Abstract
Background
Heteroplexis Chang is an endangered genus endemic to China with important ecological and medicinal value. However, due to the lack of genetic data, our conservation strategies have repeatedly been delayed by controversial phylogenetic (molecular) relationships within the genera. In this study, we reported three new Heteroplexis chloroplast (cp.) genomes (H. vernonioides, H. impressinervia and H. microcephala) to clarify phylogenetic relationships between species allocated in this genus and other related Compositae.
Results
All three new cp. genomes were highly conserved, showing the classic four regions. Size ranged from 152,984 − 153,221 bp and contained 130 genes (85 protein-coding genes, 37 tRNA, eight rRNA) and two pseudogenes. By comparative genomic and phylogenetic analyses, we found a large-scale inversion of the entire large single-copy (LSC) region in H. vernonioides, H. impressinervia and H. microcephala, being experimentally verified by PCR. The inverted repeat (IR) regions showed high similarity within the five Heteroplexis plastomes, showing small-size contractions. Phylogenetic analyses did not support the monophyly of Heteroplexis genus, whereas clustered the five species within two differentiated clades within Aster genus. These phylogenetic analyses suggested that the five Heteroplexis species might be subsumed into the Aster genus.
Conclusion
Our results enrich the data on the cp. genomes of the genus Heteroplexis, providing valuable genetic resources for future studies on the taxonomy, phylogeny, and evolution of Aster genus.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12870-022-04000-1.
Keywords: Heteroplexis, Chloroplast genome, Phylogenetic analysis
Background
Heteroplexis Chang is an oligotypic genus endemic to China, which belongs to the tribe Astereae within the Asteraceae family. It was first described as a monotypic genus (Heteroplexis vernonioide), only found in the Longzhou region, Guangxi, China (Chang, 1937). In a recent infrageneric classification of Heteroplexis, five species (H. vernonioide, H. microcephala, H. sericophylla, H. incana and H. impressinervia) were recognized on the number of hermaphroditic flower and leaf characteristics, such as glandular spots, villi, and vein [1, 2]. These genus species occur mainly in limestone areas along the valleys and mountaintops in the Guangxi Zhuang Autonomous Region of southern China. Most Heteroplexis species have a very narrow and disjunctive distribution area [3]. Such as, H. microcephala has been only recorded in four different towns in Yangshuo county (Guangxi), and H. incana has been only found in Liuzhou (Guangxi) [3]. In addition, the main effective pollinators of the Heteroplexis species are insects such as hoverflies and Vespa ducalis, which are highly susceptible to environmental and climatic factors [4]. As a result, these Heteroplexis species are threatened. Most of them have been listed on the National List of Rare and Endangered Plants in China [5]. Conservation strategies are delayed due to a lack of understanding of the interspecific relationships within this genus.
Phylogenetic studies of Heteroplexis have been underway since it was first discovered. As early as 1937, when Chang discovered Heteroplexis vernonioide for the first time, he thought that its morphological structure was similar to Brachyactis (Chang, 1937). Zhang and Kare further attributed the Heteroplexis to Erigeron - Conyza group because ‘it has more female outer florets than hermaphroditic central florets’ [6]. Later on, Shi et al. (2015), using inter simple sequence repeat (ISSR) makers, found that Heteroplexis species split into three clades H. impressinervia and H. microcephala clade, H. vernonioides and H. incana clade, and H. sericophylla clade [3]. In contrast, Hu (2015), using internal transcribed spacer (ITS) and Expressed sequence Tags (ETS) molecular markers, showed that the Heteroplexis specimens were located in the tribe Astereae, with H. sericophylla and H. vernonioides as sister species, and H. microcephala close to Aster species [7].
In higher plants, the chloroplast (cp.) is a semi-autonomous replication organelle with its own genetic material - the cp. genome [8–10]. The cp. genomes of angiosperms range from 120 to 160 kb in length and exhibit a typical quadripartite circular DNA molecule, which consists of one large single-copy (LSC) region, one small single-copy (SSC) region, and two inverted repeats (IR) [5, 11, 12]. Due to its uniparental inheritance and low recombination and substitution rate, the cp. genome (in whole or as concatenated protein-coding genes fragment) has been used for phylogenetic studies over the past decades, replacing ISSR markers [10, 13]. For example, Kim et al. (1995) constructed a phylogenetic tree of 89 species from Compositae based on chloroplast ndhF genes, identifying five major clades [14]. Whereas Panero et al. (2008) further discussed the origin, evolution, and phylogenetic relationships of Compositae using ten chloroplast-encoded genes [15]. With the advent of next-generation sequencing technology, it is more efficient and inexpensive to obtain cp. genome sequences, which generally provide more genetic information than a few gene fragments to solve the phylogenetic relationship of complicated lineages [16–18]. So far, two Heteroplexis species (H. sericophylla and H. incana) cp. genomes have been reported, and phylogenetic trees based on whole cp. genomes showed that Heteroplexis is more closely related to Aster [5, 19].
