Abstract
Resistance to colistin generated by the mcr-1 gene in Enterobacteriaceae is of great concern due to its efficient worldwide spread. Despite the fact that the Lima region has a third of the Peruvian population and more than half of the national pig and poultry production, there are no reports of the occurrence of the mcr-1 gene in Escherichia coli isolated from livestock. In the present work, we studied the occurrence of E. coli carrying the mcr-1 gene in chicken and pig farms in Lima between 2019 and 2020 and described the genomic context of the mcr-1 gene. We collected fecal samples from 15 farms in 4 provinces of Lima including the capital Lima Metropolitana and recovered 341 E. coli isolates. We found that 21.3% (42/197) and 12.5% (18/144) of the chicken and pig strains were mcr-1-positive by PCR, respectively. The whole genome sequencing of 14 mcr-1-positive isolates revealed diverse sequence types (e.g., ST48 and ST602) and the presence of other 38 genes that confer resistance to 10 different classes of antibiotics, including beta-lactamase blaCTX-M-55. The mcr-1 gene was located on diverse plasmids belonging to the IncI2 and IncHI1A:IncHI1B replicon types. A comparative analysis of the plasmids showed that they contained the mcr-1 gene within varied structures (mikB–mcr1–pap2, ISApl1–mcr1–pap2, and Tn6330). To the best of our knowledge, this is the first attempt to study the prevalence of the mcr-1 gene in livestock in Peru, revealing its high occurrence in pig and chicken farms. The genetic diversity of mcr-1-positive strains suggests a complex local epidemiology calling for a coordinated surveillance under the One-Health approach that includes animals, retail meat, farmers, hospitals and the environment to effectively detect and limit the spread of colistin-resistant bacteria.
Keywords: mcr-1 gene, colistin, chicken farm, pig farm, Escherichia coli
1. Introduction
Antimicrobial resistance (AMR) represents a growing threat to global health, principally in developing countries, where the high population density, poor medical care and unregulated use of antibiotics provide a favorable environment for the emergence and dissemination of multidrug-resistant bacteria (MDR) [1]. The increased prevalence of bacterial pathogens resistant to last-line antibiotics (carbapenems, colistin and tigecycline) raises serious concerns about our ability to treat infectious diseases in humans and animals [2]. Colistin is one of the last-resort treatments against multidrug-resistant strains of Enterobacterales, but its unregulated overuse as a therapeutic drug and growth promoter in pig and poultry farming has favored the emergence of colistin-resistant strains [3,4]. Since the discovery of the plasmid-encoded colistin resistance gene named mcr-1 in China in 2015 [5], this gene has been described in human, animal and environmental samples around the world [6]. The rapid spread of the mcr-1 gene by efficient horizontal transfer is driven by the IncI2, IncHI2 and IncX4 plasmids [4].
Escherichia coli is a commensal bacterium that inhabits the gastrointestinal tract of humans and animals and represents a major reservoir of antimicrobial resistance genes (ARGs), mostly acquired through horizontal gene transfer [7,8]. As a result of this capacity, E. coli has been commonly used as an indicator to monitor AMR in livestock, food and humans [9,10]. Even though they are normally commensal, certain strains of E. coli are associated with infections. For example, pathogenic E. coli may cause neonatal and postweaning diarrhea and edema in swine, while it may cause infections of the respiratory tract and soft tissues, resulting in colibacillosis, air sacculitis and cellulitis in chickens [11]. E. coli and Klebsiella pneumoniae carrying the mcr-1 gene have recently been reported in isolates from Peruvian hospitals [12,13,14,15], as well as in isolates from slaughtered chickens destined for human consumption [16]. Due to the recurrent detection of resistant enterobacteria in hospitals, the Peruvian government decreed in late 2019 the prohibition of the manufacture, sale and import of veterinary products containing the active compound of colistin (Polymyxin E).
The Lima region has a third of the Peruvian population and concentrates the largest animal production in the country. In fact, by 2020, 53% and 43% of the national poultry and pig production were concentrated in Lima, mainly in farms located on the outskirts of the capital Lima Metropolitana. Reporting the growing colistin resistance and mcr-1 prevalence in poultry and pig farms in low- and middle-income countries (LMIC) could be of high importance, but until now has been neglected in Peru [17]. Indeed, there is no information about the occurrence of mcr genes in livestock in Peru due to the absence of a systematic surveillance. In the present work, we investigated the occurrence of E. coli carrying the mcr-1 gene isolated from chicken and pig farms in Lima, Peru, from 2019 to 2020 and performed a genomic analysis of the isolates carrying mcr-1 to determine the genetic diversity and phylogenetic relationships of these isolates. Additionally, we characterized the virulence and ARGs profiles and explored the genomic context of the mcr-1 gene.
2. Methods
2.1. Sample Collection and Bacterial Culture
We collected 348 fecal samples from 8 chicken farms and 300 samples from 7 pig farms in 4 provinces of Lima, including the Peruvian capital, Lima Metropolitana, between 2019 and 2020 (Supplementary Table S1). All samples were later organized in pools. One pool was prepared for each chicken shed or pigpen combining 2 g of feces in a 50 mL tube; at least 5 pools per farm were obtained. Each fecal pool was diluted in buffered saline solution (0.9%) and thereafter plated onto MacConkey agar (BD Difco) and incubated overnight at 37 °C. At least five suspected colonies of E. coli (lactose-fermenting colonies, convex morphology and pinkish color appearance) were selected from each pool, inoculated on eosin methylene blue agar (EMBA) and incubated for 24 h at 37 °C, observing the growth of typical metallic green colonies. Finally, a 7-parameter biochemical test including Simmon’s citrate agar, lysine iron agar, triple sugar iron, motility–indole–lysine medium, sulfur indole motility medium, urea medium, red methyl and Voges Proskauer medium were used to confirm E. coli.
