Abstract
Leaf morphology can affect the development and yield of plants by regulating plant architecture and photosynthesis. Several factors can determine the final leaf morphology, including the leaf complexity, size, shape, and margin type, which suggests that leaf morphogenesis is a complex regulation network. The formation of diverse leaf morphology is precisely controlled by gene regulation on translation and transcription levels. To further reveal this, more and more genome data has been published for different kinds of vegetable crops and advanced genotyping approaches have also been applied to identify the causal genes for the target traits. Therefore, the studies on the molecular regulation of leaf morphogenesis in vegetable crops have also been largely improved. This review will summarize the progress on identified genes or regulatory mechanisms of leaf morphogenesis and development in vegetable crops. These identified markers can be applied for further molecular-assisted selection (MAS) in vegetable crops. Overall, the review will contribute to understanding the leaf morphology of different crops from the perspective of molecular regulation and shortening the breeding cycle for vegetable crops.
Keywords: leaf morphogenesis, vegetable crops, gene identification, molecular assisted selection (MAS), genetic map
Introduction
Since the plant leaf is an organ that grows from almost indistinguishable primordia into shapes that vary largely among genus, species, or even cultivars (Kierzkowski et al., 2019), there have been several classification methods according to different characteristics. For example, leaves can be divided into simple or complex according to the number of leaflets, lobbed or serrated according to the margin shape, and inward-curvature or outward-curvature according to the curvature direction of the leaf margin. Well-organized gene and hormone regulatory mechanisms are the prerequisites for the formation of leaf diversity.
At present, the molecular mechanisms of leaf development and morphogenesis have been well studied in the dicotyledons plant, Arabidopsis, and the monocotyledons plant, rice. Arabidopsis typically has simple and small serrated leaves and is thus the ideal plant to study leaf shape and margin formation. Previous studies have shown that leaf serration formation depends on the guidance of auxin maxima at their tips (Kawamura et al., 2010), and many proteins can regulate the leaf serrations of Arabidopsis by direct or indirect auxin process (Kong et al., 2019). Auxin biosynthesis-related proteins, YUCCA (YUC), indole-3-acetic acid carboxyl methyltransferase (IAMT1), AUXIN-RESISTANT 1 (AUX1), and PIN-FORMED 1 (PIN1) (Hay et al., 2006; Wenzel et al., 2007; Kim et al., 2011; Kasprzewska et al., 2015) can regulate the auxin maxima directly in the leaf tips for Arabidopsis. Also, the auxin and ASYMMETRIC LEAVES1 (AS1) signaling pathways converge to suppress the expression of a KNOTTED1-like homeobox (KNOX) gene, BREVIPEDICELLUS (BP), which plays a key role in generating the auxin maxima at the tips of leaf serrations (Hay et al., 2006). The mutation in INDOLE-3-BUTYRIC ACID RESPONSE 5 (IBR5) can result in changed serrations due to the disturbed auxin reporters and increased cell area (Kong et al., 2019). Besides the auxin regulatory process, other factors like gibberellins (GAs), cytokinins (CK), and microRNAs were also involved in leaf margin formation in Arabidopsis (Du et al., 2018), and these results also provide a further reference for the knowledge of leaf morphogenesis.
Unlike Arabidopsis, the research on rice leaf morphogenesis mainly focuses on its spatial extension due to the importance of ideal plant architecture, which plays a significant role in photosynthetic efficiency and grain yield (Heath and Gregory, 1938). Therefore, using molecular genetic techniques to produce proper leaf morphology and improve the photosynthesis rate can balance the relationship with the grain sink, which will effectively achieve a high yield in rice. For leaf curvature, four types of genes can control leaf curvature direction and degree according to the different gene functions, including genes regulating the adaxial/abaxial side, cell development in the adaxial side, development of sclerenchyma in the abaxial side and abnormal cuticle development (Fu et al., 2019). As for the leaf size regulation in rice, cell division-related genes, such as SEMI-ROLLED LEAF 2 (SRL2), SHALLOT-LIKE 1 (SLL1/RL9), CATION-CHLORIDE COTRANSPORTER (OsCCC1), and WUSCHEL-related homeobox 4 (OsWOX4) (Yan et al., 2006; Zhang et al., 2009; Kong et al., 2011; Liu et al., 2016; Yasui et al., 2018), transcription factors and cellulases NARROW AND ROLLED LEAF 1 (NAL2/NAL3), SLL1 (Hu et al., 2010; Zhang et al., 2014) genes related to auxin synthesis and metabolism, NAL7, TRYPTOPHAN DEFICIENT DWARF (TDD1), CONSTITUTIVELY WILTED 1 (OsCOW1), OsARF19 (Woo et al., 2007; Fujino et al., 2008; Sazuka et al., 2009; Zhang et al., 2016) have been cloned and some of these genes have been widely used in the rice molecular breeding to cultivate the proper architecture.
Besides the research on the model plants, Arabidopsis and rice, there is still a need for further understanding of the leaf development of other species which is essential to understand species-specific spatiotemporal activation of conserved processes in leaf patterning. Vegetable crops are the essential nutrient resources in our daily life so there is a demand for high-yield production and quality. Leaf size has a direct effect on the production of leafy vegetables, and leaf shape has a close relationship with fruit quality (Rowland et al., 2020). With the further discovery of high-quality genomes for vegetable crops (Hao et al., 2020), the genes regulating the leaf morphogenesis in vegetable crops have been clarified from the genome sequencing and re-sequencing data. With the highly efficient transgenic system in many vegetable crops, the functional confirmation for these identified genes also becomes more reliable. Therefore, high-density and accurate genetic maps have been constructed for many important traits in vegetable crops. In this review, the present molecular studies on leaf morphogenesis in vegetable crops are summarized which is conducive to the understanding of leaf development and contributes to molecular-assisted breeding.
The typical leaf morphology in vegetable crops
The leaf has diverse types in natural plants and can be classified according to different principles. Among them, leaf types can be generally classified into two categories according to the leaf initiation: simple or compound leaf. A simple leaf consists of a single undivided blade and a compound leaf consists of multiple leaflets, organized in pinnate, palmate, or higher-ordered structures (Champagne and Sinha, 2004). In vegetable crops, tomatoes and peas have been used as typical materials to study the formation of compound leaves. But the formation pattern of compound leaves is still quite different between these two species. Tomato has a pinnate compound leaf which is formed by three stages of leaflets, including primary (I), secondary (II), and intercalary (Int), and the leaflets have dissected margins (Blein et al., 2010). Peas have classic genetic material and some studies have indicated that their leaves are formed by several pairs of proximal leaflets (lt), distal tendrils (tdl), and leaflet-like stipules (st) (DeMason and Chetty, 2011). The complex leaf development of peas is usually divided into five stages. The P0 stage is differentiated from the apical meristem to produce the complex leaf primordium, then in the late P1 stage the primordium appears, and in the P2 stage develops the leaflet primordium. Tendril primordium develops in the P3 stage and the P4 stage forms terminal tendrils, which contribute to climbing and supporting plant growth (Gourlay et al., 2000). Therefore, due to the largely differentiated compound leaf development processes of these vegetable crops, the study of these species can contribute to a more comprehensive understanding of the process and mechanism of compound leaf development.
In addition, there is a class of important leafy plants in vegetable crops, whose leaves as the main edible organs, leaf development will greatly affect the yield and consumer choice. The proper curvature time and position of head leaves are essential for the formation of leafy heads. Among them, leafy heads vegetables include Chinese cabbage (Brassica rapa ssp. pekinensis, syn.) (Ren et al., 2020), cabbage (B. oleracea var. capitata) (Alemán-Báez et al., 2022), and lettuce (Lactuca sativa) (An et al., 2022). Leaf curvature as well as the occurrence of the leafy head is a complex biological process and also a morphological response induced by environmental factors. These factors can control the differentiation of abaxial–adaxial polarity during leaf development (Nath et al., 2003). Meanwhile, the polarity regulation process is related to cell cycle activity, cell size, cell division speed, and cell differentiation direction (Nath et al., 2003). If the cell division and growth at the edge of the leaves are slower than in the middle region, they develop into cup-like leaves with a positive Gaussian curvature value, whereas the leaves develop saddle-like with a negative Gaussian curvature value and form a leafy head (Ren et al., 2020).
