Abstract
Group A Streptococcus (GAS) expresses cell surface proteins that mediate important biological functions such as resistance to phagocytosis, adherence to plasma and extracellular matrix proteins, and degradation of host proteins. An open reading frame encoding a protein of 348 amino acid residues was identified by analysis of the genome sequence available for a serotype M1 strain. The protein has an LPATGE sequence located near the carboxy terminus that matches the consensus sequence (LPXTGX) present in many gram-positive cell wall-anchored molecules. Importantly, the central region of this protein contains 50 contiguous Gly-X-X triplet amino acid motifs characteristic of the structure of human collagen. The structural gene (designated scl for streptococcal collagen-like) was present in all 50 GAS isolates tested, which together express 21 different M protein types and represent the breadth of genomic diversity in the species. DNA sequence analysis of the gene in these 50 isolates found that the number of contiguous Gly-X-X motifs ranged from 14 in serotype M6 isolates to 62 in a serotype M41 organism. M1 and M18 organisms had the identical allele, which indicates very recent horizontal gene transfer. The gene was transcribed abundantly in the logarithmic but not stationary phase of growth, a result consistent with the occurrence of a DNA sequence with substantial homology with a consensus Mga binding site immediately upstream of the scl open reading frame. Two isogenic mutant M1 strains created by nonpolar mutagenesis of the scl structural gene were not attenuated for mouse virulence as assessed by intraperitoneal inoculation. In contrast, the isogenic mutant derivative made from the M1 strain representative of the subclone most frequently causing human infections was significantly less virulent when inoculated subcutaneously into mice. In addition, both isogenic mutant strains had significantly reduced adherence to human A549 epithelial cells grown in culture. These studies identify a new extracellular GAS virulence factor that is widely distributed in the species and participates in adherence to host cells and soft tissue pathology.
Group A Streptococcus (GAS) is a human pathogen that causes a wide variety of diseases. Although the molecular mechanisms of GAS pathogenesis are not fully understood it has been well documented that several cell surface proteins participate in host-pathogen interactions (21). Laboratory inactivation of many of these genes has been reported, and the resulting isogenic mutants have been found to be detrimentally affected in biomedically important properties such as resistance to phagocytosis (44), host cell internalization (26), and mouse virulence (7). Many of the proteins have structural features typical of the cell surface proteins of gram-positive bacteria (15), including a variable amino terminus, a central region with repeating sequences, and a cell-associated region containing a cell wall anchor motif with the amino acid sequence LPXTG(X).
Because bacterial cell surface components contribute to several phases of GAS pathogenesis, various molecular biology approaches have been used to identify previously undescribed genes encoding proteins that participate in interaction with the host (11, 48, 51). For example, a fibronectin-binding protein was found by screening a GAS expression library with antibody directed against the cell wall-associated region of streptococcal surface proteins (51). Inhibition of opsonic properties present in anti-GAS cell wall serum by fractionated streptococcal cell wall extracts allowed the identification of a new protective antigen (11). In addition, analysis of the GAS genome database permitted identification of a surface protein (designated GRAB) that regulates proteolysis at the bacterial cell surface by binding to host α2-macroglobulin (48).
Here, we report the identification and characterization of a gene encoding an extracellular protein that participates in adherence of the pathogen to host epithelial cells and contributes to virulence, as assessed by subcutaneous inoculation of mice. This gene was present in all 50 genetically diverse GAS isolates analyzed, which represent 21 distinct M protein serotypes. A noteworthy feature of the protein is the occurrence of extensive and contiguous Gly-X-X (where X is an undefined amino acid) amino acid repeats characteristic of human collagen. The gene (designated scl for streptococcal collagen-like), was transcribed abundantly in the exponential phase of growth. Isogenic strains in which the scl gene was inactivated by a nonpolar mutagenesis technique were significantly reduced in their ability to adhere to human epithelial cells grown in culture and were significantly less pathogenic in a mouse model of soft tissue infection.
MATERIALS AND METHODS
Bacterial strains and growth.
Fifty GAS strains isolated from international sources were studied. The collection had strains with 21 different M types that were verified by sequencing the emm gene fragment encoding the hypervariable amino terminus of the M protein. Two M1 strains were used for genetic manipulation, adherence assays, and animal experiments. MGAS 6708 is identical to GAS 370, the serotype M1 strain used for the Streptococcal Genome Sequencing Project (http://www.genome.ou.edu/strep.html). This strain lacks the gene (speA) encoding streptococcal pyrogenic exotoxin A. MGAS 5005 (speA positive) is representative of the M1 strains most commonly recovered from invasive infections and has been recently used in mouse model experiments (24, 30). MGAS 6708 and MGAS 5005 have been extensively characterized (23, 24, 30).
GAS strains were grown in Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) supplemented with 0.2% yeast extract (THY medium) or on tryptose agar with 5% sheep blood (Becton Dickinson, Cockeysville, Md.). Brain heart infusion agar (Difco Laboratories) supplemented with spectinomycin (150 μg/ml) was used for mutant selection. The GAS strains were incubated at 37°C in an atmosphere of 5% CO2–20% O2.
Cloning experiments were performed with Escherichia coli XL-1 Blue (Stratagene, La Jolla, Calif.) grown in Luria-Bertani medium (Difco Laboratories).
Inactivation of the scl gene in serotype M1 GAS strains.
The scl gene was inactivated in MGAS 6708 and MGAS 5005 with a nonpolar spectinomycin resistance cassette designated spc2 (30). An ∼1.9-kb chromosomal fragment containing the scl gene and flanking regions was amplified from MGAS 6708 with forward primer scl-SalI (5′-TACGCGTCGACAAGGGTAATCTGAGGCATAAG) and reverse primer scl-SmaI (5′ TCTCCCGGGTTCGCCTGACTTTCTCAC). This PCR product was digested with PstI and SmaI to generate an ∼600-bp fragment, which was cloned between the PstI-SpeI sites of the E. coli vector pFW11 (45). The SpeI site was first blunt ended with the Klenow fragment of DNA polymerase. The resulting plasmid (pSL131) contained ∼200 bp of the 3′ end coding region of scl and ∼400 bp of downstream noncoding DNA. Next, a DNA fragment of ∼700 bp was generated from the initial PCR product by digestion with SalI and XbaI. This fragment contained ∼300 bp of the 5′ end coding region of scl and ∼400 bp of the upstream noncoding region and was cloned between the SalI-HindIII site of pSL131, generating plasmid pSL132. Both XbaI and HindIII sites were first blunt-ended with the Klenow fragment. The spc2 cassette with flanking SmaI restriction sites was then inserted in frame between the SpeI (cleaves between bp 33 and 34 of the scl open reading frame [ORF], the SpeI site was first blunt-ended with the Klenow fragment) and PvuII (cleaves 41 bp from the end of the scl ORF) sites, resulting in suicide plasmid pSL134.
Electrocompetent cells made from MGAS 6708 and MGAS 5005 were electroporated with pSL134. The bacteria were plated on brain heart infusion agar containing 150 μg of spectinomycin per ml. Drug-resistant colonies were initially screened by PCR for the presence of a single amplification product arising as a result of a double recombination event. These colonies were further analyzed by Southern hybridization and DNA sequencing to confirm that the proper genetic construct had been obtained and no spurious mutations were present. Lack of scl expression by the mutants was confirmed by Northern blot and Western analyses (see Results).
RNA methods.
Total RNA was isolated and examined by Northern hybridization as described previously (30). Briefly, GAS cells grown in 10 ml of THY medium were harvested and resuspended in TE buffer (10 mM Tris [pH 7.0], 1 mM EDTA). Cells were treated at 37°C for 5 min with mutanolysin (25 U) and lysozyme (1 mg/ml) in the presence of 5 mM aurintricarboxylic acid, an RNase inhibitor. The bacteria were lysed by adding sodium dodecyl sulfate (SDS) (2% final concentration) and an equal volume of acid-phenol-chloroform at 65°C for 5 min. The samples were extracted with acid-phenol-chloroform, and contaminating DNA was removed by digestion with DNase I. The scl-specific DNA probe (whole gene) was amplified from the reference strain MGAS 6708 and biotinylated with BrightStar labeling reagents (Ambion, Austin, Tex.). A 553-bp recA-specific DNA probe was used in a control hybridization to test the integrity of the RNA. Northern blot analysis of 15 μg of RNA was performed with NorthernMax reagents (Ambion). RNA size markers (Life Technologies, Grand Island, N.Y.) were used to estimate transcript size.
DNA methods.
