Abstract
Simple Summary
This review article aims to compile the information published in the scientific literature regarding Coxiella burnetii infection in goats and their role in the epidemiology of infection, namely their association with the occurrence of Q fever in humans. Q fever presents a worldwide occurrence and the risk that it represents to humans has been recognized since its first description. The characteristics of C. burnetii justify its classification as a category B biological threat agent. International public health authorities strongly recommend global monitoring of C. burnetii, especially after large-scale Q fever epidemics occurred in The Netherlands, which originated from goat infection. An approach with the characterization of the bacterium, its strategies of infection, and clinical patterns in goats will help to understand the dynamics of infection in an epidemiological analysis and to analyze the role of goats in Q fever.
Abstract
Since its first description in the late 1930s, Q fever has raised many questions. Coxiella burnetii, the causative agent, is a zoonotic pathogen affecting a wide range of hosts. This airborne organism leads to an obligate, intracellular lifecycle, during which it multiplies in the mononuclear cells of the immune system and in the trophoblasts of the placenta in pregnant females. Although some issues about C. burnetii and its pathogenesis in animals remain unclear, over the years, some experimental studies on Q fever have been conducted in goats given their excretion pattern. Goats play an important role in the epidemiology and economics of C. burnetii infections, also being the focus of several epidemiological studies. Additionally, variants of the agent implicated in human long-term disease have been found circulating in goats. The purpose of this review is to summarize the latest research on C. burnetii infection and the role played by goats in the transmission of the infection to humans.
Keywords: zoonosis, C. burnetii, prevalence, outbreaks, genotype
1. Introduction
The history of Q fever, the disease caused by Coxiella burnetii, can be traced back to 1937, when it was described by Edward Holbrooke Derrick in Australia [1]. Almost simultaneously, in the United States, an unknown agent isolated from ticks recovered from Nine Mile Creak region, Montana, was described [2]. Australian and American teams shared their findings and concluded that they were studying the same agent and the same disease [3]. The potential risk of Q fever to public health and the large gaps in the knowledge of this disease were recognized early, namely by the World Health Organization (WHO) that, in 1950, encouraged the epidemiological research. Consequently, Q fever was reported in 51 countries from the five continents [4]. In Europe, Q fever was first reported in Greece, during the Second World War, in German soldiers who had febrile illness, the so-called “Balkan flu” [5].
Nowadays, except in New Zealand, C. burnetii is found worldwide, infecting a wide range of domestic and wildlife animals [6,7]. Q fever is listed in the Terrestrial Animal Health Code of the World Organization for Animal Health (WOAH) and all Member Countries are required to report the occurrence of the disease [8].
Since its first report, human Q fever outbreaks have been regularly reported throughout the world [9]. From 2007 until 2010, The Netherlands faced the largest Q fever outbreak ever recorded, resulting in over 4000 reported and 40,000 estimated infected people [10]. This occurrence alerted public health authorities regarding C. burnetii and the need for a harmonized monitoring of infection was highlighted [11,12,13,14]. In fact, during the last decade, the number of relevant publications on this subject increased significantly [15].
Despite the wide host range of C. burnetii, the infection is mostly recognized in domestic ruminants [7,16,17,18,19]. However, over time, human Q fever outbreaks have often been related to spill-over infection from goats to humans, as shown in Table 1.
Table 1.
Human Q fever outbreaks associated with goats.
Recently, it was observed that goats played a major role in infecting humans compared with sheep in The Netherlands Q fever outbreak, which was previously associated with small ruminants [18]. Thus, the infection patterns in goats (e.g., dynamics of infection, clinical outcomes, and shedding patterns) need clarification, and experimental studies using goats have improved our scientific knowledge on this topic. Furthermore, the effective strategy of prevention and control of Q fever, in both humans and animals, requires a One Health perspective [28,30]. This review is not to intend to give a global geographic epidemiological view of Q fever, but rather focus on the role of goats in the epidemiology of this zoonotic pathogen. First, the characteristics of the agent and its pathogenesis will be briefly described, considering its significance in understanding the clinical and excretion patterns and followed by the specific infection in goats. Then, the focus will be in the epidemiological role of goats in Q fever dissemination, finally referring to the importance of molecular tools to unravel the complex epidemiology of C. burnetii.
This review will focus on C. burnetii infection, highlighting the role of goats in the epidemiology of this zoonotic pathogen. An overview of the pathogenesis, as well as of the epidemiological characteristics influencing the dissemination of the infection, will be described referring to the importance of molecular tools to unravel the complex epidemiology of C. burnetii.
2. Coxiella burnetii: The Microorganism and Its Pathogenesis
When Q fever was first described, its causative agent was unknown. In 1948, the genus Coxiella was created and Coxiella burnetii (Philip, 1948) was listed in the 6th edition of Bergey’s Manual of Determinative Bacteriology [3,33] as the aetiological agent of Q fever.
Phylogenetic investigations based on 16S rRNA sequence analysis placed C. burnetii in the gamma group of proteobacteria, belonging to the order Legionellales, family Coxiellaceae, and genus Coxiella [34]. The first complete genome sequence of C. burnetii was published in 2003. It corresponded to the original strain (RSA 493 strain) firstly isolated from ticks in the United States, also known as the Nine Mile strain. This event led to significant advances in the knowledge of C. burnetii [35]. The genome of C. burnetii contains conserved genomic regions as well as polymorphic regions [36]. Furthermore, the insertion sequence IS1111 plays an important role in the genomic plasticity of C. burnetii. The number of IS1111 elements is highly variable between strains; many different genetic locations are described, showing a direct impact on C. burnetii genotypes [37].
C. burnetii is a small pleomorphic Gram-negative rod, presenting 0.2–0.4 μm wide and 0.4–1.0 μm long [38]. All the lipopolysaccharides (LPSs) encoding genes are in a 38 Kb region in the C. burnetii genome, and it has been observed that mutational variations in this region result in antigenic and virulence shift, termed “phase variation”. Antigenic variation results from an irreversible modification from smooth-type (phase I) to rough-type (phase II) LPS causing a dramatic reduction in virulence [39]. Thus, the avirulent rough LPS (phase II) results from a point/frameshift mutation, small deletion, or transposon insertion in a gene in the LPS biosynthetic pathway [40,41]. Therefore, the sugar composition of phase II LPS is quite different because sugars such as L-virenose dihydrohydroxystreptose and galactosamine uronyl-(1,6) glucosamine are lacking [39,42]. So, the lack of virulence is associated with a shorter LPS and not with a defect in the synthesis of other virulence factors. However, it is interesting to note that avirulent forms of other strains besides Nine Mile show different patterns of deletions/mutations, suggesting that the biosynthesis of LPS in C. burnetii is not yet completely understood [40]. The shift from virulent phase I to avirulent phase II is likely due to repeated passages of the strains in cell cultures or embryonated eggs [43].
Phase I C. burnetii can be recovered from infected hosts and the smooth-type LPS of phase I disturbs an effective immune response, giving the phase I bacterium the opportunity to survive and multiply in the host cells. Therefore, phase I C. burnetii is highly infectious [39].
C. burnetii exhibits a biphasic developmental cycle in which two main morphological forms are identified: large cell variant (LCV) and small cell variant (SCV) [44]. LCVs have a larger size (>0.5 μm), they are metabolically active, and have less electron dense forms. They have dispersed and filamentous chromatin and possess clearly distinguishable outer and cytoplasmic membranes with exposed LPS on the surface, sharing features with Gram-negative bacteria. These LCVs are sensitive to the decrease in osmotic pressure [45,46,47]. SCVs are small rod-shaped forms ranging typically from 0.2 and 0.5 μm, being filterable through 0.22 μm filters. They are very compact and present low metabolic activity [44,46]. Some structural characteristics of SCVs are the electron-dense and condensed chromatin and the unusual cell envelope characterized by a high number of cross-links in peptidoglycans, which seems to enhance environmental stability [45,48]. Thus, they are very stable in the environment, showing a high resistance to osmotic, mechanical, chemical, heat, and desiccation stresses [44,48].
The primary target cells of C. burnetii are blood-circulating monocytes, macrophages (e.g., lymph nodes, spleen, liver, and lungs) [49], and trophoblasts in pregnant females [50].
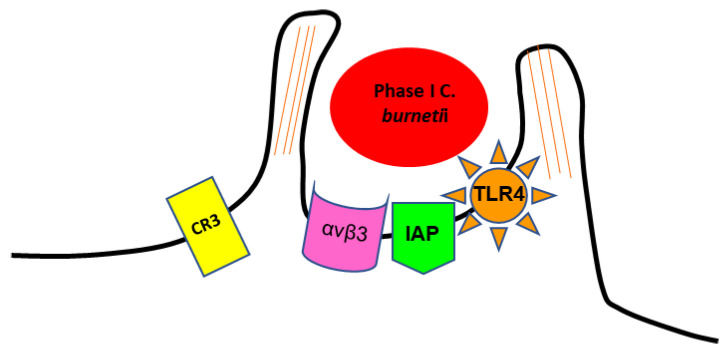
The internalisation of phase I SCV of C. burnetii in target cells involves the recognition of several receptors [51]. It is mediated by the leukocyte response integrin (LRI) (αvβ3) and an integrin-associated protein (IAP) [39,52]. The entry occurs through a microfilament-dependent endocytosis [44,51]. Phase I LPS induces a rearrangement of F-actin cytoskeleton, leading to pronounced membrane protrusions at the site of bacterial adherence. This phenomenon, called membrane ruffling, requires contact between C. burnetii and host cells, and depends on the expression of toll-like receptor type 4 (TLR4) on the host cell surface (Figure 1) [53,54,55]. The ability to use αvβ3 integrin for invasion might be exploited by C. burnetii as a mechanism to avoid the induction of an inflammatory response, as αvβ3 integrin is typically involved in the removal of apoptotic cells via phagocytosis, being generally associated with an inhibition of inflammation [52]. Thus, C. burnetii enters the cells without alerting the immune system [56].
Figure 1.
Scheme representing the internalization of phase I SCV of C. burnetii by monocyte-like cells.
