Abstract
Host-microbe interactions constitute dynamical systems that can be represented by mathematical formulations that determine their dynamic nature. Dynamical interactions can be categorized as deterministic, stochastic, or chaotic. Knowing the type of dynamical interaction is essential for understanding the system under study. Very little experimental work has been done to determine the dynamical characteristics of host-microbe interactions and its study poses significant challenges. The most straightforward experimental outcome involves an observation of time to death upon infection. However, in measuring this outcome, the internal parameters and dynamics of each host-microbe interaction are hidden from the experimentalist. To investigate whether a time-to-death (time to event) dataset provides adequate information for searching for chaotic signatures, we first determined our ability to detect chaos in simulated data sets of time-to-event measurements and successfully distinguished the time-to-event distribution of a chaotic process from a comparable stochastic one. To do so, we introduced an inversion measure to test for a chaotic signature in time-to-event distributions. Next, we searched for chaos in time-to-death of Caenorhabditis elegans and Drosophila melanogaster infected with Pseudomonas aeruginosa or Pseudomonas entomophila, respectively. We found suggestions of chaotic signatures in both systems, but caution that our results are still preliminary and highlight the need for more fine-grained and larger data sets in determining dynamical characteristics. If validated, the occurrence of chaos in host-microbe interactions would have important implications for the occurrence and outcome of infectious diseases, the reproducibility of experiments in the field of microbial pathogenesis and the prediction of future microbial threats.
Introduction
Host microbe interactions can have variable outcomes that result in at least four states, namely symbiosis, commensalism, latency, and disease (1, 2). These states can be distinguished by the amount of damage incurred on the host. When damage is sufficient to affect homeostasis disease ensues and is manifested as symptoms of distress. When damage is sufficient to cripple essential systems death ensues. Studying the dynamics of host-microbe interactions is inherently difficult, since measurements of host-microbe interactions, such as survival time post infection, are often very difficult to reproduce in a quantitative manner even when the initial conditions appear to be nearly identical. There are several non-exclusionary explanations for the lack of detailed inter-experimental reproducibility even when the overall result is reproducible. On one hand, microbial pathogenesis is dependent on a dizzying array of variables that include microbe- and host-specific factors, such as inoculum size, route of infection, host state, temperature, associated microbiota, etc (3) Even in the most carefully conducted experiment it is unlikely that all variables are perfectly controlled from experiment to experiment. This is apparent even in experiments where a qualitative variable such as host death is consistent but quantitative outcomes such as time to death can vary significantly from experiment to experiment (4). For instance, even within an experiment where a set of genetically identical hosts in the form of inbred mice are infected with the same inoculum of a bacterium or parasite, organ microbial burden across individuals can vary by 100-fold (for an example see Figure 2 in (5)). Such ranges are often attributed to experimental variation, although to our knowledge the sources of such variability have not been exhaustively investigated in any host-microbe interaction, and such studies may not be feasible with current experimental capabilities. This interpretation assumes that the system is deterministic since it operates on the implicit assumption that if an experimenter could control for all experimental variables the results would be perfectly reproducible from experiment to experiment. Yet, this assumption has never been formally tested, and it is possible that experimental noise and lack of reproducibility in host-microbe interactions also reflect the mathematical properties of the systems under study.
Dynamical systems are either stochastic or deterministic. Stochastic systems are dominated by random variables and are inherently unpredictable. Deterministic systems are predictable in that they produce the same output given the same initial conditions. Chaotic systems are a subset of deterministic systems where the long-term course of the system is highly unpredictable, even while the system is theoretically predictable. This seeming contradiction stems from the high sensitivity to initial conditions possessed by a chaotic system, popularly captured by the notion of “the butterfly effect”, where a small change to the weather system such as a butterfly flapping its wings cascades onto a large change in the system later on, potentially culminating in a tornado. In a deterministic system, the future course of the system is theoretically possible to determine, if given perfect knowledge of the current state of the system and its dynamics. But since in practice, we can never account fully and with complete precision for all the state variables, such long-term prediction is impossible. By the same token, experiments carried out in the context of a chaotic system cannot be expected to be repeatable since we cannot control for all variables with perfect precision. We emphasize to the non-specialist reader that mathematical chaos does not mean ‘chaotic ‘in the vernacular parlance. The free online dictionary defines chaotic a ‘A condition or place of great disorder or confusion’, but the mathematical definition is that of a system that is highly sensitive to the initial conditions, which is quantified by the maximal Lyapunov exponent being greater than zero, indicating that nearby trajectories at a given point in time diverge exponentially with respect to time in the future. Chaotic systems are to be contrasted with stochastic, or random, dynamical systems. In stochastic systems, neither short- nor long-term prediction is possible due to an irreducible element of randomness. For example, the outcome of a gambling game where the cards are dealt at random cannot be predicted because complete chance determines the combinations at various points.
