Abstract
Attachment to the intestinal mucosa is an essential step in the pathogenesis of diarrhea caused by enteropathogenic Escherichia coli (EPEC). Fimbriae and intimin, the outer membrane protein product of the chromosomal eae gene, contribute to this process, but their relative roles and the nature of their interaction are not known. The aim of this study was to determine the relative contribution of plasmid-encoded fimbriae, termed Ral, and intimin to the capacity of rabbit-specific EPEC (REPEC) to attach to the intestinal mucosa of rabbits. To achieve this, we constructed a series of mutants in REPEC strain 83/39 (O15:H−), in which the ralE and eae genes were insertionally inactivated. These strains were then inoculated into ligated loops of rabbit ileum, which were resected 18 h later and examined by light and electron microscopy. The results showed that intimin, but not Ral, is essential for the elicitation of attaching-effacing lesions by REPEC. Nevertheless, a Δeae Ral-bearing mutant adhered to the intestinal epithelium to the same extent as its eae-positive parent and far more extensively than an eae+ Δral strain. To examine the contribution of Ral and intimin to colonization of rabbit intestine, we fed these strains to weanling rabbits, which were killed 4 days later, so that the number of bacteria in various regions of the intestine could be determined. The results indicated that strain 83/39 requires both Ral and intimin to colonize the intestine successfully and that a Δeae ΔralE double mutant was incapable of colonizing the intestine. Taken together, these findings indicate that Ral and intimin act independently as adhesion factors of REPEC strain 83/39 and that this strain carries no other significant colonization factor. When both Ral and intimin are present, they appear to act cooperatively, with Ral-mediated adhesion preceding that mediated by intimin.
Some strains of Escherichia coli are primary intestinal pathogens which cause diarrhea (for a review see reference 18). For those varieties of diarrheagenic Escherichia coli, such as enterotoxigenic E. coli (ETEC) and enteropathogenic E. coli (EPEC), which colonize the small intestine, specific attachment factors (or adhesins) are essential virulence determinants, because they allow the bacteria to bind to the small intestinal mucosa and resist removal by peristaltic motility. In ETEC, these attachment factors are well-defined virulence determinants and include the K88 fimbriae of porcine ETEC and the various colonization factor antigens of human strains (reviewed in references 9 and 17).
The adhesins of human EPEC strains are less varied than those of ETEC. They include (i) bundle-forming pili (Bfp), which are type 4-like pili related to the toxin-coregulated pili of Vibrio cholerae (reviewed in reference 24), and (ii) intimin, which is the 94-kDa outer membrane protein product of the eae gene (6, 11). Unlike fimbrial adhesins, which typically are plasmid encoded, eae forms part of a chromosomal pathogenicity island termed the locus for enterocyte effacement (15). The latter incorporates approximately 40 open reading frames whose products act in concert to provoke the characteristic attaching and effacing (A/E) lesions, which are a distinguishing feature of intestinal infections with EPEC (reviewed in reference 8).
EPEC has several counterparts in animals, including rabbit-specific EPEC (REPEC). As for human EPEC strains, these bacteria carry the locus for enterocyte effacement, express intimin, and produce A/E lesions in the intestine of rabbits (15, 20). They differ from human EPEC, however, in that they do not carry Bfp or any other identifiable type 4-like fimbriae (20). Instead, some strains produce adhesins which closely resemble K88 fimbriae of ETEC (1).
The discovery of distinct plasmid- and chromosome-encoded adhesins in EPEC and REPEC led to the formulation of a generalized two-stage model of EPEC adhesion (5), which was supported by data derived from tissue culture assays, organ culture, and animal models (see, for example, references 3 and 12). According to this model, EPEC initially adheres to intact intestinal epithelial cells via fimbrial adhesions, after which it binds more closely via intimin. More recently, however, Hicks et al. (10) used various derivatives of a human EPEC strain and organ culture of pediatric intestine to show that intimin-mediated adherence precedes that mediated by Bfp. They concluded that Bfp are involved mainly in interbacterial binding once the bacteria have adhered to host cells. To examine the relative contribution of adhesive fimbriae and intimin to the pathogenicity of REPEC and to determine which model of EPEC adhesion applies to these bacteria, we investigated the adhesive capacity to rabbit intestine of a REPEC strain, 83/39 (serotype O15:H−), and its derivatives, which carried mutations in the genes for intimin and/or a K88-like fimbrial adhesin, known as Ral (1).
