Abstract
Atherosclerosis is a progressive cardiovascular disease in which cholesterol-rich plaques build up within arteries, increasing the risk of thrombosis, myocardial infarction, and stroke. One promising therapeutic approach is the use of high-density lipoprotein (HDL) biomimetic formulations based on ApoAI peptides that promote cholesterol efflux from plaques, ultimately leading to cholesterol excretion. Here, we describe the multivalent display of ApoAI peptides on the surface of protein nanotubes derived from the plant virus tobacco mosaic virus (TMV) and protein nanoparticles using virus-like particles from bacteriophage Qβ. Bioconjugation yielded ApoAI conjugates varying in size and morphology. We tested ABCA1-mediated cholesterol efflux using macrophage foam cells, the mitigation of reactive oxygen species in endothelial cells, and wound healing in endothelial cells. We found that the multivalent ApoAI platform, in particular the TMV-based nanotube, significantly improved the efficacy of cholesterol efflux compared to free peptides, Qβ nanoparticle formulations, and traditional HDL therapy. Finally, to better understand the mechanistic basis of enhanced cholesterol efflux, we used confocal microscopy to show that while native TMV was taken up by cells, TMV-ApoAI remained at the exterior of foam cell membranes and efflux was documented using fluorescent cholesterol. Together, these data highlight that high aspect ratio materials with multivalent display of ApoAI peptides offer unique capabilities promoting efficient cholesterol efflux and may find applications in cardiovascular therapy.
Graphical Abstract

INTRODUCTION
Atherosclerosis is a progressive cardiovascular disease (CVD) in which excess cholesterol accumulates within the arterial intima as plaques.1,2 Late-stage CVD may involve thrombosis and plaque rupture, resulting in myocardial infarction or stroke, which are the two leading causes of death worldwide.3,4 Furthermore, patients who recover from thrombotic complications or stroke can suffer long-term heart, brain, or other organ damage, the severity of which is directly related to the duration of oxygen deprivation.
The molecular basis of atherosclerotic progression is still unclear, but genetic predisposition and lifestyle can promote inflammation, which triggers endothelial cell dysfunction, proteoglycan remodeling, and a homeostatic imbalance between cholesterol deposition and cholesterol removal in the arteries.5–7 Cholesterol is mainly delivered to cells by low-density lipoprotein (LDL) via LDL receptor-mediated endocytosis and for macrophages primarily via scavenger receptor AI/AII. Cholesterol deposition and accumulation is regulated by its removal or “efflux” by high-density lipoprotein (HDL). One of the main components of HDL is apolipoprotein AI (ApoAI), which interacts with adenosine 5′-triphosphate (ATP)-binding cassette transporter A1 (ABCA1) and scavenger receptor class B type I (SR-BI) to facilitate cholesterol efflux from cholesteryl ester-loaded foam cells. There is a well-documented inverse relationship between HDL and the risk of CVD.8 However, within diseased patient population, raising HDL levels does not necessarily decrease the risk of CVD, which highlights the importance of HDL functionality. Dysfunctional ApoAI and HDL populations within atherosclerotic lesions have been reported, caused by inflammation-induced oxidative damage; dysfunctional ApoAI was found to be lipid free in these plaque regions compared to healthy vessel tissue.9 The current treatment for atherosclerosis includes preemptive and reactive changes in lifestyle, as well as surgical interventions and cholesterol-lowering drugs.
Some cholesterol-lowering drugs work indirectly. Statins block the synthesis of cholesterol and therefore reduce the accumulation of LDL cholesterol in the blood, encouraging the efflux of cholesteryl ester from foam cells within the plaque. However, long-term statin use induces the upregulation of proprotein convertase subtilisin/kexin type 9 serine protease (PCSK9), an inhibitor of the LDL receptor in the liver, which is required for cholesterol uptake.10 The FDA recently approved siRNA-based gene therapy against PCSK9 to reduce levels of LDL cholesterol.11,12 Statins can be synergized with bile acid sequestrants that prevent the absorption of cholesterol-rich bile acids from the gut and force the liver to use cholesterol in the blood, but this approach bears significant side effects.13
Other cholesterol-lowering drugs act directly, using HDL to promote cholesterol efflux and thus mediate uptake of LDL particles by interacting with foam cell receptors (such as scavenger receptor class A type I and II and ABCA1 receptor). HDL was formulated in clinical trials by combining serum-derived ApoAI with different phospholipid compositions, but the results were only moderately successful. One potential explanation is the heterogeneous nature of serum-derived ApoAI from HDL donors.8,14–17 Indeed, HDL from CVD patients was less efficient in terms of cholesterol efflux, possibly reflecting the oxidation of the ApoAI protein and lipidome.9,18,19 Interestingly, more recent clinical studies have indicated that the relationship between HDL concentration and CVD mortality is U-shaped rather than linear, suggesting that the quality of HDL is more important than the quantity for lowering LDL cholesterol.20,21
HDL biomimetic formulations of a consistent quality have been prepared using recombinant full-length (243 amino acids) ApoAI and smaller biomimetic peptides containing an amphiphilic α-helix that is functionally involved in lipid binding and cholesterol efflux. In fact, recombinantly expressed ApoAI was tested in clinical trials with promising results; however, the significance of the result was limited due to the small patient sample size and expense of therapeutic production.22 The native ApoAI structure consists of 10 amphiphilic α-helices, so ApoAI peptides have also been reengineered into multimeric forms such as tandem or branched peptides to improve their cholesterol efflux capacity and cost-efficient production. ApoAI peptides have been delivered alone or as lipid nanoparticles formulated using dimyristoyl phosphatidylcholine.23,24 Specifically, ApoAI peptide 4F has been tested in phase I clinical trials, confirming its safety but revealing issues with bioavailability (Figure S1).25,26 Of note, 4F peptide has been demonstrated to enhance protection of ApoAI from oxidative damage and efflux in a synergistic fashion, which further motivates work using the ApoAI peptide approach.27 More recently, other formulations have been tested including combinations of lipids, inorganic, and nucleic acid-based nanoparticles that present ApoAI for targeted delivery.28–30 However in these examples, low-copy-number multimeric forms were engineered, but in biology high valency may increase efficacy. Therefore, we set out to develop nanoparticle formulations with high ApoAI copy number presented on nanoparticles vs nanotubes.