In this study, we sequenced and assembled the other three Heteroplexis species whole cp. genomes and performed comprehensive structural, sequence, and phylogenetic analyses with previously published Heteroplexis spp. cp. genomes. The main goals of this study were: (1) provide new cp. genomes data and perform a comparative genomic analysis among Heteroplexis species; (2) elucidate the phylogenetic relationships of Heteroplexis species within the genus and family; and (3) identify hypervariable loci for future development of molecular markers to ascertain Heteroplexis species identification for conservation and phylogenetic studies.
Results
General CP features
Illumina HiSeq paired-end (150 pb) reads were assembled for each Heteroplexis species with NOVOPlasty software, obtaining three new full cp. genomes deposited at NCBI (H. impressinervia, MN367917; H. microcephala, MW795355; and H. vernonioides, MN462631), that were compared with the previously published two plastomes of Heteroplexis (H. incana, MN172194 and H. sericophylla, MK942054).
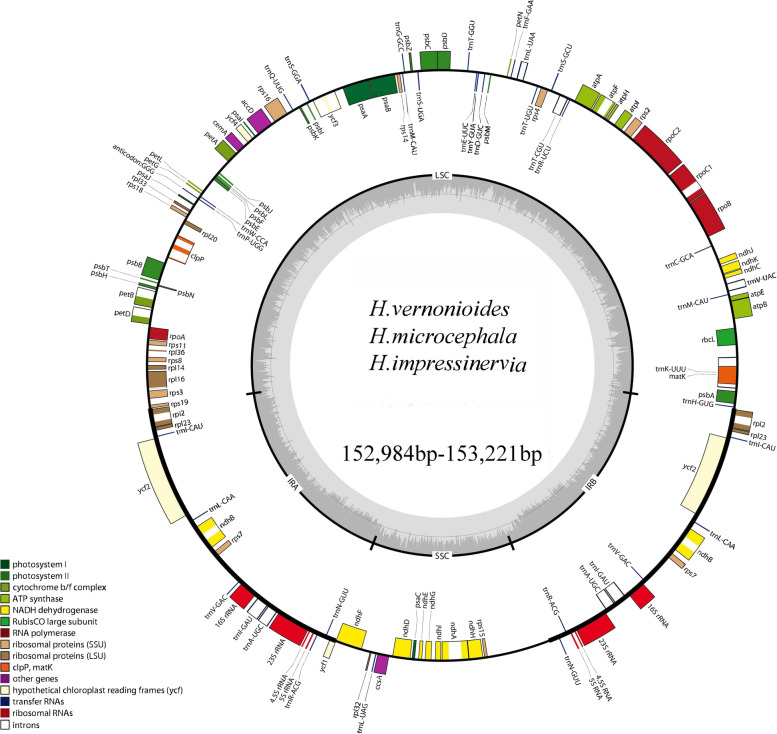
All five Heteroplexis plastomes exhibit a typical quadripartite structure (LSC-IR-SSC-IR), with a conserved gene content, relative gene position and orientation (Fig. 1; Supplementary Table S1). The cp. genome length ranged from 152,605 bp (H. incana) to 153,221 bp (H. vernonioides), with an average GC content of 37.3%, except for H. vernonioides that was 37.2%. The total number of annotated genes in the characterized Heteroplexis plastomes was of 132, distributed in 85 (79 + 6) protein-coding genes (PCGs), 37 (30 + 7) tRNAs, and 8 (4 + 4) rRNA genes, and two pseudogenes (rps19φ and ycf1φ). Six tRNA genes and 12 PCGs contained introns, of which 15 (atpF, ndhA, ndhB, rps16, rpoC1, petB, petD, rpl16, rpl2, trnA-UGC, trnI-GAU, trnG-UCC, trnK-UUU, trnL-UAA, and trnV-UAC) contained only one intron and three (clpP, rps12 and ycf3) contained two introns (Table S2).
Fig. 1.
Physical maps of three newly sequenced Heteroplexis chloroplast genomes
Repeated sequences
A total of 339 complex repeats sequences, consisting of 227 interspersed repeats (101 forward, 20 reverse, 100 palindrome, and six complementary) and 112 tandem repeats, were identified within plastome genomes using MISA, REPuter and Tandem Repeat Finder software, as described in the Material and Methods section (Figure S1A). For interspersed repeats, the sequence length is mainly concentrated in 30–60 bp, regardless of the forward or palindrome repeats. As for the tandem repeats, most were in the range of 16–40 bp, and there was a repeat over 117 bp in H. vernonioides. It is located between the gene trnT-UGU and trnG-UCC. These tandem repeats were mainly distributed in the non-coding LSC and SSC regions.
Microsatellites are small repeating units (1–6 nucleotide) within a genome nucleotide sequence [20]. The high rate of polymorphism in repeat sequences at the species level makes them one of the most common molecular markers in phylogenetic and population genetics studies [21]. The total number of SSRs range from 85 to 93 (Figure S1B), the majority of which were mononucleotide repeats (34%, with A/T showing the highest numbers), followed by trinucleotide (24%) and dinucleotides (18%), whereas other SSRS types have lower proportions (Figs. S1C, S1D). Mononucleotide repeats may play a more important role in genetic variation than other SSRs types. Our findings are similar to other studies that show that A/T repeats were the most abundant [22]. In addition, the analysis of SSR locations revealed that most SSRs were distributed in the non-coding, intergenic, and intron regions. These SSRs can be used to develop specific markers, which can be key in studying systematics and evolution of the family, with conservation purposes among others.
