Abstract
EHR-based sepsis research often uses heterogeneous definitions of sepsis leading to poor generalizability and difficulty in comparing studies to each other. We have developed OpenSep, an open-source pipeline for sepsis phenotyping according to the Sepsis-3 definition, as well as determination of time of sepsis onset and SOFA scores. The Minimal Sepsis Data Model was developed alongside the pipeline to enable the execution of the pipeline to diverse sources of electronic health record data. The pipeline’s accuracy was validated by applying it to the MIMIC-IV version 1.0 data and comparing sepsis onset and SOFA scores to those produced by the pipeline developed by the curators of MIMIC. We demonstrated high reliability between both the sepsis onsets and SOFA scores, however the use of the Minimal Sepsis Data model developed for this work allows our pipeline to be applied to more broadly to data sources beyond MIMIC.
Keywords: Sepsis, Sepsis-3, SOFA, MIMIC, critical care, phenotyping
INTRODUCTION
Sepsis is most often conceptualized as “life-threatening organ dysfunction caused by a dysregulated host response to infection (RTI).”1 Unfortunately, as no “gold standard” diagnostic test exists, identification of sepsis has relied on surrogate markers that combine suspicion of infection (SOI) with clinical markers of pathological RTI. These approaches, while clinically useful are not easily converted into a computable logic.2 Furthermore, reasonable clinicians may disagree about the nuances of the application of these definitions in specific cases. Due to these difficulties myriad definitions have evolved. Our group has previously systematically described various computational definitions of sepsis this work builds upon these efforts.3
Sepsis-3 is the most recently proposed consensus definition.4 However, to rigorously apply this definition in a computational manner requires data to be integrated harmonized and codified to make them suitable for analysis.
The Medical Information Mart for Intensive Care (MIMIC), is a frequently used source of deidentified data for studying critical care outcomes.5 MIMIC-IV version 1.0, the latest iteration of this resource, has recently been released.6 Similar to MIMIC-III the curators of MIMIC-IV developed and released a Sepsis-3 identification algorithm in SQL, hereon called the MIMIC pipeline.7 While the MIMIC-IV sepsis identification code is valuable for standardizing and expediting sepsis research using MIMIC-IV data, the pipeline is specialized for the data model of MIMIC-IV which is complex and not generalizable to other EHR datasets.
To address this gap, we have created a novel, freely available, open-source data processing pipeline, hereon called the OpenSep pipeline, to define Sepsis-3. Our intent is to produce a pipeline that can easily accept data from multiple data models with minimal transformation and to disseminate this pipeline in such a way as to enable its reproducible and rigorous use.
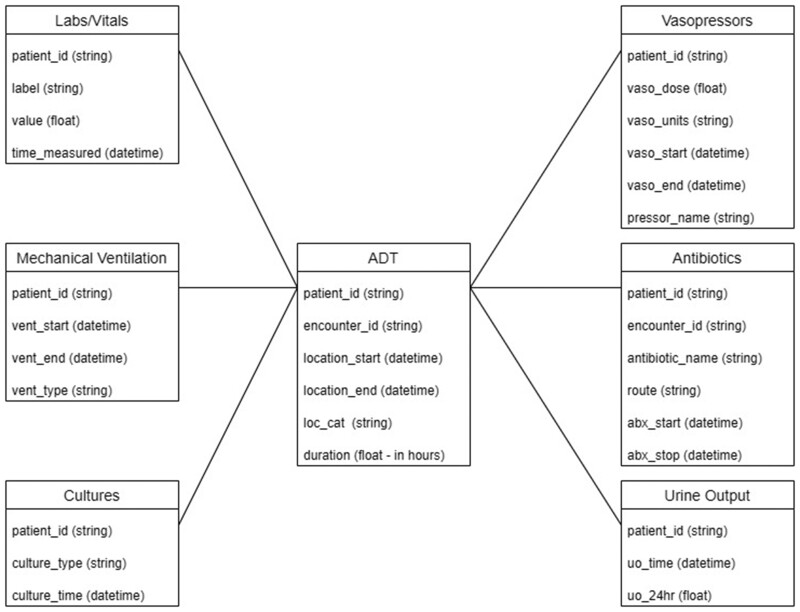
To enable the facile transformation of data, we have developed the Minimal Sepsis Data Model (MSDM) along with the pipeline to structure the data (Figure 1). The MSDM was designed to minimize the amount of data needed to assess sepsis status and illness severity, limit omission of relevant labs or measurements, and provide a simplified approach to the calculation of SOFA and determination of SOI. For instance, all patient data were related to each other by patient identifiers to avoid making arbitrary distinctions based on encounters or locations.