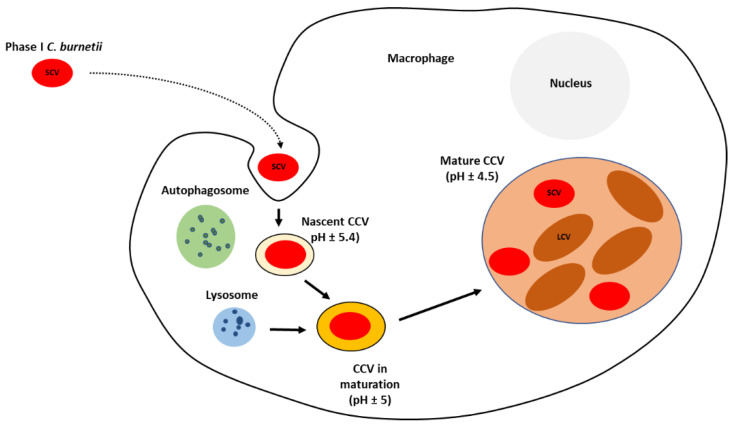
After internalization, bacteria localize within the nascent Coxiella-containing vacuole (CCV), which traffics through the endocytic cascade. It develops into an early phagosome acquiring the small GTPase RAB5. This GTPase stimulates the fusion with early endosomes, resulting in acidification of the lumen to approximately pH 5.4 and acquisition of the early-endosomal marker protein 1 (EEA1) [57,58]. Early phagosome is converted into late phagosome acquiring acid hydrolases, which are involved in pronounced degradative activity, which C. burnetii can resist [59]. This late phagosome lacks RAB5 and EEA1 but acquires lysosome-associated membrane protein 1, 2, and 3 (LAMP1, LAMP2, and LAMP3) and vacuolar ATPase, which pumps protons into the maturing phagosome to further decrease the luminal pH to about 5.0 [58,60,61]. C. burnetii persists and replicates, at a slow rate, within the large CCV with an acidic environment [39,62,63]. The process of phagosome maturation continues with its fusion with lysosomal compartments to acquire cathepsins and hydrolases. The vacuolar ATPase further reduces the pH to around 4.5 [58,64]. Phagosome maturation depends on the balance between pro-inflammatory (IFN-γ, IL-12, and IL-6) and anti-inflammatory (IL-10) cytokines [65]. C. burnetii modulates the genesis of CCV and has several strategies for adaptation to the stressful environment. It encodes a significant number of basic proteins that are probably involved in buffering the acidic environment of the CCV. Moreover, four sodium–proton exchangers and transporters for osmoprotectants are codified in its genome, allowing this bacterium to confront osmotic and oxidative stresses [35].
During its biogenesis process, CCV becomes large and contains a large number of bacteria [62]. C. burnetii does not synthesize its own CCV membrane. Multiple fusion events with autophagosomes along with endolysosomal vacuoles are essential to provide sufficient membrane to enlarge the CCV [66,67]. C. burnetii continuously directs fusion with other host cell compartments and inhibits apoptotic cell death, allowing a prolonged infectious cycle [63,68,69,70,71].
The internalised SCV, within the CCV, suffers a differentiation into replicative and metabolically active LCV (Figure 2). The low intra-phagosomal pH and perhaps enzyme system and/or nutrient sources present in the vacuole seem to trigger this differentiation. Lag phase extends to approximately two days post-infection and is composed primarily of SCV to LCV morphogenesis. The exponential phase occurs over the next four days with CCV harbouring replicating LCV almost exclusively. The LCV multiplies and persists within an expanding CCV that contains lysosomal elements, including an acid pH (5.0) and degradative proteases [44,46,59,72].
Figure 2.
Diagram of the intracellular lifecycle of C. burnetii. CCV—Coxiella-containing vacuole; SCV—small cell variant; LCV—large cell variant.
A dramatic expansion of the CCV occurs concomitantly with the appearance of replicating LCV, occupying nearly the entire cytoplasm [44,66]. These metabolically active LCVs also play an important role in cell-to-cell spread during acute infection. This process is facilitated by the display of unique LCV antigens such as a porin protein termed P1. The stationary phase begins six days post-infection, concomitantly with the re-appearance of SCV. Following the accumulation of large numbers of LCVs, C. burnetii converts back into SCVs, which are released from heavily infected cells by an undefined mechanism [44].
The resistance properties of these SCVs strongly implicate this form as responsible for long-term extracellular survival and aerosol transmission of C. burnetii [44,45].
3. Infection and Clinical Outcomes in Goats
It is globally recognized that C. burnetii infection occurs mainly by inhalation of contaminated aerosols and, because C. burnetii is a highly infective pathogen, low doses cause a high risk of illness [73,74]. So far, experimental studies on goats were not focused on estimating the infectious dose. However, in humans, it was estimated that the 50% infectious dose was around one bacterium [75].
Alveolar macrophages are the first-line defence that confronts C. burnetii [49,76]. The ability of these cells to rapidly respond recruiting additional immune cells is central for an effective antibacterial response in early stages of infection [65,77]. In primary infections, after entry into the organism, a bacteraemia occurs, leading to a systemic infection with the involvement of organs such as liver, spleen, lungs, and bone marrow [38]. The organism can subsequently disseminate to colonize and replicate in resident macrophages of different tissues and organs [78]. In pregnant goats, the main target cells are the trophoblasts in the allanthocorion, causing a placentitis and necrosis of placental tissues [79,80]. The amount of C. burnetii DNA detected increases until parturition and decreases drastically after parturition, probably by the disappearing of trophoblasts, the replication niche of C. burnetii during pregnancy [79,81]. This strong tropism of C. burnetii towards placenta does not seem to occur for other tissues of nonpregnant goats and kids, suggesting that pregnant females are more susceptible to C. burnetii infection [79,82].
Cell-mediated immunity probably plays a critical role in controlling C. burnetii infection [49,55]. Cells belonging to monocyte-macrophage lineage express polarized functional properties. This polarization seems to be closely related to the ability to control C. burnetii infection, explaining the bacterial persistence in chronic infections [83]. Classically, M1 polarized macrophages are induced by LPS, IFN-γ, and TNF-α, and participate in the resistance against intracellular pathogens involved in Th1 responses. In contrast, M2-polarized macrophages are induced by IL-4, IL-13, or IL-10 and promote Th2 responses. So, it is thought that the course of infection differs according to the macrophage polarization in response to C. burnetii infection [83]. If M1-associated molecules are expressed by macrophages, the bacterial replication will be controlled [62,83], while the stimulation of an M2 response will account for the persistence of C. burnetii in macrophages, which become highly permissive to C. burnetii replication [83,84,85].
Beyond cell-mediated response, an antibody-mediated immunity also seems to be important in C. burnetii infection [49]. Treatment of C. burnetii infection with immune sera makes the bacterium more susceptible to phagocytosis and destruction by macrophages [86]. Specific immunoglobulins are secreted following infection [38] and the infection of dendritic cells with antibody-opsonized bacteria results in increased expression of maturation markers and inflammatory cytokines in mice [49]. It can be concluded from field studies that C. burnetii antibodies are highly persistent, lasting for several months up to years [87,88]. Thus, both humoral and cellular immunity play a role in C. burnetii infection.
However, the immune control of C. burnetii might not lead to its eradication from the infected host [55]. It is also hypothesized that the uterus could be a site of latent infection, hence reactivation during pregnancy can occur [89,90].
In goats, as well as in other domestic ruminants, C. burnetii infection often goes unnoticed owing to the absence of symptoms, and the term Coxiellosis is usually used to refer this condition [8]. In the early stages after infection, C. burnetii can be detected in the blood, lungs, spleen, and liver. However, it is not clear if its presence in organs other than placenta affects the functions of these organs, as only mild lesions have been described [79,81,91,92]. Experimental infection of non-pregnant goats showed that, at late stages of infection, C. burnetii was present in mammary glands, emphasizing the milk as an important shedding route [82]. Infection of pregnant goats may cause a wide range of conditions including abortion, delivery of premature offspring, stillbirth, and weak offspring. Of these, one of the most important outcomes of the C. burnetii infection is the abortion, which occur at the end of pregnancy without premonitory signs. In dairy goat herds that experience abortions caused by C. burnetii, an increased incidence of metritis can be noticed. Notwithstanding, a clinically normal progeny, which may or may not be congenitally infected, may occur, as described in infection of non-pregnant goats [7,79,82]. However, it seems that apparently healthy kids born from infected mothers may develop respiratory and digestive tract disorders [7].
In the season that follows an abortion storm, the multiplication of the organism may be reactivated during pregnancy, leading to reproductive failures [93,94,95]. Even in asymptomatic infections, a latent infection may develop and a reactivation late in pregnancy can occur several days before parturition. Generally, when late-term abortions, stillbirths, or birth of stunted animals are observed in goat flocks, Q fever should be suspected. Usually, up to 90% of the reproductive females within the flock are infected. This is why it is mentioned that C. burnetii may cause epidemic herd outbreaks with significant animal losses owing to abortion waves and weak offspring during the parturition period [96,97].
4. Epidemiological Highlights
C. burnetii is a category B biological threat agent because of its impressive stability and resistance, its ability to aerosolize, and its virulence [43,44,45,98]. These characteristics allow the survival of this pathogen in the environment for long periods while keeping its infectivity [45,53]. In fact, viable microorganisms can be recovered after several years in dust, two years at –20 °C, seven to ten months on wool at environment temperature, 150 days in soil, for more than one month on fresh meat, and seven days in water or in milk at room temperature [17,38].
The transmission of C. burnetii may occur by direct, indirect, or vectorial transmission. The majority of natural C. burnetii infections occur by airborne transmission, resulting from the inhalation of aerosolized bacteria [79,93,96]. The resistance of C. burnetii allows it to be dispersed by wind far away from its original source. This may cause a long-distance transmission of infection leading to inter-herd transmission of C. burnetii or even to dispersion of bacteria to residential locations, causing human outbreaks [99,100,101]. Several studies on human outbreaks report this mid- to long-distance transmission [99,102,103]. Moreover, areas with high wind speed, open landscape, and high temperature increase the risk of infection [104].
Milk is considered a relevant route of bacterial shedding in the infected goats [105] and several studies evidence the presence of C. burnetii in goat milk (Table 2).
Table 2.
