Abstract
A green-based approach for the synthesis of silver nanoparticles has gained tremendous attention in biomedical applications. Fungal endophytes have been recognized as a remarkable biological source for the synthesis of potential nanodrugs. The present study focuses on the fabrication of silver nanoparticles using the fungal endophyte Penicillium oxalicum (POAgNPs) associated with the leaf of the Amoora rohituka plant. Sharp UV–visible spectra at 420 nm appeared due to the surface plasmon resonance of POAgNPs and the reduction of silver salt. FT-IR analysis revealed the presence of functional groups of bioactive compounds of P. oxalicum responsible for the reduction of silver salt and validated the synthesis of POAgNPs. A high degree of crystallinity was revealed through XRD analysis, and microscopy-based characterizations such as AFM, TEM, and FESEM showed uniformly distributed, and spherically shaped nanoparticles. Furthermore, POAgNPs showed a potential inhibitory effect against bacterial and fungal strains of pathogenic nature. POAgNPs also exhibited potential antioxidant activity against the synthetically generated free radicals such as DPPH, superoxide, hydroxyl, and nitric oxide with EC50 values of 9.034 ± 0.449, 56.378 ± 1.137, 34.094 ± 1.944, and 61.219 ± 0.69 μg/mL, respectively. Moreover, POAgNPs exhibited cytotoxic potential against the breast cancer cell lines, MDA-MB-231 and MCF-7 with IC50 values of 20.080 ± 0.761 and 40.038 ± 1.022 μg/mL, respectively. POAgNPs showed anticancer potential through inhibition of wound closure and by altering the nuclear morphology of MDA-MB-231 and MCF-7 cells. Further anticancer activity revealed that POAgNPs induced apoptosis in MDA-MB-231 and MCF-7 cells by differential expression of genes related to apoptosis, tumor suppression, and cell cycle arrest and increased the level of Caspase-3. The novel study showed that P. oxalicum-mediated silver nanoparticles exhibit potential biological activity, which can be exploited as nanodrugs in clinical applications.
Introduction
Nanotechnology has significantly emerged as an attractive technology that has gained tremendous interest in the scientific discipline due to its myriad of applications in the biomedical sector. The synthesis of small-sized particles at the nanoscale is an integrated approach combining diverse branches of science, that is, chemical, engineering, physics, electronics, and others for manufacturing nanoparticles at the nanometer scale and also allowing manipulation through biotechnological interventions. The synthesis of nanoparticles in the size range of 1–100 nm has surpassed the limitations of biological barriers and gained impetus in the advanced research field.1 The synthesis of nanoparticles, particularly metal nanoparticles, has significantly impacted the research field due to their flexibility to manipulate size, shape, structure, optical properties, and assembly and thus obtaining nanoparticles of desired physiochemical characteristics. Metallic nanoparticles including gold, copper, zinc, iron, silver, graphene, titanium, magnesium, and platinum have been used for the nanofabrication, showing their potential applications in industries, medicine, chemical engineering, bioimaging, biocatalysts, cosmetics, and antimicrobial agents.2 However, silver metal has been widely used for the synthesis of nanoparticles due to its exceptionally unique chemical stability, catalytic properties, and conductivity.3,4 Silver has long been recognized for its intrinsic antimicrobial potential and therefore, extensively employed for medicinal purposes.5,6 Overcoming the chemical- and physical-based synthesis of silver nanoparticles, biogenic synthesis of silver nanoparticles has gained momentum for the fabrication of green silver nanoparticles due to their eco-friendly nature, least generation of toxic byproducts, economically favorable characteristics, reduced side effects, and potential biological activities.7,8
The escalated demand of drugs due to the cytotoxicity and lack of tissue specificity of synthetic drugs, development of resistance by the tissues, and cost effectiveness of drugs has raised concern about maintaining the health and wellness. One of the most leading causes of death is cancer, which is considered as the deadliest disease. The global cancer statistics have shown an estimate of 19.3 million new cases of cancer (18.1 million on excluding nonmelanoma skin cancer) and about 10.0 million deaths due to cancer (9.9 million on excluding nonmelanoma skin cancer) in the year 2020 and the expected cases by 2040 are given to be 28.4 million, a 47% increase from 2020.9 Many large-sized drugs face challenges in the complete eradication of tumor cells due to their inability to penetrate into the tumor matrix. Also, the consequent use of high dose of conventional drugs causes adverse effects in the biological systems. The low bioavailability, stability, penetration, and solubility of these drugs limit their efficacy. Moreover, the major drawback exhibited by these drugs includes high systemic toxicity. Alternative to these drugs are natural-based therapies that have emerged as an effective and safer approach against diseases including cancer. Fungal endophytes are one of the most distinguished groups of the microbial community owing to their multitude of activities in biomedical and pharmaceutical industries.10−12 Many of these bioactive compounds have been identified through mass spectrometry-based techniques along with the bioinformatics tools that have revealed their chemical nature and significant biological functions.13,14 Fungal endophyte-mediated synthesis of silver nanoparticles (AgNPs) has emerged as a frontier technology to develop an effective, nontoxic, highly soluble, permeable, and budget-friendly nanodrug that provides an advanced measure for early detection, diagnostics, and treatment of deadliest diseases including cancer. The unique cellular organization of fungi aid tolerance against high mechanical and environmental pressures, easy culture and subculture, less labor and maintenance, facile synthesis protocol, less complex medium requirement, less growth time, easy scale up, plentiful extracellular enzyme secretion, and metal bioaccumulation have attracted attention toward employing fungal endophyte-mediated synthesis of silver nanoparticles over other microbes.15−17 The large size of silver metal confers extensive use of silver in the fabrication of bio-based nanoparticles. Additionally, the large surface area-to-volume ratio offers characteristic physicochemical properties that aid better biocompatibility and cellular uptake, facilitating deep penetration inside the cells, crossing the biological barriers with enhanced efficacy.18 The nontoxicity, eco-friendliness, and inexpensiveness of the fungal endophyte-mediated synthesis of silver nanoparticles have accelerated the demand of mycofabricated silver nanoparticles or nanodrugs. Recent study has shown Penicillium oxalicum- and Fusarium hainanense-derived silver nanoparticles exhibiting antimicrobial, antioxidant, larvicidal, and anticancer potency.19 In a study, P. oxalicum-mediated synthesized silver nanoparticles have shown effective cytotoxic activity against MRC-5 (human fetal lung fibroblast cells) and antimicrobial and anti-biofilm activity.20 Another finding showed Penicillium italicum-mediated synthesized silver nanoparticles exhibiting potential antimicrobial activity and cytotoxic effects against MCF-7 (human breast cancer cells).21 The fungus-based synthesis of nanoparticles using Penicillium brevicompactum (MTCC-1999) showed anticancer activity against the MCF-7 breast cancer cell line with IC50 values of 70 and 50 μg/mL after 24 and 48 h, respectively, and also inhibited the growth of several pathogenic bacterial strains.22
With this rationale, the present study has been designed to synthesize mycogenic silver nanoparticles using the fungal endophyte P. oxalicum isolated from Amoora rohituka along with their assessment for biological activities. In our previous study, we have reported the presence of unique bioactive compounds and potential biological activities of the EA extract of P. oxalicum as compared to other isolates.23 The report thus led to the further exploration of biological activities of P. oxalicum-derived bioactive compounds, and therefore, silver nanoparticles have been synthesized. This is the first study showing the biological potential of silver nanoparticles synthesized from an aqueous extract of P. oxalicum (POAgNPs). The characterization of POAgNPs was carried out by UV–vis spectroscopy, FT-IR, XRD, AFM, TEM, and FESEM, that validated the synthesis of silver nanoparticles in the nanoscale range, between 5 to 23 nm. POAgNPs have been studied for their antibacterial and antifungal activity against pathogenic bacterial strains Escherichia coli and Staphylococcus aureus and fungal strains Aspergillus flavus, A. niger, A. luchuensis, and Penicillium albicans. Green-synthesized POAgNPs showed promising antioxidant activity against DPPH, superoxide, hydroxyl, and nitric oxide free radicals. Furthermore, the cytotoxic activity of POAgNPs against the human breast cancer cells MDA-MB-231 and MCF-7 validated its significant antiproliferative potential. The anticancer activity of POAgNPs was shown through the cell migration inhibition and alteration in the nuclear morphology of breast cancer cells. Additionally, the molecular mechanism of POAgNP-mediated apoptosis in breast cancer cells was determined through differential expression of genes that regulate apoptosis. The fungal endophyte P. oxalicum-mediated synthesis of silver nanoparticles provides a novel study that elucidates its probable anticancer mechanism against human breast cancer cells. The observation revealed POAgNPs as a potential anticancer agent, and their further evaluation for anticancer activity is of considerable significance for establishing the therapeutic potential of POAgNPs in breast cancer therapy.
Results and Discussion
Visible Color Change Indicated the Synthesis of P. oxalicum-Mediated Silver Nanoparticles (POAgNPs)
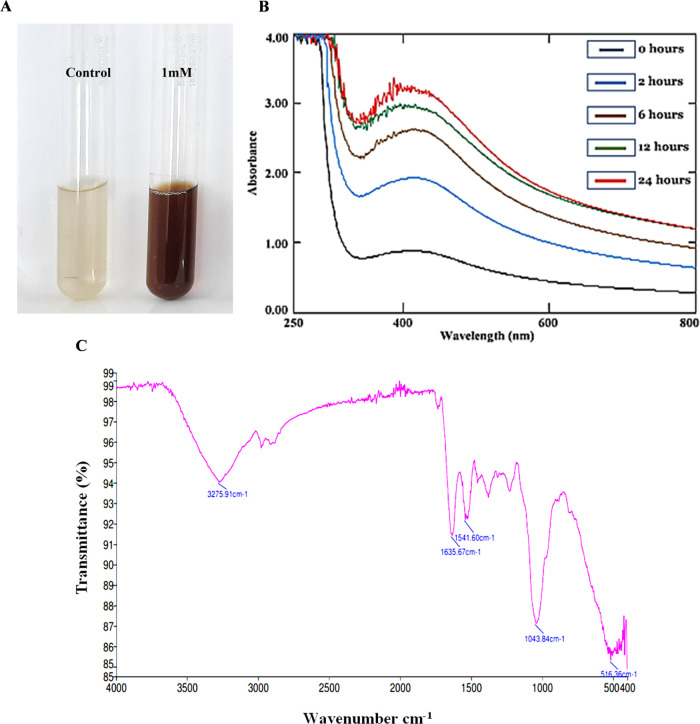
Extracellular synthesis has always been an attractive approach due to its simple downstream processing and high production rate, and therefore, fungi are considered as the most appropriate candidate for large-scale green nanoproduction. The synthesis of silver nanoparticles was marked by the change in the color of the aqueous solution from pale yellow to a brownish color (Figure 1A). The color was shifted from pale yellow to light brown to intense brown with increasing time of incubation. Researchers have reported color change as the macroscopic observation for the synthesis of silver nanoparticles.24−26 Our findings suggest that synthesis of POAgNPs is attributed to the extracellular enzymes and proteins of metabolites produced from P. oxalicum. These enzymes during the electron transfer process reduce the silver ions that gives a brown color to the solution. Many other observations have shown the involvement of NADH and NADH-dependent nitrate reductase in the biogenic synthesis of silver nanoparticles. A report suggesting the role of nitrate reductase activity in the potential biogenic synthesis of silver nanoparticles mediated by Aspergillus flavus has been shown.27 In another study, Fusarium oxysporum has been demonstrated to exhibit high potential for silver nanoparticle synthesis, which has been related to its ability to secrete the NADH-dependent nitrate reductase enzyme that causes the reduction of aqueous silver ions into AgNPs. The color alteration is due to the optical phenomenon known as surface plasmon resonance that can be explained as the oscillation of free electrons of silver nanoparticles with a specific energy, that absorbs and scatters a particular wavelength of visible light.28 The aqueous extract without silver nitrate did not show color alteration, and therefore, the appearance of brown color can be recorded as the preliminary identification for the synthesis of silver nanoparticles.
Figure 1.
Visual and IR-based characterization of silver nanoparticles synthesized by endophyte P. oxalicum (POAgNPs). (A) Visible color change after addition of 1 mM silver nitrate to the aqueous extract of P. oxalicum. (B) UV–vis spectra of POAgNPs at different time intervals showing reduction of silver nitrate and formation of silver nanoparticles. (C) FT-IR spectrum of silver nanoparticles synthesized by the endophyte P. oxalicum.
Characterization Using Different Techniques Revealed the Synthesis, Dispersity, and Size Range of POAgNPs
UV–vis Absorption Spectra (UV–vis)
The initial characterization for the biosynthesis of silver nanoparticles was determined through UV–vis spectrophotometry. The absorbance was recorded in the range of 200–800 nm at different time intervals of 0, 2, 6, 12, and 24 h. The characteristic peak was observed at a range of 400–450 nm that revealed the formation of silver nanoparticles. The value of λmax was observed to be 420 nm, which can be attributed to the phenomenon of SPR resulting from the collective oscillation of electrons in the conductive band.29 The absorbance was increased as the time for incubation was increased (Figure 1B). The observation of a peak at 420 nm indicated the release of active components from the mycelia of P. oxalicum that mediate the reduction and capping of silver nanoparticles. The enzymatic action of aqueous solution caused the bioreduction of Ag+ to Ag0 ions in the colloidal solution with the appearance of dark brown color. Earlier studies have also demonstrated the electronic excitation in AgNPs synthesized using the fungal endophyte Botryosphaeria rhodina, owing to which a characteristic peak at 450 nm was observed.30 In another study, the fungal endophytes Aspergillus tamarii, Aspergillus niger, and Penicillium ochrochloron have been used for the synthesis of silver nanoparticles that revealed the absorption at 419, 430, and 430 nm, respectively, through UV–vis absorption analysis.31
Fourier Transform Infrared Spectroscopy (FT-IR)
The FT-IR analysis showed the presence of fungal biomolecules in the synthesized silver nanoparticles. The FT-IR spectrum of silver nanoparticle is represented in Figure 1C. The bands are observed at 3295.71, 1635.67, 1541.60, 1043.84, and 516.36 cm–1 (Table S1). The bands correspond to the hydroxyl group, which arise due to the OH stretching. The involvement of the hydroxyl group as the bioreduction agent in silver nanoparticle synthesis has been previously reported.32 The band at 1635.67 is assigned to the C=C stretching vibrations of the alkenyl group. The band observed at 1541 corresponds to the aliphatic nitro compound, while the band at 1043.84 can be assigned to the C–F stretching or bending vibrations in C–H or CN stretching vibrations of aliphatic fluoro compounds or aromatic or primary amine. The observations indicated the presence of protein in the sample. Previous reports have shown that proteins stabilize the silver nanoparticles through interacting with nanoparticles by either binding through free amine groups or cysteine residues.33,34 Reports have also shown that the peptides of proteins bind to metal and cap AgNPs so as to prevent agglomeration.35 In a report, FT-IR analysis of fungal-derived silver nanoparticles showed the stretching vibration of amide groups in proteins, and this demonstrated the role of proteins as capping agents for AgNP synthesis.36 The fungal (Phoma exigua var. exigua)-mediated silver nanoparticles showed the presence of proteins as capping agents due to the presence of amide II and amide III and C=C stretching of the bonds.37 The FT-IR spectrum of Aspergillus flavus-mediated silver nanoparticles has revealed the presence of primary and secondary amines along with the aliphatic and aromatic amines that indicated the presence of proteins in the sample.38 Another band at 516.36 was identified as aliphatic iodo compounds, which arises due to the C–I stretching vibrations. In a report, aliphatic fluoro compounds, nitro groups, iodo compounds, and OH groups have been reported to be involved in the reduction and stabilization of the silver nanoparticles.39
X-ray Diffraction (XRD)
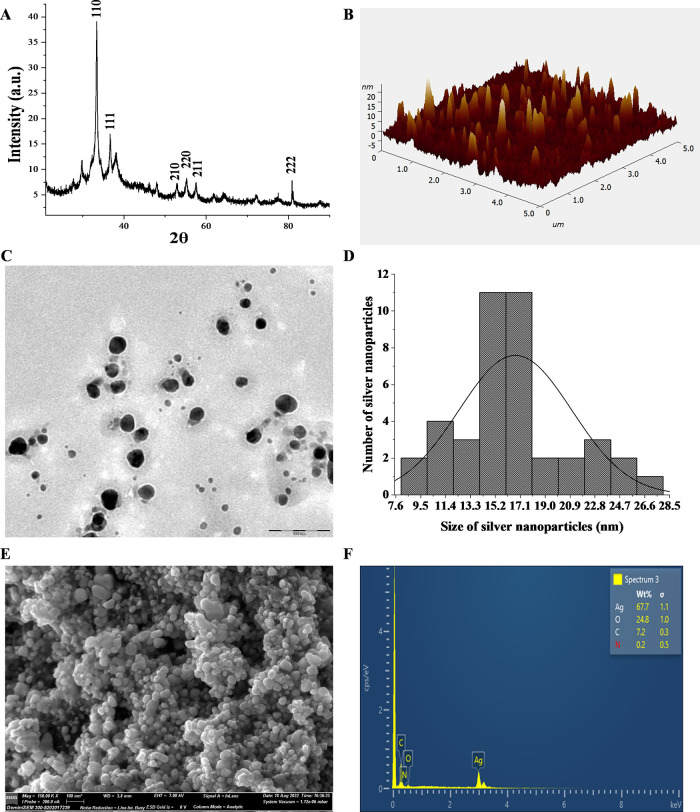
To validate the synthesis and nature of POAgNPs, powder X-ray diffraction analysis was performed. The XRD plot depicts the crystalline nature, which was shown by the intensity of the X-ray scattered pattern observed for POAgNPs at different angles. The broader peaks represent the decrease in crystallite size; however, the sharp peak shows larger crystalline domain sizes (Figure 2A). The angle between the transmitted and reflected beams is represented by 2θ, termed as diffraction angle. The diffraction peaks observed are attributed to the scattering from a specific set of parallel planes of atoms and are determined by miller indices (hkl). The miller indices (hkl) quantify the intercepts and represent the 3D orientation of the crystallographic plane. The observation showed six major peaks at 2θ values of 33.39, 36.67, 53.02, 55.21, 57.52, and 80.78 corresponding to the 110, 111, 210, 220, 211, and 222 planes of silver. After analyzing JCPDS (file no. 89-3722), the characteristic pattern of POAgNPs is observed to possess a face-centered cubic (fcc) structure. Moreover, the XRD plot also exhibited four unidentified peaks that were weaker as compared to those of silver and appeared at 2θ values of 29.76, 38.14, 47.91, and 72.06. The bioactive compounds produced from P. oxalicum may be responsible for these four unexpected peaks.40 The observation of such unpredicted peaks was corroborated with the previous findings, which identify five crystalline peaks at 2θ values of 32.28, 46.28, 54.83, 67.47, and 76.69° with the XRD pattern involving the relevant 2° range.41 In the present study, the presence of these unassigned peaks is attributed to the bioactive compounds of P. oxalicum present on the surface of POAgNPs, whereas the stronger planes imply that silver plays a key role in the biosynthesis of POAgNPs. The lattice parameters interplanar spacing (d) and full width at half maximum (FWHM) were calculated using an X’pert HighScore Plus, and the size of the synthesized silver nanoparticles was found to be in the range of 5.4–16.9 nm. A previous report has shown that silver nanoparticles synthesized using the fungus Amylomyces rouxii strain KSU-09 exhibited crystalline nature and the average size was determined to be 27 nm.42 The fungal endophyte Raphanus sativus-derived silver nanoparticles were reported to be crystalline in nature with an average particle size of 4–30 nm calculated using Scherrer’s equation (D = (kλ/β cos θ)), where D represents the crystalline size (nm), λ indicates the wavelength of X-rays (0.1541 nm), β specifies the angular line full width at half maximum (FWHM) of the peak (in radians), and θ displays the Braggs angle (in radians).43
Figure 2.
