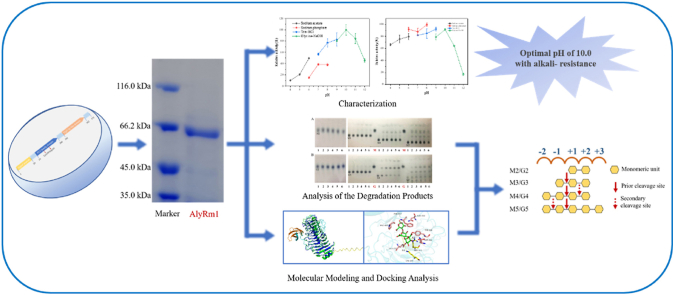
Abstract
Alginate lyase is essential for the production of alginate oligosaccharides (AOSs), which exhibit diverse bioactivities and have numerous applications in the food and pharmaceutical industries. The creation of recombinant alginate lyase by genetic engineering lays a crucial foundation for the commercialization of alginate lyase. This study cloned and expressed the polysaccharide lyase family 6 (PL6) alginate lyase gene alyrm1 from Rubrivirga marina.The optimum temperature and pH for recombinant AlyRm1 are 30 °C and 10.0, respectively. AlyRm1 shows good alkaline stability, for it remained over 80% of the enzyme activity after being incubated at pH 10.0 for 24 h AlyRm1 preferentially degrades PolyM into AOSs with degrees of polymerization (DP) 2–5 and monosaccharides as an endolytic bifunctional lyase. In addition, the analysis of degradation products toward oligosaccharides revealed that the minimal substrate of AlyRm1 is trisaccharide and clarified the degradation patterns.
Keywords: Alginate lyase, Alkaline resistant, Oligosaccharides, Molecular docking
Graphical abstract
Highlights
-
•
A novel PL6 family alginate lyase from Rubrivirga marina was characterized.
-
•
AlyRm1 is basophilic, has the highest activity at pH 10 and shows stability at a wide range of pH.
-
•
AlyRm1 is active against alginate, polyM and polyG with an endolytic activity.
-
•
Alphafold 2 is used to predict protein structure and catalytic mechanism.
1. Introduction
Alginate is a common polysaccharide exist in a variety of brown algae, constituting approximately 40% of the dry weight (Rioux and Turgeon, 2015). It is a linear unbranched polymer of homopolymeric monomers of (1 → 4)-linked β-D-mannuronic acid (M) and its C-5 epimer α-L-guluronic acid (G). Typically, there are three different types of blocks: polyguluronic acid (PolyG), polymannuronic acid (PolyM), and the heteropolymer of Ms and Gs (PolyMG or GM) (Barzkar et al., 2022). Due to its excellent physicochemical features, such as high viscosity, sol/gel transition capabilities, and water-retaining property, etc., alginate is widely employed as thickening ingredient in the food industry. (B. Zhu et al., 2019). As a matrix or carrier, alginate is predominantly used in biomedicine to prolong the drug’s release and enhance the drug's efficacy in the delivery systems. As a functional component, the use of alginate has been severely constrained refer to its polymeric structure, poor solubility, and low bioavailability (Benwei Zhu, Ni, Xiong, & Yao, 2021).
Alginate oligosaccharides have attracted great interest due to their biological activities, which include antioxidant activity, antimicrobial activity, and antitumor activity (Y. Li et al., 2021), indicating a promising application in the functional food and biopharmaceutical industries. In addition, they have been added successfully to feeds to improve aquaculture, poultry, and swine production, and they can also be utilized as elicitors of plants and microbes, and prebiotics. (J. Liu et al., 2019; Tusi et al., 2011). So far, alginate has been converted into AOSs via chemical (acid or alkali) or physical (microwave or hydrothermal treatment) or enzymatic method (alginate lyases) (M. Liu et al., 2021). Enzymatic degradation of alginate offers several benefits over physical and chemical methods, including environmental protection, energy savings, specificity, and high biological activity (Lari et al., 2019).
Alginate lyases depolymerize alginate through the β-elimination of the 1,4-glycosidic bond and subsequently generate an unsaturated C C between C4 and C5. Endolytic alginate lyases cleave long alginate chains to produce unsaturated oligosaccharides, while the exolytic alginate lyases generate unsaturated monosaccharides, which could be converted into 4-deoxy-L-erythro-5-hexoseulose uronic acid (DEH) nonenzymatically (D. M. Wang et al., 2014). Based on CAZY database (http://www.cazy.org/), alginate lyases have been classified into twelve polysaccharide lyase (PL) families (PL5, 6, 7, 14, 15, 17, 18, 31, 32, 34,36 and 39) (Jouanneau et al., 2021). In specific, alginate lyases are classified as PolyM-specific lyases (EC 4.2.2.3) which prefer degrading M blocks, and PolyG-specific lyases (EC 4.2.2.11) which prefer degrading G blocks, as well as bifunctional lyases (EC4.2.2.-) that can degrade both M blocks and G blocks (F. Xu et al., 2020). Alginate lyases are not only employed as catalysts for degrading alginate into AOSs, but also as medicines to enhance antibiotic killing of mucoid Pseudomonas aeruginosa in cystic fibrosis (Benwei Zhu, Tan, Qin, Xu, Du, & Yin, 2015). The exolytic alginate lyases could convert alginate to unsaturated monosaccharides, which split into pyruvate and glyceraldehydes-3-phosphate, and then the pyruvate can be converted into various biofuels and biochemicals (Kim et al., 2011). However, enzymes with higher stability, especially those that remain stable and highly active in industrial production scenarios, are still in shortage. In this work, a deep-sea marine bacteria Rubrivirga marina (Park et al., 2013) which harboring complete alginate metabolism pathway was chosen as the gene source for the potential enzymes with special properties, and a novel PL6 alginate lyase was characterized. Compared with other PL6 family enzymes that have been characterized, AlyRm1 has broad substrate specificity and high alkaline resistance, providing a candidate for the industrial application of alginate lyase.
2. Materials and methods
2.1. Chemicals and material
Sodium alginate (M/G ratio 77/23) was purchased from Sigma–Aldrich (St. Louis, MO, USA). Polymannuronic acid (PolyM, larger than 5 kDa) and polyguluronic acid (PolyG, larger than 5 kDa) were purchased from Carbosynth US LLC (San Diego, CA). Qingdao BZ Oligo Biotech Co., Ltd. (Qingdao, China) supplied β-D-mannuronic acid oligosaccharides (DP: 2–5, purity: over 97%) and α-L-guluronic acid oligosaccharides (DP: 2–5, purity: over 97%).
2.2. Amino acid sequence analysis of AlyRm1
The BLAST algorithm and conserved domain research on the National Center for Biotechnology Information server were applied to analyze the amino acid sequence. ExPASy ProtParam, an online analysis tool, was used to calculate some parameters of AlyRm1 including molecular weight (Mw), instability index, and isoelectric point (pI). The native signal peptide sequence of alyrm1 was predicted by SignalP 6.0 server (http://www.cbs.dtu.dk/services/SignalP/). Multi-sequence alignment was implemented by Clustal X and further analyzed through ESPript 3.0.