Specifically, we evaluated two viral nanocarriers differing in shape and size. First, bacteriophage Qβ (Qβ) is a 30 nm spherical virus-like particle (VLP) with 180 identical coat protein units arranged in T3 symmetry, allowing peptide conjugation to amine groups on the surface. Second, tobacco mosaic virus (TMV) is a rod-shaped plant virus nanoparticle (VNP) with a high aspect ratio (300 × 18 nm2) composed of 2130 coat protein units that self-assemble around a single plus-sense single-stranded RNA (Table S1). We used a TMV mutant with lysine in place of alanine at position 158 (TMV-Lys) to facilitate conjugation.31 We evaluated three biomimetic ApoAI peptides: 4FN and 4FC (based on the clinical 4F sequence with a cysteine added via an intervening GGGS linker for conjugation at the N or C terminus) as well as 23MJ (based on a preclinical multivalent formulation) (Table S2). Qβ was chosen as a model system for a nanoparticle, and TMV was chosen because we hypothesized that its high aspect ratio shape may confer unique advantages: We previously demonstrated that multivalent peptide display on TMV particles improves targeting to enhance atherosclerotic plaque imaging.32–36 Data indicate that TMV has favorable flow and margination properties that target the vessel wall. We therefore used TMV-ApoAI to probe the functional effects of multivalent ApoAI peptide display on cholesterol efflux with Qβ-ApoAI as a comparator to determine the effect of nanocarrier morphology. Our hypothesis was that multivalent TMV-ApoAI conjugates could be engineered to mimic native HDL and improve cholesterol efflux more effectively than HDL or free ApoAI peptides. We evaluated the mechanistic and functional quality of TMV-ApoAI conjugates in cholesterol-binding assays, cholesterol efflux assays, reactive oxygen species (ROS) assays, and wound healing assays.
RESULTS AND DISCUSSION
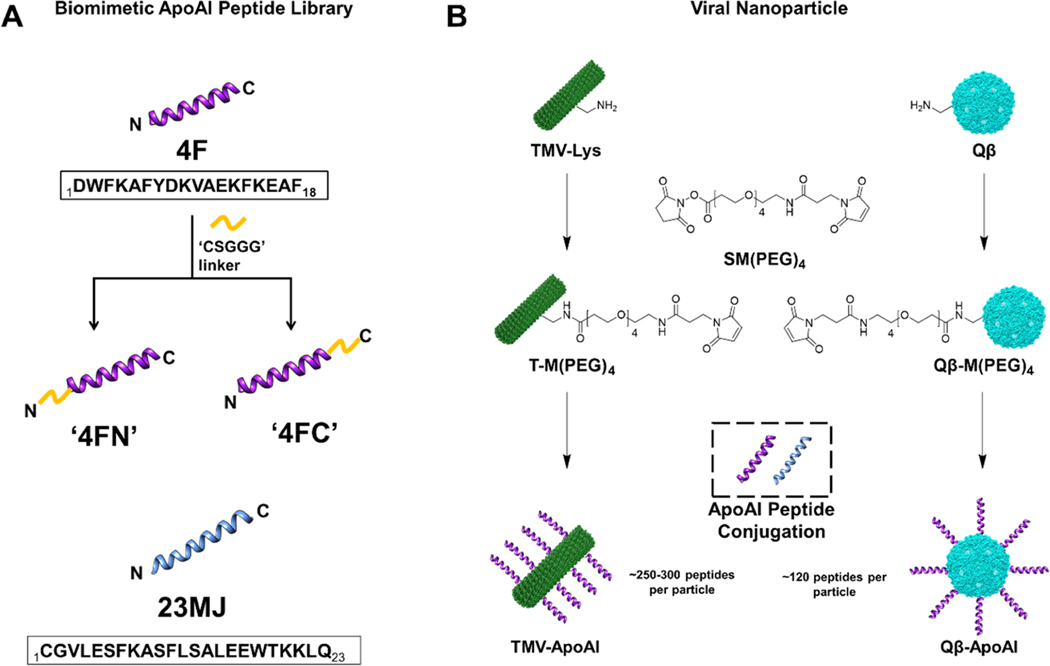
To investigate the efficacy of multivalent biomimetic ApoAI peptides displayed on the surface of viruses, we combined three model peptides (Table S1) with two carriers: a bacteriophage-derived VLP and a VNP based on a plant virus, the former serving as a model for a nanoparticle and the latter as a nanotube (Table S2). All three biomimetic ApoAI peptides are derived from functional α-helix regions (Figure S1) and have proven to be safe during clinical trials (4F) or efficacious in terms of cholesterol efflux during preclinical development (23MJ). The 4F peptide was synthesized with a CSGGG linker to attach either the N terminus (4FN) or C terminus (4FC) to the carrier, allowing us to investigate the implications of peptide orientation, whereas the 23MJ peptide contained a native N-terminal cysteine which removed the need for a linker (Figure 1A).
Figure 1.
Preparation of TMV-ApoAI and Qβ-ApoAI conjugates. (A) Biomimetic ApoAI peptides. (B) Bioconjugation of ApoAI peptides to TMV and Qβ using maleimide–thiol chemistry. TMV and Qβ were modeled in Chimera software (PDB ID: 2TMV and 1QBE, respectively). VLPs/VNPs and peptides are not drawn to scale.
Peptide conjugates (Figure 1B) were synthesized in a two-part reaction. First, heterobifunctional linker SM(PEG)4 was reacted with the VLPs/VNPs to form the maleimide intermediates T-M(PEG)4 and Qβ-M(PEG)4. Second, each intermediate was then reacted with the three ApoAI peptides and the final particles termed TMV-ApoAI and Qβ-ApoAI were purified by ultracentrifugation.
The conjugation of ApoAI peptides to TMV was confirmed by SDS-PAGE, which revealed the presence of ∼20 kDa bands in addition to ∼17.5 kDa bands corresponding to conjugated and unmodified TMV coat proteins, respectively (Figure 2A). The degree of modification was ∼12–14% as determined by densitometric analysis, agreeing with the ∼11–13% modification as determined using the bioanalyzer (Figure 2B). These values represent ∼250–300 peptides per TMV particle (Figure 2C). Dynamic light scattering (DLS) indicated a size increase for the modified TMV-ApoAI particles: TMV had a Zav of ∼203.1 nm (PDI = 0.230), a ∼1.3-fold size increase was observed for the ApoAI conjugates with a Zav of ∼255.8 nm (PDI = 0.266) for T-4FN, ∼258.7 nm (PDI = 0.223) for TMV-4FC, and ∼279.9 nm (PDI = 0.255) for TMV-23MJ (Figure 2D). Fast protein liquid chromatography (FPLC) data indicate that TMV-ApoAI peptide conjugates exhibited a typical elution profile of TMV at a retention volume of ∼7.5 mL along with a 260:280 nm absorbance ratio of ∼1.2 (Figure 2E). Additionally, the analysis of negatively stained TMV particles by TEM further confirms that the particle morphology remained intact after modification (Figure 2F). Together, these results confirm the bioconjugation of ApoAI peptides to TMV yielding intact TMV-ApoAI formulations.
Figure 2.
Characterization of TMV-ApoAI peptide conjugates. (A) SDS-PAGE with Coomassie blue (CB) stain: lane L (ladder), lane 1 unmodified TMV (TMV), lane 2 T-4FN conjugate (T-4FN), lane 3 TMV-4FC conjugate (T-4FC), and lane 4 TMV-23MJ conjugate (T-23MJ). (B) Bioanalyzer (BA) electrophoretic analysis. (C) Summarized degree of modification. (D) Dynamic light scattering intensity plots of TMV-ApoAI peptide conjugates (Zavg data are mean values, n = 3). (E) Fast protein liquid chromatograms of TMV nanoparticles; a 260:280 nm ratio at max absorption is ∼1.2. (F) TEM images of negatively stained TMV-ApoAI peptide conjugates (scale bars = 100 nm).
