Abstract
Background: Hepatic resection, radiofrequency ablation (RF), and liver transplantation (LT) represent the only available curative treatments for early stage hepatocellular carcinoma (HCC). Various studies showed that the 5-year overall survival (OS) rate reaches ∼70% after resection and ∼60% after RF. Objective: To improve the success rate of curative therapies and consequently the OS, an improvement in patients’ selection and management should be pursued. In this regard, microRNAs (miRNAs) can be helpful prognostic biomarkers. Materials and Methods: In this retrospective study, a miRNA array profiling was performed on 34 HCC blood samples which is collected before therapy (T0), 1 month (T1), and 6 months (T2) after curative treatments (resection and RF) to identify noninvasive biomarker candidates for therapy response and OS. MiRNAs were validated in 80 blood HCC samples using quantitative real-time PCR (qRT-PCR). Patients were divided into complete responder (CR) and partial responder and progressive disease (PRPD). Results: Among the selected miRNAs, miR-3201 is significantly associated with treatment response in the validation phase, showing a 23% reduction (P = .026) in CR compared to PRPD. MiR-3201 was able to distinguish CR from PRPD (area under the curve [AUC] = 0.69, 71% sensitivity, 70% specificity, P = .0036). Furthermore, lower levels of miR-3201 were associated with longer OS (hazard ratio [HR] = 2.61, P = .0006). Conclusions: Blood miR-3201 could be used as a prognostic biomarker for curative therapy response and OS in HCC.
Keywords: microRNA, blood, circulating miRNA, biomarker, hepatocellular carcinoma, liver cancer, HCC, resection, radiofrequency
Introduction
Primary liver cancer is one of the most common tumors in the world, ranking second as the most frequent cause of cancer-related death, with an estimated global incidence rate per 100,000 person-years of 9.3 and a corresponding mortality rate of 8.5 in 2018.1–3 Hepatocellular carcinoma (HCC) represents approximately 75% of the total cases of primary liver cancer, having an incidence strongly correlated to the male gender and advanced age.4,5 Almost all cases of HCC are associated with a known etiology, most frequently chronic viral hepatitis (B and C), non-alcoholic fatty liver disease (NAFLD), genetic and hereditary disorders, environmental toxins such as aflatoxin exposure, metabolic diseases including diabetes mellitus and obesity, and dietary and lifestyle factors like alcohol consumption and smoking.6,7 Despite the variety of etiologies, several factors promote the development of chronic damage during their long clinical course, often leading to liver cirrhosis, which represents one of the primary predisposing factors of HCC.8,9 Indeed, one-third of cirrhotic patients develop HCC during their lifetime.4
In contrast to the reduction of death rates observed for many cancer types, HCC mortality continues to increase by ∼2% to 3% per year due to the late diagnosis which hampers the efficacy of the currently available therapies.10,11 Only 40% of patients with HCC (Barcelona Clinic Liver Cancer [BCLC] stages 0 and A) are eligible for liver transplant, surgical resection, or tumor ablation therapies, the potentially curative treatments.12,13 Liver transplantation (LT) removes both the tumor and cirrhosis, with a 5-year overall survival (OS) rate of 70% and 6% to 18% of HCC recurrence.14,15 Hepatic resection is a recommended option in patients with small lesions and preserved liver function, providing a 5-year OS rate of 60% to 70% and median disease-free survival (DFS) of 2 years with a low risk of early recurrence in 76.5% of patients.16–19 Radiofrequency ablation (RF) is a minimally invasive therapy applicable to patients with small lesions. Around 60% of patients treated with RF can benefit from a 5-year survival, or even more patients (76%) if selected according to the BCLC guidelines.20–22 The detection of lesions at early stages coupled with a personalized therapeutical protocol for each patient may represent a strategy to improve patient survival. The assessment of blood-based biomarkers at the time of diagnosis may help clinicians in a more precise patient stratification providing at the same time additional information for better clinical management.
In this context, circulating microRNAs (miRNAs) represent a valuable tool supporting conventional clinical practices.23,24 They are small noncoding RNAs with a length of 19 to 22 nucleotides involved in the post-transcriptional gene silencing of their target genes.25,26 Besides circulating miRNAs having a value of noninvasiveness, ease of measurement, and cost-effectiveness, they are stable and not degraded in blood, which makes them suitable as diagnostic, predictive, and prognostic biomarkers.27,28 MiRNAs are generally studied in plasma and serum. However, blood can represent an interesting source since it contains additional noncoding RNAs derived from immune system cells.
