Abstract
Ornibactins are linear hydroxamate siderophores produced by Burkholderia cepacia with peptide structures similar to that of pyoverdines produced by the fluorescent pseudomonads. The gene encoding the outer membrane receptor (orbA) was identified, sequenced, and demonstrated to have significant homology with hydroxamate receptors produced by other organisms. The orbA precursor was predicted to be a protein with a molecular mass of 81 kDa. An orbA mutant was constructed and demonstrated to be unable to take up 59Fe-ornibactins or to grow in medium supplemented with ornibactins. Outer membrane protein profiles from the parent strain, K56-2, revealed an iron-regulated outer membrane protein of 78 kDa that was not detectable in the K56orbA::tp mutant. When this mutant harbored a plasmid containing the orbA gene, the 78-kDa protein was present in the outer membrane protein profiles and the mutant was able to utilize ornibactin to acquire iron. The orbA mutant was less virulent in a chronic respiratory infection model than the parent strain, indicating that ornibactin uptake and utilization are important in the pathogenesis of B. cepacia respiratory infections.
Burkholderia cepacia is an opportunistic pathogen that can cause severe respiratory infections in individuals with cystic fibrosis (CF) or chronic granulomatous disease (18). The incidence of B. cepacia infections in CF patients varies geographically, but prevalence has been reported as high as 40% in some North American centers. Approximately 20% of CF patients colonized with B. cepacia experience a rapid and often fatal pulmonary decline, sometimes associated with septicemia, even in patients with previously mild disease (reviewed in references 17 and 18). Potential virulence factors that may contribute to the severity of B. cepacia infections include siderophores.
Iron is essential for microbial growth, and bacterial pathogens must contend with an iron-restricted environment when colonizing mammalian hosts since iron is bound to transferrin and lactoferrin rendering it essentially unavailable to microbial invaders (30). Pathogenic bacteria require specialized iron acquisition systems to overcome the iron limitation imposed by the host. The most common mechanism of iron acquisition is the secretion of small chelators, called siderophores, that bind ferric iron and transport it into the cell via specific receptor-mediated membrane-associated uptake mechanisms (reviewed in references 8, 31, and 33). Siderophore-mediated iron acquisition is dependent on the activities of the TonB, ExbB, and ExbD proteins, which provide energy to the outer membrane receptor to translocate iron across the bacterial membrane (31).
B. cepacia has been reported to produce four different siderophores: ornibactins, pyochelin, salicyclic acid (SA; formerly azurechelin), and cepabactin (28, 29, 41, 43, 45, 49). Ornibactins and SA are the predominant siderophores produced by clinical isolates of B. cepacia and are produced by 87 and 92%, respectively, of B. cepacia random amplified polymorphic DNA types isolated from CF patients (9). Ornibactins are linear hydroxamate/hydroxycarboxylate siderophores composed of the conserved tetrapeptide l-Orn1(Nδ-OH,Nδ-acyl)-d - threo - Asp(β - OH) - l - Ser - l - Orn4(Nδ - OH,Nδ - formyl) - 1,4 - diaminobutane. The acyl groups of Orn1 vary in length and include 3-hydroxybutanoic acid, 3-hydroxyhexanoic acid, and 3-hydroxyoctanoic acid, forming the three different ornibactins, designated ornibactin-C4, ornibactin-C6, and ornibactin-C8 according to their acyl chain lengths (45, 46). The peptide structures of ornibactins are similar to that of the pyoverdines produced by the fluorescent pseudomonads, Pseudomonas aeruginosa and Pseudomonas fluorescens, but lack a chromophore (29, 45).
In a previous study, we used transposon mutagenesis to identify two genes required for ornibactin synthesis (42). The pvdA gene encodes the enzyme l-ornithine N5-oxygenase, which is responsible for catalyzing the hydroxylation of l-ornithine and for the formation of the hydroxamate ligands (42). The identification of the B. cepacia pvdA gene was based on its homology to the P. aeruginosa pvdA gene, which codes for the same enzyme and which is required for the synthesis of the siderophore pyoverdine (48). The B. cepacia pvdA gene product was shown to be required for both ornibactin biosynthesis and uptake, and pvdA mutants were less virulent than the parent strain in both chronic and acute models of respiratory infection (42). A pvdD homolog that demonstrated homology to peptide synthetases involved in nonribosomal peptide synthesis in a range of bacterial and fungal species was also identified (27). In the present study, we describe the identification and characterization of a gene located downstream of pvdA that codes for the outer membrane receptor for ornibactin.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. B. cepacia strain K56-2 was originally isolated from the sputum of a CF patient. This strain produces SA and ornibactins and negligible amounts of pyochelin and does not produce cepabactin (9, 23). It belongs to B. cepacia genomovar III and has the cblA gene and B. cepacia epidemic strain marker (BCESM) (25). For genetic manipulations, cultures were routinely grown at 37°C in Luria-Bertani broth (Life Technologies, Burlington, Ontario, Canada) or Bacto-Terrific broth (Difco, Detroit, Mich.). Trypticase soy agar was used to quantitate bacteria in lung homogenates. When appropriate, antibiotics were added at the following concentrations: 100 μg of ampicillin, 15 μg of tetracycline, and 1.5 mg of trimethoprim per ml for E. coli and 300 μg of tetracycline, 100 μg of streptomycin, and 100 μg of trimethoprim per ml for B. cepacia.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference(s) or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F′ Φf80dlacZΔM15Δ(lacZYA-argF)recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 deoR U169 | Life Technologies |