Therefore, leaf morphology in vegetable crops is distinct from that in Arabidopsis thaliana or monocotyledonous rice and maize, which provides the basis for studying leaf morphogenesis. According to the previous study on the model plants and the development of genome sequencing of various vegetable crops in recent years, some important markers linked with leaf morphogenesis have been identified in vegetable crops and some known genes with novel functions were also confirmed.
The regulation mechanism of leaf complexity in vegetable crops
Plant leaves initiate from the flanks of the shoot apical meristem (SAM), which are regulated by the complex genetic network (Du et al., 2018) and precise hormone concentration (Shwartz et al., 2016). When the leaf initiates from SAM, it is divided into either a simple or compound leaf and this is determined by some related genes and multiple hormones. In some vegetable crops, especially compound leaf crops such as tomatoes and peas, the molecular and physiological mechanisms also have been clarified and summarized in Table 1 .
Table 1.
Genes are involved in leaf complexity in tomato and pea.
| Gene | Species | Description | phenotype | References |
|---|---|---|---|---|
| KNOX1 | tomato | KNOX family | Overexpression of KNOX1 caused more complex leaf structure | Bharathan and Sinha, 2001 |
| PTS/TKD1 | tomato | KNOX family | 1 bp deletion in PTS/TKD1 increased leaf complexity | Kimura et al., 2008 |
| UNI | pea | LEAFY transcription factors | uni mutants produced the simple leaf | Wang et al., 2008 |
| BIP | tomato | BEL-LIKE Homeodomain gene | bip mutant showed highly complex leaf phenotype in
heirloom tomato |
Nakayama et al., 2021 |
| CRISPA | pea | MYB transcription factors | crispa mutants showed ectopic stipules, narrow leaflets, and shortened petioles with excessive adaxial expansion | DeMason and Chetty, 2014 |
| GOB | tomato | CUC subfamily protein | Primary leaflets of gob mutant are often fused, secondary leaflets and marginal serrations are absent | Blein et al., 2008; Berger et al., 2009; Ben-Gera and Ori, 2012 |
| c | tomato | RAX1 protein | c mutant reduced leaf complexity | Busch et al., 2011 |
| PIN1 | tomato and pea | Auxin efflux protein | pin1 mutants influenced leaf and leaflet initiation | Koenig et al., 2009; Zhou et al., 2011 |
| leafless | tomato | DORNRONSCHEN protein | leafless (lfs) in tomato can’t produce leaves and leaflets | Capua and Eshed, 2017 |
| IAA9 | tomato | Aux/IAA family protein | iaa9 mutants showed simplified leaves | Wang et al., 2005; Zhang et al., 2007 |
| ARF10 | tomato | Auxin response protein | The mutation in ARF10 inhibited the blade and leaflets outgrowth in tomato | Ben-Gera et al., 2016; Damodharan et al., 2016; Lin et al., 2016 |
| AGO7 | tomato | ARGONAUTE family | Overexpression of AGO7 resulted in increased leaflet numbers | Lin et al., 2016 |
| clausa | tomato | MYB transcription factor | clausa mutants elaborated compound leaves | Avivi et al., 2000; Bar et al., 2016 |
| GA2ox4 | tomato | Gibberellin 2-oxidase | Overexpression of GA2ox4 increased leaf complexity | Yanai et al., 2011 |
| PRO | tomato | DELLA protein | pro mutant showed simplified leaves | Marti et al., 2007; Bassel et al., 2008; Jasinski et al., 2008 |
In complex and fine gene regulatory networks, the class1 Knotted-like homeobox (KNOX1) gene family is essential for meristem initiation and maintenance of differentiation (Bharathan and Sinha, 2001; Wang and Jiao, 2020). When KNOX1 genes are continuously expressed, they will result in the formation of complex leaf structures in tomatoes (Bharathan and Sinha, 2001). Then, the increased expression level of KNOX1 can produce a more complex leaf structure (Hay and Tsiantis, 2010). A large-scale linkage analysis among the accessions with different leaf complexity was performed, and 1 bp deletion from the KNOX family gene (PTS/TKD1) was co-separated with the increased leaf complexity phenotype (Kimura et al., 2008). Similarly, a Potato-leaf (c) mutant showed reduced leaf complexity in tomatoes compared with the wild type, the causal gene of the c mutant was identified by the comparative genomics and showed different alleles among cultivars, which encoded Regulator of Axillary Meristem1 (RAX 1) and participated in leaf complexity patterning (Busch et al., 2011). Recently, two different Homeobox genes in tomatoes, BIPINNATA (BIP) and WUSCHEL RELATED HOMEOBOX 1 (WOX1), contribute to leaf complexity and leaflet development, respectively (Nakayama et al., 2021). Comparative genomics analysis among the wild type and domesticated tomato species revealed the bip mutation in heirloom tomatoes arose de novo during the domestication process (Nakayama et al., 2021). Therefore, such a locus identified from the natural population can be a reliable marker for the selection of tomato leaf morphology.
Unlike tomatoes, studies have found that peas belong to the legumes inverted repeat-lacking clade (IRLC) family of plants. This branch plant has a different compound leaf regulation mechanism from tomatoes and other legumes (Bar and Ori, 2015). To begin with, KNOX1 is not expressed in the leaf primordia of peas, suggesting that KNOX1 is not involved in compound leaf development in peas (Hofer et al., 2009). Instead, UNIFOLIATA (UNI) in peas encodes LEAFY (LFY)-type transcription factors and plays a similar role as KNOX1 in tomatoes (Hofer et al., 1997). UNI is initially expressed in the complex leaf primordium when the leaflets have not differentiated. With the differentiation of the leaflet primordium, the expression level of UNI was enhanced in the newly produced leaflet primordium (Hofer et al., 1997). Pea uni mutants produced simple leaves which also indicated that UNI maintains the complex leaf development state in peas (Hofer et al., 1997). Therefore, the homologous gene of LFY in peas acts as a key regulator to control the formation of compound leaves.
Besides, there are some genes regulating the leaf complexity on the transcriptional level. PHANTASTICA (SlPHAN), containing an MYB-domain protein, can regulate leaflet formation in tomatoes (Kim et al., 2003a). The expression pattern indicated that SlPHAN was expressed throughout the entire adaxial side of the leaf primordium and formed pinnate compound leaves (Kim et al., 2003a). The increased expression level of SlPHAN also can result in a variety of leaf morphologies, from simple, ternate to compound leaves (Zoulias et al., 2012). Meanwhile, SlPHAN has a complex relationship with the KNOX gene family to regulate complex formation in tomatoes. LeT6 and SlPHAN are expressed in the compound leaf and leaflet primordium (Kim et al., 2003b), but SlPHAN is down-regulated in overexpressed LeT6 mutant, which indicates that LeT6 is a negative regulator of SlPHAN (Kim et al., 2003b). There are other MYB transcription factors identified in peas. CRISPA is the ortholog of PHAN in peas, the cri-1 (CRISPA) mutant exhibits ectopic stipules, narrow leaflets, and shortened petioles with excessive adaxial expansion (Tattersall et al., 2005). CRI suppresses KNOX1 and UNI to maintain the normal development and expansion of leaves in peas (DeMason and Chetty, 2014). GOBLET (GOB) belongs to the CUP-SHAPED COTYLEDONS (CUC) subfamily, which can affect compound leaf patterning largely by its expression position, time, and level ( Blein et al., 2008; Berger et al., 2009; Ben-Gera and Ori, 2012). GOB is negatively controlled by the MYB transcription factor CLAUSA (CLUA) and microRNA miR164, whose ectopic expression in tomatoes can result in extremely elaborate compound leaves and absent secondary leaflet initiation, respectively (Berger et al., 2009; Bar and Ori, 2015).
