Abstract
Nowadays, many researchers, farmers and companies focus on the development of an environmentally friendly approach for enhancing field vegetable production and protection. Using next-generation plant biostimulants (PBs) could be effective to enhance tolerance to abiotic and biotic stresses, vegetable crop quality or nutrient efficiency which is particularly important for vegetables with a short growing season, such as Pisum sativum. Two herbal drug-containing plant conditioners Elice16Indures® (supercritical carbon dioxide extract SC-CO2) and Fitokondi® (aqueous extract) developed in the RIMPH Ltd (Hungary) were used in pea field experiments to monitor the potential of enhancing crop quality and defense response against different stress factors. Fresh leaves were collected after treatments for QuantSeq 3’ mRNA sequencing at Illumina NextSeq 550 platform and libraries were investigated by genome-wide transcriptional profiling focusing on genes associated with defense response pathways. RNA quantification datasets are presented and 86 bp long sequence reads were pre-processed and assembled that were deposited in the National Center for Biotechnology Information (NCBI), Sequence Read Archive (SRA) and Transcriptome Shotgun Assembly (TSA) databases under the BioProject PRJNA870114. Functional annotation of transcripts and pairwise differential expression with enrichment analyses are presented here to support gene expression analysis experiments.
Keywords: Illumina sequencing, Whole-genome transcriptional profiling, Defense response, Nanotechnology, Plant conditioner, RNA dataset, Elice16Indures
Specifications Table
| Subject | Plant Science: Plant Physiology |
| Specific subject area | The effect of two plant biostimulants (PBs) Elice16Indures® and Fitokondi® were examined to stimulate the enhanced normalized difference vegetation index (ENDVI) and defense response processes of pea (Pisum sativum subsp. sativum) in the field. |
| Type of data | Table Database record Figure |
| How the data were acquired | Seeds of P. sativum subsp. sativum cultivar ‘Angela’ were sown in experimental plots in four replicates and treated with two types of PBs. During the cultivation period plants were treated three times at BBCH16, BBCH51 and BBCH67 stages with Elice16Indures 20 g ha−1 (‘E20’), Elice16Indures 240 g ha−1 (‘E240’) and as a positive control, Fitokondi (‘F’) 4 l ha−1 (the normal field dose) biostimulators. Leaf samples were collected after the last treatment at BBCH74 stage. Samples were sequenced by the Illumina NextSeq550 platform and next generation sequencing (NGS) libraries were prepared using 14.6-17.5M single-end reads. Using de novo assembly a combined transcript dataset was gained (total transcripts: 7,513; total genes: 6,897) that was examined by pairwise differential expression (DEG) and gene set enrichment analysis (GSEA). Functional annotation and Gene Ontology (GO) analyses were performed according to GO terms of molecular function, cellular component and biological process. |
| Data format | Raw Analyzed Filtered |
| Description of data collection | Leaves of untreated (control) and treated (Elice16Indures 20 g ha−1, Elice16Indures 240 g ha−1 and Fitokondi 4 l ha−1) pea plants were collected at BBCH74 stages and were stored in RNA Shield at -25°C until sequencing. Sequencing was performed by a third party, Xenovea Ltd, Szeged, Hungary. |
| Data source location | EduCoMat Ltd Keszthely Hungary |
| Data accessibility | The bio project and raw reads are available in National Center for Biotechnology Information (NCBI) database under the accessions: Repository name: Plant biostimulants treatments in Pisum sativum Data identification number: PRJNA870114 Direct link to datasets: https://www.ncbi.nlm.nih.gov/search/all/?term=PRJNA870114 Repository name: Dataset of conditioning effect of herbal extract-based plant biostimulants in pea (Pisum sativum) Data identification number (DOI): 10.17632/f93mjns9t6.2 Direct link to datasets: https://data.mendeley.com/datasets/f93mjns9t6/2 |
Value of the Data
-
•
Over the past 15 years, the research and development of PBs have progressed which could rise the sustainability of agricultural systems with efficient and environmentally friendly plant-growth-promoting methods. PBs can be used in organic vegetable cultivation. Investigating the bioactivity of herbal extracts as a plant conditioner can help to improve the quality of vegetables and the priming and triggering defense responses against stress. These data may supply information about the physiological effects of herbal extracts used as PBs.
-
•
Different herbal extraction technologies have been developed to obtain the optimal amounts of active ingredients from herbs. The effect of conventional aqueous extract was compared with the effect of supercritical carbon dioxide (SC-CO2) extract with liposomal formulation. The data from RNA-seq may provide information about the effect of different extraction and formulation methods of this kind of PBs that may enhance their uptake in field-cultivated plants.
-
•
The effect of treatments was examined in a large-scale genome-wide transcriptional profiling experiment in which the collected samples were treated with different PB doses and formulations. Illumina Gene Expression Profiling (GEx) sequencing data represents the molecular genetic background of the different physiological states of the plants as a response to the two investigated formulations and dosages.