Comparative genome analysis
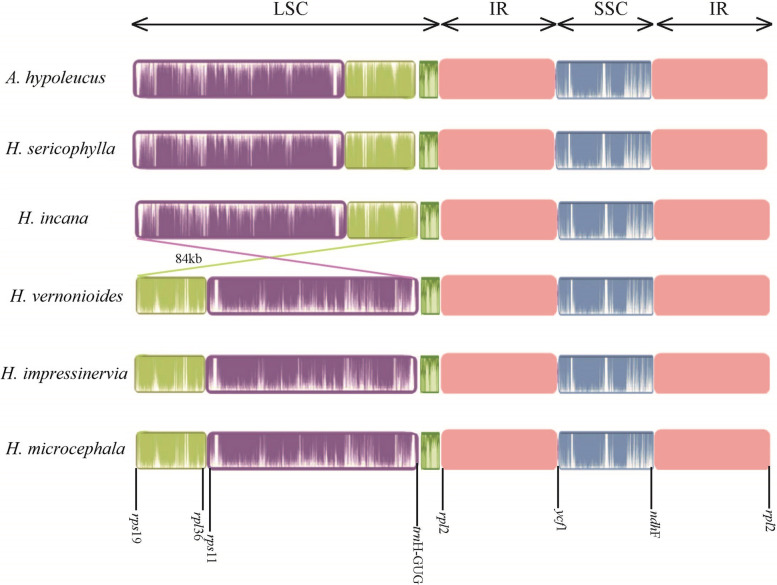
To understand the structural variation of Heteroplexis plastomes, we compared the cp. genomes of all five species and A. hersileoides with Mauve software. Our Mauve alignment (of the six cp. genomes) generated six locally collinear blocks (LCBs), each representing a homologous region without rearrangement (Fig. 2). The results revealed a large-scale inversion of the entire LSC region in H. vernonioides, H. impressinervia and H. microcephala, as shown in the altered gene positions at the two LSC/IR junctions (Fig. 3). Specific primers (Table S3) were developed to verify the authenticity of this inversion experimentally.
Fig. 2.
Plastid genome structure and gene order in the Heteroplexis chloroplast genomes compared with Aster hypoleucus
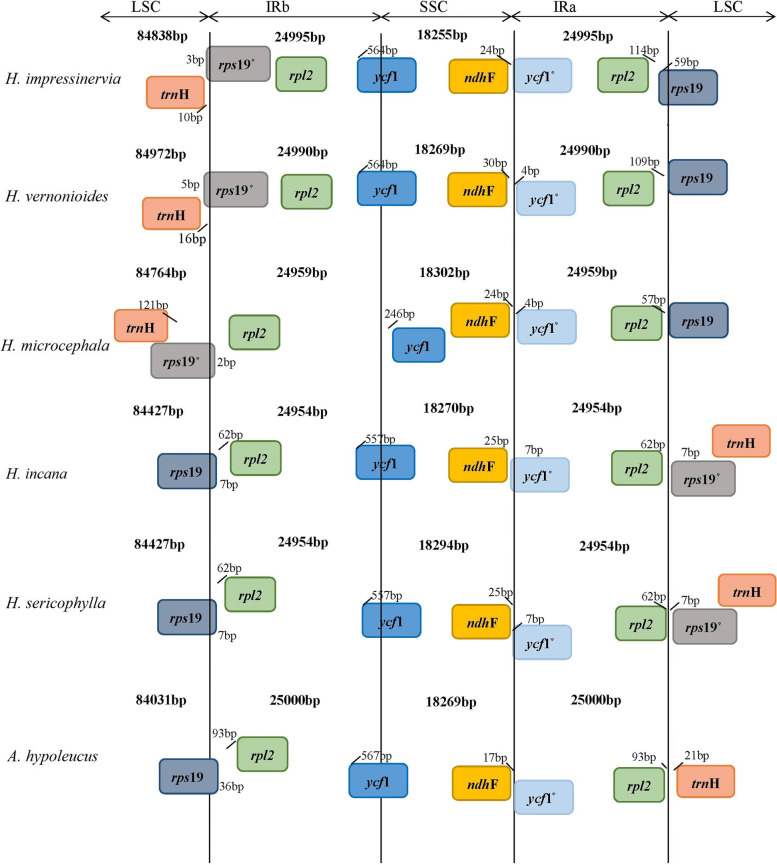
Fig. 3.