In this study, we assessed the blood miRNA expression at the time of HCC diagnosis, at 1 month and 6 months after therapy to identify miRNAs associated with a complete response to curative treatments and OS, which can help clinicians in more accurate patient management.
Materials and Methods
The reporting of this study conforms to REporting recommendations for tumour MARKer prognostic studies (REMARK) guidelines.29
Study Design
The present retrospective study was organized into 2 phases.
Discovery phase (phase 1): Thirty-four blood samples collected from 13 patients at the time of diagnosis (T0), 1 month (T1), and 6 months (T2) after curative therapy (resection and RF) were analyzed through microarray profiling. Six months after therapy, the patients were categorized, according to modified Response Evaluation Criteria In Solid Tumors (mRECIST) as complete responder (CR), partial responder (PR), and progressive disease (PD). Kruskal–Wallis test was used to determine gene expression differences in microarray analysis between CR patients versus PR + PD (PRPD) patients. In the discovery group, miRNAs associated with complete response to curative treatments were selected as blood biomarker candidates for subsequent validation.
Validation phase (phase 2): MiRNA candidates selected in the discovery phase were tested by quantitative real-time PCR (RT-qPCR) in other 80 HCC blood samples from 39 patients treated with resection and RF at the 3 considered time points. MiRNA expression determined at T0, T1, and T2 was associated with the response to therapy and patient survival.
Patients
Between 2012 and 2017, 52 consecutive patients referring to the Liver Center who were diagnosed with HCC according to the European Association for the Study of the Liver (EASL) criteria were enrolled for the study. Blood samples were collected at the time of HCC diagnosis (T0), 1 month (T1), and 6 months (T2) after curative treatments (hepatic resection [n = 18] and RF [n = 34]). Six months after therapy, 73% of patients had a complete response to treatment, while 27% did not respond. The end of the follow-up period was June 2021 (median follow-up: 48 months). The clinical and demographic features of the groups are shown in Table 1.
Table 1.
Clinical Characteristics of the Enrolled Patients.
| Discovery | Validation | |||||
|---|---|---|---|---|---|---|
| Tot (n = 13) |
CR (n = 9) |
PRPD (n = 4) |
Tot (n = 39) |
CR (n = 29) |
PRPD (n = 10) | |
| Age mean (95% CI) |
71.8 (66.2-75.1) |
73.6 (72.5-75.1) |
67.9 (55.8-80.0) |
70.4 (67.7-73.0) |
69.6 (66.3-72.9) |
72.8 (68.3-77.3) |
| Gender | ||||||
| Female | 2 | 2 | 0 | 7 | 5 | 2 |
| Male | 11 | 7 | 4 | 32 | 24 | 8 |
| Etiology | ||||||
| Alcohol or metabolic | 8 | 5 | 3 | 16 | 11 | 5 |
| Viral | 4 | 4 | 0 | 12 | 9 | 3 |
| Mixed | 1 | 0 | 1 | 11 | 9 | 2 |
| Scores | ||||||
| CTP A/B | 12/1 | 9/0 | 3/1 | 33/6 | 24/5 | 9/1 |
| BCLC 0/A | 3/10 | 2/7 | 1/3 | 6/33 | 4/25 | 2/8 |
| No. of lesions | ||||||
| Single <2cm | 2 | 2 | 0 | 4 | 4 | 0 |
| Single <5cm or 3 nodules ≤3cm |
9 | 6 | 3 | 27 | 20 | 7 |
| Large single or multi | 2 | 1 | 1 | 8 | 5 | 3 |
| AFP (ng/mL) | ||||||
| <20 | 10 | 7 | 3 | 25 | 19 | 6 |
| 20 to 400 | 3 | 2 | 1 | 6 | 3 | 3 |
| >400 | 0 | 0 | 0 | 2 | 2 | 0 |
| Treatment | ||||||
| Resection | 4 | 1 | 3 | 14 | 9 | 5 |
| RF | 9 | 8 | 1 | 25 | 20 | 5 |
Abbreviations: AFP: alpha-fetoprotein; BCLC: Barcelona Clinic Liver Cancer; CR: complete responder; CTP: Child–Turcotte-Pugh; PRPD: partial responder and progressive disease; RF: radiofrequency ablation.
All the patients provided written informed consent for the blood samples and associated clinical data collection. Patient anonymity has been preserved. The investigation was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the regional ethical committee (Comitato Etico Regionale Unico FVG, No. 14/2012 ASUITS).