Characterization of the synthesized silver nanoparticles. (A) XRD showing the crystalline nature of POAgNPs. (B) Atomic force microscopy showing homogeneously shaped silver nanoparticles. (C) TEM images showing spherical-shaped and well-dispersed silver nanoparticles. (D) Histogram of the size distribution of POAgNPs. (E) FESEM micrograph showing the surface topology of silver nanoparticles. (F) Energy dispersive X-ray spectrum of POAgNPs showing the elemental composition of POAgNPs.
Atomic Force Microscopy (AFM)
The size, morphology, surface roughness, and agglomeration of the synthesized silver nanoparticles were characterized using AFM. The randomly dispersed, homogeneously shaped, and slightly aggregated silver nanoparticles were observed (Figure 2B). The AFM analysis revealed the presence of sphere-shaped silver nanoparticles. The size of the POAgNPs was found to be in the range of 13–23 nm. Root-mean-square roughness and roughness average are crucial height parameters that reflect the synthesis of silver nanoparticles. The root-mean-square roughness (Sq) and roughness average (Sa) values of homogeneously shaped nanoparticles of POAgNPs were 2.641 and 1.737 nm, respectively. Moreover, the AFM analysis also revealed that the maximum height of the peak profile (Rmax) was 13.121 nm, which validates the particle size of POAgNPs calculated using Image J analysis. In a report, biogenically synthesized silver nanoparticles using Aspergillus tamarii have been analyzed for the particle size using AFM, which was found to be 40 nm.44 In a previous study, AFM analysis for Alternaria sp.-derived silver nanoparticles has shown the average particle size between 4 and 28 nm.43Aspergillus fumigatus-mediated synthesis of silver nanoparticles showed sphere-shaped particles with a size of around 50 nm through AFM analysis.45
Transmission Electron Microscopy (TEM)
Insights into the morphology and size of the synthesized POAgNPs were provided by TEM analysis. The microscopic observations revealed the homogeneity, good dispersion, and spherical shape of the synthesized silver nanoparticles, thus indicating their stabilization (Figure 2C). The nanoparticles were observed to be in the size range of 15–19 nm, which was determined using Image J software. The polydisperse nature of POAgNPs with variation in the particle size range has been shown through the histogram in Figure 2D, where a maximum number of nanoparticles lie in the range of 15–17 nm. A report has shown that mycogenic silver nanoparticles synthesized using Phoma exigua exhibited a size of 22 nm, analyzed through TEM.37 The morphological and size analyses of P. oxalicum-derived silver nanoparticles using TEM have revealed an average size of 6 nm.46 Another study has revealed a size range of 5.19–21.3 nm analyzed using TEM for P. oxalicum-derived silver nanoparticles.19 A size range of 10–50 nm for silver nanoparticles derived from Beauveria bassiana has been revealed by TEM analysis.47
Field Emission Scanning Electron Microscopy (FESEM)
The FESEM analysis performed for POAgNPs revealed the surface topography of the synthesized particles. The analysis showed the presence of spherical, uniformly shaped, well-distributed, and least-aggregated particles (Figure 2E). EDX analysis provided quantitative and qualitative measures of elements in nanoparticles. The sharp peak observed at 3 keV revealed the presence of silver, as it is a characteristic absorption for AgNPs due to surface plasmon resonance (Figure 2F). Elemental analysis revealed that silver was present as the main element with a percentage of 67.7%; however, other elements were also present at different percentages. Some weaker peaks corresponding to other elements were oxygen (24.8%), carbon (7.2%), and nitrogen (0.2%). The elemental analysis shows a positive correlation with the findings of FT-IR and XRD spectra that showed the presence of oxygen atoms and capping of silver through fungal metabolites, respectively. In a previous report, SEM analysis of the endophytic fungus Botryosphaeria rhodina-derived silver nanoparticles has shown the presence of variable-shaped and uniformly distributed nanoparticles.30 The endophytic fungus Cryptosporiopsis ericae PS4-mediated silver nanoparticles analyzed with SEM showed spherical nanoparticles and the EDX profile revealed the characteristic signal at approximately 3 keV, which indicated the presence of silver nanoparticles.48 The silver nanoparticles synthesized using Penicillium chrysogenum and Aspergillus oryzae have shown sphere-shaped particles, analyzed through SEM, and an absorption peak observed at 3 keV through EDS analysis indicated the presence of AgNPs.49
POAgNPs Significantly Inhibited the Growth of Pathogenic Bacterial and Fungal Strains
POAgNPs Showed Significant Antibacterial Activity against Pathogenic Bacterial Strains
The present study was performed to examine the potential of POAgNPs to inhibit the growth of bacterial strains, E. coli and S. aureus, using the agar well diffusion method. The results showed that the synthesized POAgNPs inhibited the growth of both bacteria in a concentration-dependent manner (Figure S1). The antibacterial activity of POAgNPs was observed to be highest against S. aureus, and it was comparable to that of the positive control, vancomycin. The inhibitory effect of POAgNPs against bacterial growth was calculated through measuring zones of inhibition in mm, represented in Table 1. The minimum inhibitory concentration (MIC25, MIC50, MIC70) of POAgNPs against bacterial strains was evaluated using the broth dilution method and is represented in Figure 3A, and MIC values are shown in Table S2. The MIC25, MIC50, and MIC75 values for POAgNPs against E. coli were found to be 8.710 ± 0.217, 12.369 ± 0.099, and 81.857 ± 0.453 μg/mL, respectively; for S. aureus, it was found to be 14.417 ± 0.011, 20.975 ± 0.008, and 61.614 ± 1.452 μg/mL. The effect of POAgNPs was found to have more potential against S. aureus. For the positive control (streptomycin), MIC25, MIC50, and MIC75 values have been observed to be 22.209 ± 0.090, 61.084 ± 0.392, and 90.123 ± 0.234 μg/mL against E. coli, and against S. aureus, the MIC25, MIC50, and MIC75 values for the positive control (vancomycin) were found to be 14.709 ± 0.123, 36.951 ± 0.177, and 90.476 ± 0.076 μg/mL, respectively. The negative control containing nuclease-free water did not show any inhibition in the bacterial growth.
Table 1. Diameter of Zo ne of Inhibition (mm) Induced by POAgNPs against Pathogenic Bacterial Strains and Fungal Strains.
| diameter
of zone of inhibition (mm) |
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| activity against bacterial strains | activity against fungal strains | ||||||||||||||||||||||||
| E. coli | S. aureus | A. niger | A. flavus | A. luchuensis | P. albicans | ||||||||||||||||||||
| s. no. | samples | 25 μg/mL | 50 μg/mL | 100 μg/mL | 200 μg/mL | 25 μg/mL | 50 μg/mL | 100 μg/mL | 200 μg/mL | 25 μg/mL | 50 μg/mL | 100 μg/mL | 200 μg/mL | 25 μg/mL | 50 μg/mL | 100 μg/mL | 200 μg/mL | 25 μg/mL | 50 μg/mL | 100 μg/mL | 200 μg/mL | 25 μg/mL | 50 μg/mL | 100 μg/mL | 200 μg/mL |
| 1. | POAgNPs | 6.6 | 6.6 | 7.4 | 8 | 5 | 6 | 8 | 5.1 | 9 | 13 | 6 | 8 | 7 | 9 | ||||||||||
| 2. | amphotericin B (positive control) | 11 | 13 | 14 | 14 | 6.8 | 9 | 10 | 12 | 10 | 13 | 14 | 15 | 7.2 | 8 | 9 | 9 | ||||||||
| 3. | streptomycin (positive control) | 7.2 | 8.0 | 8.0 | |||||||||||||||||||||
| 4. | vancomycin (positive control) | 6.7 | 6.8 | ||||||||||||||||||||||
| 5. | nuclease-free water (negative control) | ||||||||||||||||||||||||
Figure 3.
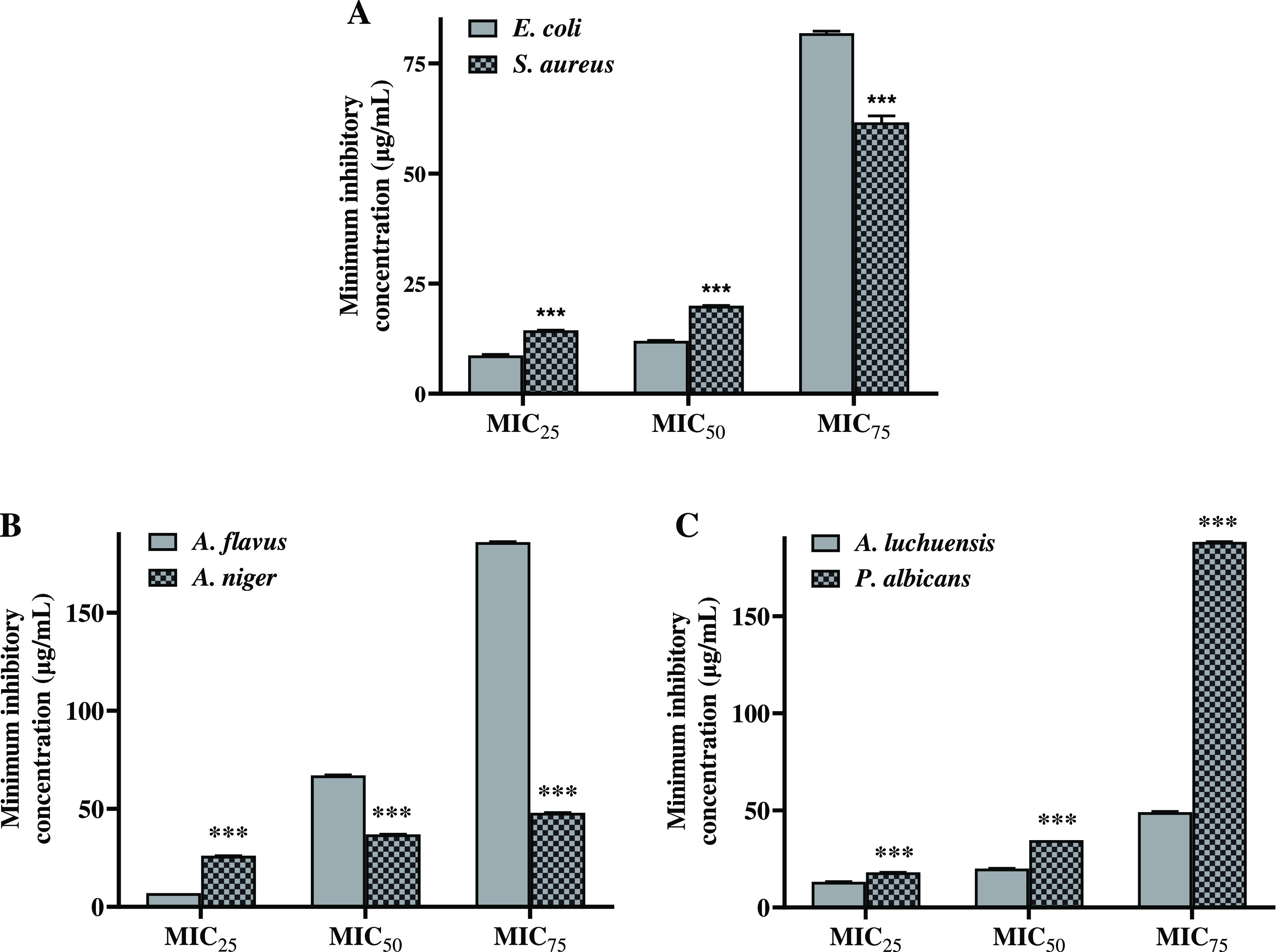
Calculation of the minimum inhibitory concentration for POAgNPs against. (A) Pathogenic bacterial strains, E. coli and S. aureus, (B) Pathogenic fungal strains, Aspergillus flavus and Aspergillus niger and (C) Aspergillus luchuensis and Penicillium albicans. All the experiments were performed in triplicate. The p-value was calculated by comparing means ± SD of percentage growth inhibition against bacteria and fungi in control and POAgNP-treated groups, using one-way ANOVA followed by Tukey to determine statistical significance values, which are as follows: ***p ≤ 0.001; ** p ≤ 0.002; and * p ≤ 0.033.