2.3. Cloning and expression of alyrm1
The alyrm1 gene (GenBank ID: WP_095509519.1) lacking the native signal peptide sequence consisting of 20 amino acids from Rubrivirga marina(GenBank ID: GCA_002283365.1) was amplified using DreamTaq Green PCR Master Mix (Thermo Fisher). The forward primer was 5′- CGGGATCCGACCGCTACGTCACGA -3′, and the reverse primer was 5′- CCCTCGAGCCGGGCCAGGGTCA -3′ (the BamHI and XhoI sites were underlined). The pET21a-AlyRm1 plasmid was transformed into E. coli host cells (DH5α and BL21) using the Ultra-Competent Cell Preps Kit (Sangon Biotech, Shanghai, China) and cultured in Luria-Berani (LB) broth with 100 mg/L of ampicillin. The cells were cultivated at 37 °C and 200 rpm till the optical density (OD) at 600 nm reached 0.8–1.0, after which the E. coli BL21(DE3) harbouring pET21a-AlyRm1 plasmid was induced with 1 mM isopropyl-D-1-thiogalactopyranoside (IPTG) at 16 °C and 180 rpm for 16 h to produce the recombinant protein. The cells were collected by centrifugation and resuspended in 20 mM sodium phosphate buffer (pH 7.4) containing 0.5 M NaCl, and broken up by using ultrasonication. The cell homogenate was centrifuged at 11000×g for 1 h at 4 °C. The supernatant was filtered through a 0.22 μm filter and then load to a HisTrap™ HP column (GE Healthcare, Uppsala, Sweden). The recombinant protein was eluted with buffer containing imidazole in gradient concentrations (50, 100, 200, 300, and 1000 mM). Protein fractions were displayed by SDS-PAGE to determine the molecular weight using Pierce™ Unstained Protein MW Marker (Thermo Fisher). The AlyRm1-containing fractions were concentrated using an Amicon ultra-centrifugal filter unit (Molecular weight cutoff value: 10000 Da; Millipore, Cork, Ireland). Bradford assay kit (Sangon Biotech, Shanghai, China) was applied to determine the concentration of the purified protein.
2.4. Enzymatic activity assay of AlyRm1
To determine the activity of AlyRm1, enzyme reaction was processed in a 200 μL 50 mM glycine-NaOH buffer (pH 10.0) with the concentration of sodium alginate at 2% (w/v). After incubated at 30 °C for 20 min, the reaction was terminated by boiling water bath for 5 min. The 3,5-dinitrosalicylic acid (DNS) assay(Bailey, 1988) was used to measure the reducing sugar concentration at 540 nm wavelength using glucose as the standard. One unit (U) was defined as the amount of enzyme capable of releasing 1 μmol of reducing sugars per minute.
2.5. Characterization of AlyRm1
The impact of temperature on AlyRm1 was investigated under different temperature intervals (20–70 °C). To test the thermostability, the enzymes were incubated at various temperatures for 1 h before the enzyme reaction. In addition, the enzymes were incubated at 30 °C for 30–120 min before the enzyme reaction to obtain the half-life time of AlyRm1 at 30 °C. After determining the optimal temperature, the effect of pH on AlyRm1 was assessed by incubating the enzyme in different buffers ranging from pH 4.0 to 12.0, including 50 mM sodium acetate buffer (pH 4.0–6.0), 50 mM sodium phosphate buffer (pH 6.0–8.0), 50 mM Tris-HCl buffer (pH 7.0–9.0), 50 mM glycine-NaOH buffer (pH 9.0–12.0) at 30 °C for 20 min. The pH stability was determined by measuring the residual activity after the enzyme was incubated in different buffers at 4 °C for 24 h. The effects of metal ions, EDTA and SDS on AlyRm1 were determined by incubating the enzyme under the optimum pH and containing different chemicals of 1 mM, and then measuring the enzymatic activity. The activity of AlyRm1 without adding metal ions was used as a control. Substrate specificity of AlyRm1 was determined using sodium alginate, PolyM and PolyG (2%, w/v) as substrates. Furthermore, the kinetic parameters of the enzyme toward these substrates were determined by measuring the initial reaction velocities under the optimal pH and temperature. The Km and Vmax values were obtained from a nonlinear regression plot using Origin 9.0 software.
2.6. Analysis of the degradation products of AlyRm1
Sodium alginate, PolyM and PolyG (2%, w/v) were digested by a 2 μM purified enzyme under optimal conditions for 0–24 h. The degradation products were firstly analyzed by thin layer chromatography (TLC) with the mobile phase of 1-butanol: acetic acid: water = 6:6:1. The developed plates were visualized Xu’s assay (X. Xu et al., 2014). The reaction products were also analyzed using High-performance liquid chromatography (HPLC) (Shimadzu LC-20A system, Kyoto, Japan) equipped with a PL aquagel-OH 20 size exclusion column (7.5 × 300 mm) (Agilent Technologies, CA) and a refractive index (RI) detector. The column was kept at 30 °C, and the mobile phase was 0.02 M NaH2PO4 with a flow rate of 0.2 mL/min. Hydrolysis reactions were conducted with β-D-mannuronic acid oligosaccharides (DP: 2–5) and α-L-guluronic acid oligosaccharides (DP: 2–5) as substrates in order to evaluate the reaction mode towards oligosaccharides and the minimal recognizable substrate of AlyRm1.
2.7. Molecular modelling and docking analysis
The structure of AlyRm1 was constructed using SWISS-MODEL online workspace using the alginate lyase AlyGC from G. chathamensis S18K6T (PDB ID: 5GKQ) with the sequence similarity of 39.81% as a template. Meanwhile, the prediction of the structure of the enzyme was performed with the program AlphaFold2 Colab [https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb]. The molecular docking of the AlyRm1 and alginate oligosaccharides (DP: 2–4) was performed using the Autodock-Vina (Trott and Olson, 2010). The ligands G4, G3, G2, M4, M3 and M2 were extracted from the AlyF-G4 complex (PDB: 6A40) (Lyu et al., 2019) and AlyGC-M4 complex (PDB: 5GKQ)(F. Xu et al., 2017) and converted to PDBQT format using by Autodock Tools (Morris et al., 2009). Analysis of the docking results and graphical presentations were performed in PyMOL (http://www.pymol.org)
