Abstract
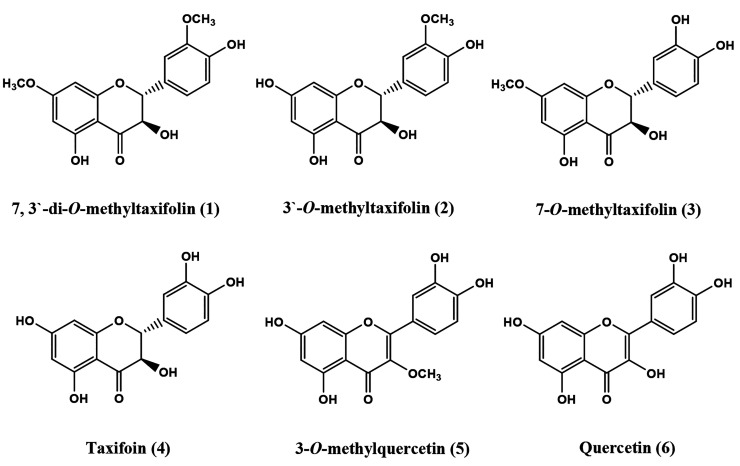
Six flavonoids present in Pulicaria jaubertii, i.e., 7,3′-di-O-methyltaxifolin (1), 3′-O-methyltaxifolin (2), 7-O-methyltaxifolin (3), taxifolin (4), 3-O-methylquercetin (5), and quercetin (6), were tested for their anticancer activities. The methylated flavonoids, compounds 1–3 and 5, were evaluated for their anticancer activities in comparison to the non-methylated parent flavonoids taxifolin (4) and quercetin (6). The structures of the known compounds were reconfirmed by spectral analyses using 1H and 13C NMR data comparisons and HRMS spectrometry. The anticancer activity of these compounds was evaluated in colon cancer, HCT-116, and noncancerous, HEK-293, cell lines using the MTT antiproliferative assays. The caspase-3 and caspase-9 expressions and DAPI (4′, 6-diamidino-2-phenylindole) staining assays were used to evaluate the apoptotic activity. All the compounds exhibited antiproliferative activity against the HCT-116 cell line with IC50 values at 33 ± 1.25, 36 ± 2.25, 34 ± 2.15, 32 ± 2.35, 34 ± 2.65, and 36 ± 1.95 μg/mL for compounds 1 to 6, respectively. All the compounds produced a significant reduction in HCT-116 cell line proliferation, except compounds 2 and 6. The viability of the HEK-293 normal cells was found to be significantly higher than the viability of the cancerous cells at all of the tested concentrations, thus suggesting that all the compounds have better inhibitory activity on the cancer cell line. Apoptotic features such as chromatin condensation and nuclear shrinkage were also induced by the compounds. The expression of caspase-3 and caspase-9 genes increased in HCT-116 cell lines after 48 h of treatment, suggesting cell death by the apoptotic pathways. The molecular docking studies showed favorable binding affinity against different pro- and antiapoptotic proteins by these compounds. The docking scores were minimum as compared to the caspase-9, caspase-3, Bcl-xl, and JAK2.
1. Introduction
Flavonoids, a vastly distributed natural product class of compounds found mostly in higher plants, have multiple benefits as food and medicines.1−3 They are also abundantly present in vegetables, fruits, and green algae. Flavonoids play a defensive role in plants against oxidative stress, which is produced in response to unfavorable environmental conditions of salinity and dryness.4,5 The polyhydroxy flavonoids exhibit protective effects against oxidative stress damage to soft tissues, i.e., the brain, heart, and liver,6 and have been used medicinally. Silymarin, a multihydroxylated flaovnolignan, is used for hepatoprotection and treatment of liver cirrhosis.7 Quercetin and taxifolin are two such polyhydroxylated flavonoids that are well-known for their potent antioxidant activity.8−10 A variety of flavonoids have also been studied for their anticancer/cytotoxic activity against various cancers and malignant cell lines.11,12 Structurally, quercetin and taxifolin have the same pattern of hydroxyl groups distribution among the three nucleus rings, i.e., A, B, and C of the flavonoid. Taxifolin, a dihydroquercetin, contains a 2,3-saturated C–C bond in the C-ring of the structure.8,9,13 Supposedly, this saturation in the taxifolin structure, as compared to the quercetin, is responsible for the lower antioxidant activity of the taxifolin in comparison to the quercetin.9 Furthermore, quercetin and taxifolin have also been shown to exhibit potential anticancer activity with definite mechanisms.13−16 One of the involved anticancer mechanisms is the inhibition of thioredoxin reductase TrxR.16 In addition, taxifolin also has significant quinone reductase activity and anticancer potential by up and down regulating several genes through an ARE-dependent mechanism.17 Some other mechanisms are also reported for taxifolin.14,18 As for the methylated flavonoids, their structures have been suggested to exhibit higher anticancer activity as compared to the corresponding hydroxylated compounds.11,19 In the current study, four methylated flavonoid aglycones, i.e., 7,3′-di-O-methyltaxifolin (1), 3′-O-methyltaxifolin (2), 7-O-methyltaxifolin (3), and 3-O-methylquercetin (5), in addition to taxifolin (4) and quercetin (6) (Figure 1), were isolated from P. jaubertii and were tested for anticancer potential against the HCT-116 cancer cell line and HEK-293 noncancer cell lines. The expression levels of caspase-3 and caspase-9 genes were measured after anticancer treatments, and DAPI staining experiments were also conducted to explore the apoptotic activity of these tested methylated compounds. The apoptotic activity of the compounds was validated through in silico receptor–ligand docking studies to provide an insight into the levels of the binding feasibility.
Figure 1.
Structures of the isolated flavonoids 1–6.
2. Materials and Methods
2.1. Reagents and Instrumentation
1H and 13C NMR measurements were carried out on a Bruker AMX-600 spectrometer operating at 600 MHz (for 1H) and 150 MHz (for 13C) in DMSO-d6 solution, and chemical shifts were expressed in δ (ppm) with reference to TMS and coupling constants (J) in hertz. The liquid chromatography quadrupole time-of-flight mass spectrometry (LC-QTOF-MS/MS) was performed in electron spray ionization (ESI) mode. The temperature of the ESI source was set at 220 °C, with a flow of dry gas at 9.0 l/min and a nebulizer pressure of 1.8 bar. The capillary voltage was set at 4500 V, and end plate offset was at 500 V. Samples were separated using a 1.7 μm 2.1 × 100 mm Acquity UPLCCSH C18 column (Waters, USA) with a 5 μL volume injection in a UPLC system (UltiMate 3000, Thermo Scientific, Germany). The separation was performed at a constant mobile phase flow rate of 0.4 mL/min, and the gradient program was 0% of solvent B at 0.01 min, ramped to 100% B in 10 min, then reduced to 0% B at 12 min, and the column equilibrated with 0% B to 14 min. The column temperature was set at 40 °C, and the temperature of the autosampler was kept at 4 °C. The MetaboScape 4.0 software (Bruker, Bremen, Germany) was used to automatically analyze the raw MS and MS/MS data. The scan EIMS-TIC, VG-ZAB-HF, X-mass (158.64, 800.00) mass spectrometer (VG Analytical, Inc.) was used for EI-MS determinations. Open column chromatography was performed using Si gel (Si gel 60, Merck), Sephadix LH-20 (Pharmacia), and SPE-C18 cartridges (Strata columns). Precoated silica gel 60 F254 plates (Merck) were used for TLC. Detection of the developed chromatograms was carried out by exposure to ammonia vapor and spraying with 1% vanillin–H2SO4, followed by heating at 100 °C for 5 min.
2.2. Plant Material
The plant material (Pulicaria jaubertii aerial parts) was collected from the Aljar region of Hajja-Yemen in August 2012 and was kindly identified by Dr. Abd El-Rahman Saeed Aldabee, Professor of Plant Taxonomy, Faculty of Science, Sana’a University, Yemen. A voucher specimen {P-01} is available in the Pharmacognosy and Medicinal Plants Department, Faculty of Pharmacy, Al-Azhar University, Cairo, Egypt.