1. Objective
Basically, there is a demand for the use of herbal drug-containing plant biostimulators during sustainable vegetable cultivation, so it is necessary to know how to apply them effectively. A revolutionary new formulation technique, the investigation of the effectiveness of the multilamellar nano-size liposome formulation provides new data on the changes in the physiological processes of the treated plants. The great value of the analysis is the whole-genome expression profiling, the published data are suitable for further comprehensive analysis.
2. Data Description
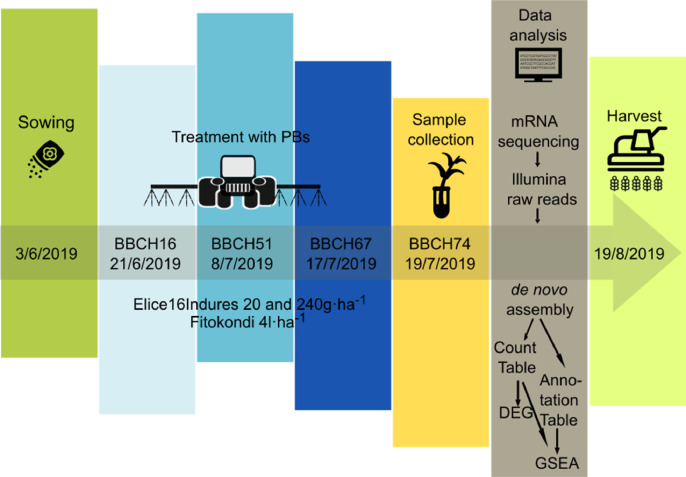
Natural PBs appear as a potential category of agricultural inputs to improve the quality of horticultural plants, as well as enhance tolerance to abiotic and biotic stresses [1,2]. To date, a few PBs have been used during the cultivation of pea (P. sativum L.) that contain marine algae extract [3], micro and macro elements [4], licorice root extract [5] and nitrophenols, nitroguaiaco [6]. Herbal drug-containing plant conditioning, general immunostimulant and yield-enhancing product Elice16Indures (https://gynki.hu/en/rimph-botanicals/products) and Fitokondi (https://www.fitokondi.hu/a-fitokondi) may have beneficial effects on plant growth and development and stress resistance. Therefore, low and high dosage treated pea plants were examined by whole-genome profiling to determine changes in the expression levels of genes involved in biochemical pathways (Fig. 1).
Fig. 1.
Timeline of PBs treatment, sample collection and data analysis process of plot pea experiment.
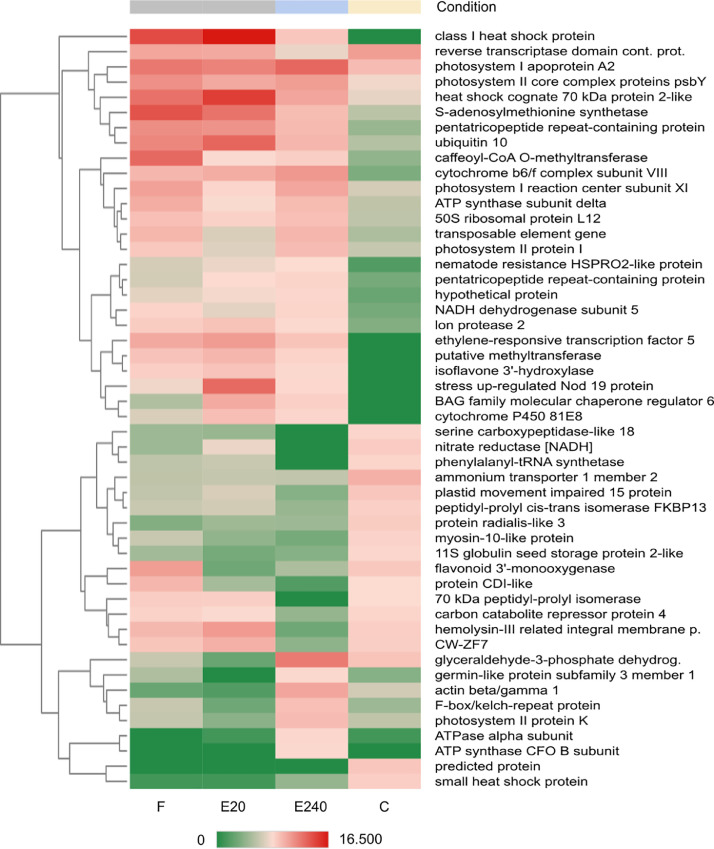
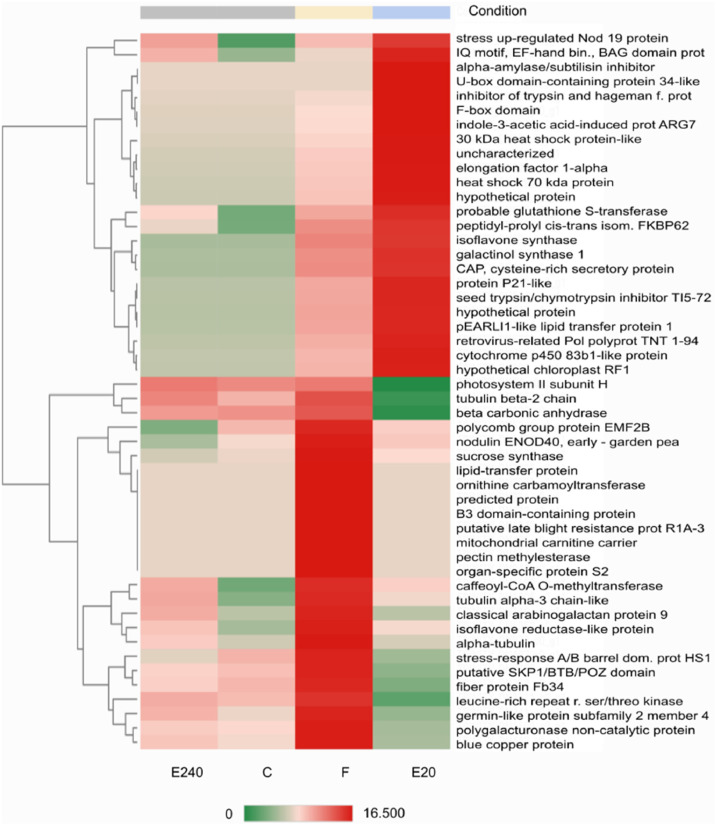
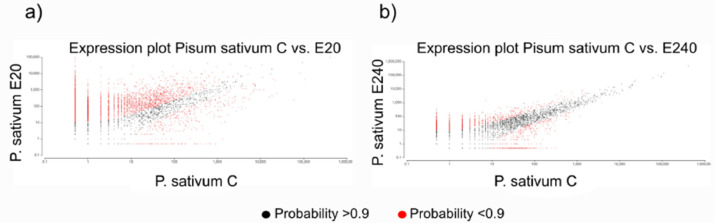
