TME) and highlight the limitations of conventional paradigms for the study of TME in EGFR-mutated tumors.
The clinical success of cancer immunotherapy has focused on tumor infiltrating lymphocytes (TILs), the crucial role in TME across cancer types, particularly CD8+T cells, which acted as soldiers to exert killing function. Specifically, the magnitude and quality of CD8+T cells were linked to the outcome of immunotherapy. Some initial studies reported that EGFR-mutated tumors presented few TILs or absence of TILs,18 however, a subsequent study indicated that there was no statistical significance in density of CD8+T cells between EGFR-mutated and wild-type tumors.19 Therefore, the deep insight into the state of T cells in cancers should be emphasized. T cell dysfunction is a hallmark of many cancers, but only parts of CD8+TILs could be effectively reinvigorated by PD-1/PD-L1 blockade.20 Recent studies have reported that CD39 was considered as the marker of exhausted T cells and also, measuring CD39 expression could be a straightforward way to isolate bystander CD8+T cells (CD39-CD8+T cells), which were not specific for tumor antigens on chronic stimulation, hence lost the ability to be reinvigorated.21 Increasing evidence has shown that patients with relatively high CD39+CD8+T cell proportions at baseline tended to have an improved response to ICI treatment which has been confirmed in colorectal cancer and hepatocellular carcinoma.21 22 However, several lines of evidence showed the opposing effect of CD39+CD8+T cells. Canale et al demonstrated that CD39+CD8+T cells exhibited an exhausted phenotype with impaired secretion of IFN-γ, TNF, IL-2 and the frequency of CD39highCD8+T cells increased with tumor growth in breast and melanoma models.23 Similarly, Sade-Feldman et al reported that CD39 expression on T cells was defined with dysfunctional states and hence, correlated with non-responders in patients with melanoma treated with ICI.24 However, unlike melanoma and breast cancer, NSCLC patients, especially those harboring EGFR mutations, were correlated with uninflamed phenotype and weak immunogenicity. In this case, the predictive value of CD39+CD8+ T cell proportions in indicating the possibility of T-cell reactivation on ICI treatment in NSCLC should be fully addressed.25 26
Here, we comprehensively analyzed the features of TME in EGFR-mutated tumors. We identified that the major distinguishing immune cells were CD8+T cells between EGFR-mutated and wild-type tumors. Additionally, impaired function of CD8+T cells were present in EGFR-mutated tumors, owing to the abundant bystander CD39-CD8+T cells without cytotoxic activities. With analysis of T-cell related cytokines to understand the regulation mechanism of CD39 expression, we found that IL-10, as the dual role of regulation in immune system,27 played pivotal roles in inducing CD8+T cells activation in CD39-dependent manner. Furthermore, we demonstrated that EGFR-mutated tumors were lack of IL-10 to induce the expression of CD39 on CD8+T cell with cytotoxic and exhausted features that could be reinvigorated by ICI treatment, supporting the poor response to anti-PD (L)1 treatment in EGFR-mutated tumors. Therefore, our data suggested that IL-10 and CD39 could be exploited for the development of novel biomarkers in EGFR-mutated tumors and also, our in vivo study showed that combining IL-10 and PD-1 blockade exerted promising antitumor effect in EGFR-mutated mouse models, inspiring the design of cytokine-based immunotherapies that exploit a specific CD8+T cells subtype to benefit EGFR-mutated NSCLC patients.
Materials and method
Gene expression datasets
To decipher the pattern of immune infiltration in EGFR-mutated and EGFR wild-type NSCLC, a total of 58 EGFR-mutated and 344 EGFR wild-type samples’ sequencing data including mutations and transcriptome profiling from The Cancer Genome Atlas (TCGA) were retrieved from the TCGA data portal in July 2019 (https://portal.gdc.cancer.gov/). To explore the correlation between genes in lung cancer, an online web server was used, named TIMER2.0 (Tumor Immune Estimation Resource, http://timer.cistrome.org/).28
Human tissue specimen and plasma collection
A total of 30 patients, diagnosed with resectable lung cancer by pathological examination were enrolled in the study for analysis of characteristics of TME in EGFR-mutated tumors. Baseline characteristics were shown in online supplemental table 1. The resected tumor specimen was kept in Tissue Storage Solution (Miltenyi Biotec) at 4°C within 48 hours for flow cytometry. For cytokine measurement in tissue specimen, 25 resected tumors among 30 cases were kept directly at −80°C until use.
jitc-2022-005436supp002.xlsx (9.6KB, xlsx)
A total of 36 patients treated with PD-1/PD-L1 inhibitors, diagnosed with advanced lung cancer were also enrolled in the study. Baseline characteristics were shown in online supplemental table 2. 5 mL blood samples within 2 days prior to initiation of ICI treatment and paired plasma samples after one cycle of ICI treatment were drawn and centrifuged at 1600 g for 10 min for plasma collection.
jitc-2022-005436supp003.xlsx (9.7KB, xlsx)
Detection of EGFR mutations in tissue samples
The EGFR mutations were detected using the Human AmoyDx EGFR Gene Mutation Diagnostic Kit (Amoy Diagnostics). The detailed procedures were performed as previously described.29 30
Isolation of human PBMC
The 5 mL Heparinized venous blood samples from patients with lung cancer were subjected to density gradient centrifugation using Ficoll-Paque plus (GE Healthcare). Peripheral blood mononuclear cells (PBMCs) were collected, washed with PBS and thawed at −80°C until use.
PBMC derived CD8+T cell activation and analysis
The 50 mL heparinized venous blood samples from 10 healthy donors were collected for isolation of PBMCs. CD8+T cells were then isolated from human PBMCs using the CD8 Microbeads (#130-045-201, Miltenyi Biotec), MS columm (Miltenyi Biotec) and MACS Seperator according to the manufacturer’s protocol. For co-culture system, isolated CD8+T cells were activated in T cell activation/expansion kit (Miltenyi Biotec), and then were replated and co-cultured with normal/tumor cells as ratio of 2:1 for 3 days. For IL-10 restimulation experiment, the CD8+T cells were initially split into four groups, control (activated with T cell activation/expansion kit) or activated in addition to different doses of recombinant human IL-10 protein (1 ng/mL, 10 ng/mL and 100 ng/mL) for 3 days. After 3 days, cells were collected, washed and immediately for further flow cytometry.
Efficacy evaluation
Progression-free survival (PFS) and ORR were used as the endpoints to analyze the correlation between the levels of cytokines or proportions of CD39+CD8+T cells and efficacy of ICI treatment in NSCLC patients. PFS was defined from the first day of ICIs treatment to the day of physician assessment of disease progression or death from any cause. ORR was calculated as percentage of patients with evaluated complete response or partial response. Objective tumor responses were assessed by investigators according to the Response Evaluation Criteria In Solid Tumors V.1.1.
Animal model
Doxycycline-inducible transgenic EGFRL858R mouse models were obtained from Dr. Liang Chen of the National Institute of Biological Sciences, Collaborative Innovation Center for Cancer Medicine. Specifically, the clara cell 10-kD protein (CC10) promoter and the reverse tetracycline transactivator (rtTA) were used to create a lung-specific, externally regulatable, overexpression transgenic system. Age-matched and sex-matched transgenic mice on FVB background were used in the experiments. The genotyping was performed at the age of 4 weeks and mice with double positive EGFRL858RCC10rtTA were administered doxycycline at a concentration of 1.0 g/L in drinking water, which was replaced twice weekly. Multifocal lung adenocarcinomas were developed after 7–8 weeks’ induction and these tumors were confirmed by CT scan after 8–10 weeks of doxycycline induction (online supplemental figure 1). The animal models were established as previously described.31 32 All the animal studies were approved by the Institutional Committee for Animal Care and Use, Shanghai Pulmonary Hospital and were performed in accordance with the institutional guidelines.
jitc-2022-005436supp004.pdf (1.4MB, pdf)
Cell lines and reagents
Beas-2B, HCC827, PC9, A549, H23, H2030, and Jurkat cell lines were maintained in DMEM or RPMI1640 (HyClone) supplemented with 10% fetal bovine serum (FBS) (Gibco) and 1% penicillin-streptomycin (Gibco). The recombinant mouse IL-10 protein (#417-ml-100CF) and recombinant human IL-10 protein (#217-IL-010) was purchased from R&D Systems (Minneapolis, MN, USA). Human EGF (# 100-15-100) was purchased from Peprotech (Cranbury, NJ, USA). Gefitinib (ZD1839) was purchased from Selleck. Anti-PD-1 mAb (#BE0146, clone: RMP1-14), IgG control (#BE0089, clone: 2A3), InVivoPure pH 7.0 Dilution Buffer (#IP0070) and InVivoPure pH 6.5 Dilution Buffer (#IP0065) were purchase from BioXcell (West Lebanon, New Hampshire, USA).
siRNA transfection
h-ENTPD1-siRNAs were commercially synthesized (Ribo-Bio Co) and transfected into cells using riboFECT CP Transfection Kit (Ribo-Bio Co). siRNA sequences targeting human genes were as below: siENTPD1-1, GCAAGGCTATCATTTCACA.
Quantitative RT-PCR
Total RNAs were isolated from cells using RNAiso Plus (TAKARA) and reverse-transcribed to cDNA using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). Diluted cDNA was subject to Quantitative RT-PCR (qRT-PCR) analysis using TB Green Premix Ex Taq II (TAKARA). Relative expression levels were calculated by the comparative Ct approach. qRT-PCR primers were listed as follows:
(1) h-ENTPD1-Forward, 5’-CTCAGGAAAAGGTGACTGAGAT-3’ and h-ENTPD1-Reverse, 5’-CTCCTTTACTCCAGCGTAAGAT-3’.