Sample Collection
Fasting whole blood samples were collected from each patient during clinical visits. Peripheral blood (3 mL per patient) was collected in Tempus™ Blood RNA Tubes containing 6 mL of Stabilizing Reagent (Thermo Fischer Scientific) and subsequently frozen at − 80 °C for long-term storage.
RNA Isolation and Quality Assessment
Total RNA was isolated from Tempus™ Blood RNA Tubes using the MagMAX™ for Stabilized Blood tubes RNA Isolation Kit (Thermo Fischer Scientific) following the manufacturer protocol. The quality of the total RNA extracted was assessed with the Agilent RNA 6000 Nano Kit (Agilent Technologies) using the 2100 Bioanalyzer Instrument (Agilent Technologies). Samples with an RNA integrity number (RIN) less than 6 were discarded from the subsequent profiling experiments.
RNA Microarray Profiling and Analysis
MiRNA profiles were analyzed through the Affymetrix GeneChip® microRNA 3.0 Array (Affymetrix®, Thermo Fischer Scientific) as previously described.30
qRT-PCR validation
Fifty nanograms of total RNA were reverse-transcribed using the qScript microRNA cDNA Synthesis Kit (QuantaBio) according to manufacturer instructions. Samples were analyzed through qRT-PCR using the PerfeCTa SYBR® Green SuperMix (QuantaBio) in a CFX-96 thermal cycler (Bio-Rad Laboratories) according to manufacturer instructions. All reactions were run in duplicate in a 25µL reaction. MiRNA primers were purchased from Metabion International AG (Metabion) or Sigma-Aldrich (Merck KGaA). MiR-486 was used as an endogenous normalizer. Expression levels were calculated using the 2−ΔΔCt formula.
Statistical analysis
The Mann–Whitney U test was used to compare the differences between the 2 independent groups. For multiple comparisons, the Kruskal–Wallis test in a one-way ANOVA procedure was used. The receiver operating characteristic (ROC) curves were plotted to estimate the discriminatory potential of the miRNAs. Survival curves were plotted according to the Kaplan–Meier method. Analyses were performed using NCSS 11 Software (2016) (NCSS, LLC, ncss.com/software/ncss).
Results
MiRNA Screening in the Discovery Cohort
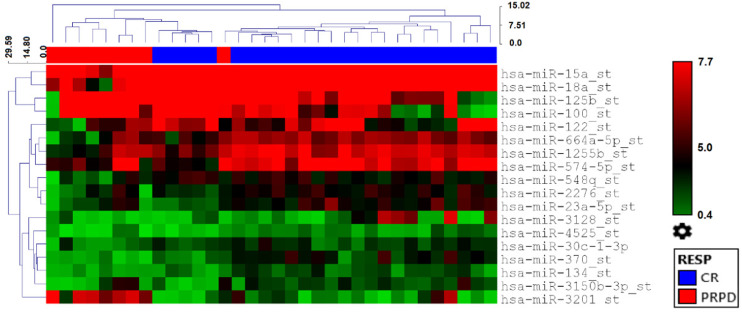
HCC blood samples collected from 13 patients at T0 (n = 13), T1 (n = 13), and T2 (n = 8) were analyzed through the Affymetrix GeneChip® miRNA 3.0 Array build on miRBase release 20. Nineteen miRNAs were differently expressed (P< .05) between CR and PRPD patients treated with curative therapies at all considered time points (Table 2 and Figure 1), with miR-370 and miR-3201 being the top-scoring miRNA in terms of both differential expression and P value (P < .001). MiR-370 showed a higher expression in CR compared to PRPD (fold change (FC) = 6.47), while miR-3201 was higher in PRPD compared to CR (FC = 5.95) (Table 2).
Table 2.
Differentially Expressed miRNAs Between CR and PRPD Patients of the Discovery Cohort at all Considered Time Points.