Precise hormone regulation is also involved in the formation of leaf complexity in vegetable crops. Firstly, the leaf development from the primordium requires the distribution of auxin in precise locations within a specific spatiotemporal developmental context (Koenig et al., 2009; Ben-Gera and Ori, 2012). The treatment of exogenous auxin or auxin transport inhibitors induces leaflet initiation or the simplification of leaf forms in peas and tomatoes, respectively (DeMason and Chawla, 2004; Koenig et al., 2009). Auxin activity maxima are also related to leaflet initiation. An auxin efflux protein PIN1 coincides with leaf and leaflet initiation in tomatoes and peas (Bai and DeMason, 2006; Koenig et al., 2009). The recombination-based mapping results showed that a severe auxin-defective tomato mutant leafless (lfs), which cannot produce leaves and leaflets, is due to the transient expression of DORNRONSCHEN (DRN) at incipient and young primordia overlapped with auxin response maxima (Capua and Eshed, 2017). Additionally, there are some auxin signaling and response proteins involved in the subsequent development of leaf dissection. The mutation in SlIAA9, an auxin response protein, exhibited simplified leaves and a similar phenotype in the tomato mutant entire (e), indicating that IAA9 was the causal gene for the e mutant (Wang et al., 2005; Zhang et al., 2007). The expression level of E/SlIAA9 has an effect on restricting lamina outgrowth between leaflets (Koenig et al., 2009; Ben-Gera and Ori, 2012). At the translation and post-translation level, E/SlIAA9 can interact with TRANSPORT INHIBITOR RESPONSE1 (SlTIR) and AUXIN F-BOX (SlAFB6), which are auxin receptors, and can be degraded by the ubiquitin 26S proteasome SCFTIR1/AFB under specific auxin level (Dharmasiri et al., 2005; Kepinski and Leyser, 2005). The auxin response protein family (ARFs) has a redundant function in leaf complexity and is always controlled by microRNAs (Ben-Gera et al., 2016; Wu et al., 2018). The repression of SlARF10 by SlmiR160 can inhibit the blade and leaflet outgrowth in tomatoes (Ben-Gera et al., 2016; Damodharan et al., 2016; Lin et al., 2016). Besides, the activity of ARF proteins is also negatively controlled by the trans-acting small interfering RNAs (ta-siRNA) and they are involved in leaf development among different plant species (Yan et al., 2010; Lin et al., 2016). In tomatoes, the tasiRNA-ARFs module was an ARGONAUTE (AGO)-dependent process. A wiry mutant with a simpler and narrow leaf in tomatoes is due to a mutation in AGO7. Conversely, the overexpression of AGO7 in tomatoes altered the production of ta-siRNA and repression of ARFs activity (Yifhar et al., 2012; Lin et al., 2016). It is demonstrated among different species that gene regulation or exogenous auxin application will cause disrupted auxin distribution and affect leaf and leaflet initiation.
Besides auxin, GAs and CK are also important for leaf morphological formation in some vegetable crops. CK usually cooperates with auxin to regulate leaf initiation. Endogenous CK can increase the expression level of CK-related genes ISOPENTENYL TRANSFERASE 7 (IPT7) and CYTOKININ OXIDASE/DEHYDROGENASE3 (CKX3) in tomatoes and then change the leaf complexity (Shwartz et al., 2016). Both local applications of auxin to develop leaf primordia and mutations in SlIAA9 can suppress the expression level of CK biosynthesis gene IPT7 and the super-compound leaf formation, suggesting that CK regulation of dissected leaf morphogenesis requires a localized auxin response (Shwartz et al., 2016). An MYB transcription factor mutant clausa showed increased leaf complexity. CK signaling was enhanced by CLAU, thus uncovering an additional instance in which heightened CK signals result in increased complexity of the tomato leaves (Avivi et al., 2000; Bar et al., 2016). Similarly, the application of exogenous GA will lead the leaves to form a simplified leaf phenotype in tomatoes (Hay et al., 2002; Fleishon et al., 2011). Some GA-related genes are also involved in the formation of leaf complexity in tomatoes and peas. GA up-regulated the expression level of UNI to make the leaves more complex in peas (DeMason and Chetty, 2011). The overexpression of a GA-responsive gene Gibberellin 2-oxidase (GA2ox4) in tomatoes and peas caused increased leaf complexity (Yanai et al., 2011). The mutation of the homologous gene PROCERA (PRO) which encodes DELLA protein in tomatoes exhibits simplified leaves (Marti et al., 2007; Bassel et al., 2008; Jasinski et al., 2008). Besides, GA application or clau pro double mutants can repress the compound leaf phenotype of the clau mutant, failing to delimit the expression domain of KNOXI, then displaying elevated KNOXI expression range and increased leaf complexity (Hay et al., 2002).
Genes related to leaf curvature in vegetable crops
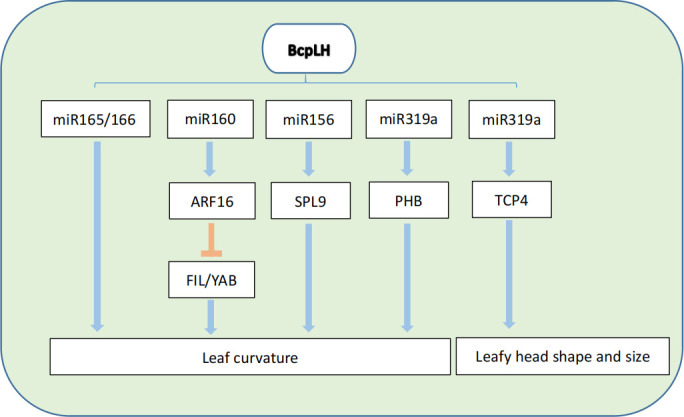
The different models of leaf adaxial-abaxial patterning can influence the leaf initiation and following development process. At the initiation stage, the defective adaxial-abaxial patterning can influence the normal initiation from the SAM and, at the development stage, the adaxial-abaxial patterning can also result in the upward- or downward-curling leaf phenotype (Yamaguchi et al., 2012). Leaf curvature is the most important factor in leafy head formation and the occurrence of the leafy head is also regulated by several adaxial-abaxial related genes and environmental factors (Ito, 1957; Liang et al., 2016). Among them, two curvature-related genes have been cloned in Chinese cabbage, Brassica rapa ssp. pekinensis LEAFY HEADS (BcpLH) and BcpLH2, which may have a redundant function in leafy head formation (Yu et al., 2000; Ren et al., 2020). BcpLH is a close homolog of Arabidopsis HYPONASTIC LEAVES 1 (HYL1) in Chinese cabbage, which participates in miRNA biogenesis. BcpLH can control curvature development and leafy head formation in Chinese cabbage by coordinating with a set of microRNAs, including miR165/166, miR160, and miR319a to regulate the time of leaf curvature ( Figure 1 ) (Ren et al., 2018; Ren et al., 2020). Conversely, these microRNAs can target some adaxial-determined genes and control the expression ratio to determine the degree and direction of leaf curvature. miR319a can positively regulate the abaxial-determined gene PHABULOSA (PHB), and ARF16 is a target gene of miR160 which can negatively regulate adaxial-determined gene FIL/YAB1 (Liu et al., 2011; Ren et al., 2018; Ren et al., 2020). SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9 (BrpSPL9) was identified by the transgenic system and miR156 can regulate BrpSPL9 to control leaf curvature, which has been considered to be a crucial gene for genetic improvement of Chinese cabbage (Wang et al., 2014). Using the recombinant inbred line (RILs) of heading and non-heading Chinese cabbage combined with QTL analysis, the miR319-targeted gene TEOSINTE BRANCHED1/CYCLOIDIA/PCF (TCP4) can regulate the shape and size of the leafy head by controlling the expression level of TCP4 ( Figure 1 ) (Yu et al., 2013; Mao et al., 2014). The other transcription factor BcMYB101 was also identified to possibly participate in the development of curly leaves in non-heading pak choi (Hou et al., 2020).