There is very little information available on the dynamical characteristics of host-microbe interactions, i.e., whether these dynamics are stochastic, predictably deterministic, or chaotic. Cryptococcus neoformans infection in the moth Galleria mellonella, followed a deterministic and predictable course, and no signatures of chaos were detected, but the inoculum used may have been too large, thereby resulting in a predictable outcome of death that overwhelmed any chaotic influences in this system (6). In contrast, analysis of influenza virus dynamics (7) and host-parasitoid systems (8) have found dynamical behavior suggestive of chaos. Chaos is widespread in the biological world and is found in predator-prey relationships (9), microbial densities in ecological systems (9), DNA content during mitosis (10), and even in processes that are considered periodic such as cardiac rhythms (11), circadian rhythms (12), gene regulation (13), human menstrual cycles (14), signaling cascades (15) and yeast budding cycles (16). Given the existence of chaotic signatures in such diverse systems it is worthwhile to consider whether it also occurs in microbial pathogenesis, a process that involves emergent properties arising from the host-microbe interaction. In this study we investigated the dynamics of Pseudomonas spp. infection in Caenorhabditis elegans and Drosophila melanogaster.
Results
Detecting chaos in time-to-death distributions
When faced with a time series sampled from some dynamical process, one might want to detect whether the underlying process is chaotic or stochastic. Of course, there could be some stochasticity on top of an underlying chaotic process, in which case one might still want to detect the presence of chaos. The difficulty arises from the fact that many statistics of a chaotic process resemble a stochastic one -- the determinism of the process rendered barely detectable due to the unstable dynamical trajectories characteristic of chaos. However, methods exist for distinguishing chaos (17, 18). One such method is the permutation spectrum test, studying the distribution of ordinal patterns in short consecutive subsequences of the time series (19, 20). In particular, the existence of forbidden ordinal patterns -- those with no or almost no occurrence -- is known as a strong indication of chaos, which distinguishes it from a truly stochastic time series. In the experiments described here, a population of hosts are exposed to pathogens, and the resulting time-to-death distribution is compared to that of a naturally occurring time-to-death distribution of a control population. In this case, the problem of detecting underlying chaos in the dynamics of host-microbe interactions, when the readout is time-to-death, is complicated by two factors. First, we do not have access to the internal dynamics of the system, only to the time at which a terminal event (death) occurs. Second, since we only gather one data point from each individual (time until death), we must study an assemblage of systems rather than a single continual system. This forces a degree of randomness on the data, especially as we cannot control the system’s internal parameters. In particular, we can no longer use the permutation spectrum test in the traditional way, since we have a distribution of time-to-deaths pulled from a sample population rather than a sample of time-series from single individual subjects.
Faced with these problems, we devised a new test that we term an “inversion measure”, which we suggest can be used in detecting chaos based only on the distribution of time-to-event in an ensemble system. This method is based on a histogram representation of the time-to-event distribution. Similarly to the permutation spectrum test, we subdivide the histogram’s bins into consecutive non-overlapping sequences of k=4 bins. For each such sequence, we check whether it is an inversion, meaning the direction of change from second to third bin is opposite to the direction of change from first to fourth. We measure the frequency of 4-bin sequences which are inversions. We base this measure on the understanding that distributions that are roughly smooth after averaging away random noise would have fewer inversions, whereas distributions that are less smooth and more irregular, which is a characteristic of chaos (17), would potentially have more inversions. Indeed, as the direction of change from the first to fourth bin is expected to indicate the local linear trend, the direction of change from the second to third bin would be expected, on average, to align with this trend as well. Our measure is inspired by the permutation spectrum test in studying the aggregate behavior of short sequences of time. We assign a p-value to the measure based on bootstrapping against a null hypothesis that the distribution is locally linear (See Methods for more details). Thus, a low p-value indicates the distribution significantly deviates from a locally linear one and may have chaotic characteristics. We note, however, that this is not a definitive indicator of chaos but rather of persistent nonlinear irregularities that could arise from chaos. In this paper, we propose this measure as a preliminary test for discriminating between truly stochastic processes and chaotic ones, based on time-to-event distributions. This is a nonparametric test as the null hypothesis makes no assumptions about the specific shape of the time-to-event distribution other than its local linearity.