The emergence of resistant pathogenic bacterial strains is responsible for causing several diseases such as respiratory, digestive, skin, and urinary tract infections. The nanosized particles bestow great advantage in combating the health conditions caused by these pathogens with reduced toxicity and side effects, stability, broad-spectrum activity, and most importantly lack of microbial resistance to them.50,51 Due to the characteristic surface-to-volume ratio of fungal endophyte-derived silver nanoparticles, they exhibit remarkable antibacterial activity even at low concentrations against a wide range of pathogenic bacterial strains.52 The antibacterial activity of silver nanoparticles can be due to their adsorption on the surface of bacteria.53 The bacterial inner wall is ruptured, and generation of reactive oxygen species leads to the dissipation of the proton motive force and cell death.54 Furthermore, the microscopic characterization of silver nanoparticles using high-resolution techniques such as TEM, SEM, and AFM has revealed the spherical nature of silver nanoparticles, which facilitates adsorption onto the bacterial surface, piercing of the bacterial cell wall, and ultimately leading to death of the microbe. The concept is supported by the previous report that has shown POAgNPs to exhibit antimicrobial activity against Gram-positive S. aureus and Gram-negative Shigella dysenteriae and Salmonella typhi.55
POAgNPs Showed Potential Antifungal Activity against Pathogenic Fungal Strains
The antifungal activity of POAgNPs was characterized against a wide range of pathogenic fungi such as A. flavus, A. niger, A. luchuensis, and P. albicans, which showed significant inhibition in a dose-dependent manner (Figure S2). The zone of inhibition was calculated for all the tested pathogenic fungi (Table 1). The MIC values for POAgNPs were calculated against the pathogenic fungal strains A. flavus and A. niger (Figure 3B) and A. luchuensis and P. albicans (Figure 3C), and the respective values are shown in Table S2. POAgNPs significantly inhibited the growth of A. niger with MIC25, MIC50, and MIC75 values of 26.964 ± 0.021, 37.692 ± 0.050, and 48.421 ± 0.079 μg/mL, respectively. Similarly, the MIC25, MIC50, and MIC75 values for A. flavus were 7.729 ± 0.032, 67.727 ± 0.193, and 186.57 ± 0.152 μg/mL, respectively; for A. luchuensis, they were 13.17 ± 0.109, 20.94 ± 0.116, and 49.08 ± 0.332 μg/mL, and for P albicans, they were 18.092 ± 0.050, 34.683 ± 0.006, and 188.22 ± 0.145 μg/mL, respectively. The MIC25, MIC50, and MIC75 values for the positive control for A. niger were observed to be 10.071 ± 0.021, 19.293 ± 0.006, and 48.437 ± 0.107 μg/mL, respectively; for A. flavus, they were 16.092 ± 0.218, 22.866 ± 0.123, and 45.801 ± 0.097 μg/mL; for A. luchuensis, they were 7.385 ± 0.009, 9.824 ± 0.009, and 12.263 ± 0.010 μg/mL; and for P albicans, they were 16.128 ± 0.074, 21.794 ± 0.195, and 95.920 ± 0.424 μg/mL, respectively. Our findings showed that fabrication of silver nanoparticles using the fungal mycelial extract is an effective approach to generate nanosized drugs, exhibiting characteristic physiochemical properties that facilitate the effective delivery of drugs inside the microbial cell, controlled release of bioactive compounds, and enhanced stability.
The nature of mycotoxins can have varying effects in humans such as carcinogenic, mutagenic, teratogenic, hepatotoxic, nephrotoxic, immunosuppressive, and embryotoxic. These pathogenic fungi have been known to adopt several strategies to evade the host immune system and therefore cause negative effects and raise major concern for human health. The limitation of existing antifungal drugs in treating the pathogenicity caused by fungal strains has been overcome by the introduction of nanosized drugs. In the present study, significant results have been obtained for the POAgNP-mediated inhibition of fungal growth. The small size and unique intrinsic surface properties of biogenic silver nanoparticles facilitate easy penetration inside the fungal cell walls through reacting with the cellular components. The silver nanoparticles get adhered to the cell membrane and interfere with the ergosterol or fatty acids and generate reactive oxygen species and alter membrane fluidity.56 The natural architecture of DNA is disrupted due to the silver nanoparticles, as they interact with DNA bases and form crosslinks and substitute hydrogen bonds adjoined to nitrogen atoms in purines and pyrimidines and also inhibit cell division.57,58 DNA replication is inhibited, ATP production is ceased, and therefore, the biochemical cycle of fungal cells is disrupted.59 The finding of the present study is in accordance with the previous findings that revealed P. oxalicum-mediated silver nanoparticles to exhibit enhanced antifungal activity in combination with antibiotics.20 Another report has shown that silver nanoparticles derived from Penicillium chrysogenum exhibited antifungal activity against a wide range of pathogenic strains, Candida tropicalis, Aspergillus niger, and Fusarium solani.60
POAgNPs Exhibited Potential Antioxidant against Synthetically Generated Free Radicals
DPPH Free Radical Scavenging Activity
The antioxidant activity of POAgNPs was evaluated against the synthetically generated DPPH free radicals (Figure 4A). The result showed that POAgNPs significantly inhibited the free radicals of DPPH with an EC50 value of 9.034 ± 0.449 μg/mL. The spectrophotometric observation at 517 nm showed a decrease in the absorbance with increasing concentration of POAgNPs that is attributed to the scavenging of free radicals. The observation suggests that POAgNPs exerted potential free radical scavenging even at a lower concentration. The scavenging ability of POAgNPs was observed to be very close to that of the well-established antioxidant agent ascorbic acid with an EC50 value of 10.5 ± 0.265 μg/mL (Table 2).
Figure 4.
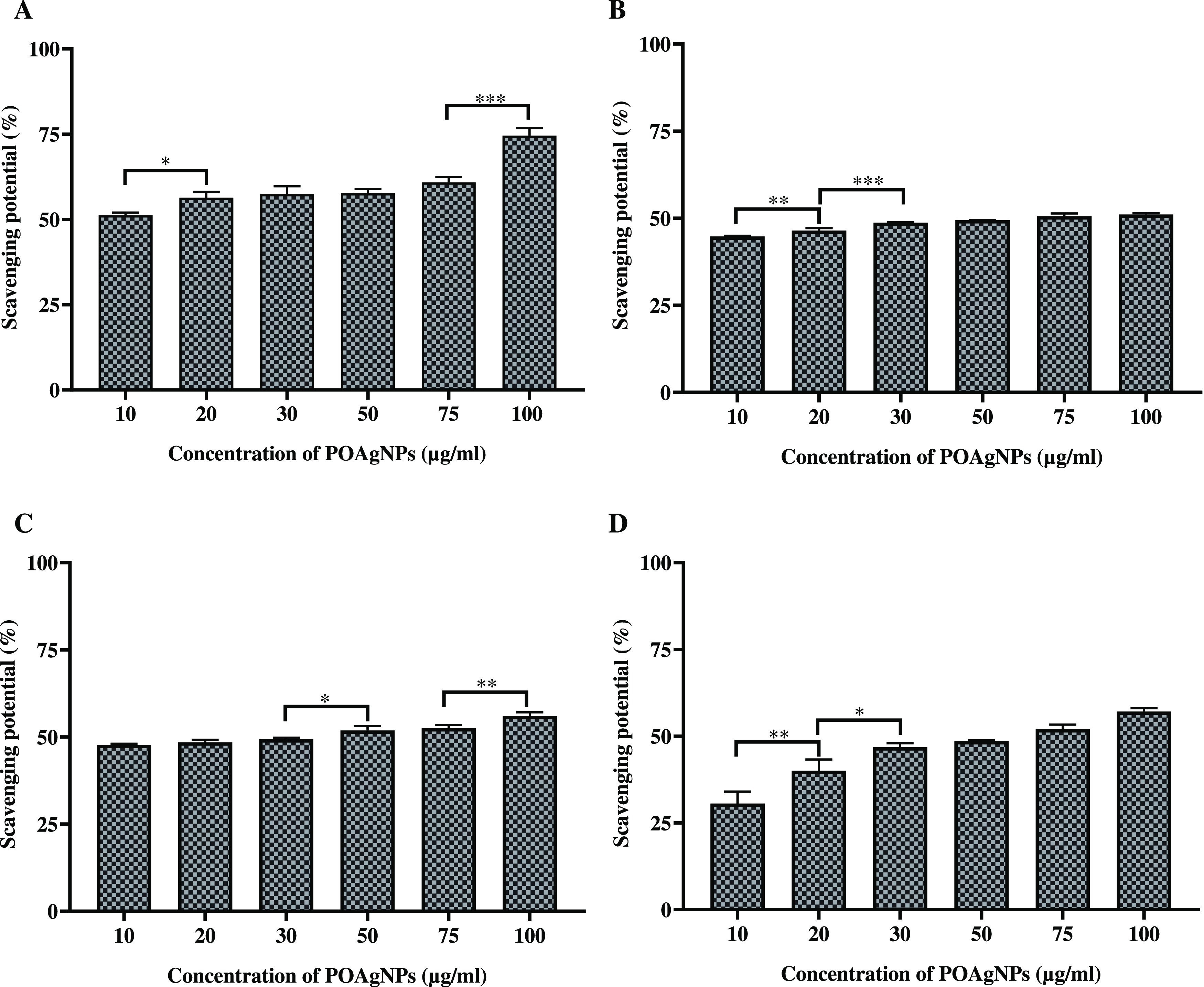
Antioxidant activity of POAgNPs against different antioxidant assays. (A) DPPH free radical scavenging activity. (B) Superoxide anion scavenging activity. (C) Hydroxyl radical scavenging activity. (D) Nitric oxide scavenging activity. All the experiments were performed in triplicate. The p-value was calculated by comparing means ± SD of the free radical scavenging potential (%), using one-way ANOVA followed by Tukey to determine statistical significance values, which are as follows: ***p ≤ 0.001; **p ≤ 0.002; and *p ≤ 0.033.
Table 2. Antioxidant and Cytotoxic Activity of POAgNPs with the Respective EC50 and IC50 Values.
| antioxidant
activity (EC50 value in μg/mL) |
cytotoxic
activity (IC50 value in μg/mL) |
||||||
|---|---|---|---|---|---|---|---|
| s. no. | samples | DPPH free radical scavenging assay | superoxide anion scavenging activity | hydroxyl radical scavenging assay | nitric oxide scavenging assay | MDA-MB-231 cells | MCF-7 cells |
| 1. | POAgNPs | 9.034 ± 0.449 | 56.378 ± 1.137 | 34.094 ± 1.944 | 61.219 ± 0.69 | 20.080 ± 0.761 | 40.038 ± 1.022 |
| 2. | ascorbic acid (positive control) | 10.5 ± 0.265 | 31.67 ± 2.867 | 19.64 ± 0.988 | 22.55 ± 0.876 | ||
| 3. | tamoxifen (positive control) | 24.572 ± 0.249 | 33.214 ± 0.824 | ||||
Superoxide Anion Scavenging Activity
The superoxide anion scavenging potential was evaluated by determining the scavenging ability of POAgNPs against superoxide anion radicals. POAgNPs exhibited potential superoxide anion scavenging activity with an EC50 value of 56.378 ± 1.137 μg/mL (Table 2). POAgNPs induced the scavenging activity against synthetically generated free radicals in a dose-dependent manner (Figure 4B). The positive control ascorbic acid showed scavenging potential against superoxide anion radicals with an EC50 value of 31.67 ± 2.867 μg/mL, which is approximately 1.5 time lower than that of POAgNPs. In the present study, the superoxide anion scavenging activity of POAgNPs has been demonstrated through the coupled reaction of PMS/NADH that is measured spectrophotometrically using NBT, which in contact with PMS/NADH forms purple-colored formazan. The oxidation of NADH in the NADH/PMS system causes reduction of NBT, which indicates the scavenging activity of POAgNPs.
Hydroxyl Radical Scavenging Activity
The assay performed in the study showed the significant scavenging potential of POAgNPs. The chemically driven generation of hydroxyl radicals was achieved through FeCl3-EDTA-H2O2, and the scavenging of radicals through POAgNPs was assessed spectrophotometrically. The scavenging potential of POAgNPs against hydroxyl radicals was in a concentration-dependent manner (Figure 4C). The activity was calculated in terms of percentage free radical scavenging potential, and the EC50 value was found to be in the significant range of 34.094 ± 1.944 μg/mL (Table 2). The EC50 value calculated for the positive control was observed to be 19.64 ± 0.988 μg/mL.
Nitric Oxide Radical Scavenging Activity
In the present study, nitric oxide free radical scavenging assay has been performed to evaluate the potential of mycogenic silver nanoparticles against synthetically generated free radicals, and significant results have been obtained. The production of nitric oxide was reduced through the activity of POAgNPs against reactive oxygen species that play an important role in elevating the levels of nitrite ions in the reaction. The quantification of nitrite ions produced was done using the Greiss reagent, and the activity of POAgNPs was observed to be in a concentration-dependent manner (Figure 4D). The EC50 value was found to be in the significant range of 61.219 ± 0.69 μg/mL (Table 2). The EC50 value for the activity of the positive control against nitric oxide radicals was found to be 22.55 ± 0.876 μg/mL.
Reactive species of oxygen and nitrogen produced through many biological reactions exert deleterious impacts on human health and contribute to various diseases. The superoxide anion radical is produced particularly in the electron-rich aerobic environment and poses serious damage to cellular components, leading to tissue damage and occurrence of several diseases such as inflammatory diseases, cancer, cardiovascular, atherosclerosis, dementia, and other pathologies. The antioxidants have the ability to neutralize the detrimental effect of free radicals by donating them electrons and thereby terminate the chain reaction and protect the vital organs. Fungal endophytes have been reported to exhibit pharmaceutically important bioactive compounds belonging to classes such as polyphenols, flavonoids, terpenoids, alkaloids, tannins, and steroids.10 Flavonoids belong to the phenolic group with potential antioxidant activities.61 Phenols have been reported to exhibit the most effective oxidative properties and therefore can be related to their antioxidant nature.62 Many reports are available, which have witnessed the correlation of the phenolic content and antioxidant property.23,63 Our previous findings have also suggested the potential antioxidant activity of P. oxalicum against chemically generated radicals and therefore validate the antioxidant activity of POAgNPs.64 Reports have suggested that mycogenic silver nanoparticles fabricated using the fungi Aspergillus versicolor and Penicillium citrinum showed potential free radical scavenging activity.65,66 Biogenically synthesized silver nanoparticles using the endophytic fungus Alternaria alternata have shown potential antioxidant activity against DPPH and nitric oxide free radicals.67 Fungus-mediated green silver nanoparticles have been reported to show potential antioxidant activity against superoxide free radicals.68
POAgNPs Exhibited Antiproliferative Activity against Breast Cancer Cells
The synthesized green silver nanoparticles have been studied for their antiproliferative activity against ER-positive MCF-7 and the triple-negative breast cancer cell line MDA-MB-231. The potential cytotoxic activity of POAgNPs was observed against both cell lines. The activity was increased as the concentration increased from 5 to 200 μg/mL. POAgNPs showed significant cytotoxic potential against the MDA-MB-231 cell line as compared to that of the positive control, tamoxifen. The spectrophotometric analysis was calculated in terms of percentage cell viability, and the IC50 value against the MDA-MB-231 cell line was observed to be 20.080 ± 0.761 and 24.572 ± 0.249 μg/mL for POAgNPs and tamoxifen, respectively. For MCF-7 cells, the IC50 value was observed to be 40.038 ± 1.022 and 33.214 ± 0.824 μg/mL for POAgNPs and tamoxifen, respectively. The viability of cells was significantly decreased at the higher dose of POAgNPs, and morphological changes were observed. A higher number of apoptotic cells were observed at higher concentrations of the drug, whereas live healthy cells were high in number at lower concentrations. The respective IC50 values observed for both cell lines are depicted in Table 2, and graphical representation for percentage cell viability at different concentrations of the drug is shown in Figure 5. The biological activity of nanoparticles is based on their shape, morphology, size, stability, and aggregation. The cytotoxic activity of silver nanoparticles has been widely studied due to their potential effects on cellular uptake, penetration, and intracellular distribution. It has been shown in previous reports that AgNPs show enhanced activity due to their characteristic property of large surface-to-volume ratio, which get easily internalized into the cells and upon interaction with the cellular constituents negatively affect the cellular signaling pathways. The assumptions made on the activity of silver nanoparticles suggest the interaction of AgNPs with mitochondria and subsequent disruption of the electron transfer chain and thereby an increase in the ROS level.69,70 The oxidative stress caused by the AgNP-mediated generation of ROS also induces toxicity against cells. The bioactive compounds of fungal endophytes and AgNPs show synergistic effects, as suggested through our previous findings,64 and therefore show enhanced cytotoxicity. The results obtained thus validate the enhanced cytotoxic potential of POAgNPs against MDA-MB-231 cancer cells as compared to that of the positive control. The altered metabolism of cancer cells and high proliferation rate make them more vulnerable for cellular uptake.71 It has been postulated in a report that polyphenols exhibit cytotoxicity against nonhealthy cells.72 Moreover, the toxic effects of silver nanoparticles increase with an increase in the surface charge of nanoparticles. The positively charged nanoparticles effectively interact with the negatively charged cell surface and penetrate deeply into the tumor matrix, evenly distribute, and accumulate in tumor cells and due to their stability pose higher cytotoxic effects.73−75 Fungus-derived silver nanoparticles have been assessed in previous reports for their cytotoxic potential against breast cancer cell lines. Trichoderma atroviride-mediated silver nanoparticles have been reported to exhibit potential cytotoxicity against MDA-MB-231 cells.76 The extracellular synthesis of silver nanoparticles using the endophytic fungus Guignardia mangiferae has revealed significant cytotoxic effects against MCF-7 cells.77 The potential cytotoxic activity of Ganoderma sessiliforme-mediated silver nanoparticles has been reported against MCF-7 & MDA-MB-231 breast cancer cell lines.78 Syed et al.79 have shown in their reports the anticancer activity of mycogenic AgNPs synthesized using the fungus Humicola sp., against the MDA-MB-231 cell line.