| SM10 | Mobilizing strain; RP4 tra genes integrated in chromosome; Kmr | 40 |
| B. cepacia | ||
| K56-2 | Clinical isolate; genomovar III | 9, 23, 25 |
| K56orbA::tp | orbA::tpr derivative of K56-2 | This study |
| Plasmids | ||
| pNOT19 | Modified pUC19 cloning vector; Apr | 37 |
| pUCP26 | Broad-host-range vector; IncP OriT; pRO1600 ori; Tcr | 38 |
| pPD519 | pNOT19 with 6.1-kb SphI fragment from K56-2 containing the pvdA, orbA, and pvdF genes | 42 |
| pEX18Tc | Suicide vector; sacB Tcr | 20 |
| pEX18Ap | Suicide vector; sacB Apr | 20 |
| p34E-Tp | Source of tp cassette; Tpr | 12 |
| pRK2013 | ColE1 Tra (RK2)+ Kmr | 14 |
| pCC524 | pEX18TC with a 3-kb EcoRI-BglII fragment containing orbA cloned into the EcoRI-BamHI sites | This study |
| pCC524T | pCC524 with a SalI fragment containing the 0.6-kb Tpr cassette from p34E-Tp inserted in the XhoI site in the orbA gene | This study |
| pPD524 | pEX18Ap with a 3-kb EcoRI-BglII fragment containing orbA cloned into the EcoRI-BamHI sites | This study |
| pPD526 | pUCP26 with a 3-kb EcoRI-HindIII fragment from pPD524 cloned into the EcoRI-HindIII sites | This study |
For siderophore-iron uptake assays, growth curves, outer membrane preparations, and animal experiments, cultures were grown at 32°C with maximum aeration in TSB-DC as previously described (35). All glassware was washed with acid and rinsed with deionized water to remove iron. All reagents were made with water purified by the Milli-Q system (Millipore, Mississauga, Ontario, Canada).
For outer membrane protein isolation, 20-ml cultures were grown for 16 h and used to inoculate 500-ml cultures that were grown to an A600 of approximately 1.0. The medium was sometimes supplemented with 20 μM ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDHA) or 50 μM FeCl3. Strains K56-2orbA::tp and K56-2orbA::tp(pUCP26) will not grow in the presence of EDDHA, so EDDHA was not added to the medium of these strains.
DNA manipulations.
Molecular biology techniques were performed as generally described by Sambrook et al. (36). Genomic DNA was isolated from K56-2 as described by Ausubel et al. (2). Recombinant plasmids were electroporated into Escherichia coli strain DH5α using a Gene Pulser (Bio-Rad, Richmond, Calif.) according to the manufacturer's recommendations or into B. cepacia K56-2orbA::tp as previously described (11).
For construction of an orbA allelic exchange mutant, a 3-kb EcoRI-BglII fragment from plasmid pPD519 containing orbA (42) was cloned into pEX18Tc (20) to construct plasmid pCC524. A 0.6-kb SalI fragment containing the Tpr cassette from p34E-Tp (12) was inserted into the XhoI site in the orbA gene. This plasmid (pCC524T) was transferred from E. coli to K56-2 by triparental mating using pRK2013 (14) as a mobilizing plasmid. Tpr transconjugants were plated on medium supplemented with 5% sucrose to select for excision of the plasmid. Insertional inactivation of orbA was confirmed by Southern hybridization and PCR using primers internal to orbA, which flanked the XhoI site. Primers orb1 (5′-TGCCGCATGTTGACCCAGTC-3′) and orb2 (5′-AACAACGACCAACGCTCGC-3′) amplified an approximately 700-bp product from K56-2 and an approximately 1,300-bp product from K56orbA::tp.
Nucleotide sequencing.
Nucleotide sequencing was performed using the ABI PRISM BigDyeTM terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase (Perkin-Elmer Corp., Mississauga, Ontario, Canada). DNA sequencing reactions were analyzed with an ABI373A DNA sequencer by the University Core DNA Services (University of Calgary). Custom oligonucleotides were synthesized by Life Technologies. Analysis of the sequence was performed with PC/Gene (Intelligenetics, Mountain View, Calif.), DNAMAN software (Lynnon Biosoft, Vaudreuil, Quebec, Canada), and ORF finder (http://www.ncbi.nlm.nih.gov/Tools/index.html). BLASTX and BLASTP programs were used to search the nonredundant sequence database for homologous sequences (1, 24). The presence of PROSITE protein patterns in the OrbA sequence was determined using the Scan-Prosite tool of the ExPASy molecular biology server of the Swiss Institute of Bioinformatics. The PSORT program on the ExPASy server was used to predict protein localization.
Iron uptake assays.
Cultures were grown to an A600 of 0.3, washed, and resuspended to a final A600 of 0.3 in TSB-DC medium (30, 44). Ornibactins (3.6 mmol) were mixed with an equal amount of 59FeCl3 in a total volume of 100 μl and equilibrated for 10 to 30 min prior to the assay. Uptake reactions were initiated by the addition of 100 μl of the 59Fe-ornibactin mixture to 10 ml of cells. One-milliliter samples of these reaction mixtures were removed at selected intervals, filtered through cellulose acetate (0.45-μM-pore-size) filters (Sartorius GmbH, Goettingen, Germany), and washed with 3 ml of 10 mM Tris (pH 7.5)–0.9% NaCl. The amount of 59Fe accumulated on the filters was measured in an LKB Compugamma counter. SA uptake assays were performed as described above except that 7.2 nmol of SA was equilibrated with 3.6 nmol of 59FeCl3 and 100 μl of 59Fe-SA was used to initiate the uptake reactions (43).