The pathogenicity for rabbits of the spontaneous rifampin-resistant mutant 83/39Rf of REPEC strain 83/39 and its nonfimbriate derivative 83/39-23 (ralE::TnphoA) have been reported previously (1, 19). An eae mutant of 83/39Rf was prepared by “reverse genetics” using a REPEC eae gene that was isolated from a cosmid library and then disrupted by insertion of a kanamycin resistance gene (kan) from the recombinant plasmid pUC4K (Pharmacia Biotech, Piscataway, N.J.) into a unique XhoI site (located 269 bp upstream of the stop codon of the REPEC eae gene [GenBank accession numbers U59502 and U59504]). The eae::kan construct together with some flanking DNA was then cloned into the suicide plasmid pJM703.1 (16). The resultant plasmid was introduced into 83/39Rf by conjugation from E. coli SM10λpir (16). A derivative of 83/39Rf, termed 39-20, in which the native eae allele had been replaced by the recombinant eae::kan gene, was isolated and characterized by Southern hybridization of EcoRV-digested genomic DNA, using DNA probes prepared from eae, kan, and pJM703.1 and by PCR using primers that flanked the XhoI insertion site (data not shown). The production of functional Ral fimbriae by strain 39-20 was established by its ability to adhere to HEp-2 cells as described previously (20).
A similar strategy was used to prepare an eae mutant of E. coli 83/39-23 (ralE::TnphoA), except that in this case, eae was inactivated by insertion of a chloramphenicol resistance (cat) gene from pBAC1 (J. Medd, unpublished data) into the EcoRV restriction site located 1 kb from the start of eae (GenBank accession number U59504). The resultant Δeae Δral mutant was designated 39-80. Both Δeae mutants were trans-complemented with pWSK29eae, which contained an intact REPEC eae gene together with approximately 1.3 kb of upstream and downstream flanking DNA in the low-copy-number plasmid, pWSK29 (25).
The ability of the bacteria to elicit A/E lesions was examined in ligated loops of rabbit ileum as described previously (19, 22). The ligated-loop model is particularly relevant when REPEC strains are tested in rabbit ileum, because the bacteria can interact with their natural host at their preferred site of attachment. As bacteria that are inoculated into ligated loops cannot be removed by peristalsis, the model is particularly useful for investigating the residual adhesive ability of bacteria from which known or suspected adhesins have been deleted.
Light-microscopic examination of sections from intestinal loops inoculated 18 h earlier with the fully virulent REPEC strain 83/39Rf (eae+ ral+) revealed extensive areas of large numbers of bacteria that were closely associated with the epithelium and chiefly within the brush border (Fig. 1A). Electron-microscopic examination of these regions revealed characteristic A/E lesions (Fig. 2A). Strain 83/39-23 (eae+ Δral) also produced A/E lesions, but the number of adherent bacteria (and consequently the extent of the A/E lesions) was far less than that found in loops inoculated with 83/39Rf (Fig. 1B). By contrast, loops inoculated with E. coli 39-20 (Δeae ral+) showed large numbers of bacteria associated with the mucosal surface (Fig. 1C) but not within the brush border as in the case of the eae-positive strains. Electron-microscopic examination of these sections confirmed that the bacteria adhered to the intact epithelium (and apparently to the overlying mucus and/or each other) and that A/E lesions were absent (Fig. 2B). The observation that 39-20 was as capable of attaching to intestinal epithelial cells as its eae+ parent indicated that this strain does not require intimin to bind to the intestinal epithelium. Moreover, the observation that strain 83/39-23 (Δral eae+) adhered less extensively than either 83/39Rf (ral+ eae+) or 39-20 (ral+ Δeae), while retaining the capacity to evoke A/E lesions, indicates that intimin-mediated binding is less efficient in this setting than that due to Ral.
FIG. 1.
Light micrographs of 0.5-μm-thick sections of rabbit ileum that were fixed in glutaraldehyde, embedded in epoxy resin, and stained with methylene blue. Each micrograph is a representation of the appearance of the mucosa in ligated loops inoculated 18 h earlier with one of the following derivatives of REPEC strain 83/39: (A) 83/39Rf (eae+ ral+), (B) 83/39-23 (eae+ Δral), (C) 39-20 (Δeae ral+), (D) 39-80 (Δeae Δral), (E) 39-20(pWSK29eae), and (F) 39-80(pWSK29eae). Arrows indicate groups of bacteria that are closely adherent to the epithelium at sites where the brush border is disrupted. Arrowheads indicate bacteria adherent to the intact brush border. Bar, 30 μm.