Comparison of the borders of LSC, SSC, and IR regions among chloroplast genomes of Heteroplexis. φ pseudogene
We compared the JL (LSC/IR) and JS (IR/SSC) boundary positions of the Heteroplexis plastomes (Fig. 3). The lengths of the IR regions were relatively uniform, ranging from 24,954 to 24,995 bp, with little contraction, especially for H. impressinervia and H. vernonioides. The JL (IR-LSC: rpl2 & rps19) boundary showed high similarity in five Heteroplexis plastomes, except for H. impressinervia and H. vernonioides. The rps19 gene crossed over the IR-LSC border and extended into the IR regions ranging for approximately 62 bp, resulting in a pseudo-copy rps19φ (62 bp in length) by the duplication of IR regions. This pseudogene rps19φ was jumped from JLB (IRb-LSC) to JLA (IRa-LSC) due to LSC region inversion in H. impressinervia, H. vernonioides and H. microcephala. In addition, the pseudogene rps19 was relocated the within the IRb region due to contraction of IR region, whereas the JLB (IRb-LSC: rps19 and rpl2) boundary genes were changed to trnH and rps19 in H. impressinervia and H. vernonioides. The JS (IR-SSC: ycf1 and ndhF) boundaries were also highly similar in Heteroplexis plastomes. The ycf1 gene crossed the IRb-SSC border and extended into the IRb region at approximately 564 bp, except in H. microcephala. This gene, like rps19 gene, also has a pseudo-copy ycf1φ (564 bp in length) due to the duplication of IR regions.
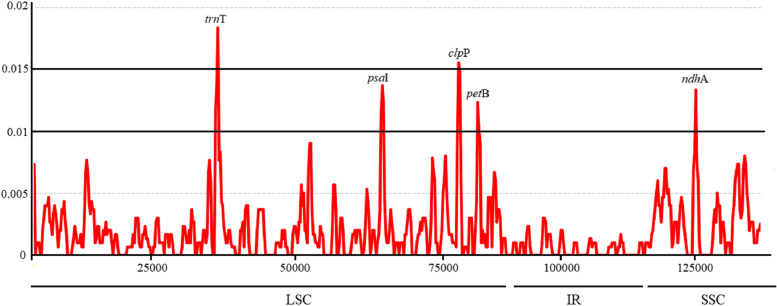
The Pi (π) value in Heteroplexis plastomes ranged from 0 to 0.0183, with an average of 0.0017. The IR regions showed low nucleotide diversity, with most of the variations occurred in the LSC and SSC regions (Fig. 4). Although the protein-coding regions were conserved, five protein-coding genes (trnT, psaI, clpP, petB and ndhA) showed significantly high π values (of > 0.01), with trnT gene showing the highest divergence value (0.0183) (Fig. 4). These polymorphic loci are candidate barcode sequences for phylogenetic inference and population genetic studies of the genus Heteroplexis.
Fig. 4.
Nucleotide diversity in the Heteroplexis chloroplast genomes
Phylogenetic analysis
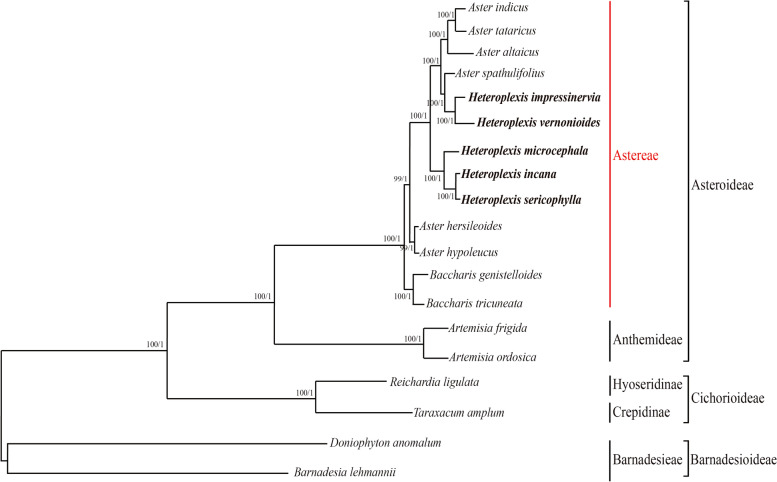
Heteroplexis genus consensus phylogeny was inferred from five data matrices (PCGs, LSC, SSC, IR and intergenic regions sequences) applying the best fit model (GTR + G) distance estimate under two different reconstruction methods (ML and BI), generating the same topology (Fig. 5 and Supplementary Figure S2). The plastid phylogenomic analysis identified a strongly supported (BS = 100, and PP = 1.0) phylogeny with two distinct not clustered clades: one harboring H. vernonioides and H. impressinervia, and the other clade with H. microcephala, H. sericophylla and H. incana (Fig. 5). This result revealed that the genus Heteroplexis was not monophyletic and embedded within the genus Aster. The clade formed by H. vernonioides and H. impressinervia was a sister clade to Aster spathulifolius, which at the same time was sister clade to other three Aster species, split from the second Heteroplexis cluster more basal to other Aster species. These results suggested that these five species should be subsumed into the genus Aster.
Fig. 5.