Ornibactin assays.
Production of ornibactins was determined using the Chrome Azurol S (CAS) assay as previously described (39, 42). SA produced by K56-2 is not detectable in the CAS assay (42).
Growth determinations.
To determine the effects of siderophores on growth, overnight cultures were subcultured into 15 ml of medium at an initial cell density corresponding to an A600 of 0.005. Pyochelin, SA, or ornibactins were added to the medium at a final concentration of 10 μg/ml. Growth experiments were performed in triplicate, and growth was measured by determining the A600 of the cultures at selected intervals.
Outer membrane protein isolation.
Outer membranes were prepared by a modification of the methods of Hancock and Nikaido (19) and Gotoh et al. (16). Cells were grown to an A600 of approximately 1.0 and washed once in 30 mM Tris-HCl (pH 8.0), and the cell pellets were frozen at −70°C. The pellets were thawed, resuspended in a solution containing 10 ml of 20% sucrose, 5 mg of DNAse/ml, and 5 mg of RNAse/ml (Sigma, St. Louis, Mo.), and disrupted by oscillation with a sonicator (Branson Cell Disruptor 350) equipped with a medium tip (continuous output; power 8 for a total time of 90 s). Cell debris was removed by centrifugation at 1,000 × g for 10 min. The supernatant was layered onto a seven-step sucrose density gradient consisting of 7 ml of 70% (wt/vol) sucrose, 7 ml of 58% (wt/vol) sucrose, 7 ml of 52% (wt/vol) sucrose, 5 ml of 48% (wt/vol) sucrose, 3 ml of 40% (wt/vol) sucrose, and 3 ml of 30% (wt/vol) sucrose prepared in 30 mM Tris-HCl (pH 8.0) and centrifuged at 100,000 × g for 10 to 15 h. The protein bands between the 52 and 58% sucrose layers and the 58 and 70% sucrose layers containing the outer membrane fraction were collected and pooled. This material was diluted with 30 mM Tris-HCl (pH 8.0), centrifuged at 17,000 × g for 15 min, and resuspended in 1 ml of 30 mM Tris-HCl (pH 8.0). Proteins were quantitated using the Bio-Rad protein assay. Ten micrograms of protein was electrophoresed on sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS–10% PAGE) gel (22).
Animal studies.
Infection experiments were performed in the chronic respiratory infection model with rats as described by Cash et al. (5). Groups of 16 male Sprague-Dawley rats weighing 150 to 170 g (Charles River Canada, Inc.) were tracheostomized under anesthesia and inoculated with the appropriate strain embedded in agar beads as previously described. On days 7 and 28 postinfection (p.i.) the lungs from four animals in each group were removed aseptically and homogenized (Polytron Homogenizer; Brinkman Instruments, Westbury, N.Y.) in 3 ml of phosphate-buffered saline (0.05 M, pH 7.2, containing 0.9% saline). Serial dilutions were plated on Trypticase soy agar and Trypticase soy agar plus the appropriate antibiotic. The lungs of four additional animals in each group were removed en bloc, fixed in 10% formalin, and examined for qualitative and quantitative pathological changes as previously described (13, 44). Infiltration of the lung with inflammatory cells and exudate was measured by the point counting method (13, 44). Briefly, with an integrating eyepiece (Zeiss, Oberkochen, Germany), the number of points overlying the surface area of the infiltrate was divided by the total number of points counted over the entire surface area of the section of the left lobe to obtain a measure of the percentage of infiltration.
Nucleotide sequence accession number.
The nucleotide sequences for the orbA and pvdF genes have been deposited in GenBank and assigned accession no. AF262994.
RESULTS
Identification of the B. cepacia ornibactin receptor gene.
In a previous study, we cloned and characterized a pvdA homolog, the gene for the enzyme l-ornithine N5-oxygenase, and determined that it was required for the biosynthesis of ornibactin in B. cepacia (42). The pvdA gene was cloned on a 6.1-kb SphI fragment from strain K56-2 (Fig. 1). To determine if this fragment contained additional genes involved in siderophore biosynthesis or uptake, the nucleotide sequence downstream of the pvdA gene was determined. An open reading frame (ORF) was identified 64 bp downstream of the stop codon of pvdA in the same frame and orientation; this ORF was predicted to code for a protein with significant homology to siderophore receptor proteins. when this ORF was used to search the GenBank database using the BlastP algorithm (1, 24), the sequences with the highest scoring alignments identified were receptors for hydroxymate type siderophores. The percent similarity between the deduced amino acid sequence encoded by this ORF and those of the most similar hydroxymate siderophores ranged from 50% for P. aeruginosa FiuA (34), a hydroxamate receptor homolog proposed to be the receptor for ferrioxamine B (47), 46 to 49% for ferrioxamine receptors such as E. coli FhuA (7), Yersinia enterocolitica FoxA (3), and Erwinia amylovora FoxR (10). Interestingly, PupB (21) and FpvA (26), the receptors for pseudobactin and pyoverdine, respectively, were only 37% similar to the putative siderophore receptor from B. cepacia although these siderophores are the most similar in structure to the ornibactins.
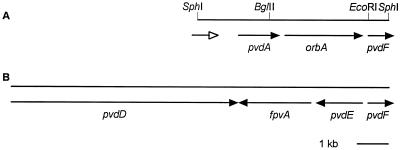
FIG. 1.