2.3. Extraction, Isolation, Purification, and Spectral Analyses of the Flavonoids
The air-dried powdered aerial parts of P. jaubertii (1.5 kg) were exhaustively extracted with methanol (7 L × 3) to yield 85 g of a gellish extract. The dried extract was suspended in 500 mL of distilled water and partitioned successively with n-hexane, ethyl acetate, and n-butanol to give 30 g of n-hexane, 12 g of ethyl acetate, and 8 g of n-butanol fractions, respectively. The ethyl acetate fraction was chromatographed on a column packed with silica gel and eluted with methylene chloride–methanol (100%: 0 to 85–15%) to obtain six fractions of A (450 mg), B (420 mg), C (950 mg), D (1.4 g), E (1.6 g), and F (2.1 g). The fraction B (420 mg) was subjected to Sephadex LH-20 column elution with 100% methanol to give two subfractions as B-1 (195 mg) and B-2 (90 mg). The subfraction B-1 was purified using Sephadex LH-20 (100% MeOH) to give compound 1 (60 mg). Fraction D (1.4 g) was applied to a Sephadex LH-20 column and eluted with 100% methanol to give three subfractions, D-1 (420 mg), D-2 (300 mg), and D-3 (330 mg). The subfractions D-1 and D-3 were separately subjected to RP-C18 columns, eluted with 90% to 20% water in methanol to give two subfractions of each, D-1a (90 mg) and D-1b (170 mg) from the subfraction D-1, and D-3a (110 mg) and D-3b (130 mg), respectively, from the subfraction D-3. The subfractions D-1a and D-3a were further subjected to Sephadex LH-20 column chromatography, eluting with 100% methanol to give compounds 2 (47 mg) and 3 (55 mg), respectively. Fraction E (1.6 g) was chromatographed on a Sephadex LH-20 column and eluted with 100% methanol to give three subfractions, E-1 (390 mg), E-2 (280 mg), and E-3 (520 mg). The subfractions E-1 and E-2 were further subjected, separately, to columns of Sephadex LH-20 and eluted with 100% methanol to afford the compounds 5 (51 mg), and 6 (43 mg), respectively. The subfraction E-3 was chromatographed on a silica gel column by eluting it with petroleum ether–ethyl acetate (80:20–70:30) to afford compound 4 (35 mg).
2.3.1. Compound 1
7,3′-Di-O-methyltaxifolin, an off-white amorphous powder; 1H NMR (DMSO-d6, 600 MHz) δ 11.89 (s, 5-OH), 9.12 (s, 4′-OH), 7.12 (d, J = 1.8 Hz, H-2′), 6.92 (dd, J = 8.0, 1.8 Hz, H-6′), 6.79 (d, J = 8.0 Hz, H-5′), 6.12 (d, J = 2.2 Hz, H-6), 6.10 (d, J = 2.2 Hz, H-8), 5.80 (d, J = 6.3 Hz, 3eq–OH), 5.10 (d, J = 11.5 Hz, H-2), 4.72 (dd, J = 11.5, 6.1 Hz, H-3), 3.79 (s, 7-OCH3), 3.78 (s, 3′-OCH3); 13C NMR (DMSO-d6, 150 MHz) δ 199.00 (C-4), 168.02 (C-7), 163.44 (C-5), 162.98 (C-9), 147.81 (C-3′), 147.53 (C-4′), 128.36 (C-1′), 121.71 (C-6′), 115.43 (C-5′), 112.67 (C-2′), 101.84 (C-10), 95.40 (C-6), 94.27 (C-8), 83.80 (C-2), 71.93 (C-3), 56.43 (7-OCH3), 56.17 (3′-OCH3); +HRESIMS m/z 333.0975 [M + H]+; −HRESIMS m/z 331.0851 [M – H]−.
2.3.2. Compound 2
3′-O-Methyltaxifolin, colorless crystals from MeOH; 1H NMR (DMSO-d6, 600 MHz) δ 11.92 (s, 5-OH), 10.84 (s, 4′-OH), 7.10 (d, J = 1.8 Hz, H-2′), 6.90 (dd, J = 8.1, 1.8 Hz, H-6′), 6.80 (d, J = 8.0 Hz, H-5′), 5.92 (d, J = 2.0 Hz, H-8), 5.87 (d, J = 2.0 Hz, H-6), 5.82 (s, 3eq–OH), 5.04 (d, J = 11.4 Hz, H-2), 4.65 (d, J = 11.4 Hz, H-3), 3.78 (s, 3′-OCH3); 13C NMR (DMSO-d6, 150 MHz) δ 198.41 (C-4), 167.24 (C-7), 163.76 (C-5), 163.02 (C-9), 147.80 (C-3′), 147.48 (C-4′), 128.52 (C-1′), 121.65 (C-6′), 115.43 (C-5′), 112.66 (C-2′), 100.93 (C-10), 96.48 (C-6), 95.48 (C-8), 83.65 (C-2), 71.84 (C-3), 56.16 (3′-OCH3); +HRESIMS m/z 319.0814 [M + H]+; −HRESIMS m/z 317.0698 [M-H]−.
2.3.3. Compound 3
7-O-Methyltaxifolin, colorless crystals from MeOH; 1H NMR (DMSO-d6, 600 MHz) δ 11.85 (s, 5-OH), 9.09, 9.03 (s, 3′, 4′-OHs), 6.88 (d, J = 1.5 Hz, H-2′), 6.75 (d, J = 1.8 Hz, H-6′), 6.74 (s, H-5′), 6.10 (d, J = 2.2 Hz, H-6), 6.07 (d, J = 2.2 Hz, H-8), 5.84 (brs, 3eq–OH), 5.02 (d, J = 11.2 Hz, H-2), 4.55 (d, J = 11.2 Hz, H-3), 3.78 (s, 7-OCH3); 13C NMR (DMSO-d6, 150 MHz) δ 198.80 (C-4), 168.02 (C-7), 163.44 (C-9), 162.94 (C-5), 146.25 (C-3′), 145.39 (C-4′), 128.30 (C-1′), 119.93 (C-6′), 115.79 (C-2′), 115.57 (C-5′), 101.81 (C-10), 95.32 (C-6), 94.22 (C-8), 83.65 (C-2), 72.08 (C-3), 56.41 (7-OCH3); +HRESIMS m/z 319.0822 [M + H]+; −HRESIMS m/z 317.0695 [M-H]−.
2.3.4. Compound 4
Dihydroquercetin (taxifolin), colorless crystals from MeOH; 1H NMR (DMSO-d6, 800 MHz) δ 11.88 (s, 5-OH), 10.80, 9.01, 8.96 (s, 7-, 3′-, 4′-OHs), 6.86 (d, J = 1.8 Hz, H-2′), 6.73 (s, H-5′, H-6′), 5.90 (d, J = 1.8 Hz, H-6), 5.89 (d, J = 2.4 Hz, H-8), 5.74 (d, J = 5.4 Hz, 3eq–OH), 4.96 (d, J = 11.4 Hz, H-2), 4.48 (dd, J = 11.4, 4.2 Hz, H-3); 13C NMR (DMSO-d6, 200 MHz) δ 197.87 (C-4), 167.27 (C-7), 163.38 (C-5), 163.06 (C-9), 146.26 (C-3′), 145.430 (C-4′), 128.08 (C-1′), 119.88 (C-6′), 115.84 (C-2′), 115.59 (C-5′), 100.97 (C-10), 96.50 (C-6), 95.03 (C-8), 83.54 (C-2), 71.94 (C-3); EIMS m/z 304 [M]+.
2.3.5. Compound 5
3-O-Methylquercetin, yellow crystals [MeOH]; 1H NMR (DMSO-d6, 600 MHz) δ 12.71 (s, 5-OH), 10.85, 9.77, 9.40 (s, 7, 3′, 4′-OHs), 7.55 (d, J = 2.2 Hz, H-2′), 7.45 (dd, J = 8.4, 2.2 Hz, H-6′), 6.91 (d, J = 8.4 Hz, H-5′), 6.41 (d, J = 1.9 Hz, H-8), 6.19 (d, J = 2.0 Hz, H-6), 3.78 (s, 3-OCH3); 13C NMR (DMSO-d6, 150 MHz) δ 178.36 (C-4), 164.68 (C-7), 161.73 (C-5), 156.79 (C-9), 156.07 (C-2), 149.17 (C-4′), 145.70 (C-3′), 138.12 (C-3), 121.25 (C-6′), 121.05 (C-1′), 116.22 (C-2′), 115.86 (C-5′), 104.64 (C-10), 99.00 (C-6), 94.05 (C-8), 60.13 (3-OCH3); +HRESIMS m/z 317.0665 [M + H]+; −HRESIMS m/z 315.0538 [M – H]−.