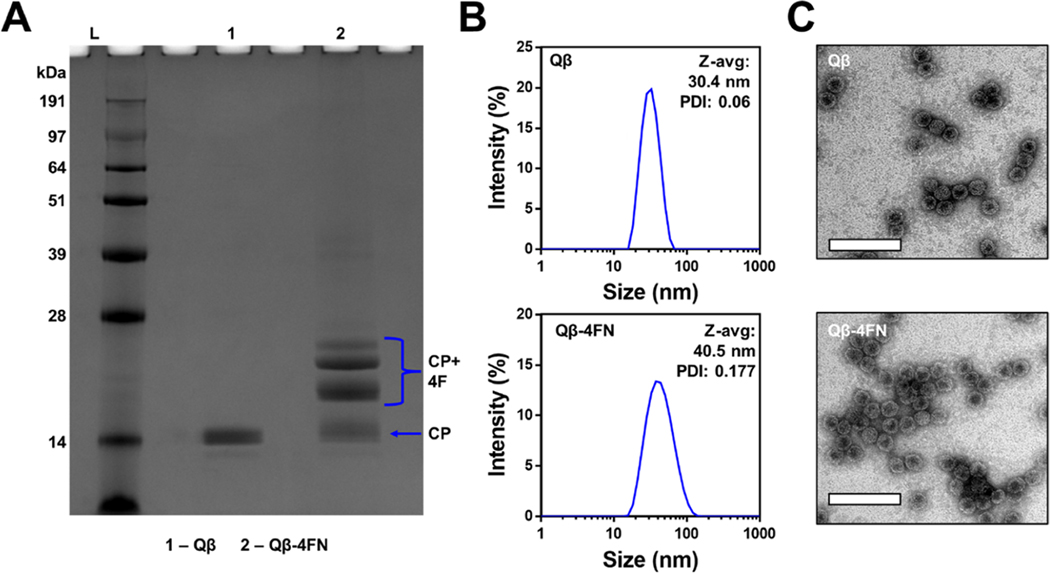
SDS-PAGE also confirmed the conjugation of Qβ to ApoAI peptide 4FN, revealing the presence of unmodified CP bands at ∼14.2 kDa plus multiple modified CP bands with higher molecular weights (Figure 3A). The degree of modification was ∼55–60% as determined by densitometric analysis. DLS indicated that the hydrodynamic diameter of Qβ−4FN was slightly larger (Zav ∼40.5 nm, PDI = 0.177) than that of unmodified Qβ (Zav ∼30.4 nm, PDI = 0.06) (Figure 3B). The structural integrity of the particles was confirmed by TEM (Figure 3C). We note that Qβ was modified only with 4FN; initial scouting experiments were performed with TMV and the three peptide candidates which revealed that 4FN was the most potent peptide (see below). Therefore, questions about the impact of the nanoparticle size/shape were only addressed using T-4FN vs Qβ−4FN. Synthesized TMV-ApoAI and Qβ-ApoAI conjugates were then employed in a cholesterol efflux assay (Figure 4A).
Figure 3.
Characterization of Qβ−4FN. (A) SDS-PAGE: lane L (ladder), lane 1 unmodified Qβ, and lane 2 Qβ−4FN. (B) Dynamic light scattering intensity plots of Qβ and Qβ−4FN (Zavg data are mean values, n = 3). (C) TEM images of negatively stained Qβ and Qβ−4FN (scale bars = 100 nm).
Figure 4.
Cholesterol efflux is improved by the multivalent display of ApoAI peptides on TMV and Qβ. (A) Schematic representation of the cholesterol efflux assay sequence. (B) Cholesterol efflux under upregulated ABCA1 conditions. Cholesterol efflux (%) = supernatant cholesterol/(supernatant cholesterol + cell lysate cholesterol) × 100. (C) Bar graph shows the efflux comparison between TMV-ApoAI conjugates and HDL (LO = particle concentration of 5.5 nM, HI = particle concentration of 0.5 μM). (D) Cholesterol efflux under homeostatic ABCA1 conditions. (E) Cholesterol efflux under homeostatic ABCA1 conditions following HI treatment with the free peptide corresponding to the peptide displayed on conjugates. (F) Bar graph shows the efflux comparison between TMV-ApoAI conjugates and corresponding free peptides at the HI particle concentration. Data are mean fluorescence intensity ± standard deviation (n = 3). Statistical analysis by one-way ANOVA with Tukey’s test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant).
Data indicate that T-4FN is a more potent cholesterol binder than TMV-4FC (∼2.5-fold), which suggests that cholesterol efflux using multivalent display of 4F peptide is highly orientational-dependent (Figure 4B,C). In addition, a spherical Qβ-ApoAI peptide conjugate does not display significant cholesterol efflux and is comparable to baseline BSA control group efflux. Therefore, a rod-shaped nanocarrier is a more suitable candidate for further investigation of multivalent peptide display effect. Structural work indicates that both the N and C termini of ApoAI peptide were involved in lipid and cholesterol binding. However, our findings demonstrate that potent efflux can still be observed in an ABCA1-dependent manner with the N terminus of the peptide reserved for conjugation (Figure 4B,D). Moreover, the terminus-specific efflux observed in our study suggest a more complex cholesterol-binding structure is at play, given that when an ABCA1 receptor is not upregulated, the observed cholesterol efflux of T-4FN drastically decreases (Figure 4D). Further structural work should be conducted on nanoparticle-ApoAI peptide conjugates to identify potential supramolecular assemblies afforded by surface engineering of ApoAI peptides that significantly boost the cholesterol efflux compared to equimolar free peptide doses (Figure 4E,F). Of note, noncovalent lipid nanoparticle formulations have been the mainstay to ApoAI peptide delivery, but our findings suggest nanocarrier-conjugated delivery is a viable, potentially more potent approach.
Our in vitro functional assays confirmed that the T-4FN candidate shows promise in the context of cardiovascular therapy. To investigate the underlying molecular mechanism, we generated a fluorescence-labeled T-4FN construct for three-dimensional (3D) confocal scanning to visualize the cellular localization of the VNPs during cholesterol efflux. This was achieved by converting internal glutamic acid residues lining the TMV inner channel (Figure S2) to alkyne groups by EDC-mediated conjugation with propargylamine, followed by the copper(I)-catalyzed azide-alkyne cycloaddition of 5FAM. The TMV-5FAM intermediate was then externally conjugated with the 4FN peptide using maleimide-thiol chemistry via an SM(PEG)4 intermediate as discussed above, producing the final fluorescent conjugate T-4FN-5FAM.