Figure 1.
Minimal Sepsis Data Model. Dataframes needed by the python pipeline to determine Sepsis-3 onset.
To validate our pipeline, we leveraged the previously available MIMIC pipeline. We describe the degree of agreement between Sepsis-3 classification, time of onset, and calculated SOFA scores between the previously developed and well validated MIMIC-pipeline and the OpenSep pipeline we developed, when used on the MIMIC-IV data.8 Finally, in cases where the 2 pipelines disagree the reasons for the disagreement were characterized.
MATERIALS AND METHODS
Study data
MIMIC-IV version 1.0 was published in March of 2021 is an update to the widely used MIMIC-III database, incorporating a greater number and variety of more recent data sources. Data were acquired from patients admitted to Beth Israel Deaconess emergency departments and intensive care units. The MIMIC-IV SQL database was reproduced on a locally hosted SQL-server as described previously.6
Data were transformed using SQL queries of from the MIMIC-IV Data Model to create dataframes conforming to the MSDM (Figure 1).
Open-source pipeline development
Suspicion of infection
SOI was defined as either cultures followed by antimicrobials in the next 72 h, or antimicrobials followed by cultures within 24 h, as consistent with the Seymour interpretation of Sepsis-3.9 Encounters meeting these criteria were identified using the Admission-Discharge-Transfer (ADT), culture, and antimicrobial data. Only the first instance of SOI was considered for each encounter. The time of SOI was defined as the earlier of antimicrobial administration or culture collection.1
Response to infection
Per the Sepsis-3 criteria, organ failure can be demonstrated by a SOFA score greater than or equal to 2 when in the ICU, or qSOFA score greater than or equal to 2 outside of the ICU.1 Given the sparsity of data outside of the ICU in the MIMIC-IV only SOFA scores were considered in the MIMIC-IV implementation. This was enforced in this pipeline to facilitate comparison, though the ability to determine Sepsis by qSOFA is also available.
For patients identified as meeting criteria for SOI, SOFA scores were calculated using the labs and vitals, ADT, mechanical ventilation, vasopressor, and urine output data (see Supplementary Table S1). Scores were calculated starting at the beginning of the encounter and then hourly, as well as at the times of ICU admission or discharge. Scores calculated outside of the ICU were discarded.
SOFA Scores were calculated as previously described.10 If no data was available for a given component or subscore at a specific time a SOFA subscore of 0 (normal) was assumed. Encounters were evaluated for whether a score greater than or equal to 2 occurred within 48 h before or 24 h after SOI. Patients who met this criterion were classified as having met Sepsis-3 criteria. Onset of sepsis was defined as the earlier of the cultures or antimicrobials.
Pipeline comparison
To ensure that scores and determinations made by each pipeline were similar, encounters were assigned an onset time and SOFA score based on both the MIMIC-IV SQL pipeline and the proposed pipeline. To account for their difference in handling the calculation interval, SOFA scores were matched by finding the nearest SOFA scores from the SQL and proposed pipelines. SOFA scores were then compared for all patients for each hour of their ICU admission.