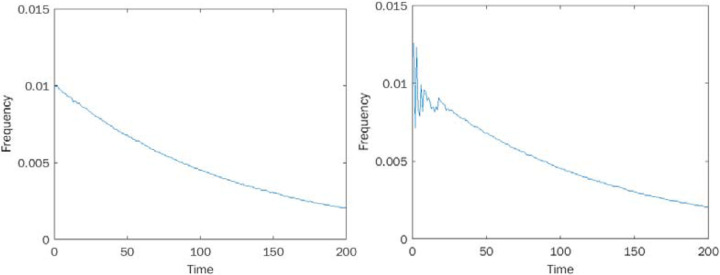
To begin validating this measure, we apply it to simulated time-to-event data from two processes, one chaotic and one stochastic. Specifically, for a chaotic process, we generate data from the logistic map (17) x → rx(1 − x), with 0 < x < 1 and parameter 0 < r < 4. The logistic map is known to be chaotic for certain parameters r. We selected r = 3.9 and measured the distribution of time-to-event, where an event is considered to occur when the dynamic variable x falls in a certain fixed small region (for us, the interval (0.2 ‥ 0.21)), and the initial distribution is uniformly sampled from the interval (0 ‥ 1). For the stochastic process, we sample from a geometric distribution with the same mean, that is the time-to-event distribution of a stochastic Bernoulli process. The resulting time distributions look qualitatively similar for sufficiently long waiting times (see Figure 1), while the distribution of short waiting times is erratic for the chaotic process.
Figure 1:
Time-to-Event distributions: A. Sampling geometric distribution. B. Sampling from the chaotic quadratic map x → rx(1 − x), r = 3.9.
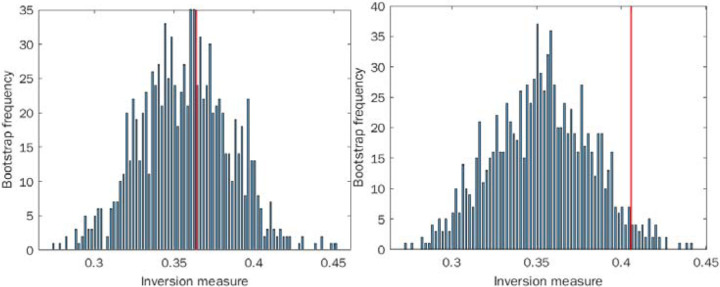
We find a higher proportion of inversions in the chaotic data as opposed to the stochastic data, and we find statistical significance for rejecting the null hypothesis of a locally linear distribution in the chaotic case (p<0.05), but not in the stochastic one (p=0.41), see Figure 2. These results suggest that our inversion measure is capable of discriminating chaos from stochastic behavior using only time-to-event distributions without access to the underlying time-series itself.
Figure 2:
Inversion method of bootstrap for locally linear approximation of the distributions (histograms): A. Sampling from a geometric distribution. B. Sampling from chaotic quadratic map. Red line indicates the actual inversion measure of the distribution.
Biological Data analysis
For the biological model systems described below, time-to-death data is obtained as described in the experimental setup in the methods section, with no preprocessing or change of units. These experiments were conducted using automated C. elegans lifespan machine analysis, which utilizes modified flatbed scanners to capture images of petri plates containing C. elegans animals feeding on a non-pathogenic bacterium (Escherichia coli) or a pathogenic bacterium (P. aeruginosa) at set intervals for processing by automated analysis software as described in (21, 22). This technology enables unbiased, continuous tracking of individual C. elegans animals throughout their adult lifespan and has the potential to determine an individual animal’s time of death to within a 15-minute interval or less. Additionally, these experiments can use relatively large populations per condition with each plate containing up to 35 worms. In the case of D. melanogaster, visual observations were made to assess survival of groups of flies (n=20–30 per vial) feeding on normal fly food or a fly food amended with a pathogenic bacterium (P. entomophila). Dead flies were counted at various time points ranging from every hour up to 48 h.
Caenorhabditis elegans
We first construct the time-to-death histograms for both non-infected and infected C. elegans data sets, to visually determine the differences between them.
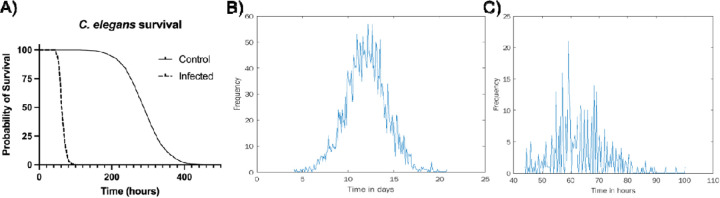
As can be observed in Figure 3, the two histograms differ substantially. While the time-to-death distribution of the non-infected individuals resembles the normal distribution, the infected individuals’ histogram is far more irregular. This irregularity may partially result from a smaller sample size of the infected population (Figure 3). The two histograms, however, come from two different underlying biological processes and so cannot be directly compared. The former results from a process of aging-related death, while the latter reflects the pathogenic process of the worm-microbe interaction and the consequent immune system response.
