Abstract
Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is associated with excessive coagulation, thrombosis, and mortality.
Objective
To provide insight into mechanisms that contribute to excessive coagulation in coronavirus 2019 (COVID-19) disease.
Patients/Methods
Blood from COVID-19 patients was investigated for coagulation–related gene expression and functional activities.
Results
Single-cell RNA sequencing (scRNA-seq) of peripheral blood mononuclear cells from severe COVID-19 patients revealed a 5.2-fold increase in tissue factor (TF [F3 gene]) transcript expression levels (P < .05), the trigger of extrinsic coagulation; a 7.7-fold increase in C1-inhibitor (SERPING1 gene; P < .01) transcript expression levels, an inhibitor of intrinsic coagulation; and a 4.4-fold increase in anticoagulant thrombomodulin (TM [THBD gene]) transcript expression levels (P < .001). Bulk RNA-seq analysis of sorted CD14+ monocytes on an independent cohort of COVID-19 patients confirmed these findings (P < .05). Indicative of excessive coagulation, 41% of COVID-19 patients’ plasma samples contained high D-dimer levels (P < .0001); of these, 19% demonstrated extracellular vesicle TF activity (P = .109). COVID-19 patients’ ex vivo plasma–based thrombin generation correlated positively with D-dimer levels (P < .01). Plasma procoagulant extracellular vesicles were elevated ∼9-fold in COVID-19 patients (P < .01). Public scRNA-seq data sets from bronchoalveolar lung fluid and our peripheral blood mononuclear cell scRNA-seq data show CD14+ monocytes/macrophages TF transcript expression levels are elevated in severe but not mild or moderate COVID-19 patients.
Conclusions
Beyond local lung injury, SARS-CoV-2 infection increases systemic TF (F3) transcript levels and elevates circulating extracellular vesicles that likely contribute to disease-associated coagulation, thrombosis, and related mortality.
Keywords: coagulation, COVID-19, thrombosis, tissue factor, transcriptome
1. Introduction
Although acute respiratory distress syndrome is the fundamental feature of severe coronavirus 2019 (COVID-19) disease, critically ill patients have a higher incidence of pulmonary thrombosis and venous thromboembolism associated with mortality [1,2]. Blood coagulation is classically divided into the tissue factor (TF)-initiated extrinsic pathway and the contact-activated intrinsic pathway that converge into a common path. For hemostasis, TF is constitutively present in the adventitial layer of blood vessels [3]. However, TF expression can be induced, leading to excessive coagulation and thrombosis [4]. Cellular damage because of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection could directly expose TF to blood and release cell debris to serve as contact surfaces for the intrinsic pathway. Cellular activation could induce TF expression locally and in blood-borne cells [5]. Consistent with local activation of coagulation, autopsied lungs from patients with COVID-19 showed severe endothelial injury, intracellular viruses, disrupted cell membranes, pulmonary vessel thrombosis, and alveolar capillary thrombosis [6]. In terms of systemic activation, Hottz et al. found that platelet activation and platelet-monocyte aggregates are elevated in severe COVID-19 disease and that TF protein is higher in these aggregates when compared to free monocytes. They also found that platelet activation and monocyte TF protein levels were associated with coagulation markers and were increased in patients on mechanical ventilation [7]. In coculture, COVID-19 patient-derived platelets induced TF protein and TF-mediated protease-activated receptors (PARs) signaling of monocytes [8].
TF requires a specific phospholipid membrane environment to become an active cofactor for factor VIIa (FVIIa) to trigger coagulation [9]. Likewise, FXa/FVa and FIXa/FVIIIa complexes require calcium and phospholipid membranes to function. Extracellular vesicles (EVs) are lipid-bound microparticles released by all types of healthy and damaged cells. Wang et al. [10] reported that, in culture, infection of macrophages with SARS-CoV-2 spike protein pseudovirus markedly increased hydrolysis of sphingomyelin and TF procoagulant activity at the cell surface and released TF-positive EVs. Several studies have shown that TF activity is elevated for EVs isolated from plasma in a subset of fatally ill COVID-19 patients suggesting a pathological role for circulating EV TF activity in COVID-19 disease [11,12].
There exist conflicting reports concerning the dysregulation of the TF (F3) gene in COVID-19 patients. Bronchoalveolar lavage fluid cells (BALFs) bulk RNA sequence data from COVID-19 patients were originally generated by Zhou et al. to define the SARS-CoV-2 genome sequence [13]. Using Zhou et al. RNA sequence data, Garvin et al. [14] reported an atypical pattern of expression for genes that would result in elevated kinin levels. In parallel with this, Mast et al. [15] reported a reduction in transcripts for the intrinsic pathway inhibitor C1-inhibitor (SERPING1) and also unchanged TF (F3) transcripts, with elevated TFPI transcripts. Their findings suggest that COVID-19 enhances intrinsic but not extrinsic coagulation in the lungs.
A study by Fitzgerald et al. analyzing publicly available RNA sequencing (RNA-seq) databases from Xiong et al. [16] and Liao et al. [17] also reported elevated TFPI transcripts in COVID-19 BALF samples. By contrast, this study found that TF (F3 gene) transcripts were significantly elevated in the lung epithelial cells of COVID-19 patients’ BALF samples, which suggests that COVID-19 enhances extrinsic coagulation locally. They also reported that TF transcript expression levels in peripheral blood mononuclear cells (PBMCs) were not elevated, concluding that it was unlikely that circulating immune cells are driving COVID-19-associated coagulopathies.
Francischetti et al. [18] reported elevated TF in COVID-19 patients’ postmortem specimens as determined by the FVIIa-ATIII complex and microparticle TF activity. TF RNA in situ hybridization studies of Subrahmanian et al. [19] reported 2-fold higher TF transcript expression in COVID-19 than in non-COVID-19 acute respiratory distress syndrome patient lungs, which coclustered with SARS-CoV-2. These data and a review of TF expression and EV activity studies strongly suggested that increased TF expression in pathogenic coronaviruses contributes to thrombosis [20].
Through a detailed analysis of samples from COVID-19 patients, we report here the influence of severe SARS-CoV-2 infection on systemic coagulation–related gene expression and functional activities. We also reanalyzed available RNA-seq data sets for BALF cells and found increased TF (F3) transcripts in severe, but not mild nor moderate, COVID-19 patients. Our findings support the hypothesis that both local lung injury with enhanced epithelial TF expression and systemic TF enhanced expression predominantly by CD14+ monocytes, contribute to COVID-19 disease-associated thrombosis and mortality.