Statistical analysis
Correlations between the SQL and proposed package sepsis labels and SOFA scores were compared using a coefficient of determination (Correlation Coefficient2). All data analysis and figure generation were performed in Python 3.9.7 (Python Software Foundation, Beaverton, OR) using Jupyter Notebook (NumFOCUS, Austin, TX). The code is freely available at https://github.com/mhofford/I2_Sepsis.11
RESULTS
SOI identification
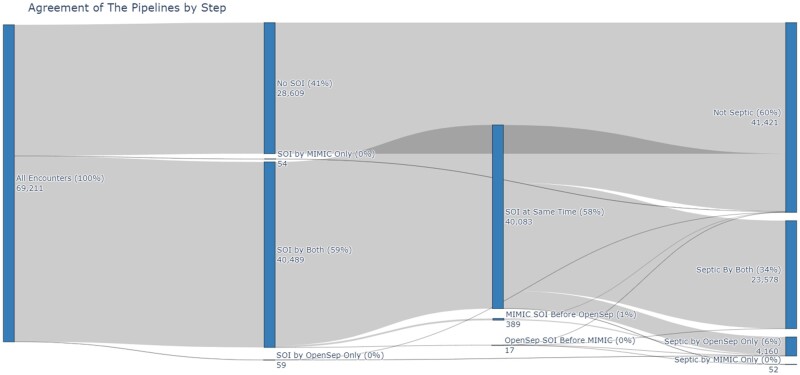
In the MIMIC-IV dataset 69 211 hospitalizations were available for analysis. 40 543 were identified by the MIMIC-pipeline and 40 548 were identified by the OpenSep-pipeline as meeting SOI. Both pipelines agreed on 40 489 of SOI determinations. Fifty-four were found to have SOI by the MIMIC-pipeline alone, and 59 were found to meet SOI by the OpenSep-pipeline alone. Three hundred eighty-nine were found to have SOI by the MIMIC-pipeline before the OpenSep-pipeline and 17 were found to have SOI by the OpenSep-pipeline before the MIMIC-pipeline (Figure 2).
Figure 2.
Agreement of the pipelines by step. Agreement of pipelines by step in determining Sepsis-3. SOI: Suspicion of Infection; RTI: Response to Infection.
Of the 519 patients that had discordant SOI assessments, 443 (85.4%) were due to the MIMIC-pipeline considering SOI that occurred greater than 24 h prior to the encounter which was not permitted in the OpenSep-pipeline as this cannot result in a classification of sepsis.1,9 Thirty-eight (7.3%) instances of discordant SOI assessments were due to an error in the MIMIC-pipeline that prevented SOI determination if antimicrobials were ordered and cultures collected at the exact same time. The final 38 (7.3%) differences in SOI emerged as the OpenSep-pipeline allows for culture collection and antimicrobial administration previous but closely timed inpatient encounters, while MIMIC-pipeline does not permit for observations that span multiple encounters (Table 1).
Table 1.
Issues identified in suspicion of infection determination
| Discrepancy | Problem identified | Number of encounters (%) |
|---|---|---|
| SOI by MIMIC not OpenSep 54 (10.4%) | SOI greater than 24 h prior to admission | 54 (100) |
| SOI by OpenSep but not MIMIC 59 (11.4%) | MIMIC does not allow for the culture and antibiotic to be at the same time | 33 (55.9) |
| MIMIC does identify SOI at the same time but for a different encounter | 26 (44.1) | |
| SOI by MIMIC before OpenSep 389 (75.0%) | MIMIC identifies SOI more than 24 h prior to the encounter | 389 (100) |
| SOI by OpenSep before MIMIC 17 (3.3%) | MIMIC does not allow for the culture and antibiotic to be at the same time | 5 (29.4) |
| MIMIC does not allow antibiotics from a different encounter | 12 (70.6) |
SOI: Suspicion of Infection.