Maximum likelihood tree and Bayesian tree were constructed based on CDS data partitions of 19 species chloroplast genomes. The number on the branches as Bayesian inference posterior probability/maximum likelihood bootstrap support values
Discussion
In this study, we reported three new plastomes of the genus Heteroplexis using Illumina sequencing and performed a series of genomic and phylogenetic analyses with previously published Heteroplexis plastomes. These new plastomes showed a typical quadripartite structure and their structure and gene content (113 unique genes) were highly conserved, as those species of the genus Aster [23]. Similarly to Aster species, 18 chloroplast protein-coding genes (six tRNA genes and 12 PCGs) were intron-containing genes [24], indicating the close relationship between the genus Heteroplexis and Aster.
The plastome of the most photosynthetic angiosperms is generally highly conserved in gene arrangement [17, 25]. However, plastome rearrangement is more common in the Compositae family. Except for Barnadesioideae, most clades have two inversions, a larger one of about 23 kb and a small one of about 3.3 kb, which may be a key feature in identifying species within the Compositae [26–28]. In Heteroplexis, a large-scale inversion of the entire LSC region was observed in H. vernonioides, H. impressinervia and H. microcephala. Although it is a common event that the cp. genome can exit in two orientations at the SSC region [29], the entire LSC region inversion is first found in Heteroplexis species.
The IR regions are an important part of the cp. genome, and its variation has been demonstrated to substantially contribute to the change in plastome size [30], and its duplication would also result in the pseudogenization of boundary genes [31]. The IR region sizes of the Heteroplexis plastomes were relatively stable, started around the rps19 gene, and terminated almost uniformly downstream of the ycf1 gene, as in Aster cp. genomes [23]. In particular, the rps19 and ycf1 genes shared the LSC/IRb and SSC/IRb boundaries, respectively. However, in the LSC/IRa and SSC/IRa boundaries, the genes were the pseudo-copy of these two parental genes. These results suggested that the duplication of the IR region resulted in the formation of partial copies of these genes, as in Aster species [23].
Previous studies of the genus Heteroplexis have consistently failed to resolve phylogenetic relationships, probably partly due to incomplete sampling or insufficient informative sites in the sampled sequences [23]. In contrast, we obtained well-supported clades, and all inter-specific relationships were well resolved (Fig. 5) by analysis of the whole plastome datasets, which was also supported by the whole genome-wide level restriction site-associated DNA sequencing (RADseq) data-sets [32]. Indicating that complete plastome data-sets have the potential to resolve the phylogenetic relationships of the genus, becoming guidance to resolve the actual taxonomic ambiguity of the Heteroplexis genus. Our strongly supported phylogeny, on which the Heteroplexis species are embedded within the genus Aster, is consistent with the results of an earlier study [7]. However, the split in two highly supported clusters (H. vernonioides and H. impressinervia; and H. microcephala, H. sericophylla and H. incana) corroborating the non-monophyly of Heteroplexis species differs from earlier studies [3, 7].
Conclusion
By chloroplast genome comparative analysis in Heteroplexis species we determined that those plastomes, although highly conserved, had undergone extensive rearrangements, including gene losses, gene duplications, relocations, pseudogenization, IR contraction and LSC inversion. These new plastid data set provided new insights into the inter-specific relationships of Heteroplexis named specimens, suggesting that the five species might be subsumed into the Aster genus and cancel the Heteroplexis genus. Our results have laid the foundation for future studies on the taxonomy, phylogeny, and evolutionary history of Aster genus.
Methods
Sampling, sequencing and annotation
All five Heteroplexis species were included in this study, among which three species were investigated for the first time, namely H. impressinervia Chang, H. vernonioides Chang and H. microcephala Y. L. Chen; the voucher specimen storage information is shown in Supplementary Table S4. All samples were identified by Prof. Yancai Shi. The sampling of three newly sequenced species was approved by the Guangxi province of China and met local policy requirements. Our experimental research, including the collection of plant materials, complies with institutional, national, or international guidelines.
The fresh leaves were obtained from the field for DNA extraction. Total genomic DNAs were extracted using the CTAB method [33] and checked by visualization after 1% agarose-gel electrophoresis. The DNA was used to construct 150 bp insert libraries per the manufacturer’s manual (Illumina Inc., San Diego, CA, USA). The libraries were then subjected to sequencing. The high-throughput sequencing was performed on the Illumina HiSeq Platform (Illumina, San Diego, CA) at Genewiz in Suzhou, China. The Illumina paired-end data were cleaned with trimmomatic version 0.36 [34] and then used for cp. genome de novo assembly by the program NOVOPlasty version 4.3 [35]. Plastome annotation was performed using Plann version 1.1 [36] and the draft annotation was inspected and corrected manually in Sequin version 16.0 (http://www.ncbi.nlm.nih.gov/) [37]. The identities of all tRNA genes were further confirmed with the tRNA-SE search server version 1.4 [38]. A physical map of the genome was generated by OGDRAW version 1.3.1 (http://ogdraw.mpimp-golm.mpg.de/) [39].