2.2. mcr-1 Gene Screening by PCR
All isolates were screened for the mcr-1 gene using the procedures described by Rebelo et al. (2018) [18]. Shortly, we used forward 5′-AGTCCGTTTGTTCTTGTGGC-3′ and reverse 5′-AGATCCTTGGTCTCGGCTTG-3′ primers with 2 µL of 10X PCR Buffer (100 mM KCl, 100 mM Tris-HCl, 20 mM MgCl2), 1.6 µL of deoxynucleotide triphosphate (dNTPs) 10 mM, 0.2 µL of DreamTaq 5 U/µL (Thermo Fisher Scientific, Waltham, MA, USA) and 2 µL of DNA in a final volume of 20 µL. The following condition were used: 1 cycle of initial denaturation at 94 °C for 15 min, followed by 25 cycles of denaturation at 94 °C for 30 s, annealing at 58 °C for 90 s and elongation at 72 °C for 60 s, with a final extension step of 72 °C for 10 min. We used the mcr-1-harboring E. coli CDC-AR-0346A reference strain (https://www.microbiologics.com/01259P) (accessed on 27 November 2022) as a positive control for all PCR runs.
2.3. Whole-Genome Sequencing and Assembly
From all mcr-1-positive E. coli, we selected 14 isolates for whole-genome sequencing (accession numbers and sequencing statistics are provided in Supplementary Table S2). The DNA was extracted from pure colonies using the PureLink™ Genomic DNA Kit (Invitrogen’, Cat. No K1820-02). DNA concentration was measured using the Qubit dsDNA HS assay (Invitrogen, Cat. No Q33230). Then 1 ng of DNA was used for Nextera XT library preparation and subsequent sequencing using 2 × 250 bp reads on the Illumina Miseq platform (Illumina, San Diego, CA, USA). The quality of the fastq files was evaluated with FastQC v0.11.9 [19], and the trimming of low-quality reads was performed with Trimmomatic v0.39 [20]. Finally, the assembly was performed with SPAdes v3.14.1 [21], and Prokka v1.14.6 [22] was used for genome annotation.
2.4. Sequence Analysis
We used the mlst v2.19.0 tool (https://github.com/tseemann/mlst) (accessed on 16 August 2022) and EzClermont v0.6.3 [23] to determine the multilocus sequence type (MLST) and phylogroup type, respectively. ARGs, virulence genes and plasmid replicon types were annotated using the Resfinder, VirulenceFinder and PlasmidFinder databases from the Center for Genomic Epidemiology with the ABRICATE v. 1.0.1 tool (https://github.com/tseemann/abricate) (accessed on 16 August 2022) using the following settings: a nucleotide identity of 80% and minimum coverage of 80%.
For the phylogenetic reconstruction, we used the following pipeline: Snippy v 4.6.0 (https://github.com/tseemann/snippy) (accessed on 15 July 2022) to generate the core-genome alignment of 14 E. coli genomes including the reference genome sequence E. coli K-12 (Genbank accession: NC_000913.3); given that recombination is widespread in bacteria genomes, Gubbins v3.2 [24] was used to detect and mask recombinant regions, and IQ-TREE v2.0 [25] to construct a maximum-likelihood tree based on a general time-reversible (GTR) nucleotide substitution model with 1000 bootstrap replicates. Tree visualization and annotation were created using the ggtree v3.0.4 [26] package in R 4.2. The genetic context of the mcr-1-encoding plasmid sequences was represented using Easyfig v2.2.2 [27].
We used a bioinformatics approach to identify the plasmid sequences. First, we used plasmidSPAdes [28] for plasmid assembly from raw data. Second, after checking if these sequences contained the mcr-1 cassette, we used plasmidfinder [29] to check if the predicted plasmid has a replicon and then we used oriTfinder [30] to identify the origin of the transfer site (oriT) and conjugative elements. Finally, we performed a search in the PLSDB database [31] to identify similar plasmid sequences. Highly similar sequences were compared with the predicted plasmids to generate a circular view using Blast Ring Image generation (BRIG) software [32].