(2) h-IL10-Forward, 5’-GTTGTTAAAGGAGTCCTTGCTG-3’ and h-IL10-Reverse, 5’-TTCACAGGGAAGAAATCGATGA-3’.
(3) h-GAPDH-Forward, 5’-GAAATCCCATCACCATCTTCCAGG-3’ and h-GAPDH-Reverse, 5’-GAGCCCCAGCCTTCTCCATG-3’.
Western blotting
Protein samples were extracted from cells using RIPA lysis buffer (Beyotime). The quantification of protein level was measured with BCA kit (Beyotime). The samples were boiled in 5X loading buffer, separated by SDS-PAGE, transferred to PVDF membranes (Millipore). After blocking, the membrane was incubated overnight at 4°C with primary antibodies as followings: p-EGFR (#3777S, Cell Signaling Technology; 1:1000 dilution), GAPDH (#5174, Cell Signaling Technology; 1:1000 dilution). The membrane was then washed with TBST and incubated with secondary antibody for 1 hour at room temperature in the dark. The targeted proteins were detected by ECL reagent (Tanon) and the exposure was performed by ChemiDoc XRS (Bio-Rad).
Cytokine measurements
Plasma samples were separated by centrifugation and then stored at −80°C until use. The concentrations of IFN-γ, IL-10, IL-12p70, IL-13, IL-1β, IL-2, IL-4, IL-6, IL-8, TNF-α in plasma were measured using the V-PLEX Proinflammatory Panel 1 (human) Kit from Meso Scale Discovery according to the manufacturer’s instructions.
Flow cytometry
The single-cell suspensions of tumor tissue were generated with tumor dissociation kit, mouse (Miltenyi Biotec) or tumor dissociation kit, human (Miltenyi Biotec) according to manufacturer’s instructions and PBMC samples were thawed for further analysis. After resuspended in FACS buffer (PBS, 2%FBS), the cells were blocked with anti-mouse CD16/CD32 (eBioscience) 10 min prior to staining with surface antibodies at 4°C. Surface markers were then added along with 10 µL Brilliant Stain Buffer (BD Bioscience) in a final staining volume of 100 µL. For intracellular staining, cells were fixed and permeabilized with Transcription Factor Buffer Set (BD Bioscience), followed by staining with intracellular antibodies. Fluorochrome-conjugated antibodies for flow cytometry analysis were listed in online supplemental table 3. Gating strategy was shown in online supplemental figure 2 and markers for cell population were defined in online supplemental table 4. The flow cytometry data were acquired on Beckman Cytoflex (Beckman Coulter) and the results were analyzed with FlowJo V.10.6.1 software (TreeStar).
jitc-2022-005436supp005.xlsx (10.7KB, xlsx)
jitc-2022-005436supp006.pdf (956.2KB, pdf)
jitc-2022-005436supp007.xlsx (10.4KB, xlsx)
Histologic analysis
The 4 µm paraffin-embedded tissue were deparaffinized and rehydrated, stained with H&E, IHC, IF following manufacturer protocol. For IHC, the tissue slides were high pressure-treated and boiled in EDTA Antigen Retrieval Solution (pH 8.0; KeyGEN BioTECH) for 5 min. The sections were kept in high-pressure for 10 min and cooled down naturally to room temperature. 3% hydrogen peroxide was used for 5 min at room temperature. After blocking the slides with 5% goat serum for 30 min, slides were incubated overnight in a humidified chamber at 4°C with primary antibodies as followings: GranzymeB (#ab4059, Abcam; 1:400 dilution). The slides were then washed with PBST and incubated with secondary antibody targeting rabbit IgG (MaxVision HRP-Polymer anti-rabbit IHC Kit, Maixim Bio) for 1 hour at room temperature. After washing with PBST, the signal was detected by DAB reagent (Maixim Bio) for 5 min at room temperature, immersed in ddH2O to stop the reaction, and subsequently counterstained with hematoxylin for 12 s. The intensities of brown-colored precipitate stained with DAB were measured and quantified with IOD or cell intensity by Image Pro Plus V.6 (Media Cybernetics). For immunofluroscence (IF), the antigen retrieval and nonspecific blocking procedure were similar to that of IHC. Sections were first incubated with primary antibody, CD8 (# 66868–1-Ig, Proteintech; 1:500 dilution) for 1 hour at room temperature and then stained with appropriate mouse fluorophore-conjugated secondary antibodies for 2 hours according to the manufacturer’s recommendations. After washing the slides with PBST, the tissues were counterstained with DAPI for cell nuclei detection. The slides were coverslipped with antifade mountant (Invitrogen) and the whole slide were scanned with Pannoramic MIDI (3D HISTECH).
RNA in situ hybridization
Chromogenic RNA in situ hybridization was performed by using ACD RNAscope 2.5 Red assays with ACD target probes: IL-10 RNAscopeProbe-Hs-IL10 (#602051). All hybridization and incubation steps were performed using the complete HybEZ Hybridization System following the manufacturer’s protocol.
Statistical analysis
Results were presented as mean±SEM. An established computational approach (CIBERSORT) to quantify cell fractions from bulk tissue gene expression profiles was applied to infer the proportions of 22 subsets of immune cells with LM22 gene signature.33 Baseline clinical characteristics of patients were listed by numbers and percentages. To analyze the difference in baseline clinical characteristics between subgroups, the χ2 or Fisher’s exact test was performed. The Kaplan-Meier method was used to analyze the survival probability and log-rank test was used to calculate the significance of differences. Cox proportional hazard model was applied for the univariate and multivariate analyses to calculate the HRs and 95% CIs. Parameters with the univariate p<0.1 were included in the multivariate model. Unpaired Student’s t-test was used to compare the statistical significance between tested and control groups. Statistical significance was determined as indicated in the figures. Significance was set to p<0.05 and represented as *p<0.05, **p<0.01, ***p<0.001. All data were analyzed by using the SPSS software (V.3.0 for Mac) and GraphPad Prism software (V.8 for Windows).
Results
EGFR-mutated lung tumors display poor response to PD-1 blockade
We explored the antitumor activity of PD-1 blockade in previously generated mouse models with autochthonous lung tumors induced by EGFR L858R .The mice were treated with IgG control or anti-PD-1 antibody at 200 µg/mouse by intraperitoneal injection every 3 days (figure 1A). Upon histological examination (figure 1B), the lungs weight was comparable between IgG control and anti-PD1 group, whereas the lungs weight was heavier than healthy group (figure 1C). In addition, the tumor invasive areas were also comparable between control and anti-PD-1 group (figure 1D), indicating that EGFR-mutated tumors were resistant to anti-PD-1 antibody which was consistent with clinical findings and confirmed that our EGFR-mutated mouse model was reliable for subsequent analyses.
Figure 1.
The efficacy of immunotherapy in EGFR-mutated lung tumors. (A) Schematic diagram depicting treatment schedule for EGFR-mutated tumors. After confirmation of developing lung tumor nodules, the mice were randomly allocated to two groups. Red arrow indicating the PD-1 blockade was administered 200 µg/mouse by intraperitoneal injection every 3 days and blue arrows indicating the IgG control was administered 200 µg/mouse by intraperitoneal injection every 3 days. Mouse were sacrificed for tumor analysis at indicated time. (B) Representative lung images of H&E staining in each group at indicated time point. Scale bars, 2.5 mm. (C) Analysis of lungs weight in each group at day 5 and day 11. (D) Analysis of tumor invasion areas in each group at Day 5 and Day 11. n=3–5 mice per group at indicated time point. Unpaired Student’s t-test was performed and results in each group were presented as mean±SEM. *p<0.05, **p<0.01, ***p<0.001, and ns represents p values with no statistical difference.
EGFR-mutated tumors show a lack of CD8+T cell infiltration and impaired CD8+T cell cytotoxic function
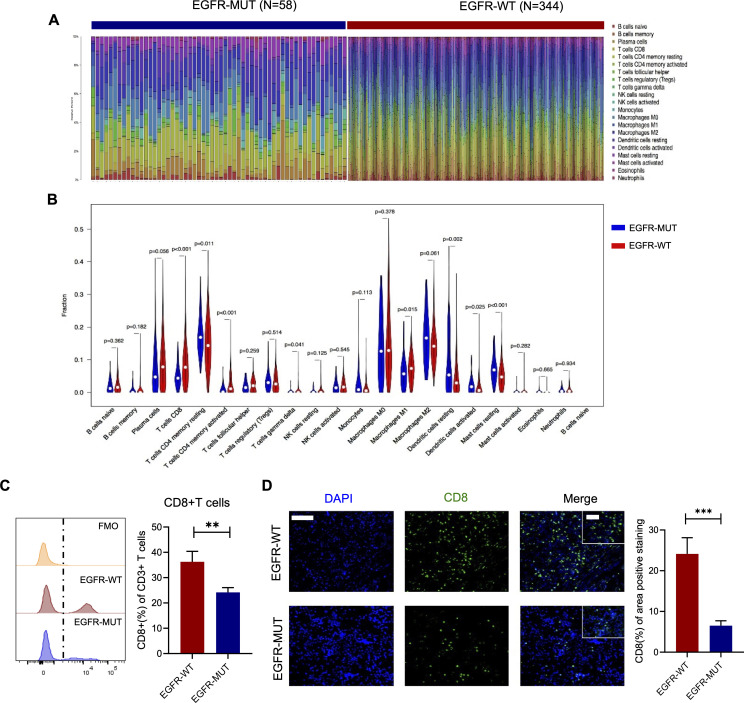
Anti-PD1 antibody appeared to be less efficacious in EGFR-mutated tumors, but the underlying mechanism is poorly understood. To uncover the underlying mechanism, we sought to decipher the unique characteristics of TME in EGFR-mutated NSCLC. First, we used the CIBERSORT to impute the fractions of 22 subsets of immune cells in EGFR-mutated and wild-type tissue samples using gene expression data from TCGA database (figure 2A). We found that EGFR-wild-type tumors were associated with higher proportions of CD8+T cells (p<0.001) and M1 macrophage (p=0.015) (figure 2B). Next, to verify the results from online database, 30 fresh primary tumor specimens and corresponding FFPE tissue samples were collected to analyze the subtypes of immune cells via flow cytometry and immunofluorescence. In general, EGFR-mutated tumors had decreased CD8+T cells infiltration compared with EGFR-wild-type tumors (figure 2C, D), whereas other immune cells including T cells, B cells, natural killer cells and myeloid cells were comparable between EGFR-mutated and EGFR-wild-type tumors (online supplemental figure 3A, B). Although EGFR-mutated tumors had more infiltration of total CD4+T cells, no significant difference was observed in numbers of T helper cells and Tregs (online supplemental figure 3C).