3. Results and discussion
3.1. Sequence analysis of the alginate lyase gene
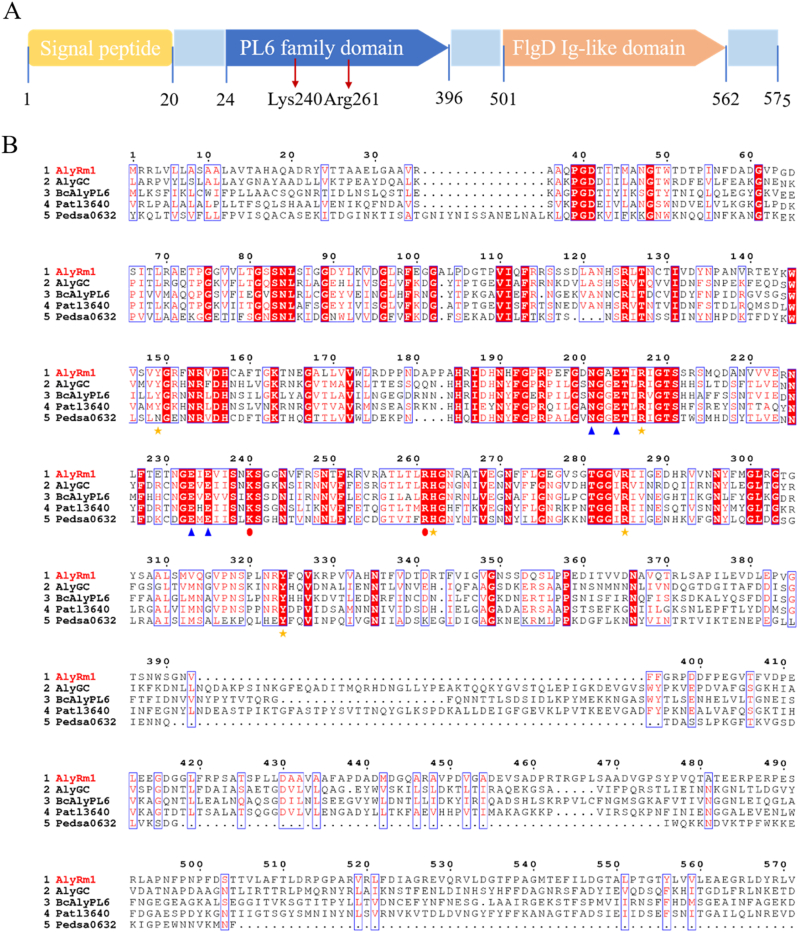
The alyrm1 gene from Rubrivirga marina is a 1728 bp open reading frame (ORF) encoding 575 amino acid residues, including a 20-amino acid signal peptide (Met1-Ala20) in the N-terminus. A conserved PL6 family domain (Val24-Gly396) suggests that AlyRm1 is a PL6 family alginate lyase. A C-terminal FlgD Ig-like domain (Phe501-Ala562) that is known to be required for hook assembly (Ohnishi et al., 1994), yet is uncommon in alginate lyase (Fig. 1A). The theoretical molecular weight (Mw) is about 61.78 kDa and the isoelectric point (pI) is 4.96. The instability index is 31.73, which classifies the protein as stable. The similarity of AlyRm1 compared to Patl3640 from Pseudoalteromonas atlantica T6c, Pedsa0632 from Pedobacter saltans (Mathieu et al., 2016), BcAlyPL6 from Bacteroides clarus(B. Wang et al., 2021), and AlyGC from G. chathamensis S18K6T (F. Xu et al., 2017) is 42.69%, 41.81%, 40.69% and 39.81%, respectively. Particularly, compared with AlgGC which is the first alginate lyase of the PL6 family whose structure was reported, those residues that are responsible for the enzymatic activity, Ca2+ coordination, and catalysis are quite conserved in AlyRm1, while residues are more variable at the C-terminal domain (Fig. 1B). Considerable PL6 enzymes have a multi-modular structure in which the PL6 family domain is coupled to additional domains, such as F5/8 type C domains, glycoside hydrolase domains, etc. BcAlyPL6, a recently discovered exolytic alginate lyase, has two domains: a PL6 family domain and an unknown C-terminal function domain. The PL6 family domain is responsible for substrate binding via the prolonged positively charged cleft, whilst the C-terminal domain is important for anchoring the substrate into the proper conformation (B. Wang et al., 2021). Little is known about a variety of other domains in the PL6 enzymes to date. This study will provide additional information about this item.
Fig. 1.
Sequence analysis of AlyRm1. (A). Conserved domain analysis (B). Sequence alignment of AlyRm1 with characterized PL6 enzymes. The residues in AlyGC whose enzymatic activity, Ca2+ coordination, and catalytic activity have been confirmed (F. Xu et al., 2017), are represented by orange stars, blue triangles, and red dots, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Biochemical characterization of AlyRm1
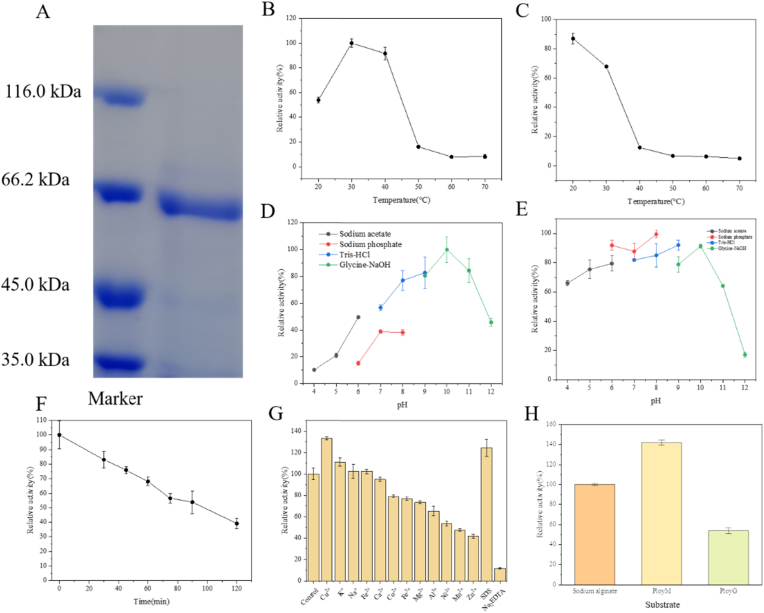
The full-length alyrm1 gene was successfully cloned and expressed in E. coli BL21(DE3). The theoretical molecular weight of AlyRm1 is 61.78 kDa, which corresponded well to the band at approximately 66.2 kDa on the SDS-PAGE (Fig. 2A). For the optimum temperature test, the results showed that AlyRm1 performed the highest activity at 30 °C (Fig. 2B). After incubation at 40 °C for 60 min, AlyRm1 lost about 80% of the enzyme activity but it preserved approximately 90% and 70% of its activity at 20 °C and 30 °C, respectively (Fig. 2C). The half-life time of AlyRm1 at 30 °C was 90 min (Fig. 2F). The optimal pH of AlyRm1 was found to be pH 10.0 (Fig. 2D). Furthermore, it was shown that more than 50% of the enzyme activity retained at pH 4.0–11.0, and over 80% of the enzyme activity retained after incubating at pH 10.0 for 24 h (Fig. 2E). As shown in Table 1, the majority of alginate lyases belonging to the PL6 family exhibited the maximum enzymatic activity at 30–50 °C and pH 7.0–9.0. AlyPL6, OUC-ScCD6, FsAlyPL6 and Alg823 all exhibited optimal activity at or above 45 °C, whereas the optimal temperature of OalS6, AlyMG, OalC6, AlyGC, AlyF and BcelPL6 was 30 or 40 °C. The majority of enzymes exhibited good thermal stability between 30 °C and 40 °C, for instance, AlyPL6 and FsAlyPL6 retained over 80% of the enzymatic activity after 1 h of incubation at 35 °C, and OUC-ScCD6 remained over 80% of the enzymatic activity after 24 h of incubation at 30 °C. Compared with other PL6 family alginate lyases, AlyRm1 showed the optimal activity at 30 °C and remained at about 90% of its activity after indicated at 20 °C for 1 h, indicating that AlyRm1 is stable at low temperature. Regarding pH, the optimal pH of AlyPL6, OUC-ScCD6, and FsAlyPL6 was 9.0 or 10.0. Nevertheless, their enzymatic activity drastically diminished when the pH of the reaction system fell below 7.0. AlyRm1 also performed the highest enzymatic activity at pH 10.0, and was stable within pH 4.0–11.0, indicating that AlyRm1 shows broad pH stability as an alkali-resistant alginate lyase. Alkali treatment usually is used to pretreat alginate for improving the saccharification biomass in enzymatic reaction (Lu et al., 2022). Thus, AlyRm1 with extensive pH performance and alkali resistance could be suitable for industrial application. In addition, the effects of metal ions, SDS and EDTA on AlyRm1 were investigated. As shown in Fig. 2G, Cu2+ and K+ enhanced the activity of AlyRm1, while some kinds of metal ions inhibited the enzyme activity, such as Co2+, Fe3+, Mg2+, Al3+, Ni2+, Mn2+ and Zn2+. 1 mM SDS significantly promoted the enzyme activity, while 1 mM metal chelating agent EDTA almost inactivated the enzyme, suggesting that metal ions have a great influence on the activity of AlyRm1.