2.3.6. Compound 6
Quercetin, yellow crystals [MeOH]; 1H NMR (DMSO-d6, 800 MHz) δ 12.47 (s, 5-OH), 10.82, 9.57, 9.34 (s, 7, 3′, 4′-OHs), 7.66 (d, J = 1.8 Hz, H-2′), 7.53 (dd, J = 8.4, 2.4 Hz, H-6′), 6.87 (d, J = 8.4 Hz, H-5′), 6.39 (d, J = 1.8 Hz, H-8), 6.17, (d, J = 2.4 Hz, H-6); 13C NMR (DMSO-d6, 200 MHz) δ 176.33 (C-4), 164.37 (C-7), 161.22 (C-5), 156.08 (C-9), 148.19 (C-4′), 147.28 (C-2), 145.55 (C-3′), 136.23 (C-3), 122.45 (C-1′), 120.46 (C-6′), 116.10 (C-2′), 115.55 (C-5′), 103.51 (C-10), 98.67 (C-6), 93.84 (C-8); EIMS m/z 302 [M]+.
2.4. In Vitro Anticancer Activity
Human colorectal carcinoma (HCT-116) and human embryonic kidney cells (HEK-293) were used to study the impact of compounds 7, 3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, taxifolin, and quercetin on cell viability, according to a previously described method.20 20,000 cells/well were seeded in 96-well plates containing DMEM, fetal bovine serum, l-glutamine, penicillin, streptomycin, and selenium chloride. They were kept at 37 °C in a CO2 incubator (Thermo-Fisher Scientific). The cells were treated with 7, 3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, taxifolin, and quercetin with 5.0 μg to 40 μg/mL dosages for 48 h and were processed for MTT assays.21 In the control group, 7,3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, taxifolin, and quercetin were not added. Cells were treated with MTT for 4 h (5.0 mg/mL) and then with DMSO (1%). The optical densities (OD) were measured with a plate reader (Biotek Instruments) at 570 nm wavelength. The statistical data presented was taken from triplicates and analyzed with GraphPad Prism 6.0 Software, USA.
2.4.1. Apoptosis Confirmation by DAPI Staining
Cancer cells were treated with the compounds for 48 h before being treated with 4% paraformaldehyde and stained with DAPI (4′,6-diamidino-2-phenylindole, 1.0 μg/mL) in the dark. DAPI stained cells were examined by using a fluorescence confocal scanning microscope (Zeiss, Germany).21
2.4.2. Detection of Caspase-3 and Caspase-9 by ELISA (Enzyme-Linked Immuno-Sorbent Assays)
To examine the impact of compounds, 7,3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, taxifolin, and quercetin on caspase-3 and caspase-9 genes expression post-treatments, cancer cells (HCT-116) were cultured at a density of 50,000 cells/well in 96-well plates in the CO2 incubator. After 24 h, if cells were 70–80% confluent, they were treated with doses of compounds, 7,3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, taxifolin, and quercetin ranging from 40 μg/mL. After 48 h, the cells were washed with phosphate-buffered saline (PBS), and 0.35% trypsin was added to remove the cells. The cells were transferred to 1.5 mL tubes and were centrifuged at 1000 rpm for 2–3 min. The supernatant was removed, pellets were resuspended in lysis buffer for 60 min, and cell lysate was transferred to another 1.5 mL tube and again centrifuged at 1000 rpm for 15 min. Then the cell lysates were transferred into fresh 1.5 mL tubes and used for the detection of caspase-3 and caspase-9, assays. Both the control and treated cells were processed with caspase-3 (catalogue no. 181418, Human Active Caspase-3 ELISA Kit) and caspase-9 (catalogue no. ab119508; caspase-9 Human ELISA kit) detection as per the protocol described by the supplier (www.ABCAM.com), and expressions of caspase-3 and caspase-9 were measured at 620 nm absorbance with a microplate reader (Bio-Tech, USA).
2.5. In Silico Studies
The crystal structures of the pro- and antiapoptotic proteins with PDB ID: 1NW9,223H0E,234HY5,244QVX,25,26 and 7Q7I,27 representing caspase-9, caspase-3, cIAP1 BIR3, Bcl-xl, STAT3, and JAK2, were downloaded from the Research Collaboratory for Structural Bioinformatics (RCSB) protein data bank. The structures of the methylated flavonoids of P. jaubertii were obtained from the PubChem database.28 The protein and ligands were prepared and optimized by using the AutoDock Tool (ADT), bundled with the MGLTools package (version 1.5.6)29 to add charges, polar hydrogen atoms, and set up the rotatable bonds. The water atoms were deleted, and the nonpolar hydrogen atoms were merged during the preparation of the protein. At that moment, PDBQT files were generated for the protein and ligands. The molecular docking was performed using AutoDock Vina v1.1.2.30 The active binding site of the proteins was chosen as the grid center and was obtained by removing the ligand. The grids were generated with the autogrid4 program distributed with AutoDock, v1.5.6.31 The center of the grid box dimensions was selected to include all atoms of the ligand set.31 The site and the coordinates of the grid box for the mentioned proteins are listed in the Supporting Information. The output docking scores were defined as affinity binding (kcal/mol). The ligand–protein interactions were analyzed using the Discovery Studio 2020 client (Dassault Systems BIOVIA, San Diego, 2020, USA).
3. Results and Discussion
3.1. Structure Confirmations of the Isolated Flavonoids
The compounds 1–4, obtained as colorless crystals, were identified as dihydroflavonols in their structures having the C-2 and C-3 single bonds, resulting in loss of the extended conjugation between the rings A/C and the ring B, as present in the quercetin molecule, the basic C2/C3 unsaturated structure (6). The compounds 1–3 were methylated derivatives of taxifolin (compound 4) and had molecular ion peaks at m/z 333.0975 [M + H]+, 319.0814 [M + H]+, and 319.0822 [M + H]+, respectively, in their HRESIMS positive ion mode. These compounds also exhibited molecular ion peaks at m/z 331.0851 [M – H]−, 317.0698 [M – H]−, and 317.0695 [M – H]−, respectively, in the HR-ESI-MS negative ion mode. Compound 4 exhibited an EIMS molecular ion peak at m/z 304 [M]+. Therefore, the molecular formulas of compounds 1–4 were determined as C17H16O7, C16H14O7, C16H14O7, and C15H12O7, respectively. As observed from the molecular ion peaks of 1–4, compound 1 has 28 mass units more than compound 4, indicating it has an additionally two methyl groups. Compounds 2 and 3 had nearly identical molecular ions with an additional 14 mass units than compound 4, indicating that these compounds have an additional methyl group. Analyses of the 1H NMR spectra of compounds 1–4 clearly showed two meta-coupled protons of H-6 and H-8, of the A-ring of the flavonoid structure, at δ 5.87 to δ 6.12 ppm. The 1H NMR spectra of compounds 1–4 also showed the typical AX system of H-2 and H-3 C-ring protons at δ 4.48 to δ 5.104 ppm with large coupling constants (11.0 to 11.5 Hz), reflecting the 2,3-trans dihydroflavonol structures.32−34 Furthermore, the B-ring protons of compounds 1–3 appeared as typical ABX systems at proton signals at δ 6.74 to δ 7.12 ppm, while those of compound 4 protons appeared as two signals at δ 6.86 ppm and δ 6.73 ppm in the ratio of 1:2, which were characteristic of H-2′ and the two accidentally equivalent H-5′ and H-6′ protons, respectively.34 The methoxyl proton signals appeared at δ 3.78 to δ 3.79 ppm in the 1H NMR spectra of the compounds 1–4. The 13C NMR spectra of compounds 1–4 displayed 15 carbon resonances consistent with flavonoid skeletons from which the two high field-signals of C-2 (δ 83.54–83.80 ppm) and C-3 (δ 71.84–72.08 ppm) of the C-ring carbon atoms characteristic of dihydroflavonols were easily identified based on reported data. The remaining carbon signals were consistent with the chemical shift values for the phloroglucinol type A-ring and catechol B-ring of dihydroflavonols.35 Furthermore, the 13C NMR spectra of compounds 1–3 revealed the presence of two additional methoxyl signals in compound 1 and one additional methoxyl signal in each of the compounds 2 and 3. The downfield shifts of the C-7 and C-10 carbons, and the upfield chemical shifts of the C-6 and C-8 carbons in compounds 1 and 3, together with the observed 1H NMR downfield shift of H-6 and H-8 protons as compared to that of compound 4 (taxifolin), indicated the presence of C-7 methoxyl groups. Furthermore, in comparison to the compound 4, the downfield shifts of C-3′, C-4′, and C-6′ carbons and the upfield shifts of C-2′ carbon in 1 and 2, in addition to the 1H NMR downfield shifts of the ortho- and para protons, H-2′ and H-6′, respectively, indicated the presence of C-3′ methoxyl groups.35,36 Additionally, the 2,3-trans dihydroflavonols of compounds 1–4 were reflected by pronounced downfield shifts of C-2 and C-4 and the upfield shift of C-3 in comparison to the corresponding chemical shifts of the 2,3-cis isomers, as reported earlier.32−34 Consequently, the compounds 1–4 were identified as 7, 3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, and dihydroquercetin (taxifolin), respectively, which were in agreement with the known data and the NMR chemical shifts values.35,37−45
Four possible stereoisomers are assumed to be present for dihydroquercetin derivatives due to the presence of two asymmetric carbon centers at C2 and C3. However, the predominant isomer in nature is the 2,3-trans isomer with the C-2 phenyl and C-3 hydroxyl groups in equatorial positions and an absolute configuration of 2R,3R.46 Compounds 1–4 exhibited distinct polarities and were isolated from different subfractions of the ethyl acetate fraction by using gradient of solvent systems (Materials and Methods). The TLC (thin layer chromatography) behavior and Rf values of all these isolates differed. The NMR chemical shifts and the coupling constants (J values) for the C2/C3 protons for each of the compounds 1–4 showed the trans configuration with coupling constant values ranging between 11.2 and 11.5 Hz, which is theoretically proposed between 10 and 15 Hz for the trans-oriented C2/C3 protons for the (isomeric) flavonoids, and that is also reported for a number of similar types of compounds.32−34 The reported coupling constant values for the cis orientation of the C2/C3 hydrogens have been reported to be approximately 3.2 Hz.32,33 In our previous work,34 the UV absorption studies with different UV shift reagents substantiated the substitution patterns of the methoxyl groups in the compounds 1–3, which were also confirmed by the NMR chemical shifts (1H and 13C) and coupling constant values of the C2/C3 protons together with 2D-NMR analyses.