Figure 3.
Survival analysis of non-infected (data from Stroustrop et al. (21), n=2992) and infected (data generated in Ausubel lab, n=407) C. elegans. A) Survival over time. Time-to-death histograms for non-infected (B) and infected (C) worms.
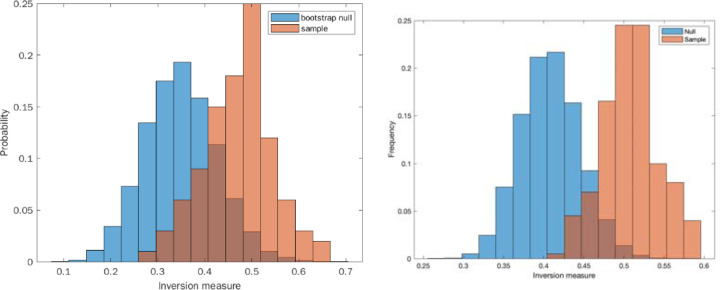
To discriminate whether our system of host-pathogen interactions exhibited chaos, we employed the inversion measure on these time distributions, as described above and in the methods section. We find statistical significance for rejecting the null hypothesis in the infected population (p<0.02), a possible indicator of chaos, and we do not find statistical significance for rejecting the null hypothesis in the uninfected population (p=0.64), see Figure 4. Here we treated the two populations similarly.
Figure 4.
Inversion method of bootstrap for locally linear approximation of the distributions (histograms): A. Sampling from non-infected C. elegans population distribution. B. Sampling from infected C. elegans population. Red line indicates the actual inversion measure of the distribution.
This observation, though not definitive due to the preliminary nature of our inversion-based test for chaos, lends credence to the hypothesis that the time-to-death data result from the infected C. elegans population is the result of an underlying chaotic dynamical process.
D. melanogaster infected with P. entomophila
We repeated a similar procedure to analyze infected and non-infected fly datasets. Here, however, we had to contend with data that was collected manually instead of by machine. While observations were mostly carried out in regular intervals, there was a specific time interval used between data collection points. This means that the number of deaths observed at the subsequent time point needed to be distributed over a wider time interval. To do this, we randomly redistributed the number of deaths observed at wider time intervals into regular subintervals. This introduces an additional element of randomness into the analysis, which we would expect to drown out the chaotic signature. Nevertheless, we still detected statistical significance (p < 0.05) in the inversion measure. In this case, rather than obtaining a single inversion measure for the histogram, we generated a distribution of inversion measures to account for the randomness of the redistribution process and compared this distribution to the null bootstrap distribution. At the same time, in the uninfected fly data we did not find a statistically significant difference between the bootstrap null and sample distribution.
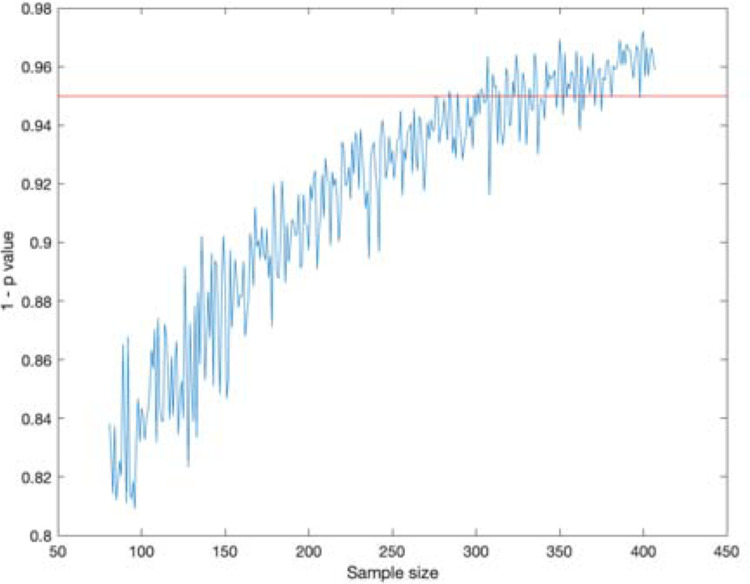
Power analysis
In order to add context to our results, we conducted a power analysis to determine how many data points can be removed while maintaining statistical significance. See Methods for more details. We conducted this analysis with the infected worm data and saw that the statistical significance threshold of p = 0.05 was reached around a sample size of 350, compared to our sample size of 407. The same analysis was performed on the data obtained from the infected fly data resulting in a required sample size of a similar order of magnitude.