Fig. 2.
(A) SDS-PAGE of purified AlyRm1. (B) Relative activities of AlyRm1 at different temperatures. (C) The thermal stability of AlyRm1. (D) Relative activities of AlyRm1 under different pHs. (E) The pH stability of AlyRm1. (F) Inactivation of AlyRm1 at 30 °C (G) The effects of metal ions, EDTA and SDS on AlyRm1. (H) Substrate specificity of AlyRm1 towards sodium alginate, PolyM and PolyG.
Table 1.
Characteristics of PL-6 alginate lyases.
| Alginate lyase | Source | Temperaturea | pHb | Metal ionsc | Action mode | Substrate Specificity | Degradation products | Reference |
|---|---|---|---|---|---|---|---|---|
| AlyPL6 | Pedobacter hainanensis NJ-02 | 45 °C, 35 °C (1 h) | 10.0, 9.0–10.0 (12 h) | Na+ (1 mM) | Endo- | Sodium alginate > polyG > polyM | 2–6 | (Q. Li et al., 2020) |
| OUC-ScCD6 | Streptomyces ecolicolor A3(2) | 50 °C, 30 °C (1 h) | 9.0, 7.0–10.0 (4 d) | Mn2+, Fe3+, Zn2+, Ba2+, Co2+ (1 mM) | Endo- | PolyM > sodium alginate > polyG | 2–4 | Cheng et al. (2020) |
| FsAlyPL6 | Flammeovirga sp. NJ-04 | 45 °C, 35 °C (1 h) | 9.0, 9.0–10.0 (20 h) | Na+, Ca2+ and Mg2+ (1 mM) | Endo- | Sodium alginate > polyM > polyG | 1–5 | (Q. Li et al., 2019) |
| BcelPL6 | Bacteroides cellulosilyticus CRE21 | 30 °C, N.D. d | 7.5, N.D. | N.D. | Endo- | Sodium alginate > polyM, polyG not detected | 2 | Stender et al. (2019) |
| AlyF | Vibrio splendidus OU02 | 30 °C, N.D. | 7.5, N.D. | Na+ (0.2 M) | Endo- | PolyG > sodium alginate, polyM not detected | 3 | Lyu et al. (2019) |
| AlyGC | Glaciecola chathamensis S18K6T | 30 °C, N.D. | 7.0, N.D. | N.D. | Exo- | PolyG > polyM > sodium alginate | 1 | (F. Xu et al., 2017) |
| OalS6 | Shewanella sp.Kz7 | 40 °C, 0–40 °C (1 h) | 7.2, 6.0–8.0 (24 h) | Na+ (1–300 Mm), K+(1–100 mM) | Exo- | PolyG > sodium alginate > polyM | 1 | (S. Li et al., 2015) |
| AlyMG | Stenotrophomas maltophilia KJ-2 | 40 °C, N.D. | 8.0, N.D. | Na+, K+ (1–100 Mm) and Ca2+ (1 mM) | Endo- | PolyMG > alginate > polyM > polyG | 2–6 | In Lee, Choi, Lee, & Kim (2012) |
| OalC6 | Cellulophaga sp.SY116 | 40 °C, 0–30 °C (1 h) | 6.6, 6.0–8.0 (6 h) | Na+ (1–500 Mm), K+ and Ca2+ (1 mM) | Exo- | PolyG > sodium alginate > polyM | 1 | (S. Li et al., 2018) |
| ALY | Pseudomonas sp.OS-ALG-9 | N.D. | N.D. | N.D. | Endo- | PolyM > polyG | N.D. | Kraiwattanapong et al. (1997) |
| Alg823 | Pseudoalteromonas carrageenovora ASY5 | 55 °C, 35–45 °C (0.5 h) | 8.0, 6.0–10.0 (24 h) | Ca2+, Mg2+, K+ and Na+ (1 mM) | Endo- | PolyM > polyG > sodium alginate | 2–3 | Zeng et al. (2019) |
| AlyRm1 | Rubrivirga marina | 30 °C, 20 °C (1 h) | 10.0, 6.0–10.0 (24 h) | Cu2+ and K+ (1 mM) | Endo- | PolyM > sodium alginate > polyG | 1–5 | This study |
The optimal temperature, the range of temperature within which the residual activity of alginate lyase was over 80% after incubated for a certain time (the incubation time).
The optimal pH, the range of pH within which the residual activity of alginate lyase was over 80% after incubated for a certain time (the incubation time).
Those metal ions increased the enzyme activity under a certain concentration (the concentration).
Not determined.
The enzyme activity of AlyRm1 was measured toward PolyM, PolyG and sodium alginate. As shown in Fig. 2H, the relative activities of AlyRm1 towards PolyM and PolyG were 141.9% and 54.0% of that for sodium alginate. Moreover, when PolyM was used as a substrate, the Km, Vmax and Kcat values were 8.1 mg/mL, 13.15 U/mg and 13.54 s−1, respectively. In contrast, the Km, Vmax and Kcat values for PolyG were 14.5 mg/mL, 9.81 U/mg and 10.10 s−1, respectively. The Km, Vmax and Kcat values for sodium alginate were 12.6 mg/mL, 10.08 U/mg and 10.38 s−1, respectively. As shown in Table 1, most PL6 alginate lyases were specific for PolyMG or PolyG, such as OalS6, AlyMG and OalC6, certain enzymes, such as AlyF, AlyGC, Patl3640, and Pedsa0640 (Violot et al., 2021) had no activity towards PolyM. In addition, BcelPL6 is a PolyM-specific lyase with negligible PolyMG activity and no PolyG activity (Stender et al., 2019). There are few PL6 alginate lyases possess a broad substrate specificity. The Vmax and Kcat values of AlyRm1 for PolyM were greater than those for PolyG and sodium alginate, indicating that its affinity for PolyM is stronger than that for PolyG. Meanwhile, AlyRm1 could also degrade sodium alginate and PolyG into AOSs, demonstrating that AlyRm1 has a broad substrate specificity, making it appropriate for industrial applications. The reducing sugar yields after 24 h were 32.6%, 23.6% and 23.8% for sodium alginate, PolyM and PolyG, respectively.