C. burnetii DNA detected in milk samples from goat herds.
| Country (Area) | Study Period | Type of Sample | Number of Samples | Test | Prevalence (%) | Reference |
|---|---|---|---|---|---|---|
| Belgium | 2009–2013 | BTM a | 1924 | Real-time PCR | 12.1 | [106] |
| France | - | BTM a | 120 | PCR | 19.0 | [105] |
| Iran | 2008 | BTM a | 110 | Nested PCR | 4.5 | [107] |
| Italy | 2018–2020 | Milk | 68 | PCR | 25.0 | [108] |
| Cheese | 15 | PCR | 6.7 | |||
| Poland | - | BTM a | 35 | Real-time PCR | 54.3 | [109] |
| Portugal | 2009–2013 | BTM a | 12 | Real-time PCR | 0.0 | [110] |
| Switzerland | 2006 | Milk | 39 | Nested PCR | 0.0 | [111] |
| The Gambia | 2012 | Milk | 33 | PCR | 2.94 | [112] |
| The Netherlands | 2008 | BTM a | 292 | Real-time PCR | 32.9 | [113] |
| Turkey | - | Milk | 50 | PCR | 4.0 | [114] |
| USA | 2012 | Milk | 387 | Real-time PCR | 2.5 | [115] |
a BTM—bulk tank milk.
Despite the knowledge that C. burnetii remains viable in unpasteurized milk, the assumption of the risk of infection by ingestion of C. burnetii milk is controversial [108,116,117]. In France, C. burnetii was detected in commercially available milk products, but, because its viability was not confirmed, the transmission by consumption of these products was not considered important [118]. Moreover, in a report concerning the public health risks related to raw drinking milk, C. burnetii is not mentioned as a biohazard to be transmitted via milk [119]. Thus, for risk assessment, it is assumed that multiplication of the pathogen in milk and milk products does not occur. Furthermore, there are insufficient data for a dose–response model for the oral route in humans [117]. It is known that the pasteurization procedure by ultra-high temperature treatment of milk (72 °C for 15 s) is adequate to eliminate viable C. burnetii from whole raw milk [120,121]. Notwithstanding, the risk of infection by consuming unpasteurized milk and raw milk dairy products may not be negligible.
Infected goats also shed high concentrations of C. burnetii in placental membranes, birth fluids, and/or abortion products [79,122]. This is an important excretion route in peri-partum period contributing to a high contamination of the environment. Furthermore, C. burnetii can also be excreted through faeces and vaginal mucus [79,113]. The shedding is normally very high at the first parturition after the infection, but occasionally, it occurs at subsequent pregnancies accompanied by a considerable number of bacteria excreted through placenta [79,94]. So, normal deliveries in infected females may contribute to the environmental contamination and should, therefore, be considered as a major zoonotic risk [79]. In the environment, bacteria can be easily aerosolized from desiccation of infected placenta and body fluids or from contaminated manure, reaching new hosts [96,101,103].
The risk of infection has been studied at the herd level and at the individual level in prevalence studies by detection of antibodies (Table 3 and Table 4, respectively).
Table 3.
Prevalence of antibodies anti-C. burnetii at goat herd level.
| Country (Area) | Study Period | Type of Sample | Sampling Method | Number | Test | Cut-Off Value | Prevalence (%) | Reference |
|---|---|---|---|---|---|---|---|---|
| Canada | 2010–2012 | Serum | Multi-stage random | 76 | ELISA | 0.4 | 63.2 | [123] |
| Great Britain | 2008 | Serum | Random stratified | 145 | ELISA | 0.4 | 3.0 | [124] |
| Ireland (Republic of) | 2005–2007 | Serum | Random | 66 | ELISA | 0.4 | 1.5 | [125] |
| Italy | 2012 | Serum | Multi-stage random | 206 | ELISA | 0.4 | 19.5 | [126] |
| Lebanon | 2014 | Serum | Random | 128 | ELISA | 0.4 | 45.3 | [127] |
| Norway | 2009 | BTM a | Random | 348 | ELISA | 0.4 | 0 | [128] |
| Portugal | 2011 | Serum | Random | 52 | ELISA | 0.30 | 28.8 | [129] |
| Spain | 2007–2008 | Serum | Random | 11 | ELISA | 0.40 | 45.0 | [130] |
| Sweden | 2010 | BTM a | Random | 58 | ELISA | 0.4 | 1.7 | [131] |
| Switzerland | 2011 | Serum | Random stratified | 72 | ELISA | 0.4 | 11.1 | [132] |
| The Netherlands | 2008 | Serum | Random | 442 | ELISA | 0.4 | 17.9 | [133] |
| USA | 2012–2014 | Serum | Random | 89 | ELISA | 0.4 | 11.5 | [115] |
a BTM—bulk tank milk.
Table 4.
Individual seroprevalence of C. burnetii in goats.
| Country (Area) | Study Period | Type of Sample | Sampling Method | Number of Samples | Test | Cut-Off Value | Prevalence (%) | Reference |
|---|---|---|---|---|---|---|---|---|
| Albania | 1995–1997 | Serum | - | 443 | ELISA | 0.4 | 8.8 | [136] |
| Bangladesh | 2009–2010 | Serum | Convenience | 529 | ELISA | 0.4 | 0.8 | [137] |
| Brazil | 2014–2015 | Serum | Convenience | 312 | ELISA | 0.4 | 55.1 | [138] |
| Canada | 2010–2012 | Serum | Multi-stage random | 2195 | ELISA | 0.4 | 32.5 | [123] |
| Ethiopia | - | Serum | Multi-stage random | 293 | ELISA | 0.4 | 35.5 | [139] |
| Great Britain | 2008 | Serum | Random stratified | 522 | ELISA | 0.4 | 0.8 | [124] |
| Greece | 2014–2015 | Serum | Convenience | 800 | ELISA | 0.4 | 14.4 | [140] |
| India | - | Serum | Convenience | 53 | ELISA | 0.4 | 5.7 | [141] |
| Iran | - | Serum | Multi-stage random | 241 | ELISA | 0.4 | 22.4 | [142] |
| Ireland (Republic of) | 2005–2007 | Serum | Random | 590 | ELISA | 0.4 | 0.3 | [125] |
| Italy | 2012 | Serum | Multi-stage random | 3185 | ELISA | 0.4 | 25.7 | [126] |
| Ivory Coast | 2012–2014 | Serum | Cluster | 622 | ELISA | 0.4 | 12.4 | [143] |
| Kenya | 2013 | Serum | Random | 280 | ELISA | 0.4 | 18.2 | [144] |
| Lebanon | 2014 | Serum | Random | 384 | ELISA | 0.4 | 17.2 | [127] |
| Reunion Island | 2011–2012 | Serum | Random | 134 | ELISA | 0.4 | 13.4 | [135] |
| Portugal | 2011 | Serum | Random | - | ELISA | 0.4 | 10.4 | [129] |
| Spain | 2007–2008 | Serum | Random | 115 | ELISA | 0.40 | 8.7 | [130] |
| Spain | 2015–2018 | Serum | Random | 135 | ELISA | 0.4 | 24.4 | [145] |
| Switzerland | 2011 | Serum | Random stratified | 321 | ELISA | 0.4 | 3.4 | [132] |
| The Gambia | 2012 | Serum | Multi-stage random | 484 | ELISA | 0.4 | 24.2 | [112] |
| The Netherlands | 2008 | Serum | Random | 3134 | ELISA | 0.4 | 7.8 | [133] |
| USA | 2012–2014 | Serum | Random | 608 | ELISA | 0.4 | 3.8 | [115] |
| Vietnam | 2016–2017 | Serum | Random | 1458 | ELISA | 0.4 | 4.1 | [146] |
At the herd level, the risk factors for C. burnetii infection are as follows: the proximity of an infected farm, the high animal density in a municipality, the high wind speed, an open landscape, the high temperature [123,134,135], the increased size of the herd [123,129,134], the poor hygiene and bio-security measures in the farm, the presence of ticks, the presence of dogs and cats in the farm [134,135], and the presence of swine on farms [123]. At an individual level, it was shown that the risk of positivity increases with age [129].
The number of Q fever cases varies geographically, and a seasonal variation is also described [100]. In the Northern Hemisphere, acute Q fever cases are more often reported in spring and early summer, showing a slow rise in reported cases in March and April, probably associated with the start of lambing/kidding, and the main peak occurs between May and July [27,147,148]. This occurs probably because of the “outside” lambing/kidding during spring associated with heavy environmental contamination with C. burnetii. It is known that the lambing season in October is not related to a higher incidence in humans, which might be owing to “indoor” lambing [96], which is also consistent with the study conducted in the South of France, showing that autumn is not a very windy season, which might explain the lower incidence of human Q fever at this time of the year [99].
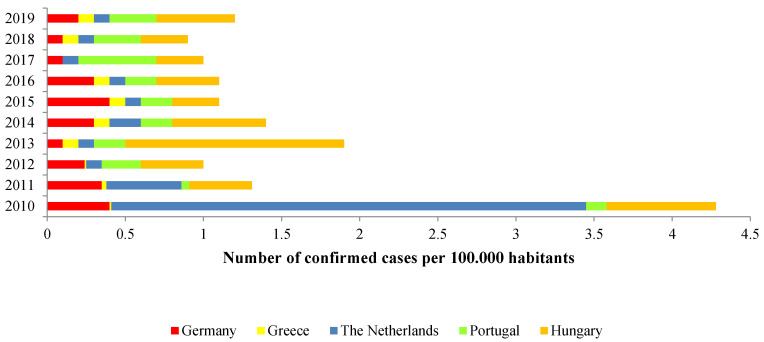
In most European countries, Q fever cases in humans and animals are reported regularly [8,148]. In humans, after the largest ever recorded outbreak in The Netherlands, the number of notified cases has suffered in general a sustained decrease in Europe and, nowadays, small outbreaks still occur, as shown on Figure 3, mainly in areas with infected livestock herds [148,149,150].
Figure 3.
Rates of confirmed human cases of Q fever in four European Countries from 2010 to 2019 [148,149,150].