Figure 5.
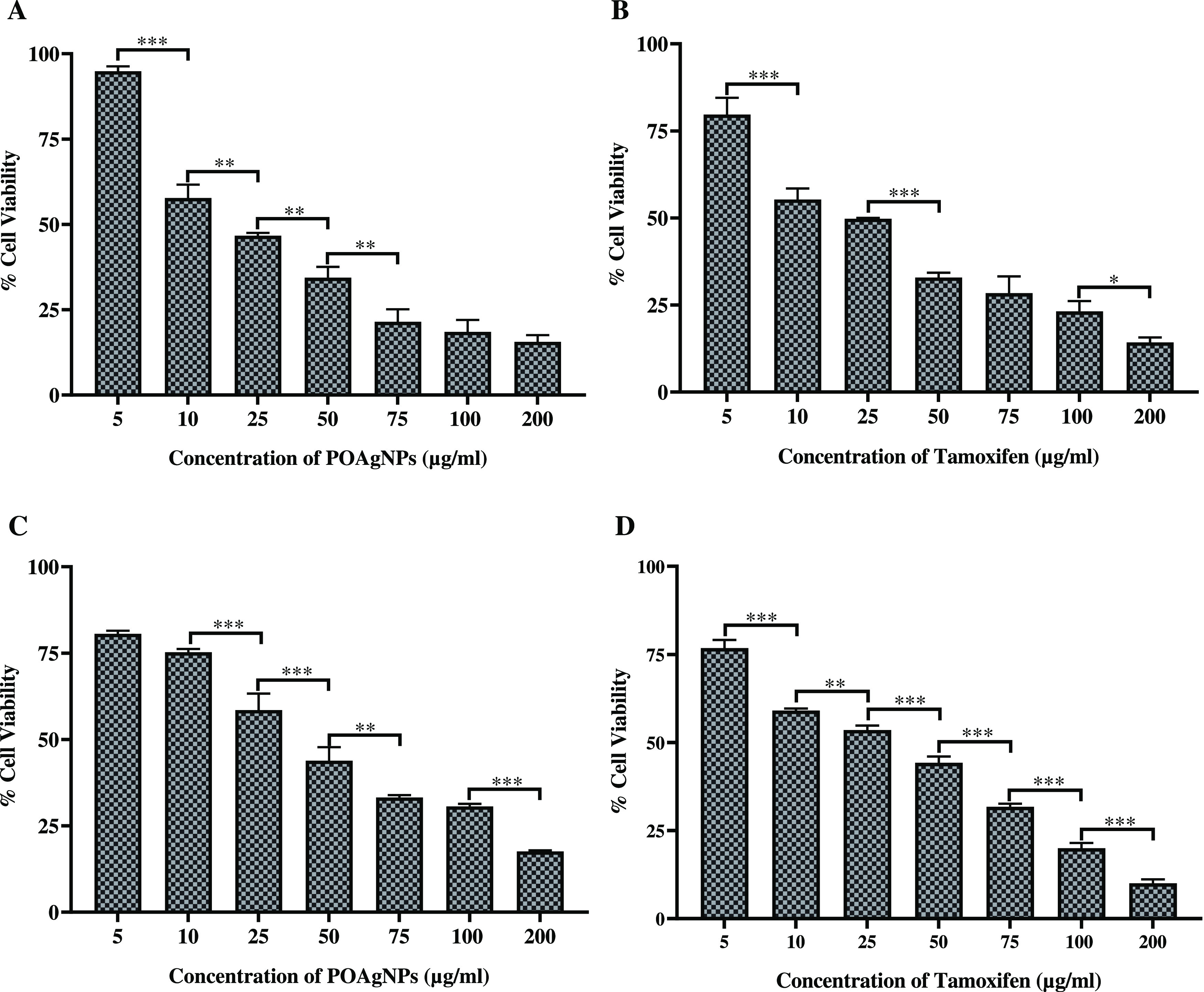
Cytotoxic activity of POAgNPs and positive control (tamoxifen) against breast cancer cell lines. (A) Cytotoxic effect of POAgNPs against the MDA-MB-231 cancer cell line. (B) Cytotoxic effect of tamoxifen against the MDA-MB-231 cancer cell line. (C) Cytotoxicity of POAgNPs against the MCF-7 cancer cell line. (D) Cytotoxicity of tamoxifen against the MCF-7 cancer cell line. All the experiments were performed in triplicate. The p-value was calculated by comparing means ± SD of percentage of cell viability of the cancer cell, using one-way ANOVA followed by Tukey to determine statistical significance values, which are as follows: ***p ≤ 0.001; **p ≤ 0.002; and *p ≤ 0.033.
POAgNPs Suppressed the Cell Migration of Breast Cancer Cells
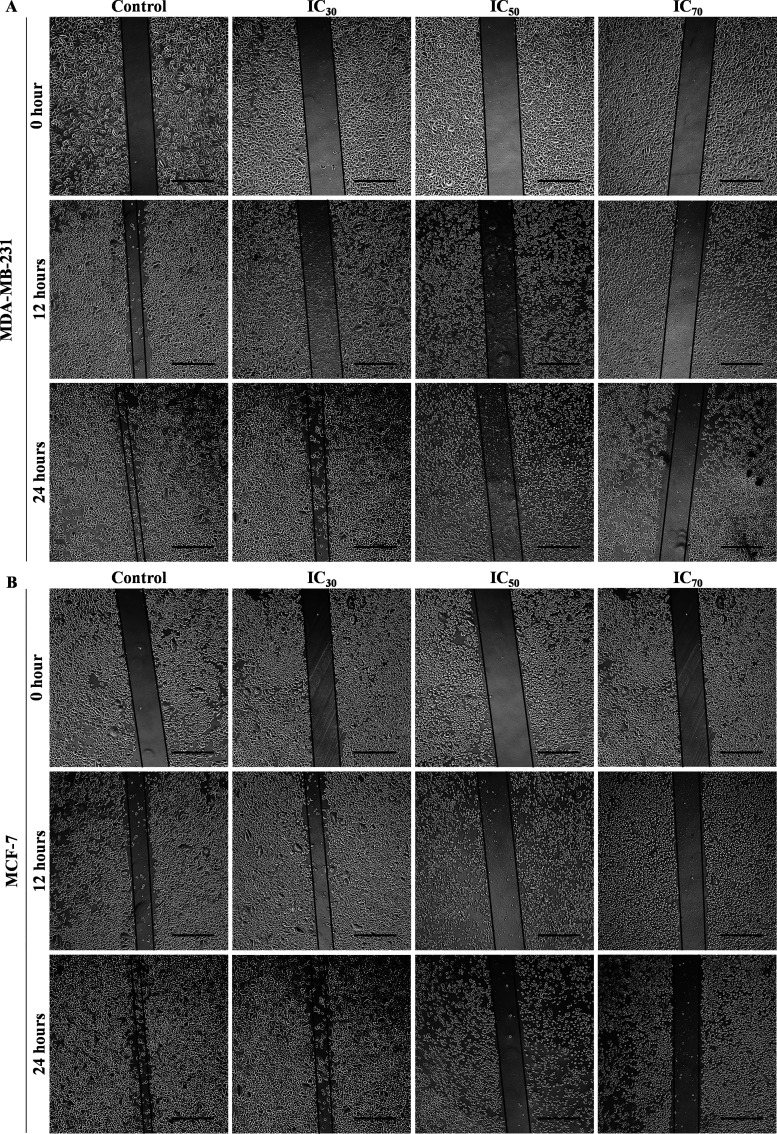
The findings showed that POAgNPs exhibit the potential to hinder cell migration in both the cell lines. The inhibition of wound healing capacity of cells was achieved in a concentration-dependent manner. The percentage wound closure was calculated using Image J software for both the cell lines at time intervals of 0, 12, and 24 h, and calculation was done with respect to the control. The cell migration was observed to be maximum in the control and decreased as the concentration of POAgNPs increased (Figure 6A,B). The % wound closure observed for MDA-MB-231 and MCF-7 cells (Figure S3) treated with POAgNPs for 12 h at the respective concentrations IC30, IC50, and IC70 was 13.87 ± 0.94, 10.56 ± 1.07, and 3.1 ± 1.49%, and 30.25 ± 1.85, 23.51 ± 1.13, and 9.10 ± 2.00%, respectively, as compared to that of the control group of 24.57 ± 2.12% (MDA-MB-231) and 46.04 ± 1.85% (MCF-7). After 24 h of treatment, the % wound closure at concentrations IC30, IC50, and IC70 was found to be 58.37 ± 2.83, 20.18 ± 0.8, and 5.16 ± 1.53%, respectively for MDA-MB-231 as compared to the control group with a % wound closure of 62.28 ± 1.06%. For MCF-7 cells, at 24 h, the % wound closure was observed as 54.185 ± 2.19, 35.35 ± 1.68, and 11.25 ± 1.41% at the respective concentrations of IC30, IC50, and IC70 as compared to the control with a value of 72.95 ± 1.03%. Several pathological processes such as metastatic dissemination, tumor invasion, and neoangiogenesis are related to the processes of cell migration. The invasive property of abnormal tumor cell migration involves signaling pathway activation that controls cytoskeletal dynamics and turnover of the cell–cell adhesions and cell matrix.80 The plasticity of cancer cells to switch between different migratory modes turns cells drug-resistant. The master regulators controlling the plasticity are targeted by chemotherapeutic drugs so as to impede tumor dissemination and metastasis. The actomyosin cytoskeleton has been reported as the minimum machinery that is required for the migratory plasticity and tumor cell invasion, obstructing which prevents cell migration and metastasis.81 The possible mechanism for achieving the suppression of cell migration could be the interaction with microtubules and altering their dynamics as presented in a previous report.82 The mechanism can be supported through previous reports that have reported the alteration in microtubule dynamics posed by chemotherapeutic drugs as a possible mechanism for inhibiting cell migration.83,84 In a previous report, the human lung carcinoma A549 cells treated with silver nanoparticles from the endophytic fungus Talaromyces purpureogenus have shown decreased ability of wound closure as compared to that of the control.30
Figure 6.
Inhibitory effect of different concentrations of POAgNPs (IC30, IC50, and IC70) on cell migration in breast cancer cell line. (A) MDA-MB-231 cancer cell line. (B) MCF-7 cancer cell line. Scale bar = 50 μm.
POAgNPs Induced Alteration of Nuclear Morphology in Breast Cancer Cells
The assessment of apoptosis and changes in the nuclear morphology can be performed using DAPI assay. In the assay, dose-dependent changes in the nuclear morphology of cancer cells were observed. The breast cancer cells MCF-7 and MDA-MB-231 were treated with IC30, IC50, and IC70 concentrations of POAgNPs, and after 24 h of incubation, significant change in the nuclear morphology was observed. The apoptotic cells were observed to be higher at the IC70 concentration of POAgNPs as compared to the IC30 concentration and control group. The control group showed an intact cell membrane with evenly shaped cells (Figure 7). while in the treated groups, a loose cell structure was observed, which indicates apoptotic events occurring in the cells induced by the drug. The preferential binding of DAPI to the minor grooves of adenine–thymine regions of DNA gives a fluorescence intensity of about 20-fold higher compared to that of unbound DAPI. The assay provides a low-cost and facile detection of apoptosis through observing the nuclear morphology. The successive stages of apoptotic events can be screened through assessing apoptosis-specific modulations of nuclear morphology. Different stages of nuclear morphological changes such as early changes (like chromatin condensation) and late stages of apoptosis (like nucleus shrinking and formation of apoptotic bodies) can be detected through DAPI assay that provides an advantage over apoptotic assays that are restricted to a single apoptotic stage.85 In one of the reports, human hepatocellular carcinoma, HCC, and human osteosarcoma, OS, treated with AgNPs synthesized using Penicillium shearii have shown morphological alterations that indicated the occurrence of apoptotic events.86 Another study has revealed mycosynthesized AgNP-treated MCF-7 cells exhibiting condensed or fragmented chromatin when stained with DAPI stain.87
Figure 7.
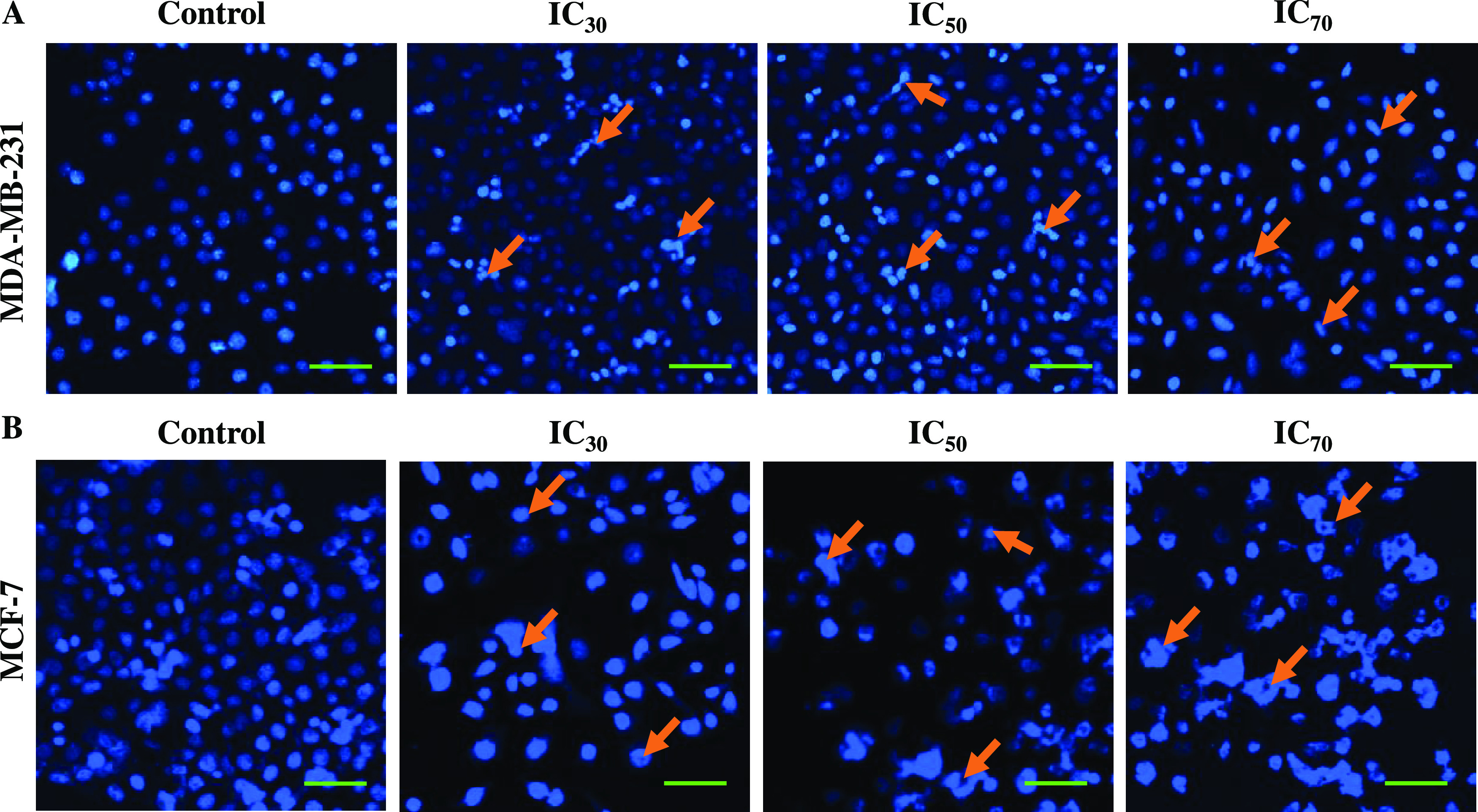
Fluorescence microscopy images of 4′,6-diamidino-2-phenylindole (DAPI)-stained breast cancer cells treated with POAgNPs in a dose-dependent manner for 24 h. (A) MDA-MB-231 cells. (B) MCF-7 cells. The images represent the morphological changes in the cells treated with IC30, IC50, and IC70 concentrations of POAgNPs as compared to those of control cells. The cells in the control group are observed to have normal rounded nuclei with normal blue color; however, the cells in the treated group are bright in color with condensed chromatin material and abnormal nuclei with an irregular cellular structure (marked through arrows in the figure) that clearly indicates apoptosis of cells. Scale bar = 10 μm.