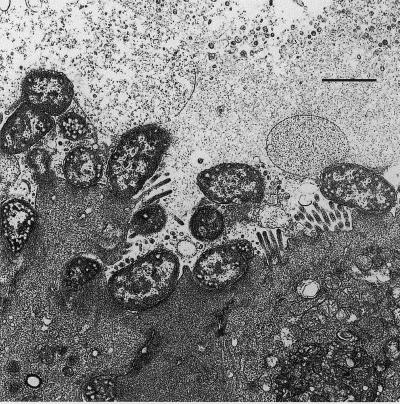
FIG. 2.
Transmission electron micrographs of mucosa from ligated ileal loops inoculated 18 h earlier with REPEC strain 83/39Rf (eae+ ral+) (A) or 39-20 (Δeae ral+) (B). Note the extensive A/E lesions in panel A and their complete absence from panel B, despite the large number of bacteria associated with the epithelium. Bar, 2 μm.
Strain 39-80 (Δeae Δral) induced no pathological changes in the intestine, which resembled that of uninoculated “control” loops (Fig. 1D). Although this strain was not observed in any section, quantitative culture of the contents of intestinal loops at the conclusion of the 18-h observation period revealed that 39-80 was no less viable than 83/39Rf or 39-20 (data not shown). Collectively, these results indicate that Ral fimbriae and intimin act independently as adhesive factors of REPEC 83/39 and that bacteria which lack both of these factors are not able to adhere to the intestinal epithelium.
When the two Δeae mutants, 39-20 and 39-80, were trans-complemented with eae on plasmid pWSK29eae, their capacity to cause A/E lesions was restored. In the case of strain 39-20(pWSK29eae), the findings resembled those in loops inoculated with 83/39Rf, except that there appeared to be more bacteria that adhered to the mucosa without inducing A/E lesions than was the case with 83/39Rf (Fig. 1E). Strain 39-80 (pWSK29eae) behaved in a manner similar to 39-20, causing A/E lesions that were sparsely distributed but otherwise indistinguishable from those induced by the fully virulent strain (Fig. 1F and 3). These results confirm the observations made using other experimental systems that intimin is essential for the A/E capacity of EPEC and related bacteria (4, 7, 14, 23). Moreover, the finding that strains 83/39Rf (ral+ eae+) and 39-20 (Δeae ral+) adhered to a similar degree to each other and more extensively than 83/39-23 (Δral eae+) suggests that binding via Ral precedes that mediated by intimin. This conclusion is in keeping with the original two-stage model of EPEC adhesion, which proposed that bacteria first adhere to the intact mucosa via fimbriae, after which they evoke A/E lesions and adhere more closely via intimin (3, 12). Accordingly, when primary adhesion via fimbriae does not occur, intimin-mediated attachment will be limited.
FIG. 3.
Transmission electron micrograph of mucosa from a ligated ileal loop inoculated 18 h earlier with REPEC strain 39-20 (Δeae ral+), which was trans-complemented with eae via plasmid pWSK29eae. Note the characteristic A/E lesions, which indicate the ability of the plasmid to restore A/E capacity to strain 39-20. Bar, 1 μm.
Our findings and those of Hicks et al. (10) regarding the sequence of events in REPEC and EPEC adherence suggest that Ral and Bfp fulfill different roles in this process. Hicks et al. (10) investigated the in vitro adhesive capacity of derivatives of a human EPEC strain, E2348/69, to human small intestine and concluded that intimin acts before Bfp, whereas our data indicate that Ral acts before intimin. The apparent discrepancy between the findings of these two studies can be attributed to the different nature of Ral and Bfp. Ral fimbriae are closely related to K88 fimbriae of ETEC (1), whose essential role in virulence by mediating bacterial attachment to the intestinal mucosa is undisputed (for a review see reference 17). By contrast, Bfp seem to be primarily involved in interbacterial attachment and detachment (2, 10, 13). In their model of EPEC adhesion, Hicks et al. (10) proposed that a factor, which remains to be identified, promotes initial contact between Bfp-bearing EPEC and the intestinal epithelium. Our data indicate that Ral fulfills this role in REPEC strain 83/39, which produces no detectable adhesins other than intimin and Ral.