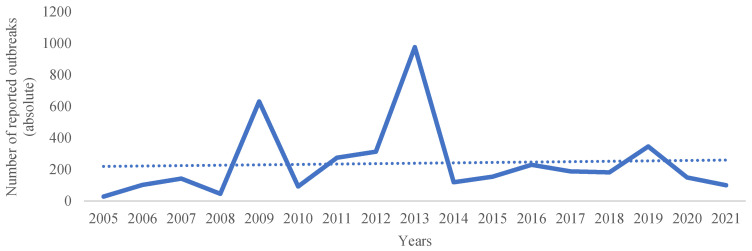
Regarding the report of infection in goats, the WAHIS Interface from the World Organization for Animal Health (WOAH) indicates the number of outbreaks reported in each country per year, which is systematized for European countries in the Figure 4. However, the analysis must be performed carefully because it is based on a notification procedure, thus it may not indicate the true prevalence as differences in the procedure of notification may differ between countries. Thus, data from epidemiological studies (Table 3 and Table 4) can be more reliable within the reality in a region/country.
Figure 4.
Number of Q fever outbreaks in goats reported in European countries from 2005 to 2021 and linear trend line [8].
C. burnetii infection in humans is usually considered an occupational threat. Some professional groups are more prone to exposure with C. burnetii. For instance, the farming workforce constitutes a relevant occupational risk group because of their contact with infected livestock, namely during breeding practices [151,152]. Moreover, veterinarians, laboratory workers, and abattoir workers are also at risk of being infected [8,153,154,155,156], as well as workers in the wool, tanneries, fur, meat, leather, and timber industries [152].
However, community outbreaks have been described very often. Factors as living on rural or sub urban areas [133] and in the proximity of positive farms [157] substantially increase the risk of Q fever.
The danger of C. burnetii being used as a biological weapon is a global concern. The low infectious dose for humans and its ability to spread over large distances on the wind would cause an enormous impact on human health. Additionally, wide-scale consequences would occur because of animal infection (i.e., domesticated, and wild animals) that could represent secondary sources of infection for humans [98] and even compromise agriculture and food production.
5. Molecular Epidemiology: An Added Value
Nowadays, molecular epidemiology is crucial in monitoring programs of C. burnetii and in investigation of Q fever outbreaks. The genetic heterogeneity of C. burnetii can be assessed by several molecular techniques. Today, the most adopted methods to define phylogeny are the multi loci variable-number tandem repeat analysis (MLVA) and the multispacer sequence typing (MST) [96,158,159]. A large number of MLVA data exist for European countries, even if the lack of consensus between scientists hampers comparison. Overall, a common pool of MLVA genotypes is present in Europe, together with novel genotypes sporadically found in specific countries [160,161,162,163]. Different MLVA genotypes could correspond to an identical MST type, indicating the MLVA method as more discriminatory than MST. Furthermore, the availability of a free-access database on the internet increased the interest in these methods to characterize C. burnetii strains circulating in a region in a normal context or in the case of outbreak [106,164,165].
A systematic genotyping provides a descriptive database, enabling to monitor the temporal and geographical evolution of strains, thus helping to trace the origins of the outbreaks and to identify interspecies transmission. These data can help to explain different scenarios of dissemination and contribute to finding efficient control measures [37,159,165,166]. For instance, in Portugal, the involvement of different genotypes in acute and in long-term infections of Q fever was found, but only one genotype found in long-term Q fever was linked to the one identified in goats from the same region [161]. Moreover, in Belgium, an emerging CbNL01-like genotype was identified in goats and this strain was isolated from half of the field samples isolated from the Dutch outbreak. However, no impact on the number of human cases was observed [106]. In Poland, examination of nine C. burnetii samples from goats revealed the presence of three MLVA genotypes (I, J, and PL1) and one sequence type (ST61). These MLVA and MST profiles were different to the strains’ profiles involved in the Q fever outbreak in the Netherlands [109]. Table 5 shows the C. burnettii genotypes identified by MST in goats and in other hosts (human, ruminants, and vectors), showing the wide transmission of specific genotypes.
Table 5.
Coxiella burnetii genotypes identified in goats and other hosts using multispacer sequence typing (MST).
| MST | Species | Country | Reference |
|---|---|---|---|
| 8 | Goat | Spain | [160,167] |
| Sheep | Spain | ||
| Human | Portugal, France, and USA | ||
| 13 | Goat | Portugal, Spain | [160,167] |
| Sheep | Spain | ||
| Cattle | Spain | ||
| Human | Portugal | ||
| Ticks | France | ||
| 18 | Goat | Germany, Spain | [160,167] |
| Sheep | Germany | ||
| Cattle | Poland | ||
| Human | France, Greece, Italy, Poland, Slovakia, and Romania | ||
| 30 | Goat | Namibia | [167] |
| 32 | Goat | Austria | [167] |
| Human | France and Germany | ||
| 53 | Goat | France | [167] |
| 58 | Goat | Libano | [167] |
| 61 | Goat | Poland | [167] |
| Cattle | Iran | [167] | |
| 62 | Goat | Iran | [167] |
| Sheep | [167] | ||
| Cattle | [167] | ||
| 66 to 70 | Goat | Thailand | [167] |
| 74 | Goat | Brazil | [167] |
| Cattle | [167] |
An analysis of MLVA and MST genotypes published in international databases showed that human isolates of C. burnetii frequently belong to the same genomic group of caprine isolates [168]. Furthermore, a geographical niche for C. burnetii genotypes was demonstrated. Although some isolates present a worldwide distribution, others show a geographic localization. Some genomic groups occur predominantly in Northern and Central Asia, Eastern and Central Europe, and Africa, while others are found mainly in central Europe or even in other areas [168]. For instance, MST8 reported in goats from Spain (Table 5) was the most found genotype in goat milk in the United States [169].
Thus, genotyping demonstrates that isolates in human Q fever are frequently genetically related to isolates circulating in goat populations, which reinforces the role of goats as important sources of infection for human populations.
6. Conclusions
In conclusion, C. burnetii presents a wide host range; however, it is mostly recognized in domestic ruminants. Q fever is not very commonly diagnosed in humans, in part because primary infection is frequently asymptomatic. The development of molecular tools has allowed to unravel the dynamic of genotypes’ circulation. Genotyping of C. burnetii indicates that, often, infected goats are the source of infection in human Q fever outbreaks. The pathogenesis is complex and not entirely understood at the host level, which highlights the requirement of future research to grasp the dynamics of the infections in specific hosts, as well as to unravel strain characteristics that may determine its virulence, affect the course of the disease and the clinical outcome, or influence the affinity to specific hosts. Yet, the association of genotypes with high virulence or host specificity remains to be demonstrated. An international genotype database facilitates the identification of emerging genotypes and their epidemiological features, also relying on a standard molecular method, which allows interlaboratory comparison. Thus, a worldwide surveillance in goats based on molecular epidemiology will be an important strategy to effective control of this zoonotic and highly resistant pathogen that fits in the One Health approach that considers animals, environment, and humans.
Author Contributions
Conceptualization and design: S.A., M.J.S. and G.J.d.S.; acquisition of data: S.A. and S.R.d.S.; analysis and interpretation of data: S.A., S.R.d.S., M.J.S. and G.J.d.S.; writing—original draft preparation: S.A. and S.R.d.S.; writing—review and editing: S.A., S.R.d.S., M.J.S. and G.J.d.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: WAHIS Interface. Available online: https://wahis.woah.org/#/home and Database on Multi Spacers Typing of Coxiella burnetii. Available online: https://ifr48.timone.univ-mrs.fr/mst/coxiella_burnetii/.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
S.A. received a Ph.D. fellowship (SFRH/BD/77823/2011) by the Portuguese Foundation for Science and Technology. Authors were supported by CNC UIDB/04539/2020; CITAB UIDB/04033/2020 and Inov4Agro LA/P/0126/2020.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Derrick E.H. “Q” fever, a new fever entity: Clinical features, diagnosis and laboratory investigation. Med. J. Aust. 1937;2:281–299. doi: 10.5694/j.1326-5377.1937.tb43743.x. [DOI] [PubMed] [Google Scholar]
- 2.Davis G.E., Cox H.R., Parker R.R., Dyer R.E. A Filter-Passing Infectious Agent Isolated from Ticks. Pub. Health Rep. 1938;53:2259–2282. doi: 10.2307/4582746. [DOI] [Google Scholar]
- 3.Bengtson I.A. Immunological relationships between the rickettsiae of Australian and American “Q” fever. Pub. Health Rep. 1941;56:272–281. doi: 10.2307/4583616. [DOI] [Google Scholar]
- 4.Kaplan M.M., Bertagna P. The geographical distribution of Q fever. Bull. World Health Organ. 1955;13:829–860. [PMC free article] [PubMed] [Google Scholar]
- 5.Caminopetros J.P. La Q-fever en Grece: Le lait source de l’infection pour l’homme et les animaux. Ann. Parasite Paris. 1948;23:107–118. doi: 10.1051/parasite/1948231107. [DOI] [PubMed] [Google Scholar]
- 6.Fox-Lewis A., Isteed K., Austin P., Thompson-Faiva H., Wolfgang J., Ussher J.E. A case of imported Q fever in New Zealand. NZMJ. 2019;132:92–94. [PubMed] [Google Scholar]
- 7.van den Brom R., van Engelen E., Roest H.I., van der Hoek W., Vellema P. Coxiella burnetii infections in sheep or goats: An opinionated review. Vet. Microbiol. 2015;181:119–129. doi: 10.1016/j.vetmic.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 8.World Organisation for Animal Health (WOAH) WAHIS Interface. [(accessed on 15 September 2022)]. Available online: https://wahis.woah.org/#/home.