3.3. Reaction mode of AlyRm1
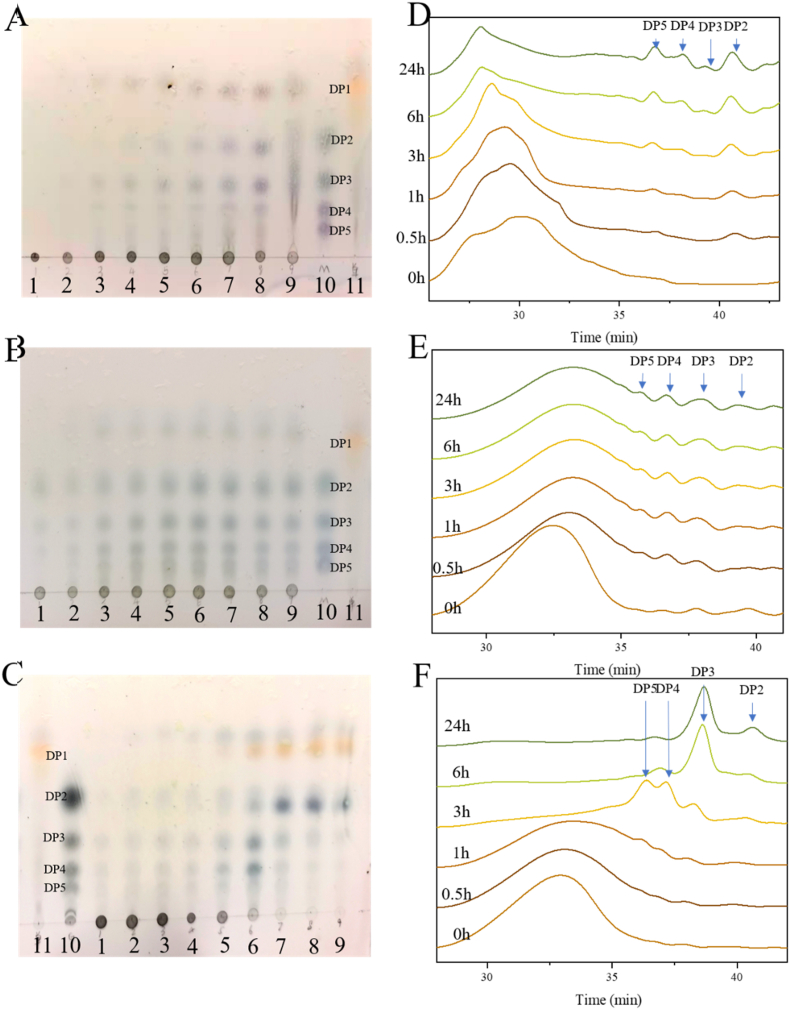
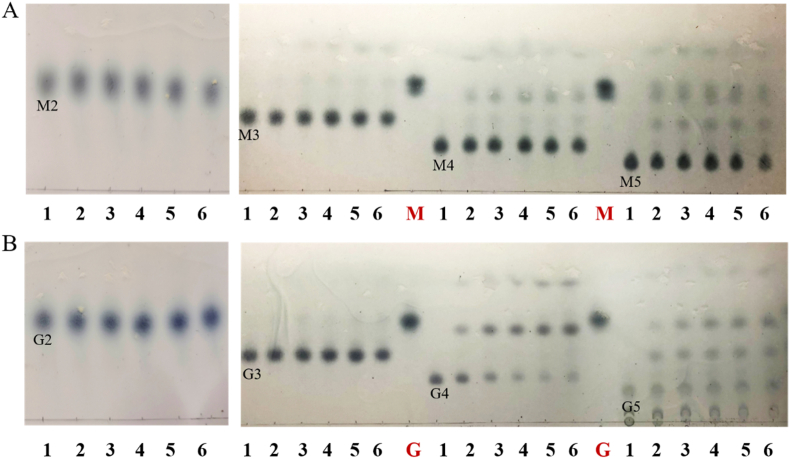
To obtain the degradation mode of AlyRm1, TLC analysis was firstly performed on the degradation products of three distinct substrates at various time points. With increasing incubation time, the length of the oligomer was seen to decrease from a relatively lengthy oligomer at the beginning of the reaction to a progressively shorter oligomer. (Fig. 3A, B, C), suggesting that AlyRm1 possesses endolytic activity. The primary products are oligosaccharides of DP 2–5, and monosaccharides were formed with the increase of incubation time. The degradation pattern was also confirmed by HPLC. It is noteworthy that DP3 was the main degradation product after a 24 h reaction when PolyG was used as the substrate (Fig. 3F). However, for PolyM and sodium alginate, the alginate oligosaccharides of DP 2–5 were uniformly distributed (Fig. 3D and E). Using oligosaccharides as substrates, we further investigated the reaction mode of AlyRm1. It demonstrated that neither M2 nor G2 can be degradated (Fig. 4). In addition, AlyRm1 could not degrade M3 and G3 efficiently, trisaccharide was still dominant after 24 h incubation for both. For M4 and G4, their main degradation products were disaccharides in the early stage, and the amount of mono- and trisaccharide increased after longer incubation times, indicating that AlyRm1 tends to degrade tetrasaccharide into disaccharides, followed by tetrasaccharide into mono- and trisaccharide. For M5 and G5, di- and trisaccharide were the main component in the early stage, and mono- and tetrasaccharide accumulated continuously, indicating that AlyRm1 tends to degrade pentasaccharide into di- and trisaccharide, followed by pentasaccharide into mono- and tetrasaccharide (Fig. 4). For alginate lyases in the PL6 family, most of them are endolytic alginate lyases, for instance, AlyPL6, OUC-SCCD6, BcelPL6 and AlyF which produce alginate oligosaccharides of DP 2–6, 2–4, 2 and 3 in the endolytic mode, respectively, whereas only a few are exolytic alginate lyases, such as AlyGC, OalS6 and OalC6.
Fig. 3.
Degradation products analysis of AlyRm1 by TLC and HPLC. (A, B, C) TLC analysis of AlyRm1 degradation products towards sodium alginate, PolyM, and PolyG. Lane 1–9: degradation products at 0 min, 5 min, 15 min, 30 min, 1 h, 3 h, 6 h, 12 h, 24 h. Lane 10: AOSs standards. Lane 11: Monosaccharide obtained by degrading sodium alginate using TcAlg1(D. Wang et al., 2018). (D, E, F) HPLC analysis of the degradation products of AlyRm1 towards sodium alginate, PolyM, and PolyG.
Fig. 4.
TLC analysis of the degradation products of AlyRm1 towards oligosaccharides. (A) β-D-mannuronic acid oligosaccharides (DP: 2–5) as substrates. (B) α-L-guluronic acid oligosaccharides (DP: 2–5) as substrates. Lane 1–6: degradation products of substrates for 0 h, 1 h, 3 h, 6 h, 12 h, 24 h. Lane M: dimannuronic acid. Lane G: diguluronic acid.
3.4. Molecular modeling and docking analysis of AlyRm1
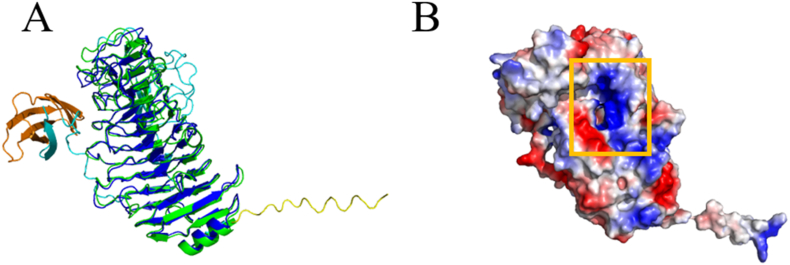
Using the SWISS-MODEL online workspace, we predicted the homology structure of AlyRm1. The GMQE and QMEAN values were 0.58 and −1.73, indicating that the homology structure of AlyRm1 is credible because GMQE and QMEAN scores should be 0–1 (Waterhouse et al., 2018) and −4–0 (Benkert et al., 2010), respectively. In addition, AlphaFold is a novel machine learning method developed by DeepMind, which has proved very successful in predicting the structure of proteins according to their amino acid sequences(Jumper et al., 2021). AlphaFold 2 was also used to predict the structure of AlyRm1. There existed a substrate-binding active cleft (Fig. 5B). The predicted structure of AlyRm1 using AlphaFold 2 and the homologous model of AlyRm1 were similar fairly with a root-mean-square deviation (RMSD) of 0.70 Å (Fig. 5A).