In an in silico analysis of the stereochemistry of the structures 1–4, the minimum energy confirmations (MEC) showed the energy requirements at 14.9810, 7.9747, 7.7360, and 0.6669 kcal/mol, respectively, for compounds 1–4. The energy requirement for the nonmethylated compound, taxifolin (4), was the minimum at 0.6669 kcal/mol, while the energy requirements for the compounds 2 and 3 were at 7.9747 and 7.7360 kcal/mol, nearly at the same level for the monomethylated compounds. The dimethylated compound 1, with both rings A and B methylated, was the highest energy requirement at 14.9810 kcal/mol. The stereochemistry at the C2/C3 carbons was predicted to be 2R,3R for all compounds 1–4, which were in conformity with the reported configuration of the dihydroquercetin derivatives.46 The structural disposition depicted in Figure 1 shows the trans dispositions of the C2/C3 hydrogen atoms of the ring C with the stereo-orientations of the other substituents, including the ring B, and the hydroxy group at the C3 position.
Compounds 5 and 6 were obtained as yellow crystals. The molecular formula of 5 was determined as C16H12O7 from the molecular ion peaks at m/z 317.0665 [M + H]+ and m/z 315.0538 [M – H]− in the HRESIMS positive and negative modes, respectively. The molecular formula of compound 6 was determined to be C15H12O7 from the molecular ion peak at m/z 302 [M]+. This reveals that compound 5 has one methyl group more than compound 6, as indicated by the additional 14 mass units in compound 5. The 1H NMR spectra of compounds 5 and 6, showed a pair of doublets at δ 6.17–6.41 ppm corresponding to the two meta-coupled protons of the A-ring and three proton resonances at δ 6.87–7.66 ppm corresponding to the three aromatic protons of the B-ring. The 1H NMR spectrum of 5 showed an additional methoxyl singlet at δ 3.78 ppm corresponding to the presence of C-3 methoxyl group. In comparison, the 13C NMR chemical shifts of compounds 5 and 6 carbon signals revealed similarities with 3-O-methylquercetin and quercetin, respectively. The placement of the methoxyl group at C-3 in compound 5 was established from the downfield shift of C-2 and C-3 carbon signals as compared to that of compound 6 (quercetin). Therefore, compounds 5 and 6 were identified as 3-O-methylquercetin and quercetin, respectively, and the obtained data were in good agreement with the reported values in the literature.35,36 These compounds, 1–6, have also been previously isolated from this plant.34,47
3.2. In Vitro Anticancer Activity
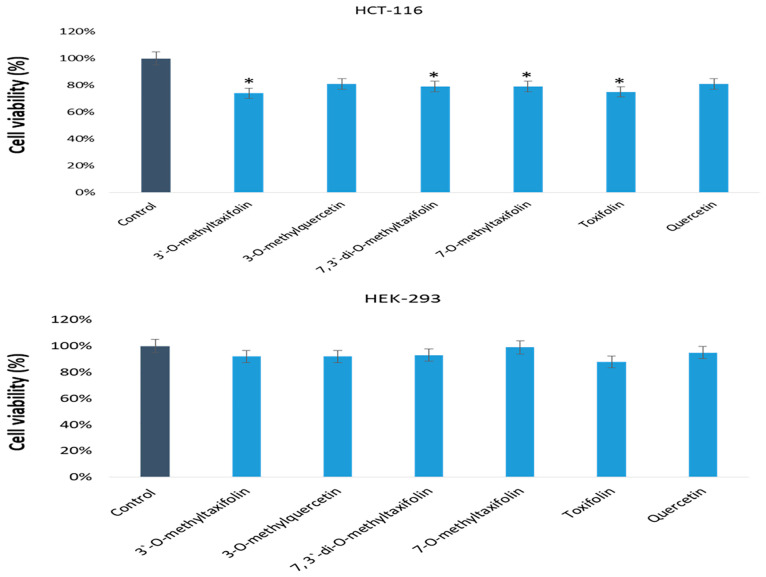
The antiproliferative potentials of 7,3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, taxifolin, and quercetin were investigated on colon cancer (HCT-116) and human embryonic kidney (HEK-293) cells. A reduction was observed in cancer cell viability after treatments with 7,3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, taxifolin, and quercetin (Figure 2). The IC50 values of these compounds, respectively, were at 33 ± 1.25, 36 ± 2.25, 34 ± 2.15, 34 ± 2.65, 32 ± 2.35, and 36 ± 1.95 μg/mL.
Figure 2.
Anticancer effects of 7,3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, taxifolin, and quercetin on HCT-16 cells and HEK-293 cells by the cell viability MTT assays after 48 h, *p < 0.05.
The treatment with 7,3′-di-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, and taxifolin produced significant reductions in the numbers of cancer cells as compared to the 3′-O-methyltaxifolin and quercetin compounds. It was also to examine whether the treatment with 7, 3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, taxifolin, and quercetin produced any cytotoxic effects on the noncancerous cells. The results showed that treatments with these compounds also reduced the HEK-293 cell numbers, but the percentage reduction was less than that observed for the cancer cell lines. This suggests that the compounds have better inhibitory action on the HCT-116 cancer cell line than on the HEK-193 noncancer cell line. This is the first study on the anticancer activity of 7,3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, taxifolin, and quercetin against HCT-116 cell lines. Previous research has also demonstrated anticancer activity of various synthetic flavonoids and other structurally related compounds on a variety of cell lines.48,49
3.3. Apoptotic Cell Death Detection by DAPI Stainings
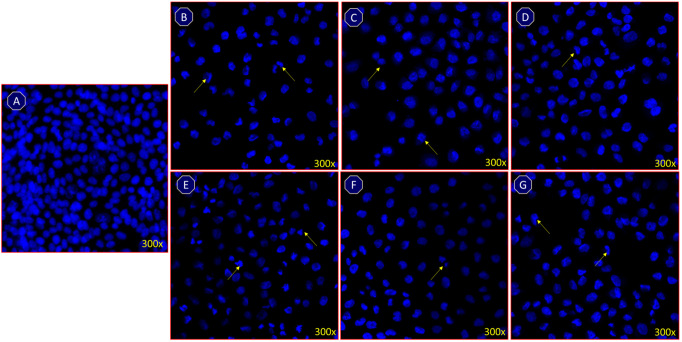
Nuclear DAPI (4′,6-diamidino-2-phenylindole) staining was used to examine whether the reduction in cancer cell numbers in response to the compounds 1–6 treatment was due to apoptosis or another form of programmed cell death.50−52 The current findings revealed that cell treatments with 7,3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, taxifolin, and quercetin induced apoptosis, and key apoptotic features, such as chromatin condensation and nuclear shrinkage, were observed (Figure 3B–G). The treatments with the compounds also caused an increase in nuclear condensation, shrinkage of the cells, and loss of shape of the cells (Figure 3 B–G) as compared to the control (Figure 3A), which suggested an increase in programmed cell death (i.e., apoptosis). In the current study, DAPI was used as an apoptotic marker, and morphological changes were observed in the cancer cells post-treatments that clearly showed that the cells were under programmed cell death.53,54
Figure 3.