QuantSeq 3′ mRNA sequencing for RNA quantification [7] of samples was performed to determine the gene expression profiling of response after treatments with PBs. GEx library construction was applied for shallow RNA-sequencing and after pre-processing Illumina RNA-seq reads of NGS libraries were deposited in the NCBI Sequence Read Archive (SRA) under the Bioproject PRJNA870114 with accession numbers SRR21124993-SRR21124996. De novo assembly of sequence reads was generated by SRA datasets to reconstruct transcripts that were deposited in Transcriptome Shotgun Assembly (TSA) database at DBJ/EMBL/GenBank under the accession GKBF01000000. Output statistics of transcriptome is summarized in Table 1. Read counts, 6,897 transcripts have been recovered from the TSA fasta file that was summarized in the CountTable (Mendeley DOI: 10.17632/f93mjns9t6.2). Using CountTable DEGs were determined to compare untreated and treated samples (Fig. 2 and Table 2.) that were visualized by heatmap (Figs. 3-5). Moreover, the DEGs of treated samples were compared (Figs. 6-8). Functional annotation and GO analysis were carried out using AnnotationTable which was performed in Mendeley database (DOI: 10.17632/f93mjns9t6.2). GO annotation was specified according to GO terms: molecular function, cellular component and biological process. The GSEA was performed with statistically significant, concordant differences between treated and untreated samples (Figs. 9-11). The whole analysis was performed by using OmicsBox.BioBam (v2.0) https://www.biobam.com/omicsbox.
Fig. 4.
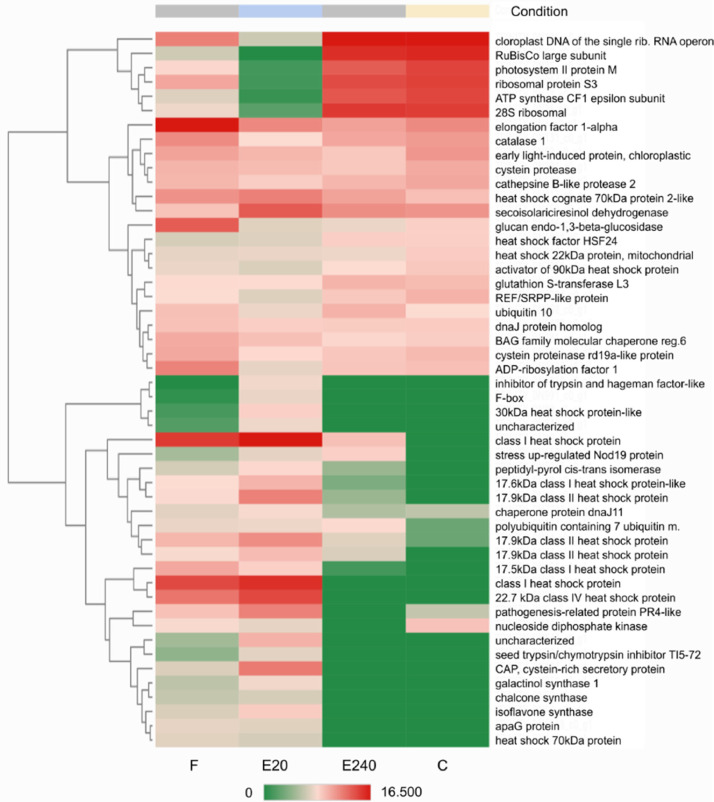
Heatmap of Top50 DEGs where untreated leaf (control ‘C’) as a reference and treated leaf by Elice16Indures 240 g ha−1 (‘E240’) as test condition was set. Samples of the treated leaf with Elice16Indures 20 g ha−1 (‘E20’) and Fitokondi 4 l ha−1 (‘F’) were shown. Annotation of transcript IDs was shown in AnnotationTable (DOI: 10.17632/f93mjns9t6.2).