Following purification, the conjugation of TMV-5FAM to 4FN peptides was confirmed by SDS-PAGE, which revealed the presence of conjugate coat protein bands at ∼20 kDa and unmodified coat protein bands at ∼17.5 kDa. Densitometric analysis indicated a ∼20% degree of modification, and the presence of 5FAM on both the modified and unmodified coat proteins was confirmed by the MultiFluor Green signal (Figure S2B). DLS indicated that the T-4FN-5FAM conjugate (Zav ∼138.5 nm, PDI = 0.232) was slightly larger than TMV-5FAM (Zav ∼120.0 nm, PDI = 0.212) (Figure S2C). UV–vis spectrophotometry indicated that ∼220–230 5FAM molecules were present per TMV particle. A260/A280 ratios were around the expected value of ∼1.2 (Figure S2D). TEM confirmed the presence of intact T-4FN-5FAM particles (Figure S2E).
A fluorescent-labeled T-4FN-5FAM was used to promote cholesterol efflux from fluorescent-labeled foam cells (Figure 5A,B). Cholesterol and T-4FN-5FAM fluorescence were quantified using Nikon AR Elements Analysis software’s 3D volume viewer tool and plotted in a bar graph (Figure 5C,D).
Figure 5.

(A and B) Cholesterol efflux measured by confocal fluorescence microscopy. Washed and fixed J774 foam cells post-12 hr treatment period: PBS only, 0.5 μM TMV-5FAM in PBS (control), and 0.5 μM T-4FN-5FAM in PBS (candidate). Green = TMV or T-4FN particles labeled with 5FAM. Red = Dil-Ac-LDL cholesterol. Blue = nuclei stained with 10 μg/mL DAPI. Purple = cell membrane stained with wheat germ agglutinin Alexa Fluor 647. Scale bars = 30 μm. Panel B shows a single-cell 3D reconstruction of cells after T-4FN-5FAM and TMV-5FAM treatment. (C) Comparison between treatment groups of total fluorescent intensity of cell-associated Dil-Ac-LDL after incubation and (D) corresponding total fluorescence intensity of cell-associated 5FAM after incubation. Data in (C) and (D) are mean ± standard deviation of n = 30 (10 cells quantified per image, three images per treatment group). Statistical analysis by one-way ANOVA with Tukey’s test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant).
The localization of T-4FN particles differed strikingly from that of unmodified TMV. Punctate foci containing TMV particles were mostly observed within the foam cells, whereas most of the T-4FN particles were extracellular, forming a dense layer on the plasma membrane (Figure 5A,B). It was evident that the formed layer was tightly associated due to its persistent membrane attachment following repeated PBS washes before fixation. Dil-LDL cholesterol fluorescence intensity was significantly lower for the T-4FN treatment compared to unmodified TMV (p < 0.01) (Figure 5C), consistent with the cholesterol efflux assay using nonfluorescent particles and cholesterol constituents. Interestingly, TMV itself promotes a baseline level of cholesterol efflux, augmented by the multivalent display of 4FN. The fluorescence intensity of 5FAM was significantly higher for T-4FN particles than unmodified TMV (p < 0.0001). There was also an inverse correlation of T-4FN cell binding and cholesterol efflux (Figure 5C,D).
To investigate the ability of the TMV particles to bind cholesterol, we mixed them with fluorescent BODIPY-cholesterol for 2 h at 37 °C in 0.01 M KP/DMSO (50:50). The cholesterol-bound nanoparticles were separated from free cholesterol by centrifugation in spin filters with a 100 kDa cutoff. Samples without cholesterol were incubated as a negative control. Purified fractions were analyzed by DLS and fluorimetry.
We observed an increase in the size of all cholesterol-incubated samples relative to cholesterol-free controls. The Zav of the HDL-positive control increased from 54.0 to 296.7 nm, while the PDI decreased from 0.421 to 0.290. However, the greatest change was observed for the T-4FN conjugate, where the Zav increased from 220.5 to 708.8 nm, while the PDI decreased from 0.193 to 0.145 (Figure 6A). Interestingly, unmodified TMV showed an inherent affinity for cholesterol, as revealed by the Zav increase from 203.1 to 293.6 nm, while the PDI decreased from 0.240 to 0.158. The lower PDI of most samples following exposure to cholesterol suggests that cholesterol binding improves order among particle populations, although TMV-4FC was an exception, showing an increase in PDI from 0.168 to 0.218 (Figure 6A). The change in fluorescence was calculated by subtracting the signal before cholesterol binding from the signal after cholesterol binding. T-4FN produced the most intense signal, consistent with the size increase observed by DLS (Figure 6B).
Figure 6.
Free BODIPY-cholesterol binding assay. (A) Dynamic light scattering intensity plots of TMV-ApoAI peptide conjugates before (black) and after (blue) incubation in vitro with BODIPY-cholesterol. (B) Fluorescence analysis of BODIPY-cholesterol binding to nanoparticles. Data are mean changes in fluorescence ± standard deviation (n = 3). Statistical analysis by one-way ANOVA with Tukey’s test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant).
Given the encouraging cholesterol efflux data confirming ABCA1-mediated cholesterol efflux as well as free cholesterol-binding mechanisms of our biomimietic multivalent ApoAI peptide nanoparticles, we carried out additional functional assays to investigate potential orthogonal benefits of our conjugates beyond cholesterol removal. HDL and the 4F peptide are known to activate endothelial nitric oxide synthase (eNOS) and upregulate nitric oxide (NO), which has atheroprotective properties. Chronic oxidative stress and inflammation are major factors driving endothelial cell dysfunction, which promotes the accumulation of oxidized LDL in the lumen, foam cell generation, and atherosclerosis. Using an SVEC endothelial cell line, which was subject to treatment and oxidative stress periods, we assessed the ability of our multivalent TMV-ApoAI to mitigate the formation of ROS (Figure 7).
Figure 7.
ROS levels were detected in SVEC cells using a 2′,7′-dichlorofluorescein diacetate fluorescent probe followed by confocal fluorescence microscopy after treatment for 8 h with the VNPs and controls followed by exposure to oxidized LDL for 4 h. (A) Representative images of two images captured from each sample well. (B) Fluorescence integrated density of intracellular ROS quantified using ImageJ. Data are mean values from the two images captured per well.
The cells were treated with 0.5 μM HDL, 0.5 μM TMV, and varying amounts of free peptide normalized depending on the TMV-ApoAI conjugates (particle treatment groups were matched on a 1:1 molar equivalent basis with HDL, whereas peptide treatment groups were matched on a 1:1 molar equivalent basis with the number of peptides displayed per TMV particle (Figure 7A)). After treatment, we induced oxidative stress by exposing the cells to 50 μg/mL ox-LDL. We found that all treatment groups were able to mitigate ROS production, with T-4FN reducing the ROS signal to near the level of the negative media control, lacking ox-LDL (Figure 7B). 23MJ peptide did not mitigate ROS; however, this could be due to the fact that it is a class Y α-helix compared to class A α-helix; class Y preferentially associates with lipids over LDL cholesterol, which in the ROS assay was the primary driver of inflammation.
The cholesterol efflux and ROS assay data suggested that T-4FN was the most promising candidate for further functional investigation. We therefore evaluated the effect of T-4FN on wound healing using SVEC cells. To better recapitulate the wound healing environment during endothelial cell dysfunction, we combined a traditional scratch wound healing assay with cellular oxidative stress.40 We compared the effects of 8 h treatments with 0.5 μM HDL, 4FN, T-4FN, or unmodified TMV followed by 4 h exposure to 50 μg/mL ox-LDL on wound closure for 12 and 24 h following 8 h (Figure 8).
