Highlights
-
•
Cuproptosis-related lncRNAs prognostic model demonstrated exceptional prognostic efficacy for osteosarcoma patients.
-
•
The biological function of identified differentially expressed genes with cuproptosis-related lncRNAs’ risk model was closely related to the PI3K-Akt pathway.
-
•
Cuproptosis-related lncRNAs were associated with osteosarcoma CAFs.
-
•
Erlotinib, MP470, and WH-4-023 might potentially effectively target the PI3K-Akt pathway drugs for OS.
Keywords: Osteosarcoma, lncRNA, Cancer-associated fibroblasts, Cuproptosis, Prognoses
Abstract
Background
Osteosarcoma is the most common primary pediatric and adolescent bone malignancy. An imbalance in copper homeostasis caused by copper ion accumulation could increase intracellular toxicity and regulate cancer cell growth. This study aimed to identify long non-coding RNAs (lncRNAs) associated with cuproptosis to predict prognosis and target drug use to improve patient survival.
Methods
RNA sequencing and relevant clinical information of ninety-three osteosarcoma patients were obtained from the TARGET database. We then identified thirteen prognostic cuproptosis-related lncRNAs(CRLncs) using coexpression and univariate Cox analyses. The prognostic risk model with three CRLncs was constructed using the least absolute shrinkage and selection operator (LASSO) and multivariate Cox regression analysis. Patients were divided into low-risk and high-risk subgroups using the median risk score. The tumor microenvironment (TME) and immune status of identified subgroups were analyzed using ESTIMATE, CIBERSORT, MCP-counter, xCELL, EPIC, and ssGSEA analyses. Functional analyses were conducted to elucidate the underlying mechanisms, including GO, KEGG, GSVA, and GSEA analyses. Also, the relationships between the model, tumor immunity, and drug sensitivity were explored. Lastly, the expression level of ZNF37BP, AL353759.1, and AC005034.5 was validated in vitro.
Results
We constructed a model containing three CRLncs (ZNF37BP, AL353759.1, and AC005034.5) and validated its excellent prognostic and predictive power. The AUC curves for 1-year, 3-year, and 5-year survival probabilities were 0.76, 0.84, and 0.89, respectively. Patients in the high-risk group had a shorter overall survival (OS) time than those in the low-risk. The stroma score and cancer-associated fibroblasts (CAFs) were significantly higher in the low-risk group. Immune cells such as T cells CD4 naive, T cells gamma delta, NK cells resting, dendritic cells resting, and mast cells activated were significantly upregulated in the high-risk group. Based on functional analyses, the PI3K-Akt pathway was identified as a critical metabolic pathway in osteosarcoma. Additionally, we obtained three potentially effective drugs for OS: erlotinib, MP470, and WH-4-023 targeting the PI3K-Akt pathway. The expression level of ZNF37BP was significantly elevated in OS cell lines than in normal osteoblast hFOB1.19 cells, and that of ATP7A, LIPT1, AL353759.1, and AC005034.5 were decreased considerably in OS cell lines.
Conclusion
Cuproptosis-related lncRNAs are correlated with the CAFs of osteosarcoma, and this could serve as a foundation for OS survival prediction and treatment.
1. Introduction
Osteosarcoma is the most common primary pediatric and adolescent bone malignancy [1]. A high mortality rate is associated with this type of cancer, especially among patients with lung metastases and resistant to treatment. Currently, neoadjuvant chemotherapy and surgery are the primary treatments for osteosarcoma [2]. However, there has been little progress in improving survival outcomes for osteosarcoma patients in the past three decades. Hence, exploring new targets and features for improving patients' clinical efficacy and survival with OS is imperative.
The tumor microenvironment (TME) plays a crucial role both in tumor progression and in therapy response [3]. Increasing evidence suggests that the bone microenvironment affects osteosarcoma metastasis. In the TME, stromal cells, especially cancer-associated fibroblasts (CAFs), which are significant components of the tumor microenvironment, play pivotal roles in tumor progression [4]. Although the CAFs are thought to be associated with osteosarcoma TME, research is lacking. Novel markers associated with CAFs characteristics of OS that could predict outcomes and assist in patient management are urgently needed.
Cuproptosis, a recently discovered mode of cell death, is distinct from ferroptosis, necroptosis, and apoptosis [5]. Excess copper in the cell binds to lipoylated components of the tricarboxylic acid(TCA) cycle, triggering proteotoxic stress and death of the cells [6]. It provides new insights into regulating intracellular copper levels in cancer treatment [7]. Meanwhile, several copper-containing coordination compounds are promising antitumor agents [8]. Emerging evidence has demonstrated that lncRNAs are crucial regulators of osteosarcoma development, progression, and invasion [9]. Deregulated expression of lncRNAs has been found to participate in the regulation of various signaling transduction pathways.
In this study, we developed a prognostic cuproptosis-related lncRNA model that could robustly predict survival time for OS patients and was associated with CAFs for osteosarcoma. We may provide new clues regarding molecular mechanisms underlying osteosarcoma, shedding new light on treatment strategies and promoting the individual-based treatment of osteosarcoma.
2. Materials and methods
2.1. Data collection and screening Cuproptosis–Related genes
RNA-sequencing (RNA-seq) data and corresponding clinical information (Table 1) were obtained from the Therapeutically Applicable Research to Generate Effective Treatments (https://ocg.cancer.gov/programs/target). The lncRNA and mRNA expression profiles were extracted from the expression data of ninety-three osteosarcoma patients. All gene expression levels were normalized using log2 (TPM + 1). Thirteen cuproptosis-related genes(CRGs)were obtained from a previous report [5] and the Molecular Signatures Database (MSigDB) gene sets [10]. OS-related CRGs were determined by intersecting the CRGs with genes from the TARGET-OS transcriptome data.
Table 1.
Characteristics of patients in TARGET-OS cohort.
| Characteristics | Alive(N = 57) | Dead(N = 36) | Total(N = 93) | p-value | FDR |
|---|---|---|---|---|---|
| Gender | 1 | 1 | |||
| Female | 25(26.88 %) | 15(16.13 %) | 40(43.01 %) | ||
| Male | 32(34.41 %) | 21(22.58 %) | 53(56.99 %) | ||
| Race | 0.24 | 0.71 | |||
| American Indian or Alaska Native | 0(0.0e + 0 %) | 1(1.08 %) | 1(1.08 %) | ||
| Asian | 4(4.30 %) | 2(2.15 %) | 6(6.45 %) | ||
| Black or African American | 5(5.38 %) | 4(4.30 %) | 9(9.68 %) | ||
| White | 38(40.86 %) | 17(18.28 %) | 55(59.14 %) | ||
| Unknown | 10(10.75 %) | 12(12.90 %) | 22(23.66 %) | ||
| Age | 0.41 | 0.81 | |||
| <18 years | 42(45.16 %) | 30(32.26 %) | 72(77.42 %) | ||
| >18 years | 15(16.13 %) | 6(6.45 %) | 21(22.58 %) | ||
| Metastasis | 1.10E-03 | 5.70E-03 | |||
| Metastatic | 7(7.53 %) | 16(17.20 %) | 23(24.73 %) | ||
| Non-metastatic | 50(53.76 %) | 20(21.51 %) | 70(75.27 %) | ||
| Primary tumor site | 0.03 | 0.14 | |||
| Arm/hand | 4(4.30 %) | 3(3.23 %) | 7(7.53 %) | ||
| Leg/foot | 53(56.99 %) | 29(31.18 %) | 82(88.17 %) | ||
| Pelvis | 0(0.0e + 0 %) | 4(4.30 %) | 4(4.30 %) | ||
2.2. Screening Cuproptosis–Related LncRNAs and Construction of the coexpression network
We used the “limma” package to perform coexpression analysis of the expression CRGs to lncRNAs to obtain the CRLncs matrix, and “ggalluvial” was used to plot the Sankey relationship between CRGs and CRLncs by Pearson's correlation analysis (|Pearson R| >0.5 and p < 0.001).