Fig. 7.
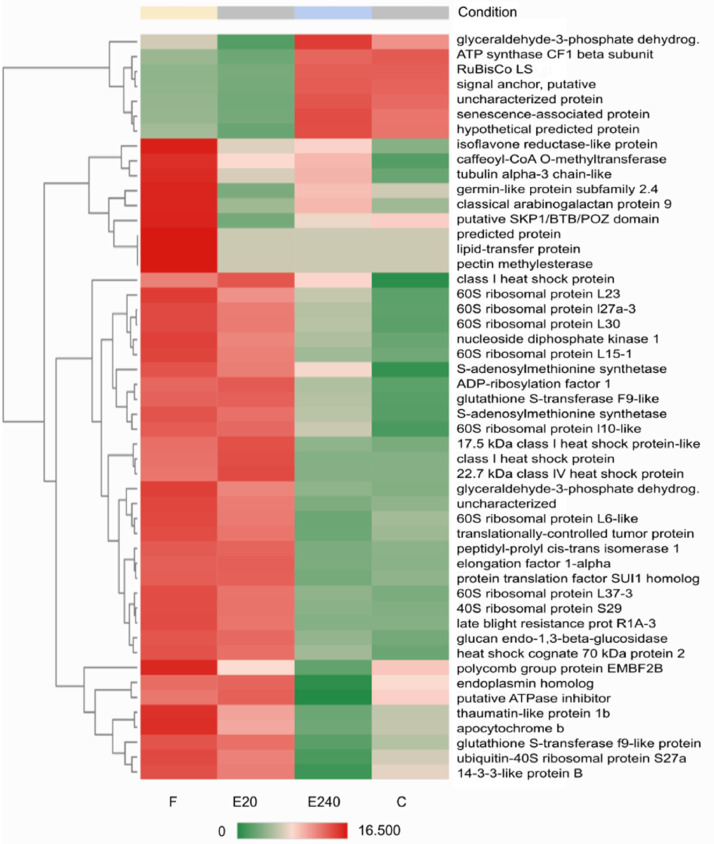
Heatmap of Top50 DEGs where treated leaf by Fitokondi 4 l ha-1 (‘F’) as a reference and treated leaf by Elice16Indures 20 g ha−1 (‘E20’) as test condition was set. Samples of the treated leaf with Elice16Indures 240 g ha−1 (‘E240’) and untreated leaf (control ‘C’) were shown. Annotation of transcript IDs was shown in AnnotationTable (DOI: 10.17632/f93mjns9t6.2).
Table 1.
RNA-Seq de novo assembly results of P. sativum samples.
| Total transcripts | 7,513 |
| Total genes | 6,897 |
| Percent GC | 35.09 |
| Total assembled bases (all transcripts) | 2,700,878 |
| Total assembled bases (longest isoform per gene) | 2,457,949 |
| Minimum length | 251 |
| Maximum length | 2,365 |
| Average length | 361.00 |
Fig. 2.
Expression plots of DEGs compared the Elice16Indures treated samples a) 'E20′ - dose 20 g ha−1 and b) 'E240′ – dose 240 g ha−1. The scatter plots show the average expression values of each condition. Differentially expressed features considering the probability threshold (0.9) are highlighted in red.
Table 2.
Statistical data of DEG in three comparisons of treated and non-treated samples. The abbreviations are as follows: ‘C’ control, untreated; ‘E20’ treated with Elice16Indures 20 g ha−1; ‘E240’ treated with Elice16Indures 240 g ha−1 and ‘F’ treated with Fitokondi 4 l ha−1.
| Pairwise comparison | Number of DE (Probability>0.9) | Up-regulated (M>0) | Down-regulated (M<0) |
|---|---|---|---|
| ‘E20’ vs. ‘C’ | 4,475 | 4,102 | 373 |
| ‘E240’ vs. ‘C’ | 1,478 | 970 | 508 |
| ‘F’ vs. ‘C’ | 4,585 | 4,353 | 232 |
Fig. 3.