Comparison of the organization of pyoverdine and ornibactin receptor genes between B. cepacia K56-2 and P. aeruginosa PAO. (A) Gene organization in K56-2. Solid arrows, genes that have been sequenced; open arrow, partial sequence of potential ORF encoding a protein with weak homology to nonribosomal peptide synthetases. (B) Gene organization in PAO (accession no. U07359).
The gene encoding the B. cepacia ferric siderophore precursor, designated orbA, is predicted to encode a 755-amino-acid polypeptide with a calculated molecular mass of 81,706 Da and an isoelectric point (pI) of 8.49. A potential signal sequence cleavage site was detected between amino acid residues 38 and 39 using the SignalP search program (33). The predicted molecular mass of the mature protein is 77,745 Da. A PROSITE database search detected a signature sequence for TonB-dependent receptor proteins at C-terminal amino acids 738 to 755, with 15 of 18 residues matching the consensus pattern ([LYGSTANE] - XXX - [GSTAENQ] - X - [PGE] - R - X - [LIVFYWA] - X - [LIVMFTA] - [STAGNQ] - [LIVMFYGTA] - X - [LIVMFYWGTADQ]-X-F) (PROSITE PDOC00354) characteristic of most TonB-dependent receptor proteins. By PSORT analysis, the predicted location of OrbA was the outer membrane.
Beginning 86 bp downstream of the orbA coding region is an ORF encoding a polypeptide with 64% identity and 74% similarity to P. aeruginosa pyoverdine synthetase F (PvdF; accession no. AAB6021), which is required for synthesis of pyoverdine, although its specific function has not be described. No other homologs were identified in the database. We have tentatively designated this gene B. cepacia pvdF.
Role of OrbA in siderophore transport.
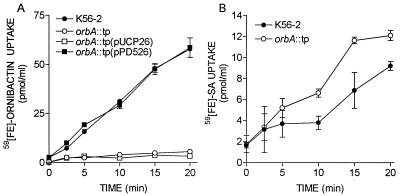
Because of its proximity to genes involved in ornibactin biosynthesis, we hypothesized that orbA was the ornibactin receptor gene. To determine if orbA was required for ornibactin uptake, an orbA mutant was constructed by inserting a trimethoprim resistance cassette into the chromosome by allelic exchange and was designated K56orbA::tp. To determine if this mutant was capable of utilizing ornibactin, its ability to take up 59Fe-ornibactins was compared to that of the parent strain. K56orbA::tp was not able to take up 59Fe-ornibactins during the 20-min assay period (Fig. 2A). When the mutant was complemented with pPD526, which contains the orbA gene, the ability of K56orbA::tp to take up 59Fe-ornibactins was restored (Fig. 2A). To determine if the iron uptake defect was specific for ferric ornibactins, the ability of K56orbA::tp to take up 59Fe-SA was also examined (Fig. 2B). The mutant was able to accumulate 59Fe-SA and interestingly, accumulated this siderophore at a slightly faster rate than K56-2, indicating that orbA was required for ornibactin but not SA uptake.
FIG. 2.
Uptake of 59Fe-siderophore complexes by B. cepacia K56-2 and K56orbA::tp. (A) Uptake assays were initiated by the addition of 59Fe-ornibactins; (B) uptake assays were initiated by the addition of 59Fe-SA. Samples of 1 ml were removed at intervals, and the amount of 59Fe accumulated was determined from a standard curve. Values are means ± standard deviations of triplicate assays. These experiments were repeated at least three times with similar results.
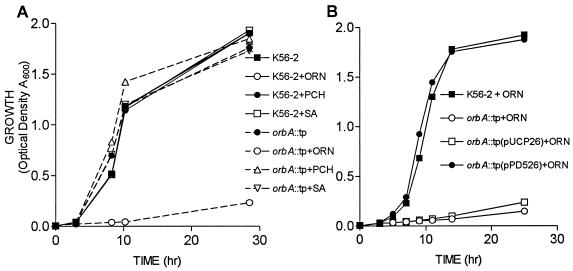
To confirm that orbA is required for ornibactin utilization, the ability of K56orbA::tp to utilize ornibactins, SA, and pyochelin for growth was examined. In TSB-DC medium K56orbA::tp and K56-2 grew at similar rates in the absence of added siderophores (Fig. 3A). No differences in growth rate were observed when either SA or pyochelin was added to the medium. When ornibactins were added to the medium, K56orbA::tp grew very poorly and did not reach an optical density greater than 0.1 (Fig. 3A). When the mutant was complemented with pPD526, the growth rate in the presence of ornibactin was restored to parental levels (Fig. 3B). K56orbA::tp was also not able to grow in the presence of the iron chelator EDDHA unless complemented with pPD526 (data not shown). The inability of K56orbA::tp to take up 59Fe-ornibactins or grow in the presence of ornibactins indicates that orbA is required for ornibactin utilization.
FIG. 3.
Effect of added siderophores on growth of B. cepacia K56-2 and K56orbA::tp. (A) Comparison of growth rates in medium containing 10 μg of either ornibactins, pyochelin, or SA/ml. K56-2, solid lines; K56orbaA::tp, dashed lines. (B) Ability of the orbA gene in trans to complement the growth of K56orbA::tp in the presence of 10 μg of ornibactins/ml. Optical densities of the cultures were determined at selected intervals. Values are means ± standard deviations of triplicate assays. These experiments were repeated twice with similar results.