SOFA score calculation
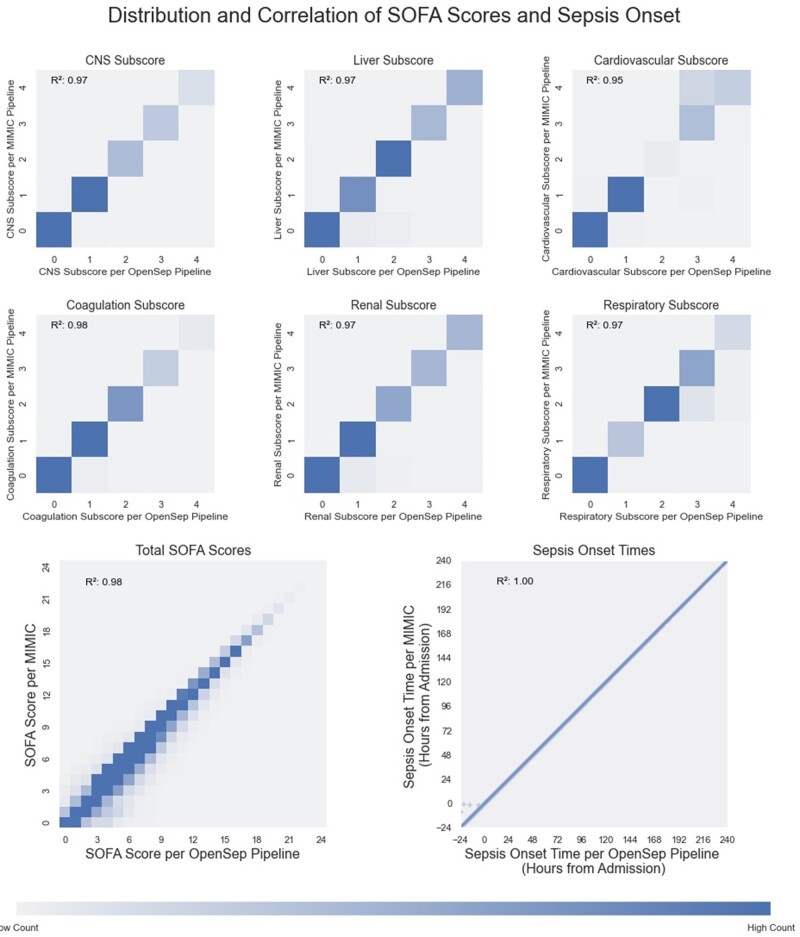
SOFA scores were compared for the 40 548 encounters that met SOI in OpenSep-pipeline. The median (IQR) number of SOFA scores per hospitalization was 54 (28–117). After matching the nearest SOFA scores from the MIMIC and OpenSep pipelines, hourly SOFA and SOFA subscore results were found to be similar between the MIMIC and OpenSep pipelines, with coefficients of determination (R2) 0.95 or greater for all SOFA subscores and 0.98 for the calculated Total SOFA score (Figure 3).
Figure 3.
Distribution and correlation of SOFA scores and Sepsis onset. Distribution and correlations of SOFA subscores, total SOFA score and Sepsis-3 onset time for SQL and python pipelines. Temporally nearest scores for all patients at each hour of the ICU admission are plotted for SOFA and SOFA sub scores. Heat maps were used to characterize the distribution for the SOFA scores as they are heavily overlapping. Maximum intensity in the heat-map was capped at the 95th percentile for each of the scores. Scatter plot was used for the onset time to display the patients that were identified earlier by the python pathway. Color bar does not relate to the scatter plot.
Sepsis-3 identification
Of the 69 211 included hospitalizations, sepsis was identified in 23 630 (34%) encounters by the MIMIC-pipeline and 27 738 (40%) by the OpenSep-pipeline. There was a high degree of overlap between the sepsis determinations with 23 578 (34.1%) hospitalizations being identified by both pipelines. The OpenSep-pipeline identified 4160 patients not identified by the MIMIC-pipeline, while the MIMIC-pipeline identified 52 patients not identified by the OpenSep-pipeline (Figure 2).
Of the 4160 patients classified as septic by the OpenSep-pipeline but not MIMIC-pipeline, 3335 (80.2%) were due to a missing or nonmatching ADT identifier (stay_id) between temporally close encounters where patients met either the SOI or RTI criteria. The remaining 541 (13.0%) were caused by a difference in SOFA score from the OpenSep-pipeline considering data from outside the ICU but within the specified timeframe. Of the 52 patients identified by MIMIC-pipeline, but not the proposed pipeline, 43 (82.7%) were due to differences in the timing of the hourly SOFA scores. The MIMIC-pipeline set time zero as the hour of ICU admission and calculated a score hourly whereas the OpenSep pipeline began at the start of the encounter and at times of location change. In 7 (13.4%) cases, the MIMIC pipeline calculated SOFA scores after the patient was discharged from the ICU and in the remaining 2 cases, the difference in sepsis determination was caused by a difference in SOI determination (Table 2).