2.3. Risk model construction and validation
By using the survival package in R, we calculated the hazard rat through univariate Cox regression(uniCox) analysis of CRGs and CRLncs associated with OS prognosis. The performed least absolute shrinkage and selection operator regression (LASSO-Cox) analysis was then conducted on the prognosis-related CRLncs screened by uniCox analysis to prevent overfitting. An optimal value was determined by the minimum lambda. In addition, an analysis of multivariate Cox regression(multiCox) was performed to determine the risk model's CRLs. The CRLncs prognostic signature score was calculated as follows: Risk Score = Σ (βi × Expi) (β: coefficients, Exp: lncRNA expression level). Since few osteosarcoma datasets contain CRLs, we randomly divided the samples with survival data into a test set and a training set using the “caret” package. We developed a risk prognosis model for the total sample, training group, and test group. According to the median CRL risk scores, we divided all samples into high- and low-risk groups. Kaplan-Meier survival analysis was performed by “survivor” and “survminer” packages to compare patients' overall survival(OS) time between high-risk and low-risk groups. Time-dependent receiver operating characteristic(ROC) analysis of OS was used to assess the sensitivity and accuracy of the predictive model by the “ timeROC ” package. The principal component analysis(PCA), t-distributed stochastic neighbor embedding(t-SNE), and uniform manifold approximation and projection(UMAP) analysis of all osteosarcoma samples were performed to observe whether the model could distinguish OS patients into high- and low-risk groups accurately.
2.4. Establishment of CRLncs prognostic nomogram for osteosarcoma
We integrated clinical characteristics(age, gender, and tumor metastasis) and the CRLncs signature risk score to investigate the independent prognostic factors via univariate and multivariate Cox regression. We then developed a prognostic nomogram based on these independent prognostic factors to predict 1-, 3-, and 5-year overall survival. Model calibration was performed using the calibration plot.
2.5. Functional enrichment analysis and KEGG pathway network construction
Differentially expressed genes(DEGs) between the high-risk and low-risk groups were identified (|log2(fold change)|>1.5 and FDR < 0.05) with the “limma” R package and functionally annotated based on the Gene Ontology(GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) with the “clusterProfiler” R package(p < 0.05, and FDR < 0.25), and was visualized in Metascape [11]. Many pathways are involved in the complex pathogenesis of DEGs; hierarchical relations and connectivity relations are among these pathways. In order to explore the possible core pathways of DEGs, we further used Cytoscape software to construct a pathway-pathway network with KEGG pathway enrichment terms.
2.6. Assessment of immune cell infiltration and immune microenvironment
In exploring the differences in immune cell infiltration in the two risk groups of osteosarcoma patients, we simultaneously utilized five algorithms(ESTIMATE, CIBERSORT, MCP-counter, xCELL, and EPIC) to estimate the immune cell infiltration. We also performed the ssGSEA algorithm to quantify the immune functions. Finally, TIDE(tumor immune dysfunction and exclusion) algorithm (https://tide.dfci.harvard.edu/) was used to predict response to immunotherapy and the CAF proportion in the low- and high-risk score subgroups [12].
2.7. Drug sensitivity prediction
R packages of “limma,” “ggpubr,” and “pRRophetic” were used to predict potential chemotherapeutic agents to treat OS on high- and low-risk OS subgroups, with an initial screening criterion of p < 0.001 [13].
2.8. Cell culture
The human fetal osteoblast cell line(hFOB1.19) and human OS cell lines(MG63 and 143B) were obtained from Shenzhen Advanced Biomechanics and Materials Lab(Shenzhen, China). A separate medium was used for each cell line (Shenzhen Advanced Biomechanics and Materials Lab; Shenzhen, China). The hFOB1.19 was used as an osteoblast cell, the MG63 was used as a non-metastatic OS cell, and 143B was used as a metastatic OS cell. We cultured human OS cells at 37 °C in an incubator with 5 % CO2 and human FOB1.19 cells at 34 °C in an incubator with 5 % CO2.
2.9. Quantitative real-time PCR(qRT-PCR)
In accordance with the manufacturer's instructions, total RNA was extracted from OS cells and hFOB1.19 by using TRIZOL(Gibco, USA); the total RNA was isolated using RNAiso Plus(AgBio. China) and then synthesized cDNA by using the cDNA Synthesis Kit(AgBio. China). qPCR was performed with an SYBR Green Real-time PCR Master Mix ki(AgBio. China). All PCR tests were performed on the StepOnePlus Real-Time PCR systems(Applied Biosystems Inc., USA). Glyceraldehyde-3-phosphate dehydrogenase(GAPDH) acted as the internal reference for normalization. All primers used for qRT–PCR were synthesized by Sangon Biotech(Sangon Biotech, Shanghai, China). Table 2 outlines the primer sequences we used.
Table 2.
Primer sequences for RT-qPCR.
| Genes | Forward | Reverse |
|---|---|---|
| GAPDH | GCACCGTCAAGGCTGAGAAC | TGGTGAAGACGCCAGTGGA |
| ATP7A | TGACCCTAAACTACAGACTCCAA | CGCCGTAACAGTCAGAAACAA |
| LIPT1 | GCTGGATGTGCAGGCTACC | GCAATGGTGATAGGCAGTAGTC |
| AC005034.5 | TGTGTGCAGTCTATTGAGGGT | TGACAAGGTAGCATCAAATCCC |
| ZNF37BP-F | ACCTTCCGTCAGAAGTCAGC | ACCCAGAGTAATTTCCCTCGTTT |
| AL353759.1 | CTCGCGCCAAAGCGAAATC | CTGCGTAGTTGCCTTTACGGA |
2.10. Statistical analysis
Statistical analyses were performed via R(version 4.2.1) and GraphPad Prism(version 8.4.3). We conducted each experiment at least three times independently. The measurement results were expressed as mean ± standard deviation (SD). Statistical analysis was conducted by Student's t-tests and one-way ANOVAs.Statistical significance is defined by *p < 0.05, **p < 0.01,and ***p < 0.001.