Fig. 5.
The overall structure of AlyRm1 (A) Overlap of the homology model structure of AlyRm1 which was obtained using the SWISS-MODEL (green) with the structure of AlyRm1 which was predicted using Alphafold 2. The color of a signal peptide fragment, PL6 family domain, and FlgD Ig-like domain are yellow, blue, and orange, respectively. The linkage between the PL6 family domain and FlgD Ig-like domain is in cyan. (B) The electrostatic surface view of AlyRm1 was predicted using Alphafold 2. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
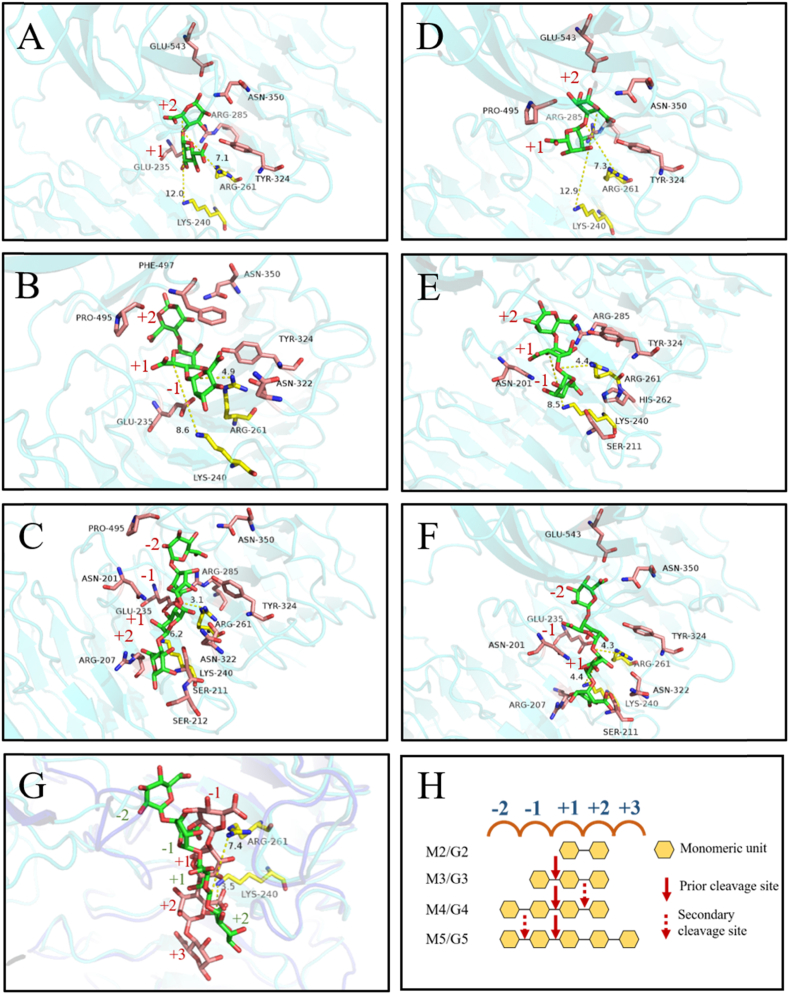
In the study, the structure of protein obtained using AlphaFold 2 was used for molecular docking analysis. The results of molecular docking with the lowest binding energy were selected for analysis. - n and + n represent the non-reducing ends and the reducing ends, respectively (Davies et al., 1997). Combining with the result that the minimum substrate of AlyRm1 is trisaccharide, the key residues for substrate binding were determined by the molecular docking analysis of AlyRm1 and M3/G3. In the AlyRm1-M3 complex, Glu235, Asn322, Tyr324, Asn350, Pro495, and Phe497 can form a hydrogen bond with the M3 molecule (Fig. 6B). For the AlyRm1-G3 complex, Asn201, Ser211, His262, Arg285, Tyr324 can form a hydrogen bond with the G3 molecule (Fig. 6E). In the AlyRm1-M4 complex, Asn201, Arg207, Ser211, Ser212, Glu235, Arg285, Asn322, Tyr324, Asn350 and Pro495 can form a hydrogen bond with the M4 molecule (Fig. 6C). For the AlyRm1-G4 complex, Asn201, Arg207, Ser211, Glu235, Asn322, Tyr324, Asn350 and Glu543 can form a hydrogen bond with the G4 molecule (Fig. 6F). Among them, Arg207, His262, Arg285 and Tyr324 are the key residues of the enzymatic activity in the corresponding position of the AlyGC, in addition, Asn201 and Glu235 are the key residues of Ca2+ coordination (F. Xu et al., 2017). BcAlyPL6 is a two-domain alginate lyase that bound one calcium ion and one melonic acid in its catalytic cleft. The calcium ion is chelated by Asn182, Glu185, Glu214 and Glu216 (corresponding to Asn201, Glu204, Glu233 and Glu235 in AlyRm1) and two water molecules. One melonic acid is mainly bound by Arg242, Arg266 and Tyr305(corresponding to Arg261, Arg285 and Tyr324 in AlyRm1) via hydrogen bonds or electrostatic interactions at the subsite −1. Asp617 and Ser619 of the C-terminal domain are involved in the formation of catalytic pockets and can interact with sugar residues at the subsite −1, suggesting that the C-terminal domain of BcAlyPL6 plays an important role in the formation of the right conformation for the substrate (B. Wang et al., 2021). For AlyRm1, Pro495 and Phe497 are located in the linkage between the PL6 family domain and the FlgD Ig-like domain, and Glu543 is located in the FlgD Ig-like domain, indicating that the FlgD Ig-like domain is crucial to the interaction of AlyRm1 with substrate. In addition, in the model of AlyRm1 complexed with M4, the distance between M4 and the catalytic residues Lys240 and Arg261 is shorter than the distance between M2 or M3 and the same residues (Fig. 6A, B, C). The same phenomenon occurs in α-L-guluronic acid oligosaccharides (Fig. 6D, E, F), validating the conclusion that AlyRm1 cannot degrade M2/G2, and cannot degrade M3/G3 effectively. Compared with the AlyGC-M4 complex, AlyGC is an exolytic alginate lyase that cleaves the subsites between – 1 and + 1 of the M4 substrate to liberate mono- and trisaccharide (F. Xu et al., 2017). M4 in the AlyRm1-M4 complex moves toward the non-reducing end, thus, AlyRm1 releases mono-, di-, and trisaccharide by cleaving the subsites between – 1 and + 2 on the M4 substrate. However, catalytic residues Lys240 and Arg261 are closer to the subsites between −1 and +1 than to the subsites between +1 and + 2 (Fig. 6C, G), indicating AlyRm1 preferentially cleaves the subsites between −1 and +1 to release disaccharide from the M4 substrate. In combination with the result of the analysis of degradation products towards oligosaccharides, we obtained the putative degradation pattern of AlyRm1 towards alginate oligosaccharides (DP: 2–5) (Fig. 6H), which provided greater insight of the action mode and substrate recognition of PL6 family alginate lyases.