The ability to utilize ornibactins is not required for production of ornibactins. Ornibactin yields in culture supernatants were measured using the CAS assay (39, 42). Culture supernatants from K56-2 and K56orbA::tp (20 μl) contained CAS activity represented by A630/A600 ratios of 0.99 ± 0.2 and 0.45 ± 0.4, respectively. Although there was a significant difference in ornibactin yields (P < 0.001; t test for unpaired observations), K56orbA::tp was able to produce ornibactins. Since the presence of ornibactins inhibits growth of K56orbA::tp, it is likely that ornibactin production is partially repressed in the mutant compared to that in the parent strain.
Identification of the ornibactin receptor.
Outer membrane preparations were isolated from K56-2 and K56orbA::tp grown in low-iron and high-iron media and analyzed for the presence of iron-regulated proteins by SDS-PAGE. K56-2 had a protein with a molecular mass of approximately 78 kDa that was expressed in low-iron medium but not in medium supplemented with 50 μM FeCl3 (Fig. 4A, compare lanes 2 and 4). In contrast, K56orbA::tp did not express this 78-kDa protein in either low- or high-iron medium (Fig. 4A, compare lanes 5 and 6). The size of this iron-regulated protein correlates with the predicted mass of 77.7 kDa for mature OrbA. Expression of the pyoverdine siderophore receptors in P. aeruginosa and Pseudomonas putida has been shown to be inducible by the presence of the cognate siderophore (15, 21). When K56-2 was grown in the presence of ornibactin, there was no apparent increase in the expression of the 78-kDa protein (Fig. 4A, compare lanes 2 and 3), suggesting that the expression of this protein is not inducible by the presence of ornibactin under these growth conditions. Outer membrane preparations from K56orbA::tp(pPD526) did contain the 78-kDa protein, indicating that the orbA gene on a plasmid was able to complement the defect in expression of this protein (Fig. 4B, compare lanes 3 and 4).
FIG. 4.
Outer membrane protein profiles of B. cepacia K56-2 and K56orbA::tp electrophoresed on SDS–10% PAGE gel and stained with Coomassie brilliant blue. (A) Lane 1, molecular mass markers (kilodaltons) indicated on the left; lanes 2 to 6, 10 μg of outer membrane protein preparations; lane 2, K56-2 grown in medium with 20 μM EDDHA; lane 3, K56-2 grown in medium with 20 μM EDDHA and 10 μg of ornibactins/ml; lane 4, K56-2 grown in medium with 50 μM FeCl3; lane 5, K56orbA::tp grown in medium with no additions; lane 6, K56orbA::tp grown in medium with 50 μM FeCl3. (B) Complementation of K56orbA::tp with the orbA gene. Lane 1, molecular mass markers (kilodaltons) indicated on the left; lane 2, K56-2 grown in medium with 20 μM EDDHA; lane 3, K56orbA::tp(pUCP26); lane 4, K56orbA::tp(pPD526).
Effect of an orbA mutation on virulence.
Previously we demonstrated that mutations in the pvdA gene markedly reduced the ability of B. cepacia to colonize and persist in acute and chronic models of respiratory infection. In addition to being unable to synthesize ornibactin the pvdA mutant was also deficient in the ability to take up Fe-ornibactins, Fe-SA, or Fe-pyochelin (42). Therefore, this mutant was restricted in iron acquisition via all known siderophore-mediated pathways. To determine the importance of ornibactin-mediated iron acquisition in a strain that could utilize SA to acquire iron, the virulence of K56orbA::tp was compared to that of the parent strain in a chronic respiratory infection model.
Rats were infected with K56-2 and K56orbA::tp, and on days 7 and 28 p.i. quantitative bacteriology and quantitative histopathological analyses were performed on lungs removed from infected animals. On day 7 p.i., there was a difference of approximately 3 log units in the number of bacteria (CFU per milliliter) recovered from the lungs between the mutant and the parent strain (Table 2). On day 28 p.i., there was a 4-log-unit difference between the mutant and the parent strain, and in fact the mutant was only recovered from the lungs of one of four animals at this time. K56-2 had similar numbers of bacteria recovered from the lungs on both days 7 and 28 p.i., indicating that the parent strain was able to persist in the lung.
TABLE 2.
Comparison of the virulence of K56-2 with that of K56orbA::tp in a chronic respiratory infection model
| Strain | Virulencea at p.i. day:
|
|||
|---|---|---|---|---|
| 7
|
28
|
|||
| CFU/ml/lung | % Pathology | CFU/ml/lung | % Pathology | |
| K56-2 | 1.5 × 106 ± 2.4 × 106 | 40.5 ± 4.2 | 2.4 × 105 ± 4.5 × 105 | 39.0 ± 4.7 |
| K56orbA::tp | 2.3 × 103 ± 1.3 × 103b | 35.7 ± 3.5 | 1.0 × 101 ± 2.1 × 101 | 11.5 ± 2.6c |
Values are means ± standard deviations for four animals.
K56orbA::tp was significantly different from K56-2 (P < 0.05) by analysis of variance (ANOVA) and Dunn's multiple comparisons test.
K56orbA::tp was significantly different from K56-2 (P < 0.05) by ANOVA.
On day 7 p.i., no significant difference in the percent lung pathology between the mutant and the parent strains was observed (Table 2). On day 28 the lungs infected with the orbA mutant had 70% less pathological involvement than did lungs infected with K56-2. The percent pathologies were nearly identical for K56-2-infected lungs on both days 7 and 28. The observed difference in lung pathology between K56-2 and K56orbA::tp on day 28 is likely due to the number of bacteria present in the lung. These data indicate that the ornibactin-mediated iron uptake pathway is important for the virulence of B. cepacia even in a strain with a functional SA-mediated iron uptake pathway.