2. Material and methods
2.1. Patient samples
Washington University-affiliated hospitals collected and made available for investigations; plasma, PBMC, and, in some cases, tissue from hospitalized COVID-19 (PCR-positive) patients. Family and patient consents were obtained per the Declaration of Helsinki. Our single-cell RNA sequencing (scRNA-seq) analysis included 6 age–matched non-COVID-19 control samples and 2 samples per patient from 12 severe (ICU) patients, of whom 6 died. Our bulk RNA-seq analysis on sorted CD14+ monocytes included 10 age–matched controls, 9 mild (nonhospitalized) and 10 severe (ICU) COVID-19 patient samples, none of whom died. Our protein and functional activities studies included 48 samples from 24 healthy controls (average age was 50.1 years) and 206 samples from 108 hospitalized, severe COVID-19 patients collected during January–May of 2021. Exclusion criteria included surgery and suspected or documented bacterial or other viral infections. Patients’ average age was 67.5 years, average body mass index was 28.9 kg/m2, 58% were males, 68% were African-American, and 31% were Caucasian. Forty-two of these patients died of COVID-19-related causes; of the 66 survivors, 43 had been in intensive care unit requiring mechanical ventilation (Figure 5A). Over 90% of patients were given anticoagulant therapies. Samples were collected at varying times relative to the onset of symptoms or times of hospital admissions. Not all samples were analyzed in every assay. Instead, in some instances, representative randomized subsets of samples were analyzed. Actual sample sizes for each study are described in the figure legends.
2.2. Reagents
Dade Innovin was from Siemens (#B4212-41, lot 549766A) and prepared as recommended, which yielded a TF concentration of 620 ± 70 pM (mean ± standard error of mean [SEM], n = 4) as measured by ELISA (Abcam; cat. # ab220653, lot GR3358225-1). Human factor VIIa (# HFVIIa), gamma carboxyglutamic acid (gla)-domainless FVIIa #(GDFVIIa), human factor X (# HFX), factor Xa, and α-thrombin (# HT), and corn trypsin inhibitor (# CTI) were from Enzyme Research Laboratories. Anti-TF antibody (anti-HuCD142, clone HTF-1) and mouse IgG isotype control (PI31903) were from Invitrogen. Factor Xa substrate was Chromogenix S-2765 (# 82141339). Pooled normal plasma (# 0010) and factor XI-deficient (# 1100) were from George King Biomedical, Inc.
2.3. Plasma sample preparations
Blood was collected from healthy controls and COVID-19 patients into EDTA (1.8 mg/mL) or into 1/9 volume of 3.8% citrate or 3.8% citrate plus 1mg/mL CTI. Plasma was prepared between 2 and 4 hours postblood collection for all samples. Blood was centrifuged at 2500 g for 10 min; plasma was collected and stored at −80 °C.
2.4. Thrombin generation assays
Fifty μL plasma was incubated with 20 μL of selective inhibitor(s) in HSAE plus 20 μL of rabbit brain cephalin (60 μg/mL; Pentapharm) at room temperature for 20 minutes. Next, the assay was initiated by adding 10 μL of 1 mM Z-GGR-AMC (Bachem, cat # 4002155) containing 50 mM CaCl2. Data were collected using a Fluoroskan Ascent instrument and analyzed as described [21].
2.5. EV preparations
EVs were prepared by the method described by Hisada and Mackman [22]. Briefly, 200 μL of plasma was thawed, diluted with 1ml of HSAE buffer (50 mM HEPES, 100 mM NaCl, 2 mg/mL bovine albumin, 1 mM EDTA; pH 7.4), and centrifuged at 23 000 g for 20 min; the supernatant was decanted and saved; EV pellet was washed with 1 mL of buffer HSAE followed by centrifugation at 23 000 g for 20 minutes. The supernatant was discarded, and the final EV pellet was resuspended in 200 μL HSAE by repeated pipetting up and down and then stored at −80 oC.
2.6. EV TF activity assays
TF activity assays were performed at 37 oC in 96-well plates as described [22]. Specifically, 40 μL of EV test sample or TF standards (1:1 serial dilution of Innovin starting at 1 pM) was added to wells containing 10 μL of 40 μg/mL anti-TF or control IgG in HSAE. Following a 20 min incubation, 50 μL of a mixture of 4.8 nM FVIIa and 150 nM FX in HSAE containing 12 mM CaCl2 was added; the plates were sealed and incubated for 2 hours. Next, to stop FXa generation, 25 μL of 30 mM EDTA in HSAE was added, followed by a 5 min incubation. Finally, 25 μL of 4 mM FXa substrate (S-2765 in H2O) was added, and absorbance at 405 nM was monitored for 15 minutes in a SpectraMax (Molecular Devices). Activity in each well was determined from the standard curve generated in the absence of anti-TF. The reported “specific TF activity” was calculated as the activity in the absence of anti-TF minus the activity in the presence of anti–TF. In cases where the calculated “specific TF activity” was <0, rather than assigning it a value of zero, it was reported as its negative value to reflect the variability associated with this assay.
2.7. EV in thrombin generation assays
Twenty microliters of EV samples were added to 50 μL of FXI-deficient plasma plus 10 μL of 100 μg/mL anti-TF; after a 10-minute incubation thrombin generation was initiated with the addition of 20 μL of calcium and substrate as described above.
2.8. Immunoassays
Plasma D-dimer protein levels were determined by immunoassays as recommended by the manufacturer (Abcam, cat. # ab260076). TFPIα plasma levels were determined with the R&D Systems ELISA assay, cat. # DY2974, however, when using the TFPIα standard supplied with the assay, plasma levels of TFPIα were calculated to be ∼5-fold higher than typically reported. Therefore, we substituted a sample of pooled normal plasma (George King, Biomedical) to generate a standard curve and defined the plasma TFPIα level as 15 ng/mL, which corresponds to the reported level of TFPIα in normal plasma [21].