- 9.de Valk H. Q fever: New insights, still many queries. Eurosurveillance. 2012;17:20062. doi: 10.2807/ese.17.03.20062-en. [DOI] [PubMed] [Google Scholar]
- 10.van Loenhout J.A.F., Paget W.J., Vercoulen J.H., Wijkmans C.J., Hautvast J.L.A., van der Velden K. Assessing the long-term health impact of Q-fever in the Netherlands: A prospective cohort study started in 2007 on the largest documented Q-fever outbreak to date. BMC Infect. Dis. 2012;12:280. doi: 10.1186/1471-2334-12-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sidi-Boumedine K., Rousset E., Henning K., Ziller M., Niemczuck K., Roest H.I.J., Thiéry R. Development of harmonised schemes for the monitoring and reporting of Q-fever in animals in the European Union. EFSA Support. Publ. 2010;7:48. doi: 10.2903/sp.efsa.2010.EN-48. [DOI] [Google Scholar]
- 12.EFSA (European Food Safety Authority) Panel on Animal Health and Welfare (AHAW); Scientific Opinion on Q Fever. EFSA J. 2010;8:1595. [Google Scholar]
- 13.European Centre for Disease Prevention and Control . Risk Assessment on Q Fever—Technical Report. ECDC; Stockholm, Sweden: 2010. p. 40. [Google Scholar]
- 14.Georgiev M., Afonso A., Neubauer H., Needham H., Thiéry R., Rodolakis A., Roest J., Stärk K.D., Stegeman J.A., Vellema P., et al. Q fever in humans and farm animals in four European countries, 1982 to 2010. Eurosurveillance. 2013;18:20407. doi: 10.2807/ese.18.08.20407-en. [DOI] [PubMed] [Google Scholar]
- 15.Farooq M., Khan A.U., El-Adawy H., Mertens-Scholz K., Khan I., Neubauer H., Ho Y.S. Research Trends and Hotspots of Q Fever Research: A Bibliometric Analysis 1990–2019. BioMed Res. Int. 2022;2022:9324471. doi: 10.1155/2022/9324471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Álvarez-Alonso R., Basterretxea M., Barandika J.F., Hurtado A., Idiazabal J., Jado I., Beraza X., Montes M., Liendo P., García-Pérez A.L. A Q Fever Outbreak with a High Rate of Abortions at a Dairy Goat Farm: Coxiella burnetii Shedding, Environmental Contamination, and Viability. Appl. Environ. Microbiol. 2018;84:e01650-18. doi: 10.1128/AEM.01650-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark N.J., Soares Magalhães R.J. Airborne geographical dispersal of Q fever from livestock holdings to human communities: A systematic review and critical appraisal of evidence. BMC Infect. Dis. 2018;18:218. doi: 10.1186/s12879-018-3135-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vellema P., Santman-Berends I., Dijkstra F., van Engelen E., Aalberts M., Ter Bogt-Kappert C., van den Brom R. Dairy Sheep Played a Minor Role in the 2005-2010 Human Q Fever Outbreak in The Netherlands Compared to Dairy Goats. Pathogens. 2021;10:1579. doi: 10.3390/pathogens10121579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byeon H.S., Nattan S., Kim J.H., Han S.T., Chae M.H., Han M.N., Ahn B., Kim Y.D., Kim H.S., Jeong H.W. Shedding and extensive and prolonged environmental contamination of goat farms of Q fever patients by Coxiella burnetii. Vet. Med. Sci. 2022;8:1264–1270. doi: 10.1002/vms3.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bond K.A., Vincent G., Wilks C.R., Franklin L., Sutton B., Stenos J., Cowan R., Lim K., Athan E., Harris O., et al. One Health approach to controlling a Q fever outbreak on an Australian goat farm. Epidemiol. Infect. 2016;144:1129–1141. doi: 10.1017/S0950268815002368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panaiotov S., Ciccozzi M., Brankova N., Levterova V., Mitova-Tiholova M., Amicosante M., Rezza G., Kantardjiev T. An outbreak of Q fever in Bulgaria. Ann. Dell’istituto Super. Sanità. 2009;45:83–86. [PubMed] [Google Scholar]
- 22.Genova-Kalou P., Vladimirova N., Stoitsova S., Krumova S., Kurchatova A., Kantardjiev T. Q fever in Bulgaria: Laboratory and epidemiological findings on human cases and outbreaks, 2011 to 2017. Eurosurveillance. 2019;24:1900119. doi: 10.2807/1560-7917.ES.2019.24.37.1900119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang M., Ma J., Jiao J., Li C., Chen L., Zhu Z., Ruan F., Xing L., Zheng X., Fu M., et al. The epidemic of Q fever in 2018 to 2019 in Zhuhai city of China determined by metagenomic next-generation sequencing. PLoS Negl. Trop. Dis. 2021;15:e0009520. doi: 10.1371/journal.pntd.0009520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fishbein D.B., Raoult D. A cluster of Coxiella burnetii infections associated with exposure to vaccinated goats and their unpasteurized dairy products. Am. J. Trop. Med. Hyg. 1992;47:35–40. doi: 10.4269/ajtmh.1992.47.35. [DOI] [PubMed] [Google Scholar]
- 25.King L.A., Goirand L., Tissot-Dupont H., Giunta B., Giraud C., Colardelle C., Duquesne V., Rousset E., Aubert M., Thiéry R., et al. Outbreak of Q fever, Florac, Southern France, Spring 2007. Vector Borne Zoonotic Dis. 2011;11:341–347. doi: 10.1089/vbz.2010.0050. [DOI] [PubMed] [Google Scholar]
- 26.Hatchette T.F., Hudson R.C., Schlech W.F., Campbell N.A., Hatchette J.E., Ratnam S., Raoult D., Donovan C., Marrie T.J. Goat-Associated Q Fever: A New Disease in Newfoundland. Emerg. Infect. Dis. 2001;7:413–419. doi: 10.3201/eid0703.017308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kovácová E., Kazár J., Simková A. Clinical and serological analysis of a Q fever outbreak in western Slovakia with four-year follow-up. Eur. J. Clin. Microbiol. Infect. Dis. 1998;17:867–869. doi: 10.1007/s100960050209. [DOI] [PubMed] [Google Scholar]
- 28.Gunther M.J., Heller J., Hayes L., Hernandez-Jover M. Dairy goat producers’ understanding, knowledge and attitudes towards biosecurity and Q-fever in Australia. Prev. Vet. Med. 2019;170:104742. doi: 10.1016/j.prevetmed.2019.104742. [DOI] [PubMed] [Google Scholar]
- 29.van der Giessen J., Vlaanderen F., Kortbeek T., Swaan C., van den Kerkhof H., Broens E., Rijks J., Koene M., De Rosa M., Uiterwijk M., et al. Signalling and responding to zoonotic threats using a One Health approach: A decade of the Zoonoses Structure in the Netherlands, 2011 to 2021. Eurosurveillance. 2022;27:2200039. doi: 10.2807/1560-7917.ES.2022.27.31.2200039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jorm L.R., Lightfoot N.F., Morgan K.L. An epidemiological study of an outbreak of Q fever in a secondary school. Epidemiol. Infect. 1990;104:467–477. doi: 10.1017/S0950268800047476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark W.H., Lennette E.H., Romer M.S. Q fever in California. IX. An outbreak aboard a ship transporting goats. Am. J. Hyg. 1951;54:35–43. [PubMed] [Google Scholar]
- 32.Bjork A., Marsden-Haug N., Nett R.J., Kersh G.J., Nicholson W., Gibson D., Szymanski T., Emery M., Kohrs P., Woodhall D., et al. First reported multistate human Q fever outbreak in the United States, 2011. Vector Borne Zoonotic Dis. 2014;14:111–117. doi: 10.1089/vbz.2012.1202. [DOI] [PubMed] [Google Scholar]
- 33.Preston W. In: Bergey’s Manual of Determinative Bacteriology. 6th ed. Bergey D.H., Breed R.S., Hitchens A.P., Murray E.G.D., editors. Williams & Wilkins; Baltimore, MD, USA: 1948. [Google Scholar]
- 34.Drancourt M., Roux V., Dang L.V., Tran-Hung L., Castex D., Chenal-Francisque V., Ogata H., Fournier P.-E., Crubézy E., Raoult D. Genotyping, Orientalis-like Yersinia pestis, and Plague Pandemics. Emerg. Infect. Dis. 2004;10:1585–1592. doi: 10.3201/eid1009.030933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seshadri R., Paulsen I.T., Eisen J.A., Read T.D., Nelson K.E., Nelson W.C., Ward N.L., Tettelin H., Davidsen T.M., Beanan M.J., et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. USA. 2003;100:5455–5460. doi: 10.1073/pnas.0931379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidi-Boumedine K., Adam G., Angen O., Aspan A., Bossers A., Roest H.J., Prigent M., Thiéry R., Rousset E. Whole genome PCR scanning (WGPS) of Coxiella burnetii strains from ruminants. Microbes Infect. 2015;17:772–775. doi: 10.1016/j.micinf.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Sidi-Boumedine K., Duquesne V., Prigent M., Yang E., Joulié A., Thiéry R., Rousset E. Impact of IS1111 insertion on the MLVA genotyping of Coxiella burnetii. Microbes Infect. 2015;17:789–794. doi: 10.1016/j.micinf.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 38.Eldin C., Mélenotte C., Mediannikov O., Ghigo E., Million M., Edouard S., Mege J.L., Maurin M., Raoult D. From Q Fever to Coxiella burnetii Infection: A Paradigm Change. Clin. Microbiol. Rev. 2017;30:115–190. doi: 10.1128/CMR.00045-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mege J.L., Maurin M., Capo C., Raoult D. Coxiella burnetii: The “query” fever bacterium—Model of immune subversion by a strictly intracellular microorganism. FEMS Microbiol. Rev. 1997;19:209–217. doi: 10.1111/j.1574-6976.1997.tb00298.x. [DOI] [PubMed] [Google Scholar]
- 40.Beare P.A., Samuel J.E., Howe D., Virtaneva K., Porcella S.F., Heinzen R.A. Genetic Diversity of the Q Fever Agent, Coxiella burnetii, Assessed by Microarray-Based Whole-Genome Comparisons. J. Bacteriol. 2006;188:2309–2324. doi: 10.1128/JB.188.7.2309-2324.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Denison A.M., Massung R.F., Thompson H.E. Analysis of the O-antigen biosynthesis regions of phase II Isolates of Coxiella burnetii. FEMS Microbiol. Lettters. 2007;267:102–107. doi: 10.1111/j.1574-6968.2006.00544.x. [DOI] [PubMed] [Google Scholar]