Figure 8.
Wound healing assay. Cells were treated with 0.5 μM HDL, 4FN, T-4FN, or unmodified TMV followed by 4 h exposure to 50 μg/mL ox-LDL to induce oxidative stress before monitoring wound healing for 12 or 24 h. Data are mean changes in fluorescence ± standard deviation (n = 3). Statistical analysis by one-way ANOVA with Tukey’s test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant).
Exposing the SVEC cells to HDL enhanced wound closure after 12 and 24 h compared with the control (serum-free medium). After 12 h, 0.5 μM HDL and 0.5 μM T-4FN increased wound closure by 8.42-fold (p < 0.0001) and 6.41-fold (p < 0.01), respectively (Figure 8A). After 24 h, 0.5 μM HDL and 0.5 μM T-4FN increased wound closure by 6.24-fold (p < 0.0001) and 4.70-fold (p < 0.001), respectively, and even the free 4FN peptide increased wound closure by 3.20-fold (p < 0.05) (Figure 7B). Unmodified TMV also improved wound closure by 2.58-fold after 12 h and 3.15-fold after 24 h. Although the difference is not statistically significant, T-4FN performed better than the 4FN peptide at both time points, suggesting that multivalent display can enhance endothelial cell wound healing. Our findings are consistent with the reported ability of HDL and the ApoAI peptide to promote wound healing. HDL stimulates endothelial cell growth, promotes wound repair in animal models of hypercholesterolemia, and promotes neovascularization. ApoAI mimetic peptides are also demonstrably pleiotropic for wound healing, reducing oxidative stress by increasing NO synthesis and converting dysfunctional, oxidized HDL to a functional form which achieves re-endothelialization.41–43
CONCLUSIONS
We have demonstrated for the first time that the multivalent display of ApoAI peptides on plant-based VNPs enhances cholesterol efflux, ROS mitigation, and wound healing compared to HDL or free ApoAI peptides. We also found that ABCA1-mediated efflux can occur without both peptide termini free. We report several critical design principles regarding VNP/VLP-ApoAI peptide conjugates, which may apply to other nanoparticle systems: (1) ABCA1-mediated cholesterol efflux is terminus-dependent (T-4FN vs TMV-4FC), (2) ABCA1-mediated cholesterol efflux is influenced by the nanoparticle shape, being more effective with TMV rods than Qβ particles, and (3) multivalent ApoAI display changes the foam cell localization of nanoparticles. Importantly, the clinical safety of several ApoAI peptides in addition to 4F has been validated, and these may also benefit from multivalent display on VNPs. Unmodified TMV also showed unanticipated inherent cholesterol-binding and ROS-mitigating properties, which should be further investigated, possibly using a library of plant viruses. The analysis of novel combinations of nanoparticles and ApoAI peptides may reinvigorate the cholesterol efflux approach to combat atherosclerosis in vivo.
MATERIALS AND METHODS
Production of TMV and Qβ.
TMV-Lys was propagated in Nicotiana benthamiana as previously described and stored in 0.01 M potassium phosphate (KP) buffer (pH 7.4) at 4 °C.31,37 TMV-Lys is described as TMV for brevity. Bacteriophage Qβ VLPs were expressed as previously reported and stored in 1× PBS.38
Peptide Library.
Peptide sequences in Table S1 were selected according to preclinical efficacy in vivo as well as phase I clinical safety and were prepared by solid-phase synthesis (GenScript). The 4F peptide was synthesized with a CSGGG linker on either the N terminus (4FN) or the C terminus (4FC), whereas the 23MJ peptide was synthesized without a linker because it contains a native N-terminal cysteine. Sequences were analyzed using PepCalc (https://pepcalc.com/) to determine characteristics such as molecular weight, isoelectric point, and hydropathicity. Peptide 3D models generated using PEP-FOLD3 were converted to the Sybyl mol2 format and imported into ALOGPS 2.1 to calculate Log P values. Peptide sequences and properties are summarized in Table S2.
Bioconjugation of ApoAI Peptides to TMV and Qβ External Lysine Residues.
The intermediate particles TMV-(PEG)4 and Qβ-(PEG)4 were produced by reacting 2 mg of TMV or Qβ (2 mg··mL–1) with SM(PEG)4 (5 molar equivalents per TMV coat protein and 17 molar equivalents per Qβ coat protein) for 2 h at 25 °C in 0.01 M KP buffer. Particles were purified by ultracentrifugation (50000 × g, 1 h, 4 °C) on a 30% (w/v) sucrose cushion in 0.01 M KP buffer. Pellets containing T-M(PEG)4 and Qβ-M(PEG)4 were washed twice and resuspended in 0.01 M KP buffer. A maleimide–thiol reaction was then carried out by adding 0.5 molar equivalents of ApoAI peptide (4FC, 4FN, or 23MJ) per TMV coat protein or 2.8 molar equivalents per Qβ.
Bioconjugation of 5-Carboxyfluorescein (5FAM) to Internal Glutamic Acid Residues of TMV.
Intermediate TMV-N3 particles were produced through a modified protocol.39 Conjugation of interior glutamic acid residues was performed by conjugation with propargylamine (25 molar equivalents per coat protein) and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide at a final concentration of 2 mg/mL TMV in 0.01 M KP buffer overnight at room temperature. Then, the intermediate was reacted via copper-(I)-catalyzed azide–alkyne cycloaddition to 5FAM (two molar equivalents per TMV coat protein) for 30 min on ice in 0.01 M KP buffer containing 2 mM aminoguanidine, 2 mM ascorbic acid sodium salt, and 1 mM copper sulfate. Particles were purified by ultracentrifugation (50000 × g, 1 h, 4 °C) on a 30% (w/v) sucrose cushion in 0.01 M KP buffer. Pellets containing TMV-5FAM were washed twice and resuspended in 0.01 M KP buffer, then dialyzed overnight against a 0.01 M KP buffer using a Spectrum Labs 10 kDa dialysis membrane (Thermo Fisher Scientific). Purified TMV-5FAM was externally conjugated to 4FN peptide as described above.
UV–vis Spectroscopy and BCA Assay.
The concentration of VLPs/VNPs in 0.01 M KP buffer was determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific). Fluorophore modification and the concentration of TMV were determined using the Beer–Lambert law. The extinction coefficient of TMV is ε(260 nm) = 3 mL·mg−1 · cm−1, molecular weight = 3.94 × 107 g·mol−1. The extinction coefficient of 5FAM is ε(493 nm) = 83,000 M−1 cm−1, molecular weight = 376 Da. The Qβ concentration was determined by measuring the total protein using a Pierce BCA assay kit (Thermo Fisher Scientific).
Dynamic Light Scattering (DLS).