DAPI stainings shows the impact of (A) control, (B) 7,3′-di-O-methyltaxifolin, (C) 3′-O-methyltaxifolin, (D) 7-O-methyltaxifolin, (E) 3-O-methylquercetin, (F) taxifolin, and (G) quercetin on colon (HCT-116) cancer cells, 48 h post-treatment.
3.4. Caspase-3 and Caspase-9 Expression
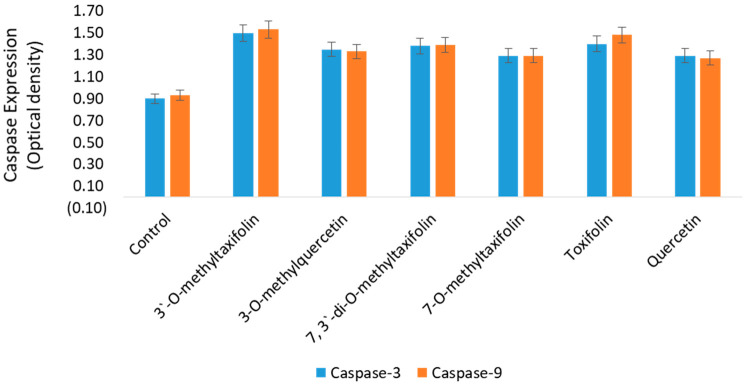
To confirm the apoptosis, expressions of caspase-3 and caspase-9 were observed in the cell lysates by ELISA. The pro-apoptotic expressions of these genes are considered the hallmark of apoptosis. After 48 h of treatment with the compounds, an increased expression of both caspase-3 and caspase-9 was observed in response to treatments by all the compounds (Figure 4). The untreated cells (control) did not show any elevation of caspase-3 and caspase-9 expressions, thus suggesting that the compounds induced cell death in the colon cancer cells were due to involvement of apoptotic pathways.
Figure 4.
Pro-apoptotic caspase-3 and caspase-9 expression in HCT-116 cells. The plot shows expression of caspase-3 and caspase-9 after 48 h treatment with 7, 3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, taxifolin, and quercetin. An increase in the caspase-3 and caspase-9 was observed post treatment. Data are presented as optical density (OD) proportional to both caspase concentrations. A high OD represents high caspase production.
According to the latest information available, this is the first report where treatments of compounds 7, 3′-di-O-methyltaxifolin, 3′-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, taxifolin, and quercetin has been shown to induce apoptotic cell death in colon cancer cells. The induction of apoptosis in the cells may cause a decrease in cytochrome C levels in the mitochondria, whereas its level increases in the cytoplasm. The cytoplasmic cytochrome C activates Apaf1, which together with pro-caspase-9 induces the formation of apoptosomes, followed by the activation of pro-caspase-3 by caspase-9.55−57
3.5. Docking Scores
To further understand the apoptosis effects of the methylated flavonoids from P. jaubertii, molecular docking studies were performed against different pro- and antiapoptotic proteins, such as the third BIR domain (BIR3) of X-linked inhibitor of apoptosis (cIAP1 BIR3), Bcl-extra-large (Bcl-xl), signal transducer and activator of transcription 3 (STAT3), and Janus kinase 2 (JAK2), including caspase-9, and caspase-3. More negative docking scores indicated more favorable binding affinity.58 Consistently, the docking studies showed that the methylated flavonoids from P. jaubertii have favorable binding affinity values against caspase-9, caspase-3, Bcl-xl and JAK2 (Table 1). The potential binding affinity varied from 7.0 to 9.5 kcal/mol, significantly, against Bcl-xl and JAK2. The highest binding affinity was detected for quercetin; the docking scores were −9.0 and −9.2 kcal/mol against Bcl-xl, and JAK2 proteins, respectively. Quercetin has been shown to directly bind and inhibit Jak2,59 and an apoptotic pathway of quercetin has been proposed to be involved in JAK2/STAT3 signaling pathway inhibition.60 Furthermore, the down-regulation of JAK2 induces the inactivation of Bcl-xl, resulting in the induction of apoptotic signals through caspase activation.61 In addition, quercetin is able to prompt caspase-dependent apoptosis by binding and inhibiting the activity of Bcl-xl.62
Table 1. Docking Scores of the Methylated Flavonoids against Various Ligands.
| Ligand Proteins | 1NW9 | 3H0E | 4HY5 | 4QVX | 6NUQ | 7Q7I |
|---|---|---|---|---|---|---|
| Compounds | Docking Scores (kcal/mol) | |||||
| 7,3′-Di-O-methyltaxifolin (1) | –6.30 | –7.00 | –5.50 | –5.90 | –5.80 | –7.20 |
| 3′-O-Methyltaxifolin (2) | –6.20 | –6.90 | –5.40 | –7.80 | –5.70 | –7.20 |
| 7-O-Methyltaxifolin (3) | –6.40 | –6.90 | –5.60 | –6.00 | –6.00 | –7.20 |
| Taxifolin (4) | –6.50 | –6.80 | –5.70 | –6.20 | –5.70 | –7.10 |
| 3-O-Methylquercetin (5) | –6.70 | –6.90 | –6.10 | –9.00 | –6.10 | –8.30 |
| Quercetin (6) | –7.10 | –7.00 | –6.30 | –9.20 | –6.00 | –9.00 |
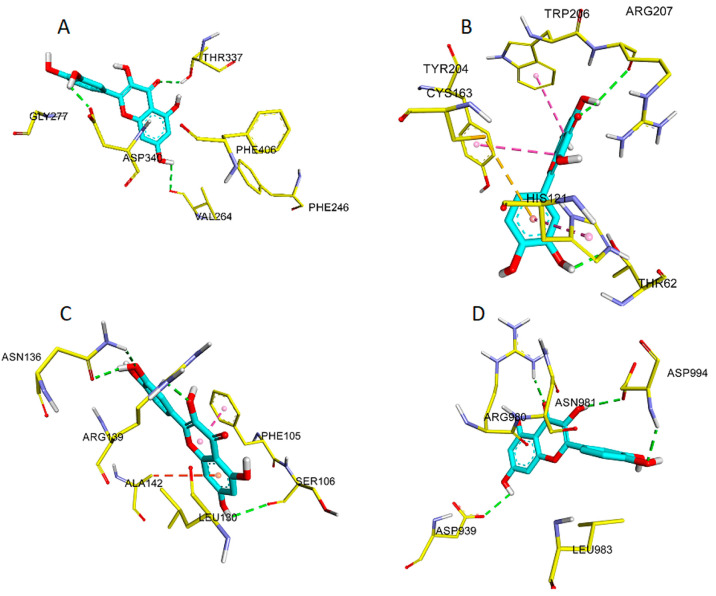
For a more detailed outlook, the binding interactions of the methylated flavonoids of P. jaubertii against pro- and antiapoptotic proteins were analyzed for their ligand–protein interactions at the binding site. The flavonoids were able to generate several hydrogen bonds (H-bonds) and π–π stackings with the caspase-9, caspase-3, Bcl-xl, and JAK2 proteins (Table 2). The quercetin participated in H-bonds with amino acids, D939, R980, and D994 in 7Q7I, while quercetin was involved in H-bonds interactions with the amino acids S106, and R139, and the π-alkyl interaction with A142, and π–π stacking with F105 in 4QVX were observed. Besides, quercetin formed H-bonds with amino acids of the protein sequence, E261, N265, Q320, and P338 in 1NW9, as well as H-bond interactions with T62, and R207, π-sulfur interaction with C163, and π–π stackings with Y204, W206, and H121 amino acids in 3H0E ligand. Figure 5 represents the binding modes of quercetin at the substrate binding sites of caspase-9, caspase-3, Bcl-xl, and JAK2. These results showed that the flavonoids of P. jaubertii possessed minimum binding affinity and favored binding interactions with Bcl-xl, JAK2, caspase-3, and caspase-9 ligands in the docking studies. Summarily, the docking scores of the methylated flavonoids against caspase-9 (PDB ID: 1NW9), caspase-3 (PDB ID: 3H0E), the third BIR domain (BIR3) of X-linked inhibitor of apoptosis (cIAP1 BIR3) (PDB ID: 4HY5), Bcl-extra-large (Bcl-xl) (PDB ID: 4QVX), signal transducer and activator of transcription 3 (STAT3) (PDB ID: 6NUQ), and Janus kinase 2 (JAK2) (PDB ID: 7Q7I) were found to confirm the apoptosis notion. Therefore, these flavonoids are suggested to be involved in the apoptotic pathway to affect the cells’ death to cause the anticancer effects.