Figure 2.
The characterizations of immune cells infiltration in EGFR-mutated tumors. (A) The overview of immune infiltrations and (B) the proportions of immune cells in EGFR-mutated and wild-type tumors (data source: TCGA, n=58 in EGFR-mutated group and n=344 in EGFR-wild-type group). (C) Flow cytometry analysis of resected lung tumors (left histogram) and ratio of CD8+ of CD3+ cells. (D) Representative immunofluorescence images (left) and analysis of CD8 positive areas (right) in EGFR-mutated and wild-type tumors (n=19 in EGFR-mutated group and n=11 in EGFR-wild-type group). Scale bars, 100 µm. Inset scale bars, 50 µm. Unpaired Student’s t-test was performed and results in each group were presented as mean±SEM. **p<0.01, ***p<0.001. TCGA, The Cancer Genome Atlas.
jitc-2022-005436supp008.pdf (23.4KB, pdf)
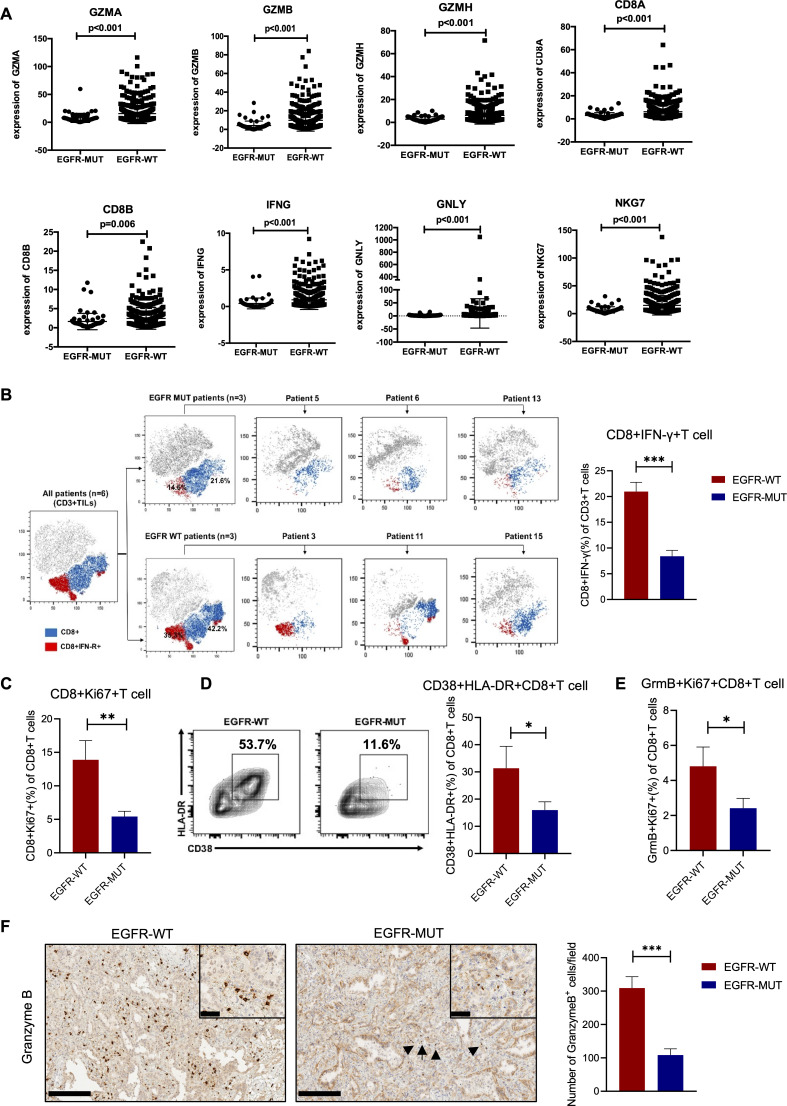
Efficacy of cancer immunotherapy depends in part on the number and properties of CD8+T cell.34 To depict the landscape of the composition, lineage and functional states of TILs in EGFR-mutated and EGFR-wild-type tumors, we extracted the RNA sequencing data from TCGA database based on the signature previously described by Guo et al.35 Expression of lymphocyte activation or functions markers, including CD8A, CD8B, and effector molecules including GZMA, GZMB, GZMH, IFNG, GNLY, NKG7 were significantly decreased in EGFR-mutated tumors (figure 3A). Additionally, as costimulatory markers, CD38 and FASLG expression was also reduced in EGFR-mutated tumors, whereas naïve markers and Treg-related makers showed no difference between two tumors subtypes (online supplemental figure 4). Similarly, in fresh NSCLC samples, EGFR-mutated tumors had lower proportion of cytotoxic (IFN-γ+) and proliferated (Ki67+) CD8+T cells (figure 3B, C). Functional or activated subsets, such as CD38+HLA-DR+ and GranzymeB+Ki67+ were also significantly less infiltrated in EGFR-mutated tumors (figure 3D, E). Similar results were observed in IHC analysis of GranzymeB+ cells, which confirmed that EGFR-mutated tumors showed less cytotoxic T cells compared with EGFR wild-type tumors (figure 3F).
Figure 3.
CD8+T cells in EGFR-mutated tumors had impaired cytotoxic function. (A) FPKM expression of GZMA, GZMB, GZMH, CD8A, CD8B, IFNG, GNLY, NKG7 comparison between EGFR-mutated and wild-type tumors (n=66 in EGFR-mutated group and n=411 in EGFR-wild type group). (B) Representative t-SNE map of CD3+T cells isolated from lung tumors and were performed simultaneously on three different patients from each cohort to explore the proportions of IFN-γ in CD8+T cells (left). FACS percentage of IFN-γ in CD8+T cells in each group. (C) Ratio of Ki67+ of CD8+T cells. (D) Representative flow cytometry images (left) and histogram showed the percentage of CD38+HLA-DR+ of CD8+ cells in each group. (E) Ratio of GranzymeB+Ki67+ of CD8+T cells. (F) Representative immunohistochemistry images (left) and numbers of GranzymeB positive cells (right). (A) Mann-Whitney U test was performed, and p value was indicated in figures. (B–F) n=19 in EGFR-mutated group and n=11 in EGFR-wild-type group. Scale bars, 200 µm. Inset scale bars, 50 µm. Unpaired Student’s t-test was performed and results in each group were presented as mean±SEM. *p<0.05, **p<0.01, ***p<0.001.
jitc-2022-005436supp009.pdf (586.5KB, pdf)
Taken together, the above findings suggested that EGFR-mutated tumors revealed a low CD8+T cell density and presented as impaired functional states.