To determine the relative contribution of Ral fimbriae and intimin to the ability of strain 83/39 to colonize rabbit intestine, we inoculated rabbits with a series of mutants in which the genes encoding these factors were inactivated. For these studies, bacterial strains were inoculated via a stomach tube into 4-week-old New Zealand White rabbits 15 min after they had received a 2-ml dose of 5% sodium bicarbonate. The inoculum comprised 2 × 108 CFU, which is approximately 100-fold greater than the number of CFU required to cause fatal diarrhea in approximately 80% of rabbits of the same age (1, 19). The colonizing ability of each test strain was determined (i) by enumerating the test bacteria in rectal swabs each day after infection (1) and (ii) by killing the rabbits on the fourth day after infection and counting the number of bacteria associated with the intestinal epithelium in the duodenum, jejunum, ileum, cecum, and colon (21). The 4-day time point was chosen because it is within the incubation period of diarrhea caused by REPEC strain 83/39 (1). Hence, bacteria recovered at this time are likely to be well established in their preferred niche in the intestine and to be replicating in the absence of disease. Once diarrhea commences, however, the distribution of bacteria in the intestine may be altered, and their opportunity to replicate is enhanced. The data obtained from quantitative bacterial cultures were log transformed and subjected to statistical analysis using Instat software (GraphPad, San Diego, Calif.) on an IBM-compatible computer. For animals that were culture negative, an arbitrary value of 10 CFU/g was used. A P value of <0.05 was taken to indicate statistical significance.
None of the rabbits given any of the test strains became ill or lost weight during the 4-day observation period. Moreover, no rabbit showed evidence of intraintestinal fluid accumulation at the time of autopsy. The numbers of bacteria recovered from different regions of the intestine of these animals are shown in Table 1. All six test strains failed to colonize the duodenum or jejunum to a notable extent, with only 2 of all 30 rabbits yielding the bacteria from these sites. By contrast, E. coli 83/39Rf (eae+ ral+) was isolated from the ileum of all five animals given this strain, in numbers ranging from 1 × 107 to 4.7 × 1010 CFU/g of mucosa (geometric mean, 1.8 × 108 CFU/g). Similar numbers of this strain were recovered from the cecum and colon, with a geometric mean of around 1.5 × 108 CFU/g for each site (Table 1). Strain 83/39-23 (Δral eae+) was recovered from only two of five rabbits given this strain. The mean number of bacteria recovered from the ileum (40 CFU/g), cecum (500 CFU/g), and colon (400 CFU/g) of these animals was significantly less than that for rabbits given the parent strain (P = 0.004, 0.03, and 0.02, respectively; Student's t test, 2-tailed). E. coli 39-20 (Δeae ral+) was recovered from three of five rabbits given this strain in numbers that were similar to those obtained from rabbits which received 83/39-23 (eae+ Δral) (geometric means for ileum, cecum, and colon: 50, 900, and 500 CFU/g, respectively), but significantly less than those obtained from rabbits given 83/39Rf (eae+ ral+) (P < 0.01; Student's t test, 2-tailed).
TABLE 1.
Mean number of bacteria in mucosal scrapings obtained from different regions of the intestine of five different rabbits inoculated with one of the test strains 4 days earlier
| Strain | Genotype | Mean no. of bacteria (CFU/g of mucosa) froma:
|
||||
|---|---|---|---|---|---|---|
| Duodenum | Jejunum | Ileum | Cecum | Colon | ||
| 83/39Rf | ral+ eae+ | 3 (1) | 5 (1) | 1.8 × 108 (5) | 1.5 × 108 (5) | 1.6 × 108 (5) |
| 83/39-23 | Δral eae+ | <102 (0) | <102 (0) | 40 (1) | 5.0 × 102 (2) | 4.0 × 102 (2) |
| 39-20 | ral+ Δeae | <102 (0) | <102 (0) | 50 (2) | 9.0 × 102 (3) | 5.0 × 102 (3) |
| 39-80 | Δral Δeae | <102 (0) | <102 (0) | <102 (0) | <102 (0) | <102 (0) |
| 39-20(pWSK29eae) | ral+ Δeae (eae) | 7 (1) | 20 (1) | 2.6 × 105 (4) | 1.6 × 105 (4) | 1.8 × 105 (4) |
| 39-80(pWSK29eae) | Δral Δeae (eae) | <102 (0) | <102 (0) | <102 (0) | <102 (0) | <102 (0) |
Data are the geometric mean CFU per gram of mucosa. The number of culture-positive animals is shown in parentheses.