The hydrodynamic diameter of VLP/VNP conjugates were probed using a Zetasizer Nano ZSP/Zen5600 instrument (Malvern Panalytical, Malvern, UK). Particle hydrodynamic diameter was calculated as the weighted mean of the intensity distribution, and samples were analyzed at 1 mg/mL concentration in 0.01 M KP buffer.
Transmission Electron Microscopy (TEM).
The VLPs/VNPs (0.1 mg/mL) in 10 μL drops of Milli-Q water (0.5 mg· mL−1) were added to Formvar FCF400-CU carbon-coated copper TEM grids (Electron Microscopy Sciences) for 2 min at room temperature. The grids were washed twice for 30 s using Milli-Q water and then stained with 10 μL of 2% (w/v) uranyl acetate for 1 mL. The samples were analyzed at 80 kV using an FEI Tecnai Spirit G2 Bio TWIN electron microscope.
SDS-PAGE and Microfluidic Gel Electrophoresis.
VLP/VNP samples (5 μg in 20 μL of reducing loading dye) were denatured at 100 °C for 5 min and loaded alongside a SeeBlue Plus2 marker onto 4–12% or 12% NuPAGE precast gels in 3-(N-morpholino)propanesulfonic acid (MOPS) buffer. After fractionation for 37 min at 200 V and 120 mA, the gels were photographed using the FluorChem R imaging system under white light for Coomassie brilliant blue detection and MultiFluor Green light for 5FAM detection. The degree of modification was determined by densitometric band analysis and the calculation of peak integration ratios using ImageJ. Coat proteins and their degree of modification was also analyzed using an Agilent Bioanalyzer 2100 and a Protein 80 Kit.
BODIPY-Cholesterol Binding Assay.
A fluorescent cholesterol, BODIPY-cholesterol, or 23-(dipyrrometheneboron difluoride)-24-norcholesterol was used to examine the ability of HDL biomimics to uptake cholesterol. VNP samples (500 μg) and equimolar HDL were incubated with 20 uM BODIPY-cholesterol for 2 h at 37 °C on a shaker at 150 rpm in 0.5 mL of 10 mM KP/DMSO. After incubation, samples were purified by centrifugation using 100 kDa molecular-weight cutoff Amicon spin filters to remove free, unbound cholesterol. Samples were analyzed for size on DLS (see above) and fluorescence before and after BODIPY-cholesterol binding using 480/508 nm excitation/emission parameters on a Tecan Plate Reader.
J774 Cholesterol Efflux Assay.
Cholesterol efflux was measured from J774 cells (ATCC) as previously described.23 Briefly, J774 cells were seeded in 96-well plates at 1 × 105 cells/100 μL in DMEM (Corning) with 10% (v/v) fetal bovine serum (FBS) and 1% (w/v) penicillin–streptomycine (P/S). Cells were incubated until they reached ∼80% confluence. After two rinses with FBS-free DMEM containing 1% (w/v) P/S, they were incubated for 48 h in 200 μL of FBS-free DMEM containing 40 μg/mL acetylated low-density lipoprotein (AcLDL), 2 μg/mL Sandoz inhibitor, 1% (w/v) P/S, and 5% (v/v) human lipoprotein-deficient serum (LPDS) to produce foam cells. Excess lipids were removed by two rinses as above, and foam cells were equilibrated for 12 h in 200 μL of FBS-free DMEM, 2 μg/mL Sandoz inhibitor, 0.3 mM CPT-cAMP, 1% (w/v) P/S, and 5% (v/v) human LPDS. After two further rinses, cholesterol efflux was initiated by adding different VLP/VNP or peptide treatments in FBS-free DMEM containing 1% (w/v) P/S and 5% (v/v) human LPDS and incubating for 15 h. The medium was then collected and centrifuged (1000 × g, 10 min, 4 °C), and the cholesterol content of the supernatant was determined using the Amplex assay kit. Cells were washed three times with ice-cold PBS and lysed using 200 μL of CytoBuster lysis buffer, and the cholesterol content of the lysate was determined using the Amplex assay kit. Cholesterol efflux was calculated by dividing the amount of cholesterol in the medium by the total cholesterol content of the medium and lysate.
ROS Assay.
SVEC cells (ATCC) were seeded in four-compartment 35 mm CELLview glass-bottom dishes or 96-well plates at 6 ×104 cells/mL in 500 μL of DMEM and were allowed to reach ∼80% confluence. The medium was then removed, and the cells were washed once with PBS before incubation with VLPs/VNPs for 8 h. We then added 100 μg/mL ox-LDL for 4 h to induce oxidative stress and detected ROS using a CellROX Green Reagent Kit. Images were captured using a Nikon A1R confocal microscope with a 20×, 0.75 numerical aperture dry objective.
Wound Healing Assay.
SVEC cell migration was investigated using a Radius 96-well kit (Cell Biolabs). We seeded 3 × 104 cells in 200 μL of DMEM per well and allowed the cells to reach ∼80% confluence. After removing the Radius Gel, cells were allowed to migrate for 24 h in the presence of various treatments. Bright-field phase contrast images were acquired after 0, 12, and 24 h using an EVOS FL microscope fitted with a 10× objective (n = 6 cells per group). Percentage wound closure was calculated by tracing the wound perimeter in ImageJ to calculate the area at t = 12 or 24 h and dividing by the initial starting area at t = 0 h.
TMV-ApoAI Distribution during Cholesterol Efflux from J774 Foam Cells.
We seeded 1 × 105 J774 cells in 500 μL of DMEM containing 10% (v/v) FBS and 1% (w/v) P/S into each zone of a four-compartment 35 mm CELLview glass-bottom dish and allowed them to reach ∼80% confluence. Cells were rinsed twice with 500 μL of FBS-free DMEM containing 1% (w/v) P/S and incubated for 48 h in 200 μL of FBS-free DMEM containing 40 μg/mL of fluorescence-labeled acetylated low-density lipoprotein (Dil-AcLDL), 2 μg/mL Sandoz inhibitor, 1% (w/v) P/S, and 5% (v/v) human LPDS to produce labeled foam cells. After two rinses as above, foam cells were equilibrated for 12 h in 200 μL of FBS-free DMEM containing 2 μg/mL Sandoz inhibitor, 0.3 mM CPT-cAMP, 1% (w/v) P/S, and 5% (v/v) human LPDS. After two further rinses, the cells were treated with 100 μL of 5FAM-labeled treatments and controls in FBS-free DMEM containing 1% (w/v) P/S and 5% (v/v) human LPDS. After 12 h, the cells were washed twice with 500 μL of PBS and then fixed for 10 min at room temperature using a PBS solution containing 10% (w/v) PFA and 25% (w/v) glutaraldehyde. Cells were then twice washed with PBS and subsequently stained for 45 min with 300uL of wheat germ agglutinin Alexa Fluor 647 (WGA-647) prepared in PBS and 5% (v/v) goat serum at a 1:500 dilution. Cells were then twice washed with PBS and then stained with 300 μL of DAPI dye (5 μg/mL in PBS) for 20 min. Cells were washed twice with PBS and then imaged using a Nikon A1R confocal microscope with a 20×, 0.75 numerical aperture dry objective.