- 42.Hoover T.A., Culp D.W., Vodkin M.H., Williams J.C., Thompson H.A. Chromosomal DNA Deletions Explain Phenotypic Characteristics of Two Antigenic Variants, Phase II and RSA 514 (Crazy), of the Coxiella burnetii Nine Mile Strain. Infect. Immun. 2002;70:6726–6733. doi: 10.1128/IAI.70.12.6726-2733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuley R., Smith H.E., Frangoulidis D., Smits M.A., Roest H.I.J., Bossers A. Cell-Free Propagation of Coxiella burnetii Does Not Affect Its Relative Virulence. PLoS ONE. 2015;10:e0121661. doi: 10.1371/journal.pone.0121661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coleman S.A., Fischer E.R., Howe D., Mead D.J., Heinzen R.A. Temporal Analysis of Coxiella burnetii Morphological Differentiation. J. Bacteriol. 2004;186:7344–7352. doi: 10.1128/JB.186.21.7344-7352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCaul T.F., Williams J.C. Developmental Cycle of Coxiella burnetii: Structure and Morphogenesis of Vegetative and Sporogenic Differentiations. J. Bacteriol. 1981;147:1063–1076. doi: 10.1128/jb.147.3.1063-1076.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heinzen R.A., Hackstadt T., Samuel J.E. Developmental biology of Coxiella burnetii. Trends Microbiol. 1999;7:149–154. doi: 10.1016/S0966-842X(99)01475-4. [DOI] [PubMed] [Google Scholar]
- 47.Seshadri R., Hendrix L.R., Samuel J.E. Differential Expression of Translational Elements by Life Cycle Variants of Coxiella burnetii. Infect. Immun. 1999;67:6026–6033. doi: 10.1128/IAI.67.11.6026-6033.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sandoz K.M., Popham D.L., Beare P.A., Sturdevant D.E., Hansen B., Nair V., Heinzen R.A. Transcriptional Profiling of Coxiella burnetii Reveals Extensive Cell Wall Remodeling in the Small Cell Variant Developmental Form. PLoS ONE. 2016;11:e0149957. doi: 10.1371/journal.pone.0149957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shannon J.G., Heinzen R.A. Adaptive immunity to the obligate intracellular pathogen Coxiella burnetii. Immunol. Res. 2009;43:138–148. doi: 10.1007/s12026-008-8059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amara A.B., Ghigo E., Le Priol Y., Lépolard C., Salcedo S.P., Lemichez E., Bretelle F., Capo C., Mege J.L. Coxiella burnetii, the agent of Q fever, replicates within trophoblasts and induces a unique transcriptional response. PLoS ONE. 2010;5:e15315. doi: 10.1371/journal.pone.0015315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Capo C., Moynault A., Collette Y., Olive D., Brown E.J., Raoult D., Mege J.L. Coxiella burnetii avoids macrophage phagocytosis by interfering with spatial distribution of complement receptor 3. J. Immunol. 2003;170:4217–4225. doi: 10.4049/jimmunol.170.8.4217. [DOI] [PubMed] [Google Scholar]
- 52.Dupuy A.G., Caron E. Integrin-dependent phagocytosis: Spreading from microadhesion to new concepts. J. Cell Sci. 2008;121:1773–1783. doi: 10.1242/jcs.018036. [DOI] [PubMed] [Google Scholar]
- 53.Meconi S., Jacomo V., Boquet P., Raoult D., Mege J.L., Capo C. Coxiella burnetii Induces Reorganization of the Actin Cytoskeleton in Human Monocytes. Infect. Immun. 1998;66:5527–5533. doi: 10.1128/IAI.66.11.5527-5533.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meconi S., Capo C., Remacle-Bonnet M., Pommier G., Raoult D., Mege J.L. Activation of protein tyrosine kinases by Coxiella burnetii: Role in actin cytoskeleton reorganization and bacterial phagocytosis. Infect. Immun. 2001;69:2520–2526. doi: 10.1128/IAI.69.4.2520-2526.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Honstettre A., Ghigo E., Moynault A., Capo C., Toman R., Akira S., Takeuchi O., Lepidi H., Raoult D., Mege J.L. Lipopolysaccharide from Coxiella burnetii is involved in bacterial phagocytosis, filamentous actin reorganization, and inflammatory responses through Toll-like receptor 4. J. Immunol. 2004;172:3695–3703. doi: 10.4049/jimmunol.172.6.3695. [DOI] [PubMed] [Google Scholar]
- 56.van Schaik E.J., Chen C., Mertens K., Weber M.M., Samuel J.E. Molecular pathogenesis of the obligate intracellular bacterium Coxiella burnetii. Nat. Rev. Microbiol. 2013;11:561–573. doi: 10.1038/nrmicro3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Heinzen R.A., Hackstadt T. A developmental stage-specific histone H1 homolog of Coxiella burnetii. J. Bacteriol. 1996;78:5049–5052. doi: 10.1128/jb.178.16.5049-5052.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kinchen J.M., Ravichandran K.S. Phagosome maturation: Going through the acid test. Nat. Rev. Mol. Cell Biol. 2008;9:781–795. doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howe D., Shannon J.G., Winfree S., Dorward D.W., Heinzen R.A. Coxiella burnetii phase I and II variants replicate with similar kinetics in degradative phagolysosome-like compartments of human macrophages. Infect. Immun. 2010;78:3465–3474. doi: 10.1128/IAI.00406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berón W., Gutierrez M.G., Rabinovitch M., Colombo M.I. Coxiella burnetii Localizes in a Rab7-Labeled Compartment with Autophagic Characteristics. Infect. Immun. 2002;70:5816–5821. doi: 10.1128/IAI.70.10.5816-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schulze-Luehrmann J., Eckart R.A., Ölke M., Saftig P., Liebler-Tenorio E., Lührmann A. LAMP proteins account for the maturation delay during the establishment of the Coxiella burnetii-containing vacuole. Cell. Microbiol. 2016;18:181–194. doi: 10.1111/cmi.12494. [DOI] [PubMed] [Google Scholar]
- 62.Ghigo E., Capo C., Tung C.H., Raoult D., Gorvel J.P., Mege J.L. Coxiella burnetii survival in THP-1 monocytes involves the impairment of phagosome maturation: IFN-g mediates its restoration and bacterial killing. J. Immunol. 2002;169:4488–4495. doi: 10.4049/jimmunol.169.8.4488. [DOI] [PubMed] [Google Scholar]
- 63.Howe D., Melnicáková J., Barák I., Heinzen R.A. Maturation of the Coxiella burnetii parasitophorous vacuole requires bacterial protein synthesis but not replication. Cell. Microbiol. 2003;5:469–480. doi: 10.1046/j.1462-5822.2003.00293.x. [DOI] [PubMed] [Google Scholar]
- 64.Flannagan R.S., Jaumouillé V., Grinstein S. The cell biology of phagocytosis. Ann. Rev. Pathol. 2012;7:61–98. doi: 10.1146/annurev-pathol-011811-132445. [DOI] [PubMed] [Google Scholar]
- 65.Barry A., Mege J., Ghigo E. Hijacked phagosomes and leukocyte activation: An intimate relationship. J. Leukoc. Biol. 2011;89:373–382. doi: 10.1189/jlb.0510270. [DOI] [PubMed] [Google Scholar]
- 66.Voth D.E., Heinzen R.A. Lounging in a lysosome: The intracellular lifestyle of Coxiella burnetii. Cell. Microbiol. 2007;9:829–840. doi: 10.1111/j.1462-5822.2007.00901.x. [DOI] [PubMed] [Google Scholar]
- 67.Pareja M.E.M., Bongiovanni A., Lafont F., Colombo M.I. Alterations of the Coxiella burnetii Replicative Vacuole Membrane Integrity and Interplay with the Autophagy Pathway. Front. Cell. Infect. Microbiol. 2017;7:112. doi: 10.3389/fcimb.2017.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lührmann A., Roy C.R. Coxiella burnetii inhibits activation of host cell apoptosis through a mechanism that involves preventing cytochrome c release from mitochondria. Infect. Immun. 2007;75:5282–5289. doi: 10.1128/IAI.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voth D.E., Howe D., Heinzen R.A. Coxiella burnetii inhibits apoptosis in human THP-1 cells and monkey primary alveolar macrophages. Infect. Immun. 2007;75:4263–4271. doi: 10.1128/IAI.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Voth D.E., Heinzen R.A. Coxiella Type IV Secretion and Cellular Microbiology. Curr. Opin. Microbiol. 2009;12:74–80. doi: 10.1016/j.mib.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vázques C.L., Colombo M.I. Coxiella burnetii modulates Beclin 1 and Bcl-2, preventing host cell apoptosis to generate a persistent bacterial infection. Cell. Death Differ. 2010;17:421–438. doi: 10.1038/cdd.2009.129. [DOI] [PubMed] [Google Scholar]
- 72.Howe D., Mallavia L.P. Coxiella burnetii Exhibits Morphological Change and Delays Phagolysosomal Fusion after Internalization by J774A. 1 Cells. Infect. Immun. 2000;68:3815–3821. doi: 10.1128/IAI.68.7.3815-3821.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jones R.M., Nicas M., Hubbard A.E., Reingold A.L. The Infectious Dose of Coxiella burnetii (Q Fever) Appl. Biosaf. 2006;11:32–41. doi: 10.1177/153567600601100106. [DOI] [Google Scholar]
- 74.Brooke R.J., Mutters N.T., Péter O., Kretzschmar M.E.E., Teunis P.F.M. Exposure to low doses of Coxiella burnetii caused high illness attack rates: Insights from combining human challenge and outbreak data. Epidemics. 2015;11:1–6. doi: 10.1016/j.epidem.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 75.Brooke R.J., Kretzschmar M.E., Mutters N.T., Teunis P.F. Human dose response relation for airborne exposure to Coxiella burnetii. BMC Infect. Dis. 2013;13:488. doi: 10.1186/1471-2334-13-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Graham J.G., MacDonald L.J., Hussain S.K., Sharma U.M., Kurten R.C., Voth D.E. Virulent Coxiella burnetii Pathotypes Productively Infect Primary Human Alveolar Macrophages. Cell. Microbiol. 2013;15:1012–1025. doi: 10.1111/cmi.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marriott H.M., Dockrell D.H. The role of the macrophage in lung disease mediated by bacteria. Exp. Lung Res. 2007;33:493–505. doi: 10.1080/01902140701756562. [DOI] [PubMed] [Google Scholar]