Standard DNA manipulation techniques were used (54). Southern blot analysis was done with a nonradioactive labeling and detection kit (ECL; Amersham Pharmacia Biotech, Inc., Piscataway, N.J.). The presence of the scl gene in GAS strains was assessed by PCR with primers that amplify the entire ORF. PCR was performed with Taq polymerase (Life Technologies) with the forward primer scl-up (5′-CTCCACAAAAGAGTGATCAGTC) and the reverse primer scl-rev (5′-TTAGTTGTTTTCTTTGCGTTT). DNA was first denatured at 94°C for 1 min. Thirty amplification cycles were performed as follows: 1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 1 min 40 s of extension at 72°C, followed by one cycle of 5 min at 72°C. The PCR products were analyzed by agarose gel electrophoresis. DNA sequencing was done with internal primers and the Taq DyeDeoxy terminator cycle sequencing kit (Applied Biosystems, Inc., Foster City, Calif.) with an ABI 377 instrument. The DNA sequence data were analyzed with Sequencher 3.1.1 (Gene Codes Corporation, Inc., Ann Arbor, Mich.) and Lasergene (DNASTAR, Inc., Madison, Wis.) software.
Protein methods.
Since the scl gene was transcribed during the log phase but not stationary phase of growth, Scl protein expression was studied in GAS cultures grown to exponential phase optical density at 600 nm ([OD600] of ∼0.5) in THY medium. On the basis of DNA sequence analysis, Scl is presumed to be a cell surface protein. Therefore, the cell wall-associated protein fractions were obtained from MGAS 5005 and MGAS 6708 wild-type strains and from their isogenic scl mutant derivatives. GAS cells harvested from 100-ml cultures were resuspended in 1 ml of 20% sucrose–10 mM Tris (pH 8.0) buffer containing 25 U of mutanolysin and 1 mg of lysozyme per ml. Cells were digested at 37°C for 1 h and pelleted by centrifugation, and the supernatants were used for subsequent analyses.
Synthetic peptide TTMTSSQRESKIKEI (corresponding to amino acid residues 42 to 56 of the Scl protein) (Fig. 1) was used to raise anti-Scl-specific antibody in rabbits (Bethyl Laboratories, Inc., Montgomery, Tex.). This peptide corresponds to amino acid residues located in the amino terminus of mature Scl from serotype M1 GAS. Immune rabbit serum had antipeptide reactivity in an enzyme-linked immunosorbent assay, whereas preimmune serum from the same rabbit did not (data not shown). Anti-Scl rabbit immunoglobulin was purified by affinity chromatography with a column made of Sepharose conjugated to the synthetic peptide.
FIG. 1.
Nucleotide and amino acid sequence of the scl gene and Scl protein in serotype M1 GAS. (A) The scl coding sequence consists of 1,047 bp (nucleotides 77 to 1123) and encodes a polypeptide of 348 amino acids. The presumed scl promoter region contains a predicted ribosome-binding site (RBS) and −10 and −35 regions. A predicted transcription site, +1, is denoted by a dot. A potential binding site for the streptococcal positive transcription regulator, Mga, overlapping with the −35 region is shown. A potential transcription terminator (tt) consisting of two inverted repeats was identified downstream of scl gene. The predicted ATG start codon and the TAA stop codon are shown in boldface type. SS, signal sequence showing the predicted cleavage site (arrowhead); V, V region of the mature Scl protein; CL, CL region consisting of 50 GXX triplets (boxed); L, L region with PGEKAPEKS repeats (underlined); WM, cell-associated Scl part containing a cell wall segment with the LPATGE motif (shaded), and cell membrane hydrophobic region with a charged tail. (B) Alignment of the consensus Mga binding site and putative promoter region of the scl gene. The consensus Mga binding site was identified on the basis of sequence alignments of several Mga-regulated genes (34). The essential bases are shown in boldface type, and essential sequences within the potential Mga binding site of the scl gene promoter which do not match with the consensus sequence are shown in lowercase type. Twenty-two of 29 (76%) essential bases are identical. Underlined bases in the putative promoter region of the scl gene promoter denote a potential −35 promoter region.
Scl production by the wild-type and mutant strains was assessed by Western blotting. Cell wall protein fractions were separated by electrophoresis with SDS–12% polyacrylamide electrophoresis and transferred to a nitrocellulose membrane (Bio-Rad, Hercules, Calif.). Immunodetection of Scl was performed with rabbit anti-Scl purified immunoglobulin G (1:500 dilution). Alkaline phosphatase-conjugated goat anti-rabbit affinity-purified immunoglobulin G (heavy and light chains) (Rockland Immunochemicals, Gilbertsville, Pa.) was used as the secondary antibody, and detection was done with a chromogenic substrate (Life Technologies). Prestained broad-range marker proteins (Bio-Rad) were used as molecular mass standards.
Mouse infection studies.
The virulence of the wild-type GAS serotype M1 strains and the scl-lacking isogenic mutants was compared with two mouse infection models as described previously (31, 32). For the intraperitoneal (i.p.) inoculation model, adult (20- to 25-g) male outbred CD-1 Swiss mice (Harlan Sprague Dawley Inc., Houston, Tex.) were used. Five-week old (20- to 30-g) outbred, immunocompetent, hairless male mice (strain Crl:SKH1-hrBR) (Charles River, Wilmington, Mass.) were used for the subcutaneous (s.c.) inoculation model. The animals were anesthetized with isoflurane (AErrane; Ohmeda Caribe Inc., Guayama, P.R.) prior to inoculation.
Inocula were prepared from GAS cells grown in THY medium. Cells harvested at an OD600 of ∼0.5 were used because the scl gene is transcribed abundantly in the exponential phase of growth (see Results). The bacteria were washed once with sterile ice-cold, pyrogen-free phosphate-buffered saline, and the OD600 was adjusted with phosphate-buffered saline to give the required inoculum. The number of CFU inoculated per mouse was verified for each experiment by colony counts on tryptose agar plates containing 5% sheep blood (Becton Dickinson). GAS cell suspensions (0.25 ml) were administered to mice i.p., and mortality was recorded every 3 h for 5 d.
Mice used in the s.c. infection model were weighed immediately before GAS inoculation. GAS cells contained in 0.1 ml were injected s.c. in the right flank of each mouse with a tuberculin syringe. Animal weight and abscess size were measured daily for the first week and twice a week thereafter for a total of 21 days. The dimensions of the abscesses were measured with a caliper. Abscess length (L) and width (W) values were used to calculate abscess volume (V = 4/3π(L/2)2 × [W/2]) and area (A = π[L/2] × [W/2]) employing equations for a spherical ellipsoid. Blood was collected from each dead animal by cardiac puncture and cultured on blood agar to confirm GAS bacteremia.
Human cell culture and adherence assay.
A549 human lung epithelial cells (ATCC CCL-185) were used to compare the level of adherence of the wild-type and scl-inactivated isogenic mutant strains. The A549 cells were cultured on glass coverslips placed in 24-well tissue culture plates with F12K nutrient mixture medium (Kaighn's modification) (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies) at 37°C in an atmosphere with 5% CO2. Cells were grown to semiconfluency (∼4 × 105 cells/well) without antibiotics.
GAS were grown in THY medium until they reached mid-log phase (OD600 of 0.3 to 0.4). The bacteria were harvested, washed once with Hanks balanced salt solution, and resuspended in Hanks balanced salt solution. The A549 human cells were inoculated with ∼108 CFU of GAS per well in F12K medium plus 10% fetal bovine serum. The tissue culture plates were centrifuged for 4 min at 700 × g to promote contact between bacteria and epithelial cells and were incubated for 1 h at 37°C with 5% CO2. The wells were washed three times to remove nonadherent bacteria. The human cells were fixed for 1 h in 1 ml of PBS–2% paraformaldehyde and stained with crystal violet.
The number of cell-associated bacteria was determined by counting 50 random A549 cells in four different fields with light microscope. The adherence experiments were performed in triplicate, and the results were expressed for each experiment as the mean number of GAS per A549 cell ± standard error of the mean.
Molecular evolutionary genetic analysis.
Multiple-sequence alignment of the inferred Scl amino acid sequences was conducted with Clustal W (59). The proportions of polymorphic synonymous (pS) and nonsynonymous (pN) sites were calculated by the method of Nei and Gojobori (39).
Statistical analysis.
Kaplan-Meier survival curves were plotted for mice inoculated with the wild-type and scl mutant strains, and the mortality rate was examined for statistical significance with the log-rank test. A t test was used to determine if the difference in adherence between wild-type and isogenic mutant strains was statistically significant.
A mixed model repeated-measures analysis was used to test for significant differences in the abscess areas and volumes, and weight loss between the groups of mice in the s.c. inoculation experiments. The repeated measures model was employed with one within-subjects and one between-subjects factor. Statistical calculations were performed with Proc mixed software (SAS Institute Inc., Cary, N.C.).