2.9. Transcript expression dataset analysis
We analyzed 3 COVID-19 RNA-seq datasets: 1) PBMC scRNA-seq data from 24 COVID-19 patient samples (mean ± SD age 70 ± 8.6 years) and 6 controls (mean ± SD age 70 ± 11.5 years) that were generated and described by Amrute et al. (database NCBI GEO accession no. GSE192391) [23]. Bulk RNA-seq data on sorted CD14+ monocytes from 10 non-COVID-19 cancer patient controls (mean ± SD age 64.9 ± 11.3 years), 9 mild (not oxygen required, non-ICU; mean ± SD age 53.4 ± 7.3 years), and 10 severe (supplemental oxygen or ICU required; mean ± SD age 60.3 ± 11 years) COVID-19 patient samples that were generated and described by Dhindsa et al. (NCBI accession no. GSE176290; note, sample GSM5362281 was removed from the study to avoid sample overlap with a scRNA-seq study) [24]. Bulk RNA-seq data were normalized to the beta-actin (ACTB) for each sample. 3) BALF scRNA-seq data from 3 moderate and 6 severe/critical COVID-19 patients and 4 controls that were generated and described by Liao et al. [17] and database NCBI GEO accession no. GSE128033. We downloaded the metadata (meta.tsv file) and the scRNA-seq data (nCoV.rds file) for COVID-19 and control samples from http://cells.ucsc.edu/covid19-balf. BALF scRNA-seq average expression for each sample was calculated using SEURAT Average Expression function.
2.10. Statistics
Statistical analyses were performed by Mann–Whitney test for unpaired data, Wilcoxon test for paired data, and Spearman test for correlations. Differences were considered statistically significant at P < .05. Analyses were performed with Prism (GraphPad Prism version 9.0). Values are expressed as mean ± SEM as indicated. ∗ denotes P < .05; ∗∗ denotes P < .01; ∗∗∗ denotes P < .001; ∗∗∗∗ denotes P < .0001.
3. Results
3.1. COVID-19 induces systemic CD14+ monocyte TF transcription
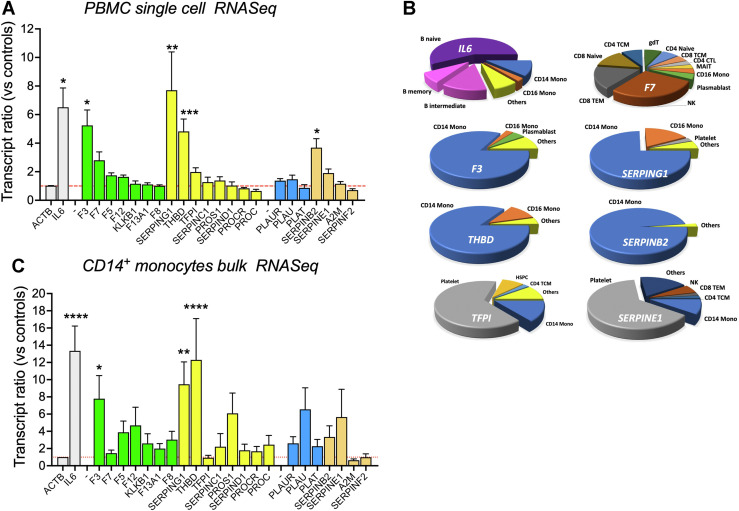
PBMC scRNA-seq analysis was conducted for coagulation–related genes for 24 PBMC samples banked at study enrollment days 0 and 7 from 12 hospitalized COVID-19 patients, 6 of whom died, and from 6 age–matched healthy controls [23]. Compared with 6 age–matched controls, the procoagulant TF (F3) transcript expression levels were elevated 5.2-fold (P < .05); the anticoagulant C1-inhibitor (SERPING1) transcripts were elevated 7.7-fold (P < .01); the anticoagulant thrombomodulin TM, (THBD) transcripts were elevated 4.4-fold (P < .001); and the fibrinolysis inhibitor PAI-2 (SERPINB2) transcripts were elevated 3.9-fold (P < .05). Transcripts that were elevated >1.75-fold, but did not achieve statistical significance included procoagulant FVII (F7), anticoagulant TFPIα (TFPI), and fibrinolysis inhibitor PAI-1 (SERPINE1). Beta-actin (ACTB) and IL-6 (IL-6) transcripts represented stable and elevated transcription controls (P < .05), respectively (Figure 1 A). The distribution of transcript expression for TF (F3), TM (THBD), C1-inhibitor (SERPING1), and PAI-2 (SERPINB2) genes across all cell types showed that CD14+ monocytes were the primary source. TFPIα (TFPI) and PAI-1 (SERPINE1) transcript expression were mainly seen in platelet-derived cells, whereas FVII (F7) transcripts showed stochastic expression (Figure 1B). Cell assignments based on transcript reads [23] showed, for COVID-19 versus controls, a 67% increase in CD14+ monocytes and 53% increase in platelets. While these increases would contribute partially to increases in transcript reads, the magnitudes of change for F3, THBD, and SERPING1 genes indicate induction of expression as the main cause for this increase. In an independent, nonoverlapping cohort of 10 COVID-19 patients and 10 age–matched controls, bulk RNA-seq analysis of sorted CD14+ monocytes [24] also showed significant increases in TF (F3, [P < .05]), TM (THBD, [P < .0001]), and C1-inhibitor (SERPING1, [P < .01]) transcript expression levels in COVID-19 cases (Figure 1C).
Figure 1.
Coagulation–related transcription changes with coronavirus 2019 (COVID-19) infection. (A) scRNA-seq analysis on PBMCs. Controls included individual samples from 6 age–matched subjects. COVID-19 samples included 2 samples per patient taken 7 days apart from 12 severe COVID-19 patients, 6 of whom died. Shown are the means and standard error of mean (SEM) vs controls for the pooled COVID-19 samples. Procoagulants in green; anticoagulants in yellow; profibrinolytic in blue; antifibrinolytics in orange. (B) Cell types proportional expression of transcripts in COVID-19 samples. (C) Bulk RNA-seq analysis on sorted CD14+ monocytes. This analysis included individual samples from 10 age–matched controls and 10 severe COVID-19 patients. Shown are the means and SEM vs controls for COVID-19 samples. Samples and patients in (A) and (C) did not overlap. Mann–Whitey test P values ∗P < .05; ∗∗P < .01; ∗∗∗P < .001; ∗∗∗∗P < .0001. Dashed red line represents the level for controls that equals 1.
Within our PBMC scRNA-seq analysis, 7629 cells (6225 COVID-19 and 1404 controls) were assigned as platelets. Among the top 10 elevated gene expression levels for these platelets, 4 (IFI27, RSAD2, CMPK2, and IFI6; data not shown) were also previously reported to be among the top 10 genes in platelets from COVID-19 patients, whereas none were in the top 10 genes reported for patients with sepsis or with H1N1 influenza [25]. These data are consistent with observed changes in transcription being attributable to COVID-19 disease and not an undetected coinfection.