Table 2. Amino Acid Residues of Caspase-9 (PDB ID: 1NW9), Caspase-3 (PDB ID: 3H0E), Bcl-extra-large (Bcl-xl) (PDB ID: 4QVX), and Janus Kinase 2 (JAK2) (PDB ID: 7Q7I) Interacting with Methylated Flavonoids.
| Ligand Proteins | 1NW9 | 3H0E | 4QVX | 7Q7I |
|---|---|---|---|---|
| Compounds | Amino acid residues | |||
| 1 | Q245, V264, Q320, T337, P338, I341 | T62, H121, W206, R207 | F105, R139, A142 | E930, R980, N981, D994 |
| 2 | H243, Q245, V264, N265, T337, P338 | T62, H121, S205, W206, R207 | F97, F105, S106, R132, R139, A142 | M929, R980, D994 |
| 3 | Q245, V264, P338, I341 | T62, H121, C163, Y204, R207 | F105, R139, A142 | M929, R980, D994 |
| 4 | E261, N265, Q320, P338 | T62, H121, C163, Y204, W206, R207 | F105, R132, R139, A142 | M929, R980, D994 |
| 5 | Q245, T337, P338 | T62, E123, C163, R207 | F105, R132, R139, A142 | K857, R980, D994 |
| 6 | V264, T337, D340 | T62, H121, C163, Y204, W206, R207 | F105, S106, N136, R139, A142 | D939, R980, D994 |
Figure 5.
Ligand-protein interactions in 1NW9 (A), 3H0E (B), 4QVX (C), and 7Q7I (D). The H-bond interactions are shown as green dotted lines, and the π–π stacking interactions are shown as lavender dotted lines. π–sulfur interactions are shown as orange dotted lines, and π–alkyl interactions are shown as red dotted lines.
4. Conclusions
Colon cancer cells (HCT-116) and noncancer cells (HEK-293) showed reductions in cell viability after treatments with the compounds 7,3′-di-O-methyltaxifolin, 7-O-methyltaxifolin, 3-O-methylquercetin, and taxifolin, which produced more significant reductions in cancer cell numbers as compared to the 3′-O-methyltaxifolin and quercetin. Compound treatment induced apoptotic features, such as chromatin condensation and nuclear shrinkage. An increase in both the caspase-3, and caspase-9 expressions occurred post-treatments in the HCT-116 cell lines, suggesting the apoptotic cells’ death. The molecular docking studies of the parent flavonoids and the methylated flavonoids against different pro- and antiapoptotic proteins demonstrated the favorable bindings of caspase-9, caspase-3, Bcl-xl, and JAK2, thereby supporting the notion of induced cells death by an apoptotic pathway to produce the anticancer effects.
Acknowledgments
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education and, Saudi Arabia for funding this research work through the project number (QU-IF-1-2-2). The authors also thank the technical support of Qassim University
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c05565.
Figure S1: 1H NMR spectrum of compound 1 (DMSO-d6, 600 MHz). Figure S2: 13C NMR spectrum of compound 1 (DMSO-d6, 150 MHz). Figure S3: + ESIMS of compound 1. Figure S4: −ESIMS of compound 1. Figure S5: 1H NMR spectrum of compound 2 (DMSO-d6, 600 MHz). Figure S6: 13C NMR spectrum of compound 2 (DMSO-d6, 150 MHz). Figure S7: +ESIMS of compound 2. Figure S8: −ESIMS of compound 2. Figure S9: 1H NMR spectrum of compound 3 (DMSO-d6, 600 MHz). Figure S10: 13C NMR spectrum of compound 3 (DMSO-d6, 150 MHz). Figure S11: +ESIMS of compound 3. Figure S12: −ESIMS of compound 3. Figure S13: 1H NMR spectrum of compound 4 (DMSO-d6, 600 MHz). Figure S14: 13C NMR spectrum of compound 4 (DMSO-d6, 600 MHz). Figure S15: 1H NMR spectrum of compound 5 (DMSO-d6, 600 MHz). Figure S16: 13C NMR spectrum of compound 5 (DMSO-d6, 150 MHz). Figure S17: +ESIMS of compound 5. Figure S18: −ESIMS of compound 5. Figure S19: 1H NMR spectrum of compound 6 (DMSO-d6, 600 MHz). Figure S20: 13C NMR spectrum of compound 6 (DMSO-d6, 600 MHz) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Miyazawa T.; Nakagawa K.; Asai A. Biodynamics of Natural Antioxidants in Humans. Kagaku to Seibutsu 2000, 38 (2), 104–114. 10.1271/kagakutoseibutsu1962.38.104. [DOI] [Google Scholar]
- Obakan-Yerlikaya P.; Arisan E. D.; Coker-Gurkan A.; Palavan-Unsal N.. Breast Cancer and Flavonoids as Treatment Strategy. In Breast Cancer - From Biology to Medicine; InTech, 2017. 10.5772/66169. [DOI] [Google Scholar]
- Kumar S.; Pandey A. K. Chemistry and Biological Activities of Flavonoids: An Overview. Sci. world J. 2013, 2013, 1. 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers T. J.; Colmer T. D. Plant Salt Tolerance: Adaptations in Halophytes. Ann. Bot. 2015, 115 (3), 327–331. 10.1093/aob/mcu267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flowers T. J.; Hajibagheri M. A.; Clipson N. J. W. Halophytes. Q. Rev. Biol. 1986, 61 (3), 313–337. 10.1086/415032. [DOI] [Google Scholar]
- Mohammed H.Natural and Synthetic Flavonoid Derivatives with Potential Antioxidant and Anticancer Activities. PhD Dissertation, 2009. [Google Scholar]
- Flora K.; Hahn M.; Rosen H.; Benner K. Milk Thistle (Silybum Marianum) for the Therapy of Liver Disease. American Journal of Gastroenterology 1998, 93, 139–143. 10.1111/j.1572-0241.1998.00139.x. [DOI] [PubMed] [Google Scholar]
- Vladimirov Y. A.; Proskurnina E. V.; Demin E. M.; Matveeva N. S.; Lubitskiy O. B.; Novikov A. A.; Izmailov D. Y.; Osipov A. N.; Tikhonov V. P.; Kagan V. E. Dihydroquercetin (Taxifolin) and Other Flavonoids as Inhibitors of Free Radical Formation at Key Stages of Apoptosis. Biochem. 2009, 74 (3), 301–307. 10.1134/S0006297909030092. [DOI] [PubMed] [Google Scholar]
- Silva M. M.; Santos M. R.; Caroço G.; Rocha R.; Justino G.; Mira L. Structure-Antioxidant Activity Relationships of Flavonoids: A Re-Examination. Free Radic. Res. 2002, 36 (11), 1219–1227. 10.1080/198-1071576021000016472. [DOI] [PubMed] [Google Scholar]
- Akinmoladun A. C.; Oladejo C. O.; Josiah S. S.; Famusiwa C. D.; Ojo O. B.; Olaleye M. T. Catechin, Quercetin and Taxifolin Improve Redox and Biochemical Imbalances in Rotenone-Induced Hepatocellular Dysfunction: Relevance for Therapy in Pesticide-Induced Liver Toxicity?. Pathophysiology 2018, 25 (4), 365–371. 10.1016/j.pathophys.2018.07.002. [DOI] [PubMed] [Google Scholar]
- Mohammed H. A.; El-Wahab A.; Mohammed F.; Shaheen U.; Mohammed A. E.-S. I.; Abdalla A. N.; Ragab E. A. Isolation, Characterization, Complete Structural Assignment, and Anticancer Activities of the Methoxylated Flavonoids from Rhamnus Disperma Roots. Molecules 2021, 26 (19), 5827. 10.3390/molecules26195827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H. A.; Al-Omar M. S.; Khan R. A.; Mohammed S. A. A.; Qureshi K. A.; Abbas M. M.; Al Rugaie O.; Abd-Elmoniem E.; Ahmad A. M.; Kandil Y. I. Chemical Profile, Antioxidant, Antimicrobial, and Anticancer Activities of the Water-Ethanol Extract of Pulicaria Undulata Growing in the Oasis of Central Saudi Arabian Desert. Plants 2021, 10 (9), 1811. 10.3390/plants10091811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makena P. S.; Pierce S. C.; Chung K.; Sinclair S. E. Comparative Mutagenic Effects of Structurally Similar Flavonoids Quercetin and Taxifolin on Tester Strains Salmonella Typhimurium TA102 and Escherichia Coli WP-2 UvrA. Environ. Mol. Mutagen. 2009, 50 (6), 451–459. 10.1002/em.20487. [DOI] [PubMed] [Google Scholar]