The incompetent CD8+T cells in EGFR-mutated tumors are characterized as absence of CD39 expression
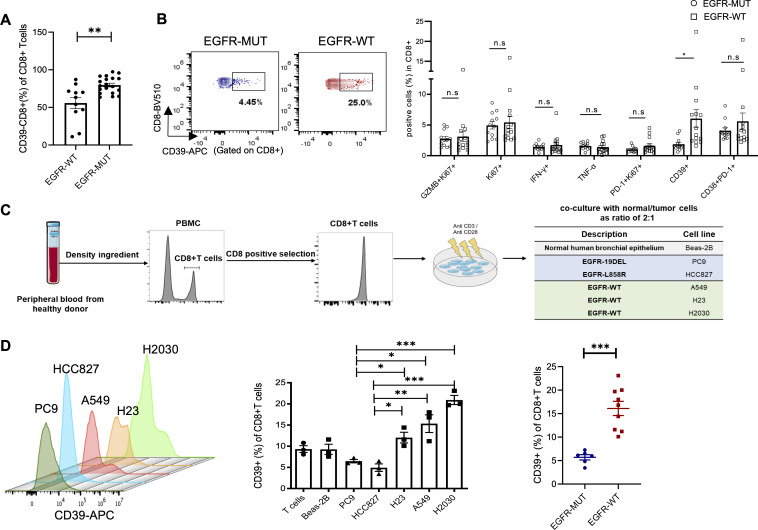
Having identified that the impaired functional CD8+T cells were highly presented in EGFR-mutated TME, we speculated that these incompetent CD8+T cells had a specific signature to be quantified. Simoni et al demonstrated that in human cancers, bystander infiltrating CD8+T cells, characterized as the absence of CD39, lacked the hallmarks of chronic antigen stimulation at the tumor site and hence, lost the ability to exert the T-cell-mediated killing function.21 Therefore, we next investigated whether CD39 by CD8+TILs were linked with EGFR mutation status. Interestingly, the frequency of CD39-CD8+T cells were significantly higher in EGFR-mutated tumors in our cohort (figure 4A). We also obtained the peripheral blood from treatment-naïve, advanced NSCLC patients (n=26). Of note, the CD39+CD8+T cells were enriched in EGFR-wild-type (n=14) tumors instead of EGFR-mutated tumors (n=12), whereas no significant difference was observed in proportions of GranzymB+Ki67+, Ki67+, IFN-γ+, TNF-α+ in CD8+T cells. Several predictive biomarkers for anti-PD-1 treatment, such as PD-1+Ki67+CD8+T cells36 and CD38+PD-1+CD8+T cells37 were also similar in EGFR-mutated and wild-type patients (figure 4B). Since these results demonstrated abundant bystander CD39-CD8+T cells were present in EGFR-mutated tumors, we asked whether the expression of CD39 on CD8+T cells was regulated by the activation of EGFR signaling. We cocultured isolated human CD8+T cells with lung epithelial normal cells or lung tumor cells in vitro. Isolated human CD8+T cells were activated and then cocultured with normal (Beas-2B) cells, EGFR-mutated tumor cells (PC9, HCC827) and EGFR-wild-type tumors (A549, H23, H2030) (figure 4C). After co-cultured for 72 hours, the expression of CD39 on CD8+T cells was tested. We found that CD8+T cells co-cultured with EGFR-wild-type tumor cells had significantly increased CD39 expression compared with those co-cultured with EGFR-mutated tumor cells (figure 4D), suggesting that EGFR-wild-type tumor cell had direct impact on CD39 expression on CD8+T cells.
Figure 4.
EGFR-mutated tumors revealed abundant CD39-CD8+T cells. (A) FACS percentage of CD39-CD8+T cells in tissue specimen (n=19 in EGFR-mutated group and n=11 in EGFR-wild-type group). (B) Flow cytometry analysis of functional markers in CD8+T cells in PBMC of advanced lung cancer patient (n=12 in EGFR-mutated group and n=14 in EGFR-wild-type group). (C) Schematic of in vitro co-culture experiments. Human CD8+T cells were isolated and activated with anti CD3/CD28 beads, and then were replated and co-cultured with normal/tumor cells as ratio of 2:1 for 72 hours. (D) FACS percentage of CD39+ in CD8+T cells after co-cultured with different cells. Unpaired Student’s t-test was performed and results in each group were presented as mean±SEM. *p<0.05, **p<0.01, ***p<0.001, and ns represents p values with no statistical difference. PBMC, peripheral blood mononuclear cell.
Thus, these data showed that EGFR-mutated tumors presented abundant bystander CD39-CD8+T cells and CD39 expression on CD8+T cells were closely linked with EGFR mutational status.
CD39 expression determines CD8+T cell cytotoxic function and predicts ICI efficacy
The results described in the previous section suggested a striking lack of CD39 expression in CD8+T cells of EGFR-mutated tumors. Hence, to better depict the characteristics of CD39+CD8+T cells and bystander CD39-CD8+T cells, we performed flow cytometry on human PBMCs. Compared with CD39-CD8+ counterparts, CD39+CD8+T cells showed hallmarks of functional markers (CD38+HLA-DR+, Ki67+, GZMB+Ki67+) and exhausted phenotypes (PD-1+Ki67+) (figure 5A). Transcriptome profiling data from TCGA indicated that ENTPD1, encoding CD39, was in positive proportion to expression of genes related to cytotoxic and functional signatures, including IFNG, GZMB, IL2RB, PRF1, GZMA, and TNF (figure 5B). In line with these results, flow cytometry analysis of tumor tissues indicated that the frequency of CD39 expression was positively correlated to the proportions of CD38+HLA-DR+T cells (r=0.68, p<0.001) (figure 5C). To further assess the impact of CD39 expression on the function of CD8+T cells, we tested the proliferated or functional state of Jurkat cell with or without knocking down ENTPD1. With 70% interference efficacy of ENTPD1 (figure 5D), the percentage of Ki67+ and GranzymeB+ cells were strikingly reduced (figure 5E). Next, we investigated whether CD39+CD8+T cells were linked to anti-PD1-treatment efficacy. We obtained peripheral blood from 11 patients with advanced NSCLC before and after one-cycle of ICI treatment. As expected, the proportions of PD-1+CD8+T cells were significantly decreased after anti-PD-1 treatment, in accordance with the principles of ICI therapy (figure 5F). We found that patients with rapid radiological response (n=5) were characterized by high baseline CD39+population in CD8+T cells compared with non-responders (n=6), suggesting a potential role of CD39+CD8+T cells in predicting ICI efficacy (figure 5G).
Figure 5.
CD39 expression determined CD8+T cell cytotoxic function and predicts ICI efficacy. (A) Expression of functional and exhausted markers by CD39+ and CD39- subpopulation of CD8+T cells from human PBMCs. (B) Correlations between expression of cytotoxic markers and ENTPD1 (encoding CD39) (data source: TCGA). (C) Correlations between percentage of CD38+HLA-DR+ and CD39+ in CD8+T cells infiltrated in lung tumor tissues (n=30). (D) Interference efficacy of si-ENTPD1 in Jurkat cells. (E) FACS percentage of Ki67 and GranzymeB+ in Jurkat cells. (F) Schematic for blood collection time point in lung cancer patients treated with PD-1 blockade (left). FACS analysis of PD-1+ in CD8+T cells at pretreatment and after one-cycle of treatment (right) (n=11). (G) The numbers of responders (PR) and non-responders (PD+SD) and FACS percentage of baseline CD39+ in CD8+T cells in peripheral blood. Unpaired Student’s t-test was performed and results in each group were presented as mean±SEM. *p<0.05, ***p<0.001. ICI, immune checkpoint inhibitor; TCGA, The Cancer Genome Atlas. PBMC, peripheral blood mononuclear cell.
Together, these results demonstrated that the presence of CD39 determines the CD8+T cells to possess the cytotoxic and exhausted features and correlated with effective ICI response, supporting the notion that the poor response to anti-PD-1 treatment in EGFR-mutated tumors could be attributed to the abundance of bystander CD39-CD8+T cells and lack of CD39+CD8+T cells in EGFR-mutated tumors.
IL-10 is downregulated in EGFR-mutated tumors
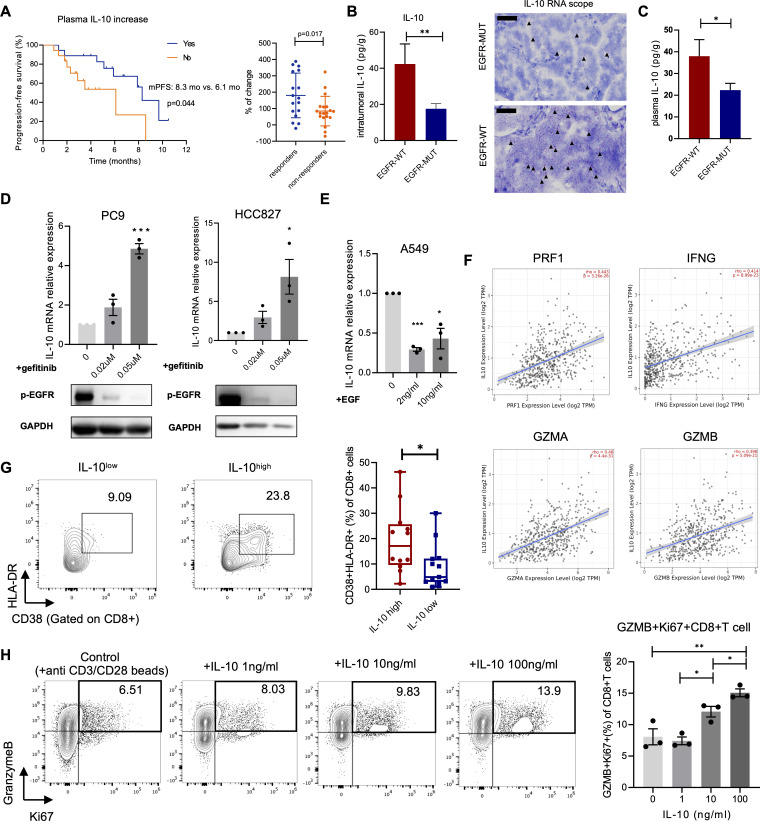
To understand the reason behind the distinct CD39 expression in EGFR-mutated and wild-type tumors, cytokine networks raised our research interest. Cytokines are potent regulators secreted by diverse cell types and act as soluble protein mediators of cell communication in complex TME.38 39 Given the roles of cytokines in activating, maintaining, and involving in adaptive immune response to contribute to antitumor immunity, we hypothesized that the expression of CD39 on CD8+T cells was regulated by a certain T cell-related cytokine which could also predict ICI efficacy.40 Hence, concentrations of 8 T-cell-related cytokines, including IL-2, IL-4, IL-6, IL8, IL-10, IL-12p70, IFN-γ, and TNF-α levels were measured in plasma of 36 patients diagnosed with advanced NSCLC treated with ICIs at baseline, and at 2–3 weeks after initiating treatment (online supplemental figure 5). Interestingly, only changes in plasma IL-10 reflected tumor response, patients with increased plasma IL-10 had longer PFS (mPFS: 8.3 vs 6.1 months, p=0.044) and better objective response (p=0.017) (figure 6A, online supplemental figure 6). Multivariate analysis showed that patients who received subsequent treatment line of immunotherapy had inferior PFS (HR=15.671, p=0.011). Except for treatment-line, only increased plasma IL-10 was linked with better PFS instead of other cytokines (p=0.052 and p=0.050, respectively) (online supplemental table 5). Based on these data, we sought to investigate whether poor response to ICI treatment was attributed to the distinct level of IL-10 in EGFR-mutated tumors compared with EGFR-wild-type tumors. As expected, intratumoral and plasma levels of IL-10 were significantly lower in EGFR-mutated tumors (figure 6B–C). IL-10 is predominately produced by leukocytes, including macrophages, myeloid-derived suppressor cells (MDSCs) and T helper (Th) cells.41 Since the immune cells, which were the conventional source of IL-10 showed no difference in proportions between EGFR-mutated tumors or wild-type, we tested whether the intrinsic EGFR signaling pathway in tumor cells contributed to different secretion levels of IL-10. In vitro study showed that with EGFR-TKI (gefitinib) to block EGFR signaling pathway in EGFR-mutated tumor cells (PC9, HCC827), IL-10 expression was significantly increased in dose-dependent manner (figure 6D). Conversely, with the addition of EGF to restimulate EGFR signaling pathway in EGFR-wild-type cells (A549), IL-10 expression was dramatically decreased (figure 6E), suggesting that different levels of IL-10 in EGFR-mutated and wild-type tumors were likely to be associated with the activation of EGFR signaling in tumor cells. To further investigate the impact of IL-10 on CD8+T cells, we firstly extracted transcriptome profiling data from TCGA and found a positive correlation between IL-10 expression and activated markers of T cells, such as PRF1, IFNG, GZMA and GZMB (figure 6F). Also, CD8+T cells in tIL-10high (tissue-IL-10) patients had higher expression of T cell activated markers (figure 6G). To establish the direct link between IL-10 and function of CD8+T cells, we performed the in vitro restimulation model with human CD8+T cells. We found that IL-10 induced the activated phenotype of CD8+T cells with elevated expression of GZMB+Ki67+ (figure 6H).