Discussion
In this study we first established that a time-to-death distribution provided adequate information to analyze dynamical signatures in C. elegans infected with P. aeruginosa and D. melanogaster infected with P. entomophila, in search for evidence of chaotic behavior. While we found evidence for chaos in both models with the infection time-to-death series, no such evidence was found in the non-infected populations (normal aging). To our knowledge, this is the first evidence that host-microbe interactions may be subject to chaotic behavior. Knowledge of whether virulence is deterministic, stochastic, or chaotic, and the types of host-microbe interactions that manifest those properties is critically important for understanding the fundamental attributes of virulence. Identifying the relevant variables that potentially drive the dynamical behavior in such interactions could have profound implications for public health, vaccine design, and preparedness against emerging pathogens.
The observation of chaotic signatures in infected animals has important implications for the predictability of microbial pathogenesis. Both models are invertebrates, which lack adaptive immunity. We don’t know the generalizability of these observations to other host-microbe systems, but the occurrence of chaotic dynamics in vertebrates would affect our ability to predict future threats. For example, the COVID-19 pandemic has highlighted the importance in identifying other potential threats in nature. The challenge of virulence forecasting is highlighted by the fact that there are more than 300,000 viruses among the 5,500 known mammalian species (23). Could the subset of mammalian viruses that threaten humans be predicted? Some may argue that the answer to this question must await additional mechanistic information that could be used to model human infections. However, even if we knew all the steps by which a single virus could interact with host cells, the immune response to that virus, the potential for transmissibility, prediction of virulence in a population, etc., it would not be possible if the overall process is stochastic or chaotic because small differences in the host-microbe interaction would have major effects, as manifested by a range of outcomes from asymptomatic infection to pathogenicity, which in the extreme results in the death of the host. An example of this phenomenon is the unpredictability of infection at the level of the individual, as evident in the current SARS-CoV-2 pandemic where outcomes range from asymptomatic infection to death and are determined by a set of variables that in combination produce uncertain outcomes (3). Chaotic systems are inherently unpredictable although it is possible to generate probabilistic models for short term prediction of such systems, as is done in applications of stochastic processes and machine learning to weather forecasting. Hence, our finding that some host-microbe interactions may manifest as chaotic dynamical systems implies that absolute predictability with regards to the outcome of these interactions may be impossible if these dynamics are widespread in nature. Nevertheless, knowledge of whether some host-microbe interactions are chaotic could inform efforts to control such diseases including the possibility of exploiting properties of chaos, itself, to our advantage. For example, once the relevant driving forces have been identified in the analysis of chaotic systems, tools from the field of control theory in general, and chaos control theory in particular, can be applied to hypothesize perturbations that could be applied to stabilize a chaotic system (24).
In our prior exploration of chaos with C. neoformans infected Galleria melonella (6), we found that the outcome of infection was deterministic and predictable without evidence of chaos. In the course of this work, we have learned that the time-to-death data must be obtained as precisely as possible for each individual host. Prior work evaluated time-to-death measured at large intervals (e.g., daily), but this is inadequate for detecting dynamical fluctuations on a finer scale, as is the case in our experimental analysis. Many studies in the field of microbial virulence rely on measuring the percentage of animals that survive daily and round to nearest 24 h interval irrespective of the fact that each individual animal succumbed at a different time. This is a form of averaging that is done for experimental convenience since it is often not practical to monitor the experiment continuously to record the exact time of death. While exploring methods to study chaos in host-microbe interactions we found that such averaging could reduce or abolish chaotic signatures. In light of the current analysis, our inability to find chaotic signatures in our prior study may have been a false-negative result stemming from death sampling at large intervals and low power, since it analyzed only 240 events (6). Hence, detection of chaos in host-microbe interactions is highly dependent on experimental design and the common practice for making measurements at large time intervals produces data that is unsuitable for ascertaining the dynamical nature of the system. Fortunately, with the availability of continuous monitoring devices such as cameras it is possible to obtain accurate survival data for each individual in the study and thus generate a fine-grained time-to-death histogram that can be analyzed by the methods described here. Though our results provide statistically significant conclusions, a larger sample size and fine-grained time-to-death measurements are required to solidify our conclusions.