3. Results
3.1. Identification of Prognosis-Related CRGs and Co-expressed CRLncs in osteosarcoma
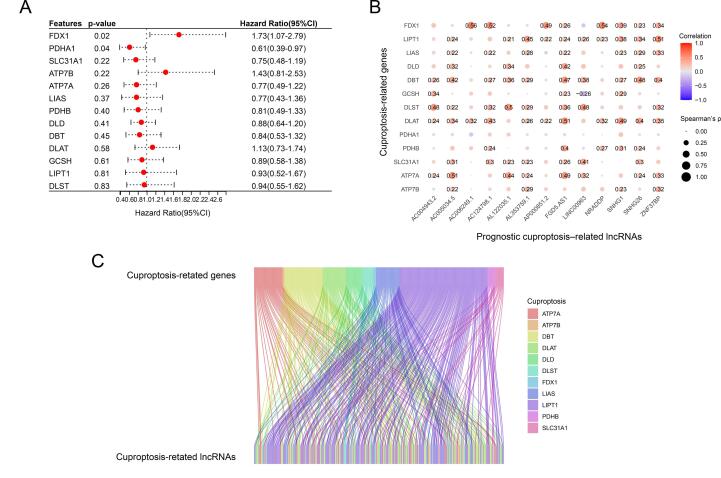
Thirteen OS-related CRGs were identified. By univariate Cox analysis, two prognosis-related CRGs (FDX1 and PDHA1) were identified(p < 0.05) (Fig. 1A). We identified 264 lncRNAs (|Coefficient| > 0.5 and p < 0.001) that were co-expressed in OS(Fig. 1B). By univariate Cox analysis, thirteen prognosis-related lncRNAs were identified(p < 0.05), of which six lncRNAs were identified as protective factors with a hazard ratio(HR) < 1, while the other seven lncRNAs were considered as risk factors. The correlation between CRGs and prognostic CRLncs in the TARGET-OS cohort was visualized through the Sankey diagram by packages of “ggalluvial” [14].
Fig. 1.
Identification of prognosis-related CRGs and co-expressed CRLncs in Osteosarcoma. (A) Univariate cox analysis of the survival rate of OS using the thirteen CRGs. (B) Correlation between CRGs and co-expressed prognostic CRLncs. (C) Sankey diagram plot of CRGs and CRLncs.
3.2. Construction and validation of the prognostic signature
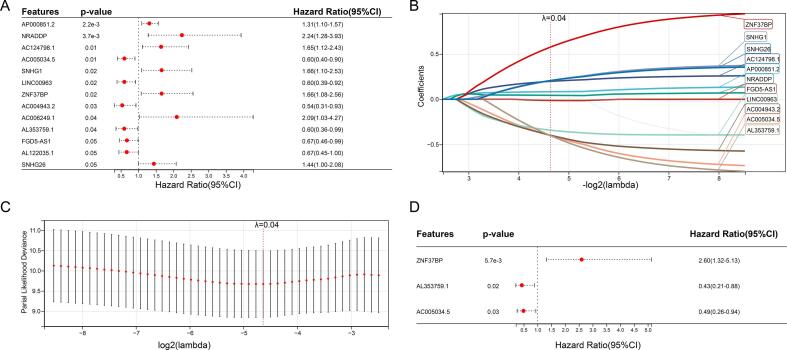
Subsequently, we performed LASSO analysis to avoid overfitting and construct the best prognostic signature. According to the optimal penalty parameter(λ = 0.0402642963670375) value, eleven CRLncs participated in further multivariate Cox regression analysis to determine the optimal number of CRLncs used to establish the risk model (Fig. 2A–D). Finally, we identified three lncRNAs(ZNF37BP, AC005034.5, and AL353759.1). Here are the calculations used to calculate the risk score: Risk score =(0.956917270253414 * ZNF37BP exp.) + (-0.707198563219447 * AC005034.5 exp.) + (-0.833381321267523 * AL353759.1 exp.). A median risk score of each patient was used to divide them into high-risk and low-risk groups. Using the “caret” package, the total sample group was divided into low-risk and high-risk groups(N = 46 and 47, respectively). In the training group, there were 24 participants at high risk(N = 24) and 23 participants at low risk(N = 23). Twenty-three participants were assigned to the high-risk and 23 to the low-risk group of the test group.
Fig. 2.
Construction of prognostic risk model. (A) Univariate Cox analysis with CRLncs. (B-C) LASSO analysis with optimal penalty lambda value, (D) Three prognostic CRLncs (ZNF37BP, AC005034.5, and AL353759.1) was identified under multiCox analysis.
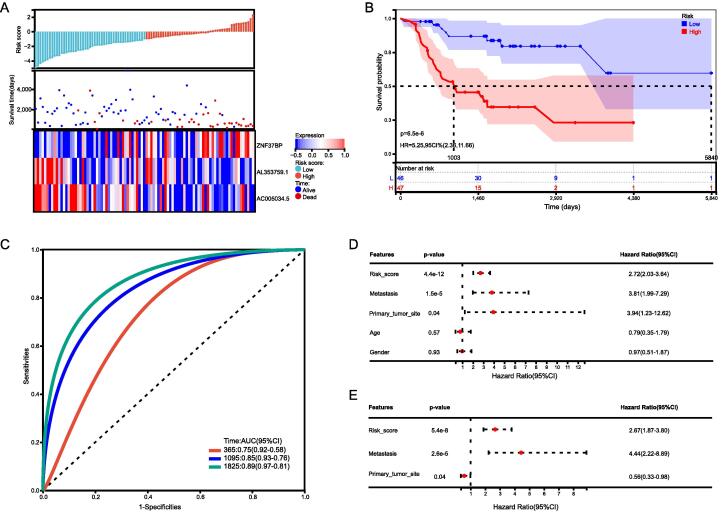
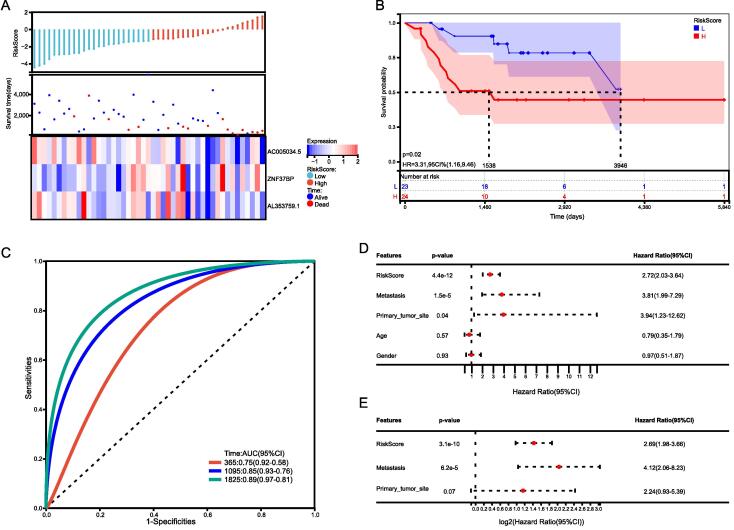
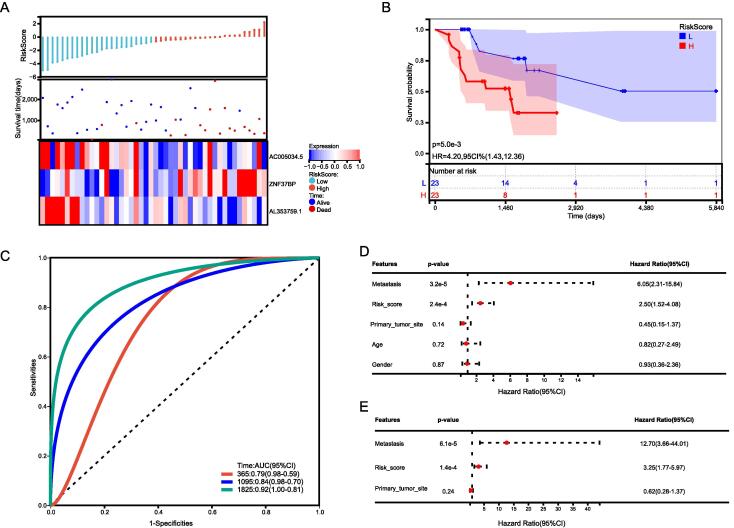
Using three CRLncs, we compared the expression, distribution of risk scores, and survival status between two risk groups in the total group, the training group, and the test group, to assess the predictive power of the risk model. The established risk model successfully classified the patients into high-risk and low-risk groups. We observed a significant decrease in survival as the risk score increased; patients in the low-risk group had better overall survival than those in the high-risk group (Fig. 4, Fig. 5, Fig. 6). As expected, AC005034.5 and AL353759.1 are protective factors that tended to be downregulated as the risk score increased. ZNF37BP was a risk factor, and the expression showed an upregulated trend with the increase in risk score.