Nucleotide sequence accession number.
The scl sequence reported here has been deposited in GenBank under accession number 330137.
RESULTS
Identification and analysis of the scl gene and inferred Scl protein in serotype M1 GAS.
A common theme of the cell-associated carboxy-terminal part of surface proteins from gram-positive cocci is the presence of a charged tail of up to 7 amino acid residues preceded by a hydrophobic transmembrane domain (15). The cell-wall domain is located amino terminal to this hydrophobic segment and contains a highly conserved cell anchor LPXTG(X) motif. The protein sequence corresponding to the hydrophobic cell membrane domain of the cell surface protein M6 (FFTAAALTVMATAGVAAVV) (14) was used to search the Streptococcal Genome Sequencing Project database (http://www.genome.ou.edu/strep.html), and several sequences with similarity to the query were identified. One of these ORFs (designated scl for streptococcal collagen-like) is shown in Fig. 1A. The scl ORF is 1,047 bp long (nucleotide 77 to 1123) and would encode a protein with 348 amino acid residues. A potential promoter located upstream of this ORF includes a −10 region (tataat; perfect match of the consensus sequence) and a −35 region (tttaca; five of six bases identical [in boldface type] to the consensus sequence ttgaca (52). A potential ribosome-binding site (gaaagaga; the consensus sequence is taaggagg [identical residues shown in boldface type]) is located 9 nucleotides upstream from the ATG-Met start codon (20, 57).
The predicted molecular mass of the Scl protein is ∼36.3 kDa. As expected for an exported GAS cell surface protein, the amino terminus (encompassing amino acid residues 1 to 37) (Fig. 1A) has structural features characteristic of signal sequences (42), such as a short amino-terminal hydrophilic region followed by a hydrophobic transmembrane segment. The signal peptidase cleavage site usually has a small amino acid residue, and Ala37 satisfies this criterion in Scl. The predicted molecular mass of the mature protein (residues 38 to 348) is ∼32.4 kDa, and the predicted isoelectric point is 5.54. The amino-terminal segment of the inferred mature Scl protein (amino acid residues 38 to 105) is hydrophilic and has a predicted α-helical structure.
A noteworthy structural feature of Scl is a collagen-like region consisting of 50 Gly-X-X (GXX) triplet repeats that encompass amino acid residues 106 to 255 (designated CL, for collagen-like region) (47). The GXX sequence in collagen forms a right-handed triple helix with three molecular chains. The inferred bacterial cell-associated part of Scl (designated the WM region, encoded by amino acid residues 267 to 348) has an apparent cell wall domain with a cell anchor motif (LPATGE) and a hydrophobic transmembrane segment followed by a charged tail at the carboxy terminus. The CL and WM regions are connected with a linker sequence (L) consisting of repeats with a PGEKAPEKS core sequence. In M1 GAS there are two of these repeats.
scl gene expression in M1 GAS strains.
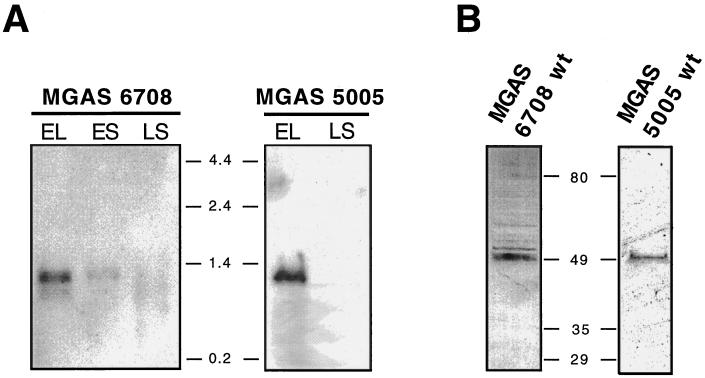
Northern blot analysis was performed to determine if the scl gene was transcribed (Fig. 2A). Total RNA was isolated from strain MGAS 6708 grown to logarithmic (OD600 ≈ 0.5) or stationary (OD600 ≈ 0.8 and overnight) phase and probed with the entire scl gene amplified by PCR. A single transcript of ∼1.2 kb corresponding to the predicted length of a single-gene scl transcript was identified. The same results were obtained with strain MGAS 5005. A predicted +1 transcription start site is shown in Fig. 1A. In addition, a potential transcription terminator with two inverted repeat sequences is present 40 bp downstream from the TAA stop codon. When these upstream and downstream putative regulatory regions are considered, the calculated 1,193-bp size of the scl transcript agrees with the size of the hybridizing band identified by Northern analysis (Fig. 2A).
FIG. 2.
scl gene expression in M1 GAS strains. (A) Northern blot analysis of the total RNA isolated from cultures of MGAS 6708 and MGAS 5005 harvested during the logarithmic (EL [early logarithmic] [OD600 ≈ 0.5]) or stationary (ES [early stationary] [OD600 ≈ 0.8] or LS [late stationary, overnight]) phase of growth. RNA (15 μg) was loaded in each lane. A DNA probe consisting of the entire scl gene was amplified by PCR from the MGAS 6708 source strain and labeled by biotinylation. The blot was developed with streptavidin-peroxidase conjugate and a chemiluminescence detection method was used to visualize hybridizing bands. A single transcript of ∼1.2 kb was identified in log-phase cultures. scl gene transcription decreased markedly during the stationary phase of growth. RNA size markers (in kilobases) were used to estimate the size of the scl transcript. (B) Western blot analysis of the cell wall-associated protein fractions from exponential GAS cultures. Both wild-type (wt) M1 serotype MGAS 6708 and MGAS 5005 strains have an immunoreactive cell wall-associated protein migrating at ∼49 kDa. Molecular mass standards (in kilodaltons) are shown.
Only weak scl transcription occurred in the early stationary phase of growth, and no scl-specific mRNA was detected in the overnight cultures of both M1 strains tested. Together, this pattern of scl transcription is characteristic of genes regulated by Mga (multiple gene regulator), a positive transcriptional regulator of several virulence genes (4, 6, 43). In this regard, we note that upstream of the scl ORF there is a DNA sequence homologous to the consensus Mga binding site (34) (Fig. 1B). This potential binding site for Mga overlaps with the predicted −35 promoter region, a molecular arrangement consistent with the characteristics of Mga binding sites (43). Northern blot analysis of isogenic M1 strains (wild type and mga mutant) confirmed our prediction that scl1 is under mga control (unpublished data).
The presence of the Scl protein in the cell wall-associated fraction was confirmed by analysis of two genetically distinct GAS strains harvested in the exponential phase of growth (Fig. 2B). Both wild-type MGAS 5005 and MGAS 6708 strains expressed a protein with an approximate molecular mass of 49 kDa that reacted with rabbit antisera raised against an M1-specific Scl peptide. Although the predicted molecular mass of the mature Scl protein is ∼32 kDa, it is a common observation that cell surface proteins from gram-positive bacteria migrate aberrantly slowly in SDS-polyacrylamide electrophoresis gels (1, 27, 48). It is also well known that proteins with repetitive motifs tend to migrate aberrantly. Taken together, the data indicate that the scl gene is transcribed and translated by GAS serotype M1 strains.
scl gene variation among genetically diverse GAS strains.
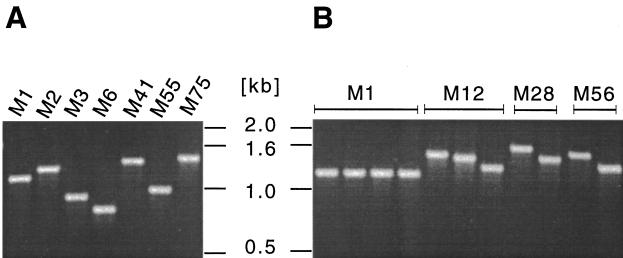
Inasmuch as GAS isolates can have extensive chromosomal structural heterogeneity, we next screened 50 genetically diverse and epidemiologically unrelated strains by PCR with scl-specific primers to assess the distribution of the scl gene. A PCR product was obtained from all 50 strains, but considerable size variation was identified between strains expressing different M protein serotypes (Fig. 3A). In addition, scl size variation was identified among serotype M12, M28, and M56 organisms (Fig. 3B).
FIG. 3.
Size variation in the scl gene. The complete scl gene and putative promoter region were amplified by PCR and analyzed by agarose gel electrophoresis. All 50 strains tested yielded a single PCR product, and representative examples of the size variants identified are shown. (A) Size variation in the scl gene in different M-type GAS strains. (B) scl gene size variation among strains of certain M types, including M12, M28, and M56.