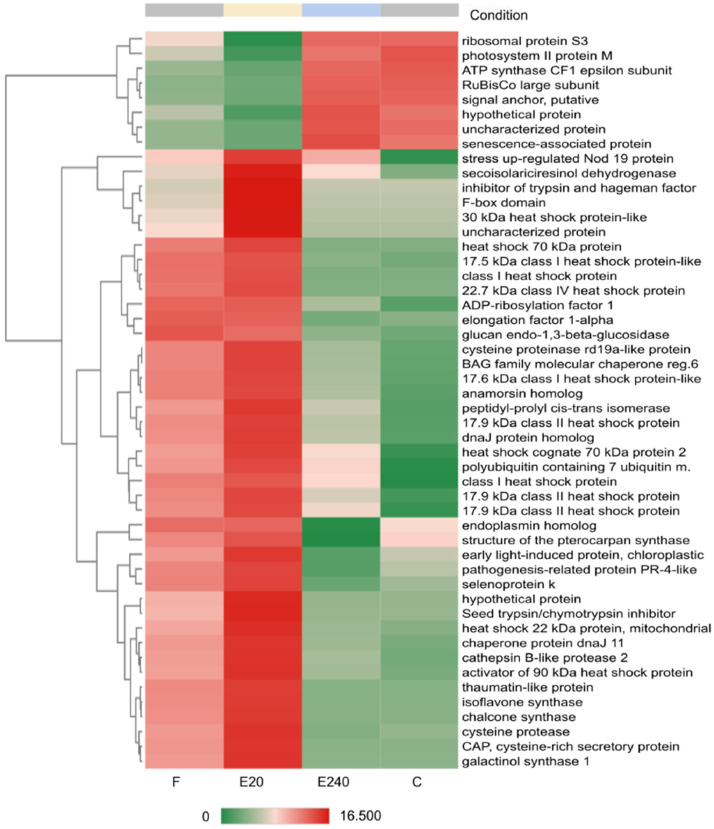
Heatmap of Top50 DEGs where untreated leaf (control ‘C’) as a reference and treated leaf by Elice16Indures 20 g ha−1 (‘E20’) as test condition was set. Samples of the treated leaf with Elice16Indures 240 g ha−1 (‘E240’) and Fitokondi 4 l ha−1 (‘F’) were shown. Annotation of transcript IDs was shown in AnnotationTable (DOI: 10.17632/f93mjns9t6.2).
Fig. 5.
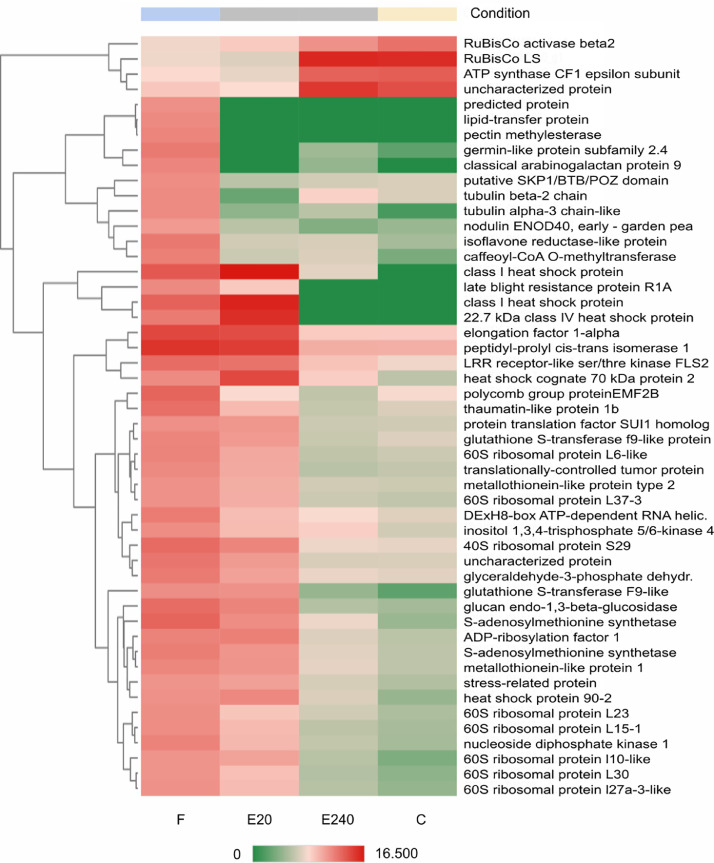
Heatmap of Top50 DEGs where untreated leaf (control ‘C’) as a reference and treated leaf by Fitokondi 4 l ha−1 (‘F’) as test condition was set. Samples of the treated leaf with Elice16Indures 20 g ha−1 (‘E20’) and Elice16Indures 240 g ha−1 (‘E240’) were shown. Annotation of transcript IDs was shown in AnnotationTable (DOI: 10.17632/f93mjns9t6.2).
Fig. 6.
Heatmap of Top50 DEGs where treated leaf by Elice16Indures 20 g ha−1 (‘E20’) as a reference and treated leaf by Elice16Indures 240 g ha−1 (‘E240’) as test condition was set. Samples of the treated leaf with Fitokondi 4 l ha−1 (‘F’) and untreated leaf (control ‘C’) were shown. Annotation of transcript IDs was shown in AnnotationTable (DOI: 10.17632/f93mjns9t6.2).
Fig. 8.
Heatmap of Top50 DEGs where treated leaf by Fitokondi 4 l ha-1 (‘F’) as a reference and treated leaf by Elice16Indures 240 g ha−1 (‘E240’) as test condition was set. Samples of the treated leaf with Elice16Indures 20 g ha−1 (‘E20’) and untreated leaf (control ‘C’) were shown. Annotation of transcript IDs was shown in AnnotationTable (DOI: 10.17632/f93mjns9t6.2).