Figure 1.
Genetic regulation of leafy head development in Chinese Cabbage.
In order to further investigate the molecular mechanism of leafy head formation, segregated populations were constructed using non-heading pak choi (PC-101) with Chinese cabbage, then a high-density DH-88 genetic map for leafy head formation-related QTLs was generated, which was located on the chromosomes A03, A05, and A08. These QTLs related to heading traits were identified to contribute the breeding selection in Chinese cabbage (Sun et al., 2018). Another recombination of pak choi (PC-101) and Chinese cabbage (CC-48) was also constructed and 22 QTLs for leafy head-related traits were discovered. Among them, five QTLs were located on chromosomes A04 and A05 linked with head leaf shape (Sun et al., 2021). In lettuce, two cultivars including the heading lettuce (PI536839) and the non-heading lettuce (PI344074) were used to generate a genetic population. After BSR-seq and complementation test, a homologous gene of knotted 1 (KN1) in maize was the responsible gene for heading in lettuce. KN1 protein in lettuce (LsKN1) has conserved KNOX and homeobox domains (Yu et al., 2020). Besides the regulation at the genetic level, the plant hormones and environmental conditions are also critical for leaf curvature. Previous physiological experiments have confirmed that the formation of the leafy head of cabbage is due to the growth of the unbalanced distribution of auxin on both sides of the leafy head (Dong and Huang, 2018). External environmental factors can influence leafy head development including the photosynthesis period, temperature, and fertilizer application (Ito, 1957). Thus, the cooperation between genetic background and proper growth environment can promote the good growth of leafy vegetable crops.
Genes related to leaf shape and size formation in vegetable crops
After leaf initiation from SAM, determinate cells are induced to change division patterns along three axes: proximal/distal, adaxial/abaxial, and medial/lateral. These affect the later cell division and extension, thus controlling the final leaf size and the formation of the leaf margin (Dkhar and Pareek, 2014). The complicated process of leaf shape and size also indicates they are quantitative traits and are controlled by multiple genes. Therefore, several QTLs or genes linked with leaf shape have been identified in vegetable crops by QTL mapping combined with other sequencing methods. For instance, two QTLs (qLeaf_or-1, qLeaf_or-2) with a high effect on leaf traits were mapped between two pea lines with tendrilless and afila leaf types and combined with specific locus amplified fragment sequencing (SLAF-seq), a high-density genetic map was constructed containing many SNP markers linked to leaf traits (Zheng et al., 2019). A comparative mapping and gene cloning approach among bio, ele1 (larger leaf size), and lath (smaller leaf size) identified a KIX domain protein and an ortholog of Arabidopsis PEAPOD (PPD) which were involved in organ size determination in peas (Li et al., 2019). A WD40 repeat domain-containing protein was mapped to regulate the leaf size in cucumbers and the mutation site W264 in this protein was also conserved in the diverse natural cucumber population. Besides genetic analysis, the transcriptome analysis also provided evidence that LL might regulate or interact with some known organ size genes including AP2-like ethylene-responsive transcription factor (ANT-like), GROWTH-REGULATING FACTOR (GRF-1), GRF-4, GIF-1, TCP4, TCP7, and TCP15 to regulate the leaf and lateral organ size together in cucumbers (Yang et al., 2018). The characteristics of the leaf margin and the underlying mechanisms also confer additional complexity which results in diverse leaf shapes. The main types of leaf margins are entire, serrated, or lobed. Recent genetic analysis and mapping of the genes related to the lobed-leaf trait in various vegetable crops showed that the lobed-leaf is controlled by a single dominant gene, LOBED LEAF1 (LL1), in watermelons (Citrullus lanatus L.) (Wei et al., 2017), but by a single recessive gene in melons (Cucumis melo L.) (Gao et al., 2014). On the contrary, it was believed that the lobed leaf was a quantitative trait and it was speculated that four candidate genes were responsible in ornamental kale (Ren et al., 2019). Later, a fine mapping and BSA-seq result from two inbred lines with different leaf phenotypes in ornamental kale indicated the candidate gene for the feathered-leaved trait (BoFL) was located on chromosome C9 and co-segregated with a CAPS marker (CAPS4610) (Feng et al., 2020). Some known proteins performed a novel function in the leaf margin development of vegetable crops. The SNP in CsPID identified by MutMap+ analysis caused the palmate cucumber leaves to become round with a lobed margin (Zhang et al., 2018; Liu et al., 2019; Song et al., 2019). PINOID (PID) encodes a serine/threonine kinase and catalyzes the phosphorylation of auxin efflux transporter PIN to regulate the cellular auxin efflux, which has a function in inflorescence and root development in Arabidopsis, maize, and rice (Benjamins et al., 2003; McSteen et al., 2007; Morita and Kyozuka, 2007). But in cucumbers, CsPID not only participates in floral organ development but also regulates leaf margin development (Zhang et al., 2018; Liu et al., 2019; Song et al., 2019).
There are also other genes regulating the leaf shape and size formation in the transcriptional process. YABBY family genes have been reported to be involved in auxin flux and auxin response in Arabidopsis (Sarojam et al., 2010). And in tomatoes, the co-expression of YABBY1 and YABBY3 can regulate the leaf size both in cultivated and wild species, and the evolutionary analysis suggests that YABBY1 emerged earlier than YABBY3, evolving from a common ancestor before the divergence of dicotyledonous plants into Rosids and Asterids (Filyushin et al., 2018). The down-regulation of ARGONAUTE1 (AGO1) in tomatoes resulted in morphological defects in leaf adaxial-abaxial patterning, meanwhile, the expression level of some adaxial-abaxial domain formation-related genes also significantly changes, such as ARF4 and NPR5 (Wang et al., 2015). In tomatoes, driving the miR164-resistant form of GOBm via the FIL promoter in tomatoes produced rounded, rumpled, and deeply lobed leaves (Berger et al., 2009). Besides, the ectopic expression of HANABA TARANU (CsHAN1) in cucumbers produced highly lobed leaves (Ding et al., 2015). The cooperation between these genes can determine the type of leaf margin.
Summary and outlook
The previous studies on dicots Arabidopsis and monocots rice have revealed the formation process of leaf morphogenesis (Vercruysse et al., 2020; Wang et al., 2020), and provided further reference for the leaf morphogenesis of other plant species. Leaf morphology plays a vital role in some vegetable crops, especially, since leaves are the edible organs for some leafy vegetable crops including Chinese cabbage and lettuce. Therefore, the research on leaf morphology of vegetable crops can largely contribute to a high yield and quality which are the main goals of breeding. At present, MAS breeding has become a major method to improve breeding efficiency and shorten the breeding cycle. However, due to gene evolution and leaf characters, many genes showed new functions or mechanisms in vegetable crops that were species-specific. Understanding the molecular mechanism in different species also can provide more knowledge about leaf morphogenesis. This review summarized the identified genes and molecular mechanisms for leaf morphology in vegetable crops and provided a reference for the application of some reliable markers in vegetable crops. For example, heading is a typical trait for some leafy vegetable crops and there is always different demand for heading and non-heading cultivars according to the consumers’ preferences. Several heading-related markers (BrpSPL9, BcpLH, BcpLH2) have been identified in Chinese cabbage which can be applied for genotyping among different cultivars at an early seeding stage (Wang et al., 2014; Ren et al., 2018; Ren et al., 2020). Besides MAS, the transgenetic system has also become more reliable in some vegetable crops (Kim et al., 2021), thus, transformation by precisely regulating the target gene could be a possible approach for high-efficiency breeding. For instance, leaf size is often precisely controlled by the expression level of some leaf-size-related genes, including TCP4, GRF, and some microRNAs (Yu et al., 2013; Mao et al., 2014; Yang et al., 2018). The vector construction by overexpression, knockout, or knockdown of the target gene and transformation into certain cultivars can then largely proceed in the breeding process. With the improvement of genome information and the maturity of the genetic transformation system of various vegetable crops, the problems in transformation, such as the off-target and redundant functions of several genes, could be avoided as much as possible. Overall, due to the high consumption of vegetable crops, a reduced breeding cycle by screening at the seedling stage or genetic transformation could be conducive to breeding ideal cultivars for vegetable crops.