- 78.Stein A., Louveau C., Lepidi H., Ricci F., Baylac P., Davoust B., Raoult D. Q fever pneumonia: Virulence of C. burnetii pathovars in a murine model of aerosol infection. Infect. Immun. 2005;73:2469–2477. doi: 10.1128/IAI.73.4.2469-2477.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roest H.I.J., van Gelderen B., Dinkla A., Frangoulidis D., van Zijderveld F., Rebel J., van Keulen L. Q fever in pregnant goats: Pathogenesis and excretion of C. burnetii. PLoS ONE. 2012;7:e48949. doi: 10.1371/journal.pone.0048949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ammerdorffer A., Roest H.I., Dinkla A., Post J., Schoffelen T., van Deuren M., Sprong T., Rebel J.M. The effect of C. burnetii infection on the cytokine response of PBMCs from pregnant goats. PLoS ONE. 2014;9:e109283. doi: 10.1371/journal.pone.0109283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sánchez J., Souriauy A., Buendía A.J., Arricau-Bouvery N., Matínez C.M., Salinas J., Rodolakis A., Navarro J.A. Experimental Coxiella burnetii Infection in Pregnant Goats: A Histopathological and Immunohistochemical Study. J. Comp. Path. 2006;135:108–115. doi: 10.1016/j.jcpa.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 82.Roest H.I.J., Dinkla A., Koets A.P., Post J., van Keulen L. Experimental Coxiella burnetii infection in non-pregnant goats and the effect of breeding. Vet. Res. 2020;51:74. doi: 10.1186/s13567-020-00797-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Benoit M., Barbarat B., Bernard A., Olive D., Mege J.L. Coxiella burnetii, the agent of Q fever, stimulates an atypical M2 activation program in human macrophages. Eur. J. Immunol. 2008;38:1065–1070. doi: 10.1002/eji.200738067. [DOI] [PubMed] [Google Scholar]
- 84.Ghigo E., Capo C., Raoult D., Mege J.L. Interleukin-10 stimulates Coxiella burnetii replication in human monocytes through tumor necrosis factor down-modulation: Role in microbicidal defect of Q fever. Infect. Immun. 2001;69:2345–2352. doi: 10.1128/IAI.69.4.2345-2352.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghigo E., Imbert G., Capo C., Raoult D., Mege J.L. Interleukin-4 induces Coxiella burnetii replication in human monocytes but not in macrophages. Ann. New York Acad. Sci. 2003;990:450–459. doi: 10.1111/j.1749-6632.2003.tb07410.x. [DOI] [PubMed] [Google Scholar]
- 86.Zhang G., Russell-Lodrigue K.E., Andoh M., Zhang Y., Hendrix L.R., Samuel J.E. Mechanisms of vaccine-induced protective immunity against Coxiella burnetii infection in BALB/c mice. J. Immunol. 2007;179:8372–8380. doi: 10.4049/jimmunol.179.12.8372. [DOI] [PubMed] [Google Scholar]
- 87.Roest H.I.J., Bossers A., Rebel J.M.J. Q Fever Diagnosis and Control in Domestic Ruminants. Dev. Biol. 2013;135:183–189. doi: 10.1159/000188081. [DOI] [PubMed] [Google Scholar]
- 88.Teunis P.F.M., Schimmer B., Notermans D.W., Leenders A.C.A.P., Wever P.C., Kretzschmar M.E.E., Schneeberger P.M. Time-course of antibody responses against Coxiella burnetii following acute Q fever. Epidemiol. Infect. 2013;141:62–73. doi: 10.1017/S0950268812000404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Langley J.M., Marrie T.J., Leblanc J.C., Almudevar A., Resch L., Raoult D. Coxiella burnetii seropositivity in parturient women is associated with adverse pregnancy outcomes. Am. J. Obstet. Gynecol. 2003;189:228–232. doi: 10.1067/mob.2003.448. [DOI] [PubMed] [Google Scholar]
- 90.Carcopino X., Raoult D., Bretelle F., On Boubli L., Stein A. Managing Q Fever during Pregnancy: The Benefits of Long-Term Cotrimoxazole Therapy. Clin. Infect. Dis. 2007;45:548–555. doi: 10.1086/520661. [DOI] [PubMed] [Google Scholar]
- 91.Lennette E.H., Holmes M.A., Abinanti F.R. Remove from marked Records, Q. Fever Studies. XIV. Observations on the Pathogenesis of the Experimental Infection induced in Sheep by the Intravenous Route. Am. J. Hyg. 1952;55:254–267. [PubMed] [Google Scholar]
- 92.Martinov S.P., Neikov P., Popov G.V. Experimental Q fever in sheep. Eur. J. Epidemiol. 1989;5:428–431. doi: 10.1007/BF00140134. [DOI] [PubMed] [Google Scholar]
- 93.Berri M., Rousset E., Hechard C., Champion J.L., Dufour P., Russo P., Rodolaskis A. Progression of Q Fever and Coxiella burnetii shedding in milk after an outbreak of enzootic abortion in a goat herd. Vet. Rec. 2005;156:548–549. doi: 10.1136/vr.156.17.548. [DOI] [PubMed] [Google Scholar]
- 94.Berri M., Rousset E., Champion J.L., Russo P., Rodolaskis A. Goats may experience reproductive failures and shed Coxiella burnetii at two successive parturitions after a Q fever infection. Res. Vet. Sci. 2007;83:47–52. doi: 10.1016/j.rvsc.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 95.Van den Brom R., Vellema P. Q fever outbreaks in small ruminants and people in the Netherlands. Small Rum. Res. 2009;86:74–79. doi: 10.1016/j.smallrumres.2009.09.022. [DOI] [Google Scholar]
- 96.Arricau-Bouvery N., Rodolakis A. Is Q fever an emerging or re-emerging Zoonosis? Vet. Res. 2005;36:327–349. doi: 10.1051/vetres:2005010. [DOI] [PubMed] [Google Scholar]
- 97.Eibach R., Bothe F., Runge M., Fischer S.F., Philipp W., Ganter M. Q fever: Baseline monitoring of a sheep and a goat flock associated with human infections. Epidemiol. Infect. 2012;140:1939–1949. doi: 10.1017/S0950268811002846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roest H.I.J., Bossers A., van Zijderveld F.R., Rebel J.M.L. Clinical microbiology of Coxiella burnetii and relevant aspects for the diagnosis and control of the zoonotic disease Q fever. Vet. Q. 2014;33:148–160. doi: 10.1080/01652176.2013.843809. [DOI] [PubMed] [Google Scholar]
- 99.Tissot-Dupont H., Amadei M.A., Nezri M., Raoult D. Wind in November, Q fever in December. Emerg. Infect. Dis. 2004;10:1264–1269. doi: 10.3201/eid1007.030724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.de Rooij M.M.T., Borlée F., Smit L.A.M., de Bruin A., Janse I., Heederik D.J.J., Wouters I.M. Detection of Coxiella burnetii in Ambient Air after a Large Q Fever Outbreak. PLoS ONE. 2016;11:e0151281. doi: 10.1371/journal.pone.0151281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hermans T., Jeurissen L., Hackert V., Hoebe C. Land-applied goat manure as a source of human Q-fever in the Netherlands, 2006–2010. PLoS ONE. 2014;9:e96607. doi: 10.1371/journal.pone.0096607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hawker J.I., Ayres J.G., Blair I., Evans M.R., Smith D.L., Smith E.G., Burge P.S., Carpenter M.J., Caul E.O., Coupland B., et al. A large outbreak of Q fever in the West Midlands: Windborne spread into a metropolitan area? Commun. Dis. Public Health. 1998;1:180–187. [PubMed] [Google Scholar]
- 103.Schimmer B., Schegget R., Wegdam M., Züchner L., de Bruin A., Schneeberger P.M., Veenstra T., Vellema P., van der Hoek W. The use of a geographic information system to identify a dairy goat farm as the most likely source of an urban Q-fever outbreak. BMC Infect. Dis. 2010;10:69. doi: 10.1186/1471-2334-10-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nusinovici S., Frössling J., Widgren S., Beaudeau F., Lindberg A. Q fever infection in dairy cattle herds: Increased risk with high wind speed and low precipitation. Epidemiol. Infect. 2015;143:3316–3326. doi: 10.1017/S0950268814003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rodolakis A., Berri M., Héchard C., Caudron C., Souriau A., Bodier C.C., Blanchard B., Camuset P., Devillechaise P., Natorp J.C., et al. Comparison of Coxiella burnetii shedding in milk of dairy bovine, caprine and ovine herds. J. Dairy Sci. 2007;90:5352–5360. doi: 10.3168/jds.2006-815. [DOI] [PubMed] [Google Scholar]
- 106.Boarbi S., Mori M., Rousset E., Sidi-Boumedine K., Van Esbroeck M., Fretin D. Prevalence and molecular typing of Coxiella burnetii in bulk tank milk in Belgian dairy goats, 2009–2013. Vet. Microbiol. 2014;170:117–124. doi: 10.1016/j.vetmic.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 107.Rahimi E., Ameri M., Karim G., Doosti A. Prevalence of Coxiella burnetii in bulk milk samples from dairy bovine, ovine, caprine, and camel herds in Iran as determined by polymerase chain reaction. Foodborne Pathog. Dis. 2011;8:307–310. doi: 10.1089/fpd.2010.0684. [DOI] [PubMed] [Google Scholar]
- 108.Basanisi M.G., La Bella G., Nobili G., Raele D.A., Cafiero M.A., Coppola R., Damato A.M., Fraccalvieri R., Sottili R., La Salandra G. Detection of Coxiella burnetii DNA in sheep and goat milk and dairy products by droplet digital PCR in south Italy. Int. J. Food Microbiol. 2022;366:109583. doi: 10.1016/j.ijfoodmicro.2022.109583. [DOI] [PubMed] [Google Scholar]
- 109.Jodełko A., Szymańska-Czerwińska M., Rola J.G., Niemczuk K. Molecular detection of Coxiella burnetii in small ruminants and genotyping of specimens collected from goats in Poland. BMC Vet. Res. 2021;17:341. doi: 10.1186/s12917-021-03051-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Anastácio S., Carolino N., Sidi-Boumedine K., da Silva G.J. Q fever dairy herd status determination based on serological and molecular analysis of bulk tank milk. Transbound. Emerg. Dis. 2016;63:293–300. doi: 10.1111/tbed.12275. [DOI] [PubMed] [Google Scholar]