POAgNPs Induced Differential Gene Expression in Breast Cancer Cells
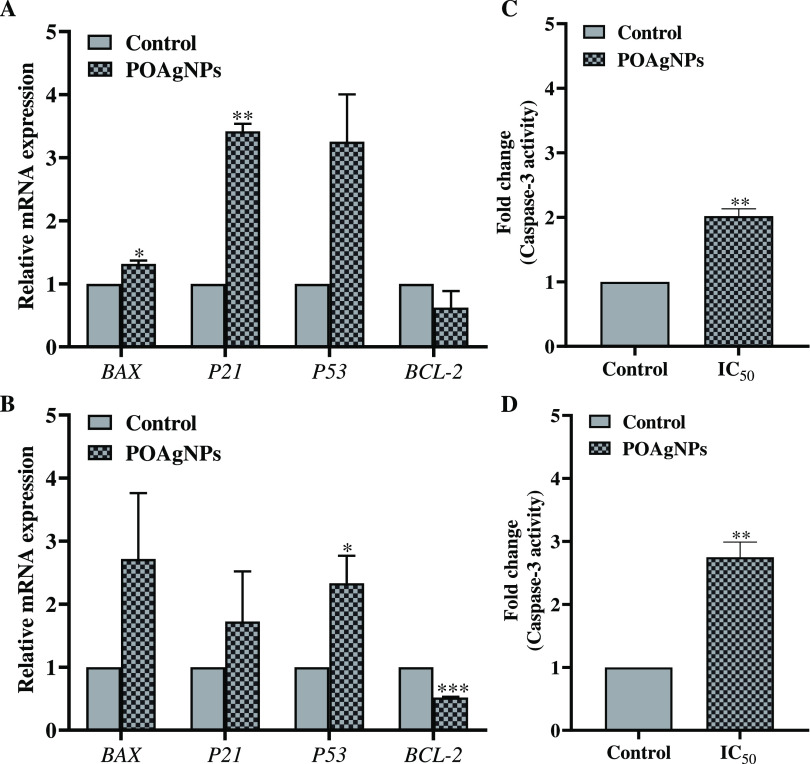
The cytotoxic effect of anticancer drugs is majorly achieved through the pathway of programmed cell death (PCD). The apoptotic pathway follows the simultaneous upregulation and downregulation of several apoptosis-related genes. The anticancer mechanism of POAgNPs was screened through the differential gene expression. The present study showed the upregulation of apoptotic genes, P21, P53, and BAX, whereas downregulation of the anti-apoptotic gene BCl-2 was observed for both cell lines (Figure 8A,B). The relative gene expression of the proapoptotic gene BAX in MDA-MB-231 showed a 1.315 ± 0.055-fold increase; however, gene responsible for the cell cycle arrest, P21 was found to be upregulated by 3.419 ± 0.120-fold. The gene expression for the tumor suppressor gene P53 was found to be in the order of 3.253 ± 0.752-fold increase as compared to that of the control. The relative gene expression for the anti-apoptotic gene BCL-2 revealed downregulation by 0.622 ± 0.264-fold as compared to that of the control. A similar pattern was observed for MCF-7 cells with the fold change of 2.718 ± 1.044, 1.725 ± 0.795, 2.331 ± 0.437, and 0.518 ± 0.013 for BAX, P21, P53, and BCL-2, respectively, as compared to the control. The transcription factors including p53 and p21 play a major role in cell cycle arrest and sequentially regulate the mitochondrial-dependent apoptotic pathway.88 p53 is considered as the sequence-specific and potential transcription factor that causes induction of cell cycle arrest and apoptosis.89 The transcriptional target of p53 is p21 that plays a role in cell cycle inhibition and therefore acts either as a positive or negative regulator of p53 for inducing apoptosis.90 p53 on interaction with Bax causes permeabilization of mitochondria. Bax-induced apoptosis can be a p53-dependent or p53-independent pathway. Both the genes, Bax and p53, cooperatively induce apoptosis in cancer cells.91 Bax is the proapoptotic member of the Bcl-2 family and is the key inducer of the mitochondrial-mediated intrinsic apoptotic pathway.92 Bcl-2 is involved in the suppression of cytochrome c from mitochondria and blocks caspase activation that is a core part of the intrinsic apoptotic pathway. The decreased level of the anti-apoptotic gene BCL-2 thus reveals the adoption of intrinsic apoptotic activity by POAgNPs through the suppression of anti-apoptotic gene expression. The altered gene expression of BCl-2 (downregulated) and BAX (upregulated) causes the release of cytochrome c from the mitochondria into the cytosol, promoting caspase activation (Caspase-3 and -9) and finally cell death. The regulation of apoptosis through the intrinsic pathways is crucially determined through the ratio of proapoptotic to anti-apoptotic Bcl-2 family genes. The present study showed the significant increase in the P53 expression, which showed its primal role in the apoptosis pathway. Previous findings show that p53 induces aminoterminal conformational alteration that indicates the release of cytochrome c from the mitochondria and sequentially driven process of mitochondrial-mediated intrinsic apoptotic pathway93,94 The present study thus gained evidence of the differential gene expression of POAgNP-treated cancer cells and intrinsic pathway adopted by POAgNPs to induce apoptosis in cancer cells. In a previous report, mycogenic silver nanoparticles synthesized using Penicillium aurantiogresium have been shown to exhibit cytotoxic effects against colon (HCT) and breast (MCF-7) human cancer cell lines and Vero (normal cell lines). Caspase-3-induced apoptosis was observed, which was further evidenced through the upregulation of Caspase-3 and Bax genes and downregulation of the Bcl-2 gene.95 The apoptotic mechanism adopted by Pleurotus ostreatu-derived AgNPs has shown a decreased level of Bcl-2 expression and increased levels of p53, Bax, and cytochrome c expression.96
Figure 8.
Representation of differential gene expressions and enhanced Caspase-3 activation induced after treatment of breast cancer cells with POAgNPs. Upregulation of the apoptotic genes BAX, P21, and P53 and downregulation of the anti-apoptotic gene BCL-2 after treatment with the IC50 concentration of POAgNPs (A). MDA-MB-231 cancer cell line (B). MCF-7 cancer cell line. Fold change in Caspase-3 activity after treatment of breast cancer cells with the IC50 concentration of POAgNPs (C). MDA-MB-231 cancer cell line (D). MCF-7 cancer cell line. All the experiments were performed in triplicate. The p-value was calculated by comparing means ± SD of relative mRNA expression using the unpaired Student t-test (*p < 0.05, **p < 0.01, ***p < 0.001) and fold change in caspase-3 activity, using one-way ANOVA followed by Tukey to determine statistical significance values, which are as follows: ***p ≤ 0.001; **p ≤ 0.002; and *p ≤ 0.033.
Expression of Caspase-3 Induced in Breast Cancer Cells Treated with POAgNPs
The assay performed in the study shows the significant fold increase in the Caspase-3 activity in both cell lines MDA-MB-231 and MCF-7 after treatment with POAgNPs (Figure 8C,D). The fold change observed was 2.018 ± 0.114 and 2.748 ± 0.240416 for MDA-MB-231 and MCF-7 cells, respectively. Caspase-3 is one of the effector caspases that execute apoptosis. They are crucial mediators of programmed cell death that catalyze the specific cleavage of many cellular proteins which are responsible for cell survival and maintenance.97,98 Caspase-3 is an intracellular cysteine protease that occurs as a proenzyme and gets activated during the cascade of events linked with apoptosis. The calorimetric assay is based on the cleavage of the substrate in the presence of Caspase-3, and dissociated pNA gives the measure of Caspase-3 activity. Previous reports have also shown similar findings, in which MCF-7 cells treated with AgNPs derived from the fungus Trichoderma viride showed Caspase-3 activation.99 The colon cancer cell line HT-29 treated with mycogenic silver nanoparticles synthesized using Aspergillus niger also showed an increased level of Caspase-3 activity as compared to that of the control.100 Another report showed that Penicillium shearii-mediated silver nanoparticles also triggered an increase in the level of Caspase-3 activity in human hepatocellular carcinoma, HCC, and human osteosarcoma, OS, treated with the IC50 concentration of mycogenic nanoparticles.86 The present study thus supports the Caspase-3-dependent mitochondrial apoptotic pathway.
Conclusions
The novel approach of biogenic synthesis of silver nanoparticles using the fungal endophytes P. oxalicum isolated from A. rohituka has shown wide-spectrum biological activity of POAgNPs. UV–vis analysis revealed the characteristic peak in the range of 400–450 nm obtained for the synthesized POAgNPs. The microscopic analysis showed the presence of monodispersed, sphere-shaped, nanosized particles and crystalline nature of nanoparticles, as revealed by XRD analysis. The biologically important active compounds present in the aqueous extract of P. oxalicum responsible for the reduction of silver were confirmed through FT-IR analysis. The synthesized POAgNPs were found to be effective against pathogenic bacterial and fungal strains and also possessed antioxidant activity against the free radicals generated through in vitro antioxidant assays. Furthermore, the in vitro cytotoxic potential of POAgNPs was also screened against the breast cancer cell lines MDA-MB-231 and MCF-7, and significant results were obtained. The inhibition of cell migration in the treated cells as compared to that in the control cells was observed through wound healing assay. The anticancer mechanism was confirmed through visualizing the apoptosis-specific modulations of the nuclear morphology using DAPI staining. The occurrence of apoptotic events was further confirmed through qRT-PCR, which revealed the expression of genes responsible for apoptosis, tumor suppression, and cell cycle arrest. Moreover, enhanced Caspase-3 activity in POAgNP-treated cells as compared to that of the control indicated adoption of a caspase-3-mediated mitochondrial apoptotic pathway. The study provided a facile, budget-friendly, and effective means to fabricate mycogenic silver nanoparticles. The extracellular synthesis method provides a great opportunity for the large-scale production of pharmaceutically important silver nanoparticles. The synthesis protocol shows a great prospect for the formation of potential AgNPs, as the characteristic property of AgNPs was observed at a AgNO3 concentration of 1 mM, which is of great interest as a low concentration of silver poses the least toxic effects to the health and environment. The synergistic effect of bioactive compounds and silver exhibits enhanced biological compatibility and potentially targets tumor cells. In this way, this report evidently shows scalability, applicability, and biocompatibility of the synthesized silver nanoparticles, and therefore, mycogenic silver nanoparticles can serve as nanodrugs and can be explored for their biomedical applications.
Materials and Methods
Fermentation and Biogenic Synthesis of AgNPs
The fermentation of the fungal endophyte P. oxalicum was done using 100 mL of potato dextrose broth and incubated for 7 days at 26 ± 2 °C in a shaking incubator at a speed of 100 rpm. After a week of incubation, the mycelial mat was harvested through filtration and washed twice with sterile double-distilled water to remove traces of medium components. Furthermore, 10 g of the fungal biomass was added to 100 mL of SDDW in a 250 mL Erlenmeyer flask and incubated for 24 h at 28 ± 4 °C in a shaking incubator at the speed of 100 rpm. The filtrate containing a cocktail of fungal bioactive compounds was filtered using Whatmann No.1 filter paper, retaining the aqueous solution and discarding the mycelial mass. The aqueous extract was then added to 100 mL of 1 mM silver nitrate and incubated under dark conditions for 24 h at 28 ± 4 °C. The aqueous extract was centrifuged at 15,000 rpm for 15 min twice or thrice followed by washing with nuclease-free water to separate the synthesized silver nanoparticle in the form of pellets. The supernatant was discarded, and the pellet was retained following drying in a hot air oven at 50 °C for 24 h and thereafter crushed for characterization. For subsequent experiments, nuclease-free water was used as a solvent to dissolve powdered silver nanoparticles.
Characterization and Evaluation of Physicochemical Properties of Fungal Endophyte-Mediated Green Silver Nanoparticles
UV–vis Absorption Spectra
The preliminary detection for the synthesis of silver nanoparticles was done on the basis of visible color change from light yellow to dark brown, which indicated the conversion of Ag+ ions into AgNPs. The synthesized nanoparticles were observed through an UV–Vis spectrophotometer (Shimadzu-UV–vis spectrophotometer; Kyoto, Japan) to measure the surface plasmon resonance of green nanoparticles, with the spectrum recorded between 200 and 800 nm.
Fourier Transform Infrared Spectroscopy Analysis
Further Fourier transform infrared spectroscopy (FT-IR) was performed to detect the functional groups corresponding to the fungal compounds responsible for the reduction of silver to fabricate silver nanoparticles. The analysis was performed in the range of 500–4000 cm–1 by the attenuated total reflectance (ATR) method using an IR spectrophotometer (PerkinElmer).
X-ray Diffraction Analysis
The crystalline nature of green nanoparticles was determined using X-Ray diffraction (XRD) using the model Bruker, Model-D8 Advance (Eco) at a voltage of 40 keV with Cu-Ka radiation and a wavelength of 1.5418 Å. The diffractograms were recorded over the angle range of 20–80. The lyophilized sample of silver nanoparticle was placed on a glass grid containing the silicon substrate for XRD analysis. The analysis was done using Origin 2022b software, and diffraction peaks were identified with the miller indices. The calculation of average size was done using software X’pert HighScore Plus.
Atomic Force Microscopy
The AFM analysis allows the determination of particle size, shape, surface morphology, and also the surface roughness of the synthesized silver nanoparticles. AFM offers 3D characterization of AgNPs with sub-nanometer resolution. Nanoparticle size distributions and variable geometry can be analyzed through AFM. To prepare a thin film of silver nanoparticles, POAgNPs, for AFM analysis, a homogeneous solution (diluted in methanol) of POAgNPs was dropped onto a sterile thin glass slide. The air-dried glass slides having a thin film were subjected to AFM analysis. Images were captured at a resolution of 256 × 256 pixels, with a constant nominal force of 40 N/m at room temperature and a cantilever frequency of 300 kHz. The surface roughness of POAgNPs in the 2D and 3D modes was estimated using Nova Px 3.2.5, NT-MDT spectrum software, whereas the measurement of particle size was done using Image J software.
Transmission Electron Microscopy
The analysis of size, shape, and morphology of the nanoparticles was done using a transmission electron microscope (Tecnai G2 20 S-TWIN TEM), which was operated at an accelerating voltage of 200 kV. The analysis was performed by taking a drop of POAgNP solution on a carbon grid followed by drying at room temperature; images were captured, and the size measurement was done using Image J software.
Field Emission Scanning Electron Microscopy
The surface morphology, size, and three-dimensional image of the synthesized silver nanoparticles were analyzed using a scanning electron microscope (Zeiss). The dried, crystalline nanoparticle was taken over carbon tape, and analysis was done with an accelerating voltage of 20 kV. The elemental composition of the mycogenic silver nanoparticle was confirmed through energy dispersive analysis of X-rays (EDAX).
Antimicrobial Activity
Antibacterial Assay
The mycosynthesized silver nanoparticles were evaluated for their antibacterial activity against the bacterial pathogens Escherichia coli (Gram negative) and Staphylococcus aureus (Gram positive) using the agar well diffusion method. For the assay, 30 mL of Muller Hinton agar was poured into the sterilized Petri plates and left for some time. Upon solidification of media, a bacterial lawn was prepared using 120 μL of freshly prepared bacterial broth culture. The assay was performed according to the previous report with some minor modifications.20 Agar wells of 8 mm diameter were formed, and 75 μL of different concentrations (25, 50, 100, and 200 μg/ mL) of the synthesized POAgNPs was loaded into the wells. The antibacterial activity of the synthesized POAgNPs was compared with streptomycin and vancomycin, positive controls against E. coli and S. aureus, respectively, and nuclease-free water as a negative control. After inoculation, the plates were incubated at 37 °C for 24 h, and the zone of inhibition was measured in mm.