Author contributions
NH and TW wrote the article. JC, CW, YZ and YD summarized the references. All authors contributed to the article and approved the submitted version.
Funding
This review is supported by the National Natural Science Foundation of China (31972407, 31972429, 32011540003), and the Natural Science Foundation of Hunan Province (2020JJ4363 and 2021JJ10032).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Alemán-Báez J., Qin J., Cai C., Zou C., Bucher J., Paulo M., et al. (2022). Genetic dissection of morphological variation in rosette leaves and leafy heads in cabbage (Brassica oleracea var. capitata). Theor. Appl. Genet. 135, 3611–3628. doi: 10.1007/s00122-022-04205-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- An G., Yu C., Yan C., Wang M., Zhang W., Jia Y., et al. (2022). Loss-of-function of SAWTOOTH 1 affects leaf dorsiventrality genes to promote leafy heads in lettuce. Plant Cell 34, 4329–4327. doi: 10.1093/plcell/koac234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avivi Y., Lev-Yadun S., Morozova N., Libs L., Williams L., Zhao J., et al. (2000). Clausa, a tomato mutant with a wide range of phenotypic perturbations, displays a cell type-dependent expression of the homeobox gene LeT6/TKn2 . Plant Physiol. 124, 541–552. doi: 10.1104/pp.124.2.541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F., DeMason D. (2006). Hormone interactions and regulation of unifoliata, PsPK2, PsPIN1 and LE gene expression in pea (Pisum sativum) shoot tips. Plant Cell Physiol. 47, 935–948. doi: 10.1093/pcp/pcj066 [DOI] [PubMed] [Google Scholar]
- Bar M., Israeli A., Levy M., Ben Gera H., Jimenez-Gomez J. M., Kouril S., et al. (2016). CLAUSA is a MYB transcription factor that promotes leaf differentiation by attenuating cytokinin signaling. Plant Cell 28, 1602–1615. doi: 10.1105/tpc.16.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M., Ori N. (2015). Compound leaf development in model plant species. Curr. Opin. Plant Biol. 23, 61–69. doi: 10.1016/j.pbi.2014.10.007 [DOI] [PubMed] [Google Scholar]
- Bassel G. W., Mullen R. T., Bewley J. D. (2008). procera is a putative DELLA mutant in tomato (Solanum lycopersicum): Effects on the seed and vegetative plant. J. Exp. Bot. 59, 585–593. doi: 10.1093/jxb/erm354 [DOI] [PubMed] [Google Scholar]
- Ben-Gera H., Dafna A., Alvarez J. P., Bar M., Mauerer M., Ori N. (2016). Auxin-mediated lamina growth in tomato leaves is restricted by two parallel mechanisms. Plant J. 86, 443–457. doi: 10.1111/tpj.13188 [DOI] [PubMed] [Google Scholar]
- Ben-Gera H., Ori N. (2012). Auxin and LANCEOLATE affect leaf shape in tomato via different developmental processes. Plant Signal. Behav. 7, 1255–1257. doi: 10.4161/psb.21550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamins R., Ampudia C. S., Hooykaas P. J., Offringa R. (2003). PINOID-mediated signaling involves calcium-binding proteins. Plant Physiol. 132, 1623–1630. doi: 10.1104/pp.103.019943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger Y., Harpaz-Saad S., Brand A., Melnik H., Sirding N., Alvarez J. P., et al. (2009). The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 136, 823–832. doi: 10.1242/dev.031625 [DOI] [PubMed] [Google Scholar]
- Bharathan G., Sinha N. R. (2001). The regulation of compound leaf development. Plant Physiol. 127, 1533–1538. doi: 10.1104/pp.010867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blein T., Hasson A., Laufs P. (2010). Leaf development: what it needs to be complex. Curr. Opin. Plant Biol. 13, 75–82. doi: 10.1016/j.pbi.2009.09.017 [DOI] [PubMed] [Google Scholar]
- Blein T., Pulido A., Vialette-Guiraud A., Nikovics K., Morin H., Hay A., et al. (2008). A conserved molecular framework for compound leaf development. Science 322, 1835–1839. doi: 10.1126/science.1166168 [DOI] [PubMed] [Google Scholar]
- Busch B. L., Schmitz G., Rossmann S., Piron F., Ding J., Bendahmane A., et al. (2011). Shoot branching and leaf dissection in tomato are regulated by homologous gene modules. Plant Cell 23, 3595–3609. doi: 10.1105/tpc.111.087981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capua Y., Eshed Y. (2017). Coordination of auxin-triggered leaf initiation by tomato LEAFLESS. proc. Natl. Acad. Sci. U.S.A. 114, 3246–3251. doi: 10.1073/pnas.1617146114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champagne C., Sinha N. (2004). Compound leaves: Equal to the sum of their parts? Development 131, 4401–4412. doi: 10.1242/dev.01338 [DOI] [PubMed] [Google Scholar]
- Damodharan S., Zhao D., Arazi T. (2016). A common miRNA160-based mechanism regulates ovary patterning, floral organ abscission and lamina outgrowth in tomato. Plant J. 86, 458–471. doi: 10.1111/tpj.13127 [DOI] [PubMed] [Google Scholar]
- DeMason D. A., Chawla R. (2004). Roles for auxin during morphogenesis of the compound leaves of pea (Pisum sativum). Planta 218, 435–448. doi: 10.1007/s00425-003-1100-x [DOI] [PubMed] [Google Scholar]
- DeMason D. A., Chetty V. J. (2011). Interactions between GA, auxin, and UNI expression controlling shoot ontogeny, leaf morphogenesis, and auxin response in Pisum sativum (Fabaceae): or how the uni-tac mutant is rescued. Am. J. Bot. 98, 775–791. doi: 10.3732/ajb.1000358 [DOI] [PubMed] [Google Scholar]
- DeMason D. A., Chetty V. (2014). Phenotypic characterization of the CRISPA (ARP gene) mutant of pea (Pisum sativum, fabaceae): a reevaluation. Am. J. Bot. 101, 408–427. doi: 10.3732/ajb.1300415 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N., Dharmasiri S., Estelle M. (2005). The f-box protein TIR1 is an auxin receptor. Nature 435, 441–445. doi: 10.1038/nature03543 [DOI] [PubMed] [Google Scholar]
- Ding L., Yan S., Jiang L., Liu M., Zhang J., Zhao J., et al. (2015). HANABA TARANU regulates the shoot apical meristem and leaf development in cucumber (Cucumis sativus l.). J. Exp. Bot. 66, 7075–7087. doi: 10.1093/jxb/erv409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dkhar J., Pareek A. (2014). What determines a leaf’s shape? Evo. Devo. 5, 47. doi: 10.1186/2041-9139-5-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Huang H. (2018). Auxin polar transport flanking incipient primordium initiates leaf adaxial-abaxial polarity patterning. J. Integr. Plant Biol. 60, 455–464. doi: 10.1111/jipb.12640 [DOI] [PubMed] [Google Scholar]
- Du F., Guan C., Jiao Y. (2018). Molecular mechanisms of leaf morphogenesis. Mol. Plant 11, 1117–1134. doi: 10.1016/j.molp.2018.06.006 [DOI] [PubMed] [Google Scholar]
- Feng X., Li X., Yang X., Zhu P. (2020). Fine mapping and identification of the leaf shape gene BoFL in ornamental kale. Theor. Appl. Genet. 133, 1303–1312. doi: 10.1007/s00122-020-03551-x [DOI] [PubMed] [Google Scholar]