| ID | CR Avg. (log2) | PRPD Avg. (log2) | FC (linear) |
P value |
|---|---|---|---|---|
| hsa-miR-370 | 4.34 | 1.65 | 6.47 | .0003 |
| hsa-miR-769-3p | 4.72 | 2.54 | 4.52 | .0174 |
| hsa-miR-574-5p | 7.52 | 5.76 | 3.39 | .0441 |
| hsa-miR-134 | 3.41 | 1.87 | 2.89 | .0272 |
| hsa-miR-664a-5p | 6.42 | 4.93 | 2.81 | .0318 |
| hsa-miR-30c-1-3p | 4.05 | 2.62 | 2.7 | .0133 |
| hsa-miR-18a | 9.09 | 7.68 | 2.67 | .0175 |
| hsa-miR-1255b | 7.21 | 5.81 | 2.64 | .0121 |
| hsa-miR-15a | 10.98 | 9.58 | 2.64 | .0420 |
| hsa-miR-3128 | 2.64 | 1.26 | 2.61 | .0101 |
| hsa-miR-122 | 6.55 | 5.25 | 2.45 | .0438 |
| hsa-miR-2276 | 5.12 | 3.93 | 2.28 | .0258 |
| hsa-miR-4525 | 2.42 | 1.23 | 2.28 | .0281 |
| hsa-miR-23a-5p | 4.91 | 3.78 | 2.19 | .0405 |
| hsa-miR-548q | 5.2 | 4.17 | 2.05 | .0021 |
| hsa-miR-3150a-5p | 1.01 | 2.11 | −2.15 | .0482 |
| hsa-miR-125b | 7.82 | 9.8 | −3.95 | .0316 |
| hsa-miR-100 | 7.04 | 9.04 | −4.02 | .0353 |
| hsa-miR-3201 | 1.62 | 4.2 | −5.95 | .0009 |
Abbreviations: CR: complete responder; FC: fold change; PRPD: partial responder and progressive disease.
Figure 1.
Heatmap with the pseudo-color scale underneath the differentially expressed miRNAs between CR (blue) and PRPD (red) in the group of patients treated with curative therapies, at all considered time points (T0, T1, and T2). Unsupervised hierarchical clustering was used to order samples and miRNAs and the log2-transformed microarray signal was considered. The sample tree with optimized leaf ordering was drawn using Euclidean distances and average linkages for cluster-to-cluster distance.
Abbreviations: CR: complete responder; PRPD: partial responder and progressive disease.
Identification of miRNA Biomarker Candidates for Curative Treatments
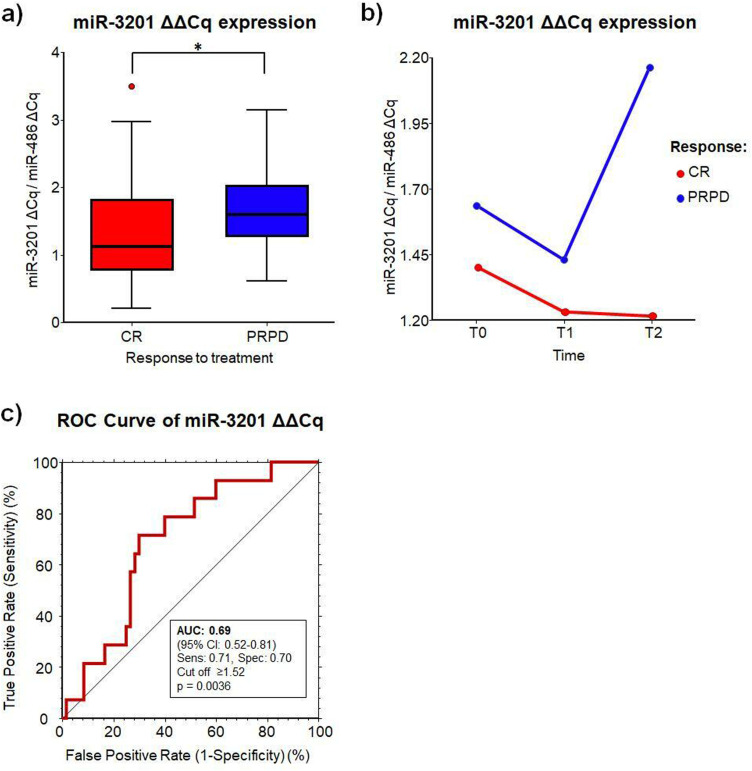
We used the qRT-PCR assays to confirm the expression of 2 selected miRNA candidates from the miRNA array study (miR-370 and miR-3201). We assessed the miRNA expression in other 80 blood samples from HCC patients collected at T0 (n = 35), T1 (n = 24), and T2 (n = 21). Among the selected miRNAs, only miR-3201 was confirmed as statistically significant in the differential expression between CR and PRPD at all considered time points with a reduction of 23% in CR (P = .026) (Table 3 and Figure 2a). Indeed, the expression of miR-3201 was always higher in patients not responding to therapy compared to responders, although not significant (P = .06) (Figure 2b), with the highest difference observed at T2. In addition, there was no difference in the miRNA expression when comparing resection versus RF at all considered time points (resection: 1.45 ± 0.82 vs RF: 1.30 ± 0.72, P = .59), before (T0: 1.21 ± 0.63 vs 1.44 ± 0.76, P = .35), and after the treatments (T1 + T2: 1.68 ± 0.95 vs 1.20 ± 0.68, P = .15).