Fig. 6.
Molecular docking analysis of AlyRm1. The ligands in Figures A, B, C, D, E, and F were M2, M3, M4, G2, G3, and G4, respectively. Lys240 and Arg261 labelled yellow are the catalytic base and acid of the reaction, respectively. (G) Comparison of the M4 position in the AlyGC-M4 complex and the AlyRm1-M4 complex. The M4 are shaped in red and green in the AlyGC-M4 and AlyRm1-M4 complex, respectively. (H) Putative degradation pattern of AlyRm1 towards alginate oligosaccharides (DP: 2–5). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Conclusion
In conclusion, a new PL6 family alginate lyase AlyRm1 from Rubrivirga marina was cloned, purified, and characterized. AlyRm1 shows good alkaline stability, for it retained over 80% of the enzyme activity after being incubated at pH 10.0 for 24 h. In addition, AlyRm1 preferred to degrade PolyM into AOSs (DP: 2–5) and monosaccharides, suggesting that AlyRm1 possesses endolytic activity, yet it could digest PolyG and sodium alginate. More importantly, the interaction of substrate binding was obtained by the molecular docking analysis. It was shown that Asn201, Arg207, Ser211, Ser212, Glu235, His262, Arg285, Asn322, Tyr324, Asn350, Pro495, Phe497 and Glu543 are crucial substrate binding residues. Combining with the result of the analysis of degradation products towards oligosaccharides, we obtained the putative degradation pattern of AlyRm1 towards alginate oligosaccharides (DP: 2–5) (Fig. 6H) These unique properties enabled a more demanding application of AlyRm1 in the functional food and biopharmaceutical industries for the production of AOSs.
CRediT authorship contribution statement
Yuting Zheng: Writing – original draft, Methodology, Formal analysis. Yujie Wang: Methodology, Formal analysis. Meiling Dan: Methodology, Formal analysis. Yanping Li: Methodology, Formal analysis. Guohua Zhao: Conceptualization, Supervision. Damao Wang: Writing – review & editing, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by Chongqing Science and Technology Bureau (cstc2020jcyj-msxmX0463), Guangxi Key Laboratory of Agricultural Resources Chemistry and Biotechnology Open Fund (2021KF04), “the Fundamental Research Funds for the Central Universities, Southwest University” (No. SWU019034), Chongqing Key Laboratory of Speciality Food Co-Built by Sichuan and Chongqing, Postgraduate mentor team-building program of Southwest University (XYDS201905) .
Data availability
No data was used for the research described in the article.
References
- Bailey M.J. A note on the use of dinitrosalicylic acid for determining the products of enzymatic reactions. Appl. Microbiol. Biotechnol. 1988;29(5):494–496. [Google Scholar]
- Barzkar N., Sheng R., Sohail M., Jahromi S.T., Babich O., Sukhikh S., Nahavandi R. Alginate lyases from marine bacteria: an enzyme ocean for sustainable future. Molecules. 2022;27(11):3375. doi: 10.3390/molecules27113375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P., Biasini M., Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2010;27(3):343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D., Liu Z., Jiang C., Li L., Xue C., Mao X. Biochemical characterization and degradation pattern analysis of a novel PL-6 alginate lyase from Streptomyces coelicolor A3(2) Food Chem. 2020;323 doi: 10.1016/j.foodchem.2020.126852. [DOI] [PubMed] [Google Scholar]
- Davies G.J., Wilson K.S., Henrissat B. Nomenclature for sugar-binding subsites in glycosyl hydrolases. Biochem. J. 1997;321(Pt 2):557–559. doi: 10.1042/bj3210557. Pt 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- In Lee S., Choi S.H., Lee E.Y., Kim H.S. Molecular cloning, purification, and characterization of a novel polyMG-specific alginate lyase responsible for alginate MG block degradation in Stenotrophomas maltophilia KJ-2. Appl. Microbiol. Biotechnol. 2012;95(6):1643–1653. doi: 10.1007/s00253-012-4266-y. [DOI] [PubMed] [Google Scholar]
- Jouanneau D., Klau L.J., Larocque R., Jaffrennou A., Duval G., Le Duff N., Roret T., Jeudy A., Aachmann F.L., Czjzek M., Thomas F. Structure–function analysis of a new PL17 oligoalginate lyase from the marine bacterium Zobellia galactanivorans DsijT. Glycobiology. 2021;31(10):1364–1377. doi: 10.1093/glycob/cwab058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A., Bridgland A., Meyer C., Kohl S.A.A., Ballard A.J., Cowie A., Romera-Paredes B., Nikolov S., Jain R., Adler J., Back T., Petersen S., Reiman D., Clancy E., Zielinski M., Steinegger M., Pacholska M., Berghammer T., Bodenstein S., Silver D., Vinyals O., Senior A.W., Kavukcuoglu K., Kohli P., Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.S., Lee C.-G., Lee E.Y. Alginate lyase: structure, property, and application. Biotechnol. Bioproc. Eng. 2011;16(5):843. [Google Scholar]
- Kraiwattanapong J., Ooi T., Kinoshita S. Cloning and sequence analysis of the gene (alyII) coding for an alginate lyase of Pseudomonas sp. OS-ALG-9. Biosci., Biotechnol., Biochem. 1997;61(11):1853–1857. doi: 10.1271/bbb.61.1853. [DOI] [PubMed] [Google Scholar]
- Lari Z., Ahmadzadeh H., Hosseini M. In: Advances in Feedstock Conversion Technologies for Alternative Fuels and Bioproducts. Hosseini M., editor. Woodhead Publishing; 2019. Chapter 2 - cell wall disruption: a critical upstream process for biofuel production; pp. 21–35. [Google Scholar]
- Li Q., Hu F., Wang M., Zhu B., Ni F., Yao Z. Elucidation of degradation pattern and immobilization of a novel alginate lyase for preparation of alginate oligosaccharides. Int. J. Biol. Macromol. 2020;146:579–587. doi: 10.1016/j.ijbiomac.2019.12.238. [DOI] [PubMed] [Google Scholar]
- Li Q., Hu F., Zhu B., Sun Y., Yao Z. Biochemical characterization and elucidation of action pattern of a novel polysaccharide lyase 6 family alginate lyase from marine bacterium flammeovirga sp. NJ-04. Mar. Drugs. 2019;17(6) doi: 10.3390/md17060323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Wang L., Chen X., Zhao W., Sun M., Han Y. Cloning, expression, and biochemical characterization of two new oligoalginate lyases with synergistic degradation capability. Mar. Biotechnol. 2018;20(1):75–86. doi: 10.1007/s10126-017-9788-y. [DOI] [PubMed] [Google Scholar]