Repeat sequence analysis
Simple-sequence repeats were analyzed using MIcroSAtellite version 2.1 (MISA, https://webblast.ipk-gatersleben.de/misa) [40], with parameters set to 10, 5, 4, 3, 3, and 3 for mono-, di-, tri-, tetra-, penta-, and hexa-nucleotides respectively. The long repetitive sequences containing forward, palindromic, reverse, and complementary repeats were analyzed using the software REPuter version 1.1 (https://bibiserv.techfak.uni-bielefeld.de/reputer) [41] with a 30 bp minimum repeat size and a Hamming distance of 3. Tandem Repeats Finder version 4.04 (http://tandem.bu.edu/trf/trf.html) [42] was used to detect tandem repeats, with parameters set to 2 for the alignment parameter match and 7 for mismatches and indels.
Comparative analyses and identification of highly divergent regions
All five Heteroplexis plastomes of H. impressinervia (MN367917), H. vernonioides (MN462631), H. microcephala (MW795355), H. sericophylla (MK942054) and H. incana (MN172194) were included in a comparative analysis. Firstly, the general features (including the genome structure, size, GC content, and gene content) of the five Heteroplexis cp. genomes were characterized using Geneious version 10.2.6 [43]. Secondly, Gene arrangements were further analyzed using Mauve alignment version 1.2 [44] with default parameters. The plastome of Aster hersileoides (NC_042944), which has a typical Astereae tribe plastome organization, was used as the reference in Mauve alignments. Thirdly, the junction of the plastomes was analyzed with IRscope version 3.1 (https://irscope.shinyapps.io/irapp/) [45]. Lastly, to further identify the hypervariable regions and the nucleotide diversity Pi (π), all five Heteroplexis chloroplast genome sequences were aligned using the MAFFT version 7.450 [46] with default parameters. The Pi (π) values were calculated using sliding window analysis (window length = 600 bp and step size = 200 bp excluding sites with alignment gaps) to detect highly divergent regions (i.e., mutation hotspots) in DnaSP version 6.0 [47].
Phylogenetic analysis
To reconstruct the phylogenetic relationships, we complemented our data set of three Heteroplexis plastomes with two previously published Heteroplexis plastomes and 14 plastomes from Compositae (Supplementary Table S5), which included ten species from the Asteroideae subfamily, two species from Cichorioideae subfamily and two species from Barnadesioideae subfamily. The species Barnadesia lehmannii (MH341582) and Doniophyton anomalum (NC_048450) from the Barnadesioideae subfamily were used as an outgroup in the phylogenetic inference. We aligned the plastome sequences of all 19 selected species using MAFFT version 7.450 [46], with manual inspection and correction. We extracted three datasets from the finally aligned plastome matrix to assess the consistency of phylogenetic reconstructions based on different plastome regions. These included sequences of: (a) the protein-coding genes (PCGs), (b) LSC regions, (c) SSC regions, (d) IR regions, and (e) intergenic regions. We used maximum likelihood (ML) and Bayesian inference (BI) methods for phylogenetic analyses. Model Finder version 2.4 [48] was used to find the best model. The best-scoring ML tree was inferred using PhyML version 3.0 [49] with the GTRGAMMAX model for each partition, and clade support was assessed using the rapid bootstrap algorithm with 1,000 replicates. Four parallel Markov chain Monte Carlo (MCMC) runs were performed for the BI method using MrBayes version 3.1.2 [49, 50]. A total of 1,000,000 generations were run with sampling every 500 generations, and the first 25% of samples were discarded as burn-in. The consensus trees were finally edited using Figtree version 1.4.4 [51].
Supplementary Information
Additional file 1: Supplementary Material 1.
Additional file 2: Supplementary Material 2.
Acknowledgements
We are grateful to the Guangxi Institute of Botany for its support. We also thank the personnel of the International Science Editing company for their services in editing this manuscript.
Abbreviations
- IR
Inverted repeat
- LSC
Large single copy
- SSC
Small single copy
- rRNA
Ribosomal rNA
- tRNA
Transfer rNA
- CTAB
Cetyltrimethylammonium bromide
- PCR
Polymerase chain reaction
- SSR
Simple sequence repeat
- BI
Bayesian inference
- ML
Maximum likelihood
- BS
Branch support
Authors’ contributions
DN analyzed the data and wrote the first draft. DLL assisted with the experiments. ZY analyzed the data. SYC provided materials and secured funding. LBB planned and directed the study and revised the manuscript. It is to mention that all authors read and approved the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (Grant Nos. 31960276).
Availability of data and materials
All annotated chloroplast genomes have been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/), accession numbers are provided in Supplementary Table S5.
Declarations
Ethics approval and consent to participate
The sampling of three newly sequenced Heteroplexis species was approved by Guangxi province of China and met local policy requirements. Our experimental research, including the collection of plant materials, are complies with institutional, national or international guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Na Duan and Lili Deng contributed equally to this work.
Contributor Information
YanCai Shi, Email: shiyancainan@163.com.
Bingbing Liu, Email: lbb2015@sxu.edu.cn.