DISCUSSION
In this study we have identified a gene that encodes the outer membrane receptor for the siderophore ornibactin. The gene, designated orbA, is predicted to encode a precursor protein with a molecular mass of 81,706 Da. The predicted mass of the mature protein is 77,745 Da. Examination of the outer membrane protein profiles revealed that a 78-kDa iron-regulated protein was present in the parent strain but not present in an orbA mutant. The orbA mutant was not able to utilize ornibactin for iron transport or growth.
Previously, we demonstrated that B. cepacia pvdA mutants were markedly less virulent than the parent strain, K56-2, in a chronic respiratory infection model (42). These mutants were cleared from the lungs of the majority of animals examined on day 28 p.i. In addition to not producing ornibactin these mutants were also reduced in their ability to take up iron complexes to either ornibactin or SA (42). The orbA mutant showed a reduction in virulence similar to that of the pvdA mutants in the respiratory infection model in that it was also cleared from the lungs of animals examined on day 28 p.i. Although K56orbA::tp produced SA and could take up 59Fe-SA at a higher rate than the parent, the SA-mediated iron acquisition system was not able to compensate for the defect in ornibactin utilization. These studies indicate that, although SA can function as a siderophore in in vitro assays (43, 49), it is not able to compete with host iron binding proteins in vivo and promote iron acquisition. SA was also not able to compete with ornibactin in the in vitro growth assay, since addition of exogenous ornibactin almost completely inhibited growth of K56orbA::tp (Fig. 3). Previously, a mutant with a Tn5 insertion in the pvdA gene was shown to hyperproduce SA (42). This mutant did not produce detectable zones on CAS agar (39), however, indicating that SA also does not compete effectively with the CAS dye for iron in the medium used for these assays. K56-2 does not produce cepabactin and produces only negligible amounts of pyochelin (9). It would be interesting to determine the effects of an ornibactin uptake mutation in strains that produce pyochelin and/or cepabactin to determine if these siderophore-mediated iron acquisition systems could compensate for a defect in ornibactin utilization in vivo.
Infection with K56orbA::tp resulted in lung histopathologic changes similar to those produced by the parent strain on day 7 p.i., although on day 28 p.i. the histopathologic changes were 70% less in lungs infected with K56orbA::tp than in lungs infected with K56-2 (Table 2). Animals infected with pvdA mutants; however, had 50 to 60% less histopathologic changes in the lungs on day 7 p.i. than animals infected with K56-2 (42). The differences in pathology observed on day 7 p.i. between the pvdA and orbA mutants cannot be attributed to differences in the number of bacteria since the CFUs of K56orbA::tp and K56orbA::tp recovered from the lungs were not significantly different and in both cases were approximately 3 log units lower than the number of bacteria recovered from the lungs of K56-2-infected animals. Since K56orbA::tp produces approximately 45% of parental ornibactin levels and since the pvdA mutant does not produce ornibactins, it is possible that ornibactin produced by K56orbA::tp contributes to lung damage. P. aeruginosa pyochelin, when loaded with iron, has been shown to generate hydroxyl radicals in the presence of neutrophil sources of superoxide and hydrogen peroxide (4, 6). Ferripyochelin was also shown to promote hydroxyl radical-mediated damage to airway epithelial cells (4). Ferripyoverdine, which is more related to ornibactins in terms of structure than pyochelin, did not catalyze the generation of hydroxyl radicals (6). It has not been reported if ornibactins have properties similar to those of pyochelin in terms of hydroxyl radical generation.
The orbA gene is located between pvdA, which is required for ornibactin synthesis, and a pvdF homolog, which is reportedly required for pyoverdine synthesis in P. aeruginosa. The orbA mutant produced ornibactin (data not shown), suggesting that the insertion of the trimethoprim cassette did not affect the expression of either of the flanking genes. Expression of the ornibactin receptor was restored to K56orbA::tp by introduction of pPD526 containing the orbA gene on a 3-kb fragment. There were no obvious −35 and −10 consensus sequences identified upstream of orbA; however, these results suggest that orbA may have its own promoter. We were unable to identify any consensus promoter sequences upstream of pvdA (42). Few promoter sequences have been determined in Burkholderia spp. for comparison with consensus promoter sequences.
The genes involved in ornibactin synthesis and uptake in B. cepacia K56-2 are arranged quite differently from the corresponding genes in P. aeruginosa PAO (Fig. 1). In P. aeruginosa, the pyoverdine receptor gene, fpvA, is located between pvdE and pvdD, which are divergently transcribed. pvdF is located upstream of pvdD and transcribed in the opposite orientation. The pvdA gene is located approximately 11.9 kb downstream of pvdF (Pseudomonas Genome Project; www.pseudomonas.com). At least seven ORFs have been identified between pvdA and pvdF, and these ORFs are presumably not involved in iron transport mechanisms. In B. cepacia and orbA receptor is located between pvdA and pvdF and all three ORFs are in the same orientation. An ORF which encodes a protein that exhibits weak homology to nonribosomal peptide synthetases was identified in the same orientation upstream of pvdA. This ORF, which is only partially contained on the 6.1-kb SphI fragment, may be involved in the synthesis of ornibactins. The differences in gene organization between B. cepacia and P. aeruginosa may indicate that there are differences in regulation or possibly function between these two iron transport systems. Further studies on both the ornibactin and pyoverdine biosynthesis and uptake systems are needed to explore this possibility.