The scl gene in these 50 strains was sequenced to determine the molecular basis of size variation. No allelic variation was found among strains expressing serotype M1 (n = 7 isolates), M2 (n = 2), M3 (n = 5), M6 (n = 4), or M18 (n = 3). Three mature Scl variants each were identified among six M12 isolates and four M28 GAS strains (Table 1). In addition, two Scl variants each were found among M52 (n = 3), M56 (n = 2), and M77 (n = 2) organisms. Two Scl proteins from M49 GAS strains differed by one amino acid residue within the signal peptide. scl gene variation among isolates of serotypes M4, M9, M22, M41, M55, M57, M75, M76, ST2035, and ST2967 could not be assessed, because only one isolate of each of these M types was sequenced.
TABLE 1.
Size variation in the scl gene GAS strains expressing 21 M types
| M type | No. of strains sequenced | scl patterna (no. of strains) | No. of amino acidis in V region | No. of GXY motifs in CL region | No. of PGEKAPEKS repeats in L region |
|---|---|---|---|---|---|
| 1 | 7 | 67 | 50 | 2 | |
| 2 | 2 | 85 | 46 | 3 | |
| 3 | 5 | 70 | 10, 25b | 2 | |
| 4 | 1 | 70 | 42 | 6 | |
| 6 | 4 | 70 | 14 | 2 | |
| 9 | 1 | 69 | 41 | 6 | |
| 12 | 6 | I (1) | 75 | 38 | 7 |
| II (4) | 75 | 34 | 7 | ||
| III (1) | 75 | 21 | 7 | ||
| 18 | 3 | 67 | 50 | 2 | |
| 22 | 1 | 72 | 45 | 5 | |
| 28 | 4 | I (2) | 75 | 48 | 7 |
| II (1) | 75 | 38 | 7 | ||
| III (1) | 75 | 34 | 7 | ||
| 41 | 1 | 83 | 62 | 2 | |
| 49c | 2 | 71 | 46 | 5 | |
| 52 | 3 | I (1) | 68 | 60 | 5 |
| II (2) | 68 | 59 | 5 | ||
| 55 | 1 | 70 | 25 | 3 | |
| 56 | 2 | I (1) | 71 | 55 | 5 |
| II (1) | 71 | 38 | 5 | ||
| 57 | 1 | 75 | 30 | 2 | |
| 75 | 1 | 69 | 44 | 5 | |
| 76 | 1 | 67 | 52 | 4 | |
| 77 | 2 | I (1) | 73 | 55 | 5 |
| II (1) | 73 | 47 | 5 | ||
| ST2035 | 1 | 85 | 43 | 5 | |
| ST2967 | 1 | 66 | 45 | 6 |
Refers to PCR product size and DNA sequence data.
The scl gene in M3 isolates has a mutation which creates an in-frame TAA stop codon within the 11th GXX triplet motif.
The two isolates of M49 differed at amino acid position 7 (H or Y) in the signal sequence.
When comparing the 37 distinct Scl variants identified, well-conserved and highly polymorphic regions were found. The amino-terminal region of the mature Scl proteins (amino acids 38 to 105 [Fig. 1A]) was highly polymorphic among these strains. For purposes of description and discussion, the amino-terminal portion of the mature Scl protein is designated as the V (variable) region. When comparing different M types, the V region was polymorphic in number of amino acids and primary sequence (Table 1). The CL region, located adjacent to the V region, also was highly variable in length as a consequence of the different number of GXX motifs. The CL region in Scl from all M1 strains (Fig. 1A) has 50 GXX triplets. However, 14 GXX motifs were identified in the four M6 organisms studied, whereas one M41 isolate had 62 GXX motifs. The GXX motifs present in the 50 GAS isolates also differed in primary amino acid sequence. For example, 21 distinct GXX amino acid motif sequences were identified among the 50 GXX triplets in the CL region of the M1 strains. In the aggregate, 50 distinct GXX amino acid motif sequences were identified in the CL regions of the 50 strains studied. Thus, the CL region was characterized by variation in number and primary amino acid sequence of the GXX amino acid motifs, thereby resulting in a highly polymorphic segment of Scl.
The carboxy-terminal end of the CL region (the segment located closer to the cell wall) was less polymorphic than the amino-terminal end of the CL region located more distant from the cell wall anchor. Between the CL region and the conserved LPATGE cell wall anchor is a segment of the protein referred to as the L region. When comparing the sequences from the 50 strains analyzed, the L region was composed of two to seven conserved repeats, each containing an identical 9-amino-acid core sequence (PGEKAPEKS) (Table 1).
Two observations warrant special comment. First, serotype M1 and M18 strains had the identical scl allele, a result indicative of very recent horizontal gene transfer. Second, serotype M3 organisms had a nucleotide substitution creating a stop codon that would truncate the Scl protein at amino acid 139 of the Scl protein. Truncation of Scl at this location would result in an exported protein with an inferred molecular mass of 11 kDa, containing 10 GXX triplets.
Nonpolar inactivation of the scl gene in serotype M1 GAS.
To facilitate assessment of the role of Scl in host-pathogen interaction, isogenic nonpolar scl mutant derivatives were constructed from serotype M1 strains MGAS 6708 and MGAS 5005. We focused the analysis on M1 strains because this is the most common M type recovered from invasive infections in many case series. MGAS 6708, a speA-lacking strain, is identical to the strain whose chromosome was recently sequenced (59), and MGAS 5005 (speA positive) is representative of organisms causing most cases of human invasive disease and pharyngitis (24, 36). Two M1 organisms were used because considerable evidence has accrued in recent years demonstrating that chromosomal variation occurs among M1 strains (23, 36). Hence, not all M1 strains are genetically equivalent. In addition, although a chromosomal sequence is available for MGAS 6708, this strain is not genetically representative of the vast majority of M1 organisms recovered from invasive and pharyngitis cases worldwide (23).
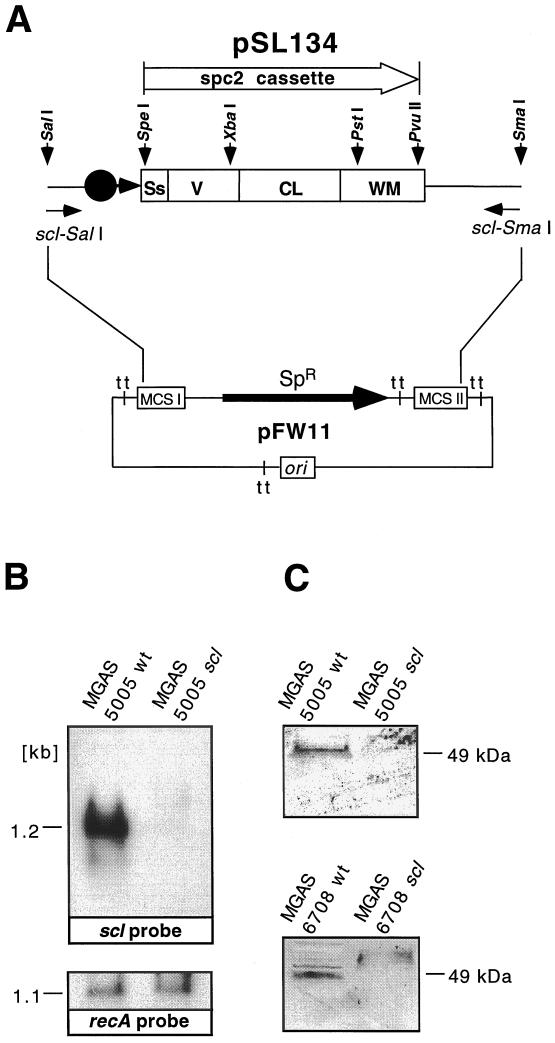
Each strain yielded numerous spectinomycin-resistant colonies after electroporation with suicide plasmid pSL134 (Fig. 4A). One scl mutant derivative of MGAS 6708 and one of MGAS 5005 were selected for further studies after analysis by PCR, Southern hybridization, and sequencing documented that the correct molecular genetic constructs had been generated (data not shown). These genetic analyses found that the mutants of the scl gene in both strains were identical and contained the expected deletion of nucleotides 114 to 1082 and replacement with the spc2 nonpolar cassette (Fig. 4A). Also as expected, scl-specific transcripts were not detected in the mutant strains during exponential growth, a time when abundant scl-specific mRNA is synthesized by the parental organisms (Fig. 4B). These results were further supported by the lack of the Scl protein in the periplasmic fraction of the scl mutants, whereas the parental wild-type isolates produced Scl (Fig. 4C). The isogenic mutants had the same colony morphology and identical growth curves as the parental isolates (data not shown).