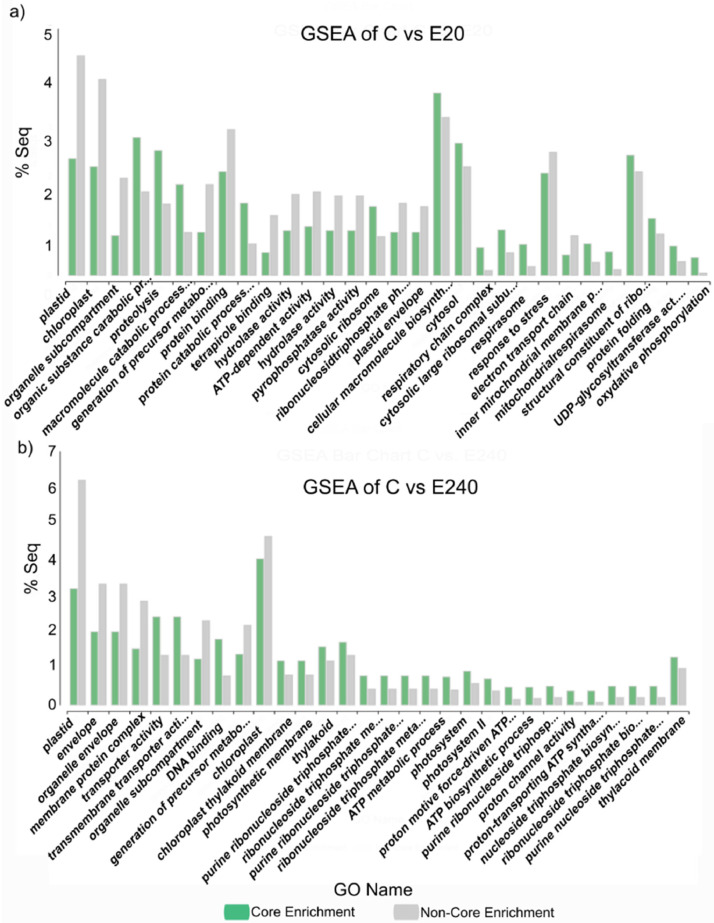
Fig. 9.
Most specific upregulated, annotated genes related GO categories that were examined by GSEA using Fisher exact test in the sample treated leaf by a) Elice16Indures 20 g ha−1 (‘E20’) and b) Elice16Indures 240 g ha−1 (‘E240’). The reference was untreated (‘C’) sample, respectively. The sample mark Core Enrichment means genes with the leading-edge subset within the gene set that contributes most to the enrichment result.
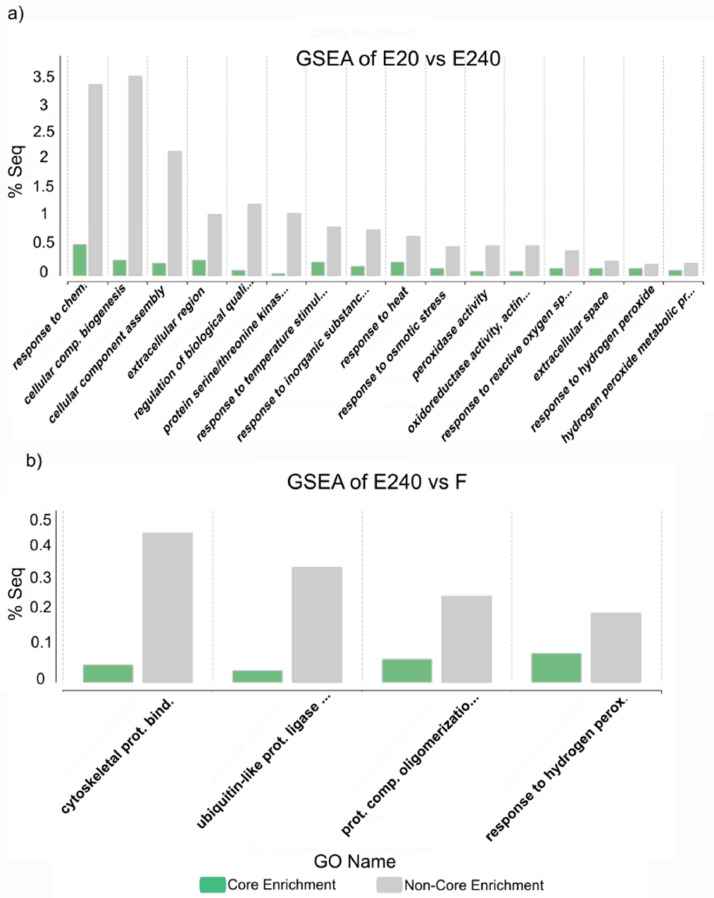
Fig. 11.
Most specific upregulated, annotated genes related GO categories that were examined by GSEA using Fisher exact test in the sample treated leaf by a) Elice16Indures 240 g ha−1 (‘E240’) with the Elice16Indures 20 g ha−1 (‘E20’) treated sample, as the reference. b) Fitokondi 4 l ha-1 (‘F’) was compared with the Elice16Indures 240 g ha-1 (‘E240’) treated sample, as the reference. The sample mark Core Enrichment means genes with the leading-edge subset within the gene set that contributes most to the enrichment result.