B. cepacia produces three different ornibactins, ornibactin-C4, ornibactin-C6, and ornibactin-C8 (45, 46). The preparation of purified ornibactins used in the growth experiments and the 59Fe-ornibactin uptake assays contained a mixture of these three ornibactin molecules purified from strain K56-2. The fact that K56orbA::tp was not able to take up the 59Fe-ornibactin mixture or grow in the presence of ornibactins added to the culture medium suggests that OrbA is the receptor for all three ornibactin molecules, regardless of the acyl side chain.
B. cepacia produces at least four siderophores, ornibactins, pyochelin, SA, and cepabactin. Little is known about the genes or their products involved in the biosynthesis and uptake of these siderophores in this organism. In this study we have reported the first identification of a siderophore receptor in B. cepacia and have extended our studies which indicate that the ornibactin-mediated iron acquisition system is required for virulence of B. cepacia in chronic respiratory infections.
ACKNOWLEDGMENTS
This study was supported by a grant from the Canadian Cystic Fibrosis Foundation. S.L. is the recipient of an Alberta Heritage Foundation for Medical Research Studentship award. C.C. was the recipient of an Alberta Heritage Foundation for Medical Research Summer Studentship award.
We thank Donald E. Woods for quantitative analyses of histopathologic changes in the lungs.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 3.Baumler A J, Hantke K. Ferrioxamine uptake in Yersinia enterocolitica: characterization of the receptor protein FoxA. Mol Microbiol. 1992;6:1309–1321. doi: 10.1111/j.1365-2958.1992.tb00852.x. [DOI] [PubMed] [Google Scholar]
- 4.Britigan B E, Rasmussen G T, Cox C D. Augmentation of oxidant injury to human pulmonary epithelial cells by the Pseudomonas aeruginosa siderophore pyochelin. Infect Immun. 1997;65:1071–1076. doi: 10.1128/iai.65.3.1071-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cash H A, Woods D E, McCullough B, Johanson W G, Jr, Bass J A. A rat model of chronic respiratory infection with Pseudomonas aeruginosa. Am Rev Respir Dis. 1979;119:453–459. doi: 10.1164/arrd.1979.119.3.453. [DOI] [PubMed] [Google Scholar]
- 6.Coffman T J, Cox C D, Edeker B L, Britigan B E. Possible role of bacterial siderophores in inflammation. Iron bound to the Pseudomonas siderophore pyochelin can function as a hydroxyl radical catalyst. J Clin Investig. 1990;86:1030–1037. doi: 10.1172/JCI114805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coulton J W, Mason P, Cameron D R, Carmel G, Jean R, Rode H N. Protein fusions of beta-galactosidase to the ferrichrome-iron receptor of Escherichia coli K-12. J Bacteriol. 1986;165:181–192. doi: 10.1128/jb.165.1.181-192.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crosa J H. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol Rev. 1989;53:517–530. doi: 10.1128/mr.53.4.517-530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darling P, Chan M, Cox A D, Sokol P A. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect Immun. 1998;66:874–877. doi: 10.1128/iai.66.2.874-877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dellagi A, Reis D, Vian B, Expert D. Expression of the ferrioxamine receptor gene of Erwinia amylovora CFBP 1430 during pathogenesis. Mol Plant-Microbe Interact. 1999;12:463–466. doi: 10.1094/MPMI.1999.12.5.463. [DOI] [PubMed] [Google Scholar]
- 11.Dennis J J, Sokol P A. Electrotransformation of Pseudomonas. In: Nickoloff J A, editor. Electroporation and electrofusion of microorganisms. Clifton, N.J: Humana Press; 1995. pp. 125–133. [DOI] [PubMed] [Google Scholar]
- 12.DeShazer D, Woods D E. Broad-host-range cloning and cassette vectors based on the R388 trimethoprim resistance gene. BioTechniques. 1996;20:762–764. doi: 10.2144/96205bm05. [DOI] [PubMed] [Google Scholar]
- 13.Dunnil M S. Quantitative methods in the study of pulmonary pathology. Thorax. 1962;17:320–328. doi: 10.1136/thx.17.4.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figuski D H, Helenski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gensberg K, Hughes K, Smith A W. Siderophore-specific induction of iron uptake in Pseudomonas aeruginosa. J Gen Microbiol. 1992;138:2381–2387. doi: 10.1099/00221287-138-11-2381. [DOI] [PubMed] [Google Scholar]
- 16.Gotoh N, White N J, Chaowagul W, Woods D E. Isolation and characterization of the outer-membrane proteins of Burkholderia (Pseudomonas) pseudomallei. Microbiology. 1994;140:797–805. doi: 10.1099/00221287-140-4-797. [DOI] [PubMed] [Google Scholar]
- 17.Govan J R, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govan J R, Hughes J E, Vandamme P. Burkholderia cepacia: medical, taxonomic and ecological issues. J Med Microbiol. 1996;45:395–407. doi: 10.1099/00222615-45-6-395. [DOI] [PubMed] [Google Scholar]