FIG. 4.
scl gene expression in wild-type and mutant M1 GAS. (A) Schematic representation of suicide plasmid pSL134 used for nonpolar inactivation of the scl gene in serotype M1 GAS. Most of the scl gene sequence (969 of 1,047 bp) was replaced with a nonpolar spectinomycin resistance cassette (spc2) inserted in frame. To enhance the likelihood of homologous recombination, ∼400 bp of DNA upstream and downstream of the scl ORF also was cloned. pSL134 is a suicide plasmid that cannot replicate in GAS. Ss, signal sequence; V, variable region; CL, collagen-like region; WM, cell wall and cell membrane region; tt, transcription terminator; MCS, multiple cloning site. (B) Northern blot analysis of the total RNA from logarithmic-phase GAS cultures. Samples (15 μg) were blotted and hybridized with scl- and recA-specific biotinylated DNA probes and detected by chemiluminescence. The scl-specific transcript is detected in the wild-type (wt) but not mutant sample. Control hybridization with the recA probe showed a positive signal for both the wild-type and scl mutants. Identical results were obtained for the MGAS 6708 isogenic pair (data not shown). (C) Western blot analysis of the cell wall-associated protein fractions obtained from the wild-type and scl mutant strains. Scl is detected in the samples prepared from the wild-type but not mutant strains.
Scl and GAS virulence in mice.
Genetic inactivation of expression of several GAS extracellular products has been reported to significantly decrease virulence in mice (7, 32, 35). In addition, we have shown in the present study that the scl gene was transcribed in the phase of growth when several known GAS virulence factors are made. As a consequence, we compared the ability of the wild-type parental organisms and isogenic mutant strains to cause mouse death after i.p. inoculation.
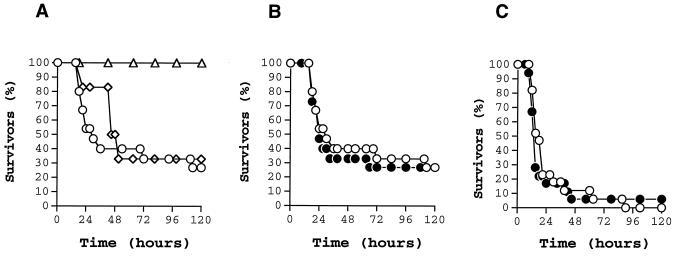
Previous experiments demonstrated that i.p. inoculation of 2 × 107 CFU of wild-type MGAS 5005 (speA positive) caused the death of 90% of mice after 5 days (data not shown). To determine the inoculum of MGAS 6708 (speA lacking) required to cause equivalent mortality, mice were inoculated i.p. with twofold serial dilutions ranging from ∼107 to ∼109 CFU. Interestingly, at the highest inoculum used (∼109 CFU), only 70% of mice died (Fig. 5A), which means that MGAS 6708 is significantly less virulent than MGAS 5005. This result is consistent with the rarity of recovery of this M1 subclone from human infections (23).
FIG. 5.
Kaplan-Meier survival curves showing mouse lethality caused by the wild type (open symbols) and isogenic scl-inactivated mutants (closed symbols). Experiments were performed with CD-1 Swiss mice inoculated i.p. with M1 type GAS. (A) Determination of the inoculum size for MGAS 6708 wild-type GAS. Inocula used were 1.1 × 109 CFU (circles), 7.5 × 108 CFU (diamonds), and 3.3 × 108 CFU or less (triangle). (B) MGAS 6708 wild-type strain (1.1 × 109 CFU [n = 15 mice/group]) and MGAS 6708 mutant (1.2 × 109 CFU [n = 15 mice/group]). (C) MGAS 5005 wild-type (2.2 × 107 CFU [n = 17 mice/group]) and MGAS 5005 mutant (2.3 × 107 CFU [n = 18 mice/group]). The differences in mouse mortality between the wild-type and mutant strains were not statistically significant.
We next inoculated mice i.p. with the wild-type and scl mutant strains (∼2.2 × 107 CFU was used for the MGAS 5005 strain pair and ∼109 CFU was used for the MGAS 6708 strain pair). No significant difference in mouse mortality was observed for either pair of isogenic GAS strains (P < 0.539 [log-rank test] for the MGAS 5005 strain pair; P < 0.821 [log-rank test] for the MGAS 6708 strain pair) (Fig. 5B and C). The same results were obtained when the i.p. inoculation experiments were repeated (data not shown). Hence, we conclude that Scl does not participate significantly in virulence in this mouse model.
Certain extracellular GAS products also have been reported to participate in the pathogenesis of invasive skin disease when mice are injected s.c. with GAS (2, 28, 31). We next investigated the effect of inactivation of the scl gene on mouse mortality and skin pathology after s.c. inoculation with the wild-type MGAS 5005 and isogenic mutant derivative. The MGAS 6708 strain pair was not studied in this model due to its relative lack of virulence in the i.p. model. There was no significant difference between MGAS 5005 mutant and wild-type strains in mouse mortality (P < 0.3279 [log-rank test]). Mice inoculated with both wild-type and mutant strains developed skin lesions that healed over 3 weeks of the experiment (Table 2). Similarly, after the initial weight loss, mice in both experimental groups regained weight. When weight loss and severity of the skin lesions in two experimental groups were compared on days 2 and 4 after the inoculation, the differences were not statistically significant (t test), probably due to the large variation within animal groups. However, when those parameters were compared over the time of the experiment, the differences in both weight loss (P < 0.0001) and the severity of soft-tissue pathology (area, P < 0.0001; volume, P < 0.0001) between mice injected with the wild-type GAS and scl isogenic mutant were statistically significant (mixed model repeated measures). Hence, inactivation of the scl gene was associated with a decreased morbidity in this model of GAS pathogenesis.
TABLE 2.
Skin pathology in mice infected s.c. with the wild-type and scl isogenic mutant GAS strains
| Animal group infected with GAS | Severity of skin lesions
|
|||
|---|---|---|---|---|
| Mean area (cm2) (range) at:
|
Mean volume (cm3) (range) at:
|
|||
| 48 h | 96 h | 48 h | 96 h | |
| 5005 wild type | 152 (43–549) | 192 (39–901) | 2,150 (234–12,828) | 3,437 (261–24,644) |
| 5005 scl | 110 (31–234) | 158 (31–706) | 1,167 (167–3,600) | 2,485 (167–16,964) |
Scl and GAS adherence to human epithelial cells.
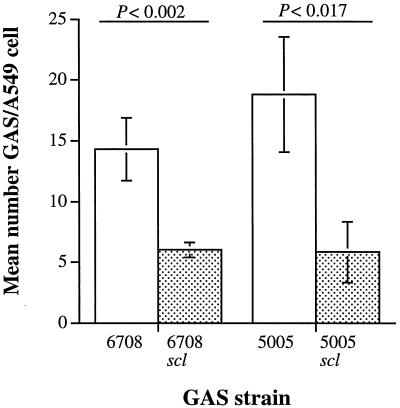
Attachment of GAS to host cells via adhesins is considered to be an important step in successful colonization. We hypothesized that Scl may act as an adhesin as a consequence of its extracellular location and collagen-like structure. To test this hypothesis, we compared the ability of wild-type MGAS 5005 and MGAS 6708 and their isogenic mutant derivatives to adhere to human A549 epithelial cells grown in vitro. There was no significant difference in the number of wild-type MGAS 5005 and MGAS 6708 that adhered to the A549 epithelial cells. In contrast, compared to the wild-type parental organisms, significantly fewer isogenic mutant bacteria adhered to the host cells (Fig. 6). The growth indices in the tissue culture medium were identical for the parental and mutant bacterial strains (data not shown). Taken together, the data are consistent with the hypothesis that the lack of Scl expression caused by nonpolar inactivation of the scl gene results in a detrimental effect on GAS adherence to host cells in vitro.
FIG. 6.
Adherence of isogenic M1 GAS strains to cultured human epithelial cells. Mid-log phase cultures of the wild-type strains (open bars) and scl-inactivated mutants (filled bars) were incubated with A549 epithelial cells for 1 h. Unattached bacteria were removed by washing. The attached GAS were stained and counted microscopically. The results are presented as mean numbers of GAS cells attached to each A549 cell ± standard error of the mean (error bars). The difference in adherence between the wild type and mutant derivatives was significant by t test for both MGAS 5005 and MGAS 6708 strain pairs.
Molecular evolutionary genetic analysis.