3.2. TF (F3) elevated transcript expression in monocytes and BALF cells is associated with disease severity
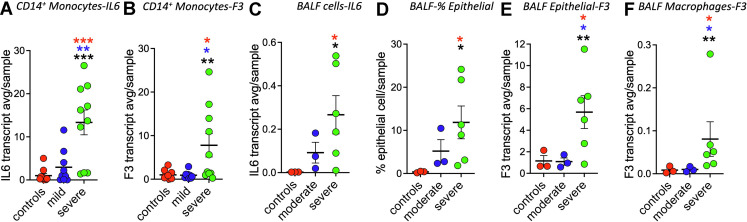
Our bulk RNA-seq study also included a cohort of 9 samples from COVID-19 patients who were not hospitalized and were classified as having a mild disease. For these samples, IL-6 transcript levels were less elevated than in the severe cohort, defined as being in the intensive care unit (Figure 2 A). TF (F3) transcript expression for patients with mild disease was similar to controls rather than those with severe disease (P < .05; [Figure 2B]). These data support that increases in TF (F3) transcript expression levels depend on disease severity. The scRNA-seq study performed by Liao et al. included 3 control, 3 moderate, and 6 severe/critical COVID-19 BALF samples. Using these data, Fitzgerald et al. reported TF (F3) to be differentially enriched in lung epithelial cells in COVID-19 samples. We separated Liao et al. BALF data samples based on their classification into healthy controls, moderate and severe/critical COVID-19 infection for further analysis. As expected, IL-6 average transcript expression levels correlated with disease severity (Figure 2C). Consistent with COVID-19 causing lung damage the fraction of epithelial cells, as identified by Liao et al., increased ∼15-fold in the moderate samples and ∼33 fold in the severe/critical samples (Figure 2D). For these cells, the average transcript expression levels for TF (F3) in controls and moderates were similar but significantly elevated 4.9-fold in severe disease (P < .05) [Figure 2E]). For cells identified by Liao et al. as macrophages, controls and patients with the moderate disease showed similar levels of TF (F3) of transcript expression, whereas severe/critical patient samples were significantly elevated 9.5-fold (P < .05; [Figure 2F]). The Liao et al. BALF data and our PBMC data are consistent with severe COVID-19 disease increasing TF transcript expression levels in macrophages/monocytes.
Figure 2.
Tissue factor (TF) (F3) transcription is dependent on disease severity. (A and B) Bulk RNA sequencing analysis of sorted CD14+ monocytes. (A) CD14+ monocytes IL-6 gene transcripts are elevated in severe but not mild COVID-19 disease patients. (B) CD14+ monocytes TF (F3 gene) transcripts are elevated in severe but not mild COVID-19 disease patients. (C) Bronchoalveolar lavage fluid (BALF) IL-6 gene transcripts are elevated in severe but not moderate COVID-19 disease patients. (D) Epithelial cell numbers are increased in BALF from patients with severe COVID-19 disease. (E) Epithelial cell TF (F3 gene) transcripts are elevated in severe but not moderate COVID-19 disease patient plasmas. (F) Macrophage TF (F3 gene) transcripts are elevated in severe but not moderate COVID-19 disease. Mann–Whitney test P values as indicated in Figure 1, with red denoting vs controls, blue denoting vs mild or moderate, and black denoting vs combined controls and mild or moderate.
3.3. COVID-19 treatment impacts TFPI
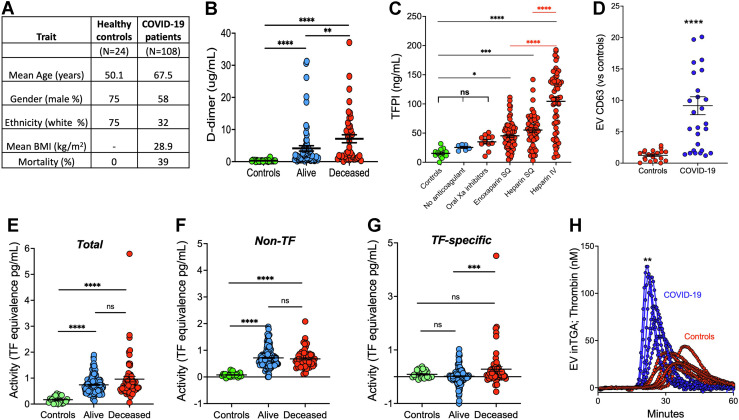
Consistent with the observed coagulation–related gene transcriptional changes, coagulation biomarker D-dimer levels were elevated in COVID-19 patient samples (P < .0001, [Figure 3 A, B]). Forty-one percent of the COVID-19 patients’ samples had D-dimer levels > 2 μg/mL, 4 times the normal upper limit (P < .0001). D-dimer levels were elevated with disease and increased levels correlated with mortality (P < .05, [Figure 3B]).
Figure 3.
Coagulation activity in COVID-19 plasma. (A) Patient characteristics. (B) Highest plasma level of D-dimer achieved over the course of disease for each patient versus outcomes. Healthy controls (n = 24); Alive (n = 66): Deceased (n = 42). (C) COVID-19 patients’ anticoagulant treatments versus plasma TFPIα levels. TFPIα levels for multiple samples from healthy controls (n = 12), and for COVID-19 patients on no anticoagulant (n = 6), an oral Xa inhibitor (n = 11), enoxaparin SQ (n = 72), heparin SQ (n = 54), or heparin IV (n = 58). (D) Extracellular vesicle CD63 levels for control and COVID-19 samples. (E–G) Extracellular vesicle tissue factor activity assays. (E) Total, (F) nonspecific, and (G) tissue factor-specific activity for healthy controls (n = 23) and COVID-19 patient samples from alive (n = 151 samples from 66 survivors) and deceased (n = 60 samples from 42 deceased) patients. (H) COVID-19 extracellular vesicles increase thrombin generation. Mann–Whitney test P values for COVID-19 vs controls as indicated in Figure 1.