- Filyushin M. A., Slugin M. A., Dzhos E. A., Kochieva E. Z., Shchennikova A. V. (2018). Coexpression of YABBY1 and YABBY3 genes in lateral organs of tomato species (Solanum, section lycopersicon). Dokl. Biochem. Biophys. 478, 50–54. doi: 10.1134/S160767291801012X [DOI] [PubMed] [Google Scholar]
- Fleishon S., Shani E., Ori N., Weiss D. (2011). Negative reciprocal interactions between gibberellin and cytokinin in tomato. New Phytol. 190, 609–617. doi: 10.1111/j.1469-8137.2010.03616.x [DOI] [PubMed] [Google Scholar]
- Fujino K., Matsuda Y., Ozawa K., Nishimura T., Koshiba T., Fraaije M. W., et al. (2008). NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genom. 279, 499–507. doi: 10.1007/s00438-008-0328-3 [DOI] [PubMed] [Google Scholar]
- Fu X., Xu J., Zhou M., Chen M., Shen L., Li T., et al. (2019). Enhanced expression of QTL qLL9/DEP1 facilitates the improvement of leaf morphology and grain yield in rice. Int. J. Mol. Sci. 20, 866. doi: 10.3390/ijms20040866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Ning X., Wang Y., Wang X., Yan W., Zhang Z., et al. (2014). Fine mapping of a gene that confers palmately lobed leaf (pll) in melon (Cucumis melo l.). Euphytica 200, 337–347. doi: 10.1007/s10681-014-1151-z [DOI] [Google Scholar]
- Gourlay C. W., Hofer J. M., Ellis T. H. N. (2000). Pea compound leaf architecture is regulated by interactions among the genes UNIFOLIATA, COCHLEATA, AFILA, and TENDRIL-LESS . Plant Cell 12, 1279–1294. doi: 10.1105/tpc.12.8.1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N., Han D., Huang K., Du Y., Yang J., Zhang J., et al. (2020). Genome-based breeding approaches in major vegetable crops. Theor. Appl. Genet. 133, 1739–1752. doi: 10.1007/s00122-019-03477-z [DOI] [PubMed] [Google Scholar]
- Hay A., Barkoulas M., Tsiantis M. (2006). ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis . Development 133, 3955–3961. doi: 10.1242/dev.02545 [DOI] [PubMed] [Google Scholar]
- Hay A., Kaur H., Phillips A., Hedden P., Hake S., Tsiantis M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12, 1557–1565. doi: 10.1016/S0960-9822(02)01125-9 [DOI] [PubMed] [Google Scholar]
- Hay A., Tsiantis M. (2010). KNOX genes: Versatile regulators of plant development and diversity. Development 137, 3153–3165. doi: 10.1242/dev.030049 [DOI] [PubMed] [Google Scholar]
- Heath D. V., Gregory F. G. (1938). The constancy of the mean net assimilation rate and its ecological importance. Ann. Bot. Lond. 2, 811–818. doi: 10.1093/oxfordjournals.aob.a084036 [DOI] [Google Scholar]
- Hofer J., Turner L., Hellens R., Ambrose M., Matthews P., Michael A., et al. (1997). UNIFOLIATA regulates leaf and flower morphogenesis in pea. Curr. Biol. 7, 581–587. doi: 10.1016/S0960-9822(06)00257-0 [DOI] [PubMed] [Google Scholar]
- Hofer J., Turner L., Moreau C., Ambrose M., Isaac P., Butcher S., et al. (2009). Tendril-less regulates tendril formation in pea leaves. Plant Cell 21, 420–428. doi: 10.1105/tpc.108.064071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H., Zhang C., Hou X. (2020). Cloning and functional analysis of BcMYB101 gene involved in leaf development in pak choi (Brassica rapa ssp. Chinensis). Int. J. Mol. Sci. 21, 2750. doi: 10.3390/ijms21082750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Zhu L., Zeng D. L., Gao Z. Y., Guo L. B., Fang Y. X., et al. (2010). Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol. Biol. 73, 283–292. doi: 10.1007/s11103-010-9614-7 [DOI] [PubMed] [Google Scholar]
- Ito H., Kato T. (1957). Studies on the head formation of chinese cabbage. J. Jpn. Soc. Hortic. Sci. 26, 154–162. doi: 10.2503/jjshs.26.154 [DOI] [Google Scholar]
- Jasinski S., Tattersall A., Piazza P., Hay A., Martinez-Garcia J. F., Schmitz G., et al. (2008). PROCERA encodes a DELLA protein that mediates control of dissected leaf form in tomato. Plant J. 56, 603–612. doi: 10.1111/j.1365-313X.2008.03628.x [DOI] [PubMed] [Google Scholar]
- Kasprzewska A., Carter R., Swarup R., Bennett M., Monk N., Hobbs J. K., et al. (2015). Auxin influx importers modulate serration along the leaf margin. Plant J. 83, 705–718. doi: 10.1111/tpj.12921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura E., Horiguchi G., Tsukaya H. (2010). Mechanisms of leaf tooth formation in arabidopsis. Plant J. 62, 429–441. doi: 10.1111/j.1365-313X.2010.04156.x [DOI] [PubMed] [Google Scholar]
- Kepinski S., Leyser O. (2005). The arabidopsis f-box protein TIR1 is an auxin receptor. Nature 435, 446–451. doi: 10.1038/nature03542 [DOI] [PubMed] [Google Scholar]
- Kierzkowski D., Runions A., Vuolo F., Strauss S., Lymbouridou R., Routier-Kierzkowska A. L., et al. (2019). A growth-based framework for leaf shape development and diversity. Cell 177, 1405–1418. doi: 10.1016/j.cell.2019.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kang Y., Yang E., Cho M., Schafleitner R., Lee J., et al. (2021). Applications and major achievements of genome editing in vegetable crops: A review. Front. Plant Sci. 12, 688980. doi: 10.3389/fpls.2021.688980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., McCormick S., Timmermans M., Sinha N. (2003. a). The expression domain of PHANTASTICA determines leaflet placement in compound leaves. Nature 424, 438–443. doi: 10.1038/nature01820 [DOI] [PubMed] [Google Scholar]
- Kim J. I., Murphy A. S., Baek D., Lee S. W., Yun D. J., Bressan R. A., et al. (2011). YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in arabidopsis thaliana. J. Exp. Bot. 62, 3981–3992. doi: 10.1093/jxb/err094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Pham T., Hamidi A., McCormick S., Kuzoff R. K., Sinha N. (2003. b). Reduced leaf complexity in tomato wiry mutants suggests a role for PHAN and KNOX genes in generating compound leaves. Development 130, 4405–4415. doi: 10.1242/dev.00655 [DOI] [PubMed] [Google Scholar]
- Kimura S., Koenig D., Kang J., Yoong F., Sinha N. (2008). Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr. Biol. 9, 672–677. doi: 10.1016/j.cub.2008.04.008 [DOI] [PubMed] [Google Scholar]
- Koenig D., Bayer E., Kang J., Kuhlemeier C., Sinha N. (2009). Auxin patterns Solanum lycopersicum leaf morphogenesis. Development 136, 2997–3006. doi: 10.1242/dev.033811 [DOI] [PubMed] [Google Scholar]
- Kong X. Q., Gao X. H., Sun W., An J., Zhao Y. X., Zhang H. (2011). Cloning and functional characterization of a cation–chloride cotransporter gene OsCCC1. plant mol. Biol. 75, 567–578. doi: 10.1007/s11103-011-9744-6 [DOI] [PubMed] [Google Scholar]