3. Results
3.1. Prevalence of the mcr-1 Gene in Poultry and Pig Farms
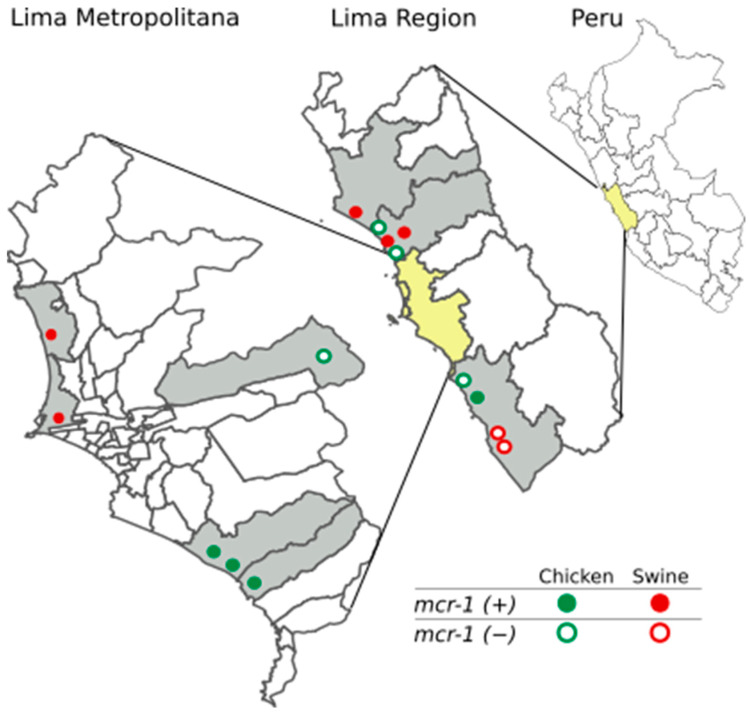
A total of 15 farms located in Lima were investigated in this study. We collected 648 fecal samples and recovered 197 E. coli isolates from 8 chicken farms and 144 E. coli isolates from 7 pig farms. The mcr-1 gene was identified in four of eight chicken farms in three Lima provinces, and in five of seven pig farms located in three Lima provinces (Figure 1). The occurrence of the mcr-1 gene was variable: 81% of the isolates were mcr-1-positive in the AV8 farm (26/32 isolates), while 52.2% (12/23) were positive in the AV4 farm, and just 10.7% (3/28) were positive in the AV5 farm (see Supplementary Table S1). Overall, the mcr-1-specific PCR identified the gene in 21.3% (42/197) of the isolates from poultry and in 12.5% (18/144) of the isolates from pigs.
Figure 1.
Geographical distribution of pig and poultry farms sampled in Lima, Peru. The location of the farms with the presence of mcr-1-positive E. coli isolates from chicken and swine is represented by circles filled in green and red, respectively. The location of the farms without the presence of mcr-1-positive E. coli isolates from chicken and swine is represented by green and red circles, respectively.
3.2. Genetic Characterization of E. coli Harboring mcr-1 and Resistome
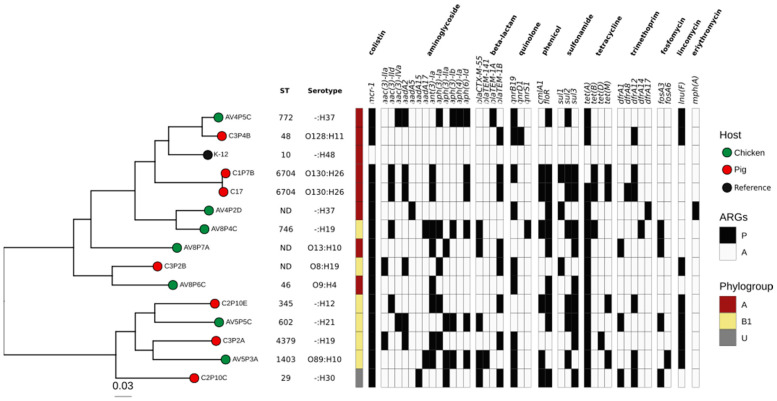
The whole genome of 14 mcr-1-positive E. coli was sequenced (7 genomes from pigs and 7 from chicken). The sequence size varied from 4.75 to 5.90 Mb. A total of 10 different MLSTs were identified in 11 isolates, while 3 were not determined (Supplementary Table S2). Most isolates were identified as phylogenetic groups A (n = 7) and B1 (n = 6), while one isolate was typed as an unknown phylogroup (U). There was no differential clustering between the isolates from poultry and porcine sources (Figure 2).
Figure 2.
Resistome of 14 mcr-1-harboring E. coli isolated from pig and poultry farms in Lima, Peru. From left to right: phylogenomic tree based on SNPs of 14 E. coli genomes from chicken (circle filled with green) and pig (circle filled with red) farms. The E. coli K-12 strain was used as the reference. First column indicates the phylogroups A (red), B (yellow) or U (gray). The heatmap shows the presence/absence (P/A) of ARGs detected in mcr-1-positive E. coli genomes.
Interestingly, the genome sequence analysis showed a high number of resistance genes. We detected 39 different genes that confer resistance to 10 different classes of antibiotics, including mcr-1 (Table 1 and Figure 2). Five isolates contained the blaCTX-M-55 gene for resistance to extended-spectrum beta-lactamase (ESBL). At least 70% of the isolates contained a gene for resistance to ampicillin, chloramphenicol, kanamycin and trimethoprim, and at least 90% of the isolates contained a gene for resistance to streptomycin, sulfisoxazole and tetracycline.
Table 1.
ARGs profile of 14 E. coli genomes carrying mcr-1.