Table 3.
Differentially Expressed miRNAs Between CR and PRPD Patients of the Validation Cohort at all Considered Time Points
| ID | CR Avg. ± SD (linear) | PRPD Avg. ± SD (linear) | FC (linear) |
P value |
|---|---|---|---|---|
| hsa-miR-370 | 0.58 ± 0.42 | 0.54 ± 0.49 | 1.07 | .450 |
| hsa-miR-3201 | 1.30 ± 0.73 | 1.69 ± 0.64 | 0.77 | .026 |
Abbreviations: CR, complete responder; FC: fold change;PRPD: partial responder and progressive disease.
Figure 2.
MiR-3201 expression in HCC blood samples. (a) Mean ΔΔCq expression of miR-3201 in CR and PRPD. (b) Mean ΔΔCq expression of miR-3201 in CR and PRPD versus time. (c) Receiver operating curve (ROC) analysis of miR-3201 when considering all times.
Abbreviations: CR: complete responder; PRPD: partial responder and progressive disease.
To validate the discriminatory potential of miR-3201 between CR and PRPD at all considered times, the area under the curve (AUC) was determined with a ROC analysis (Figure 2c). MiR-3201 showed an AUC = 0.69 (95% CI: 0.52-0.81, P = .0036), with a sensitivity and specificity of 71% and 70%, respectively, at a cut-off determined at ≥1.52.
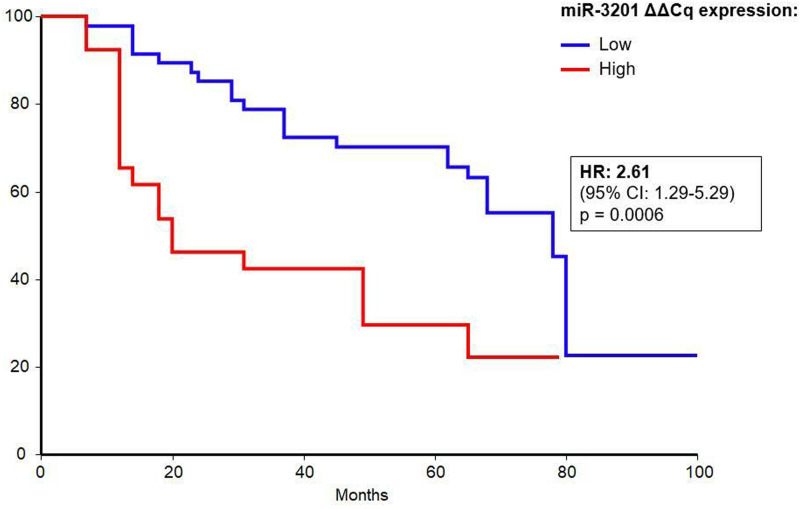
Higher Levels of miR-3201 are Associated With longer Survival in HCC Patients
Cut-off value determined by ROC curve analysis (1.52) was used to compare the survival time of patients with low versus high miRNA expression. Kaplan–Meier survival analysis demonstrated that low expression of miR-3201 was significantly associated with longer OS in patients, at all considered time points (P = .0006), with a hazard ratio (HR) of 2.61 (95% CI: 1.29-5.29) (Figure 3), corresponding to CR patients that have lower levels of the circulating miRNA. Indeed, the OS rates at 24 months were 85% for CR and 46% for PRPD patients, 70% and 42% at 48 months, and 55% and 22% at 72 months.
Figure 3.
Kaplan–Meier survival analysis by log-rank test for blood miR-3201.
Abbreviation: CI: confidence interval; HR: hazard ratio.
Discussion
Hepatic resection and RF are considered curative treatments for HCC, showing a 5-year OS rate ranging from 60% to 70% for resection and ∼60% for RF.16,18–22 However, there is a portion of patients that have a relapse. In this context, MiRNAs can represent helpful prognostic biomarkers.