Fig. 4.
Total sample group. (A) Distribution of survival status and risk score and the heatmap illustrating the expression of the three candidate CRLncs in the high- and low-risk groups. (B) Survival curve of the osteosarcoma patients in the two groups, (C) the time-dependent ROC curve of the risk model. (D) UniCox regression analysis (E) and multiCox regression analysis of the clinicopathological characteristics in the total sample cohort.
Fig. 5.
Training group. (A) Distribution of survival status and risk score and the heatmap of the three candidate CRLncs. (B) K-M plot in the two groups. (C) ROC plot of the risk model. (D) UniCox regression analysis (E) and multiCox regression analysis of the clinicopathological characteristics in the training cohort.
Fig. 6.
Test group. (A) Distribution of survival status and risk score and the heatmap of the three candidate CRLncs. (B) K-M plot in the two groups. (C) ROC plot of the risk model. (D) UniCox regression analysis (E) and multiCox regression analysis of the clinicopathological characteristics in the test cohort.
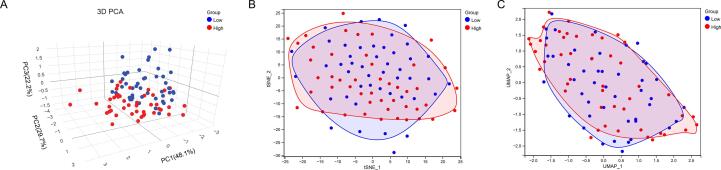
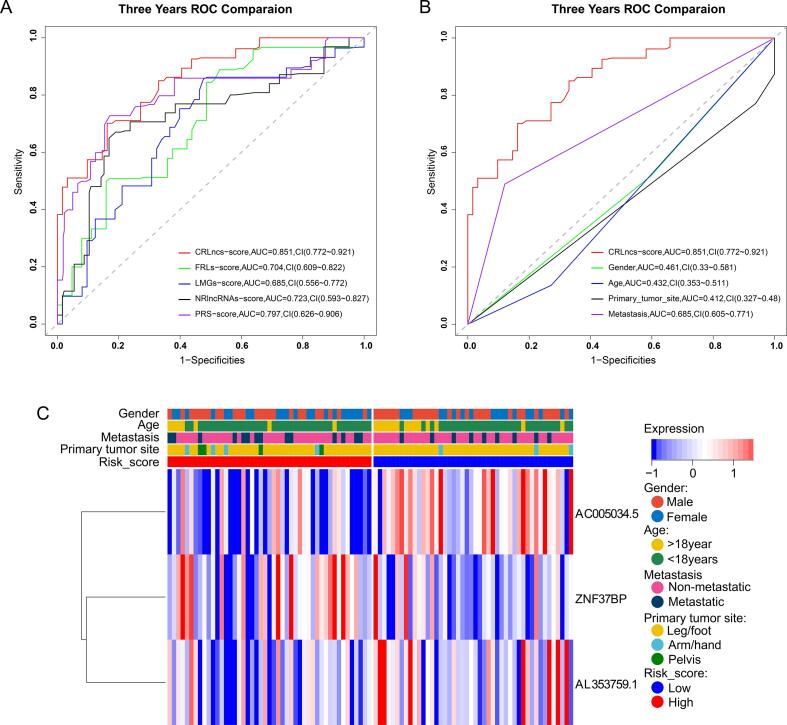
The PCA, t-SNE, and UMAP analyses revealed that all TARGET OS cohort patients were separated into clusters depending on high- and low-risk scores, illustrating the model's accuracy(Fig. 3A–C). We obtained the risk scores based on the risk score calculation formulas provided in the literature and compared the different receiver operating characteristic(ROC) curves for predicting OS survival in the TARGET-OS cohort. We found that our CRLncs score had a 3-year area under the curve (AUC) = 0.851, which was higher than FRLncs score (AUC = 0.704, P = 0.124, deLong's test), LMGs score (AUC = 0.685, P = 0.074, deLong's test), NRLncs score (AUC = 0.723, P = 0.009, deLong's test), PRS score (AUC = 0.797, P = 0.299, deLong's test) (Fig. 7A). We assessed the quality of each five models by the Akaike information criterion (AIC), and with the AIC values of 251.0236(the lowest value of our CRLncs score), 282.7302 (FRLncs score), 284.6824(the highest value of LMGs score), 279.9217(NRLncs score), and 265.0673(PRS score). Also, the improvement in the prediction of prognostic outcomes was evaluated by calculating the net reclassification index(NRI). As a whole, our CRLncs model yielded an NRI of 64.4 % (compared to the FRLncs model), 55.8 %(compared to the LMGs model), 46.3 %(compared to the NRLncs model), and 34.2 %(compared to the PRS model). The NRI statistic suggests that our CRLncs model better predicts OS survival than the FRLncs score, LMGs score, NRLncs score, and PRS score models.
Fig. 3.
(A-C) The PCA, t-SNE, and UMAP analyses revealed that all TARGET OS cohort patients were separated into clusters depending on high- and low-risk scores, illustrating the model's accuracy.
Fig. 7.
Three years ROC comparison of five prognostic risk models and clinicopathological characteristics in the TARGET-OS cohort. (A-B) Comparison ROC plot of five prognostic risk models and clinicopathological characteristics. (C) A heatmap plot of three CRLncs expression levels and a correlation between CRLncs scores and clinicopathological factors.
In comparing the survival rates of high-risk and low-risk patients, the Kaplan–Meier survival analysis revealed a relatively high survival rate in low-risk patients in the total sample group, training group, and test group (Fig. 4, Fig. 5, Fig. 6). In the total sample group, the AUCs at one year were 0.76, 3 years were 0.84, and 5 years were 0.89 (Fig. 4C). ROC curves for the training group showed a higher AUC at one year (0.76), three years (0.84), and five years (0.89) (Fig. 5C). ROC curves for the test group had higher AUCs at one year (AUC = 0.79), three years (AUC = 0.84), and five years (AUC = 0.92) (Fig. 6C).