Table 2.
Issues identified in Sepsis-3 determination
| Discrepancy | Problem identified | Number of encounters (%) |
|---|---|---|
| Sepsis by MIMIC but not OpenSep 49 (1.2%) | MIMIC finds sepsis due to slight difference in timing | 42 (85.7) |
| MIMIC uses a SOFA score outside of the ICU time | 7 (14.3) | |
| Sepsis by OpenSep not MIMIC 3876 (98.8%) | Information documented prior to the ICU is not considered by MIMIC | 541 (12.2) |
| MIMIC join fails because stay_id is null | 3188 (82.3) | |
| MIMIC join fails because stay_id does not match qualifying SOFA stay_id | 147 (3.8) |
Sepsis-onset time
For the 23 578 patients that met sepsis criteria using both the MIMIC and the OpenSep pipelines, the sepsis onset times matched nearly perfectly, with a correlation coefficient (R2) close to 1.0, with only 5 (0.02%) patients having discordant times due to the proposed pipeline considering cultures obtained in prior encounters (Figure 3).
DISCUSSION
In this report, we describe the MSDM and OpenSep an open-source, highly generalizable approach to sepsis labeling and assessment of illness severity. The proposed pipeline identified septic patients with a high degree of correlation with the well-validated SQL approach of the MIMIC developers. Investigation into discordant cases revealed that the majority were the result of the proposed pipeline identifying patients as septic which the MIMIC/SQL method did not. This emerged due to the fact that the SQL-based code did not look for SOI or RTI criteria outside of the ICU and used encounter level criteria rather than time-based as proposed by the Sepsis-3 guidelines. The proposed pipeline, facilitated by the MSDM, ran agnostic to location and encounter-level identifiers and used the time-based criteria outlined in the Sepsis-3 consensus statement to look for RTI and SOI criteria, leading a higher sensitivity. To illustrate an example, consider a patient that presented to the emergency department and had blood cultures taken and was discharged home. If that patient returned the following day and received antimicrobial therapy and met RTI criteria, they would be included in the proposed pipeline, but not in the SQL-based approach.
Another source of discordance between the 2 methods arises from the interval at which they calculated SQL scores. In the SQL pipeline time is rounded down and then counted in whole hours from the time of ICU admission. The proposed pipeline considers the beginning and end of each ICU admission and each hour in a stepwise manner from admission without rounding. This was implemented this way to allow the proposed pipeline to consider qSOFA scores outside of the ICU in other datasets that might have more data from non-ICU stays. This accounted for only minor differences in SOFA scores and rarely changed the ultimate characterization of sepsis (Tables 1 and 2).
The pipeline described here is a useful tool however, several limitations remain. There is room for performance improvement throughout the codebase, which was traded for readability of the code in order to prioritize usability and ease of review. Additionally, there is no widely agreed upon consensus for what cultures and antimicrobial should be included in defining sepsis. The proposed pipeline currently allows for the user to determine which antimicrobials and cultures should be included. Finally, while the MSDM was designed to be facile when working with EHR derived data, heterogeneity in EHR data models and implementations necessitates that the user transformed their data to the MSDM for the pipeline to function. Additionally, some user manipulation of the data may be necessary if patients included in the cohort have multiple unique identifiers.
EHR-based research in Sepsis is currently limited by the nonuniform application of definitions across the field. The MIMIC team has provided a standard computational definition for Sepsis-3 however their work cannot easily be applied other datasets as the code used to identify sepsis in their cohort is complex and specific to their difficult to generalize data model. We have successfully created an open source pipeline to more easily apply the Sepsis-3 definition to a broad array of EHR derived data in a simplified data model and validated that pipeline by comparing results to those obtained using the MIMIC-pipeline on MIMIC data. We hope this tool can be used to expedite and harmonize clinical outcomes research across diverse datasets.
Supplementary Material
Contributor Information
Mackenzie R Hofford, Department of Medicine, Institute for Informatics, Washington University School of Medicine in St. Louis, St. Louis, Missouri, USA; Division of General Medicine, Department of Medicine, Washington University School of Medicine in St. Louis, St. Louis, Missouri, USA.