Active TF in the blood is expected to form a clot and thus be short-lived unless it is in an inhibitory complex with endogenous TFPI. Heparin treatment is known to elevate circulating TFPIα levels [26,27]. TFPIα levels were not elevated in the few nonanticoagulated patients who declined treatment. Samples from patients placed on oral inhibitors showed a nonsignificant elevation in TFPIα levels. However, it is important to note that the oral inhibitor treatments were predominantly used to wean recovering patients off of heparins for eventual discharge from the hospital. By contrast, TFPIα levels were significantly elevated in patients given subcutaneous or intravenous heparin-like treatment with the highest TFPIα levels associated with intravenous infusion of heparin (P < .0001, [Figure 3C]). The lower D-dimer levels in the oral inhibitor group versus the other anticoagulant groups are consistent with these samples being obtained from recovering patients. Although TFPIα levels correlated with D-dimer and mortality, these data demonstrate that TFPIα is a marker of heparin treatment, not a marker of disease.
3.4. COVID-19 patient plasma EVs enhance coagulation
It has been reported that SARS-CoV-2 infection increases EV TF activity [11,12]. We used the procedures of Hisada et al. [22] to isolate EVs and assay for EV TF activity in our COVID-19 patients’ samples. We assayed EVs for CD63, an established biomarker for EVs [28], and found CD63 to be ∼9-fold elevated in COVID-19 EV samples (P < .0001; [Figure 3D]). To determine TF activity, we ran 2 assays, one without (total activity) and one with an anti-TF antibody (nonspecific activity), and TF-specific activity was calculated as the difference between the 2 assays (Figure 3E–G). For the COVID-19 samples, 19% demonstrated EV TF activity. EV TF activity showed a trend toward correlating with D-dimer levels (P = .109). Similar to previous reports [11,12] for a subset of patients, elevated TF-specific activity was associated with mortality (Figure 3G). Of interest, COVID-19 samples showed significantly greater non-TF-specific activity than controls (P < .0001) (Figure 3F). Consistent with this non-TF activity being a membrane-based phenomenon, rabbit brain cephalin membranes provided similar non-TF activity in this assay, and gamma-carboxyglutamic acid (gla)-domainless FVIIa, which does not bind to membranes, did not substitute for intact FVIIa, which requires membrane binding to function (data not shown). Additionally, we found that isolated COVID-19 EVs significantly increased thrombin generation of anti-TF-treated FXI-deficient plasma, which suggests the EV contains a TF-independent procoagulant membrane activity (Figure 3H; [P < .01]). CD63 levels correlated linearly with EV nonspecific thrombin generation potential (R2 = 0.83, P < .001 [data not shown]). The source(s) of the excessive procoagulant EVs in COVID-19 patient samples could not be determined.
3.5. COVID-19 patient plasma samples appear hypo-coagulant because of heparin treatment
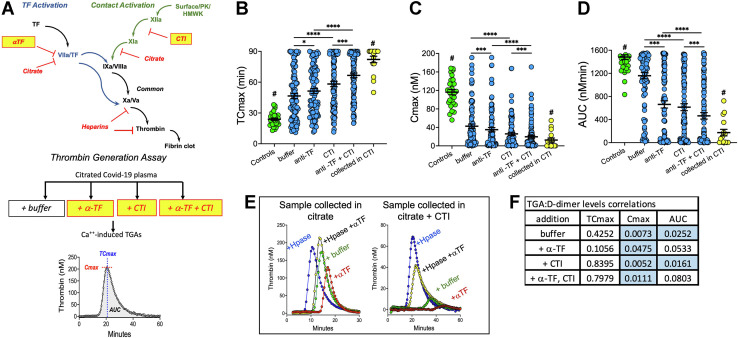
In an attempt to distinguish the relative contributions of TF-mediated extrinsic and contact-activated intrinsic coagulation in COVID-19 infection, thrombin generation assays (TGA) were performed on citrate or EDTA-collected blood samples that were processed into plasma samples. These chelators disrupt the initial step of TF-induced coagulation but do not block the contact activation system until FXIa has been produced. Assays triggered with Ca++ were run in the presence and absence of a blocking anti-TF monoclonal antibody and/or CTI, which inhibits the contact pathway by inhibiting FXIIa ( Figure 4 A). Compared to controls, COVID-19 samples showed less thrombin generation, defined by lower Cmax (maximal thrombin concentration), delayed TCmax (time to Cmax), and lower area under the curve (AUC; [Figure 4B–D]). The hypo-coagulant status of COVID-19 plasma samples was attributed to heparin treatments (unpublished and Figure 4E). In a limited study, we found that collecting blood directly into citrate plus CTI dramatically reduced thrombin generation (Figure 4E). Despite the confounding influence of sample processing and heparin treatments, several thrombin generation parameters correlated with D-dimer plasma protein levels (Figure 4F).
Figure 4.
Ca++–induced thrombin generation in COVID-19. (A) Schematic for conducting thrombin generation assays (TGAs). (B–D) TGA outcomes for controls (n = 30, green) and COVID-19 samples (n = 92, blue) collected in EDTA or citrate and COVID-19 samples collected in EDTA + CTI (n = 16, yellow). Shown are individual data points, and means ± standard error of mean for: (B) TCmax; (C) Cmax; and (D) AUC. (E) Influences of heparin and processing on TGAs. Comparison of one COVID-19 patient’s blood collected in citrate (left panel) vs citrate + CTI (right panel). CTI was added to the citrated–only sample after processing it to plasma. Samples were preincubated for 10 min with buffer (+ buffer; green), anti-TF (+ α-TF; red), heparinase (+ Hpase, blue), or both heparinase I and anti-TF (+ Hpase + α -TF; yellow). Thrombin generation parameters correlation with D-dimer levels, Spearman test (F). Wilcoxon test for (B), (C) and (D); Spearman test for (F); P values are as indicated in Figure 1.
4. Discussion
The relative contributions of localized lung versus systemic responses to COVID-19-associated thrombosis are unclear. Likewise, whether TF triggers this excessive coagulation, contact activation, or both are not fully resolved. Bulk RNA-seq data from BALF cells [13] analyzed by Mast et al. on 9 COVID-19 patients and 3 healthy controls did not reveal differences in TF (F3) gene expression [15]. By contrast, Fitzgerald et al. [17] found TF (F3) to be significantly differentially expressed in BALF cells of 2 COVID-19 patients versus 3 controls for bulk RNA-seq data from Xiong et al. [29] and to be increased in BALF epithelial cells of 9 COVID-19 patients versus 4 controls using scRNA-seq data from Liao et al. [16]. Fitzgerald et al. also found that PBMC TF (F3) gene expression levels were not changed in 3 COVID-19 patients versus 3 controls of bulk RNA-seq data also from Xiong et al. Our analysis focused on circulating PBMCs responses to COVID-19 and included scRNA-seq and bulk RNA-seq analysis that combined for a total of 34 severe COVID-19, 9 mild patients and 16 control samples recruited at Washington University in St. Louis. Our 2 independent cohorts of patients showed TF(F3) transcript expression levels to be significantly elevated 5- and 7-fold, respectively. Our further evaluation of the scRNA-seq data from Liao et al. showed an increase in TF (F3) transcript expression in BALF macrophage cells. Our datasets and those of Liao et el. also demonstrated that TF (F3) transcript expression levels depend on disease severity. Noteworthy, the disease status of the data sets of Zhou et al. analyzed by Mast et al. was not provided. If these samples were from patients with less severe diseases, TF (F3) gene expression changes would not be expected.