3.3. Relationship between the CRLncs signature and the clinicopathological characteristics
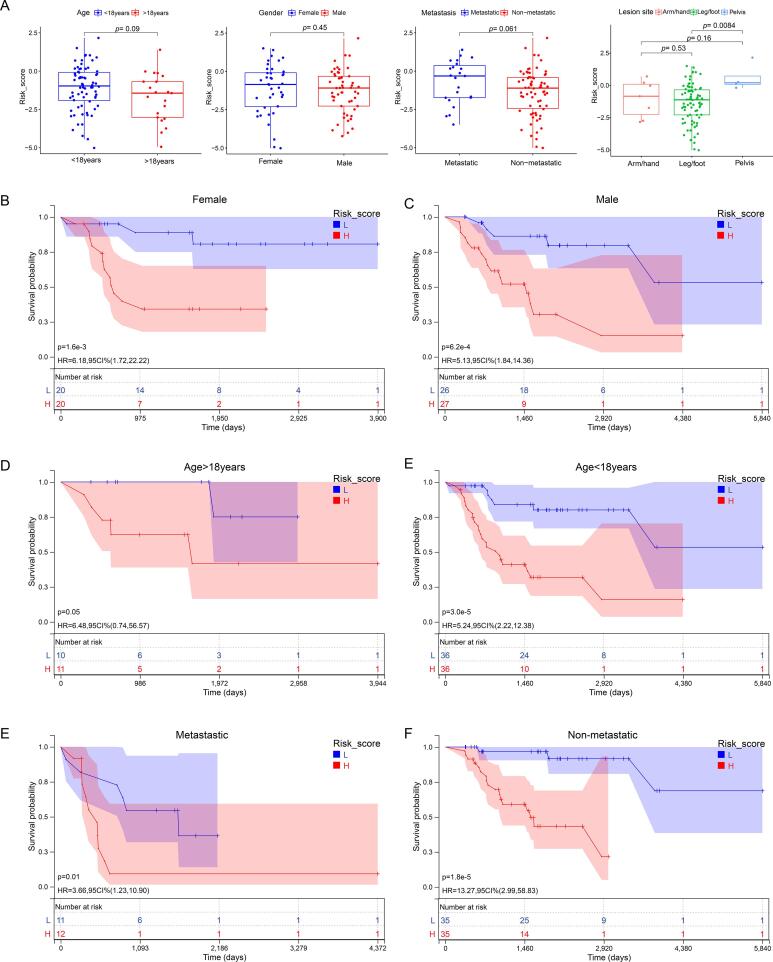
A heatmap was used to illustrate the relationships between the clinicopathological characteristics of the two risk subgroups (Fig. 7C). There was no significant difference between patients of different ages, sexes, primary tumor sites, or metastatic status in terms of risk score, indicating no association between risk score and clinical characteristics (Fig. 8A). However, in different sex, age, and metastasis subgroups, patients with high-risk scores had worse OS than patients with low-risk scores (Fig. 8B–F).
Fig. 8.
Association of risk score and clinical characteristics. (A) No significant difference was identified in OS patients with different ages, sex, primary tumor sites, and metastases. (B-F) Independence analysis and K-M plot of the CRLncs model in different sex(B, C), age(D, E), and metastasis(F, G) subgroups.
We used univariate and multivariate Cox regression analyses to test the independent prognostic value of CRLncs-scores under the influence of other clinical factors. As shown in Fig. 4, Fig. 5, Fig. 6, CRLncs-score and metastasis were independent prognostic indicators for patients with osteosarcoma. The ROC curves show that the CRLncs-score(AUC = 0.851) has a higher prediction accuracy than age(AUC = 0.0.461, P = 1.249e-07, deLong's test), gender(AUC = 0.461, P = 8.384e-07, deLong's test), primary tumor site(AUC = 0.412, P = 2.449e-07, deLong's test) and metastasis(AUC = 0.685, P = 0.008474, deLong's test), as shown in Fig. 7B. As a result of these findings, the CRLncs risk model appears to have high predictive value in the TARGET cohort.
3.4. Identification of the composite prognostic nomogram
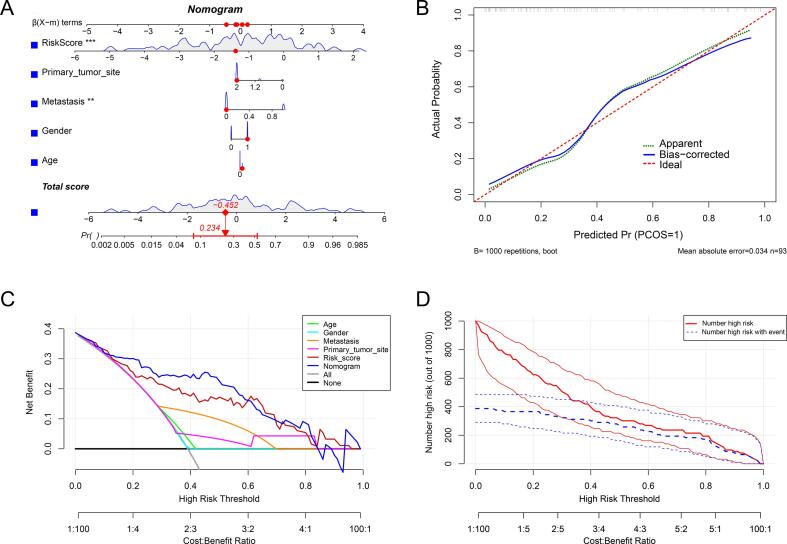
We integrated the CRLncs-score with clinical variables to improve the accuracy of predicting the prognosis of osteosarcoma patients. As shown in(Fig. 9A), the composite nomogram improved significantly in assessing survival relative to the clinical model. Then, the calibration curve was used to evaluate the predictive power of the nomogram model. The calibration curve indicated that the error between the actual risk and the predicted risk is minimal, suggesting the nomogram model owns high accuracy in predicting OS (Fig. 9B). Decision curve analysis (DCA) indicated that the “nomogram” curve was higher than the gray line, “Age” curve, “Gender” curve, “ Metastasis ” curve, and “CRLncs-score” curve, suggesting that the patients could benefit from the nomogram model at a high-risk threshold from 0 to 1, and the clinical benefit of the nomogram model was higher than the “ Age ” “ Gender ” “ Metastasis, ”and “ CRLncs-score ” curve (Fig. 9C). The clinical impact curve on the ground of the DCA curve shows the “Number high risk” curve was close to the “Number high risk with event” curve at the high-risk threshold from 0.3 to 1, which indicated that the nomogram model has an excellent predictive power (Fig. 9D).
Fig. 9.
Construction and calibration of the nomogram. (A) Nomogram integrated the risk score and clinical characteristics. (B) The calibration curve to assess the predictive power of the nomogram model. (C) DCA curve to evaluate the clinical value of the nomogram model. (D) Clinical impact curve based on the DCA curve to assess the nomogram model.