- Li S., Wang L., Han F., Gong Q., Yu W. Cloning and characterization of the first polysaccharide lyase family 6 oligoalginate lyase from marine Shewanella sp. Kz7. J. Biochem. 2015;159(1):77–86. doi: 10.1093/jb/mvv076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zheng Y., Zhang Y., Yang Y., Wang P., Imre B., Wong A.C.Y., Hsieh Y.S.Y., Wang D. Brown algae carbohydrates: structures, pharmaceutical properties, and research challenges. Mar. Drugs. 2021;19(11) doi: 10.3390/md19110620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yang S., Li X., Yan Q., Reaney M.J.T., Jiang Z. Alginate oligosaccharides: production, biological activities, and potential applications. Compr. Rev. Food Sci. Food Saf. 2019;18(6):1859–1881. doi: 10.1111/1541-4337.12494. [DOI] [PubMed] [Google Scholar]
- Liu M., Liu L., Zhang H.-f., Yi B., Everaert N. Alginate oligosaccharides preparation, biological activities and their application in livestock and poultry. J. Integr. Agric. 2021;20(1):24–34. [Google Scholar]
- Lu Y., Zhou J., Gu Q., Yang W., Yang L., Yu X. Cloning, expression and improvement of catalytic activity of alginate lyase by site-directed mutation. Syst. Microbiol. Biomanufact. 2022;2(3):555–567. [Google Scholar]
- Lyu Q., Zhang K., Shi Y., Li W., Diao X., Liu W. Structural insights into a novel Ca(2+)-independent PL-6 alginate lyase from Vibrio OU02 identify the possible subsites responsible for product distribution. Biochim. Biophys. Acta Gen. Subj. 2019;1863(7):1167–1176. doi: 10.1016/j.bbagen.2019.04.013. [DOI] [PubMed] [Google Scholar]
- Mathieu S., Henrissat B., Labre F., Skjak-Braek G., Helbert W. Functional exploration of the polysaccharide lyase family PL6. PLoS One. 2016;11(7) doi: 10.1371/journal.pone.0159415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K., Ohto Y., Aizawa S., Macnab R.M., Iino T. FlgD is a scaffolding protein needed for flagellar hook assembly in Salmonella typhimurium. J. Bacteriol. 1994;176(8):2272–2281. doi: 10.1128/jb.176.8.2272-2281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Song J., Yoshizawa S., Choi A., Cho J.-C., Kogure K. Rubrivirga marina gen. nov., sp. nov., a member of the family Rhodothermaceae isolated from deep seawater. Int. J. Syst. Evol. Microbiol. 2013;63(Pt_6):2229–2233. doi: 10.1099/ijs.0.046318-0. [DOI] [PubMed] [Google Scholar]
- Rioux L.-E., Turgeon S.L. In: Seaweed Sustainability. Tiwari B.K., Troy D.J., editors. Academic Press; San Diego: 2015. Chapter 7 - seaweed carbohydrates; pp. 141–192. [Google Scholar]
- Stender E.G.P., Dybdahl Andersen C., Fredslund F., Holck J., Solberg A., Teze D., Peters G.H.J., Christensen B.E., Aachmann F.L., Welner D.H., Svensson B. Structural and functional aspects of mannuronic acid-specific PL6 alginate lyase from the human gut microbe Bacteroides cellulosilyticus. J. Biol. Chem. 2019;294(47):17915–17930. doi: 10.1074/jbc.RA119.010206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusi S.K., Khalaj L., Ashabi G., Kiaei M., Khodagholi F. Alginate oligosaccharide protects against endoplasmic reticulum- and mitochondrial-mediated apoptotic cell death and oxidative stress. Biomaterials. 2011;32(23):5438–5458. doi: 10.1016/j.biomaterials.2011.04.024. [DOI] [PubMed] [Google Scholar]
- Violot S., Galisson F., Carrique L., Jugnarain V., Conchou L., Robert X., Thureau A., Helbert W., Aghajari N., Ballut L. Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity. Glycobiology. 2021;31(11):1557–1570. doi: 10.1093/glycob/cwab073. [DOI] [PubMed] [Google Scholar]
- Wang B., Dong S., Li F.L., Ma X.Q. Structural basis for the exolytic activity of polysaccharide lyase family 6 alginate lyase BcAlyPL6 from human gut microbe Bacteroides clarus. Biochem. Biophys. Res. Commun. 2021;547:111–117. doi: 10.1016/j.bbrc.2021.02.040. [DOI] [PubMed] [Google Scholar]
- Wang D., Aarstad O.A., Li J., McKee L.S., Saetrom G.I., Vyas A., Srivastava V., Aachmann F.L., Bulone V., Hsieh Y.S. Preparation of 4-Deoxy-L-erythro-5-hexoseulose uronic acid (DEH) and guluronic acid rich alginate using a unique exo-alginate lyase from thalassotalea crassostreae. J. Agric. Food Chem. 2018;66(6):1435–1443. doi: 10.1021/acs.jafc.7b05751. [DOI] [PubMed] [Google Scholar]
- Wang D.M., Kim H.T., Yun E.J., Kim D.H., Park Y.-C., Woo H.C., Kim K.H. Optimal production of 4-deoxy-l-erythro-5-hexoseulose uronic acid from alginate for brown macro algae saccharification by combining endo- and exo-type alginate lyases. Bioproc. Biosyst. Eng. 2014;37(10):2105–2111. doi: 10.1007/s00449-014-1188-3. [DOI] [PubMed] [Google Scholar]
- Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Chen X.-L., Sun X.-H., Dong F., Li C.-Y., Li P.-Y., Ding H., Chen Y., Zhang Y.-Z., Wang P. Structural and molecular basis for the substrate positioning mechanism of a new PL7 subfamily alginate lyase from the arctic. J. Biol. Chem. 2020;295(48):16380–16392. doi: 10.1074/jbc.RA120.015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Dong F., Wang P., Cao H.-Y., Li C.-Y., Li P.-Y., Pang X.-H., Zhang Y.-Z., Chen X.-L. Novel molecular insights into the catalytic mechanism of marine bacterial alginate lyase AlyGC from polysaccharide lyase family 6. J. Biol. Chem. 2017;292(11):4457–4468. doi: 10.1074/jbc.M116.766030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Wu X., Wang Q., Cai N., Zhang H., Jiang Z., Wan M., Oda T. Immunomodulatory effects of alginate oligosaccharides on murine macrophage RAW264.7 cells and their structure–activity relationships. J. Agric. Food Chem. 2014;62(14):3168–3176. doi: 10.1021/jf405633n. [DOI] [PubMed] [Google Scholar]
- Zeng J., An D., Jiao C., Xiao Q., Weng H., Yang Q., Xiao A. Cloning, expression, and characterization of a new pH- and heat-stable alginate lyase from Pseudoalteromonas carrageenovora ASY5. J. Food Biochem. 2019;43(7) doi: 10.1111/jfbc.12886. [DOI] [PubMed] [Google Scholar]
- Zhu B., Ni F., Sun Y., Ning L., Yao Z. Elucidation of degrading pattern and substrate recognition of a novel bifunctional alginate lyase from Flammeovirga sp. NJ-04 and its use for preparation alginate oligosaccharides. Biotechnol. Biofuels. 2019;12:13. doi: 10.1186/s13068-019-1352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B., Ni F., Xiong Q., Yao Z. Marine oligosaccharides originated from seaweeds: source, preparation, structure, physiological activity and applications. Crit. Rev. Food Sci. Nutr. 2021;61(1):60–74. doi: 10.1080/10408398.2020.1716207. [DOI] [PubMed] [Google Scholar]
- Zhu B., Tan H., Qin Y., Xu Q., Du Y., Yin H. Characterization of a new endo-type alginate lyase from Vibrio sp. W13. Int. J. Biol. Macromol. 2015;75:330–337. doi: 10.1016/j.ijbiomac.2015.01.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.