In addition, further validation of our inversion measure and the bootstrap method for obtaining the statistical significance are necessary to solidify our conclusions, including an understanding of how the choice of number (individuals infected) and size of bins (time intervals) affect the results, when a finer time-resolution of the data sampling will become available. In fact, this analysis indicates the need to develop new methods for following the dynamics of host-microbe interactions inside the host since the currently used variables to measure outcome such as time-to-death, weight loss, fever, etc., do not have sufficient resolution to provide a conclusive result.
In recent years there has been great concern about the reproducibility of biomedical research (25, 26). The detection of chaotic signatures in host-microbe interactions implies that these systems may have inherent challenges to reproducibility. Experimenters studying virulence or the efficacy of a therapeutic intervention in an animal model often choose an inoculum that rapidly kills all members of the infected group, leaving no time for the host-pathogen dynamics to present itself. Such doses can ensure conclusive results with relatively few animals but translate into making the experimental system deterministic. Consequently, experiments that force an outcome are not informative of the dynamical nature of natural host-microbe interactions. However, even when using a large inoculum to ensure mortality, there is considerable variation in such variables as time-to-death. In this regard, our observation of chaotic signatures suggests that differences in the microbial load administered and/or the site of infection could translate into large differences in effect size, negating the consistency and reproducibility of inter-experimental comparisons.
In summary, our work shows that a time-to-death distribution provides adequate information to analyze for chaotic signatures in host-microbe interactions and that such signatures exist in C. elegans-P. aeruginosa and D. melanogaster-P. entomophila infection systems. The methods described here provide a suitable approach for exploring other host-microbe interactions, which will expand our understanding of the fundamental dynamics of underlying infectious diseases.
Methods and Materials
C. elegans assays
Wild type (N2) C. elegans were maintained using standard methods. All lifespan and killing assays were performed using the C. elegans Lifespan Machine (21, 22). For lifespan survival of uninfected worms (N2 worms feeding on E. coli OP50), we used the publicly available original data set from Stroustrup et al. 2013 (19). For infection data, assays were performed in the Ausubel Lab using N2 worms and P. aeruginosa strain PA14 prepared as described (27). Survival data is available on figshare - https://figshare.com/s/0c8fdeccef1098972b73.
D. melanogaster assays
Pseudomonas entomophila was streaked from lab glycerol stocks onto LB supplemented with 10% milk and incubated at 29°C overnight (26). Isolated colonies showing lytic activity on milk plates were then inoculated into LB medium and cultured at 29°C for 18 h. Pellets from concentrated cultures (5000 RPM for 20 min at 4°C, OD600 = 200) were mixed 1:1 with 2.5% sucrose; for the negative control 2.5% sucrose was mixed 1:1 with LB. Adult wild-type (Canton-S) flies (4–6 days old, n=25–35 flies/vial) were starved in empty vials for 2 h at 29°C and then transferred to infection vials that consisted of our normal fly food (28) covered with a Whatman filter disc onto which the prepared inoculum or media control was applied (140 μl of the 1:1 bacteria/media: sucrose mix). Flies were kept at 29°C for the duration of the assay but were flipped into new vials without pathogen/infection filters on day 3 post-infection. Pathogen-independent mortality was recorded at 2 h post-infection. Thereafter, the number of dead flies per vial was recorded by visual observation at different (random) time intervals for approximately seven days after infections. For lifespan survival, cohorts of wild-type (Canton-S) female flies (4–6 days old, n=25–35 flies/vial) were monitored every 1–3 days for survival. Flies were flipped into new media tubes every 3 days. Cohorts were monitored for up to 8 days for infected flies or for non-infected flies until all flies in the vial were dead. Survival data is available on figshare - https://figshare.com/s/0c8fdeccef1098972b73.
The inversion measure on a time distribution
Given a distribution of times, we first convert the distribution into a histogram. In the case when the time points are naturally discrete, such as in our simulated data of waiting times sampled from discrete chaotic or stochastic processes, then we use each discrete time point as a bin. In the case of a continuous process, then we pick a parameter n and divide the time distribution into n bins.
Given a histogram with n bins, where each bin contains a whole number, we first add a random noise, distributed uniformly at random between 0 and epsilon, where epsilon<1, to the histogram so as to break ties, ensuring each bin has a unique value while preserving the order of the unequal counts. Then we subdivide the bins into consecutive nonoverlapping sequences of 4 bins each, with the remainder discarded. For each such sequence x1, x2, x3, x4, it is countertrend, or an inversion, if (x4 − x1) and (x3 − x2) have the same signs (positive or negative). We then calculated the frequency of sequences which are inversions. Since this frequency depends on the randomness of the tie-breaking procedure, we rerun this process 1000 times with different randomizations and take the average.