3.5. Functional analysis of DEGs based on CRLncs-score
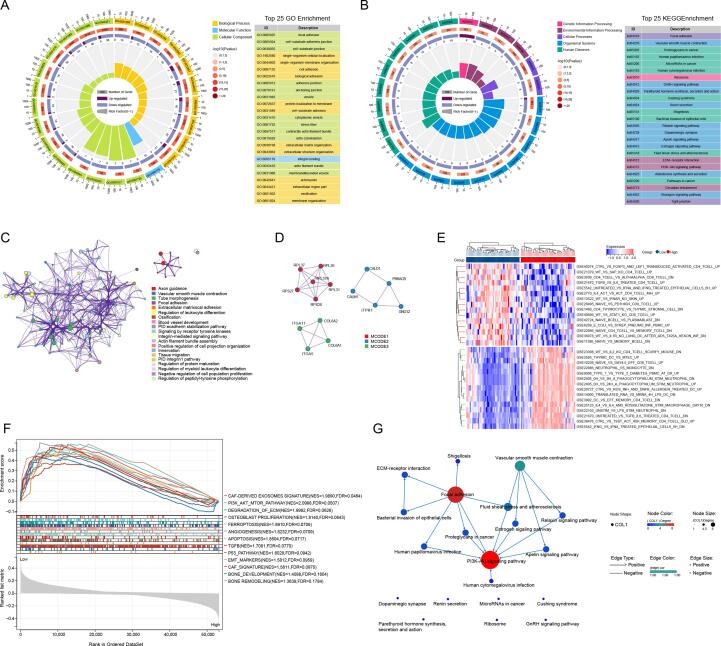
To further explore and investigate the underlying biological processes and signaling pathways, we identified a total of 167 DEGs by the cut-off of log2|FC| > 1.5 and FDR < 0.05, of which 129 genes were down-regulated, and 38 genes were upregulated in the TARGET OS cohort between two CRLncs risk score subgroups. GO analysis revealed that the 167 DEGs were mainly involved in focal adhesion, extracellular matrix organization, blood vessel development, ossification, and regulation of leukocyte differentiation (Fig. 10A). In the KEGG pathway analysis, these DEGs were primarily associated with pathways in cancer, ECM-receptor interaction, PI3K-Akt signaling pathway, tight junction, and microRNAs in cancer (Fig. 10B). The PPI analysis further screened three submodels, mainly involved in ECM-receptor interaction, immune response, and tumor development(Fig. 10 C-D). Additionally, we constructed the KEGG pathway interaction network to reveal DEGs' hierarchical and connectivity relations. As shown in (Fig. 10F), the PI3K-Akt signaling pathway, focal adhesion, ECM-receptor interaction, and vascular smooth muscle contraction could be the critical biological processes that affect the prognosis of osteosarcoma patients. As indicated, these DEGs were closely associated with immunity disorders in osteosarcoma patients, which may be the underlying mechanism for predicting the prognosis of osteosarcoma patients.
Fig. 10.
Functional analysis of DEGs based on CRLncs-score. The circular plot and visualization of the biological process were enriched by GO analysis(A), and the signaling pathways were enriched by KEGG analysis(B). The PPI network construction(C, D) and heatmap illustrating the GSVA analysis (E) result of DEGs were shown. (F) GSEA analysis visualization. (G) KEGG pathway interaction network revealed the hierarchical and connectivity relations of the DEGs'.
3.6. Differences in the tumor microenvironment
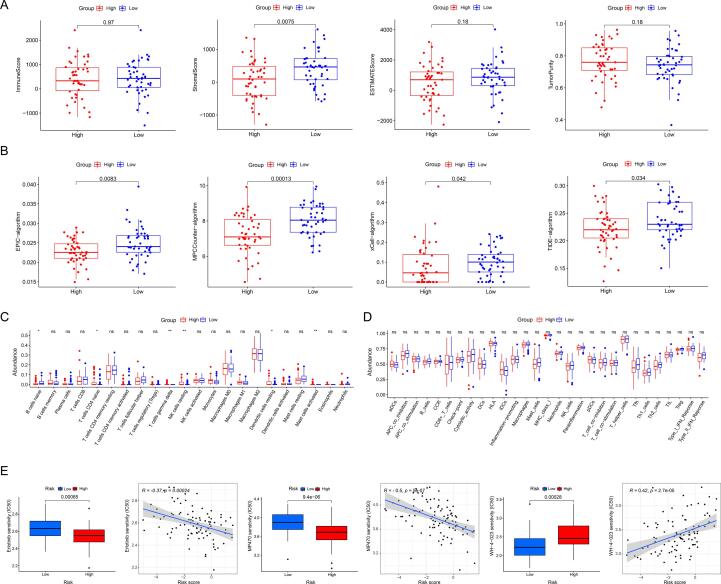
The ESTIMATE algorithm showed that the immune score(p = 0.97), estimate score(p = 0.18), and tumor purity (p = 0.18) did not differ significantly between the two subgroups. In contrast, the stroma score(p = 0.0075) was lower in the high-risk group(Fig. 11A). With the MCPCounter, xCELL, EPIC, and TIDE algorithms, we revealed that the abundance of fibroblasts or CAFs significantly differed between the two subgroups (Fig. 11B). The difference in immune cell infiltration was calculated using CIBERSORT and ssGSEA algorithms (Supplementary Fig. 1). We observed that the two groups significantly differed between B cells naive, T cells CD4 naive, T cells gamma delta, NK cells resting, dendritic cells resting, and mast cells activated. Notably, the immune cells and function did not differ significantly between the two types(Fig. 11C, D). Moreover, the subsequent GSVA and GSEA analysis showed that the CAF-derived exosomes signature, PI3K-AKT-MTOR pathway, degradation of ECM, osteoblast proliferation, TGF-β, bone development, and bone remodeling were enriched (Fig. 10E, F).
Fig. 11.
TME and TIME analysis based on CRLncs-score. (A) Differential analysis of tumor microenvironment by ESTIMATE algorithm. (B). Inferred CAF proportion scores of OS patients by four algorithms (see Methods). EPIC, MCP-Counter, xCell, and TIDE indicated the assumed CAF proportion. The stroma score (A) and CAFs(B) were significantly higher in the low-risk group. Immune cell(C) and immune function(D) differential analysis were shown using ssGSEA algorithms. (E) Selecting suitable targeting drugs for different risk- groups of patients.
3.7. Drugs with potential efficacy in OS
A sensitivity analysis revealed significant sensitivity to erlotinib(EGFR inhibitor, p = 0.00065), MP470(multitarget tyrosine kinase inhibitors, p = 9.4e − 06), and WH40222(potent and selective Lck and Src inhibitor, p = 0.00028) in both high- and low-risk populations. Patients with low risk responded better to erlotinib and MP470, while those with high risk responded better to WH4-023. (Fig. 11E). The predicted sensitive drugs were closely correlated to the above-enriched pathways. Our results demonstrate that the CRLncs-score can be used to predict chemotherapy drug sensitivity in osteosarcoma patients.
3.8. Expression of CRGs and CRLncs validation in OS
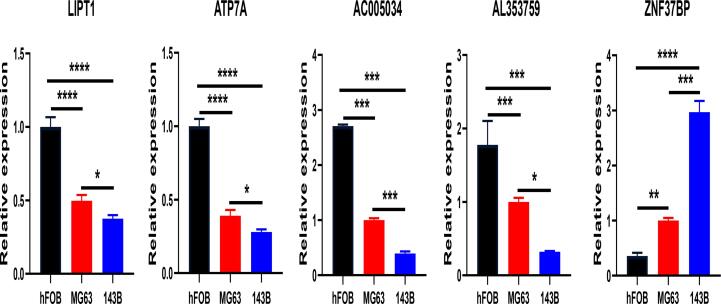
We selected three OS cell lines to assess the expression levels of CRGs and CRLncs. The control group was the normal osteoblast hFOB1.19, the MG63 was used as a non-metastatic OS cell, and 143B was used as a metastatic OS cell. The expression levels of ATP7A, LIPT1, AL353759.1, and AC005034.5 were significantly lower in MG63 and 143B cells than in normal osteoblast hFOB1.19 cells. Conversely, ZNF37BP expression levels were significantly increased in MG63 and 143B cell lines (Fig. 12).
Fig. 12.