Antifungal Assay
The antifungal activity of POAgNPs was assessed using methods described earlier with some minor modifications.20 The panel of pathogenic fungal strains, namely Aspergillus flavus, Aspergillus niger, Aspergillus luchuensis, and Penicillium albicans, was tested for the assay. A volume of 30 mL of sterilized potato dextrose agar was poured into the sterilized Petri plates and left until the medium solidifies. A volume of 120 μL of fresh broth culture of fungal strains was spread over the surface of the agar plate, and 8 mm of agar wells was formed. The wells were then loaded with 75 μL of mycosynthesized POAgNPs with different concentrations (25, 50, 100, and 200 μg/mL), and plates were then incubated for 24–48 h at 25 °C, and the zone of inhibition was then measured in mm. Amphotericin B was used as a positive control, and nuclease-free water was used as a negative control.
Determination of Minimum Inhibitory Concentration (MIC) of POAgNPs
The MIC value represents the minimum concentration of drug that inhibits the microbial growth as compared to that of the control. The MIC determination of POAgNPs against pathogenic bacterial strains, E. coli and S. aureus, and pathogenic fungal strains, A. flavus, A. niger, A. luchuensis, and P. albicans, was done using the broth dilution method as described in the guidelines of Clinical Laboratory Standards Institute (CLSI). For the test, an overnight-cultured bacterial inoculum was adjusted to the concentration of 2 × 105 CFU/mL in phosphate buffer saline (PBS), and for the fungal inoculum, spore suspension was used to adjust the concentration at 2 × 105 spores/mL using PBS. Using a 96-well microtiter plate, 100 μL of the Luria Bertani broth and potato dextrose broth for bacteria and fungi, respectively, was transferred to each well, and further 100 μL of the POAgNPs was added to one of the wells. Consequently, the test sample was serially diluted up to twofold in media as 100 μL of the test sample from the previous well was added to the second well, and then, 100 μL was again transferred from the second well to the third well, and this was followed for all the wells to be tested, leaving the last well as a negative control. Thereafter, 100 μL of the bacterial/fungal inoculum was added to each well with the control group containing media and microbial inoculum only. Eventually, after adding all the reaction components, the plate was incubated at 37 °C and 25 °C for bacteria and fungi, respectively. After incubation, the plates were observed for the turbidity, and minimum inhibitory concentrations (MIC25, MIC50, and MIC75) were calculated through recording absorbance at 600 nm using a microplate reader (Thermo Fisher Scientific).
Antioxidant Activity
DPPH Free Radical Scavenging Assay
The free radical scavenging activity of the synthesized POAgNPs was determined against stable DPPH• (2,2-diphenyl-2-picrylhydrazyl hydrate, SRL, India) spectrophotometrically.101 The free radical scavenging assay was performed using 50 μg/mL concentration of DPPH solution in methanol using the method described earlier.102 To the different concentrations of POAgNPs (10, 20, 30, 50, 75, and 100 μg/mL), 1 mL of freshly prepared DPPH solution was added. The reaction samples were incubated in a dark place for 30 min, and absorbance was recorded at 517 nm using an UV spectrophotometer (Shimadzu-UV–vis spectrophotometer; Kyoto, Japan). DPPH solution (50 μg/mL) was used as a control, and methanol was used as a blank. Different concentrations of L-ascorbic acid (SRL, India) (10–100 μg/mL) was used as a positive control. The calculation for the percentage of radical scavenging activity was measured using the formula
Superoxide Anion Scavenging Assay
The potential of POAgNPs to scavenge synthetically produced superoxide anions was evaluated using PMS–NADH-NBT. The reaction mixture containing varied concentrations of POAgNPs (10, 20, 30, 50, 75, and 100 μg/mL) were added with 1 mL of 150 μM nitroblue tetrazolium (NBT) solution (SRL, India) (dissolved in 1X phosphate buffer saline, pH 7.4) and 1 mL of 468 μM NADH solution (dissolved in 1× PBS, pH 7.4). The reaction was initiated with the addition of 100 μL of 60mM PMS (SRL, India) (dissolved in 1× PBS, pH 7.4); furthermore, the reaction mixture was incubated for 5 min at 25 °C. The activity of POAgNPs was assayed spectrophotometrically through the decreased reduction of NBT, inhibiting the formation of violet-colored formazan.103 Absorbance was recorded at 560 nm taking PMS–NADH-NBT as a control, methanol as a blank, and L-ascorbic acid at different concentrations (10–100 μg/mL) as a positive control. The percentage superoxide anion scavenging potential of POAgNPs was evaluated using the formula
Hydroxyl Radical Scavenging Assay
The scavenging potential of POAgNPs against the generated hydroxyl radicals was evaluated spectrophotometrically. Briefly, the reaction mixture was prepared using 3.75mM 2-deoxyribose (SRL, India), 1 mM H2O2 (Qualigens Fine Chemicals, India), 100 μM FeCl3 (Merck), and 100 mM EDTA. Different concentrations (10, 20, 30, 50, 75, and 100 μg/mL) of POAgNPs were added to the reaction mixture and incubated for 1 h at 37 °C. After incubation, 1 mL of 1% thiobarbituric acid (TBA) (SD Fine Chemicals, India) and 1 mL of 2% trichloro-acetic acid (TCA) (SRL, India) were added to the reaction mixture and incubated in a water bath for 20 min at 90 °C. The mixture was then left to cool, and absorbance was recorded at 417 nm using an UV–vis spectrophotometer (Shimadzu-UV–vis spectrophotometer; Kyoto, Japan). The reaction without a sample was used as a control, with methanol as a blank and l-ascorbic acid in varied concentrations (10–100 μg/mL) as a positive control.104 The calculation for the hydroxyl radical scavenging of POAgNPs was calculated using the following formula
Nitric Oxide Scavenging Assay
Briefly, different concentrations of POAgNPs (10, 20, 30, 50, 75, and 100 μg/mL) were added to 150 μL of sodium nitroprusside (SRL, India) (10 mM in 1× PBS, pH 7.4). The mixture was then incubated for 150 min. After incubation, 200 μL of Griess reagent (SRL, India) was added to the reaction mixture and left for 30 min. Furthermore, absorbance was measured for the chromophore that formed during diazotization of nitrite with sulfanilamide, and further coupling with 1-naphthylethyelenediamine dihydrochloride was observed at 546 nm using an UV spectrophotometer (Shimadzu-UV–vis spectrophotometer; Kyoto, Japan).105 Methanol was used as a blank, and L-ascorbic acid in varied concentrations (10–100 μg/mL) was used as a positive control. The percentage nitric oxide radical scavenging activity was calculated using the formula
Cell Viability Assay
Cell Growth and Maintenance
The breast cancer cell lines MDA-MB-231 and MCF-7 were grown in DMEM (Dulbecco′s modified Eagle medium) containing 10% fetal bovine serum and 1% antibiotic solution (penicillin/streptomycin) and were maintained at 37 °C in 5% CO2. The cells were grown until the monolayer adherent cells were observed with 80–90% confluency.106
The cell viability assay was done according to the method described earlier.107 The monolayer adherent cells were trypsinized on sub-confluency, and cells were then counted using trypan blue. An equal number of cells, 5 × 104 cells/well, were seeded in 96 wells and left overnight at 37 °C in a 5% CO2 incubator (Panasonic, Sakata, Japan) for cell adherence. Furthermore, treatment with different concentrations of POAgNPs (5, 10, 25, 50, 75, 100, and 200 μg/mL) was given to the cells and incubated for 24 h at 37 °C in a 5% CO2 incubator. After incubation, 20 μL of MTT (SRL) (5 mg/mL in 1X PBS, pH 7.4) was added in each well, and after incubation for 4 h, the plate was centrifuged for 20 min at 3000 rpm. The medium containing MTT was removed from the well, and the formazan crystals formed were dissolved with 100 μL of DMSO (100%) and mixed properly by pipetting up and down. Absorbance for the formation of violet-colored formazan crystals was read using a microplate reader (Thermo Fisher Scientific) at 570 nm. The cytotoxic effect of medium control containing only media, vehicle control (nuclease-free water), and positive control (tamoxifen with the same concentration as that of POAgNPs) was also checked. The calculation for the percentage cell viability was done using the formula
Wound Healing Assay
The wound healing assay is an in vitro technique for examining the cell migration or wound healing property of cancer cells. The cell migration assay is based on the creation of a cell-free area in a confluent monolayer through physical exclusion. The cell migration is induced for the exposure of the cell-free area, and the gap is filled. However, in the presence of an anticancer agent cell, migration or wound closure is inhibited. Upon 80–90% confluency, cells were harvested through trypsinization and were seeded at a density of 2 × 105 cells for MCF-7 and MDA-MB-231 and incubated for 24 h at 37 °C in a 5% CO2 incubator. Cells were grown to a complete monolayer, and then, the cell-free area was created with the breadth of the scratch maintained uniformly. Fresh medium was added to the well, and treatment with IC30, IC50, and IC70 concentration (Table S3) was given to MDA-MB-231 and MCF-7 cells. The migration of cells or wound closure was photographed over several hours, 0, 12, and 24 h. The percentage wound closure was calculated for the respective cell lines. Experiments were performed in triplicate, and the data were recorded and analyzed statistically.
Nuclear Staining with the DAPI Fluorescent Dye
The altered nuclear morphology of cancer cells treated with POAgNPs was evaluated using DAPI assay. DAPI is a fluorescent dye that is used for the quantitation of nuclei and to assess the gross cell morphology. It is associated with the minor groove of the dsDNA and preferably binds to the adenine–thymine cluster. For the assay, breast cancer cells (MDA-MB-231 and MCF-7) were seeded at a density of 2 × 105 cells/well for the control and treated groups. Once the cells get confluent, treatment with different concentrations of POAgNPs, IC30, IC50, and IC70 (Table S3), was provided. The cells were then incubated for 24 h at 37 °C in a 5% CO2 incubator. After incubation, cells were washed with 4% paraformaldehyde followed by phosphate buffer saline (PBS). Furthermore, permeabilization was done by adding 70% methanol to the cells and further incubated in −20 °C for 30 min. Again, the cells were washed with PBS, and staining was done using 1 μg/mL DAPI and left for 15 min, after which images were recorded using a fluorescence microscope.
Quantitative Real-Time PCR (qRT-PCR) for Assessing Differential Gene Expressions
The breast cancer cells, MDA-MB-231 and MCF-7, were seeded at a density of 2 × 105 cells per well. On attaining confluency, the cells were treated with the IC50 concentration of POAgNPs and incubated at 37 °C and 5% CO2 concentration for 24 h. After incubation, total mRNA isolation was done from the cells using the TRIzol reagent (TRI reagent, Invitrogen) as directed by the manufacturer’s protocol. The concentration and purity of isolated RNA was measured using a Nanodrop (Thermo Scientific). The purified RNA was then subjected to cDNA synthesis using a cDNA synthesis kit (Thermo Scientific) following the manufacturer’s protocol with some minor modifications.108 Furthermore, qRT-PCR was performed by following the standard guideline of MIQE109 using a Thermo Fisher Quantstudio 5 real-time PCR system and PowerUp SYBR Green Master Mix (Thermo). The gene expression was determined using different sets of apoptotic and anti-apoptotic genes, represented in Table S4 with their sequences. Mean Ct values were calculated and normalized to β-actin (internal control). The analysis was done through the comparison of the ΔΔCT value of the vehicle control with the POAgNP-treated group, and calculation of relative mRNA was done in terms of fold change using the 2–ΔΔCT method. Experiments were performed in triplicate.
Caspase-3 Activity Assay
Caspase-3 plays a significant role in the induction of cell apoptosis. During apoptosis, caspase-3 is cleaved into the active form of caspase-3 and induces apoptosis through a chain reaction. The caspase activity assay is a quantitative enzymatic assay that was performed according to the manufacturer’s protocol (Elabscience). The cancer cells (MDA-MB-231 and MCF-7) grown in DMEM media were treated with the IC50 concentration of POAgNPs. After 24 h of incubation, cell lysate was prepared, and cellular protein was collected after centrifugation at 12,000 rpm for 20 min at 4 °C. The control and treated samples of MDA-MB-231 and MCF-7 cells were then analyzed for caspase-3 activity with DEVD-pNA that acts as a caspase-3-specific substrate, and absorbance was measured after 24 h using a microplate reader (Thermo Fisher Scientific) at 405 nm. The reaction mixture without the sample was used as a blank, and calculation of the caspase activity was done using the formula
Statistical Analysis
The experiments of minimum inhibitory concentration determination, antioxidant activity cytotoxic activity, and caspase-3 activity were performed in triplicate (n = 3). Data are presented as mean ± S.D. in the histogram. For the statistical significance, one-way ANOVA (analysis of variance) followed by Tukey was performed using Graph Pad Prism 8.0.2 software, and mean ± S.D. values of all groups were compared. Statistical significance of qRT-PCR data was evaluated by comparing the means ± SEM of both control and treated groups by unpaired Student t-test (*p < 0.05, **p < 0.01, ***p < 0.001) using Graph Pad Prism 8.0.2 software.
Acknowledgments
P.G. would like to thank the Science and Engineering Research Board (SERB), India, for Junior Research Fellowship under Empowerment and Equity Opportunities for Excellence in Science (EMEQ) scheme. N.R. would like to thank the University Grants Commission, New Delhi, India, for Junior and Senior Research Fellowship. A.V. would like to thank the Council of Scientific and Industrial Research, New Delhi, India, for Junior Research Fellowship. R.K. acknowledges the internal funding from Banaras Hindu University, Varanasi, India, (under the Institute of Eminence Scheme). The corresponding author on behalf of all the authors thanks the Interdisciplinary School of Life Sciences (ISLS), Banaras Hindu University, Varanasi, India, Central Instrumental Facility (CIF), IIT (BHU), Varanasi, India, and Sophisticated Central Instrumentation Facility (SCIF), IIT Dharwad, India. The VG laboratory is funded by the Science and Engineering Research Board (SERB)-EMEQ grant (EEQ/2019/000025), University Grants Commission, New Delhi, India, and internal funding from Banaras Hindu University, Varanasi, India (under the Institute of Eminence Scheme).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c05605.
FTIR analysis of POAgNPs, representation of the calculated values for MIC25, MIC50, and MIC75 determined for POAgNPs and the positive control against pathogenic bacterial and fungal strains, IC30, IC50, and IC70 for both cancer cell line, list of primers with their sequences used in qRT-PCR analysis, antibacterial activity of POAgNPs represented through zone of inhibition against pathogenic bacterial strains, and representation of POAgNP-induced growth inhibition against pathogenic fungal strains
(PDF)
Author Contributions
∥ N.R. and A.V. contributed equally to this work. P.G.: data curation; formal analysis; investigation; methodology; writing—original draft; writing—review and editing; and visualization. N.R.: data curation; formal analysis; investigation; and writing—review and editing. A.V.: data curation; formal analysis; investigation; writing—review and editing. D.S.: FE-SEM. S.P.S.: FE-SEM. R.K.: writing—review and editing. S.K.S.: writing—review and editing. D.K.: writing—review and editing. V.G.: conceptualization; methodology; formal analysis; funding acquisition; investigation; project administration; resources; software; and and writing—review and editing.
The authors declare no competing financial interest.