In 2017, Cho et al. suggested circulating miR-26a and miR-29a as prognostic biomarkers predicting poor DFS and LT-free survival in Hepatitis B virus (HBV)-related HCC patients undergoing curative treatments (resection or RF).31 Indeed, pre-treatment low plasma levels of miR-26a and miR-29a were significantly associated with poor LT-free survival.31 More recently, Yokota et al. identified exosomal miR-638 as a prognostic biomarker for resected patients.32 Patients with low levels of miR-638 showed longer DFS, having a 2-year DFS rate of 77.4% when compared with 47.1% of high miRNA expressing group.32 In our recent study, we identified serum miR-4443, miR-4530, and miR-4454 as biomarkers to predict therapy response to curative treatments with a sensitivity and specificity of 72% and 75%, respectively.33 In addition, higher serum levels of miR-4454 (HR = 2.79, P = .029) and miR-4530 (HR = 2.97, P = .011) were associated with longer OS. All these studies investigated circulating miRNAs in serum and plasma, providing evidence to the potential use of circulating miRNAs as valuable biomarkers for HCC patients’ management. However, to our knowledge, there is a lack of studies exploring the value of circulating miRNAs in whole blood, which may provide additional information to clinicians.
In this study, we investigated the potential of blood miRNAs as biomarkers of therapy response in a group of 52 patients treated with hepatic resection or RF. The blood samples were collected at different time points: at the time of diagnosis (T0), 1 month after therapy (T1), and 6 months after therapy (T2). The initial discovery phase, performed on 13 patients, identified 19 differently expressed miRNAs between CR and PRPD patients with miR-370 and miR-3201 being the top-scoring miRNAs. The subsequent validation confirmed the potential of miR-3201 as a biomarker of response to hepatic resection and RF.
The blood levels of miR-3201 were significantly decreased in patients responding to therapy at all the considered time points (P = .026), showing an increasing trend of expression over time in PRPD patients. Moreover, ROC curve analysis evidenced the interesting role of miR-3201 in discriminating CR from PRPD patients with a sensitivity and specificity of 71% and 70%, respectively (AUC = 0.69), thus suggesting its potential use as a noninvasive biomarker to assess the response to curative treatments. In addition, blood miR-3201 showed a relevant performance in identifying patients with longer OS. Indeed, in the Kaplan–Meier analysis, patients with low expression of miR-3201 have a significantly higher OS, at all the considered time points, thus suggesting the potential of miR-3201 as a noninvasive prognostic biomarker for the survival of patients treated with hepatic resection and RF.
The function of miR-3201 is still controversial. MiR-3201 expression was significantly reduced in tissues of recurrent epithelial ovarian cancer,34 while it was significantly increased in the serum of melanoma patients when compared with healthy volunteers.35 In addition Su et al. showed a correlation between serum miR-3201 upregulation and higher OS in pancreatic cancer suggesting the involvement of miR-3201 in different pro-carcinogenic pathways.36 However, all these studies underline the still unclear role of this miRNA in the tumor. Indeed, both the biological function of miR-3201 and its role as a biomarker need to be deeply studied.
Conclusions
Considering the percentage of patients not reaching a complete response at 6 months after therapy, there is still a margin for the improvement of the patient selection and management during the follow-up to increase patients’ life expectancy. In this regard, biomarkers in biofluids may be of particular interest for the stratification of patients responding to curative therapies. In our study, we showed a possible role for blood miR-3201 as a biomarker for HCC patients undergoing curative treatments.
The limitation of our study consists of the restricted number of enrolled patients. However, the repeated measurements of this miRNA at 3 different time points showed a consistency in the differential expression between CR and PRPD patients, as well as in the identification of patients with longer OS. Despite this limitation, the study provides new hints for an extensive study confirming the potential of miR-3201 as a blood biomarker in HCC.
Abbreviations
- AUC
area under the curve
- BCLC
Barcelona Clinic Liver Cancer
- CR
complete responder
- DFS
disease-free survival
- EASL
European Association for the Study of the Liver
- FC
fold change
- HBV
Hepatitis B virus
- HCC
hepatocellular carcinoma
- HR
hazard ratio
- LT
liver transplantation
- miRNAs
microRNAs
- mRECIST
modified Response Evaluation Criteria In Solid Tumors
- NAFLD
non-alcoholic fatty liver disease
- OS
overall survival
- PRPD
partial responder and progressive disease
- REMARK
REporting recommendations for tumour MARKer prognostic studies
- RF
radiofrequency ablation
- RIN
RNA integrity number
- ROC
receiver operating characteristic
- RT-qPCR
quantitative real-time PCR
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study was partially supported by D. NAMICA, PORFESR 2007 to 2013 of the region Friuli Venezia Giulia, Progetti Internazionali 2020 (DGR 2195, December 20, 2019) from Regione Autonoma FVG, Programma di cooperazione Interreg V-A Italia-Slovenia 2014-2020 - Bando mirato per progetti standard n. 07/2019 - Progetto “C3B” CUP J95F19000490006, and an intramural grant from the Italian Liver Foundation.