Q-PCR analysis for the expression of CRGs and CRLncs in osteoblast hFOB1.19 and OS cell lines. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, each experiment was repeated three times.
4. Discussion
Osteosarcoma is the most common primary pediatric and adolescent bone malignancy [1]. This type of cancer is characterized by a high mortality rate, especially for patients with resistant lung metastases. In osteosarcoma, the bone microenvironment plays a crucial role in its onset and development [15]. Increasing evidence suggests that the bone microenvironment affects osteosarcoma metastasis [16]. CAFs are essential stromal components of the TME that regulate the antitumor activity of tumor-infiltrating immune cells, including innate and adaptive immune cells [17]. Many studies have shown that interactions between immune cells and other immune components can modulate the tumor immune microenvironment(TIME) and thus inhibit the antitumor immune response [18]. Although CAFs are thought to be associated with osteosarcoma TME, research is lacking [19], [20], [21]. Consequently, a novel marker associated with CAFs characteristics of OS is urgently needed for predicting outcomes and assisting in patient management.
Copper is an essential nutrient for many cellular functions. Multiple proteins are also regulated by it allosterically in many signaling pathways. Recently, researchers have found that tumors require a higher level of copper than healthy tissues [15]. And a copper-dependent cell death known as “cuproptosis” was recently reported by Tsvetkov et al. [5]. Excess copper in the cell binds to lipoylated components of the TCA cycle, triggering proteotoxic stress and death of the cells. It provides new insights into regulating intracellular copper levels in cancer treatment. In a recent study, copper-targeting drugs may help improve anti-cancer immunotherapies. LncRNAs are crucial to osteosarcoma development, progression, and invasion [22]. Deregulated expression of lncRNAs has been found to participate in the regulation of various signaling transduction pathways in osteosarcoma [23]. In light of this, we investigate the correlation between the tumor microenvironment, immune microenvironment, and CRLncs to improve prognosis and treatment for OS.
The current study focused on the relationships between CRLncs, CAFs, the immune microenvironment, chemotherapy sensitivity, and prognosis in osteosarcomas. Using univariate Cox regression, Lasso Cox regression, and multivariate Cox regression, we developed a unique CRLncs risk model including three lncRNAs(ZNF37BP, AC005034.5, and AL353759.1). The CRLncs model appeared to be a reliable predictor of OS patients' prognosis. A high-risk score was indicative of a poorer prognosis than a low-risk score. Many risk models have been developed for OS currently, such as necroptosis-associated lncRNA signature [24]), ferroptosis-related lncRNA signature [25], lipid-metabolism gene signature [26], and pyroptosis-related gene signature [27], however, our study arrived at a unique conclusion compared to the above studies. The results of our 3-year ROC analysis (AUC = 0.84) confirmed that the signature could have accurate and sensitive predictive efficacy. Furthermore, we calculated and compared the AIC values and NRIs, which allowed us to comprehensively compare the predictive effectiveness of different models. Our CRLncs model exhibited the lowest AIC value and yielded an increase in NRI. In multivariate cox regression analysis combined with clinical characteristics, both the CRLncs risk score and tumor metastasis were independent prognostic factors for the overall survival of osteosarcoma patients in both the training and validation sets. We constructed a nomogram including clinicopathological variables and risk scores. We found that the CRLncs risk score was the highest weighted score in this nomogram, followed by metastasis. The nomogram showed perfect agreement between observed and predicted rates for 1-, 3-, and 5-year overall survival. The CRLncs risk score has only three lncRNAs included; however, it has a higher and more stable predictive capacity; we believe it is more likely to be applied to clinical practice.
To further evaluate how the CRLncs regulates OS, we then compared the gene expression levels between high-risk and low-risk CRLncs scores. A total of 167 essential genes were identified. GO, KEGG and PPI analysis of the identified DEGs implied that dysregulation of the extracellular matrix organization, immunity, and bone remodeling might mediate tumorigenesis and progression of osteosarcoma by cuproptosis-related lncRNAs. Additionally, the KEGG pathway interaction network result revealed that the PI3K-Akt signaling pathway, focal adhesion, ECM-receptor interaction, and vascular smooth muscle contraction might be the critical biological processes that affect the prognosis of osteosarcoma patients. Further, the subsequent GSVA and GSEA analysis showed that the CAF-derived exosomes signature, the PI3K-Akt signaling pathway, degradation of ECM, osteoblast proliferation, angiogenesis, TGF-β, bone development, and bone remodeling ranks high in the low-risk score group. Synthesizing the above findings, we could reasonably assume that the PI3K-Akt signaling pathway may be the central pathway associated with osteosarcoma development, and the dysregulation crosstalk between the cancer-associated fibroblasts and immune cells in the TME resulted in the impairment of TIME and bone remodeling, thereby leading to a poor prognosis in osteosarcoma.
The heterogeneity of the TME plays an essential role in tumor development, including infiltrating immune cells, extracellular matrix, tumor purity, and noncellular components [3]. A variety number of mechanisms are involved in how CAFs promote the growth and invasion of cancer cells in the tumor microenvironment [28]. Consequently, we studied the role of the CRLncs risk score in predicting the CAFs and immune microenvironment landscape of osteosarcoma. The ESTIMATE algorithm showed that neither immune score, estimate score, nor tumor purity significantly differed between the two subgroups. However, our investigation revealed that the stroma score and the abundance of fibroblasts or CAFs were significantly higher in the low-risk group. Using the CIBERSORT algorithm, we revealed a higher level of tumor infiltration in the high-risk group for T cells CD4 naive, T cells gamma delta, NK cells resting, dendritic cells resting, and mast cells activated. However, the ssGSEA algorithm did not show significant differences in immune cells or function between the two subgroups. Combined with the functional and previous survival analyses, we could reasonably assume that CAFs may contribute to the worse prognosis among osteosarcoma patients.
The combination of modern surgery and systemic chemotherapy has improved osteosarcoma treatment dramatically, but survival has not changed significantly [29]). Apoptotic escape and chemoresistance have been a concern in osteosarcoma [30]. Oncologists have seen better outcomes with targeted chemotherapy instead of cytotoxic chemotherapy. Target inhibitors, like erlotinib and MP470, inhibit the growth of various cancer cell lines. Researchers found that a combination of MP470 and erlotinib inhibits tumor growth by inhibiting the HER family/PI3K/Akt pathway. However, there is little evidence of experience with these two drugs in patients with osteosarcoma. Several new Src inhibitors have been introduced to combat OS; however, the results seem promising [31]. Erlotinib and MP470 were more sensitive in the low-risk group in this study; patients with a high-risk score, however, were more susceptible to WH − 4–023. This study suggests that erlotinib, MP470, and WH − 4–023 may be potential therapeutic drugs for OS and provide a direction for further research.
Finally, it was experimentally verified in vitro that the expression levels of ATP7A, LIPT1, AL353759.1, and AC005034.5 were significantly down-regulated in MG63 and 143B OS cells than in normal osteoblast hFOB1.19 cells. On the contrary, the expression level of ZNF37BP was elevated in OS cells than in normal osteoblast cells. Furthermore, we have found that the expression levels of ATP7A, LIPT1, AL353759.1, AC005034.5, and ZNF37BP were significant between MG63 and 143B OS cells. Based on the above findings, we could reasonably assume that these five genes were associated with tumor progress and prognosis. To a certain extent, this corresponds to the reliability of our bioinformatics analysis.