| Class | Antibiotics | Number of mcr-1 + E. coli (%) | Gene Name (n) |
|---|---|---|---|
| Aminoglycoside | gentamicin | 8 (57) | aac(3)-IIa (2), aac(3)-IId (4), aac(3)-IVa (1), aac(3)-VIa (1) |
| hygromycin B | 1 (7) | aph(4)-Ia (1) | |
| kanamycin | 11 (79) | aph(3’)-Ia (5), aph(3’)-IIa (4), aph(6)-Id (6) | |
| streptomycin | 13 (93) | aadA2 (6), aadA5 (1), aadA15 (1), aadA17 (2), ant(3’’)-Ia (8), aph(3’’)-Ib (4) | |
| Beta-lactam | ampicillin | 10 (71) | blaTEM-1A (1), blaTEM-1B (8), blaTEM-141 (1) |
| ceftriaxone | 5 (36) | blaCTX-M-55 (5) | |
| Quinolone | ciprofloxacin | 10 (71) | qnrB19 (9), qnrD1 (1), qnrS2 (1) |
| Folate pathway antagonist | trimethoprim | 10 (71) | dfrA1 (3), dfrA8 (1), dfrA12 (6), dfrA14 (1), dfrA17 (1) |
| Fosfomycin | fosfomycin | 5 (36) | fosA3 (4), fosA6 (1) |
| Glycylcycline | tetracycline | 14 (100) | tet(A) (13), tet(B) (3), tet(D) (1), tet(M) (4) |
| lincomycin | 7 (50) | Inu(F) (7) | |
| Macrolide | erythromycin | 1 (7) | mph(A) (1) |
| Phenicol | chloramphenicol | 11 (79) | cmlA1 (6), floR (10) |
| Sulphonamide | sulfamethoxazole | 13 (93) | sul1 (3), sul2 (7), sul3 (9) |
Forty-two genes encoding virulence factors were detected in all E. coli genomes using the VirulenceFinder tool v2.0 (see Supplementary Figure S1 and Table S3), including genes related to evasion/invasion (capU, kpsE, kpsMII_K5, gad, iss, ompT, sepA, traT), toxins (astA, cea, cib, hlyF, stb, toxB), secretion system (cif, espABFJ, nleABC, terC, tir), adherence (eae, lpfA, perA, tsh) and iron uptake (fyuA, ireA, irp2, iucC, iutA, sitA). The AV5P5C isolate from chicken was classified as APEC because it presented genes encoding outer membrane protein (ompT), hemolysin (hlyF), increased serum survival (iss), aerobactin siderophore receptor (iutA), temperature-sensitive hemagglutinin (tsh) and siderophores (IucC, sitA) [33,34].
Additionally, 33 types of plasmid replicons were identified in the analyzed genomes; the most overrepresented was IncFIB, followed by IncX1, ColRNAI, IncFIC(FII) and IncFII(pHN7A8) (Supplementary Figure S1 and Table S3).
3.3. Characterization of the Genetic Context of the mcr-1 Gene
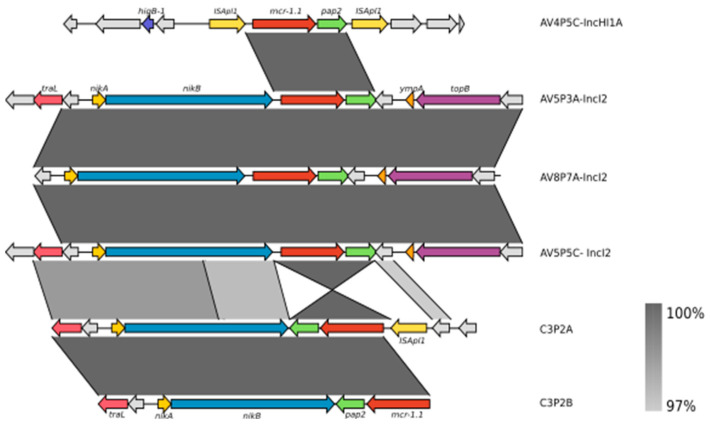
The mcr-1 sequence of all E. coli genomes was 100% identical to the mcr-1 sequence from the Resfinder database. The exploration of the genetic context of mcr-1 allowed us to identify three different types of mcr-1-containing cassettes suggesting a diverse genetic context of mcr-1-harboring E. coli in the farms of Lima (Figure 3). Two different context structures were identified in chicken farms. The ISApl1–mcr1–pap2–ISApl1 was identified in the AV4P5C isolate, which belongs to ST48. This cassette shows a structure called Tn6330 inserted into an IncHI1A:IncHI1B hybrid plasmid. Tn6330 is a composite transposon that improves the transmission of the mcr-1 gene [35]. The second structure shows the nikB–mcr1–pap2 composition that has lost ISApl1 both upstream and downstream. This structure was found within IncI2 plasmids in the AV4P2D, AV5P5C, AV5P3A and AV8P7A isolates (Figure 3). Interestingly, a BLASTN search of the AV5P3A plasmid carrying mcr-1 showed a high similarity of this plasmid to the IncI2 plasmid pkpCOL17 (99% of identity and 99.97% of coverage) identified in K. pneumoniae isolated from a patient in a Peruvian hospital [15] (Supplementary Figure S2). In a pig isolate (C3P2A), we observed the presence of a downstream copy of ISApl1 only. Due to short-read sequencing, we were not able to determine the genetic context of the mcr-1 gene for six isolates because of incomplete assembly.
Figure 3.
Genetic context of the mcr-1 gene in E. coli genomes isolated from chicken and pig farms in Lima. Six representative sequences show the diversity of the structural context of the mcr-1 gene from chicken and pig E. coli isolates in this study. The mcr-1 gene is marked in red. ISApl1 transposase, pap2 and nickB genes are marked in yellow, green and yellow, respectively. Regions of homology between sequences (>97%) are indicated by the graded shading.