References
- 1.Chen YL. Endemic and retrospectively endangered Heteroplexis genus in Guangxi. Guihaia. 1985;5(4):337–43. [Google Scholar]
- 2.Liang JY. Two new species of Heteroplexis CHANG (Compositae) Guihaia. 1994;14(2):126–9. [Google Scholar]
- 3.Shi YC, Zhou R, Fan JS, Lu ZY, Wei JQ, Jiang YS. Analysis of genetic relationships between Heteroplexis by ISSR. Seed. 2015;34(4):5–7. doi: 10.16590/j.cnki.1001-4705.2015.04.036. [DOI] [Google Scholar]
- 4.Shi YC, Zhou R, Tang JM, Lu ZY, Chai SF, Wei X. Study on the floral biology and breeding system of Heteroplexis plants. Acta Bot Boreali-Occidentalia Sinica. 2015;35(4):824–9. [Google Scholar]
- 5.Shi YC, Zhang Y, Liu BB. Complete chloroplast genome sequence of Heteroplexis sericophylla (Asteraceae), a rare and vulnerable species endemic to China. Mitochondrial DNA B Resour. 2019;4(2):3719–20. doi: 10.1080/23802359.2019.1680323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang XP, Bremer K. A cladistic analysis of the tribe Astereae (Asteraceae) with notes on their evolution and subtribal classification. Plant Syst Evol. 1993;184(3–4):259–83. [Google Scholar]
- 7.Hu HH. A systematic study of the genus Heteroplexis (Asteraceae). A dissertation submitted to University of Chinese academy of Sciences. 2015.
- 8.Gao LZ, Liu YL, Zhang D, Li W, Gao J, Liu Y, Eichler EE. Evolution of Oryza chloroplast genomes promoted adaptation to diverse ecological habitats. Commun Biol. 2019;2(2):278. doi: 10.1038/s42003-019-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li YT, Dong Y, Liu YC, Yu XY, Yang MS, Huang YR. Comparative analyses of Euonymus chloroplast genomes: genetic structure, screening for loci with suitable polymorphism, positive selection genes, and phylogenetic relationships within Celastrineae. Front Plant Sci. 2021;11:2307. doi: 10.3389/fpls.2020.593984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang H, Zhang L, Xie HH, Liu JQ, Xi ZX, Xu XT. The conservation of chloroplast genome structure and improved resolution of infrafamilial relationships of Crassulaceae. Front Plant Sci. 2021;12:631884. doi: 10.3389/fpls.2021.631884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan N, Deng Y, Liu Y, Zhang Y, Zhang L, Wang C, Liu BB. The complete chloroplast genome of Sophora alopecuroides (Fabaceae) Mitochondrial DNA B Resour. 2019;4(1):1336–7. doi: 10.1080/23802359.2019.1596760. [DOI] [Google Scholar]
- 12.Zhao YB, Yin JL, Guo HY, Zhang YY, Xiao W, Sun C, Wu JY, Qu XB, Yu J, Wang XM, Xiao JF. The complete chloroplast genome provides insight into the evolution and polymorphism of Panax ginseng. Front Plant Sci. 2015;5:696. doi: 10.3389/fpls.2014.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao KH, Li LQ, Quan H, Yang JB, Zhang ZR, Liao ZH, Lan XZ. Comparative analyses of chloroplast genomes from 14 Zanthoxylum species: identification of variable DNA markers and phylogenetic relationships within the genus. Front Plant Sci. 2021;11:605793. doi: 10.3389/fpls.2020.605793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim KJYU, Jansen RK. The ndhF sequence evolution and the major clades in the sunflower family. Proc Natl Acad Sci U S A. 1995;92(22):10379–83. doi: 10.1073/pnas.92.22.10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panero JL, Funk VA. The value of sampling anomalous taxa in phylogenetic studies: major clades of the Asteraceae revealed. Mol Phylogenet Evol. 2008;47(2):757–82. doi: 10.1016/j.ympev.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Zang M, Su Q, Weng Y, Lu L, Zheng X, Ye D, Zheng R, Cheng T, Shi J, Chen J. Complete chloroplast genome of Fokienia hodginsii (Dunn) Henry et Thomas: insights into repeat regions variation and phylogenetic relationships in Cupressophyta. Forests. 2019;10(7):528. doi: 10.3390/f10070528. [DOI] [Google Scholar]
- 17.Henriquez CL, Abdullah, Ahmed I, Carlsen MM, Zuluaga A, Croat TR, Mckain MR. Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae) Genomics. 2020;112(3):2349–60. doi: 10.1016/j.ygeno.2020.01.006. [DOI] [PubMed] [Google Scholar]
- 18.Li HW, Liu B, Davis CC, Yang Y. Plastome phylogenomics, systematics, and divergence time estimation of the Beilschmiedia group (Lauraceae) Mol Phylogenet Evol. 2020;151:106901. doi: 10.1016/j.ympev.2020.106901. [DOI] [PubMed] [Google Scholar]
- 19.Qin F, Shi YC, Zhang Y, Wei X, Liu BB. Complete chloroplast genome sequence of Heteroplexis incana (Asteraceae), a rare species endemic to China. Mitochondrial DNA B Resour. 2019;4(2):3031–2. doi: 10.1080/23802359.2019.1666685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ping JY, Feng PP, Li JY, Zhang RJ, Wang T. Molecular evolution and SSRs analysis based on the chloroplast genome of Callitropsis funebris. Ecol Evol. 2021;11(9):4786–802. doi: 10.1002/ece3.7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng G, Wei LL, Ma L, Wu ZQ, Gu CH, Chen K. Comparative analyses of chloroplast genomes from 13 Lagerstroemia (Lythraceae) species: identification of highly divergent regions and inference of phylogenetic relationships. Plant Mol Biol. 2020;102(6):659–76. doi: 10.1007/s11103-020-00972-6. [DOI] [PubMed] [Google Scholar]