3. Experimental Design, Materials and Methods
3.1. Plant materials and Treatments
P. sativum subsp. sativum cultivar ‘Angela’ plants were cultivated on experimental plots with four repeats that different treatments were distributed randomly in each block in Tata, Hungary (GPS coordinates: 47 deg37′58.30"N, 18 deg15′54.36"E). Plants were treated at three phenological development stages BBCH16 (21/6/2019), BBCH51 (8/7/2019) and BBCH67 (17/7/2019) with Elice16Indures 20 g ha−1, Elice16Indures 240 g ha−1 and Fitokondi 4 l ha−1 (the normal field dose) biostimulators using Euro Pulvé plot sprayer with TeeJet 11004 AIXR nozzle. Leaf samples were collected two days after the last treatment at BBCH74 stage (19/7/2019) for NGS libraries such as control sample (non-treated) and treated samples ‘E20’ (treated with Elice16Indures 20 g ha−1), ‘E240’ (treated with Elice16Indures 240 g ha−1) and ‘F’ (treated with Fitokondi 4 l ha−1). Moreover, untreated pods were collected for NGS library ‘P’. Elice16Indures plant conditioner contains a high amount of SC-CO2 garlic cloves extract (71.5 percent) and other 10 different herbal extracts encapsulated in 150-200 nm size multilamellar liposomes using sunflower lecithin. Fitokondi contains aqueous extract of medicinal plants.
3.2. Sequencing and Bioinformatics
3.2.1. NGS Library Preparation and Sequencing
Approximately 30 mg of plant tissues were homogenized with 100 µl of TRI-Reagent (Zymo Research, Irvine, US) using SILAMAT S5 vibrator (Ivoclar Vivadent, Schaan, Liechtenstein). Total RNA was extracted using Direct-zol™ RNA MiniPrep System (Zymo Research, Irvine, US) and GEx library construction, QuantSeq 3‘ mRNA-Seq Library Prep Kit FWD for Illumina (Lexogen GmbH, Wien, Austria) was practiced conforming protocol proposed by manufacturer's, respectively. Pooled libraries were diluted to 1.8 pM for 1 × 86 bp single-end sequencing with 75-cycle High Output v2 Kit on the NextSeq 550 Sequencing System (Illumina, San Diego, US).
3.2.2. Pre-processing, Assembly and Gene-Level Quantification
During the pre-processing Trimmomatic software was applied for trimming reads, clipping adapters and removing contamination sequences [8]. De novo assembly of full-length transcripts of single-end read sets was performed by using Trinity method [9]. The report of RNA-seq de novo assembly is summarized in Table 1. To evaluate gene expression of RNA-sequencing CountTables were constructed with OmixBox.BioBam (https://www.biobam.com/omicsbox) using the HTseq package. The CountTable was created using the treated leaf samples (‘E20’, ‘E240’ and ‘F’) with the control, non-treated leaf (‘C’) sample. CountTables were deposited to Mendeley dataset (DOI: 10.17632/f93mjns9t6.2) and provided the basis for the following analyses.
3.2.3. Analyses of Count Data
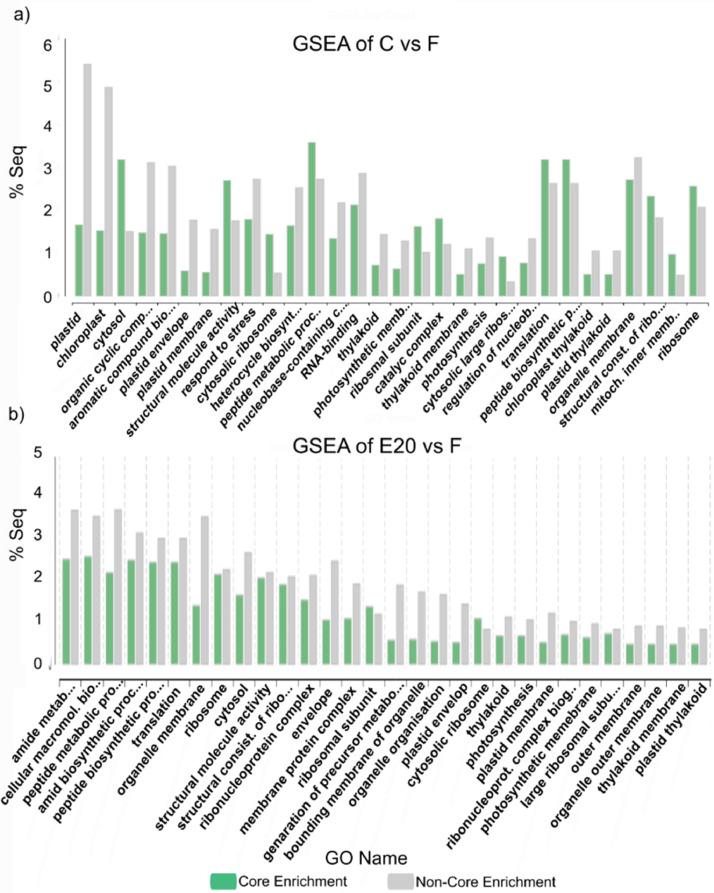
Pairwise differential expression analysis of Omicsbox (software package NOISeq v2.40.0) which belongs to the Bioconductor project [10,11] was used as an application of pairwise comparison of two different experimental conditions. The method was used to identify classes of transcripts that were over-represented using CountTable and AnnotationTable. Differentially expressed genomic features/genes (number of total features: 6,897) were expressed in pairwise comparisons of treated and non-treated samples (‘E20’ vs. ‘C’; ‘E240’ vs. ‘C’ and ‘F’ vs. ‘C’). The distribution of three DEGs was shown in heatmap (Figs. 3-5) and in Table 2.). Moreover, the DEGs of treated samples (‘E20’ vs. ‘E240’; ‘E20’ vs. ‘F’; ‘E240’ vs. ‘F’) were compared (Figs. 6-8). The functional annotation of DEGs was determined by Fisher's Exact Test and the overexpressed genes of GSEA [12] indicated in the bar chart showing GO categories (Fig. 9, Fig. 10, Fig. 11).