Strain 39-80 (Δeae Δral) was not obtained from any of the five rabbits given this strain. This finding was subsequently confirmed in a larger sample of 10 rabbits (data not shown). The eae-trans-complemented mutant 39-80(pWSK29eae) was also not obtained from any animal. By contrast, trans-complementation of 39-20(Δeae ral+) with pWSK29eae led to a significant increase in the number of bacteria recovered, from geometric means of 50, 900, and 500 CFU/g for the ileum, cecum, and colon, respectively, to 2.6 × 105, 1.6 × 105, and 1.8 × 105 CFU/g for the same three sites (P ≤ 0.05; Student's t test, 2-tailed).
These investigations have shown that REPEC strain 83/39 requires both Ral fimbriae and intimin to colonize rabbit intestine at 4 days. As in the ileal loop model, Ral and intimin appeared to act independently of each other, insofar as the numbers of 83/39-23 (eae+ Δral) and 39-20 (Δeae ral+) bacteria recovered from the ileum, cecum, and colon were similar. Strain 39-80, the Δral Δeae double mutant, was not isolated from any rabbit, confirming that strain 83/39 does not produce any significant independently acting adhesin other than intimin and Ral. The finding that 83/39-23 (eae+ Δral) and 39-20 (Δeae ral+) were recovered in similar numbers from the intestines of orally inoculated rabbits appears to contradict the ileal loop data, which indicated that Ral-bearing strains 83/39Rf and 39-20 were present in greater numbers than their Δral derivatives. The reason for this difference may lie in the nature of the experimental models, in that the ileal loop assay is useful for demonstrating relatively early events in adhesion, whereas enumeration of intraintestinal bacteria 4 days after peroral inoculation provides an indication of overall bacterial infectivity. In the future, it would be useful to compare the kinetics of infection of the two categories of mutant by examining more time points after infection.
Although pWSK29eae was able to restore the A/E phenotype to the Δeae mutants 39-20 and 39-80, the eae-trans-complemented derivatives of these strains were present in the intestine in lower numbers than their respective eae+ homologs, namely, 83/39Rf and 83/39-23. Inefficient trans-complementation suggests that eae was not expressed in normal amounts by these strains, as is often the case when chromosomal mutations are rectified by trans-complementing plasmids. This finding could also be explained by relative in vivo instability of pWSK29eae in derivatives of REPEC 83/39. To investigate this possibility, we replica plated the bacterial isolates from rabbits given 39-20(pWSK29eae) on media designed to select for 39-20 alone or 39-20(pWSK29eae). These studies showed that more than 99% of all isolates obtained from rectal swabs and the intestines of rabbits given 39-20(pWSK29eae) retained the plasmid throughout the observation period, notwithstanding the lack of specific selection.
Taken together, the results of the experiments reported here suggest that during the early stages of infection, REPEC 83/39 adheres to intestinal epithelial cells via Ral and then more closely via intimin as part of the A/E process. In the absence of intimin, REPEC strains can attach via Ral, but they are unable to adhere firmly and by 4 days will have attained significantly lower numbers than strains bearing both Ral and intimin. On the other hand, strains which lack Ral but express intimin are able to colonize the intestine to a limited extent, but once they have attached, they do so firmly and are able to persist for longer periods than bacteria which have attached via Ral alone.
Acknowledgments
We are indebted to J. E. Peeters, National Institute of Veterinary Research, Brussels, Belgium, for the gift of E. coli 83/39. We also gratefully acknowledge assistance provided by Louise Taylor.
This work was supported in part by grants from the Australian National Health and Medical Research Council and the Murdoch Children's Research Institute.
