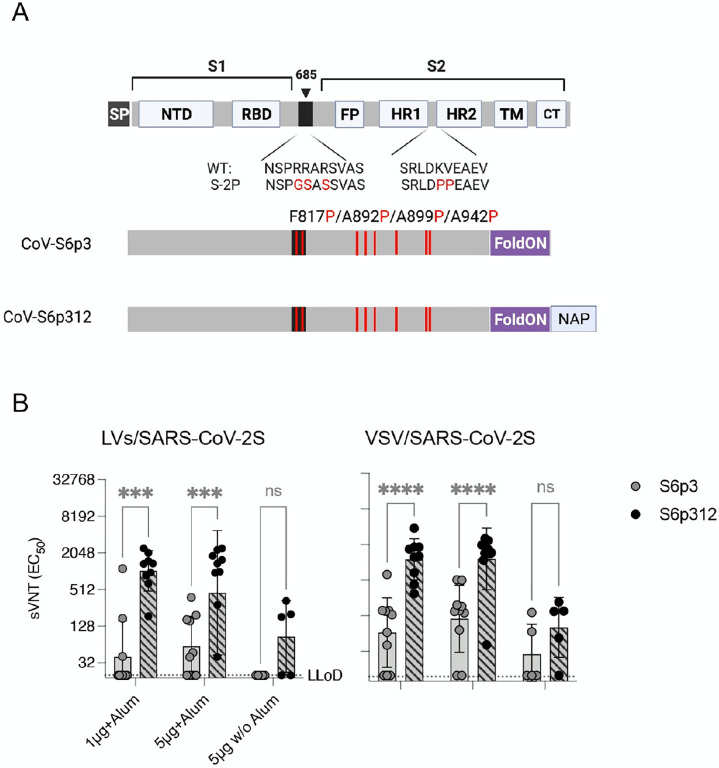
Figure 2. Multimerization of SARS-CoV-2 spike enhances neutralizing antibody responses.
(A) Schematic diagram of the full-length SARS-CoV-2 spike and engineered full-length ectodomain spikes. Some of the structural domains shown include the cleavable signal peptide (SP), N-terminal domain (NTD), receptor-binding domain (RBD), S2 cleavage site (685, black), fusion peptide (FP), heptad repeats 1 and 2 (HR1 and HR2), transmembrane domain (TM) and cytoplasmic tail (CT). The native furin cleavage site was altered (RRAR→GSAS) to inhibit proteolytic cleavage, and six prolines, noted in red text, were introduced to further increase stability. Further modifications include the C-terminal domain of the T4 fibritin (foldON, purple) placed at the C-terminus of the spike and the H. pylori neutrophil-activating protein (NAP, blue) preceded by a GlySer linker. (B) Pseudovirus-neutralizing antibody responses. Mice were vaccinated once with either 1 or 5 μg of alum-adjuvanted proteins, and neutralizing antibodies in serum samples collected 21 days post-vaccination were quantified using LV-SARS-CoV-2 pseudoviruses (left panel) and VSV-SARS-CoV-2-S pseudoviruses (right panel). Antibody titers below the lower limit of detection (LLoD) were replaced with 0.5xLLoD. Black dots represent individual mice, and bars and error bars depict the geometric mean ± geometric standard deviation, respectively. Statistical analysis among groups was calculated by two-way ANOVA with Bonferroni’s post test (ns, p>0.05; ****, p<0.0001).