To obtain a p-value for the inversion measure, we used a bootstrap against the null hypothesis that the histogram is locally linear, as follows. For each consecutive sequence of 4 bins, let x1, x2, x3, x4 be the corresponding sequence of whole numbers, totaling to N. We calculate a line of best fit for xi as a function of i, and let this line of best fit be the null distribution for this sequence. If the line goes below 0, we reset the negative numbers to 0’s. We then resample N points according to the null distribution. We repeat this process for each sequence of 4 bins, and then recalculate the inversion measure. We do this 1,000 times to generate a bootstrapped null distribution of the inversion measure. We then calculate a t-statistic and its p-value by comparing the sample inversion measure to this null distribution.
For the simulated data, we simulated 100,000 time points for each process (chaotic and stochastic).
For the fly datasets, in which some observations were not made in regular intervals, we redistributed events along gaps as follows. Whenever one or more observations were missed, we randomly uniformly distributed the next-observed number of events along the interval between the given observation and the prior observation. We then formed a histogram from this redistributed data and proceeded as before to calculate the inversion measure. Due to the randomness of redistribution, we carried this process out 1,000 times, thus generating a sample distribution of inversion measures. For each of these 1,000 histograms, we ran the bootstrap process with a sample size of 1,000, thereby generating a total of 1,000,000 inversion measures that make up the null distribution. We used these two distributions to generate a p value by estimating the probability that the sample inversion measure is greater than or equal to the null inversion measure.
For the uninfected flies, we combined two distinct datasets. In this case, we redistributed each dataset along gaps as mentioned above, and only then combined the two histograms into one and proceeded with the analysis as just described
Power analysis
For the power analysis, we varied a parameter n starting from 20% of the full sample size (n = 82 for the worm data) to 100% of the full sample size (n = 407 for the worm data). For each n, we randomly sampled with replacement n data points (i.e. “events”) from the given distribution. We used this reduced sample to recalculate a p-value for the inversion measure relative to a null distribution, by repeating the same analysis on this reduced sample. We repeated this procedure 100 times for each n-value and took the average of the p-value.
Figure 5.
Survival analysis of non-infected (n=366) and infected (n=673) D. melanogaster. A) Survival over time. Time-to-death histograms for non-infected (B) and infected (C) flies.
Figure 6:
Comparing the null and sample distributions of the inversion measure. A: non-infected. B: infected.
Figure 7:
Power analysis for the C. elegans dataset
Importance.
Is microbial pathogenesis a predictable scientific field? At a time when we are dealing with Coronavirus Disease 2019 (COVID-19) there is intense interest in knowing about the epidemic potential of other microbial threats and new emerging infectious diseases. To know whether microbial pathogenesis will ever be a predictable scientific field requires knowing whether a host-microbe interaction follows deterministic, stochastic, or chaotic dynamics. If randomness and chaos are absent from virulence, there is the hope for prediction in the future regarding the outcome of microbe-host interactions. Chaotic systems are inherently unpredictable although it is possible to generate short-term probabilistic models, as is done in applications of stochastic processes and machine learning to weather forecasting. Information on the dynamics of a system is also essential for understanding the reproducibility of experiments, a topic of great concern in biological sciences. Our study finds preliminary evidence for chaotic dynamics in infectious diseases.
Acknowledgements.
NAB is supported in part by R35GM128871. AC is supported in part by R01 AI152078, R01 HL059842, and AI052733. AB was supported in part by NIH R01-CA164468 and R01-DA033788 to AB.