Luca Grisetti is supported by a PhD scholarship from the University of Trieste.
Institutional Review Board Statement: The study was approved by the regional ethical committee (Comitato Etico Regionale Unico FVG, No. 14/2012 ASUITS).
Informed Consent Statement: Informed consent was obtained from all subjects involved in the study.
Data Availability: The datasets used during the current study are available from the corresponding author upon reasonable request.
ORCID iD: Luca Grisetti https://orcid.org/0000-0002-0243-9290
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Colombet M, Soerjomataram Iet al. et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941-1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Castet F, Heikenwalder Met al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol . 2022;19(3):151-172. doi: 10.1038/s41571-021-00573-2. [DOI] [PubMed] [Google Scholar]
- 4.Galle PR, Forner A, Llovet JMet al. EASL Clinical practice guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69(1):182-236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 5.Petrick JL, Florio AA, Znaor Aet al. International trends in hepatocellular carcinoma incidence, 1978-2012. Int J Cancer. 2020;147(2):317-330. doi: 10.1002/ijc.32723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawla P, Sunkara T, Muralidharan P, Raj JP. Update in global trends and aetiology of hepatocellular carcinoma. Contemp Oncol. 2018;22(3):141-150. doi: 10.5114/wo.2018.78941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.European Association for Study of Liver, European Organisation for Research and Treatment of Cancer. EASL-EORTC clinical practice guidelines: Management of hepatocellular carcinoma. Eur J Cancer. 2012;48(5):599-641. doi: 10.1016/j.ejca.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 8.Pinzani M, Rosselli M, Zuckermann M. Liver cirrhosis. Best Pract Res Clin Gastroenterol. 2011;25(2):281-290. doi: 10.1016/j.bpg.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Zhou W-C, Zhang Q-B, Qiao L. Pathogenesis of liver cirrhosis. WJG. 2014;20(23):7312-7324. doi: 10.3748/wjg.v20.i23.7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W, Wei C. Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 2020;7(3):308-319. doi: 10.1016/j.gendis.2020.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren Z, Ma X, Duan Z, Chen X. Diagnosis, therapy, and prognosis for hepatocellular carcinoma. Anal Cell Pathol. 2020;2020:e8157406. doi: 10.1155/2020/8157406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llovet JM, Zucman-Rossi J, Pikarsky Eet al. et al. Hepatocellular carcinoma. Nature Reviews Disease Primers. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 13.Song P, Cai Y, Tang H, Li C, Huang J. The clinical management of hepatocellular carcinoma worldwide: A concise review and comparison of current guidelines from 2001 to 2017. Biosci Trends. 2017;11(4):389-398. doi: 10.5582/bst.2017.01202. [DOI] [PubMed] [Google Scholar]
- 14.Clavien P-A, Lesurtel M, Bossuyt PMM, Gores GJ, Langer B, Perrier A. Recommendations for liver transplantation for hepatocellular carcinoma: An international consensus conference report. Lancet Oncol. 2012;13(1):e11-e22. doi: 10.1016/S1470-2045(11)70175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Au KP, Chok KSH. Multidisciplinary approach for post-liver transplant recurrence of hepatocellular carcinoma: A proposed management algorithm. World J Gastroenterol. 2018;24(45):5081-5094. doi: 10.3748/wjg.v24.i45.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Xie H, Hu Met al. et al. Recent progress in treatment of hepatocellular carcinoma. Am J Cancer Res. 2020;10(9):2993-3036. [PMC free article] [PubMed] [Google Scholar]
- 17.Cucchetti A, Zhong J, Berhane S, et al. The chances of hepatic resection curing hepatocellular carcinoma. J Hepatol. 2020;72(4):711-717. doi: 10.1016/j.jhep.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 18.Hong SK, Lee K-W, Hong S. Young, et al. Efficacy of Liver Resection for Single Large Hepatocellular Carcinoma in Child-Pugh A Cirrhosis: Analysis of a Nationwide Cancer Registry Database. Frontiers in Oncology. 2021 [accessed July 28, 2021]; 0. https://www.frontiersin.org/articles/10.3389/fonc.2021.674603/full. doi:10.3389/fonc.2021.674603.