Figure 6.
IL-10 was correlated with ICI response and downregulated in EGFR-mutated tumors. (A) Kaplan-Meier curve for PFS according to the change of IL-10 level in plasma of NSCLC patients treated with anti-PD-1(L1) antibodies (left). Percentage of IL-10 level changes in responders and non-responders. (B) Concentration of IL-10 in tumor tissues (left) (n=14 in EGFR-mutated group and n=7 in EGFR-wild-type group) and images for IL-10 RNA in situ hybridization (right). IL-10 mRNA signals (red dots with black arrows) were indicated. Scale bars, 50 µm. (C) Concentration of IL-10 in plasma of lung cancer patients (n=25 in EGFR-mutated group and n=12 in EGFR-wild-type group). (D) IL-10 mRNA relative expression on different doses of gefitinib treated with PC9 and HCC827 (upper). Western blot showed the expression of p-EGFR on gefitinib treatment (lower). (E) IL-10 mRNA relative expression on different doses of EGF treated with A549. (F) Correlations between expression of cytotoxic T-cell related markers and IL10 (data source: TCGA). (G) FACS percentage of CD38+HLA-DR+ in CD8+T cells infiltrated in tumor tissues with IL-10high or IL-10low level (n=12 in IL-10high group and n=13 in IL-10low group). (H) FACS percentage of GZMB+Ki67+ in CD8+T cells isolated from human peripheral blood on different doses of IL-10 restimulation. Unpaired Student’s t-test was performed and results in each group were presented as mean±SEM. *p<0.05, **p<0.01, ***p<0.001. ICI, immune checkpoint inhibitor; TCGA, The Cancer Genome Atlas.
jitc-2022-005436supp010.pdf (66KB, pdf)
jitc-2022-005436supp011.xlsx (10.8KB, xlsx)
These results confirmed that (1) increased IL-10 was associated with ICI response and could directly activate CD8+T cells and (2) IL-10 expression was directly regulated by EGFR signaling pathway and poor response to anti-PD-1 treatment in EGFR-mutated tumors could be possibly attributed to the absence of IL-10.
IL-10 upregulates CD39 expression and thus enhances T cell-receptor-mediated CD8+T cell function
The data outlined earlier indicated that compared with EGFR-wild-type tumors, EGFR-mutated tumors showed low IL-10 level and lack of CD39+CD8+T cells, which were both significantly correlated with ICI efficacy. To investigate the intrinsic correlations behind these two parameters, we profiled ENTPD1 and IL-10 expression using TIMER database. Clearly, ENTPD1 expression was positively related to IL-10 expression and the proportions of CD39+CD8+T cells were higher in tIL-10-high patients (figure 7A, B). Moreover, IL-10 directly induced CD39 expression on CD8+T cells in dose-dependent manner in vitro (figure 7C). Since IL-10 stimulated cytotoxicity of CD8+T cells and upregulated CD39 expression, also, CD39+CD8+T cells possessed the cytotoxic features, we next asked whether the functional states of T cells depended on CD39 expression after IL-10 stimulation. Jurkat cells with or without ENTPD1 knockdown, were stimulated with anti-CD3/CD28 beads and with presence of different doses of IL-10 in vitro for 3 days. With knockdown of ENTPD1, the proliferated and cytotoxic ability of Jurkat cells were not increased whereas IL-10 induced the proportions of GranzymeB+ and Ki67+ cells without ENTPD1 knockdown (figure 7D). These results supported the concept that IL-10 induced cytotoxic activity of CD8+T cells on account of CD39 expression.
Figure 7.
IL-10 upregulated CD39 expression and thus enhances T cell-receptor-mediated CD8+T cell function. (A) Correlations between expression of IL-10 and ENTPD1 (encoding CD39) (data source: TCGA). (B) FACS percentage of CD39+ in CD8+T cells infiltrated in tumor tissues with IL-10high or IL-10low level (n=12 in IL-10high group and n=13 in IL-10low group). (C) FACS percentage of CD39+ in CD8+T cells isolated from human peripheral blood on different doses of IL-10 restimulation. (D) FACS percentage of Ki67 and GranzymeB+ in Jurkat cells with or without knockdown of ENTPD1 on different doses of IL-10 restimulation. Unpaired Student’s t-test was performed and results in each group were presented as mean±SEM. *p<0.05, **p<0.01, ***p<0.001, and ns represents p values with no statistical difference. TCGA, The Cancer Genome Atlas.
Collectively, the above findings further suggested that EGFR-mutated tumors were lack of adequate IL-10 to stimulate CD8+T cells expressing CD39, thus abundant bystander CD39-CD8+T cells without potential ability to be reinvigorated on ICI treatment resulted in poor response to immunotherapy.
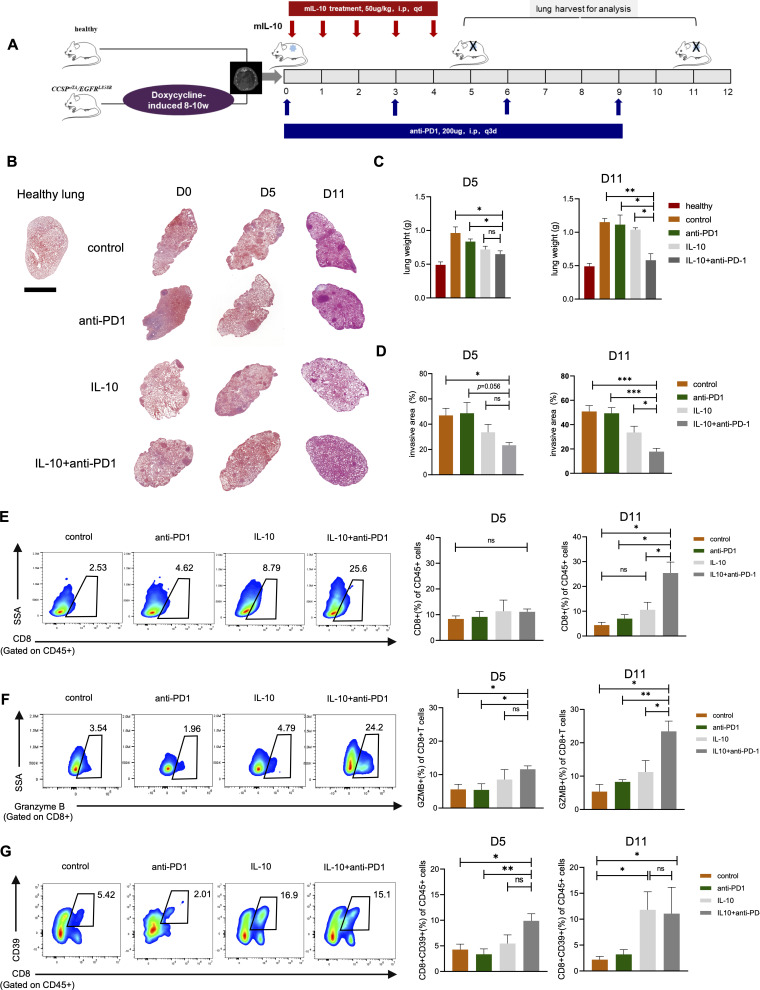
Combining rIL-10 and PD-1 blockade optimized antitumor effects in EGFR-mutated lung tumors
Since IL-10 was downregulated in EGFR-mutated tumors to inadequately induce abundant cytotoxic CD39+CD8+T cells, which possessed activated but exhausted feature, hence responded poorly to anti-PD-1 treatment, we next investigated whether combing IL-10 and PD-1 blockade could optimize antitumor activity in EGFR-mutated autochthonous lung tumors mouse model. After confirming the formation of autochthonous lung tumors, the mice were randomly divided into four groups as schedules shown in figure 8A, namely control group, anti-PD-1 antibody group, IL-10 group, and IL-10 plus anti-PD-1 antibody group. Upon histological examination (figure 8B), the lungs weight was comparable in the lungs of recombinant IL-10 (rIL-10) monotherapy and combination group, whereas was lighter than the lungs of control and anti-PD-1 group on day 5. Surprisingly, the lungs weight in combination group was continually decreased and lighter than rIL-10 monotherapy group on day11 (figure 8C). Similar results were also observed in tumor invasive areas (figure 8D), thus suggesting the synergetic antitumor effect of rIL-10 combining with PD-1 blockade in EGFR-mutated lung tumors. We then investigated the impact of different treatment regimens on the TME. On day5, the proportions of CD8+T cells were comparable in four groups, but the proportions of GranzymB+CD8+T cells in combination group were higher than other groups. On Day 11, the combination groups had the highest numbers of CD8+T cells infiltration with cytotoxicity (figure 8E, F). To test whether IL-10 regulated CD39 expression on CD8+T cells in vivo, we analyzed the proportions of CD39+CD8+T cells in four groups. As expected, rIL-10 monotherapy and combination group had elevated numbers of CD39+CD8+T cells compared with control and anti-PD-1 group (figure 8G).