- 19.Hancock R E, Nikaido H. Outer membranes of gram-negative bacteria. XIX. Isolation from Pseudomonas aeruginosa PAO1 and use in reconstitution and definition of the permeability barrier. J Bacteriol. 1978;136:381–390. doi: 10.1128/jb.136.1.381-390.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoang T T, Karkhoff-Schweizer R R, Kutchma A J, Schweizer H P. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 21.Koster M, van de Vossenberg J, Leong J, Weisbeek P J. Identification and characterization of the pupB gene encoding an inducible ferric-pseudobactin receptor of Pseudomonas putida WCS358. Mol Microbiol. 1993;8:591–601. doi: 10.1111/j.1365-2958.1993.tb01603.x. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lewenza S, Conway B, Greenberg E P, Sokol P A. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J Bacteriol. 1999;181:748–756. doi: 10.1128/jb.181.3.748-756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madden T L, Tatusov R L, Zhang J. Applications of network BLAST server. Methods Enzymol. 1996;266:131–141. doi: 10.1016/s0076-6879(96)66011-x. [DOI] [PubMed] [Google Scholar]
- 25.Mahenthiralingam E, Coenye T, Chung J W, Speert D P, Govan J R W, Taylor P, Vandamme P. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol. 2000;38:910–913. doi: 10.1128/jcm.38.2.910-913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMorran B J, Merriman M E, Rombel I T, Lamont I L. Characterisation of the pvdE gene which is required for pyoverdine synthesis in Pseudomonas aeruginosa. Gene. 1996;176:55–59. doi: 10.1016/0378-1119(96)00209-0. [DOI] [PubMed] [Google Scholar]
- 27.Merriman T R, Merriman M E, Lamont I L. Nucleotide sequence of pvdD, a pyoverdine biosynthetic gene from Pseudomonas aeruginosa: PvdD has similarity to peptide synthetases. J Bacteriol. 1995;177:252–258. doi: 10.1128/jb.177.1.252-258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer J M, Hohnadel D, Halle F. Cepabactin from Pseudomonas cepacia, a new type of siderophore. J Gen Microbiol. 1989;135:1479–1487. doi: 10.1099/00221287-135-6-1479. [DOI] [PubMed] [Google Scholar]
- 29.Meyer J M, Van V T, Stintzi A, Berge O, Winkelmann G. Ornibactin production and transport properties in strains of Burkholderia vietnamiensis and Burkholderia cepacia (formerly Pseudomonas cepacia) Biometals. 1995;8:309–317. doi: 10.1007/BF00141604. [DOI] [PubMed] [Google Scholar]
- 30.Mietzner T, Morse S. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu Rev Nutr. 1994;14:471–493. doi: 10.1146/annurev.nu.14.070194.002351. [DOI] [PubMed] [Google Scholar]
- 31.Moeck G S, Coulton J W. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol Microbiol. 1998;28:675–681. doi: 10.1046/j.1365-2958.1998.00817.x. [DOI] [PubMed] [Google Scholar]
- 32.Neilands J B. Microbial iron compounds. Annu Rev Biochem. 1981;50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 34.Ochsner U A, Vasil M L. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci USA. 1996;93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohman D E, Sadoff J C, Iglewski B H. Toxin A-deficient mutants of Pseudomonas aeruginosa PA103: isolation and characterization. Infect Immun. 1980;28:899–908. doi: 10.1128/iai.28.3.899-908.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 37.Schweizer H P. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol Microbiol. 1992;6:1195–1204. doi: 10.1111/j.1365-2958.1992.tb01558.x. [DOI] [PubMed] [Google Scholar]
- 38.Schweizer H P, Klassen T, Hoang T. Improved methods for gene analysis and expression in Pseudomonas spp. In: Nakazawa T, Furukawa K, Hass D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C.: ASM Press; 1996. pp. 229–237. [Google Scholar]
- 39.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 40.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 41.Sokol P A. Production and utilization of pyochelin by clinical isolates of Pseudomonas cepacia. J Clin Microbiol. 1986;23:560–562. doi: 10.1128/jcm.23.3.560-562.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sokol P A, Darling P, Woods D E, Mahenthiralingam E, Kooi C. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding l-ornithine N5-oxygenase. Infect Immun. 1999;67:4443–4455. doi: 10.1128/iai.67.9.4443-4455.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokol P A, Lewis C J, Dennis J J. Isolation of a novel siderophore from Pseudomonas cepacia. J Med Microbiol. 1992;36:184–189. doi: 10.1099/00222615-36-3-184. [DOI] [PubMed] [Google Scholar]
- 44.Sokol P A, Woods D E. Relationship of iron and extracellular virulence factors to Pseudomonas aeruginosa lung infections. J Med Microbiol. 1984;18:125–133. doi: 10.1099/00222615-18-1-125. [DOI] [PubMed] [Google Scholar]
- 45.Stephan H, Freund S, Beck W, Jung G, Meyer J M, Winkelmann G. Ornibactins—a new family of siderophores from Pseudomonas. Biometals. 1993;6:93–100. doi: 10.1007/BF00140109. [DOI] [PubMed] [Google Scholar]
- 46.Stephan H, Freund S, Meyer J, Winklemann G, Gung G. Structure elucidation of the gallium-ornibactin complex by 2D-NMR spectroscopy. Liebigs Ann Chem. 1993;1993:43–48. [Google Scholar]
- 47.Vasil M L, Ochsner U A. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol Microbiol. 1999;34:399–413. doi: 10.1046/j.1365-2958.1999.01586.x. [DOI] [PubMed] [Google Scholar]
- 48.Visca P, Ciervo A, Orsi N. Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdin biosynthetic enzyme l-ornithine N5-oxygenase in Pseudomonas aeruginosa. J Bacteriol. 1994;176:1128–1140. doi: 10.1128/jb.176.4.1128-1140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Visca P, Ciervo A, Sanfilippo V, Orsi N. Iron-regulated salicylate synthesis by Pseudomonas spp. J Gen Microbiol. 1993;139:1995–2001. doi: 10.1099/00221287-139-9-1995. [DOI] [PubMed] [Google Scholar]