4. Discussion
We studied the occurrence of E. coli carrying the mcr-1 gene in 15 livestock farms in Lima, Peru, from 2019 to 2020. The results of the PCR showed that E. coli was positive for mcr-1 in 9 of the 15 farms evaluated in this study at different rates, i.e., in 21% (42/197) of the isolates from poultry and in 12% (18/144) of the isolates from pigs. To the best of our knowledge, this is the first study to investigate the occurrence of mcr-1-positive E. coli isolates in farms in Peru. Previous works reported E. coli isolates carrying the mcr-1 gene in samples of clinical and food origin in Peru [12,13,14,16]. In December 2019, the Ministry of Agriculture and Irrigation of Peru (MINAGRI) published a resolution prohibiting the use of colistin in food-producing animals [36]. The effect of the ban was not evaluated in this study because all samples were collected before the application of the resolution in March 2020. We expect a reduction in mcr-1 prevalence as was observed in China. After the implementation of the colistin prohibition for veterinary use in China, the prevalence of E. coli carrying mcr-1 decreased from 45% to 19% between 2016 and 2018 in pig farms [37]. In South America, the circulation of mcr-1-harboring Enterobacteriaceae isolates has a higher prevalence in animals (8.7%) than in food (5.4%) or humans (2.0%), mainly in Brazil, Bolivia and Argentina [38].
All isolates sequenced in this study belong to different sequence types, suggesting an important diversity in mcr-1-positive E. coli. These isolates were classified as belonging to phylogroup A or B1, with commensal E. coli usually found in humans and animal hosts [39,40]. The clones found in our study, ST48, ST602, ST746, ST46, ST345, were previously reported in clinical isolates from humans and other hosts; ST602 is widely distributed internationally [41,42]. This information is concerning, since it suggests that these strains have the ability to move and proliferate in different ecological niches, which may facilitate the genetic exchange of the mcr-1 gene and other antibiotic resistance genes between a wide range of bacterial species. A highly diverse resistome was revealed, with 39 different genes conferring resistance to 15 different antibiotics including ESBL, chloramphenicol, ciprofloxacin, tetracycline and sulfamethoxazole, indicating an extensive circulation of E. coli carrying multiple antibiotic-resistant genes in livestock in Lima; in fact, Peru is considered one of the countries with a high projected increase of antimicrobial consumption by livestock [43]. We detected that mcr-1 was associated with resistance mechanisms to beta-lactams; five mcr-1-positive E. coli also encoded blaCTX-M-55, while other eight isolates produced the blaTEM-1B gene. The co-occurrence of mcr-1 and beta-lactam genes was also reported previously in South America, in samples from chicken meat in Brazil and in E. coli isolated from pig farms and companion animals in Argentina [44,45,46].
The composite transposon Tn6330 (ISApl1–mcr1–pap2–ISApl1) is considered the main vehicle for mcr-1 mobilization [6,35]. Only one out of fourteen mcr-1-positive E. coli sequenced in this work contained Tn6330 with both copies of ISApl1 within an IncHI1A:IncHI1B plasmid, while in another isolate, we noted the presence of an upstream copy of ISApl1. Cassettes with the Tn6330 structure are generally mobilized by IncHI2 plasmids; however, E. coli has been reported harboring mcr-1 into hybrid plasmids containing the incompatible types IncHI1A and IncHI1B in Asia [47,48]. Up to now, only four plasmids have been described to carry the mrc-1 gene in Latin America: IncX4, IncP, IncI2 and IncHI2 [49]. On other hand, some E. coli genomes from chicken and pigs presented mcr-1 carried by the IncI2 plasmid lacking the ISApl1 copies. The plasmid IncI2 has already been described to spread different mcr genes variants in Latin American countries such as Argentina, Brazil and Uruguay [50]. According to global genomic studies, mcr-1 sequences with two copies of ISApl1 are found in lesser frequency than sequences with only a single copy of ISApl1, while the majority of positive mcr-1 isolates do not present the ISApl1 sequence [4,6]. Due to the short-read sequencing method used in this work, we were not able to determine the genetic context of the mcr-1 gene for six mcr-1-positive genomes because of the incomplete assembly of the (plasmid) sequences. Therefore, we cannot exclude the presence of other plasmid types that mobilize the mcr-1 gene in our isolates.
In conclusion, we determined the occurrence of mcr-1-harboring E. coli in chicken farms (21.3%) and pig farms (12.5%) in Lima. The genomic analysis showed diverse lineages of E. coli carrying the mcr-1 gene mobilized by the IncI2 and IncHI1A:IncHI1B plasmids, including the presence of ISApl1 copies enhancing the dissemination of mcr-1. The elevated prevalence of multidrug-resistant strains in farms in Lima could serve as a reservoir of ARGs that can be disseminated by farmers or food, impacting public health. We need to expand the genomic and epidemiological surveillance of colistin resistance in farmers, livestock, the environment, wastewater and hospitals to understand the dynamic of mcr-1 transmission in Peru.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/antibiotics11121781/s1, Figure S1: Virulome and plasmid replicons of 14 mcr-1 E. coli.; Figure S2: Genetic characteristics of the Inc2 mcr-1-carrying plasmid identified in this study. Circular view and alignment comparison of closely related IncI2 plasmids carrying the mcr-1 gene.; Supplementary Table S1: Sampling information and mcr-1 positivity by PCR in the farms sampled in this study.; Supplementary Table S2: Accession numbers and sequencing statistics of 14 E. coli genomes carrying the mcr-1 gene.; Supplementary Table S3: Virulence factors and plasmid types predicted by abricate using VFDB (Virulence Factor Database) and Plasmidfinder. Values represent the gene coverage.