References
- 1.Casadevall A, Pirofski L. 1999. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect Immun 67:3703–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casadevall A, Pirofski L. 2003. The damage-response framework of microbial pathogenesis. Nature Microbiol Rev 1:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadevall A, Pirofski LA. 2018. What Is a Host? Attributes of Individual Susceptibility. Infect Immun 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailoo JD, Reichlin TS, Wurbel H. 2014. Refinement of experimental design and conduct in laboratory animal research. ILAR J 55:383–391. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, Schmidt RE, Mizushima N, Deretic V, Sibley LD, Virgin HW. 2008. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4:458–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garcia-Solache MA, Izquierdo-Garcia D, Smith C, Bergman A, Casadevall A. 2013. Fungal virulence in a lepidopteran model is an emergent property with deterministic features. mBio 4:e00100–e00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts MG, Hickson RI, McCaw JM, Talarmain L. 2019. A simple influenza model with complicated dynamics. J Math Biol 78:607–624. [DOI] [PubMed] [Google Scholar]
- 8.Preedy KF, Schofield PG, Chaplain MA, Hubbard SF. 2007. Disease induced dynamics in host-parasitoid systems: chaos and coexistence. J R Soc Interface 4:463–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becks L, Hilker FM, Malchow H, Jurgens K, Arndt H. 2005. Experimental demonstration of chaos in a microbial food web. Nature 435:1226–1229. [DOI] [PubMed] [Google Scholar]
- 10.Macieira-Coelho A. 1995. Chaos in DNA partition during the last mitoses of the proliferative life-span of human fibroblasts. FEBS Lett 358:126–128. [DOI] [PubMed] [Google Scholar]
- 11.Wu GQ, Arzeno NM, Shen LL, Tang DK, Zheng DA, Zhao NQ, Eckberg DL, Poon CS. 2009. Chaotic signatures of heart rate variability and its power spectrum in health, aging and heart failure. PLoS ONE 4:e4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leloup JC, Goldbeter A. 1999. Chaos and birhythmicity in a model for circadian oscillations of the PER and TIM proteins in drosophila. J Theor Biol 198:445–459. [DOI] [PubMed] [Google Scholar]
- 13.Rodrigo G, Kirov B, Shen S, Jaramillo A. 2013. Theoretical and experimental analysis of the forced LacI-AraC oscillator with a minimal gene regulatory model. Chaos 23:025109. [DOI] [PubMed] [Google Scholar]
- 14.Derry G, Derry P. 2010. Characterization of chaotic dynamics in the human menstrual cycle. Nonlinear Biomed Phys 4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zumsande M, Gross T. 2010. Bifurcations and chaos in the MAPK signaling cascade. J Theor Biol 265:481–491. [DOI] [PubMed] [Google Scholar]
- 16.Sriram K, Bernot G, Kepes F. 2007. A minimal mathematical model combining several regulatory cycles from the budding yeast cell cycle. IET Syst Biol 1:326–341. [DOI] [PubMed] [Google Scholar]
- 17.Strogatz SH. 2018. Nonlinear dynamics and chaos with student solutions manual: With applications to physics, biology, chemistry, and engineering. CRC press. [Google Scholar]
- 18.Grassberger P, Procaccia I. 2004. Measuring the strangeness of strange attractors, p 170–189, The theory of chaotic attractors. Springer. [Google Scholar]
- 19.Kulp CW, Zunino L. 2014. Discriminating chaotic and stochastic dynamics through the permutation spectrum test. Chaos 24:033116. [DOI] [PubMed] [Google Scholar]
- 20.Kulp CW. 2013. Detecting chaos in irregularly sampled time series. Chaos 23:033110. [DOI] [PubMed] [Google Scholar]
- 21.Stroustrup N, Ulmschneider BE, Nash ZM, López-Moyado IF, Apfeld J, Fontana W. 2013. The Caenorhabditis elegans Lifespan Machine. Nat Methods 10:665–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McEwan DL, Feinbaum RL, Stroustrup N, Haas W, Conery AL, Anselmo A, Sadreyev R, Ausubel FM. 2016. Tribbles ortholog NIPI-3 and bZIP transcription factor CEBP-1 regulate a Caenorhabditis elegans intestinal immune surveillance pathway. BMC Biol 14:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A, Ojeda-Flores R, Arrigo NC, Islam A, Ali KS, Hosseini P, Bogich TL, Olival KJ, Sanchez-Leon MD, Karesh WB, Goldstein T, Luby SP, Morse SS, Mazet JA, Daszak P, Lipkin WI. 2013. A strategy to estimate unknown viral diversity in mammals. mBio 4:e00598–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schöll E, Schuster HG. 2007. Handbook of Chaos Control. Wiley-VCH, Weinheim. [Google Scholar]
- 25.Pusztai L, Hatzis C, Andre F. 2013. Reproducibility of research and preclinical validation: problems and solutions. Nat Rev Clin Oncol 10:720–724. [DOI] [PubMed] [Google Scholar]
- 26.Casadevall A, Fang FC. 2010. Reproducible science. Infect Immun 78:4972–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McEwan DL, Kirienko NV, Ausubel FM. 2012. Host translational inhibition by Pseudomonas aeruginosa Exotoxin A Triggers an immune response in Caenorhabditis elegans. Cell Host Microbe 11:364–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lesperance DNA, Broderick NA. 2020. Meta-analysis of Diets Used in Drosophila Microbiome Research and Introduction of the Drosophila Dietary Composition Calculator (DDCC). G3 (Bethesda) 10:2207–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]