Mast et al. and Fitzgerald et al. BALF bulk RNA-seq data analyses found enhanced TFPI transcript expression levels and decreased C-1 inhibitor (SERPING1) transcript expression levels in the lungs of COVID-19 patients, which would favor the intrinsic coagulation pathway [15,17]. Our PBMC scRNA-seq analyses showed nonsignificant changes in TFPI transcript expression and significant increases in SERPING1 transcript levels of expression along with elevated TF transcript expression levels. Our findings support a systemic response of circulating CD14+ monocytes to SARS-CoV-2 infection, which favors activation of the TF extrinsic pathway and inhibition of the intrinsic pathway of coagulation. Together, these studies suggest that the coagulopathy induced by COVID-19 in the lung differs (at least partly) from that in the circulation.
In addition to initiating coagulation, TF enhances inflammatory responses through cellular effects mediated by PARs. These cell-signaling effects of TF are direct, through TF/FVIIa and TF-/FVIIa/FXa cell surface complexes to activate PAR-2 [30,31], and indirect, through the actions of thrombin and FXa generated down-stream in the coagulation process, which activate PARs 1, 3, and 4 [32]. It has been reported that TF (F3) expression is increased in inflammatory monocytes from patients with HIV infection [33] and, in a baboon model of Escherichia coli-induced sepsis, inhibition of the TF pathway not only prevented disseminated intravascular coagulation but reduced inflammation and promoted survival [34]. Our findings support that increases in TF expression in response to SARS-CoV-2 infection are similar to other viral and bacterial infections. It is probable that increases in CD14+ monocyte TF expression also contribute to COVID-19 systemic inflammation.
In this study, we also found a significant elevation of thrombomodulin TM (THBD) transcripts in CD14+ monocytes. As an anticoagulant, TM complexes with thrombin to activate protein C, which then proteolytically inactivate cofactors FVIIIa and FVa to inhibit coagulation. TM also regulates inflammation via its lectin binding domain by inhibiting leukocyte-mediated vessel injury, neutralizing cell-damage-mediated inflammation, and suppressing the complement system [35,36]. It is possible that CD14+ monocytes’ increase in THBD transcript expression also dampens the excessive coagulation and inflammatory responses to SARS-CoV-2 infections. In summary, we found significant transcriptional changes in CD14+ monocytes from COVID-19 patients, with simultaneous increases in procoagulant and proinflammatory TF (F3 gene), anticoagulant and anti-inflammatory C1-inhibitor (SERPING1 gene), and anticoagulant thrombomodulin TM (THBD gene) transcript expression.
Several laboratories have reported TGA results for COVID-19 patient samples, but each used Ca++ with TF to trigger the assays, which negates the ability to quantify endogenous triggering activity if present [[37], [38], [39]]. Only Bouck et al. conducted TGAs triggered with Ca++ and without TF [40]. They found no difference between control and COVID-19 samples but did show that anti-TF antibodies impacted these assays. We found in our TGAs that COVID-19 samples were overall hypo-coagulated because of heparin treatment. Our investigations also revealed that the relative impact of COVID-19 disease on Ca++-induced TGAs was less than what occurs during sample processing via contact activation. Collecting blood samples directly into citrate plus CTI prevented the contact activation, but this precluded our ability to define any impact COVID-19 might exert via triggering by the intrinsic coagulation pathway. Similar to Bouck et al., we also found that anti-TF antibody impacted these assays, and, in addition, we found TGA data to correlate with COVID-19 samples’ D-dimer protein levels. If one is to use TGAs to evaluate COVID-19 patient samples, it is recommended that blood be collected directly into citrate (or EDTA) containing CTI and plasma be treated with heparinase prior to analysis. However, any contribution of SARs-CoV-2 infection to contact activation will no longer be observable.
The isolation of EV from plasma by centrifugation and washing eliminates both heparin and contact activation factors, allowing for EV to be investigated for procoagulant potential. Similar to other reports [11,12], for a subset of critically ill COVID-19 patients, we found EV TF activity was elevated. In our study, most critically ill patients with high D-dimer did not show EV TF activity, perhaps because of the assay limit of detection. EV TF activity has been considered a potential biomarker for COVID-19 prognosis [20], although currently, D-dimer is a simpler assay and a more predictable biomarker. Conducting EV TF activity assays also revealed an elevated level of non–TF procoagulant potential in COVID-19 samples that we identified as an increase in EV phospholipid membranes. Though the sources of this EV could not be identified, it is likely that multiple sources, including local lung injury, are likely to contribute to the circulating EV.
In conclusion, we demonstrate significant upregulation of F3 transcripts in circulating CD14+ monocytes and BALF cells from COVID-19 patients. These data support a critical role of the TF-triggered extrinsic pathway in COVID-19-associated systemic thrombosis. Interestingly, SARS-CoV-2 infection upregulates SERPING1 and THBD transcript expression in CD14+ monocytes, which could inhibit the intrinsic and common pathways of coagulation, respectively. In addition, we found that severe SARS-CoV-2 infection increases circulating EV levels and corresponding membrane coagulant activity. Together, the increases in both TF transcriptional expression and elevated procoagulant EV membranes likely contribute to thrombosis. To reduce COVID-19-associated thrombosis, selective targeting of the factor VIIa/TF coagulation triggering complex warrants investigation.