The observations that Scl is an extracellular protein potentially subjected to selective pressure in the host suggested that molecular evolutionary genetic analysis would provide insight into the processes shaping variation at the scl locus. Multiple-sequence alignment of 50 inferred Scl amino acid sequences by the Clustal W method required introduction of numerous alignment gaps, particularly in the V and CL domains. To determine if the level of selective constraint varies in different regions of Scl, we calculated the proportion of nonsynonymous and synonymous nucleotide substitutions (pN and pS, respectively) for the four Scl domains among the unique scl alleles (the L region was analyzed together with the CL region). The proportions, calculated as pN − pS ± standard error, for the following domains were as indicated: signal sequence, −0.0999 ± 0.0243; V region, −0.1785 ± 0.0304; CL region (the L region was analyzed together with the CL region), −0.1887 ± 0.0261; wall membrane region, −0.0151 ± 0.0092; overall, −0.0819 ± 0.0087. The difference, pN − pS, is a measure of the degree of selective constraint. The more negative the value, the less the contribution of amino acid replacements and the greater the contribution of synonymous (silent, not resulting in amino acid replacements) nucleotide substitutions. A difference of zero indicates selectively neutral variation, where the per site rates of pS and pN are equal. A positive difference (amino acid replacements exceed silent substitutions) suggests the action of diversifying (positive) selection. In general, the degree of selective constraint is low for scl as a whole, with the value of pN − pS approaching zero. The level of constraint is not uniform, however. The region of highest constraint spans the V and CL domains, with the two values differing by 0.01. In contrast, the values for the V and CL domains differ from the signal sequence and wall membrane domains by an average of 0.1261. The pattern of nonsynonymous and synonymous changes across the gene suggests the locus is responding to selective pressure, and the elevated constraint of the V and CL domains demonstrates the functional importance of this region.
DISCUSSION
Bacterial pathogenesis is a complex process that can involve attachment, colonization, internalization by epithelial and endothelial cells, local tissue destruction, and dissemination to distant anatomic sites. GAS express virulence factors that mediate one or more of these processes, including cell surface adhesins that participate in attachment to host cells (21). In this study we identified a new streptococcal cell surface molecule with the following characteristics: (i) the structural gene is commonly found in GAS strains and is expressed in a growth phase-dependent fashion, (ii) the protein contains a large region composed of GXX repeats with structural homology to human collagen, (iii) the protein participates in GAS attachment to human epithelial cells, and (iv) the protein participates in soft-tissue pathogenesis in the mouse.
The scl gene is common among GAS strains and is expressed in the logarithmic phase of growth.
GAS is composed of a heterogeneous array of chromosomal genotypes. Many genes encoding cell surface or other extracellular proteins have considerable allelic variation (25, 37), and many are not present in all GAS isolates. We found that the scl gene is widely distributed among 50 GAS strains that together represent the breadth of genomic diversity in the species. However, extensive size and protein sequence variation was identified (Fig. 7). The comparison of sequence data indicated that the amino-terminal signal sequence and the carboxy-terminal cell wall and membrane regions were largely conserved, whereas the central V and CL regions varied between the strains. This result suggested that the Scl protein might be under positive selection for amino acid replacements. To test this hypothesis, we measured the level of selective constraint, an indicator of the functional importance of the molecule and the degree of selective pressure present (38). Overall, constraint is low, with the value of pN − pS approaching zero. However, the degree of constraint is not uniform, for the central region of the gene (encoding V and CL) possesses an elevated negative value of pN − pS. Variation of pN − pS across a protein has been used by Reid et al. (49) to assess the relative functional importance of specific domains in an extracellular antigen of pathogenic Escherichia coli. The high level of codon bias present in the CL region (characterized by an increased proline and lysine content) and the requirement that glycine occupy every third residue demonstrate an increased level of selective constraint as indicated by the change in pN − pS. Presumably, the maintenance of the GXX motif in Scl is essential for the formation of a structure potentially required for interaction with host molecules such as extracellular matrix proteins and integrins. Hence, the level of constraint in the CL region restricts the nature of the changes such that the GXX motif is maintained. The molecular nature and biological importance of the constraint acting on the V region is yet to be elucidated.
FIG. 7.
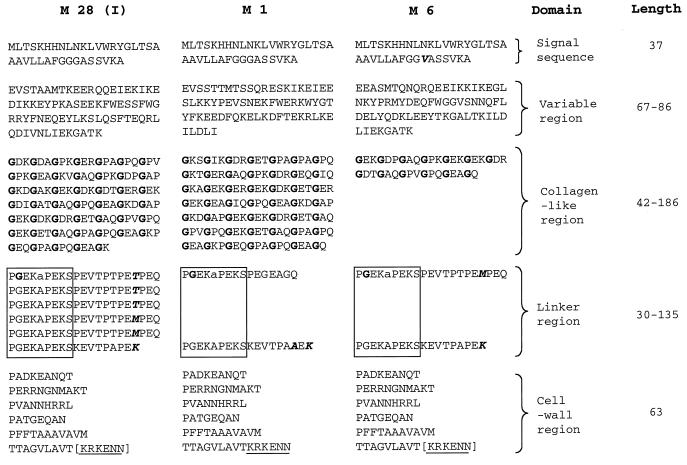
Schematic representation of variation in the Scl protein sequence among three different M-type GAS strains. The amino acid sequence of the Scl protein from M1 (source strain), M6, and M28 (scl pattern I [Table 1]) GAS strains is shown. The length of each Scl domain among all 50 GAS strains tested is shown as the number of amino acid residues. Glycine residues within the CL region are shown in boldface type. Polymorphic sites within the signal sequence and linker region are in boldface type and italicized. The signal sequence consists of 37 amino acids in all strains tested. In addition to the G31V substitution in the Scl signal sequence of the M6 strain, the following amino acid replacements were found in other GAS strains: H7Y, L9I, L12Q, and T19I. The alanine (A256 in M1 strain) residue (shown in lowercase type) located after the last GEK triplet motif is counted as the first residue of the linker region. The cell wall region contains the charged-tail sequence KRKENN (underlined). The KRKEEN motif in the M6 and M28 strains is shown in brackets because DNA sequence analysis did not identify all of the gene due to primer placement. The following polymorphic changes were identified in the cell wall region in all strains tested (amino acid residue numbers correspond to the sequence in the M1 strain): N292S and T335I.
The scl gene is abundantly transcribed in logarithmic growth but not in the stationary growth phase. Temporal variation in the expression of proven and presumed GAS virulence factors has been well described. Several complex gene regulatory circuits are known, including the Mga regulon (6, 43, 56); the CovR-CovS two-component regulatory system (13, 22, 29); a positive regulator, RofA (16); and the negative regulator Nra (46). Genes controlled by Mga are expressed in the exponential phase of growth in a process that involves binding of Mga to promoter regions. Mga binding sites generally overlap with the −35 regulatory sequence (34, 43). Our analysis identified a sequence in the presumed scl promoter with homology with the consensus Mga binding site sequence (34) (Fig. 1B). Taken together, these data indicate that the scl gene is present in most if not all GAS strains and is coexpressed with other virulence factors under Mga regulation.
Scl contains a unique GXX triplet repeat region.
The Scl protein contains a region consisting of extensive GXX triplet repeats characteristic of human collagen. CL repeats are exceedingly rare in prokaryotic pathogens. Interestingly, a region of 30 amino acid with 10 GXX repeats was described previously in the GAS bacteriophage-encoded hyaluronidase (33). In addition, Erickson et al. (12) reported that a platelet aggregation-associated protein of Streptococcus sanguis has a sequence (PGEQGPK) with two GXX repeats. However, the roles of these proteins in virulence were not assessed with isogenic mutant strains and model systems.
In human collagen, three chains entwine to form a right-handed superhelix (47). Three characteristics of the GXX region in Scl suggest that it is capable of forming a similar quaternary structure. First, Scl fulfills the absolute requirement that glycine occupy every third residue (only glycine can fit into the center of the triple helix without distorting it). Second, on the basis of computer modeling, the GXX region of Scl is predicted to form a coiled structure. Third, lysines and prolines essential for triple-helix stabilization together comprise ∼31% (percent by frequency) of the nonglycine residues.
Scl and host interactions.