Author Contributions
L.M.H., L.L.E., D.C. and C.G.D.G. conceived the idea and designed the experiments. L.L.E., G.M.M. and L.M.H. coordinated the sample collection. D.C., C.G.D.G., Y.I.L., T.S.E. and C.L.R.C. performed bacteria isolation and sequencing. D.C., C.G.D.G., C.L.R.C., Y.I.L. and T.S.E. analyzed the data. D.C., C.G.D.G., C.L.R.C., Y.I.L. and T.S.E. wrote the manuscript. D.C., C.G.D.G., C.L.R.C., L.L.E., R.H.R.A., N.L. and L.M.H. performed the manuscript review and editing. L.L.E. and L.M.H. supervised this work. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Genome sequence data analyzed in this study can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA892251.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was funded by PROCIENCIA-CONCYTEC, through its executing unit ProCiencia—Proyectos de Investigación Básica 2018-01, Contract number No. 127-2018-FONDECYT. CD and YI were supported by Vicerrectorado de Investigación y Postgrado—Universidad Nacional Mayor de San Marcos (Project Number A21081091).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sulis G., Gandra S. Access to Antibiotics: Not a Problem in Some LMICs. Lancet Glob. Health. 2021;9:e561–e562. doi: 10.1016/S2214-109X(21)00085-1. [DOI] [PubMed] [Google Scholar]
- 2.WHO Critically Important Antimicrobials for Human Medicine: 6th Revision. [(accessed on 20 October 2022)]. Available online: https://www.who.int/publications-detail-redirect/9789241515528.
- 3.Kempf I., Jouy E., Chauvin C. Colistin Use and Colistin Resistance in Bacteria from Animals. Int. J. Antimicrob. Agents. 2016;48:598–606. doi: 10.1016/j.ijantimicag.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 4.Matamoros S., van Hattem J.M., Arcilla M.S., Willemse N., Melles D.C., Penders J., Vinh T.N., Thi Hoa N., Bootsma M.C.J., van Genderen P.J., et al. Global Phylogenetic Analysis of Escherichia coli and Plasmids Carrying the mcr-1 Gene Indicates Bacterial Diversity but Plasmid Restriction. Sci. Rep. 2017;7:15364. doi: 10.1038/s41598-017-15539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y.-Y., Wang Y., Walsh T.R., Yi L.-X., Zhang R., Spencer J., Doi Y., Tian G., Dong B., Huang X., et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016;16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 6.Wang R., van Dorp L., Shaw L.P., Bradley P., Wang Q., Wang X., Jin L., Zhang Q., Liu Y., Rieux A., et al. The Global Distribution and Spread of the Mobilized Colistin Resistance Gene mcr-1. Nat. Commun. 2018;9:1179. doi: 10.1038/s41467-018-03205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L., Madec J.-Y., Lupo A., Schink A.-K., Kieffer N., Nordmann P., Schwarz S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018;6:4. doi: 10.1128/microbiolspec.ARBA-0026-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leekitcharoenphon P., Johansson M.H.K., Munk P., Malorny B., Skarżyńska M., Wadepohl K., Moyano G., Hesp A., Veldman K.T., Bossers A., et al. Genomic Evolution of Antimicrobial Resistance in Escherichia coli. Sci. Rep. 2021;11:15108. doi: 10.1038/s41598-021-93970-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brisola M.C., Crecencio R.B., Bitner D.S., Frigo A., Rampazzo L., Stefani L.M., Faria G.A. Escherichia coli Used as a Biomarker of Antimicrobial Resistance in Pig Farms of Southern Brazil. Sci. Total Environ. 2019;647:362–368. doi: 10.1016/j.scitotenv.2018.07.438. [DOI] [PubMed] [Google Scholar]
- 10.Vu Thi Ngoc B., Le Viet T., Nguyen Thi Tuyet M., Nguyen Thi Hong T., Nguyen Thi Ngoc D., Le Van D., Chu Thi L., Tran Huy H., Penders J., Wertheim H., et al. Characterization of Genetic Elements Carrying mcr-1 Gene in Escherichia coli from the Community and Hospital Settings in Vietnam. Microbiol. Spectr. 2022;10:e0135621. doi: 10.1128/spectrum.01356-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang H., Chen S., White D.G., Zhao S., McDermott P., Walker R., Meng J. Characterization of Multiple-Antimicrobial-Resistant Escherichia coli Isolates from Diseased Chickens and Swine in China. J. Clin. Microbiol. 2004;42:3483–3489. doi: 10.1128/JCM.42.8.3483-3489.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ugarte Silva R.G., Olivo López J.M., Corso A., Pasteran F., Albornoz E., Sahuanay Blácido Z.P. Anales de la Facultad de Medicina. Volume 79. UNMSM; Lima, Peru: 2018. Resistencia a Colistín Mediado por el gen mcr-1 Identificado en Cepas de Escherichia coli y Klebsiella pneumoniae: Primeros Reportes en el Perú; pp. 213–217. [DOI] [Google Scholar]
- 13.Deshpande L.M., Hubler C., Davis A.P., Castanheira M. Updated Prevalence of mcr-like Genes among Escherichia coli and Klebsiella pneumoniae in the SENTRY Program and Characterization of mcr-1.11 Variant. Antimicrob. Agents Chemother. 2019;63:e02450-18. doi: 10.1128/AAC.02450-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yauri-Condor K., Zavaleta Apestegui M., Sevilla-Andrade C.R., Sara J.P., Villoslado Espinoza C., Taboada W.V., Gonzales-Escalante E. Extended-Spectrum Beta-Lactamase-Producing Enterobacterales Carrying the mcr-1 Gene in Lima, Peru. Rev. Peru. Med. Exp. Salud Pública. 2021;37:711–715. doi: 10.17843/rpmesp.2020.374.5832. [DOI] [PubMed] [Google Scholar]