Supplementary Material
References
- Blanco E.; Shen H.; Ferrari M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015, 33, 941–951. 10.1038/nbt.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M.; Fawcett D.; Sharma S.; Tripathy S. K.; Poinern G. E. J. Green synthesis of metallic nanoparticles via biological entities. Materials 2015, 8, 7278–7308. 10.3390/ma8115377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhah Ridzuan A.; Ibrahim S.; Karman S.; Ab Karim M. S.; Zaman W. S. W. K.; Khuen C. C. Study on Electrical Conductivity of Graphene Oxide Decorated with Silver Nanoparticle for Electrochemical Sensor Development. Int. J. Electrochem. Sci. 2021, 16, 210557. 10.20964/2021.05.03. [DOI] [Google Scholar]
- Yaqoob A. A.; Ahmad H.; Parveen T.; Ahmad A.; Oves M.; Ismail I. M.; Qari H. A.; Umar K.; Mohamad Ibrahim M. N. Recent advances in metal decorated nanomaterials and their various biological applications: a review. Front. Chem. 2020, 8, 341. 10.3389/fchem.2020.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim W.; Barnard R. T.; Blaskovich M.; Ziora Z. M. Antimicrobial silver in medicinal and consumer applications: a patent review of the past decade (2007–2017). Antibiotics 2018, 7, 93. 10.3390/antibiotics7040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman G.; Ali E.; Jabbar I.; Saleem A. Synthesis, characterization, antibacterial and cytotoxic effects of silver nanoparticles. Dig. J. Nanomater. Biostructures. 2014, 9, 787–796. [Google Scholar]
- Hawar S. N.; Al-Shmgani H. S.; Al-Kubaisi Z. A.; Sulaiman G. M.; Dewir Y. H.; Rikisahedew J. J. Green synthesis of silver nanoparticles from Alhagi graecorum leaf extract and evaluation of their cytotoxicity and antifungal activity. J. Nanomater. 2022, 2022, 1–8. 10.1155/2022/1058119. [DOI] [Google Scholar]
- Khane Y.; Benouis K.; Albukhaty S.; Sulaiman G. M.; Abomughaid M. M.; Al Ali A.; Aouf D.; Fenniche F.; Khane S.; Chaibi W. J. N.; et al. Green synthesis of silver nanoparticles using aqueous Citrus limon zest extract: Characterization and evaluation of their antioxidant and antimicrobial properties. Nanomaterials 2022, 12, 2013. 10.3390/nano12122013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H.; Ferlay J.; Siegel R. L.; Laversanne M.; Soerjomataram I.; Jemal A.; Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J. Clin. 2021, 71, 209–249. 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Rai N.; Kumari Keshri P.; Verma A.; Kamble S. C.; Mishra P.; Barik S.; Kumar Singh S.; Gautam V. Plant associated fungal endophytes as a source of natural bioactive compounds. Mycology 2021, 12, 139–159. 10.1080/21501203.2020.1870579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A.; Singh R. K.; Khare S.; Tripathi R.; Pandey R. K.; Singh A. K.; Gautam V.; Tripathi J. S.; Singh S. K. Characterization and evaluation of mycosterol secreted from endophytic strain of Gymnema sylvestre for inhibition of α-glucosidase activity. Sci. Rep. 2019, 9, 17302 10.1038/s41598-019-53227-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai N.; Gupta P.; Keshri P. K.; Verma A.; Mishra P.; Kumar D.; Kumar A.; Singh S. K.; Gautam V. Fungal Endophytes: an Accessible Source of Bioactive Compounds with Potential Anticancer Activity. Appl. Biochem. Biotechnol. 2022, 194, 3296–3319. 10.1007/s12010-022-03872-1. [DOI] [PubMed] [Google Scholar]
- Gupta P.; Verma A.; Rai N.; Singh A. K.; Singh S. K.; Kumar B.; Kumar R.; Gautam V. Mass Spectrometry-Based Technology and Workflows for Studying the Chemistry of Fungal Endophyte Derived Bioactive Compounds. ACS Chem. Biol. 2021, 16, 2068–2086. 10.1021/acschembio.1c00581. [DOI] [PubMed] [Google Scholar]
- Barik S.; Rai N.; Mishra P.; Singh S. K.; Gautam V. Bioinformatics: How it helps to boost modern biological research. Curr. Sci. 2020, 118, 698–699. [Google Scholar]
- Mandal D.; Bolander M. E.; Mukhopadhyay D.; Sarkar G.; Mukherjee P. The use of microorganisms for the formation of metal nanoparticles and their application. Appl. Microbiol. Biotechnol. 2006, 69, 485–492. 10.1007/s00253-005-0179-3. [DOI] [PubMed] [Google Scholar]
- Rahimi G.; Alizadeh F.; Khodavandi A. Mycosynthesis of silver nanoparticles from Candida albicans and its antibacterial activity against Escherichia coli and Staphylococcus aureus. Trop. J. Pharm. Res. 2016, 15, 371–375. 10.4314/tjpr.v15i2.21. [DOI] [Google Scholar]
- van den Hondel C. A. M. J. J.; Punt P. J.; van Gorcom R. F. Production of extracellular proteins by the filamentous fungus Aspergillus. Antonie van Leeuwenhoek 1992, 61, 153–160. 10.1007/BF00580623. [DOI] [PubMed] [Google Scholar]
- Ingle A.; Gade A.; Pierrat S.; Sonnichsen C.; Rai M. Mycosynthesis of silver nanoparticles using the fungus Fusarium acuminatum and its activity against some human pathogenic bacteria. Curr. Nanosci. 2008, 4, 141–144. 10.2174/157341308784340804. [DOI] [Google Scholar]
- Thakor R.; Mistry H.; Patel H.; Jhala D.; Parmar N.; Bariya H. Biogenic synthesis of silver nanoparticles mediated by the consortium comprising the marine fungal filtrates of Penicillium oxalicum and Fusarium hainanense along with their antimicrobial, antioxidant, larvicidal and anticancer potency. J. Appl. Microbiol. 2022, 133, 857–869. 10.1111/jam.15611. [DOI] [PubMed] [Google Scholar]
- Baker A.; Iram S.; Syed A.; Elgorban A. M.; Al-Falih A. M.; Bahkali A. H.; Khan M. S.; Kim J. Potentially bioactive fungus mediated silver nanoparticles. Nanomaterials 2021, 11, 3227. 10.3390/nano11123227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha Z. K.; Hawar S. N.; Sulaiman G. M. Extracellular biosynthesis of silver nanoparticles from Penicillium italicum and its antioxidant, antimicrobial and cytotoxicity activities. Biotechnol. Lett. 2019, 41, 899–914. 10.1007/s10529-019-02699-x. [DOI] [PubMed] [Google Scholar]
- Majeed S.; Abdullah M. S. b.; Nanda A.; Ansari M. T. J. J. o. T. U. f. S. In vitro study of the antibacterial and anticancer activities of silver nanoparticles synthesized from Penicillium brevicompactum (MTCC-1999). J. Taibah Univ. Sci. 2016, 10, 614–620. 10.1016/j.jtusci.2016.02.010. [DOI] [Google Scholar]
- Rai N.; Keshri P. K.; Gupta P.; Verma A.; Kamble S. C.; Singh S. K.; Gautam V. Bioprospecting of fungal endophytes from Oroxylum indicum (L.) Kurz with antioxidant and cytotoxic activity. PLoS One 2022, 17, e0264673 10.1371/journal.pone.0264673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabadi H.; Honary S. Biofabrication of gold and silver nanoparticles for pharmaceutical applications. Pharm. Biomed. Res. 2016, 2, 1–7. 10.18869/acadpub.pbr.2.1.1. [DOI] [Google Scholar]
- Mistry H.; Thakor R.; Patil C.; Trivedi J.; Bariya H. Biogenically proficient synthesis and characterization of silver nanoparticles employing marine procured fungi Aspergillus brunneoviolaceus along with their antibacterial and antioxidative potency. Biotechnol. Lett. 2021, 43, 307–316. 10.1007/s10529-020-03008-7. [DOI] [PubMed] [Google Scholar]
- Danagoudar A.; Pratap G.; Shantaram M.; Ghosh K.; Kanade S. R.; Sannegowda L. K.; Joshi C. G. Antioxidant, cytotoxic and anti-choline esterase activity of green silver nanoparticles synthesized using Aspergillus austroafricanus CGJ-B3 (endophytic fungus). Anal. Chem. Lett. 2021, 11, 15–28. 10.1080/22297928.2021.1883477. [DOI] [Google Scholar]
- Zomorodian K.; Pourshahid S.; Sadatsharifi A.; Mehryar P.; Pakshir K.; Rahimi M. J.; Arabi Monfared A. Biosynthesis and characterization of silver nanoparticles by Aspergillus species. Biomed Res. Int. 2016, 2016, 1–6. 10.1155/2016/5435397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A.; Mukherjee P.; Senapati S.; Mandal D.; Khan M. I.; Kumar R.; Sastry M. Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium oxysporum. Colloids Surf. B 2003, 28, 313–318. 10.1016/S0927-7765(02)00174-1. [DOI] [Google Scholar]
- Ashraf H.; Anjum T.; Riaz S.; Naseem S. Microwave-assisted green synthesis and characterization of silver nanoparticles using Melia azedarach for the management of Fusarium wilt in tomato. Front. Microbiol. 2020, 11, 238. 10.3389/fmicb.2020.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akther T.; Mathipi V.; Kumar N. S.; Davoodbasha M.; Srinivasan H. Fungal-mediated synthesis of pharmaceutically active silver nanoparticles and anticancer property against A549 cells through apoptosis. Environ. Sci. Pollut. Res. 2019, 26, 13649–13657. 10.1007/s11356-019-04718-w. [DOI] [PubMed] [Google Scholar]
- Joshi S. R.; Devi L. S. Ultrastructure, Ultrastructures of silver nanoparticles biosynthesized using endophytic fungi. J. Microsc. Ultrastruct. 2015, 3, 29. 10.1016/j.jmau.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A.; Arya A.; Chundawat T. S. Green synthesis of silver nanoparticles using green alga (Chlorella vulgaris) and its application for synthesis of quinolines derivatives. Synth. Commun. 2019, 49, 1926–1937. 10.1080/00397911.2019.1610776. [DOI] [Google Scholar]
- Gole A.; Dash C.; Ramakrishnan V.; Sainkar S.; Mandale A.; Rao M.; Sastry M. Pepsin– gold colloid conjugates: preparation, characterization, and enzymatic activity. Langmuir 2001, 17, 1674–1679. 10.1021/la001164w. [DOI] [Google Scholar]
- Mandal S.; Phadtare S.; Sastry M. Interfacing biology with nanoparticles. Curr. Appl. Phys. 2005, 5, 118–127. 10.1016/j.cap.2004.06.006. [DOI] [Google Scholar]
- Balaji D.; Basavaraja S.; Deshpande R.; Mahesh D. B.; Prabhakar B.; Venkataraman A. Extracellular biosynthesis of functionalized silver nanoparticles by strains of Cladosporium cladosporioides fungus. Colloids Surf. B 2009, 68, 88–92. 10.1016/j.colsurfb.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Selvi K. V.; Sivakumar T. Isolation and characterization of silver nanoparticles from Fusarium oxysporum. Int. J. Curr. Microbiol. Appl. Sci. 2012, 1, 56–62. [Google Scholar]
- Shende S.; Gade A.; Rai M. Large-scale synthesis and antibacterial activity of fungal-derived silver nanoparticles. Environ. Chem. Lett. 2017, 15, 427–434. 10.1007/s10311-016-0599-6. [DOI] [Google Scholar]
- Vigneshwaran N.; Ashtaputre N.; Varadarajan P.; Nachane R.; Paralikar K.; Balasubramanya R. Biological synthesis of silver nanoparticles using the fungus Aspergillus flavus. Mater. Lett. 2007, 61, 1413–1418. 10.1016/j.matlet.2006.07.042. [DOI] [Google Scholar]
- Krishnan V.; Bupesh G.; Manikandan E.; Thanigai A.; Magesh S.; Kalyanaraman R.; Maaza M. Green synthesis of silver nanoparticles using Piper nigrum concoction and its anticancer activity against MCF-7 and Hep-2 cell lines. J. Antimicro. 2016, 2, 2472. 10.4172/2472-1212.1000123. [DOI] [Google Scholar]
- Suvith V.; Philip D. Catalytic degradation of methylene blue using biosynthesized gold and silver nanoparticles. Spectrochim. Acta, Part A 2014, 118, 526–532. 10.1016/j.saa.2013.09.016. [DOI] [PubMed] [Google Scholar]
- Jeeva K.; Thiyagarajan M.; Elangovan V.; Geetha N.; Venkatachalam P. Caesalpinia coriaria leaf extracts mediated biosynthesis of metallic silver nanoparticles and their antibacterial activity against clinically isolated pathogens. Ind. Crops Prod. 2014, 52, 714–720. 10.1016/j.indcrop.2013.11.037. [DOI] [Google Scholar]
- Musarrat J.; Dwivedi S.; Singh B. R.; Al-Khedhairy A. A.; Azam A.; Naqvi A. Production of antimicrobial silver nanoparticles in water extracts of the fungus Amylomyces rouxii strain KSU-09. Bioresour. Technol. 2010, 101, 8772–8776. 10.1016/j.biortech.2010.06.065. [DOI] [PubMed] [Google Scholar]
- Singh T.; Jyoti K.; Patnaik A.; Singh A.; Chauhan R.; Chandel S. Biosynthesis, characterization and antibacterial activity of silver nanoparticles using an endophytic fungal supernatant of Raphanus sativus. J. Genet. Eng. Biotechnol. 2017, 15, 31–39. 10.1016/j.jgeb.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda A.; Nayak B.; Krishnamoorthy M. Antimicrobial properties of biogenic silver nanoparticles synthesized from phylloplane fungus, Aspergillus tamarii. Biocatal. Agric. Biotechnol. 2018, 16, 225–228. 10.1016/j.bcab.2018.08.002. [DOI] [Google Scholar]
- Vijayan S.; Divya K.; George T. K.; Jisha M. Biogenic synthesis of silver nanoparticles using endophytic fungi Fusarium oxysporum isolated from Withania somnifera (L.), its antibacterial and cytotoxic activity. J. Bionanosci. 2016, 10, 369–376. 10.1166/jbns.2016.1390. [DOI] [Google Scholar]
- Du L.; Xu Q.; Huang M.; Xian L.; Feng J.-X. Synthesis of small silver nanoparticles under light radiation by fungus Penicillium oxalicum and its application for the catalytic reduction of methylene blue. Mater. Chem. Phys. 2015, 160, 40–47. 10.1016/j.matchemphys.2015.04.003. [DOI] [Google Scholar]
- Tyagi S.; Tyagi P. K.; Gola D.; Chauhan N.; Bharti R. K. Extracellular synthesis of silver nanoparticles using entomopathogenic fungus: characterization and antibacterial potential. SN Appl. Sci. 2019, 1, 1545. 10.1007/s42452-019-1593-y. [DOI] [Google Scholar]
- Devi L. S.; Joshi S. R. Evaluation of the antimicrobial potency of silver nanoparticles biosynthesized by using an endophytic fungus, Cryptosporiopsis ericae PS4. J. Microbiol. 2014, 52, 667–674. 10.1007/s12275-014-4113-1. [DOI] [PubMed] [Google Scholar]
- Pereira L.; Dias N.; Carvalho J.; Fernandes S.; Santos C.; Lima N. J. J. o. a. m. Synthesis, characterization and antifungal activity of chemically and fungal-produced silver nanoparticles against T richophyton rubrum. J. Appl. Microbiol. 2014, 117, 1601–1613. 10.1111/jam.12652. [DOI] [PubMed] [Google Scholar]
- Lara H. H.; Ayala-Núnez N. V.; Ixtepan Turrent L. d. C.; Rodríguez Padilla C. Bactericidal effect of silver nanoparticles against multidrug-resistant bacteria. World J. Microbiol. Biotechnol. 2010, 26, 615–621. 10.1007/s11274-009-0211-3. [DOI] [Google Scholar]
- Sondi I.; Salopek-Sondi B. Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria. J. Colloid Interface Sci. 2004, 275, 177–182. 10.1016/j.jcis.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Oves M.; Aslam M.; Rauf M. A.; Qayyum S.; Qari H. A.; Khan M. S.; Alam M. Z.; Tabrez S.; Pugazhendhi A.; Ismail I. M. Antimicrobial and anticancer activities of silver nanoparticles synthesized from the root hair extract of Phoenix dactylifera. Mater. Sci. Eng. C 2018, 89, 429–443. 10.1016/j.msec.2018.03.035. [DOI] [PubMed] [Google Scholar]
- Cho K.-H.; Park J.-E.; Osaka T.; Park S.-G. The study of antimicrobial activity and preservative effects of nanosilver ingredient. Electrochim. Acta 2005, 51, 956–960. 10.1016/j.electacta.2005.04.071. [DOI] [Google Scholar]