- Kong X., Huang G., Xiong Y., Zhao C., Wang J., Song X., et al. (2019). IBR5 regulates leaf serrations development via modulation of the expression of PIN1. Int. J. Mol. Sci. 20, 4429. doi: 10.3390/ijms20184429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Liu B., Wu J., Cheng F., Wang X. (2016). Genetic variation and divergence of genes involved in leaf adaxial-abaxial polarity establishment in Brassica rapa. front. Plant Sci. 7, 94. doi: 10.3389/fpls.2016.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Liu W., Zhuang L., Zhu Y., Wang F., Chen T., et al. (2019). BIGGER ORGANS and ELEPHANT EAR-LIKE LEAF1 control organ size and floral organ internal asymmetry in pea. J. Exp. Bot. 70, 179–191. doi: 10.1093/jxb/ery352 [DOI] [PubMed] [Google Scholar]
- Lin D., Xiang Y., Xian Z., Li Z. (2016). Ectopic expression of SlAGO7 alters leaf pattern and inflorescence architecture and increases fruit yield in tomato. Physiol. Plant 157, 490–506. doi: 10.1111/ppl.12425 [DOI] [PubMed] [Google Scholar]
- Liu X., Hao N., Li H., Ge D., Du Y., Liu R., et al. (2019). PINOID is required for lateral organ morphogenesis and ovule development in cucumber. J. Exp. Bot. 70, 5715–5730. doi: 10.1093/jxb/erz354 [DOI] [PubMed] [Google Scholar]
- Liu Z., Jia L., Wang H., He Y. (2011). HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. J. Exp. Bot. 62, 4367–4381. doi: 10.1093/jxb/err167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Li M., Liu K., Tang D., Sun M., Li Y., et al. (2016). Semi-rolled Leaf2 modulates rice leaf rolling by regulating abaxial side cell differentiation. J. Exp. Bot. 67, 2139–2150. doi: 10.1093/jxb/erw029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Wu F., Yu X., Bai J., Zhong W., He Y. (2014). MicroRNA319a-targeted brassica rapa ssp. pekinensis TCP genes modulate head shape in chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 164, 710–720. doi: 10.1104/pp.113.228007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti C., Orzaez D., Ellul P., Moreno V., Carbonell J., Granell A. (2007). Silencing of DELLA induces facultative parthenocarpy in tomato fruits. Plant J. 52, 865–876. doi: 10.1111/j.1365-313X.2007.03282.x [DOI] [PubMed] [Google Scholar]
- McSteen P., Malcomber S., Skirpan A., Lunde C., Wu X., Kellogg E., et al. (2007). barren inflorescence2 encodes a co-ortholog of the PINOID serine/threonine kinase and is required for organogenesis during inflorescence and vegetative development in maize. Plant Physiol. 144, 1000–1011. doi: 10.1104/pp.107.098558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y., Kyozuka J. (2007). Characterization of OsPID, the rice ortholog of PINOID, and its possible involvement in the control of polar auxin transport. Plant Cell Physiol. 48, 540–549. doi: 10.1093/pcp/pcm024 [DOI] [PubMed] [Google Scholar]
- Nakayama H., Rowland S., Cheng Z., Zumstein K., Kang J., Kondo Y., et al. (2021). Leaf form diversification in an ornamental heirloom tomato results from alterations in two different HOMEOBOX genes. Curr. Biol. 31, 4788–4799. doi: 10.1016/j.cub.2021.08.023 [DOI] [PubMed] [Google Scholar]
- Nath U., Crawford B. C., Carpenter R., Coen E. (2003). Genetic control of surface curvature. Science 299, 1404–1407. doi: 10.1126/science.1079354 [DOI] [PubMed] [Google Scholar]
- Ren J., Liu Z., Du J., Fu W., Hou A., Feng H. (2019). Fine-mapping of a gene for the lobed leaf, BoLl, in ornamental kale (Brassica oleracea l. var. acephala). Mol. Breed. 39, 40. doi: 10.1007/s11032-019-0944-0 [DOI] [Google Scholar]
- Ren W., Wang H., Bai J., Wu F., He Y. (2018). Association of microRNAs with types of leaf curvature in Brassica rapa . Front. Plant Sci. 9, 73. doi: 10.3389/fpls.2018.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W., Wu F., Bai J., Li X., Yang X., Xue W., et al. (2020). BcpLH organizes a specific subset of microRNAs to form a leafy head in Chinese cabbage (Brassica rapa ssp. pekinensis). Hortic. Res. 7, 1. doi: 10.1038/s41438-019-0222-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland S. D., Zumstein K., Nakayama H., Cheng Z., Flores A. M., Chitwood H., et al. (2020). Leaf shape is a predictor of fruit quality and cultivar performance in tomato. New Phytol. 226, 851–865. doi: 10.1111/nph.16403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarojam R., Sappl P. G., Goldshmidt A., Efroni I., Floyd S. K., Eshed Y., et al. (2010). Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell 22, 2113–2130. doi: 10.1105/tpc.110.075853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazuka T., Kamiya N., Nishimura T., Ohmae K., Sato Y., Imamura K., et al. (2009). A rice tryptophan deficient dwarf mutant, tdd1, contains a reduced level of indole acetic acid and develops abnormal flowers and organless embryos. Plant J. 60, 227–241. doi: 10.1111/j.1365-313X.2009.03952.x [DOI] [PubMed] [Google Scholar]
- Shwartz I., Levy M., Ori N. (2016). Hormones in tomato leaf development. Dev. Biol. 419, 132–142. doi: 10.1016/j.ydbio.2016.06.023 [DOI] [PubMed] [Google Scholar]
- Song M., Cheng F., Wang J., Wei Q., Fu W., Yu X., et al. (2019). A leaf shape mutant provides insight into PINOID Serine/Threonine kinase function in cucumber (Cucumis sativus l.). J. Integr. Plant Biol. 61, 1000–1014. doi: 10.1111/jipb.12739 [DOI] [PubMed] [Google Scholar]
- Sun X., Gao Y., Lu Y., Zhang X., Luo S., Li X., et al. (2021). Genetic analysis of the “head top shape” quality trait of Chinese cabbage and its association with rosette leaf variation. Horti. Res. 8, 106. doi: 10.1038/s41438-021-00541-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Luo S., Luo L., Wang X., Chen X., Lu Y., et al. (2018). Genetic analysis of Chinese cabbage reveals correlation between rosette leaf and leafy head variation. Front. Plant Sci. 9, 1455. doi: 10.3389/fpls.2018.01455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattersall A., Turner L., Knox M., Ambrose M., Ellis N., Hofer J. (2005). The mutant crispa reveals multiple roles for PHANTASTICA in pea compound leaf development. Plant Cell 17, 1046–1060. doi: 10.1105/tpc.104.029447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui Y., Ohmori Y., Takebayashi Y., Sakakibara H., Hirano H. (2018). WUSCHELRELATED HOMEOBOX4 acts as a key regulator in early leaf development in rice. PLoS Genet 14, e1007365. doi: 10.1371/journal [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercruysse J., Baekelandt A., Gonzalez N., Inzé D. (2020). Molecular networks regulating cell division during arabidopsis leaf growth. J. Exp. Bot. 23, 2365–2378. doi: 10.1093/jxb/erz522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jiao Y. (2020). Keeping leaves in shape. Nat. Plants 6, 436–437. doi: 10.1038/s41477-020-0660-0 [DOI] [PubMed] [Google Scholar]