- 15.Gonzales-Escalante E., Ruggiero M., Cerdeira L., Esposito F., Fontana H., Lincopan N., Gutkind G., Di Conza J. Whole-Genome Analysis of a High-Risk Clone of Klebsiella pneumoniae ST147 Carrying Both mcr-1 and blaNDM-1 Genes in Peru. Microb. Drug Resist. 2022;28:171–179. doi: 10.1089/mdr.2021.0128. [DOI] [PubMed] [Google Scholar]
- 16.Murray M., Salvatierra G., Dávila-Barclay A., Ayzanoa B., Castillo-Vilcahuaman C., Huang M., Pajuelo M.J., Lescano A.G., Cabrera L., Calderón M., et al. Market Chickens as a Source of Antibiotic-Resistant Escherichia coli in a Peri-Urban Community in Lima, Peru. Front. Microbiol. 2021;12:635871. doi: 10.3389/fmicb.2021.635871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Boeckel T.P., Pires J., Silvester R., Zhao C., Song J., Criscuolo N.G., Gilbert M., Bonhoeffer S., Laxminarayan R. Global Trends in Antimicrobial Resistance in Animals in Low- and Middle-Income Countries. Science. 2019;365:eaaw1944. doi: 10.1126/science.aaw1944. [DOI] [PubMed] [Google Scholar]
- 18.Rebelo A.R., Bortolaia V., Kjeldgaard J.S., Pedersen S.K., Leekitcharoenphon P., Hansen I.M., Guerra B., Malorny B., Borowiak M., Hammerl J.A., et al. Multiplex PCR for Detection of Plasmid-Mediated Colistin Resistance Determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for Surveillance Purposes. Euro Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2018;23:17-00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews S. FastQC. A Quality Control Tool for High Throughput Sequence Data. [(accessed on 14 July 2022)]. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 20.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seemann T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinform. Oxf. Engl. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 23.Waters N.R., Abram F., Brennan F., Holmes A., Pritchard L. Easy Phylotyping of Escherichia coli via the EzClermont Web App and Command-Line Tool. Access Microbiol. 2020;2:acmi000143. doi: 10.1099/acmi.0.000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croucher N.J., Page A.J., Connor T.R., Delaney A.J., Keane J.A., Bentley S.D., Parkhill J., Harris S.R. Rapid Phylogenetic Analysis of Large Samples of Recombinant Bacterial Whole Genome Sequences Using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu G., Smith D.K., Zhu H., Guan Y., Lam T.T.-Y. Ggtree: An r Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data. Methods Ecol. Evol. 2017;8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 27.Sullivan M.J., Petty N.K., Beatson S.A. Easyfig: A Genome Comparison Visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antipov D., Hartwick N., Shen M., Raiko M., Lapidus A., Pevzner P.A. PlasmidSPAdes: Assembling Plasmids from Whole Genome Sequencing Data. Bioinformatics. 2016;32:3380–3387. doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]
- 29.Carattoli A., Hasman H. PlasmidFinder and In Silico PMLST: Identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS) In: de la Cruz F., editor. Horizontal Gene Transfer: Methods and Protocols. Springer US; New York, NY, USA: 2020. pp. 285–294. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 30.Li X., Xie Y., Liu M., Tai C., Sun J., Deng Z., Ou H.-Y. OriTfinder: A Web-Based Tool for the Identification of Origin of Transfers in DNA Sequences of Bacterial Mobile Genetic Elements. Nucleic Acids Res. 2018;46:W229–W234. doi: 10.1093/nar/gky352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmartz G.P., Hartung A., Hirsch P., Kern F., Fehlmann T., Müller R., Keller A. PLSDB: Advancing a Comprehensive Database of Bacterial Plasmids. Nucleic Acids Res. 2022;50:D273–D278. doi: 10.1093/nar/gkab1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alikhan N.-F., Petty N.K., Ben Zakour N.L., Beatson S.A. BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genom. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbieri N.L., Pimenta R.L., de Melo D.A., Nolan L.K., de Souza M.M.S., Logue C.M. mcr-1 Identified in Fecal Escherichia coli and Avian Pathogenic E. coli (APEC) From Brazil. Front. Microbiol. 2021;12:799. doi: 10.3389/fmicb.2021.659613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarowska J., Futoma-Koloch B., Jama-Kmiecik A., Frej-Madrzak M., Ksiazczyk M., Bugla-Ploskonska G., Choroszy-Krol I. Virulence Factors, Prevalence and Potential Transmission of Extraintestinal Pathogenic Escherichia coli Isolated from Different Sources: Recent Reports. Gut Pathog. 2019;11:10. doi: 10.1186/s13099-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snesrud E., McGann P., Chandler M. The Birth and Demise of the ISApl1-mcr-1-ISApl1 Composite Transposon: The Vehicle for Transferable Colistin Resistance. mBio. 2018;9:e02381-17. doi: 10.1128/mBio.02381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El Peruano Resolucion Directoral-No 0091-2019-MINAGRI-SENASA-DIAIA. Disponen Prohibir la Importación, Comercialización, Fabricación o Elaboración de Productos Veterinarios que Contengan el Principio Activo Colistina (Polimixina E) o Cualquiera de sus Sales y Dictan Diversas Disposiciones. [(accessed on 29 April 2022)]. Available online: http://busquedas.elperuano.pe/normaslegales/disponen-prohibir-la-importacion-comercializacion-fabricac-resolucion-directoral-no-0091-2019-minagri-senasa-diaia-1832393-1/