Figure 8.
Combining rIL-10 and PD-1 blockade reduced tumor growth in EGFR-mutated tumors. (A) Schematic diagram depicting treatment schedule for EGFR-mutated tumors. After confirmation of developing lung tumor nodules, the mice were randomly allocated to four groups. Control group: mice were intraperitoneally injected with 100 µL PBS for 5 days, followed by IgG control at 200 µg/mouse by intraperitoneal injection every 3 days. Anti-PD1 group: mice were intraperitoneally injected with 100 µL PBS for 5 days, followed by anti-PD-1 antibody at 200 µg/mouse by intraperitoneal injection every 3 days. IL-10 group: rmIL-10 was dissolved in PBS and administered to mice at a dose of 50 mg/kg via intraperitoneal injection for 5 days. IL-10+ anti-PD1 group: rmIL-10 was administered to mice at a dose of 50 mg/kg via intraperitoneal injection for 5 days, followed by anti-PD-1 antibody at 200 µg/mouse by intraperitoneal injection every 3 days. Mice were anesthetized and euthanized at indicated time and lung tissue were harvested for analysis. (B) Representative lung images of H&E staining in each group at indicated time point. Scale bars, 2.5 mm. (C) Analysis of lungs weight in each group at day 5 and day 11. (D) Analysis of tumor invasion areas in each group at day 5 and day 11. (E) FACS percentage of CD8+ in CD45+ cells at day 5 and day 11. (F) FACS percentage of GranzymeB+ in CD8+ cells at day 5 and day 11. (G) FACS percentage of CD39+CD8+ in CD45+ cells at day 5 and day 11. n=3–5 mice per group at indicated time point. Unpaired Student’s t-test was performed and results in each group were presented as mean±SEM. *p<0.05, **p<0.01, ***p<0.001, and ns represents p values with no statistical difference.
Thus, in vivo data demonstrated that IL-10 could potentiate the antitumor effect when used in combination with anti-PD-1 antibody in EGFR-mutated lung cancer mouse models.
Discussion
The failure of PD-1 blockade therapy in EGFR-mutated NSCLC emphasized the demand to decipher the immense complexity of the TME and uncover the current therapeutic vulnerabilities. In this study, we identified that owing to low level of IL-10 to induce the expression of CD39 on CD8+T cells, EGFR-mutated tumors presented fewer phenotypically cytotoxic and exhausted CD39+CD8+T cells that could be potentially reinvigorated by ICI treatment. Instead, abundant bystander CD39-CD8+T cells with impaired cytotoxic functions could be associated with poor observed response to anti-PD-1 treatment in EGFR-mutated tumors. With rIL-10 treatment in EGFR mutated mouse models, the CD8+T cells in lung tissues were characterized by high expression of CD39 and functional features, confirming the synergetic antitumor effect of rIL-10 and PD-1 blockade to drive extinction of EGFR-mutated tumor cells. Our insights might inspire the design of improved immunotherapies that exploit a specific CD8+T cells subtype to benefit EGFR-mutated NSCLC patients.
With the burgeoning use of ICI for NSCLC, these immunotherapies fail to benefit the EGFR-mutated patients, hence, there has been a great need to understand which immune cell functions in the TME were impaired to be perturbed to inhibit tumor growth. The favorable prognostic significance of TILs, which acted as soldiers to exert killing functions, has been unequivocally established in NSCLC.42 Abundant CD8+T cells with cytotoxic functions infiltrated in the TME were the key determinants of success on ICI treatment. Combing TCGA data and analyses of resected lung tumors in our study cohort, we found that CD8+T cells were less infiltrated in EGFR-mutated tumor, which were consistent with other most studies.18 25 However, a previous study showed there was no statistical significance in CD8+T cells density between EGFR-mutated and wild-type tumors.19 In addition, Toki et al reported that although abundant CD8+T cells were present in EGFR-mutated tumors, high proportions of inactivated status were observed (low expression of Ki67+ and GranzymeB+) compared with EGFR-wild-type or KRAS-mutated tumors.43 In line with these findings, our resulted also suggested impaired CD8+T cell cytotoxic states were detected in the TME of EGFR-mutated tumors. Collectively, we proposed that difference in CD8+T cells between EGFR-mutated and wild-type tumors limited the immuno-response and additionally, to overcome the critical barrier to effective immunotherapy in EGFR-mutated tumors, increasing the proportions of CD8+T cells with cytotoxic functions were more rational than merely facilitating infiltration of CD8+T cells regardless of this functional status.
The presence of CD39 on CD8+T cells defined the population with cytotoxic and proliferated features that could be reinvigorated by ICI treatment.44 45 Based on this notion, we found that bystander CD39-CD8+T cells were abundant in EGFR-mutated tumors, whereas CD39+CD8+T cells with cytotoxic and exhausted markers were less infiltrated compared with EGFR-wild-type tumors. Additionally, patients with high baseline CD39+CD8+ T cell proportions had an improved response to ICI treatment in NSCLC which was supported by a previous study.21 Therefore, our findings showed that EGFR-mutated tumors were a lack of activated CD8+T cells, mostly were owing to the abundant bystander CD8+T cells, supporting that the poor observed anti-PD-1 treatment in EGFR-mutated tumors could be attributed to abundance of bystander CD39-CD8+T cells with impaired cytotoxic functions.
On this basis, approaches to expand the population of CD39+CD8+T cells might be an important component to improve the immunotherapy response in EGFR-mutated patients. Therefore, to increase the proportions of CD39+CD8+T cells with cytotoxic features pre-existing in EGFR-mutated tumors before ICI treatment, we explored the molecular mechanism linking with regulation of CD39 expression on CD8+T cells. Here, we demonstrated that IL-10, which was downregulated in EGFR-mutated tumors might be the key cytokine to upregulate CD39 expression, promote CD8+T cells to possess activated features. IL-10 has been known as immune suppressive cytokines and deficiency in IL-10 was correlated with autoimmune and inflammatory disease.41 46 Recently, IL-10 has gained master switch from tumor-promoting inflammation to antitumor immunity. Increasing evidence showed that pharmacological administration of IL-10 induced tumor rejections by enhancing CD8+T cell-mediated antitumor immunity with high expression of IFN-γ and granzymes.47 In line with this, our in vitro study found that IL-10 stimulated T-cell receptor-mediated CD8+T cell activation and importantly, IL-10 has been shown to induce activation of CD8+T cells in a CD39-dependent manner, supporting the notion that owing to insufficient IL-10 in EGFR-mutated tumors, CD8+T cells were present as bystanders in the TME. Meanwhile, Khalil et al identified eight genes associated with long DFS and inflamed TME in EGFR-mutated tumors via analyzing data from TCGA and PANCAN database, namely IL-10, BTLA, CD8A, CD39, CCR2, CSF-1R, ICOS, CD4, further corroborating that the key role of IL-10 and CD39 in EGFR-mutated tumors and their value in predicting ICI efficacy.48 As previously described, in EGFRL858R -driven mouse models, both gefitinib and osimertinib treatment significantly increased the levels of IL-10 in serum, suggesting that blocking EGFR signaling pathway, which mimicked EGFR-wild-type tumors resulted in increased levels of IL-1031. Similarly, our in vitro study also provided direct evidence that EGFR signaling pathway in tumor cells was associated with IL-10 expression. However, the regulatory mechanisms require more investigations.