- Wang H., Jones B., Li Z., Frasse P., Delalande C., Regad F., et al. (2005). The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 17, 2676–2692. doi: 10.1105/tpc.105.033415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Li R., Wen L., Fu D., Zhu B., Luo Y., et al. (2015). Functional analysis and RNA sequencing indicate the regulatory role of argonaute1 in tomato compound leaf development. PloS One 10, e0140756. doi: 10.1371/journal.pone.0140756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wu F., Bai J., He Y. (2014). BrpSPL9 (Brassica rapa ssp. pekinensis SPL9) controls the earliness of heading time in Chinese cabbage. Plant Biotechnol. J. 12, 312–321. doi: 10.1111/pbi.12138 [DOI] [PubMed] [Google Scholar]
- Wang J., Xu J., Qian Q., Zhang G. (2020). Development of rice leaves: how histocytes modulate leaf polarity establishment. Rice Sci. 27, 468–479. doi: 10.1016/j.rsci.2020.09.004 [DOI] [Google Scholar]
- Wang H., Chen J., Wen J., Tagede M., Li G., Liu Y., et al. (2008). Control of compound leaf development by FLORICAULA/LEAFY ortholog SINGLE LEAFLET1 in Medicago truncatula . Plant Physiol. 146, 1759–1772. doi: 10.1104/pp.108.117044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Chen X., Wang Z., Liu Q., Li H., Zhang Y., et al. (2017). Genetic mapping of the LOBED LEAF 1 (ClLL1) gene to a 127.6-kb region in watermelon (Citrullus lanatus l.). PloS One 12, e0180741. doi: 10.1371/journal.pone.0180741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel C. L., Schuetz M., Yu Q., Mattsson J. (2007). Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in arabidopsis thaliana. Plant J. 49, 387–398. doi: 10.1111/j.1365-313X.2006.02977.x [DOI] [PubMed] [Google Scholar]
- Woo Y. M., Park H. J., Su’udi M., Yang J. I., Park J. J., Back K., et al. (2007). Constitutively wilted 1, a member of the rice YUCCA gene family, is required for maintaining water homeostasis and an appropriate root to shoot ratio. Plant Mol. Biol. 65, 125–136. doi: 10.1007/s11103-007-9203-6 [DOI] [PubMed] [Google Scholar]
- Wu L., Tian Z., Zhang J. (2018). Functional dissection of auxin response factors in regulating tomato leaf shape development. Front. Plant Sci. 9, 957. doi: 10.3389/fpls.2018.00957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Nukazuka A., Tsukaya H. (2012). Leafadaxial-abaxial polarity specification and lamina outgrowth: evolution and development. Plant Cell Physiol. 53, 1180–1194. doi: 10.1093/pcp/pcs074 [DOI] [PubMed] [Google Scholar]
- Yan J., Cai X., Luo J., Sato S., Jiang Q., Yang J., et al. (2010). The REDUCED LEAFLET genes encode key components of the trans-acting small interfering RNA pathway and regulate compound leaf and flower development in Lotus japonicus. Plant Physiol 152, 797–807. doi: 10.1104/pp.109.140947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai O., Shani E., Russ D., Ori N. (2011). Gibberellin partly mediates LANCEOLATE activity in tomato. Plant J. 68, 571–582. doi: 10.1111/j.1365-313X.2011.04716.x [DOI] [PubMed] [Google Scholar]
- Yang L., Liu H., Zhao J., Pan Y., Cheng S., Lietzow C. D., et al. (2018). LITTLELEAF (LL) encodes a WD40 repeat domain-containing protein associated with organ size variation in cucumber. Plant J. 95, 834–847. doi: 10.1111/tpj.13991 [DOI] [PubMed] [Google Scholar]
- Yan C. J., Yan S., Zhang Z. Q., Liang G. H., Lu J. F., Gu M. H. (2006). Genetic analysis and gene fine mapping for a rice novel mutant (rl9(t)), with rolling leaf character. Chin. Sci. Bull. 51, 63–69. doi: 10.1007/s11434-005-1142-5 [DOI] [Google Scholar]
- Yifhar T., Pekker I., Peled D., Friedlander G., Pistunov A., Sabban M., et al. (2012). Failure of the tomato trans-acting short interfering RNA program to regulate AUXIN RESPONSE FACTOR3 and ARF4 underlies the wiry leaf syndrome. Plant Cell 24, 3575–3589. doi: 10.1105/tpc.112.100222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Peng J., Feng X., Yang S., Zheng Z., Tang X., et al. (2000). Cloning and structural and expressional characterization of BcpLH gene preferentially expressed in folding leaf of Chinese cabbage. Sci. China Ser. C. Life Sci. 43, 321–329. doi: 10.1007/BF02879292 [DOI] [PubMed] [Google Scholar]
- Yu X., Wang H., Zhong W., Bai J., Liu P., He Y. (2013). QTL mapping of leafy heads by genome resequencing in the RIL population of Brassica rapa . PloS One 8, e76059. doi: 10.1371/journal.pone.0076059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Yan C., Liu Y., Liu Y., Jia Y., Lavelle D., et al. (2020). Upregulation of a KN1 homolog by transposon insertion promotes leafy head development in lettuce. Proc. Natl. Acad. Sci. U.S.A. 117, 33668–33678. doi: 10.1073/pnas.2019698117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Chen R., Xiao J., Qian C., Wang T., Li H., et al. (2007). A single-base deletion mutation in SlIAA9 gene causes tomato (Solanum lycopersicum) entire mutant. J. Plant Res. 120, 671–678. doi: 10.1007/s10265-007-0109-9 [DOI] [PubMed] [Google Scholar]
- Zhang C., Chen F., Zhao Z., Hu L., Liu H., Cheng Z., et al. (2018). Mutations in CsPID encoding a Ser/Thr protein kinase are responsible for round leaf shape in cucumber (Cucumis sativus l.). Theor. Appl. Genet. 131, 1379–1389. doi: 10.1007/s00122-018-3084-8 [DOI] [PubMed] [Google Scholar]
- Zhang G. H., Li S. Y., Wang L., Ye W. J., Zeng D. L., Rao Y. C., et al. (2014). LSCHL4 from japonica cultivar, which is allelic to NAL1, increases yield of indica super rice 93-11. Mol. Plant 7, 1350–1364. doi: 10.1093/mp/ssu055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S. Z., Wu T., Liu S. J., Liu X., Jiang L., Wan J. M. (2016). Disruption of OsARF19 is critical for floral organ development and plant architecture in rice (Oryza sativa l.). Plant Mol. Biol. Rep. 34, 748–760. doi: 10.1007/s11105-015-0962-y [DOI] [Google Scholar]
- Zhang G. H., Xu Q., Zhu X. D., Qian Q., Xue H. W. (2009). SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21, 719–735. doi: 10.1105/tpc.108.061457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Xu F., Li Q., Wang G., Liu N., Gong Y., et al. (2019). QTL mapping combined with bulked segregant analysis identify SNP markers linked to leaf shape traits in pisum sativum using SLAF sequencing. Front. Genet. 9. doi: 10.3389/fgene.2018.00615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Han L., Hou C., Metelli A., Qi L., Tadege M., et al. (2011). Developmental analysis of a Medicago truncatula smooth leaf margin1 mutant reveals context-dependent effects on compound leaf development. Plant celll 23, 2106–2124. doi: 10.1105/tpc.111.085464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulias N., Koenig D., Hamidi A., McCormick S., Kim M. (2012). A role for PHANTASTICA in medio-lateral regulation of adaxial domain development in tomato and tobacco leaves. Ann. Bot. 109, 407–418. doi: 10.1093/aob/mcr295 [DOI] [PMC free article] [PubMed] [Google Scholar]