- Baghel S. S.; Shrivastava N.; Baghel R. S.; Agrawal P.; Rajput S. A Review of Quercetin: Antioxidant and Anticancer Properties. World J. Pharm. Pharm. Sci. 2012, 1 (1), 146–160. [Google Scholar]
- Asmi K. S.; Lakshmi T.; Balusamy S. R.; Parameswari R. Therapeutic Aspects of Taxifolin–An Update. J. Adv. Pharm. Educ. Res. Jul-Sep 2017, 7 (3), 187. [Google Scholar]
- Lu J.; Papp L. V.; Fang J.; Rodriguez-Nieto S.; Zhivotovsky B.; Holmgren A. Inhibition of Mammalian Thioredoxin Reductase by Some Flavonoids: Implications for Myricetin and Quercetin Anticancer Activity. Cancer Res. 2006, 66 (8), 4410–4418. 10.1158/0008-5472.CAN-05-3310. [DOI] [PubMed] [Google Scholar]
- Lee S. B.; Cha K. H.; Selenge D.; Solongo A.; Nho C. W. The Chemopreventive Effect of Taxifolin Is Exerted through ARE-Dependent Gene Regulation. Biol. Pharm. Bull. 2007, 30 (6), 1074–1079. 10.1248/bpb.30.1074. [DOI] [PubMed] [Google Scholar]
- Sunil C.; Xu B. An Insight into the Health-Promoting Effects of Taxifolin (Dihydroquercetin). Phytochemistry 2019, 166, 112066. 10.1016/j.phytochem.2019.112066. [DOI] [PubMed] [Google Scholar]
- Walle T.Methoxylated Flavones, a Superior Cancer Chemopreventive Flavonoid Subclass? In Seminars in cancer biology; Elsevier, 2007; Vol. 17, pp 354–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almessiere M. A.; Slimani Y.; Rehman S.; Khan F. A.; Polat E. G.; Sadaqat A.; Shirsath S. E.; Baykal A. Synthesis of Dy-Y Co-Substituted Manganese-zinc Spinel Nanoferrites Induced Anti-Bacterial and Anti-Cancer Activities: Comparison between Sonochemical and Sol-Gel Auto-Combustion Methods. Mater. Sci. Eng., C 2020, 116, 111186. 10.1016/j.msec.2020.111186. [DOI] [PubMed] [Google Scholar]
- Almessiere M. A.; Slimani Y.; Rehman S.; Khan F. A.; Güngüneş Ç. D.; Güner S.; Shirsath S. E.; Baykal A. Magnetic Properties, Anticancer and Antibacterial Effectiveness of Sonochemically Produced Ce3+/Dy3+ Co-Activated Mn-Zn Nanospinel Ferrites. Arab. J. Chem. 2020, 13 (10), 7403–7417. 10.1016/j.arabjc.2020.08.017. [DOI] [Google Scholar]
- Shiozaki E. N.; Chai J.; Rigotti D. J.; Riedl S. J.; Li P.; Srinivasula S. M.; Alnemri E. S.; Fairman R.; Shi Y. Mechanism of XIAP-Mediated Inhibition of Caspase-9. Mol. Cell 2003, 11 (2), 519–527. 10.1016/S1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Havran L. M.; Chong D. C.; Childers W. E.; Dollings P. J.; Dietrich A.; Harrison B. L.; Marathias V.; Tawa G.; Aulabaugh A.; Cowling R. 3, 4-Dihydropyrimido (1, 2-a) Indol-10 (2H)-Ones as Potent Non-Peptidic Inhibitors of Caspase-3. Bioorg. Med. Chem. 2009, 17 (22), 7755–7768. 10.1016/j.bmc.2009.09.036. [DOI] [PubMed] [Google Scholar]
- Hashimoto K.; Saito B.; Miyamoto N.; Oguro Y.; Tomita D.; Shiokawa Z.; Asano M.; Kakei H.; Taya N.; Kawasaki M. Design and Synthesis of Potent Inhibitor of Apoptosis (IAP) Proteins Antagonists Bearing an Octahydropyrrolo [1, 2-a] Pyrazine Scaffold as a Novel Proline Mimetic. J. Med. Chem. 2013, 56 (3), 1228–1246. 10.1021/jm301674z. [DOI] [PubMed] [Google Scholar]
- Bai L.; Zhou H.; Xu R.; Zhao Y.; Chinnaswamy K.; McEachern D.; Chen J.; Yang C.-Y.; Liu Z.; Wang M. A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression in Vivo. Cancer Cell 2019, 36 (5), 498–511. 10.1016/j.ccell.2019.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z.-F.; Hasvold L.; Wang L.; Wang X.; Petros A. M.; Park C. H.; Boghaert E. R.; Catron N. D.; Chen J.; Colman P. M. Discovery of a Potent and Selective BCL-XL Inhibitor with in Vivo Activity. ACS Med. Chem. Lett. 2014, 5 (10), 1088–1093. 10.1021/ml5001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellaway C. R.; Baldwin I. R.; Bamborough P.; Barker D.; Bartholomew M. A.; Chung C.; Dümpelfeld B.; Evans J. P.; Fazakerley N. J.; Homes P. Investigation of Janus Kinase (JAK) Inhibitors for Lung Delivery and the Importance of Aldehyde Oxidase Metabolism. J. Med. Chem. 2022, 65, 633. 10.1021/acs.jmedchem.1c01765. [DOI] [PubMed] [Google Scholar]
- Kim S.; Chen J.; Cheng T.; Gindulyte A.; He J.; He S.; Li Q.; Shoemaker B. A.; Thiessen P. A.; Yu B. PubChem 2019 Update: Improved Access to Chemical Data. Nucleic Acids Res. 2019, 47 (D1), D1102–D1109. 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M.; Huey R.; Lindstrom W.; Sanner M. F.; Belew R. K.; Goodsell D. S.; Olson A. J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30 (16), 2785–2791. 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2009, 31 (2), 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowsky L.; Weber J.; Balasupramaniam S.; Baumann K.; Proschak E. A Diverse Benchmark Based on 3D Matched Molecular Pairs for Validating Scoring Functions. ACS omega 2018, 3 (5), 5704–5714. 10.1021/acsomega.7b01194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofazzal Islam M.; Tahara S. Dihydroflavonols from Lannea Coromandelica. Phytochemistry 2000, 54 (8), 901–907. 10.1016/S0031-9422(00)00048-0. [DOI] [PubMed] [Google Scholar]
- Clark-Lewis J. W. Flavan Derivatives. XXI. Nuclear Magnetic Resonance Spectra, Configuration, and Conformation of Flavan Derivatives. Aust. J. Chem. 1968, 21 (8), 2059–2075. 10.1071/CH9682059. [DOI] [Google Scholar]
- Ragab E. A.; Raafat M. A New Monoterpene Glucoside and Complete Assignments of Dihydroflavonols of Pulicaria Jaubertii: Potential Cytotoxic and Blood Pressure Lowering Activity. Nat. Prod. Res. 2016, 30 (11), 1280–1288. 10.1080/14786419.2015.1055492. [DOI] [PubMed] [Google Scholar]
- Agrawal P. K.Carbon-13 NMR of Flavonoids; Elsevier, 2013. [Google Scholar]
- Harborne J. B.; Mabry T. J. The Flavonoids: Advances in Research. J. Chem. Educ. 2013, 72 (3), A73. 10.1021/ed072pA73.11. [DOI] [Google Scholar]
- Markham K. R.; Ternai B.; Stanley R.; Geiger H.; Mabry T. J. Carbon-13 NMR Studies of Flavonoids—III: Naturally Occurring Flavonoid Glycosides and Their Acylated Derivatives. Tetrahedron 1978, 34 (9), 1389–1397. 10.1016/0040-4020(78)88336-7. [DOI] [Google Scholar]