Given the fact that EGFR-mutated tumors were lack of IL-10 to induce adequate CD39+CD8+T cells in tumors that could be reinvigorated by ICI treatment, we next utilized the EGFR L858R -driven mouse model to investigate the efficacy of rIL-10 combining with PD-1 blockade in vivo. Combining rIL-10 with PD-1 blockade significantly decreased the lungs weight and tumor invasion areas. Moreover, rIL-10 expanded the proportions of CD39+CD8+T cells and on this basis, cytotoxic CD8+T cells were proliferated, hence exerted killing function via secreting high levels of granzymeB. Our findings provided the preliminary evidence that expanding a specific and cytotoxic CD8+T cell subpopulation boost ICI efficacy in EGFR-mutated tumors. Indeed, rIL-10, used in the current mouse model had a very short plasma half-life, which would limit its clinical use.49 Therefore, to overcome the short-life of IL-10, Pegilodecakin (pegylated IL-10) a long-acting IL-10 receptor agonist that induces oligoclonal T-cell expansion was designed. Moreover, a phase Ib trial was performed in patients with advanced solid tumors treated with Pegilodecakin combined with pembroizumab or nivolumab. The results reported that the combination of PEG-IL10 and pembroizumab resulted in an overall response rate of 43% (12/28) in NSCLC.50 This promising data initiated the phase II clinical trials in NSCLC (CYPRESS1 and CYPRESS2) patients treated with PEG-IL10 combining with anti-PD-1 treatment in first or second-line treatment. However, the results were dismal with no improved PFS (CYPRESS1: 6.28 vs 6.08 months; CYPRESS2: 1.87 vs 1.94 months) and OS (CYPRESS2: 16.33 months versus not reached; 6.7 versus 7.0 months) and overall higher toxicities compared with ICI alone.51 The negative results from clinical trials were not supposed to deny the future explorations of this combination in EGFR-mutated tumors. First, the trials excluded the patients with EGFR mutations and admittedly, the TME components are distinct in EGFR-mutated and wild-type tumors.11 Based on our findings, IL-10 were limited in EGFR-mutated tumors instead of EGFR-wild-type tumors. In EGFR-mutated tumors, additional IL-10 acted as supplementary mediator whereas, in EGFR-wild-type tumors, whether additional administration of IL-10 going beyond physiological dose resulted in high toxicity needs further investigation. Second, evidence showed that even PEG-IL10 which was specially designed to prolong its circulation time in serum was still detected lower than 10% of the initial dose.52 Therefore, many challenges remain to harness cytokines for cancer therapy, especially delivery methods, context dependencies and short half-life. Lastly, the source of IL-10 might act as different functions. Previous studies demonstrated that macrophage and MDSC secreted IL-10 inhibited CD8+T cell killer function.53 54 However, Cetuximab-based IL-10 fusion protein (CmAb-(IL10)2) allowed tumor-targeted delivery of IL-10 and demonstrated potent antitumor effects in preclinical study.55 The mechanism of different cell-originated IL-10-mediated tumor-promoting or tumor-suppressing effects awaits further investigations. Collectively, although many challenges remain, our research provided the frame of mind that the combination of IL-10 and PD-1 blockade still be worthy explored in EGFR-mutated tumors.
Our study is somewhat limited by short-life of rmIL-10 applied in mouse models and the IL-10 level in serum was not dynamically detected. Owing to the long period of reproduction of EGFRL858R -driven mouse, the sample size in each group was relatively small, further preclinical studies involving modified IL-10 are warranted. Additionally, we analyzed the TME components with samples of resected lung tumors and of note, the TME features may be distinct between early stage and late-stage lung tumors. More investigations are needed to confirm the findings in biopsy samples in patients with advanced lung cancer. Moreover, to emphasize the crucial role of CD39 in the process of IL-10 inducing activation of CD8+T cells, only in vitro experiments were performed. In vivo study should also be performed with the specific depletion of CD39 on CD8+T cells and tested the tumor invasion area and cytotoxic functions of CD8+T cells after administration of IL-10 in EGFR-mutated tumors.
In summary, we demonstrated that IL-10 and CD39+CD8+T cells played pivotal roles in predicting ICI treatment response. Our findings provided evidence that EGFR-mutated tumors were lack of IL-10 to induce the expression of CD39 on CD8+T cell and IL-10 enhanced CD8+T cell-mediated cytotoxic function in a CD39-dependent manner, hence abundant bystander CD39-CD8+T cells were present in TME which were associated with poor response to ICI treatment. Combining IL-10 and PD-1 blockade showed promising antitumor effect in EGFR-mutated mouse models. Therefore, our data suggested that IL-10 and CD39 could be exploited for the development of novel biomarkers in weak immunogenicity tumors and also, it will be interesting to explore whether modified IL-10 with minimizing toxicity can improve therapeutic efficacy when combined with ICI in EGFR-mutated tumors.
jitc-2022-005436supp001.pdf (845.3KB, pdf)
jitc-2022-005436supp012.pdf (29.1KB, pdf)
jitc-2022-005436supp013.pdf (80.8KB, pdf)
Acknowledgments
We thank Jiang Fan, Yong Yang and Jie Dai for clinical specimen collection. We thank Hongbin Ji, Jun Zhang, Fei Li and Haipeng Liu for helpful insight for this project. We thank Renhong Tang, Jianquan Wu, Jun Jiang and Simeng Chen for providing research platform and technical support.
Footnotes
Contributors: All authors contributed to this study. MQ, FZ, HL, SR and CZhou conceived and designed the study; MQ, HL, XLiu, TJ and HW carried out the experiments; YJ, XLi, CZhao, LC and XC contributed to data analysis and interpreted the results; MQ and FZ drafted the manuscript. SR and CZhou revised the manuscript. CZhou acted as the guarantor of the manuscript. All authors reviewed the manuscript and approved the version to be submitted to journal.
Funding: This work was supported by Natural Science Foundation of China (grant 81871865 to CaZ, 82172869 to SR), Shanghai Collaborative Innovation Program (grant 2020CXJQ02 to CaZ), Shanghai Technology Innovative Project (grant 19411950300, 19411950301 to CaZ) and Shanghai Sailing Program (grant 22YF1425900 to MQ).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as online supplemental information.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by the Institutional Ethics Committee of Shanghai Pulmonary Hospital(K20-288). Participants gave informed consent to participate in the study before taking part.
References
- 1. Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced Nonsquamous non-small-cell lung cancer. N Engl J Med 2015;373:1627–39. 10.1056/NEJMoa1507643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med 2016;375:1823–33. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 3. Herbst RS, Baas P, Kim D-W, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540–50. 10.1016/S0140-6736(15)01281-7 [DOI] [PubMed] [Google Scholar]
- 4. Reck M, Rodríguez-Abreu D, Robinson AG, et al. Updated Analysis of KEYNOTE-024: Pembrolizumab Versus Platinum-Based Chemotherapy for Advanced Non-Small-Cell Lung Cancer With PD-L1 Tumor Proportion Score of 50% or Greater. J Clin Oncol 2019;37:537–46. 10.1200/JCO.18.00149 [DOI] [PubMed] [Google Scholar]
- 5. Shi Y, Au JS-K, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 2014;9:154–62. 10.1097/JTO.0000000000000033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mazieres J, Drilon A, Lusque A, et al. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol 2019;30:1321–8. 10.1093/annonc/mdz167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016;387:1837–46. 10.1016/S0140-6736(16)00587-0 [DOI] [PubMed] [Google Scholar]
- 8. Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): a phase 3, open-label, multicentre randomised controlled trial. Lancet 2017;389:255–65. 10.1016/S0140-6736(16)32517-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee CK, Man J, Lord S, et al. Checkpoint inhibitors in metastatic EGFR-mutated non-small cell lung cancer-a meta-analysis. J Thorac Oncol 2017;12:403–7. 10.1016/j.jtho.2016.10.007 [DOI] [PubMed] [Google Scholar]
- 10. Lee CK, Man J, Lord S, et al. Clinical and molecular characteristics associated with survival among patients treated with checkpoint inhibitors for advanced non-small cell lung carcinoma: a systematic review and meta-analysis. JAMA Oncol 2018;4:210–6. 10.1001/jamaoncol.2017.4427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Qiao M, Jiang T, Liu X, et al. Immune checkpoint inhibitors in EGFR-mutated NSCLC: Dusk or dawn? J Thorac Oncol 2021;16:1267–88. 10.1016/j.jtho.2021.04.003 [DOI] [PubMed] [Google Scholar]
- 12. Boussiotis VA. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N Engl J Med 2016;375:1767–78. 10.1056/NEJMra1514296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Akbay EA, Koyama S, Carretero J, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov 2013;3:1355–63. 10.1158/2159-8290.CD-13-0310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garassino MC, Cho B-C, Kim J-H, et al. Durvalumab as third-line or later treatment for advanced non-small-cell lung cancer (ATLANTIC): an open-label, single-arm, phase 2 study. Lancet Oncol 2018;19:521–36. 10.1016/S1470-2045(18)30144-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carbone DP, Reck M, Paz-Ares L, et al. First-Line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med 2017;376:2415–26. 10.1056/NEJMoa1613493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015;348:69–74. 10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- 17. Vokes N, Jimenez Alguilar E, Adeni A, et al. MA19.01 efficacy and genomic correlates of response to Anti-PD1/PD-L1 blockade in non-small cell lung cancers harboring targetable oncogenes. Journal of Thoracic Oncology 2018;13:S422. 10.1016/j.jtho.2018.08.474 [DOI] [Google Scholar]