- 22.Munyao JN, Dong X, Yang JX, Mbandi M, Hu GW. Complete chloroplast genomes of Chlorophytum comosum and Chlorophytum gallabatense: genome structures, comparative and phylogenetic analysis. Plants. 2020;9(3):296. doi: 10.3390/plants9030296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tyagi S, Jung J, Kim JS, Won SY. Comparative analysis of the complete chloroplast genome of mainland Aster spathulifolius and other Aster Species. Plants. 2020;9(5):568. doi: 10.3390/plants9050568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen XF, Guo S, Yin Y, Zhang JJ, Yin XM, Zhu GW. Complete chloroplast genome sequence and phylogenetic analysis of. Aster tataricus Molecules. 2018;23(10):2426. doi: 10.3390/molecules23102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmer JD, Thompson WF. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell. 1982;29(2):537–50. doi: 10.1016/0092-8674(82)90170-2. [DOI] [PubMed] [Google Scholar]
- 26.Ki-Joong K, Keung-Sun C, Jansen RK. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol Biol Evol. 2005;(9):1783–1792. [DOI] [PubMed]
- 27.Timme RE, Kuehl JV, Boore JL, Jansen RK. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: identification of divergent regions and categorization of shared repeats. Am J Bot. 2007;94(3):302–12. doi: 10.3732/ajb.94.3.302. [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, Hahn FM, Mcmahan CM, Cornish K, Whalen MC. Comparative analysis of the complete sequence of the plastid genome of Parthenium argentatum and identification of DNA barcodes to differentiate Parthenium species and lines. BMC Plant Biol. 2009;9(1):131. doi: 10.1186/1471-2229-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doyle JJ, Davis JI, Soreng RJ, Garvin D, Anderson MJ. Chloroplast DNA inversions and the origin of the grass family (Poaceae) Proc Natl Acad Sci U S A. 1992;89(16):7722–6. doi: 10.1073/pnas.89.16.7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jansen RK, Ruhlman TA. Plastid genomes of seed plants. In. Dordrecht: Springer Netherlands. 2012;103–126.
- 31.Xie JB, Chen SS, Xu WJ, Zhao YY, Zhang DQ. Origination and function of plant pseudogenes. Plant Signal Behav. 2019;14(8):1625698. doi: 10.1080/15592324.2019.1625698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu X, Liang H, Jiang H, Kang M, Wei X, Deng L, Shi YC. Phylogeographic structure of Heteroplexis (Asteraceae), an endangered endemic genus in the limestone karst regions of southern China. Front Plant Sci. 2022;13:999964. doi: 10.3389/fpls.2022.999964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doyle JJ. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19(1):11–5. [Google Scholar]
- 34.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dierckxsens N, Mardulyn P, Smits G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017;45(4):e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang DI, Cronk QCB, Plann A command-line application for annotating plastome sequences. Appl Plant Sci. 2015;3(8):1500026. doi: 10.3732/apps.1500026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dennis AB, Ilene KM, David JL, James O, David LW. GenBank: update. Nucleic Acids Res. 2004;32:D23–6. doi: 10.1093/nar/gkh045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greiner S, Lehwark P, Bock R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 2019;47:W59–64. doi: 10.1093/nar/gkz238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiel T, Michalek W, Varshney RK, Graner A. Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.) Theor Appl Genet. 2003;106:411–22. doi: 10.1007/s00122-002-1031-0. [DOI] [PubMed] [Google Scholar]
- 41.Stefan K, Choudhuri JV, Enno O, Chris S, Jens S, Robert G. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29(22):4633–42. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gary B. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–80. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. <background-color:#CCFF99;uvertical-align:sub;>Bioinformatics</background-color:#CCFF99;uvertical-align:sub;>. 2012;28(12):1647–1649. [DOI] [PMC free article] [PubMed]
- 44.Darling AE, Mau B, Perna NT, Stajich JE. Progressive Mauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5(6):e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali A, Jaakko H, Peter P. IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics. 2018;34(17):3030–1. doi: 10.1093/bioinformatics/bty220. [DOI] [PubMed] [Google Scholar]
- 46.Kazutaka K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Julio R, Albert FM, Carlos SJ, Sara GR, Pablo L, Ramos-Onsins SE, Alejandro SG. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. 2017;34(12):3299–302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 48.Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler AV, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14(6):587–9. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stéphane G, Franck L, Patrice D, Olivier G. PHYML Online - a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–9. doi: 10.1093/nar/gki352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ronquist F, Huelsenbeck JP. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 51.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29(8):1969–73. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Material 1.
Additional file 2: Supplementary Material 2.
Data Availability Statement
All annotated chloroplast genomes have been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/), accession numbers are provided in Supplementary Table S5.