- Krenn L.; Miron A.; Pemp E.; Petr U.; Kopp B. Flavonoids from Achillea Nobilis L. Zeitschrift für Naturforsch. C 2003, 58 (1–2), 11–16. 10.1515/znc-2003-1-202. [DOI] [PubMed] [Google Scholar]
- Wang X.; Zhou H.; Zeng S. Identification and Assay of 3′-O-Methyltaxifolin by UPLC–MS in Rat Plasma. J. Chromatogr. B 2012, 911, 34–42. 10.1016/j.jchromb.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Ternai B.; Markham K. R. Carbon-13 NMR Studies of Flavonoids—I: Flavones and Flavonols. Tetrahedron 1976, 32 (5), 565–569. 10.1016/S0040-4020(01)93772-X. [DOI] [Google Scholar]
- Marco J. A.; Sanz-Cervera J. F.; Yuste A.; Oriola M. C. Sesquiterpene Lactones and Dihydroflavonols from Andryala and Urospermum Species. Phytochemistry 1994, 36 (3), 725–729. 10.1016/S0031-9422(00)89805-2. [DOI] [Google Scholar]
- Herz W.; Gibaja S.; Bhat S. V.; Srinivasan A. Dihydroflavonols and Other Flavonoids of Eupatorium Species. Phytochemistry 1972, 11 (9), 2859–2863. 10.1016/S0031-9422(00)86525-5. [DOI] [Google Scholar]
- Harborne J. B.The Flavonoids: Advances in Research since 1980; Springer, 2013. [Google Scholar]
- Grade M.; Piera F.; Cuenca A.; Torres P.; Bellido I. S. Flavonoids from Inula Viscose. Planta Med. 1985, 39, 414–419. 10.1055/s-2007-969536. [DOI] [PubMed] [Google Scholar]
- Balza F.; Towers G. H. N. Dihydroflavonols of Artemisia Dracunculus. Phytochemistry 1984, 23 (10), 2333–2337. 10.1016/S0031-9422(00)80546-4. [DOI] [Google Scholar]
- Lundgren L. N.; Theander O. Cis-and Trans-Dihydroquercetin Glucosides from Needles of Pinus Sylvestris. Phytochemistry 1988, 27 (3), 829–832. 10.1016/0031-9422(88)84101-3. [DOI] [Google Scholar]
- El-Ghaly E.-S. M.; Shaheen U.; Ragab E.; El-hila A. A.; Abd-Allah M. R.. Bioactive Constituents of Pulicaria Jaubertii: A Promising Antihypertensive Activity. Pharmacogn. J. 2016, 8 ( (1), ). [Google Scholar]
- El Rayes S. M.; Aboelmagd A.; Gomaa M. S.; Ali I. A. I.; Fathalla W.; Pottoo F. H.; Khan F. A. Convenient Synthesis and Anticancer Activity of Methyl 2-[3-(3-Phenyl-Quinoxalin-2-Ylsulfanyl) Propanamido] Alkanoates and N-Alkyl 3-((3-Phenyl-Quinoxalin-2-Yl) Sulfanyl) Propanamides. ACS omega 2019, 4 (20), 18555–18566. 10.1021/acsomega.9b02320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamim S.; Khan K. M.; Salar U.; Ali F.; Lodhi M. A.; Taha M.; Khan F. A.; Ashraf S.; Ul-Haq Z.; Ali M. 5-Acetyl-6-Methyl-4-Aryl-3, 4-Dihydropyrimidin-2 (1H)-Ones: As Potent Urease Inhibitors; Synthesis, in Vitro Screening, and Molecular Modeling Study. Bioorg. Chem. 2018, 76, 37–52. 10.1016/j.bioorg.2017.10.021. [DOI] [PubMed] [Google Scholar]
- Mou L.; Liang B.; Liu G.; Jiang J.; Liu J.; Zhou B.; Huang J.; Zang N.; Liao Y.; Ye L. Berbamine Exerts Anticancer Effects on Human Colon Cancer Cells via Induction of Autophagy and Apoptosis, Inhibition of Cell Migration and MEK/ERK Signalling Pathway. J. BUON 2019, 24 (5), 1870–1875. [PubMed] [Google Scholar]
- Khorsandi L.; Orazizadeh M.; Niazvand F.; Abbaspour M. R.; Mansouri E.; Khodadadi A. Quercetin Induces Apoptosis and Necroptosis in MCF-7 Breast Cancer Cells. Bratislava Med. J. 2017, 118 (2), 123–128. 10.4149/BLL_2017_025. [DOI] [PubMed] [Google Scholar]
- Mettu A.; Talla V.; Thumma S.; Prameela S. N. J. Mechanistic Investigations on Substituted Benzene Sulphonamides as Apoptosis Inducing Anticancer Agents. Bioorg. Chem. 2020, 95, 103539. 10.1016/j.bioorg.2019.103539. [DOI] [PubMed] [Google Scholar]
- Bi Y.; Min M.; Shen W.; Liu Y. Genistein Induced Anticancer Effects on Pancreatic Cancer Cell Lines Involves Mitochondrial Apoptosis, G0/G1cell Cycle Arrest and Regulation of STAT3 Signalling Pathway. Phytomedicine 2018, 39, 10–16. 10.1016/j.phymed.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Mettu A.; Talla V.; Naikal S. J. P. Novel Anticancer Hsp90 Inhibitor Disubstituted Pyrazolyl 2-Aminopyrimidine Compound 7t Induces Cell Cycle Arrest and Apoptosis via Mitochondrial Pathway in MCF-7 Cells. Bioorg. Med. Chem. Lett. 2020, 30 (20), 127470. 10.1016/j.bmcl.2020.127470. [DOI] [PubMed] [Google Scholar]
- Guadagno J.; Xu X.; Karajgikar M.; Brown A.; Cregan S. Microglia-Derived TNFα Induces Apoptosis in Neural Precursor Cells via Transcriptional Activation of the Bcl-2 Family Member Puma. Cell Death Dis. 2013, 4 (3), e538–e538. 10.1038/cddis.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larasati V. A.; Lembang G. V.; Tjahjono Y.; Winarsih S.; Ana I. D.; Wihadmadyatami H.; Kusindarta D. L. In Vitro Neuroprotective Effect of the Bovine Umbilical Vein Endothelial Cell Conditioned Medium Mediated by Downregulation of IL-1β, Caspase-3, and Caspase-9 Expression. Vet. Sci. 2022, 9 (2), 48. 10.3390/vetsci9020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’amelio M.; Cavallucci V.; Cecconi F. Neuronal Caspase-3 Signaling: Not Only Cell Death. Cell Death Differ. 2010, 17 (7), 1104–1114. 10.1038/cdd.2009.180. [DOI] [PubMed] [Google Scholar]
- Sabbah D. A.; Vennerstrom J. L.; Zhong H. Docking Studies on Isoform-Specific Inhibition of Phosphoinositide-3-Kinases. J. Chem. Inf. Model. 2010, 50 (10), 1887–1898. 10.1021/ci1002679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin E. J.; Lee J. S.; Hong S.; Lim T.-G.; Byun S. Quercetin Directly Targets JAK2 and PKCδ and Prevents UV-Induced Photoaging in Human Skin. Int. J. Mol. Sci. 2019, 20 (21), 5262. 10.3390/ijms20215262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.; Li J.; Liu T.; Li S.; Feng J.; Yu Q.; Zhang J.; Chen J.; Zhou Y.; Ji J. Quercetin Shows Anti-tumor Effect in Hepatocellular Carcinoma LM3 Cells by Abrogating JAK2/STAT3 Signaling Pathway. Cancer Med. 2019, 8 (10), 4806–4820. 10.1002/cam4.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozgit J. M.; Bebernitz G.; Patil P.; Ye M.; Parmentier J.; Wu J.; Su N.; Wang T.; Ioannidis S.; Davies A. Effects of the JAK2 Inhibitor, AZ960, on Pim/BAD/BCL-XL Survival Signaling in the Human JAK2 V617F Cell Line SET-2. J. Biol. Chem. 2008, 283 (47), 32334–32343. 10.1074/jbc.M803813200. [DOI] [PubMed] [Google Scholar]
- Primikyri A.; Chatziathanasiadou M. V.; Karali E.; Kostaras E.; Mantzaris M. D.; Hatzimichael E.; Shin J.-S.; Chi S.-W.; Briasoulis E.; Kolettas E. Direct Binding of Bcl-2 Family Proteins by Quercetin Triggers Its pro-Apoptotic Activity. ACS Chem. Biol. 2014, 9 (12), 2737–2741. 10.1021/cb500259e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.