- 18. Gainor JF, Shaw AT, Sequist LV, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: a retrospective analysis. Clin Cancer Res 2016;22:4585–93. 10.1158/1078-0432.CCR-15-3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mansuet-Lupo A, Alifano M, Pécuchet N, et al. Intratumoral immune cell densities are associated with lung adenocarcinoma gene alterations. Am J Respir Crit Care Med 2016;194:1403–12. 10.1164/rccm.201510-2031OC [DOI] [PubMed] [Google Scholar]
- 20. Miller BC, Sen DR, Al Abosy R, et al. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 2019;20:326–36. 10.1038/s41590-019-0312-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Simoni Y, Becht E, Fehlings M, et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 2018;557:575–9. 10.1038/s41586-018-0130-2 [DOI] [PubMed] [Google Scholar]
- 22. Liu T, Tan J, Wu M, et al. High-affinity neoantigens correlate with better prognosis and trigger potent antihepatocellular carcinoma (HCC) activity by activating CD39+CD8+ T cells. Gut 2021;70:1965–77. 10.1136/gutjnl-2020-322196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Canale FP, Ramello MC, Núñez N, et al. CD39 Expression Defines Cell Exhaustion in Tumor-Infiltrating CD8+ T Cells. Cancer Res 2018;78:115–28. 10.1158/0008-5472.CAN-16-2684 [DOI] [PubMed] [Google Scholar]
- 24. Sade-Feldman M, Yizhak K, Bjorgaard SL, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 2018;175:998–1013. 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dong Z-Y, Zhang J-T, Liu S-Y, et al. EGFR mutation correlates with uninflamed phenotype and weak immunogenicity, causing impaired response to PD-1 blockade in non-small cell lung cancer. Oncoimmunology 2017;6:e1356145. 10.1080/2162402X.2017.1356145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yeong J, Suteja L, Simoni Y, et al. Intratumoral CD39+CD8+ T Cells Predict Response to Programmed Cell Death Protein-1 or Programmed Death Ligand-1 Blockade in Patients With NSCLC. J Thorac Oncol 2021;16:1349–58. 10.1016/j.jtho.2021.04.016 [DOI] [PubMed] [Google Scholar]
- 27. Oft M. Immune regulation and cytotoxic T cell activation of IL-10 agonists - Preclinical and clinical experience. Semin Immunol 2019;44:101325. 10.1016/j.smim.2019.101325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res 2020;48:W509–14. 10.1093/nar/gkaa407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He Y, Song L, Wang H, et al. Mutational profile Evaluates response and survival to first-line chemotherapy in lung cancer. Adv Sci 2021;8:2003263. 10.1002/advs.202003263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu S, Wu F, Li X, et al. Patients with short PFS to EGFR-TKIs predicted better response to subsequent anti-PD-1/PD-L1 based immunotherapy in EGFR common mutation NSCLC. Front Oncol 2021;11:639947. 10.3389/fonc.2021.639947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jia Y, Li X, Jiang T, et al. EGFR-targeted therapy alters the tumor microenvironment in EGFR-driven lung tumors: implications for combination therapies. Int J Cancer 2019;145:1432–44. 10.1002/ijc.32191 [DOI] [PubMed] [Google Scholar]
- 32. Jia Y, Zhao S, Jiang T, et al. Impact of EGFR-TKIs combined with PD-L1 antibody on the lung tissue of EGFR-driven tumor-bearing mice. Lung Cancer 2019;137:85–93. 10.1016/j.lungcan.2019.09.016 [DOI] [PubMed] [Google Scholar]
- 33. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568–71. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guo X, Zhang Y, Zheng L, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat Med 2018;24:978–85. 10.1038/s41591-018-0045-3 [DOI] [PubMed] [Google Scholar]
- 36. Kamphorst AO, Pillai RN, Yang S, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc Natl Acad Sci U S A 2017;114:4993–8. 10.1073/pnas.1705327114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Verma V, Shrimali RK, Ahmad S, et al. PD-1 blockade in subprimed CD8 cells induces dysfunctional PD-1+CD38 hi cells and anti-PD-1 resistance. Nat Immunol 2019;20:1231–43. 10.1038/s41590-019-0441-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin J-X, Leonard WJ. Fine-tuning cytokine signals. Annu Rev Immunol 2019;37:295–324. 10.1146/annurev-immunol-042718-041447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Briukhovetska D, Dörr J, Endres S, et al. Interleukins in cancer: from biology to therapy. Nat Rev Cancer 2021;21:481–99. 10.1038/s41568-021-00363-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol 2009;9:480–90. 10.1038/nri2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ouyang W, O'Garra A. Il-10 family cytokines IL-10 and IL-22: from basic science to clinical translation. Immunity 2019;50:871–91. 10.1016/j.immuni.2019.03.020 [DOI] [PubMed] [Google Scholar]
- 42. Brambilla E, Le Teuff G, Marguet S, et al. Prognostic effect of tumor lymphocytic infiltration in resectable non-small-cell lung cancer. J Clin Oncol 2016;34:1223–30. 10.1200/JCO.2015.63.0970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Toki MI, Mani N, Smithy JW, et al. Immune marker profiling and programmed death ligand 1 expression across NSCLC mutations. J Thorac Oncol 2018;13:1884–96. 10.1016/j.jtho.2018.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu W, Zhao Z, Feng B, et al. CD8+CD39+ T cells mediate anti-tumor cytotoxicity in bladder cancer. Onco Targets Ther 2021;14:2149–61. 10.2147/OTT.S297272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lubbers JM, Ważyńska MA, van Rooij N, et al. Expression of CD39 identifies activated intratumoral CD8+ T cells in mismatch repair deficient endometrial cancer. Cancers 2022;14. 10.3390/cancers14081924. [Epub ahead of print: 11 04 2022]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Glocker E-O, Kotlarz D, Boztug K, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med 2009;361:2033–45. 10.1056/NEJMoa0907206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Naing A, Infante JR, Papadopoulos KP, et al. PEGylated IL-10 (Pegilodecakin) Induces Systemic Immune Activation, CD8+ T Cell Invigoration and Polyclonal T Cell Expansion in Cancer Patients. Cancer Cell 2018;34:775–91. 10.1016/j.ccell.2018.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Khalil M, Elkhanany A, Yang Y, et al. The tumor microenvironment in EGFR-driven loco-regional lung adenocarcinoma can predict higher risk of recurrence. Journal of Clinical Oncology 2019;37:8521. 10.1200/JCO.2019.37.15_suppl.8521 [DOI] [Google Scholar]
- 49. Huhn RD, Radwanski E, Gallo J, et al. Pharmacodynamics of subcutaneous recombinant human interleukin-10 in healthy volunteers. Clin Pharmacol Ther 1997;62:171–80. 10.1016/S0009-9236(97)90065-5 [DOI] [PubMed] [Google Scholar]
- 50. Naing A, Wong DJ, Infante JR, et al. Pegilodecakin combined with pembrolizumab or nivolumab for patients with advanced solid tumours (IVY): a multicentre, multicohort, open-label, phase 1B trial. Lancet Oncol 2019;20:1544–55. 10.1016/S1470-2045(19)30514-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spigel D, Jotte R, Nemunaitis J, et al. Randomized phase 2 studies of checkpoint inhibitors alone or in combination with Pegilodecakin in patients with metastatic NSCLC (cypress 1 and cypress 2). J Thorac Oncol 2021;16:327–33. 10.1016/j.jtho.2020.10.001 [DOI] [PubMed] [Google Scholar]
- 52. Mattos A, de Jager-Krikken A, de Haan M, et al. PEGylation of interleukin-10 improves the pharmacokinetic profile and enhances the antifibrotic effectivity in CCl₄-induced fibrogenesis in mice. J Control Release 2012;162:84–91. 10.1016/j.jconrel.2012.05.041 [DOI] [PubMed] [Google Scholar]
- 53. Koh J, Kim Y, Lee KY, et al. MDSC subtypes and CD39 expression on CD8+ T cells predict the efficacy of anti-PD-1 immunotherapy in patients with advanced NSCLC. Eur J Immunol 2020;50:1810–9. 10.1002/eji.202048534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang B, Vogelzang A, Miyajima M, et al. B cell-derived GABA elicits IL-10+ macrophages to limit anti-tumour immunity. Nature 2021;599:471–6. 10.1038/s41586-021-04082-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qiao J, Liu Z, Dong C, et al. Targeting Tumors with IL-10 Prevents Dendritic Cell-Mediated CD8+ T Cell Apoptosis. Cancer Cell 2019;35:901–15. 10.1016/j.ccell.2019.05.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005436supp002.xlsx (9.6KB, xlsx)
jitc-2022-005436supp003.xlsx (9.7KB, xlsx)
jitc-2022-005436supp004.pdf (1.4MB, pdf)
jitc-2022-005436supp005.xlsx (10.7KB, xlsx)
jitc-2022-005436supp006.pdf (956.2KB, pdf)
jitc-2022-005436supp007.xlsx (10.4KB, xlsx)
jitc-2022-005436supp008.pdf (23.4KB, pdf)
jitc-2022-005436supp009.pdf (586.5KB, pdf)
jitc-2022-005436supp010.pdf (66KB, pdf)
jitc-2022-005436supp011.xlsx (10.8KB, xlsx)
jitc-2022-005436supp001.pdf (845.3KB, pdf)
jitc-2022-005436supp012.pdf (29.1KB, pdf)
jitc-2022-005436supp013.pdf (80.8KB, pdf)
Data Availability Statement
Data are available on reasonable request. Data are available on reasonable request. All data relevant to the study are included in the article or uploaded as online supplemental information.