- Shahverdi A. R.; Fakhimi A.; Shahverdi H. R.; Minaian S. Synthesis and effect of silver nanoparticles on the antibacterial activity of different antibiotics against Staphylococcus aureus and Escherichia coli. Nanomedicine 2007, 3, 168–171. 10.1016/j.nano.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Feroze N.; Arshad B.; Younas M.; Afridi M. I.; Saqib S.; Ayaz A. Fungal mediated synthesis of silver nanoparticles and evaluation of antibacterial activity. Microsc. Res. Tech. 2020, 83, 72–80. 10.1002/jemt.23390. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan V. S.; Mudiam M. K. R.; Kumar M.; Dwivedi S. P.; Singh S. P.; Prasad T. Silver nanoparticles induced alterations in multiple cellular targets, which are critical for drug susceptibilities and pathogenicity in fungal pathogen (Candida albicans). Int. J. Nanomed. 2018, 13, 2647. 10.2147/IJN.S150648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.; Xue B.; Wang L.; Zhang Y.; Liu L.; Zhou Y. Fungus-mediated green synthesis of nano-silver using Aspergillus sydowii and its antifungal/antiproliferative activities. Sci. Rep. 2021, 11, 10356 10.1038/s41598-021-89854-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorzoni L.; de Paula e Silva A. C.; Marcos C. M.; Assato P. A.; de Melo W. C.; de Oliveira H. C.; Costa-Orlandi C. B.; Mendes-Giannini M. J.; Fusco-Almeida A. M. J. F. i. m. Antifungal therapy: new advances in the understanding and treatment of mycosis. Front. Microbiol. 2017, 08, 36. 10.3389/fmicb.2017.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K. B.; Bard A. J. Interaction of silver (I) ions with the respiratory chain of Escherichia coli: an electrochemical and scanning electrochemical microscopy study of the antimicrobial mechanism of micromolar Ag+. Biochemistry 2005, 44, 13214–13223. 10.1021/bi0508542. [DOI] [PubMed] [Google Scholar]
- Abd El Aty A. A.; Mohamed A. A.; Zohair M. M.; Soliman A. A. Statistically controlled biogenesis of silver nano-size by Penicillium chrysogenum MF318506 for biomedical application. Biocatal. Agric. Biotechnol. 2020, 25, 101592 10.1016/j.bcab.2020.101592. [DOI] [Google Scholar]
- Taruscio T. G.; Barney D. L.; Exon J. Content and profile of flavanoid and phenolic acid compounds in conjunction with the antioxidant capacity for a variety of northwest Vaccinium berries. J. Agric. Food Chem. 2004, 52, 3169–3176. 10.1021/jf0307595. [DOI] [PubMed] [Google Scholar]
- Moskovitz J.; Yim M. B.; Chock P. B. Free radicals and disease. Arch. Biochem. 2002, 397, 354–359. 10.1006/abbi.2001.2692. [DOI] [PubMed] [Google Scholar]
- Rai N.; Keshri P. K.; Gupta P.; Verma A.; Kamble S. C.; Singh S. K.; Gautam V. J. P. o. Bioprospecting of fungal endophytes from Oroxylum indicum (L.) Kurz with antioxidant and cytotoxic activity. PLoS One 2022, 17, e0264673 10.1371/journal.pone.0264673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A.; Gupta P.; Rai N.; Tiwari R. K.; Kumar A.; Salvi P.; Kamble S. C.; Singh S. K.; Gautam V. J. J. o. F. Assessment of biological activities of fungal endophytes derived bioactive compounds Isolated from Amoora rohituka. J. Fungi 2022, 8, 285. 10.3390/jof8030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netala V. R.; Kotakadi V. S.; Bobbu P.; Gaddam S. A.; Tartte V. Endophytic fungal isolate mediated biosynthesis of silver nanoparticles and their free radical scavenging activity and anti microbial studies. 3 Biotech 2016, 6, 132. 10.1007/s13205-016-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danagoudar A.; Pratap G.; Shantaram M.; Ghosh K.; Kanade S. R.; Joshi C. G. Characterization, cytotoxic and antioxidant potential of silver nanoparticles biosynthesised using endophytic fungus (Penicillium citrinum CGJ-C1). Mater. Today Commun. 2020, 25, 101385 10.1016/j.mtcomm.2020.101385. [DOI] [Google Scholar]
- Govindappa M.; Manasa D. J.; Vinaykiya V.; Dutta S.; Pawar R.; Raghavendra V. B.; et al. Screening of Antibacterial and Antioxidant Activity of Biogenically Synthesized Silver Nanoparticles from Alternaria alternata, Endophytic Fungus of Dendrophthoe falcata-a Parasitic Plant. Bionanoscience 2022, 12, 128–141. 10.1007/s12668-021-00932-4. [DOI] [Google Scholar]
- Madhanraj R.; Eyini M.; Balaji P. Antioxidant assay of gold and silver nanoparticles from edible basidiomycetes mushroom fungi. Free Radic. Antioxid. 2017, 7, 137–142. 10.5530/fra.2017.2.20. [DOI] [Google Scholar]
- Park E.-J.; Yi J.; Kim Y.; Choi K.; Park K. Silver nanoparticles induce cytotoxicity by a Trojan-horse type mechanism. Toxicol. in Vitro 2010, 24, 872–878. 10.1016/j.tiv.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Bressan E.; Ferroni L.; Gardin C.; Rigo C.; Stocchero M.; Vindigni V.; Cairns W.; Zavan B. Silver nanoparticles and mitochondrial interaction. Int. J. Dent. 2013, 2013, 1–8. 10.1155/2013/312747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns R. A.; Harris I. S.; Mak T. W. Regulation of cancer cell metabolism. Nat. Rev. Cancer 2011, 11, 85–95. 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Li L.; Tsao R.; Yang R.; Liu C.; Zhu H.; Young J. C. J. J. o. a.; Polyphenolic profiles and antioxidant activities of heartnut (Juglans ailanthifolia var. cordiformis) and Persian walnut (Juglans regia L.). J. Agric. Food Chem. 2006, 54, 8033–8040. 10.1021/jf0612171. [DOI] [PubMed] [Google Scholar]
- Foroozandeh P.; Aziz A. A. Insight into cellular uptake and intracellular trafficking of nanoparticles. Nanoscale Res. Lett. 2018, 13, 339. 10.1186/s11671-018-2728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auría-Soro C.; Nesma T.; Juanes-Velasco P.; Landeira-Viñuela A.; Fidalgo-Gomez H.; Acebes-Fernandez V.; Gongora R.; Almendral Parra M. J.; Manzano-Roman R.; Fuentes M. Interactions of nanoparticles and biosystems: microenvironment of nanoparticles and biomolecules in nanomedicine. Nanomaterials 2019, 9, 1365. 10.3390/nano9101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeevanandam J.; Barhoum A.; Chan Y. S.; Dufresne A.; Danquah M. K. Review on nanoparticles and nanostructured materials: history, sources, toxicity and regulations. Beilstein J. Nanotechnol. 2018, 9, 1050–1074. 10.3762/bjnano.9.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saravanakumar K.; Wang M.-H. Trichoderma based synthesis of anti-pathogenic silver nanoparticles and their characterization, antioxidant and cytotoxicity properties. Microb. Pathog. 2018, 114, 269–273. 10.1016/j.micpath.2017.12.005. [DOI] [PubMed] [Google Scholar]
- Balakumaran M.; Ramachandran R.; Kalaichelvan P. Exploitation of endophytic fungus, Guignardia mangiferae for extracellular synthesis of silver nanoparticles and their in vitro biological activities. Microbiol. Res. 2015, 178, 9–17. 10.1016/j.micres.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Mohanta Y. K.; Nayak D.; Biswas K.; Singdevsachan S. K.; Abd_Allah E. F.; Hashem A.; Alqarawi A. A.; Yadav D.; Mohanta T. K. Silver nanoparticles synthesized using wild mushroom show potential antimicrobial activities against food borne pathogens. Molecules 2018, 23, 655. 10.3390/molecules23030655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed A.; Saraswati S.; Kundu G. C.; Ahmad A. Biological synthesis of silver nanoparticles using the fungus Humicola sp. and evaluation of their cytoxicity using normal and cancer cell lines. Spectrochim. Acta, Part A 2013, 114, 144–147. 10.1016/j.saa.2013.05.030. [DOI] [PubMed] [Google Scholar]
- Friedl P.; Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 2011, 147, 992–1009. 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Pandya P.; Orgaz J. L.; Sanz-Moreno V. Modes of invasion during tumour dissemination. Mol. Oncol. 2017, 11, 5–27. 10.1002/1878-0261.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Decker C. C.; Zechner L.; Krstin S.; Wink M. In vitro wound healing of tumor cells: inhibition of cell migration by selected cytotoxic alkaloids. BMC Pharmacol. Toxicol. 2019, 20, 1–12. 10.1186/s40360-018-0284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.; Ganguly A.; Cabral F. Inhibition of cell migration and cell division correlates with distinct effects of microtubule inhibiting drugs. J. Biol. Chem. 2010, 285, 32242–32250. 10.1074/jbc.M110.160820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijman M. N.; van Nieuw Amerongen G. P.; Laurens N.; van Hinsbergh V. W.; Boven E. Microtubule-targeting agents inhibit angiogenesis at subtoxic concentrations, a process associated with inhibition of Rac1 and Cdc42 activity and changes in the endothelial cytoskeleton. Mol. Cancer Ther. 2006, 5, 2348–2357. 10.1158/1535-7163.MCT-06-0242. [DOI] [PubMed] [Google Scholar]
- Mandelkow R.; Guembel D.; Ahrend H.; Kaul A.; Zimmermann U.; Burchardt M.; Stope M. B. Detection and quantification of nuclear morphology changes in apoptotic cells by fluorescence microscopy and subsequent analysis of visualized fluorescent signals. Anticancer Res. 2017, 37, 2239–2244. 10.21873/anticanres.11560. [DOI] [PubMed] [Google Scholar]
- Fageria L.; Pareek V.; Dilip R. V.; Bhargava A.; Pasha S. S.; Laskar I. R.; Saini H.; Dash S.; Chowdhury R.; Panwar J. Biosynthesized protein-capped silver nanoparticles induce ros-dependent proapoptotic signals and prosurvival autophagy in cancer cells. ACS Omega 2017, 2, 1489–1504. 10.1021/acsomega.7b00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BalaKumaran M.; Ramachandran R.; Balashanmugam P.; Jagadeeswari S.; Kalaichelvan P. J. M. T. Comparative analysis of antifungal, antioxidant and cytotoxic activities of mycosynthesized silver nanoparticles and gold nanoparticles. Mater. Technol. 2022, 37, 411–421. 10.1080/10667857.2020.1854518. [DOI] [Google Scholar]
- Zhang Z.; Wang C.-Z.; Du G.-J.; Qi L.-W.; Calway T.; He T.-C.; Du W.; Yuan C.-S. Genistein induces G2/M cell cycle arrest and apoptosis via ATM/p53-dependent pathway in human colon cancer cells. Int. J. Oncol. 2013, 43, 289–296. 10.3892/ijo.2013.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B.; Lane D.; Levine A. J. Surfing the p53 network. Nature 2000, 408, 307–310. 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Abbas T.; Dutta A. J. N. R. C. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer 2009, 9, 400–414. 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu J.-J.; Dumont P.; Hafey M.; Murphy M. E.; George D. L. Mitochondrial p53 activates Bak and causes disruption of a Bak–Mcl1 complex. Nat. Cell Biol. 2004, 6, 443–450. 10.1038/ncb1123. [DOI] [PubMed] [Google Scholar]
- Jendrossek V. Targeting apoptosis pathways by Celecoxib in cancer. Cancer Lett. 2013, 332, 313–324. 10.1016/j.canlet.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Thangam R.; Sathuvan M.; Poongodi A.; Suresh V.; Pazhanichamy K.; Sivasubramanian S.; Kanipandian N.; Ganesan N.; Rengasamy R.; Thirumurugan R.; Kannan S. Activation of intrinsic apoptotic signaling pathway in cancer cells by Cymbopogon citratus polysaccharide fractions. Carbohydr. Polym. 2014, 107, 138–150. 10.1016/j.carbpol.2014.02.039. [DOI] [PubMed] [Google Scholar]
- Henry H.; Thomas A.; Shen Y.; White E. Regulation of the mitochondrial checkpoint in p53-mediated apoptosis confers resistance to cell death. Oncogene 2002, 21, 748–760. 10.1038/sj.onc.1205125. [DOI] [PubMed] [Google Scholar]
- Elshawy O. E.; Helmy E. A.; Rashed L. A. Preparation, characterization and in vitro evaluation of the antitumor activity of the biologically synthesized silver nanoparticles. Adv. Nanopart. 2016, 5, 149. 10.4236/anp.2016.52017. [DOI] [Google Scholar]
- Ismail A. F.; Ahmed M. M.; Salem A. A. Biosynthesis of silver nanoparticles using mushroom extracts: induction of apoptosis in HepG2 and MCF-7 cells via caspases stimulation and regulation of BAX and Bcl-2 gene expressions. J. Pharm. biomed. sci. 2015, 5, 1–9. [Google Scholar]
- Boyce M.; Degterev A.; Yuan J. Caspases: an ancient cellular sword of Damocles. Cell Death Differ. 2004, 11, 29–37. 10.1038/sj.cdd.4401339. [DOI] [PubMed] [Google Scholar]
- Crawford E. D.; Wells J. A. Caspase substrates and cellular remodeling. Annu. Rev. Biochem. 2011, 80, 1055–1087. 10.1146/annurev-biochem-061809-121639. [DOI] [PubMed] [Google Scholar]
- Kulandaivelu B.; Gothandam K.. Cytotoxic effect on cancerous cell lines by biologically synthesized silver nanoparticles Braz Arch. Biol. Technol. 2016, 59, 10.1590/1678-4324-2016150529. [DOI]
- Chengzheng W.; Jiazhi W.; Shuangjiang C.; Swamy M. K.; Sinniah U. R.; Akhtar M.; Umar A. Biogenic synthesis, characterization and evaluation of silver nanoparticles from Aspergillus niger JX556221 against human colon cancer cell line HT-29. J. Nanosci. Nanotechnol. 2018, 18, 3673–3681. 10.1166/jnn.2018.15364. [DOI] [PubMed] [Google Scholar]
- Contreras-Guzmán E. S.; Strong F. C. III Determination of tocopherols (Vitamin E) by reduction of cupric ion. J. AOAC Int. 1982, 65, 1215–1221. 10.1093/jaoac/65.5.1215. [DOI] [Google Scholar]
- Brand-Williams W.; Cuvelier M.-E.; Berset C. J. L.-F. s. Technology, Use of a free radical method to evaluate antioxidant activity. LWT - Food Sci. Technol. 1995, 28, 25–30. 10.1016/S0023-6438(95)80008-5. [DOI] [Google Scholar]
- Gülçin İ.; Alici H. A.; Cesur M. Determination of in vitro antioxidant and radical scavenging activities of propofol. Chem. Pharm. Bull. 2005, 53, 281–285. 10.1248/cpb.53.281. [DOI] [PubMed] [Google Scholar]
- Saeed N.; Khan M. R.; Shabbir M. Antioxidant activity, total phenolic and total flavonoid contents of whole plant extracts Torilis leptophylla L. BMC Complement Altern. Med. 2012, 12, 12. 10.1186/1472-6882-12-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcocci L.; Maguire J. J.; Droylefaix M. T.; Packer L. The nitric oxide-scavenging properties of Ginkgo biloba extract EGb 761. Biochem. Biophys. Res. Commun. 1994, 201, 748–755. 10.1006/bbrc.1994.1764. [DOI] [PubMed] [Google Scholar]
- Rose D. P.; Connolly J. M. Effects of fatty acids and inhibitors of eicosanoid synthesis on the growth of a human breast cancer cell line in culture. Cancer Res. 1990, 50, 7139–7144. [PubMed] [Google Scholar]
- Kaur N.; Arora D. S.; Kalia N.; Kaur M. Antibiofilm, antiproliferative, antioxidant and antimutagenic activities of an endophytic fungus Aspergillus fumigatus from Moringa oleifera. Mol. Biol. Rep. 2020, 47, 2901–2911. 10.1007/s11033-020-05394-7. [DOI] [PubMed] [Google Scholar]
- Gautam V.; Singh A.; Yadav S.; Singh S.; Kumar P.; Sarkar Das S.; Sarkar A. K. J. D. Conserved LBL1-ta-siRNA and miR165/166-RLD1/2 modules regulate root development in maize. Development 2021, 148, dev190033 10.1242/dev.190033. [DOI] [PubMed] [Google Scholar]
- Bustin S. A.; Benes V.; Garson J. A.; Hellemans J.; Huggett J.; Kubista M.; Mueller R.; Nolan T.; Pfaffl M. W.; Shipley G. L.; et al. The MIQE Guidelines: M inimum I nformation for Publication of Q uantitative Real-Time PCR E xperiments. Clin. Chem. 2009, 55, 611–622. 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.