- 19.Liu W, Wang K, Bao Q, Sun Y, Xing B-C. Hepatic resection provided long-term survival for patients with intermediate and advanced-stage resectable hepatocellular carcinoma. World J Surg Oncol. 2016;14:62. doi: 10.1186/s12957-016-0811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiina S, Tateishi R, Arano T, et al. Radiofrequency ablation for hepatocellular carcinoma: 10-year outcome and prognostic factors. Am J Gastroenterol. 2012;107(4):569-577. doi: 10.1038/ajg.2011.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee DH, Lee JM, Lee JYet al. et al. Radiofrequency ablation of hepatocellular carcinoma as first-line treatment: Long-term results and prognostic factors in 162 patients with cirrhosis. Radiology. 2014;270(3):900-909. doi: 10.1148/radiol.13130940. [DOI] [PubMed] [Google Scholar]
- 22.Schullian P, Laimer G, Putzer Det al. et al. Stereotactic radiofrequency ablation as first-line treatment of recurrent HCC following hepatic resection. Eur J Surg Oncol. 2020;46(8):1503-1509. doi: 10.1016/j.ejso.2020.03.207. [DOI] [PubMed] [Google Scholar]
- 23.Shen S, Lin Y, Yuan Xet al. et al. Biomarker MicroRNAs for diagnosis, prognosis and treatment of hepatocellular carcinoma: A functional survey and comparison. Sci Rep. 2016;6:38311. doi: 10.1038/srep38311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin Y, Wong YS, Goh BKP, et alet al. Circulating microRNAs as Potential Diagnostic and Prognostic Biomarkers in Hepatocellular Carcinoma. Scientific Reports. 2019 [accessed May 20, 2020]; 9. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6639394. doi: 10.1038/s41598-019-46872-8. [DOI]
- 25.Cui M, Wang H, Yao Xet al. et al. Circulating MicroRNAs in cancer: Potential and challenge. Front Genet. 2019;10:626. doi: 10.3389/fgene.2019.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwak PB, Iwasaki S, Tomari Y. The microRNA pathway and cancer. Cancer Sci. 2010;101(11):2309-2315. doi: 10.1111/j.1349-7006.2010.01683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grasedieck S, Sorrentino A, Langer Cet al. et al. Circulating microRNAs in hematological diseases: Principles, challenges, and perspectives. Blood . 2013;121(25):4977-4984. doi: 10.1182/blood-2013-01-480079. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Peng R, Wang J, Qin Z, Xue L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin Epigenetics. 2018;10(1):59. doi: 10.1186/s13148-018-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumour MARKer prognostic studies (REMARK). Br J Cancer. 2005;93(4):387-391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pascut D, Krmac H, Gilardi Fet al. et al. A comparative characterization of the circulating miRNome in whole blood and serum of HCC patients. Sci Rep. 2019;9:8265. doi: 10.1038/s41598-019-44580-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho HJ, Kim SS, Nam JS, et al. Low levels of circulating microRNA-26a/29a as poor prognostic markers in patients with hepatocellular carcinoma who underwent curative treatment. Clin Res Hepatol Gastroenterol. 2017;41(2):181-189. doi: 10.1016/j.clinre.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 32.Yokota Y, Noda T, Okumura Y, et al. Serum exosomal miR-638 is a prognostic marker of HCC via downregulation of VE-cadherin and ZO-1 of endothelial cells. Cancer Sci. 2021;112(3):1275-1288. doi: 10.1111/cas.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pratama MY, Visintin A, Crocè LS, Tiribelli C, Pascut D. Circulatory miRNA as a biomarker for therapy response and disease-free survival in hepatocellular carcinoma. Cancers (Basel). 2020;12(10):E2810. doi: 10.3390/cancers12102810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chong GO, Jeon H-S, Han HSet al. et al. Differential MicroRNA expression profiles in primary and recurrent epithelial ovarian cancer. Anticancer Res. 2015;35(5):2611-2617. [PubMed] [Google Scholar]
- 35.Margue C, Reinsbach S, Philippidou Det al. Comparison of a healthy miRNome with melanoma patient miRNomes: Are microRNAs suitable serum biomarkers for cancer? Oncotarget. 2015;6(14):12110-12127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su Q, Zhu EC, Qu Yet al. et al. Serum level of co-expressed hub miRNAs as diagnostic and prognostic biomarkers for pancreatic ductal adenocarcinoma. J Cancer. 2018;9(21):3991-3999. doi: 10.7150/jca.27697. [DOI] [PMC free article] [PubMed] [Google Scholar]