Acknowledgments
This study utilized samples obtained from the Washington University School of Medicine’s COVID-19 biorepository; which is supported by the Barnes-Jewish Hospital Foundation, the Siteman Cancer Center grant P30 CA091842 from the National Cancer Institute of the National Institutes of Health, and the Washington University Institute of Clinical and Translational Sciences grant UL1TR002345 from the National Center for Advancing Translational Sciences of the National Institutes of Health. The BJC Foundation and ICTS provided additional support for these studies to A.L.S. and G.J.R. Additional support was provided by NIH R37 AI049653-20S2 (G.L.R.), the Burroughs Wellcome Fund (A.L.S.), R01 HL120728, and R01 HL141794 (J.D.P.). The content is solely the responsibility of the authors and does not necessarily represent the view of the NIH. We thank Jane O'Halloran, MD, PhD; Rachel Presti, MD, PhD; Charles Goss, PhD; Mansi Agarwal, PhD; Mark Watson, MD, PhD; and Phillip Mudd, MD, PhD, who developed and maintained the biorepository.
Author contributions
T.J.G., N.Z., J.M.A., K.E.R., A.L.S., G.J.R, and J.D.P. designed research; T.J.G., L.A.H., N.Z., J.M.A., R.S., I.E., M.M., and E.C.E. performed research; T.J.G., L.A.H., N.Z., J.M.A., I.E., C.C., A.L.S., G.J.R., and J.D.P. analyzed data and contributed analytical tools; T.J.G., L.A.H., N.Z., I.E., A.L.S., G.J.R., and J.D.P. wrote the article.
Declaration of competing interest
There are no competing interests to disclose.
Footnotes
Manuscript handled by: P. Liaw
Final decision: P. Liaw, 01 November 2022
Supplementary material
References
- 1.Cui S., Chen S., Li X., Liu S., Wang F. Prevalence of venous thromboembolism in patients with severe novel coronavirus pneumonia. J Thromb Haemost. 2020;18:1421–1424. doi: 10.1111/jth.14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helms J., Tacquard C., Severac F., Leonard-Lorant I., Ohana M., Delabranche X., Merdji H., Clere-Jehl R., Schenck M., Gandet F.F., Fafi-Kremer S., Castelain V., Schneider F., Grunebaum L., Anglés-Cano E., Sattler L., Mertes P.-M. High risk of thrombosis in patients with severe SARS-CoV-2 infection: a multicenter prospective cohort study. Intensive Care Med. 2020;46:1089–1098. doi: 10.1007/s00134-020-06062-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drake T.A., Ruf W., Morrissey J.H., Edgington T.S. Functional tissue factor is entirely cell surface expressed on lipopolysaccharide-stimulated human blood monocytes and a constitutively tissue factor-producing neoplastic cell line. J Cell Biol. 1989;109:389–395. doi: 10.1083/jcb.109.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grover S.P., Mackman N. Tissue factor: an essential mediator of hemostasis and trigger of thrombosis. Arterioscler Thromb Vasc Biol. 2018;38:709–725. doi: 10.1161/ATVBAHA.117.309846. [DOI] [PubMed] [Google Scholar]
- 5.Mackman N., Antoniak S., Wolberg A.S., Kasthuri R., Key N.S. Coagulation abnormalities and thrombosis in patients infected with SARS-CoV-2 and other pandemic viruses. Arterioscler Thromb Vasc Biol. 2020;40:2033–2044. doi: 10.1161/ATVBAHA.120.314514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ackermann M., Verleden S.E., Kuehnel M., Haverich A., Welte T., Laenger F., Arno V., Christopher W., Helge S., Alexandar T., William W.L., Vincent W.L., Steven J.M., Danny J. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in COVID-19. N Engl J Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hottz E.D., Azevedo-Quintanilha I.G., Palhinha L., Teixeira L., Barreto E.A., Pão C.R.R., Cassia R., Sérgio F., Thiago M.L.S., Pedro K., Fernando A.B., Patrícia T.B. Platelet activation and platelet-monocyte aggregate formation trigger tissue factor expression in patients with severe COVID-19. Blood. 2020;136:1330–1341. doi: 10.1182/blood.2020007252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hottz E.D., Martins-Gonçalves R., Palhinha L., Azevedo-Quintanilha I.G., de Campos M.M., Sacramento C.Q., Temerozo J.R., Soares V.C., Dias S.S., Teixeira L., Castro Í. Platelet-monocyte interaction amplifies thromboinflammation through tissue factor signaling in COVID-19. Blood Adv. 2022;6:5085–5099. doi: 10.1182/bloodadvances.2021006680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ansari S.A., Pendurthi U.R., Rao L.V.M. Role of cell surface lipids and thiol-disulphide exchange pathways in regulating the encryption and decryption of tissue factor. Thromb Haemost. 2019;119:860–870. doi: 10.1055/s-0039-1681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J., Pendurthi U.R., Yi G., Rao L.V.M. SARS-CoV-2 infection induces the activation of tissue factor-mediated coagulation via activation of acid sphingomyelinase. Blood. 2021;138:344–349. doi: 10.1182/blood.2021010685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosell A., Havervall S., von Meijenfeldt F., Hisada Y., Aguilera K., Grover S.P., Ton L., Nigel M., Charlotte T. Patients with COVID-19 have elevated levels of circulating extracellular vesicle tissue factor activity that is associated with severity and mortality-brief report. Arterioscler Thromb Vasc Biol. 2021;41:878–882. doi: 10.1161/ATVBAHA.120.315547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krishnamachary B., Cook C., Kumar A., Spikes L., Chalise P., Dhillon N.K. Extracellular vesicle-mediated endothelial apoptosis and EV-associated proteins correlate with COVID-19 disease severity. J Extracell Vesicles. 2021;10 doi: 10.1002/jev2.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garvin M.R., Alvarez C., Miller J.I., Prates E.T., Walker A.M., Amos B.K., Mast A.E., Justice A., Aronow B., Jacobson D. A mechanistic model and therapeutic interventions for COVID-19 involving a RAS-mediated bradykinin storm. Elife. 2020;9:e6. doi: 10.7554/eLife.59177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mast A.E., Wolberg A.S., Gailani D., Garvin M.R., Alvarez C., Miller J.I., Aronow B., Jacobson D. SARS-CoV-2 suppresses anticoagulant and fibrinolytic gene expression in the lung. Elife. 