Statistical Methods.
Samples were prepared in triplicate unless otherwise stated. Data were processed by one-way analysis of variance (ANOVA) with Tukey’s test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns = not significant) using GraphPad Prism 6.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by a grant from NIH (R01-HL137674 to N.F.S.) and M.D.S. was supported in part by CBI training program NIH T32-GM112584. We acknowledge the UCSD Cancer Center Microscopy Shared Facility Specialized Support Grant P30 CA23100-28 for access to confocal microscopy
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.bioconjchem.2c00371.
Nanoparticle and peptide properties and structural models (Tables S1 and S2 and Figure S1) and synthesis and characterization of TMV-5FAM (Figure S2) (PDF)
The authors declare the following competing financial interest(s): Dr. Steinmetz is a co-founder of, has equity in, and has a financial interest with Mosaic ImmunoEngineering Inc. Dr. Steinmetz serves as Director, Board Member, and Acting Chief Scientific Officer, and paid consultant to Mosaic. The other authors declare no conflicts of interest.
Contributor Information
Matthew D. Shin, Department of NanoEngineering and, Center for Nano-ImmunoEngineering, University of, California San Diego, La Jolla, California 92039, United, States
Oscar A. Ortega-Rivera, Department of NanoEngineering and Center for Nano-ImmunoEngineering, University of California San Diego, La Jolla, California 92039, United, States
Nicole F. Steinmetz, Department of NanoEngineering, Center for Nano-ImmunoEngineering, Department of Bioengineering, Department of Radiology, Moores Cancer, Center, and Institute for Materials Discovery and Design, University of California San Diego, La Jolla, California, 92039, United States
REFERENCES
- (1).Lusis AJ Atherosclerosis. Nature 2000, 407, 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Libby P; Buring JE; Badimon L; Hansson GK; Deanfield J; Bittencourt MS; Tokgözoğlu L; Lewis EF Atherosclerosis. Nat. Rev. Dis. Primers 2019, 5, No. 56. [DOI] [PubMed] [Google Scholar]
- (3).Virani SS; Alonso A; Aparicio HJ; Benjamin EJ; Bittencourt MS; Callaway CW; Carson AP; Chamberlain AM; Cheng S; Delling FN; et al. Heart Disease and Stroke Statistics2021 Update. Circulation 2021, 143 (8), e254–e743. [DOI] [PubMed] [Google Scholar]
- (4).Dagenais GR; Leong DP; Rangarajan S; Lanas F; LopezJaramillo P; Gupta R; Diaz R; Avezum A; Oliveira GBF; Wielgosz A; et al. Variations in Common Diseases, Hospital Admissions, and Deaths in Middle-Aged Adults in 21 Countries from Five Continents (PURE): A Prospective Cohort Study. Lancet 2020, 395, 785–794. [DOI] [PubMed] [Google Scholar]
- (5).Weber C; Noels H. Atherosclerosis: Current Pathogenesis and Therapeutic Options. Nat. Med 2011, 17, 1410–1422. [DOI] [PubMed] [Google Scholar]
- (6).Wolf D; Ley K. Immunity and Inflammation in Atherosclerosis. Circulation Research 2019, 124 (2), 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Soehnlein O; Libby P. Targeting Inflammation in Atherosclerosis from Experimental Insights to the Clinic. Nat. Rev. Drug Discovery 2021, 20, 589–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kingwell BA; Chapman MJ Future of High-Density Lipoprotein Infusion Therapies. Circulation 2013, 128, 1112–1121. [DOI] [PubMed] [Google Scholar]
- (9).Huang Y; DiDonato JA; Levison BS; Schmitt D; Li L; Wu Y; Buffa J; Kim T; Gerstenecker G; et al. An Abundant Dysfunctional Apolipoprotein A1 in Human Atheroma. Nat. Med 2014, 20, 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Reiner Ž Resistance and Intolerance to Statins. Nutr., Metab. Cardiovasc. Dis 2014, 24, 1057–1066. [DOI] [PubMed] [Google Scholar]
- (11).Sinning D; Landmesser U. Low-Density LipoproteinCholesterol Lowering Strategies for Prevention of Atherosclerotic Cardiovascular Disease: Focus on SiRNA Treatment Targeting PCSK9 (Inclisiran). Curr. Cardiol. Rep 2020, 22, No. 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Novartis receives EU approval for Leqvio* (inclisiran), a firstin-class siRNA to lower cholesterol with two doses a year**. Novartis. https://www.novartis.com/news/media-releases/novartis-receives-euapproval-leqvio-inclisiran-first-class-sirna-lower-cholesterol-two-dosesyear (accessed Jun 22, 2022).
- (13).Hegele RA 7 - Lipoprotein and Lipid Metabolism. In Emery and Rimoin’s Principles and Practice of Medical Genetics and Genomics, Seventh Edition; Pyeritz RE, Korf BR, Grody WW, Eds.; Academic Press, 2021; pp 235–278. [Google Scholar]
- (14).Takata K; Di Bartolo BA; Nicholls SJ High-Density Lipoprotein Infusions. Cardiol. Clin 2018, 36, 311–315. [DOI] [PubMed] [Google Scholar]
- (15).Rohatgi A; Khera A; Berry JD; Givens EG; Ayers CR; Wedin KE; Neeland IJ; Yuhanna IS; Rader DR; et al. HDL Cholesterol Efflux Capacity and Incident Cardiovascular Events. N. Engl. J. Med 2014, 371, 2383–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).He H; Hong K; Liu L; Schwendeman A. Artificial High-Density Lipoprotein-Mimicking Nanotherapeutics for the Treatment of Cardiovascular Diseases. WIREs Nanomed. Nanobiotechnol 2021, 13, No. e1737. [DOI] [PubMed] [Google Scholar]
- (17).Bonizzi A; Piuri G; Corsi F; Cazzola R; Mazzucchelli S. HDL Dysfunctionality: Clinical Relevance of Quality Rather Than Quantity. Biomedicines 2021, 9, 729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Martínez-López D; Camafeita E; Cedó L; Roldan-Montero R; Jorge I; García-Marqués F; Gómez-Serrano M; BonzonKulichenko E; Blanco-Vaca F; Blanco-Colio LM; et al. APOA1 Oxidation Is Associated to Dysfunctional High-Density Lipoproteins in Human Abdominal Aortic Aneurysm. eBioMedicine 2019, 43, 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Javaheri A; Rader DJ Apolipoprotein A-I and Cholesterol Efflux. Circ. Res 2014, 114, 1681–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hamer M; O’Donovan G; Stamatakis E. High-Density Lipoprotein Cholesterol and Mortality. Arterioscler., Thromb., Vasc. Biol 2018, 38, 669–672. [DOI] [PubMed] [Google Scholar]