Fig. 10.
Most specific upregulated, annotated genes related GO categories that were examined by GSEA using Fisher exact test in the sample treated leaf by a) Fitokondi 4 l ha-1 (‘F’) with the reference untreated (‘C’) sample, as the reference. b) Fitokondi 4 l ha-1 (‘F’) sample was compared with Elice16Indures 20 g ha−1 (‘E20’) sample, as the reference. The sample mark Core Enrichment means genes with the leading-edge subset within the gene set that contributes most to the enrichment result.
Ethics Statements
Not relevant for the data.
CRediT Author Statement
Barbara Kutasy: Investigation, Methodology, Validation, Writing – original draft preparation; Visualization; Kincső Decsi: Methodology, Validation; Géza Hegedűs: Software, Investigation; Eszter Virág: Conceptualization, Methodology, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The work was founded by the KFI_16-1-2017-0457 - Development and production of a plant-based pesticide-plant conditioner for use in organic farming – the project of the Hungarian Government. The authors wish to acknowledge the support of József Péter Pallos, executive director, RIMPH Ltd. for the project finance. We express our thanks to Dr. Márta Kiniczky to the PBs formulation, Plant-Art Research Ltd. to release available plant growing and treatment and Xenovea Ltd, Hungary to perform NGS library preparation and sequencing. We are grateful to the editor and reviewers for the valuable comments that have helped to improve the manuscript. Project no. TKP2021- 665 EGA-20 (Biotechnology) has been implemented with the support provided by the National Research, Development and Innovation Fund of Hungary, financed under the TKP2021-EGA funding scheme. The work is supported by the GINOP-2.3.4-15-2020-00008 project. The project is co-financed by the European Union and the European Regional Development Fund.
Data Availability
References
- 1.Rouphael Y., Colla G. Biostimulants in agriculture. Front. Plant Sci. 2020;11:40. doi: 10.3389/fpls.2020.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du Jardin P. Plant biostimulants: definition, concept, main categories and regulation. Sci. Horticult. 2015;196:3–14. doi: 10.1016/j.scienta.2015.09.021. [DOI] [Google Scholar]
- 3.Duarte I.J., et al. Macroalgae as soil conditioners or growth promoters of Pisum sativum (L) Annual Res. Rev. Biol. 2018:1–8. doi: 10.9734/ARRB/2018/43272. [DOI] [Google Scholar]
- 4.Sulewska H., et al. Changes in Pisum sativum L. plants and in soil as a result of application of selected foliar fertilizers and biostimulators. Agronomy. 2020;10(10):1558. doi: 10.3390/agronomy10101558. [DOI] [Google Scholar]
- 5.Desoky E.-S.M., et al. Stimulating antioxidant defenses, antioxidant gene expression, and salt tolerance in Pisum sativum seedling by pretreatment using licorice root extract (LRE) as an organic biostimulant. Plant Physiol. Biochem. 2019;142:292–302. doi: 10.1016/j.plaphy.2019.07.020. [DOI] [PubMed] [Google Scholar]
- 6.Klimek-Kopyra A., et al. Application of biostimulants influences shoot and root characteristics of seedlings of winter pea (Pisum sativum L.) Acta Agrobotanica. 2019;72(2) doi: 10.5586/aa.1771. [DOI] [Google Scholar]
- 7.Moll P., et al. QuantSeq 3′ mRNA sequencing for RNA quantification. Nat. Methods. 2014;11(12):i–iii. doi: 10.1038/nmeth.f.376. [DOI] [Google Scholar]
- 8.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grabherr M.G., et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011;29(7):644. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tarazona S., et al. NOIseq: a RNA-seq differential expression method robust for sequencing depth biases. EMBnet J. 2011;17(B):18–19. doi: 10.14806/ej.17.B.265. [DOI] [Google Scholar]
- 11.Tarazona S., et al. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucl. Acids Res. 2015;43(21) doi: 10.1093/nar/gkv711. e140-e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.