Our studies indicated that Scl contributes significantly to adherence of GAS to human epithelial cells grown in culture, thereby adding to the relatively large number of surface-associated bacterial molecules that contribute to this process (21). The scl gene is transcribed in the logarithmic phase of growth, suggesting that the adhesin is made early in the course of host-pathogen interaction. Other molecules that contribute to host adherence that are made early in the growth phase include M protein (5), M-like proteins (17), and serum opacity factor (7). Among nonprotein streptococcal adhesins, hyaluronic acid capsule (55) and lipoteichoic acid (21, 40) also participate in adherence of GAS. Although our studies show that Scl participates in adherence in vitro, we did not address the molecular mechanism whereby Scl contributes to GAS-host cell interaction. Inasmuch as many eukaryotic cells bind collagen through receptors expressed on cell surfaces (3, 61, 62), Scl may participate in colonization of the host by docking GAS to host cell receptors (60). One biomedically important example of a collagen ligand is integrins, a class of transmembrane proteins located on the surface of mammalian cells that participate in cell-cell adhesion and attachment to the extracellular matrix (53). Interactions between GAS products and integrins via the RGD sequence have recently been reported (9, 41, 58). Integrins also bind to collagen through a different mechanism based on the triple helical structure of collagen (50). Since all 50 strains of 21 different M types had the scl gene, this mechanism could be widespread in GAS. This type of interaction between the CL region of the Scl protein and extracellular matrix and integrins could promote adherence of GAS to host tissue and trigger host-cell signal transduction.
Our data also showed that Scl contributes to soft tissue pathogenesis in a mouse model of inoculation, but they did not reveal a molecular mechanism to explain this observation. We favor the idea that the Scl protein is not directly toxic to the host but, rather, participates in soft tissue disease as a consequence of delayed clearance by the host. Under this hypothesis, Scl-positive organisms are cleared more slowly than the mutant because wild-type bacteria are not recognized as foreign as rapidly as mutant cells. An alternative hypothesis is that wild-type organisms are more resistant to phagocytosis and subsequent killing by host cells.
Autoimmunity issues.
Infection with GAS can result in sequelae such as acute rheumatic fever, rheumatic heart disease, glomerulonephritis, and other manifestations. It is widely believed that some of these postinfection sequelae have an autoimmune component, and evidence has been presented to support this idea (10). There has also been considerable speculation that GAS-induced autoantibodies play a role in many other human diseases, although conclusive evidence has not been obtained (18). The common theme in these and other autoimmune diseases including rheumatoid arthritis and systemic lupus erythematosus is the pathology of collagen within diseased sites and the presence of anticollagen antibodies in patient sera (19). Moreover, autoimmune diseases can be induced in experimental animals after exposure to collagen (8). The discovery of a widely distributed extracellular GAS protein with structural features similar to human collagen may add another dimension to GAS-induced autoimmunity considerations. In addition, our study found that the scl gene in M3 organisms had a mutation that introduced a stop codon. This polymorphism would create a truncated protein that lacks the cell wall anchor motif and, hence, would be secreted free into the extracellular milieu. It is possible that the Scl protein made by the M3 strains would interact with the host in a fundamentally different fashion than GAS cell wall-anchored Scl. Additional experiments will be required to test this idea.
ACKNOWLEDGMENTS
We thank N. P. Hoe for helpful discussion.
This research was supported by Public Health Service grant AI-33119 to J.M.M., and by funds from the Moran Foundation to S.L.
REFERENCES
- 1.Akesson P, Sjoholm A G, Bjorck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 2.Ashbaugh C D, Warren H B, Carey V J, Wessels M R. Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J Clin Investig. 1998;102:550–560. doi: 10.1172/JCI3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camper L, Hellman U, Lundgren-Akerlund E. Isolation, cloning, and sequence analysis of the integrin subunit α10, a β1-associated collagen binding integrin expressed on chondrocytes. J Biol Chem. 1998;273:20383–20389. doi: 10.1074/jbc.273.32.20383. [DOI] [PubMed] [Google Scholar]
- 4.Caparon M G, Scott J R. Identification of a gene that regulates expression of M protein, the major virulence determinant of group A streptococci. Proc Natl Acad Sci USA. 1987;84:8677–8681. doi: 10.1073/pnas.84.23.8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caparon M G, Stephens D S, Olsen A, Scott J R. Role of M protein in adherence of group A streptococci. Infect Immun. 1991;59:1811–1817. doi: 10.1128/iai.59.5.1811-1817.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, Bormann N, Cleary P P. VirR and Mry are homologous trans-acting regulators of M protein and C5a peptidase expression in group A streptococci. Mol Gen Genet. 1993;241:685–693. doi: 10.1007/BF00279912. [DOI] [PubMed] [Google Scholar]
- 7.Courtney H S, Chiang H C, Thacker J L, Dale J B. Serum opacity factor is a major fibronectin-binding protein and a virulence determinant of M type 2 Streptococcus pyogenes. Mol Microbiol. 1999;32:89–98. doi: 10.1046/j.1365-2958.1999.01328.x. [DOI] [PubMed] [Google Scholar]
- 8.Cremer M A, Rosloniec E F, Kang A H. The cartilage collagens: a review of their structure, organization, and role in the pathogenesis of experimental arthritis in animals and in human rheumatic disease. J Mol Med. 1998;76:275–288. doi: 10.1007/s001090050217. [DOI] [PubMed] [Google Scholar]
- 9.Cue D, Dombek P E, Lam H, Cleary P P. Streptococcus pyogenes serotype M1 encodes multiple pathways for entry into human epithelial cells. Infect Immun. 1998;66:4593–4601. doi: 10.1128/iai.66.10.4593-4601.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cunningham M W. Cross-reactive antigens of group A streptococci. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: American Society for Microbiology Press; 2000. pp. 66–77. [Google Scholar]
- 11.Dale J B, Chiang E Y, Liu S, Courtney H S, Hasty D L. New protective antigen of group A streptococci. J Clin Investig. 1999;103:1261–1268. doi: 10.1172/JCI5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erickson P R, Herzberg M C. The Streptococcus sanguis platelet aggregation-associated protein. Identification and characterization of the minimal platelet-interactive domain. J Biol Chem. 1993;268:1646–1649. [PubMed] [Google Scholar]
- 13.Federle M J, McIver K S, Scott J R. A response regulator that represses transcription of several virulence operons in the group A Streptococcus. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischetti V A. Streptococcal M protein: molecular design and biological behavior. Clin Microbiol Rev. 1989;2:285–314. doi: 10.1128/cmr.2.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischetti V A. Surface proteins on gram-positive bacteria. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: American Society for Microbiology Press; 2000. pp. 11–24. [Google Scholar]
- 16.Fogg G C, Gibson C M, Caparon M G. The identification of rofA, a positive-acting regulatory component of prtF expression: use of an mγδ-based shuttle mutagenesis strategy in Streptococcus pyogenes. Mol Microbiol. 1994;11:671–684. doi: 10.1111/j.1365-2958.1994.tb00345.x. [DOI] [PubMed] [Google Scholar]
- 17.Frick I M, Crossin K L, Edelman G M, Bjorck L. Protein H—a bacterial surface protein with affinity for both immunoglobulin and fibronectin type III domains. EMBO J. 1995;14:1674–1679. doi: 10.1002/j.1460-2075.1995.tb07156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibofsky A, Kerwar S, Zabriskie J B. Rheumatic fever. Rheum Dis Clin N Am. 1998;24:237–259. doi: 10.1016/s0889-857x(05)70007-7. [DOI] [PubMed] [Google Scholar]
- 19.Gioud M, Meghlaoui A, Costa O, Monier J C. Antibodies to native type I and II collagens detected by an enzyme linked immunosorbent assay (ELISA) in rheumatoid arthritis and systemic lupus erythematosus. Collagen Rel Res. 1982;2:557–564. doi: 10.1016/s0174-173x(82)80009-5. [DOI] [PubMed] [Google Scholar]