- (21).Kosmas CE; Martinez I; Sourlas A; Bouza KV; Campos FN; Torres V; Montan PD; Guzman E. High-Density Lipoprotein (HDL) Functionality and Its Relevance to Atherosclerotic Cardiovascular Disease. DIC 2018, 7, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Nissen SE; Tsunoda T; Tuzcu EM; Schoenhagen P; Cooper CJ; Yasin M; Eaton GM; Lauer MA; Sheldon WS; Grines CL; et al. Effect of Recombinant ApoA-I Milano on Coronary Atherosclerosis in Patients With Acute Coronary Syndromes: A Randomized Controlled Trial. JAMA 2003, 290, 2292–2300. [DOI] [PubMed] [Google Scholar]
- (23).Zhao Y; Imura T; Leman LJ; Curtiss LK; Maryanoff BE; Ghadiri MR Mimicry of High-Density Lipoprotein: Functional Peptide–Lipid Nanoparticles Based on Multivalent Peptide Constructs. J. Am. Chem. Soc 2013, 135, 13414–13424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Zhao Y; Black AS; Bonnet DJ; Maryanoff BE; Curtiss LK; Leman LJ; Ghadiri MR In Vivo Efficacy of HDL-like Nanolipid Particles Containing Multivalent Peptide Mimetics of Apolipoprotein A-I[S]. J. Lipid Res 2014, 55, 2053–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bloedon LT; Dunbar R; Duffy D; Pinell-Salles P; Norris R; DeGroot BJ; Movva R; Navab M; Fogelman AM; Rader DJ Safety, Pharmacokinetics, and Pharmacodynamics of Oral ApoA-I Mimetic Peptide D-4F in High-Risk Cardiovascular Patients. J. Lipid Res 2008, 49, 1344–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Dunbar RL; Movva R; Bloedon LT; Duffy D; Norris RB; Navab M; Fogelman AM; Rader DJ Oral Apolipoprotein A-I Mimetic D-4F Lowers HDL-Inflammatory Index in High-Risk Patients: A First-in-Human Multiple-Dose, Randomized Controlled Trial. Clin. Transl. Sci 2017, 10, 455–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).White CR; Datta G; Wilson L; Palgunachari MN; Anantharamaiah GM The ApoA-I Mimetic Peptide 4F Protects Apolipoprotein A-I from Oxidative Damage. Chem. Phys. Lipids 2019, 219, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Nandwana V; Ryoo S-R; Kanthala S; McMahon KM; Rink JS; Li Y; Venkatraman SS; Thaxton CS; Dravid VP High-Density Lipoprotein-like Magnetic Nanostructures (HDLMNS): Theranostic Agents for Cardiovascular Disease. Chem. Mater 2017, 29, 2276–2282. [Google Scholar]
- (29).Henrich SE; Hong BJ; Rink JS; Nguyen ST; Thaxton CS Supramolecular Assembly of High-Density Lipoprotein Mimetic Nanoparticles Using Lipid-Conjugated Core Scaffolds. J. Am. Chem. Soc 2019, 141, 9753–9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kuai R; Li D; Chen YE; Moon JJ; Schwendeman A. High-Density Lipoproteins: Nature’s Multifunctional Nanoparticles. ACS Nano 2016, 10, 3015–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Geiger FC; Eber FJ; Eiben S; Mueller A; Jeske H; Spatz JP; Wege C. TMV Nanorods with Programmed Longitudinal Domains of Differently Addressable Coat Proteins. Nanoscale 2013, 5, 3808–3816. [DOI] [PubMed] [Google Scholar]
- (32).Pitek AS; Wang Y; Gulati S; Gao H; Stewart PL; Simon DI; Steinmetz NF Elongated Plant Virus-Based Nanoparticles for Enhanced Delivery of Thrombolytic Therapies. Mol. Pharmaceutics 2017, 14, 3815–3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Park J; Gao H; Wang Y; Hu H; Simon DI; Steinmetz NF S100A9-Targeted Tobacco Mosaic Virus Nanoparticles Exhibit High Specificity toward Atherosclerotic Lesions in ApoE–/– Mice. J. Mater. Chem B 2019, 7, 1842–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Park J; Wen AM; Gao H; Shin MD; Simon DI; Wang Y; Steinmetz NF Designing S100A9-Targeted Plant Virus Nanoparticles to Target Deep Vein Thrombosis. Biomacromolecules 2021, 22, 2582–2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Bruckman MA; Jiang K; Simpson EJ; Randolph LN; Luyt LG; Yu X; Steinmetz NF Dual-Modal Magnetic Resonance and Fluorescence Imaging of Atherosclerotic Plaques in Vivo Using VCAM-1 Targeted Tobacco Mosaic Virus. Nano Lett. 2014, 14, 1551–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Wen AM; Wang Y; Jiang K; Hsu GC; Gao H; Lee KL; Yang AC; Yu X; Simon DI; Steinmetz NF Shaping BioInspired Nanotechnologies to Target Thrombosis for Dual OpticalMagnetic Resonance Imaging. J. Mater. Chem. B 2015, 3, 6037–6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Wen AM; Shukla S; Saxena P; Aljabali AAA; Yildiz I; Dey S; Mealy JE; Yang AC; Evans DJ; Lomonossoff GP; Steinmetz NF Interior Engineering of a Viral Nanoparticle and Its Tumor Homing Properties. Biomacromolecules 2012, 13, 3990–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Ortega-Rivera OA; Shin MD; Chen A; Beiss V; MorenoGonzalez MA; Lopez-Ramirez MA; Reynoso M; Wang H; Hurst BL; Wang J; et al. Trivalent Subunit Vaccine Candidates for COVID-19 and Their Delivery Devices. J. Am. Chem. Soc 2021, 143, 14748–14765. [DOI] [PubMed] [Google Scholar]
- (39).Bruckman MA; Steinmetz NF Chemical Modification of the Inner and Outer Surfaces of Tobacco Mosaic Virus (TMV). Methods Mol. Biol 2014, 1108, 173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Liu D; Ding Z; Wu M; Xu W; Qian M; Du Q; Zhang L; Cui Y; Zheng J; Chang H; et al. The Apolipoprotein A-I Mimetic Peptide, D-4F, Alleviates Ox-LDL-Induced Oxidative Stress and Promotes Endothelial Repair through the ENOS/HO-1 Pathway. J. Mol. Cell. Cardiol 2017, 105, 77–88. [DOI] [PubMed] [Google Scholar]
- (41).He D; Zhao M; Wu C; Zhang W; Niu C; Yu B; Jin J; Ji L; Willard B; Mathew AV; et al. Apolipoprotein A-1 Mimetic Peptide 4F Promotes Endothelial Repairing and Compromises Reendothelialization Impaired by Oxidized HDL through SR-B1. Redox Biol. 2018, 15, 228–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Rosenbaum MA; Chaudhuri P; Abelson B; Cross BN; Graham LM Apolipoprotein A-I Mimetic Peptide Reverses Impaired Arterial Healing after Injury by Reducing Oxidative Stress. Atherosclerosis 2015, 241, 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Bastaki KM; Tarlton JMR; Lightbody RJ; Graham A; Martin PE Homo Sapiens (Hsa)-MicroRNA (MiR)-6727–5p Contributes to the Impact of High-Density Lipoproteins on Fibroblast Wound Healing In Vitro. Membranes 2022, 12, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.