2021;10 doi: 10.7554/eLife.64330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 17.FitzGerald E.S., Chen Y., Fitzgerald K.A., Jamieson A.M. Lung epithelial cell transcriptional regulation as a factor in COVID-19-associated coagulopathies. Am J Respir Cell Mol Biol. 2021;64:687–697. doi: 10.1165/rcmb.2020-0453OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Francischetti I.M.B., Toomer K., Zhang Y., Jani J., Siddiqui Z., Brotman D.J., Hooper J.E., Kickler T.S. Upregulation of pulmonary tissue factor, loss of thrombomodulin and immunothrombosis in SARS-CoV-2 infection. EClinMed. 2021;39 doi: 10.1016/j.eclinm.2021.101069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subrahmanian S., Borczuk A., Salvatore S., Fung K.M., Merrill J.T., Laurence J., Ahamed J. Tissue factor upregulation is associated with SARS-CoV-2 in the lungs of COVID-19 patients. J Thromb Haemost. 2021;19:2268–2274. doi: 10.1111/jth.15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackman N., Grover S.P., Antoniak S. Tissue factor expression, extracellular vesicles, and thrombosis after infection with the respiratory viruses influenza A virus and coronavirus. J Thromb Haemost. 2021;19:2652–2658. doi: 10.1111/jth.15509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girard T.J., Grunz K., Lasky N.M., Malone J.P., Broze G.J., Jr. Re-evaluation of mouse tissue factor pathway inhibitor and comparison of mouse and human tissue factor pathway inhibitor physiology. J Thromb Haemost. 2018;16:2246–2257. doi: 10.1111/jth.14288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hisada Y., Mackman N. Cancer cell-derived tissue factor-positive extracellular vesicles: biomarkers of thrombosis and survival. Curr Opin Hematol. 2019;26:349–356. doi: 10.1097/MOH.0000000000000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amrute J.M., Perry A.M., Anand G., Cruchaga C., Hock K.G., Farnsworth C.W., Randolph G.J., Lavine K.J., Steed A.L. Cell specific peripheral immune responses predict survival in critical COVID-19 patients. Nat Commun. 2022;13:882. doi: 10.1038/s41467-022-28505-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhindsa S., Zhang N., McPhaul M.J., Wu Z., Ghoshal A.K., Erlich E.C., Mani K., Randolph G.J., Edwards J.R., Mudd P.A., Diwan A. Association of circulating sex hormones with inflammation and disease severity in patients with COVID-19. JAMA Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manne B.K., Denorme F., Middleton E.A., Portier I., Rowley J.W., Stubben C., Petrey A.C., Tolley N.D., Guo L., Cody M., Weyrich A.S. Platelet gene expression and function in patients with COVID-19. Blood. 2020;136:1317–1329. doi: 10.1182/blood.2020007214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandset P.M., Abildgaard U., Larsen M.L. Heparin induces release of extrinsic coagulation pathway inhibitor (EPI) Thromb Res. 1988;50:803–813. doi: 10.1016/0049-3848(88)90340-4. [DOI] [PubMed] [Google Scholar]
- 27.Novotny W.F., Palmier M., Wun T.C., Broze G.J., Miletich J.P. Purification and properties of heparin-releasable lipoprotein-associated coagulation inhibitor. Blood. 1991;78:394–400. [PubMed] [Google Scholar]
- 28.Andreu Z., Yanez-Mo M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Jiang A., Guo D., Hu W., Yang J., Tang Z., Wu H. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camerer E., Huang W., Coughlin S.R. Tissue factor- and factor X-dependent activation of protease-activated receptor 2 by factor VIIa. Proc Natl Acad Sci USA. 2000;97:5255–5260. doi: 10.1073/pnas.97.10.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riewald M., Ruf W. Mechanistic coupling of protease signaling and initiation of coagulation by tissue factor. Proc Natl Acad Sci USA. 2001;98:7742–7747. doi: 10.1073/pnas.141126698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coughlin S.R. Thrombin signalling and protease-activated receptors. Nature. 2000;407:258–264. doi: 10.1038/35025229. [DOI] [PubMed] [Google Scholar]
- 33.Schechter M.E., Andrade B.B., He T., Richter G.H., Tosh K.W., Policicchio B.B., Singh A., Raehtz K.D., Sheikh V., Ma D., Brocca-Cofano E. Inflammatory monocytes expressing tissue factor drive SIV and HIV coagulopathy. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aam5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Creasey A.A., Chang A.C., Feigen L., Wun T.C., Taylor F.B., Hinshaw L.B. Tissue factor pathway inhibitor reduces mortality from Escherichia coli septic shock. J Clin Invest. 1993;91:2850–2860. doi: 10.1172/JCI116529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang J.C. Acute respiratory distress syndrome as an organ phenotype of vascular microthrombotic disease: based on hemostatic theory and endothelial molecular pathogenesis. Clin Appl Thromb Hemost. 2019;25 doi: 10.1177/1076029619887437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foley J.H., Conway E.M. Cross talk pathways between coagulation and inflammation. Circ Res. 2016;118:1392–1408. doi: 10.1161/CIRCRESAHA.116.306853. [DOI] [PubMed] [Google Scholar]
- 37.Campello E., Bulato C., Spiezia L., Boscolo A., Poletto F., Cola M., Gavasso S., Simion C., Radu C.M., Cattelan A., Tiberio I. Thrombin generation in patients with COVID-19 with and without thromboprophylaxis. Clin Chem Lab Med. 2021;59:1323–1330. doi: 10.1515/cclm-2021-0108. [DOI] [PubMed] [Google Scholar]
- 38.de la Morena-Barrio M.E., Bravo-Pérez C., Miñano A., de la Morena-Barrio B., Fernandez-Perez M.P., Bernal E., Gómez-Verdu J.M., Herranz M.T., Vicente V., Corral J., Lozano M.L. Prognostic value of thrombin generation parameters in hospitalized COVID-19 patients. Sci Rep. 2021;11:7792. doi: 10.1038/s41598-021-85906-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polimeni A., Leo I., Spaccarotella C., Mongiardo A., Sorrentino S., Sabatino J., De Rosa S., Indolfi C. Differences in coagulopathy indices in patients with severe versus non-severe COVID-19: a meta-analysis of 35 studies and 6427 patients. Sci Rep. 2021;11 doi: 10.1038/s41598-021-89967-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouck E.G., Denorme F., Holle L.A., Middelton E.A., Blair A.M., de Laat B., Schiffman J.D., Yost C.C., Rondina M.T., Wolberg A.S., Campbell R.A. COVID-19 and sepsis are associated with different abnormalities in plasma procoagulant and fibrinolytic activity. Arterioscler Thromb Vasc Biol. 2021;41:401–414. doi: 10.1161/ATVBAHA.120.315338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.