- 20.Gold L, Pribnow D, Schneider T, Shinedling S, Singer B W, Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- 21.Hasty D L, Ofek I, Courtney H S, Doyle R J. Multiple adhesins of streptococci. Infect Immun. 1992;60:2147–2152. doi: 10.1128/iai.60.6.2147-2152.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heath A, DiRita V J, Barg N L, Engleberg N C. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect Immun. 1999;67:5298–5305. doi: 10.1128/iai.67.10.5298-5305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoe N, Nakashima K, Grigsby D, Pan X, Dou S J, Naidich S, Garcia M, Kahn E, Bergmire-Sweat D, Musser J M. Rapid molecular genetic subtyping of serotype M1 group A Streptococcus strains. Emerg Infect Dis. 1999;5:254–263. doi: 10.3201/eid0502.990210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoe N P, Nakashima K, Lukomski S, Grigsby D, Liu M, Kordari P, Dou S-J, Pan X, Vuopio-Varkila J, Salmenlinna S, McGeer A, Low D E, Schwartz B, Schuchat A, Naidich S, De Lorenzo D, Fu Y-X, Musser J M. Rapid selection of structural variants of group A Streptococcus complement-inhibiting protein in serotype M1 epidemic waves. Nat Med. 1999;5:924–929. doi: 10.1038/11369. [DOI] [PubMed] [Google Scholar]
- 25.Hollingshead S K, Readdy T L, Yung D L, Bessen D E. Structural heterogeneity of the emm gene cluster in group A streptococci. Mol Microbiol. 1993;8:707–717. doi: 10.1111/j.1365-2958.1993.tb01614.x. [DOI] [PubMed] [Google Scholar]
- 26.Jadoun J, Ozeri V, Burstein E, Skutelsky E, Hanski E, Sela S. Protein F1 is required for efficient entry of Streptococcus pyogenes into epithelial cells. J Infect Dis. 1998;178:147–158. doi: 10.1086/515589. [DOI] [PubMed] [Google Scholar]
- 27.Kihlberg B-M, Collin M, Olsen A, Bjorck L. Protein H, an antiphagocytic surface protein in Streptococcus pyogenes. Infect Immun. 1999;67:1708–1714. doi: 10.1128/iai.67.4.1708-1714.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuo C-F, Wu J-J, Lin K-Y, Tsai P-J, Lee S-C, Jin Y-T, Lei H-Y, Lin Y-S. Role of streptococcal pyrogenic exotoxin B in the mouse model of group A streptococcal infection. Infect Immun. 1998;66:3931–3935. doi: 10.1128/iai.66.8.3931-3935.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levin J C, Wessels M R. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- 30.Lukomski S, Hoe N P, Abdi I, Rurangirwa J, Kordari P, Liu M, Shu-Jun D, Adams G G, Musser J M. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000;68:535–542. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukomski S, Montgomery C A, Rurangirwa J, Geske R S, Barrish J P, Adams G J, Musser J M. Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect Immun. 1999;67:1779–1788. doi: 10.1128/iai.67.4.1779-1788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Woischnik M, Podbielski A, Musser J M. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Investig. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marciel A M, Kapur V, Musser J M. Molecular population genetic analysis of a Streptococcus pyogenes bacteriophage-encoded hyaluronidase gene: recombination contributes to allelic variation. Microb Pathog. 1997;22:202–217. doi: 10.1006/mpat.1996.9999. [DOI] [PubMed] [Google Scholar]
- 34.McIver K S, Heath A S, Green B D, Scott J R. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. J Bacteriol. 1995;177:6619–6624. doi: 10.1128/jb.177.22.6619-6624.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moses A E, Wessels M R, Zalcman K, Alberti S, Natanson-Yaron S, Menes T, Hanski E. Relative contribution of hyaluronic acid capsule and M protein to virulence in a mucoid strain of the group A Streptococcus. Infect Immun. 1997;65:64–71. doi: 10.1128/iai.65.1.64-71.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musser J M, Krause R M. The revival of group A streptococcal diseases with a commentary on staphylococcal toxic shock syndrome. In: Krause R M, Fauci A, editors. Emerging infections. San Diego, Calif: Academic Press; 1998. pp. 185–218. [Google Scholar]
- 37.Natanson S, Sela S, Moses A E, Musser J M, Caparon M G, Hanski E. Distribution of fibronectin-binding proteins among group A streptococci of different M types. J Infect Dis. 1995;171:871–878. doi: 10.1093/infdis/171.4.871. [DOI] [PubMed] [Google Scholar]
- 38.Nei M. Molecular evolutionary genetics. New York, N.Y: Columbia University Press; 1987. [Google Scholar]
- 39.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 40.Ofek I, Simpson W A, Beachey E H. Formation of molecular complexes between a structurally defined M protein and acylated or deacylated lipoteichoic acid of Streptococcus pyogenes. J Bacteriol. 1982;149:426–433. doi: 10.1128/jb.149.2.426-433.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozeri V, Rosenshine I, Mosher D F, Fassler R, Hanski E. Roles of integrins and fibronectin in the entry of Streptococcus pyogenes into cells via protein F1. Mol Microbiol. 1998;30:625–637. doi: 10.1046/j.1365-2958.1998.01097.x. [DOI] [PubMed] [Google Scholar]
- 42.Perlman D, Halvorson H O. A putative signal recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983;167:391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- 43.Podbielski A, Flosdorff A, Weber-Heynemann J. The group A streptococcal virR49 gene controls expression of four structural vir regulon genes. Infect Immun. 1995;63:9–20. doi: 10.1128/iai.63.1.9-20.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Podbielski A, Schnitzler N, Beyhs P, Boyle M D P. M-related protein (Mrp) contributes to group A streptococcal resistance to phagocytosis by human granulocytes. Mol Microbiol. 1996;19:429–441. doi: 10.1046/j.1365-2958.1996.377910.x. [DOI] [PubMed] [Google Scholar]
- 45.Podbielski A, Spellerberg B, Woischnik M, Pohl B, Lutticken R. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS) Gene. 1996;177:137–147. doi: 10.1016/0378-1119(96)84178-3. [DOI] [PubMed] [Google Scholar]
- 46.Podbielski A, Woischnik M, Leonard B A B, Schmidt K-H. Characterization of nra, a global negative regulator gene in group A streptococci. Mol Microbiol. 1999;31:1051–1064. doi: 10.1046/j.1365-2958.1999.01241.x. [DOI] [PubMed] [Google Scholar]
- 47.Ramachandran G N. Stereochemistry of collagen. Int J Peptide Protein Res. 1988;31:1–16. [PubMed] [Google Scholar]
- 48.Rasmussen M, Muller H-P, Bjorck L. Protein GRAB of Streptococcus pyogenes regulates proteolysis at bacterial surface by binding α2-macroglobulin. J Biol Chem. 1999;274:15336–15344. doi: 10.1074/jbc.274.22.15336. [DOI] [PubMed] [Google Scholar]
- 49.Reid S D, Selander R K, Whittam T S. Sequence diversity of flagellin (fliC) alleles in pathogenic Escherichia coli. J Bacteriol. 1999;181:153–160. doi: 10.1128/jb.181.1.153-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rich R L, Deivanayagam C C S, Owens R T, Carson M, Hook A, Moore D, Yang V W-C, Narayana S V L, Hook M. Trench-shaped binding sites promote multiple classes of interactions between collagen and the adherence receptors, α1β1 integrin and Staphylococcus aureus Cna MSCRAMM. J Biol Chem. 1999;274:24906–24913. doi: 10.1074/jbc.274.35.24906. [DOI] [PubMed] [Google Scholar]
- 51.Rocha C L, Fischetti V A. Identification and characterization of a novel fibronectin-binding protein on the surface of group A streptococci. Infect Immun. 1999;67:2720–2728. doi: 10.1128/iai.67.6.2720-2728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosenberg D E, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 53.Ruoslahti E. RGD and other recognition sequences for integrins. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 54.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 55.Schrager H M, Alberti S, Cywes C, Dougherty G J, Wessels M R. Hyaluronic acid capsule modulates M protein-mediated adherence and acts as a ligand for attachment of group A Streptococcus to CD44 on human keratinocytes. J Clin Investig. 1998;101:1708–1716. doi: 10.1172/JCI2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scott J R, Fischetti V A. Expression of streptococcal M protein in Escherichia coli. Science. 1983;221:758–760. doi: 10.1126/science.6192499. [DOI] [PubMed] [Google Scholar]
- 57.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stockbauer K E, Magoun L, Liu M, Burns E H, Jr, Gubba S, Renish S, Pan X, Bodary S C, Baker E, Coburn J, Leong J M, Musser J M. A natural variant of the cysteine protease virulence factor of group A Streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins αvβ3 and αIIbβ3. Proc Natl Acad Sci USA. 1999;96:242–247. doi: 10.1073/pnas.96.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vercellotti G M, McCarthy J B, Lindholm P, Peterson P K, Jacob H S, Furcht L T. Extracellular matrix proteins (fibronectin, laminin, and type IV collagen) bind and aggregate bacteria. Am J Pathol. 1985;120:13–21. [PMC free article] [PubMed] [Google Scholar]
- 61.Walzog B, Schuppan D, Heimpel C, Hafezi-Moghadam A, Gaehtgens P, Ley K. The leukocyte integrin Mac-I (CD11b/CD18) contributes to binding of human granulocytes to collagen. Exp Cell Res. 1995;218:28–38. doi: 10.1006/excr.1995.1127. [DOI] [PubMed] [Google Scholar]
- 62.Wayner E A, Carter W G. Identification of multiple cell adhesion receptors for collagen and fibronectin in human fibrosarcoma cells possessing unique α and β subunits. J Cell Biol. 1987;105:1873–1884. doi: 10.1083/jcb.105.4.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]