- 37.Shen C., Zhong L.-L., Yang Y., Doi Y., Paterson D.L., Stoesser N., Ma F., Ahmed M.A.E.-G.E.-S., Feng S., Huang S., et al. Dynamics of mcr-1 Prevalence and mcr-1-Positive Escherichia coli after the Cessation of Colistin Use as a Feed Additive for Animals in China: A Prospective Cross-Sectional and Whole Genome Sequencing-Based Molecular Epidemiological Study. Lancet Microbe. 2020;1:e34–e43. doi: 10.1016/S2666-5247(20)30005-7. [DOI] [PubMed] [Google Scholar]
- 38.Mendes Oliveira V.R., Paiva M.C., Lima W.G. Plasmid-Mediated Colistin Resistance in Latin America and Caribbean: A Systematic Review. Travel Med. Infect. Dis. 2019;31:101459. doi: 10.1016/j.tmaid.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 39.Duriez P., Clermont O., Bonacorsi S., Bingen E., Chaventré A., Elion J., Picard B., Denamur E. Commensal Escherichia coli Isolates Are Phylogenetically Distributed among Geographically Distinct Human Populations. Microbiology. 2001;147:1671–1676. doi: 10.1099/00221287-147-6-1671. [DOI] [PubMed] [Google Scholar]
- 40.Madoshi B.P., Kudirkiene E., Mtambo M.M.A., Muhairwa A.P., Lupindu A.M., Olsen J.E. Characterisation of Commensal Escherichia coli Isolated from Apparently Healthy Cattle and Their Attendants in Tanzania. PLoS ONE. 2016;11:e0168160. doi: 10.1371/journal.pone.0168160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fuentes-Castillo D., Esposito F., Cardoso B., Dalazen G., Moura Q., Fuga B., Fontana H., Cerdeira L., Dropa M., Rottmann J., et al. Genomic Data Reveal International Lineages of Critical Priority Escherichia coli Harbouring Wide Resistome in Andean Condors (Vultur Gryphus Linnaeus, 1758) Mol. Ecol. 2020;29:1919–1935. doi: 10.1111/mec.15455. [DOI] [PubMed] [Google Scholar]
- 42.Aworh M.K., Abiodun-Adewusi O., Mba N., Helwigh B., Hendriksen R.S. Prevalence and Risk Factors for Faecal Carriage of Multidrug Resistant Escherichia coli among Slaughterhouse Workers. Sci. Rep. 2021;11:13362. doi: 10.1038/s41598-021-92819-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Boeckel T.P., Brower C., Gilbert M., Grenfell B.T., Levin S.A., Robinson T.P., Teillant A., Laxminarayan R. Global Trends in Antimicrobial Use in Food Animals. Proc. Natl. Acad. Sci. USA. 2015;112:5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monte D.F., Mem A., Fernandes M.R., Cerdeira L., Esposito F., Galvão J.A., Franco B.D.G.M., Lincopan N., Landgraf M. Chicken Meat as a Reservoir of Colistin-Resistant Escherichia coli Strains Carrying mcr-1 Genes in South America. Antimicrob. Agents Chemother. 2017;61:e02718-16. doi: 10.1128/AAC.02718-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Faccone D., Moredo F.A., Giacoboni G.I., Albornoz E., Alarcón L., Nievas V.F., Corso A. Multidrug-Resistant Escherichia coli Harbouring mcr-1 and blaCTX-M Genes Isolated from Swine in Argentina. J. Glob. Antimicrob. Resist. 2019;18:160–162. doi: 10.1016/j.jgar.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 46.Rumi M.V., Mas J., Elena A., Cerdeira L., Muñoz M.E., Lincopan N., Gentilini É.R., Di Conza J., Gutkind G. Co-Occurrence of Clinically Relevant β-Lactamases and MCR-1 Encoding Genes in Escherichia coli from Companion Animals in Argentina. Vet. Microbiol. 2019;230:228–234. doi: 10.1016/j.vetmic.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 47.Li R., Zhang P., Yang X., Wang Z., Fanning S., Wang J., Du P., Bai L. Identification of a Novel Hybrid Plasmid Coproducing MCR-1 and MCR-3 Variant from an Escherichia coli Strain. J. Antimicrob. Chemother. 2019;74:1517–1520. doi: 10.1093/jac/dkz058. [DOI] [PubMed] [Google Scholar]
- 48.Li R., Du P., Zhang P., Li Y., Yang X., Wang Z., Wang J., Bai L. Comprehensive Genomic Investigation of Coevolution of Mcr Genes in Escherichia coli Strains via Nanopore Sequencing. Glob. Chall. 2021;5:2000014. doi: 10.1002/gch2.202000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lentz S.A.M., Dalmolin T.V., Barth A.L., Martins A.F. mcr-1 Gene in Latin America: How Is It Disseminated Among Humans, Animals, and the Environment? Front. Public Health. 2021;9:648940. doi: 10.3389/fpubh.2021.648940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papa-Ezdra R., Grill Diaz F., Vieytes M., García-Fulgueiras V., Caiata L., Ávila P., Brasesco M., Christophersen I., Cordeiro N.F., Algorta G., et al. First Three Escherichia coli Isolates Harbouring mcr-1 in Uruguay. J. Glob. Antimicrob. Resist. 2020;20:187–190. doi: 10.1016/j.jgar.2019.07.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequence data analyzed in this study can be found here: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA892251.