Sean C Yu, Department of Medicine, Institute for Informatics, Washington University School of Medicine in St. Louis, St. Louis, Missouri, USA; Department of Biomedical Engineering, School of Engineering, Washington University School in St. Louis, St. Louis, Missouri, USA.
Alistair E W Johnson, Program in Child Health Evaluative Sciences, Peter Gilgan Centre for Research and Learning, The Hospital for Sick Children, Toronto, ON, Canada.
Albert M Lai, Department of Medicine, Institute for Informatics, Washington University School of Medicine in St. Louis, St. Louis, Missouri, USA.
Philip R O Payne, Department of Medicine, Institute for Informatics, Washington University School of Medicine in St. Louis, St. Louis, Missouri, USA.
Andrew P Michelson, Department of Medicine, Institute for Informatics, Washington University School of Medicine in St. Louis, St. Louis, Missouri, USA; Division of Pulmonary and Critical Care, Department of Medicine, Washington University School of Medicine in St. Louis, St. Louis, Missouri, USA.
AUTHOR CONTRIBUTIONS
MRH, APM, SCY, and AEWJ conceived of the study and developed the study design. MRH and SCY created and the OpenSep pipeline. All authors assisted in drafting the work. All authors contributed to data analysis and interpretation. All authors have approved its submission and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy and integrity of any part of the work are appropriately investigated and resolved.
SUPPLEMENTARY MATERIAL
Supplementary material is available at JAMIA Open online.
CONFLICT OF INTEREST STATEMENT
None declared.
DATA AVAILABILITY
The data underlying this work are available in physio-net.6 The MIMIC derived schemas are also publically available at https://github.com/MIT-LCP/mimic-code/tree/main/mimic-iv.8 The code for the OpenSep pipeline is available at https://github.com/mhofford/I2_Sepsis.11
References
- 1. Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016; 315 (8): 801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yu SC, Betthauser KD, Gupta A, et al. Comparison of sepsis definitions as automated criteria. Crit Care Med 2021; 49 (4): e433–43. [DOI] [PubMed] [Google Scholar]
- 3. Angus DC, Seymour CW, Coopersmith CM, et al. A framework for the development and interpretation of different sepsis definitions and clinical criteria. Crit Care Med 2016; 44 (3): e113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Coopersmith CM, De Backer D, Deutschman CS, et al. Surviving sepsis campaign: research priorities for sepsis and septic shock. Intensive Care Med 2018; 44 (9): 1400–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rogers P, Wang D, Lu Z.. Medical information mart for intensive care: a foundation for the fusion of artificial intelligence and real-world data. Front Artif Intell 2021; 4: 691626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson A, Bulgarelli L, Pollard T, et al. MIMIC-IV (Version 1.0). PhysioNet2020. 10.13026/77z6-9w59. [DOI] [Google Scholar]
- 7. Johnson AEW, Aboab J, Raffa JD, et al. A comparative analysis of sepsis identification methods in an electronic database. Crit Care Med 2018; 46 (4): 494–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Johnson AE, Stone DJ, Celi LA, et al. The MIMIC Code Repository: enabling reproducibility in critical care research. J Am Med Inform Assoc 2018; 25 (1): 32–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seymour CW, Liu VX, Iwashyna TJ, et al. Assessment of clinical criteria for sepsis: for the Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016; 315 (8): 762–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lambden S, Laterre PF, Levy MM, et al. The SOFA score-development, utility and challenges of accurate assessment in clinical trials. Crit Care 2019; 23 (1): 374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hofford M, Yu SC, Johnson AEW, et al. A Python Pipeline for SOFA Score Calculation and Sepsis-3 Classification—v1.0. 2022. 10.5281/zenodo.6799609. Accessed December 9, 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this work are available in physio-net.6 The MIMIC derived schemas are also publically available at https://github.com/MIT-LCP/mimic-code/tree/main/mimic-iv.8 The code for the OpenSep pipeline is available at https://github.com/mhofford/I2_Sepsis.11