This study has some limitations. First, the mechanisms underlying CRLncRNAs in OS remain unclear and need further investigation. Despite having been internally validated by the whole group, the training group, and the testing group, the external validation was not performed in other databases due to a lack of lncRNA expression profiles and OS datasets. Thirdly, the expression levels of two CRGs and three CRLncs were validated by qRT–PCR in two OS cell lines and one normal osteoblast hFOB1.19 cell; more samples, more OS cell lines would be helpful for a more solid analysis of the evidence. Further research is needed to verify these lncRNAs' prognostic utility in animals and clinical trials.
5. Conclusion
Three cuproptosis-related lncRNAs were used to construct a robust prognostic, predictive model, which showed remarkable prognostic value for OS patients. Based on the tumor microenvironment and immune-related analyses, these three CRLncs can guide the CAFs of OS, and three potential OS-targeted drugs were identified as well. Our study offers valuable insight into predicting the OS patients' prognoses and may even assist their treatment.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This study is financially supported by the Natural Science Foundation of Guangdong Guangdong Province (Grant no. 2022A1515010293).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbo.2022.100463.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zhao X., Wu Q., Gong X., Liu J., Ma Y. Osteosarcoma: a review of current and future therapeutic approaches. Biomed. Eng. 2021;20(1):24. doi: 10.1186/s12938-021-00860-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gill J., Gorlick R. Advancing therapy for osteosarcoma. Nat. Rev. Clin. Oncol. 2021;18(10):609–624. doi: 10.1038/s41571-021-00519-8. [DOI] [PubMed] [Google Scholar]
- 3.Ugel S., Cane S., De Sanctis F., Bronte V. Monocytes in the tumor microenvironment. Annu. Rev. Pathol. 2021;16:93–122. doi: 10.1146/annurev-pathmechdis-012418-013058. [DOI] [PubMed] [Google Scholar]
- 4.Cangkrama M., Liu H., Whipman J., Zubair M., Matsushita M., Di Filippo M., et al. A protumorigenic mDia2-MIRO1 axis controls mitochondrial positioning and function in cancer-associated fibroblasts. Cancer Res. 2022;82(20):3701–3717. doi: 10.1158/0008-5472.CAN-22-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsvetkov P., Coy S., Petrova B., Dreishpoon M., Verma A., Abdusamad M., et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375(6586):1254–1261. doi: 10.1126/science.abf0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S.R., Bu L.L., Cai L. Cuproptosis: lipoylated TCA cycle proteins-mediated novel cell death pathway. Signal Transduct. Target. Ther. 2022;7(1):158. doi: 10.1038/s41392-022-01014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen X., Zhang X., Chen J., Yang Q., Yang L., Xu D., et al. Hinokitiol copper complex inhibits proteasomal deubiquitination and induces paraptosis-like cell death in human cancer cells. Eur. J. Pharmacol. 2017;815:147–155. doi: 10.1016/j.ejphar.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Veiga N., Alvarez N., Castellano E.E., Ellena J., Facchin G., Torre M.H. Comparative study of antioxidant and pro-oxidant properties of homoleptic and heteroleptic copper complexes with amino acids, dipeptides and 1,10-phenanthroline: the quest for antitumor compounds. Molecules. 2021;26(21) doi: 10.3390/molecules26216520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park E.G., Pyo S.J., Cui Y., Yoon S.H., Nam J.W. Tumor immune microenvironment lncRNAs. Brief. Bioinform. 2022;23(1) doi: 10.1093/bib/bbab504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24(10):1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geeleher P., Cox N., Huang R.S. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9):e107468. doi: 10.1371/journal.pone.0107468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunson J.C. Ggalluvial: layered grammar for alluvial plots. J. Open Source Softw. 2020;5(49):2017. doi: 10.21105/joss.02017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blockhuys S., Wittung-Stafshede P. Roles of copper-binding proteins in breast cancer. Int. J. Mol. Sci. 2017;18(4) doi: 10.3390/ijms18040871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang C., Tian Y., Zhao F., Chen Z., Su P., Li Y., et al. Bone microenvironment and osteosarcoma metastasis. Int. J. Mol. Sci. 2020;21(19) doi: 10.3390/ijms21196985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao X., Xu J., Wang W., Liang C., Hua J., Liu J., et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol. Cancer. 2021;20(1):1–30. doi: 10.1186/s12943-021-01428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binnewies M., Roberts E.W., Kersten K., Chan V., Fearon D.F., Merad M., et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corre I., Verrecchia F., Crenn V., Redini F., Trichet V. The osteosarcoma microenvironment: a complex but targetable ecosystem. Cells. 2020;9(4) doi: 10.3390/cells9040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahai E., Astsaturov I., Cukierman E., DeNardo D.G., Egeblad M., Evans R.M., et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer. 2020;20(3):174–186. doi: 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X., Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov. 2019;18(2):99–115. doi: 10.1038/s41573-018-0004-1. [DOI] [PubMed] [Google Scholar]
- 22.Ghafouri-Fard S, Shirvani-Farsani Z, Hussen BM, Taheri M. The critical roles of lncRNAs in the development of osteosarcoma. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2021;135:111217. [DOI] [PubMed]
- 23.Liu H., Zong C., Sun J., Li H., Qin G., Wang X., et al. Bioinformatics analysis of lncRNAs in the occurrence and development of osteosarcoma. Transl. Pediatr. 2022;11(7):1182–1198. doi: 10.21037/tp-22-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G., Zhang X., Feng W., Wang J. Prediction of PROGNOSIS AND IMMUNOTHERAPY OF OSTEOSARCOMA BASED ON NECROPTOSIS-RELATEd lncRNAs. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.917935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., He R., Lei X., Mao L., Yin Z., Zhong X., et al. Comprehensive analysis of a ferroptosis-related lncRNA signature for predicting prognosis and immune landscape in osteosarcoma. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.880459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian H., Lei T., Hu Y., Lei P. Expression of lipid-metabolism genes is correlated with immune microenvironment and predicts prognosis in osteosarcoma. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.673827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., He R., Lei X., Mao L., Jiang P., Ni C., et al. A novel pyroptosis-related signature for predicting prognosis and indicating immune microenvironment features in osteosarcoma. Front. Genet. 2021;12 doi: 10.3389/fgene.2021.780780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park D., Sahai E., Rullan A. SnapShot: cancer-associated fibroblasts. Cell. 2020;181(2):486–e1. doi: 10.1016/j.cell.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Chen C., Xie L., Ren T., Huang Y., Xu J., Guo W. Immunotherapy for osteosarcoma: Fundamental mechanism, rationale, and recent breakthroughs. Cancer Lett. 2021;500:1–10. doi: 10.1016/j.canlet.2020.12.024. [DOI] [PubMed] [Google Scholar]
- 30.Fernandes I., Melo-Alvim C., Lopes-Bras R., Esperanca-Martins M., Costa L. Osteosarcoma pathogenesis leads the way to new target treatments. Int. J. Mol. Sci. 2021;22(2) doi: 10.3390/ijms22020813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akiyama T., Dass C.R., Choong P.F. Novel therapeutic strategy for osteosarcoma targeting osteoclast differentiation, bone-resorbing activity, and apoptosis pathway. Mol. Cancer Ther. 2008;7(11):3461–3469. doi: 10.1158/1535-7163.MCT-08-0530. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.