REFERENCES
- 1.Adams L M, Simmons C, Rezmann L, Strugnell R A, Robins-Browne R. Identification and characterization of a K88- and CS31A-like operon of a rabbit enteropathogenic Escherichia coli strain which encodes fimbriae involved in the colonization of rabbit intestine. Infect Immun. 1997;65:5222–5230. doi: 10.1128/iai.65.12.5222-5230.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieber D, Ramer S W, Wu C Y, Murray W J, Tobe T, Fernandez R, Schoolnik G K. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science. 1998;280:2114–2118. doi: 10.1126/science.280.5372.2114. [DOI] [PubMed] [Google Scholar]
- 3.Boedeker E C, Cheney C P. RDEC-1 enteric infection of rabbits: a model for epidemic infantile diarrhea caused by enteropathogenic Escherichia coli (EPEC) strains. In: Keusch G, Wadström T, editors. Experimental bacterial and parasitic infections. New York, N.Y: Elsevier Biomedical Press; 1983. pp. 203–211. [Google Scholar]
- 4.Dean-Nystrom E A, Bosworth B T, Moon H W, O'Brien A D. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect Immun. 1998;66:4560–4563. doi: 10.1128/iai.66.9.4560-4563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnenberg M S, Kaper J B. Enteropathogenic Escherichia coli. Infect Immun. 1992;60:3953–3961. doi: 10.1128/iai.60.10.3953-3961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnenberg M S, Tacket C O, James S P, Losonsky G, Nataro J P, Wasserman S S, Kaper J B, Levine M M. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J Clin Investig. 1993;92:1412–1417. doi: 10.1172/JCI116717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donnenberg M S, Tzipori S, McKee M L, O'Brien A D, Alroy J, Kaper J B. The role of the eae gene of enterohemorrhagic Escherichia coli in intimate attachment in vitro and in a porcine model. J Clin Investig. 1993;92:1418–1424. doi: 10.1172/JCI116718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frankel G, Phillips A D, Rosenshine I, Dougan G, Kaper J B, Knutton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol Microbiol. 1998;30:911–921. doi: 10.1046/j.1365-2958.1998.01144.x. [DOI] [PubMed] [Google Scholar]
- 9.Gaastra W, Svennerholm A M. Colonization factors of human enterotoxigenic Escherichia coli (ETEC) Trends Microbiol. 1996;4:444–452. doi: 10.1016/0966-842x(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 10.Hicks S, Frankel G, Kaper J B, Dougan G, Phillips A D. Role of intimin and bundle-forming pili in enteropathogenic Escherichia coli adhesion to pediatric intestinal tissue in vitro. Infect Immun. 1998;66:1570–1578. doi: 10.1128/iai.66.4.1570-1578.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jerse A E, Yu J, Tall B D, Kaper J B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knutton S, Lloyd D R, McNeish A S. Adhesion of enteropathogenic Escherichia coli to human intestinal enterocytes and cultured human intestinal mucosa. Infect Immun. 1987;55:69–77. doi: 10.1128/iai.55.1.69-77.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knutton S, Shaw R K, Anantha R P, Donnenberg M S, Zorgani A A. The type IV bundle-forming pilus of enteropathogenic Escherichia coli undergoes dramatic alterations in structure associated with bacterial adherence, aggregation and dispersal. Mol Microbiol. 1999;33:499–509. doi: 10.1046/j.1365-2958.1999.01495.x. [DOI] [PubMed] [Google Scholar]
- 14.Marchès O, Nougayrede J P, Boullier S, Mainil J, Charlier G, Raymond I, Pohl P, Boury M, De Rycke J, Milon A, Oswald E. Role of Tir and intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect Immun. 2000;68:2171–2182. doi: 10.1128/iai.68.4.2171-2182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy B, Fekete P Z. Enterotoxigenic Escherichia coli (ETEC) in farm animals. Vet Res. 1999;30:259–284. [PubMed] [Google Scholar]
- 18.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robins-Browne R M, Tokhi A M, Adams L M, Bennett-Wood V. Host-specificity of enteropathogenic Escherichia coli from rabbits: lack of correlation between adherence in vitro and pathogenicity for laboratory animals. Infect Immun. 1994;62:3329–3336. doi: 10.1128/iai.62.8.3329-3336.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robins-Browne R M, Tokhi A M, Adams L M, Bennett-Wood V, Moisidis A V, Krejany E O, O'Gorman L E. Adherence characteristics of attaching and effacing strains of Escherichia coli from rabbits. Infect Immun. 1994;62:1584–1592. doi: 10.1128/iai.62.5.1584-1592.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robins-Browne R M, Tzipori S, Gonis G, Hayes J, Withers M, Prpic J K. The pathogenesis of Yersinia enterocolitica infection in gnotobiotic piglets. J Med Microbiol. 1985;19:297–308. doi: 10.1099/00222615-19-3-297. [DOI] [PubMed] [Google Scholar]
- 22.Robins-Browne R M, Yam W C, O'Gorman L E, Bettelheim K A. Examination of archetypal strains of enteropathogenic Escherichia coli for properties associated with bacterial virulence. J Med Microbiol. 1993;38:222–226. doi: 10.1099/00222615-38-3-222. [DOI] [PubMed] [Google Scholar]
- 23.Schauer D B, Falkow S. The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect Immun. 1993;61:4654–4661. doi: 10.1128/iai.61.11.4654-4661.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tennent J M, Mattick J S. Type 4 fimbriae. In: Klemm P, editor. Fimbriae: aspects of adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press; 1994. pp. 127–146. [Google Scholar]
- 25.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]