- 111.Fretz R., Schaeren W., Tanner M., Baumgartner A. Screening of various foodstuffs for occurrence of Coxiella burnetii in Switzerland. Int. J. Food Microbiol. 2007;116:414–418. doi: 10.1016/j.ijfoodmicro.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 112.Klaasen M., Roest H.J., van der Hoek W., Goossens B., Secka A., Stegeman A. Coxiella burnetii seroprevalence in small ruminants in The Gambia. PLoS ONE. 2014;9:e85424. doi: 10.1371/journal.pone.0085424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van den Brom R., van Engelen E., Luttikholt S., Moll L., van Maanen K., Vellema P. Coxiella burnetii in bulk tank milk samples from dairy goat and sheep farms in The Netherlands. Vet. Rec. 2012;170:310. doi: 10.1136/vr.100304. [DOI] [PubMed] [Google Scholar]
- 114.Can H.Y., Elmali E., Karagöz A. Detection of Coxiella burnetii in cows’, goats’, and ewes’ bulk milk samples using polymerase chain reaction (PCR) Mljekarstvo. 2015;65:26–31. doi: 10.15567/mljekarstvo.2015.0104. [DOI] [Google Scholar]
- 115.Bauer A.E., Hubbard K.R., Johnson A.J., Messick J.B., Weng H.Y., Pogranichniy R.M. A cross sectional study evaluating the prevalence of Coxiella burnetii, potential risk factors for infection, and agreement between diagnostic methods in goats in Indiana. Prev. Vet. Med. 2016;126:131–137. doi: 10.1016/j.prevetmed.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 116.Loftis A.D., Priestley R.A., Massung R.F. Detection of Coxiella burnetii in Commercially Available Raw Milk from the United States. Foodborne Pathog. Dis. 2010;7:1453–1456. doi: 10.1089/fpd.2010.0579. [DOI] [PubMed] [Google Scholar]
- 117.Gale P., Kelly L., Mearns R., Duggan J., Snary E.L. Q fever through consumption of unpasteurised milk and milk products—A risk profile and exposure assessment. J. Appl. Microbiol. 2015;118:1083–1095. doi: 10.1111/jam.12778. [DOI] [PubMed] [Google Scholar]
- 118.Eldin C., Angelakis E., Renvoisé A., Raoult D. Coxiella burnetii DNA, but not viable bacteria, in dairy products in France. Am. J. Trop. Med. Hyg. 2013;88:765–769. doi: 10.4269/ajtmh.12-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.EFSA BIOHAZ Panel (EFSA Panel on Biological Hazards) Scientific Opinion on the public health risks related to the consumption of raw drinking milk. EFSA J. 2015;13:3940. doi: 10.2903/j.efsa.2015.3940. [DOI] [Google Scholar]
- 120.Enright J.B., Sadler W.W., Thomas R.C. Pasteurization of milk containing the organism of Q Fever. Am. J. Public Health. 1957;47:695–700. doi: 10.2105/AJPH.47.6.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wittwer M., Hammer P., Runge M., Valentin-Weigand P., Neubauer H., Henning K., Mertens-Scholz K. Inactivation Kinetics of Coxiella burnetii During High-Temperature Short-Time Pasteurization of Milk. Front. Microbiol. 2022;12:753871. doi: 10.3389/fmicb.2021.753871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Arricau-Bouvery N., Souriau A., Lechopier P., Rodolakis A. Experimental Coxiella burnetii infection in pregnant goats: Excretion routes. Vet. Res. 2003;34:423–433. doi: 10.1051/vetres:2003017. [DOI] [PubMed] [Google Scholar]
- 123.Meadows S., Jones-Bitton A., McEwen S., Jansen J., Menzies P. Coxiella burnetii seropositivity and associated risk factors in goats in Ontario, Canada. Prev. Vet. Med. 2015;121:199–205. doi: 10.1016/j.prevetmed.2015.06.014. [DOI] [PubMed] [Google Scholar]
- 124.Lambton S.L., Smith R.P., Gillard K., Horigan M., Farren C., Pritchard G.C. Serological survey using ELISA to determine the prevalence of Coxiella burnetii infection (Q fever) in sheep and goats in Great Britain. Epidemiol. Infect. 2016;144:19–24. doi: 10.1017/S0950268815000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ryan E., Kirby M., Clegg T., Collins D.M. Seroprevalence of Coxiella burnetii antibodies in sheep and goats in the Republic of Ireland. Vet. Rec. 2011;169:280. doi: 10.1136/vr.d5208. [DOI] [PubMed] [Google Scholar]
- 126.Rizzo F., Vitale N., Ballardini M., Borromeo V., Luzzago C., Chiavacci L., Mandola M.L. Q fever seroprevalence and risk factors in sheep and goats in northwest Italy. Prev. Vet. Med. 2016;130:10–17. doi: 10.1016/j.prevetmed.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 127.Dabaja M.F., Greco G., Villari S., Vesco G., Bayan A., Bazzal B.E., Ibrahim E., Gargano V., Sciacca C., Lelli R., et al. Occurrence and risk factors of Coxiella burnetii in domestic ruminants in Lebanon. Comp. Immunol. Microbiol. Infect. Dis. 2019;64:109–116. doi: 10.1016/j.cimid.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 128.Kampen A.H., Hopp P., Groneng G.M., Melkild I., Urdahl A.M., Karlsson A.C., Tharaldsen J. No indication of Coxiella burnetii infection in Norwegian farmed ruminants. BMC Vet. Res. 2012;8:59. doi: 10.1186/1746-6148-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Anastacio S., Tavares N., Carolino N., Sidi-Boumedine K., da Silva G.J. Serological evidence of exposure to Coxiella burnetii in sheep and goats in central Portugal. Vet. Microbiol. 2013;167:500–505. doi: 10.1016/j.vetmic.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 130.Ruiz-Fons F., Astobiza I., Barandika J.F., Hurtado A., Atxaerandio R., Juste R.A., Garcia-Perez A.L. Seroepidemiological study of Q fever in domestic ruminants in semi-extensive grazing systems. BMC Vet. Res. 2010;6:3. doi: 10.1186/1746-6148-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ohlson A., Malmsten J., Frössling J., Bölske G., Aspán A., Dalin A.M., Lindberg A. Surveys on Coxiella burnetii infections in Swedish cattle, sheep, goats and moose. Acta Vet. Scand. 2014;56:39. doi: 10.1186/1751-0147-56-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Magouras I., Hunninghaus J., Scherrer S., Wittenbrink M.M., Hamburger A., Stärk K.D., Schüpbach-Regula G. Coxiella burnetii Infections in Small Ruminants and Humans in Switzerland. Transbound. Emerg. Dis. 2017;64:204–212. doi: 10.1111/tbed.12362. [DOI] [PubMed] [Google Scholar]
- 133.van den Brom R., Moll L., van Schaik G., Vellema P. Demography of Q fever seroprevalence in sheep and goats in The Netherlands in 2008. Prev. Vet. Med. 2013;109:76–82. doi: 10.1016/j.prevetmed.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 134.Schimmer B., Luttikholt S., Hautvast J.L.A., Graat E.A.M., Vellema P., van Duynhoven Y.T.H.P. Seroprevalence and risk factors of Q fever in goats on commercial dairy goat farms in the Netherlands, 2009-2010. BMC Vet. Res. 2011;7:81. doi: 10.1186/1746-6148-7-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cardinale E., Esnault O., Beral M., Naze F., Michault A. Emergence of Coxiella burnetii in Ruminants on Reunion Island? Prevalence and Risk Factors. PLoS Neglect. Trop. Dis. 2014;8:e3055. doi: 10.1371/journal.pntd.0003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cekani M., Papa A., Kota M., Velo E., Berxholi K. Report of a serological study of Coxiella burnetii in domestic animals in Albania. Vet. J. 2008;175:276–278. doi: 10.1016/j.tvjl.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 137.Haider N., Rahman M.S., Khan S.U., Mikolon A., Osmani M.G., Gurley E.S., Shanta I.S., Paul S.K., Macfarlane-Berry L., Islam A., et al. Serological Evidence of Coxiella burnetii Infection in Cattle and Goats in Bangladesh. Ecohealth. 2015;12:354–358. doi: 10.1007/s10393-015-1011-x. [DOI] [PubMed] [Google Scholar]
- 138.de Oliveira J.M.B., Rozental T., de Lemos E.R.S., Forneas D., Ortega-Mora L.M., Porto W.J.N., da Fonseca Oliveira A.A., Mota R.A. Coxiella burnetii in dairy goats with a history of reproductive disorders in Brazil. Acta Trop. 2018;183:19–22. doi: 10.1016/j.actatropica.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 139.Tesfaye A., Sahele M., Sori T., Guyassa C., Garoma A. Seroprevalence and associated risk factors for chlamydiosis, coxiellosis and brucellosis in sheep and goats in Borana pastoral area, southern Ethiopia. BMC Vet. Res. 2020;16:145. doi: 10.1186/s12917-020-02360-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Filioussis G., Theodoridis A., Papadopoulos D., Gelasakis A.I., Vouraki S., Bramis G., Arsenos G. Serological prevalence of Coxiella burnetii in dairy goats and ewes diagnosed with adverse pregnancy outcomes in Greece. Ann. Agric. Environ. Med. 2017;24:702–705. doi: 10.26444/aaem/80706. [DOI] [PubMed] [Google Scholar]
- 141.Vaidya V.M., Malik S.V.S., Bhilegaonkar K.N., Rathore R.S., Kaur S., Barbuddhe S.B. Prevalence of Q fever in domestic animals with reproductive disorders. Comp. Immunol. Microbiol. Infect. Dis. 2010;33:307–321. doi: 10.1016/j.cimid.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 142.Ezatkhah M., Alimolaei M., Khalili M., Sharifi H. Seroepidemiological study of Q fever in small ruminants from Southeast Iran. J. Infect. Public Health. 2015;8:170–176. doi: 10.1016/j.jiph.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 143.Kanouté Y.B., Gragnon B.G., Schindler C., Bonfoh B., Schelling E. Epidemiology of brucellosis, Q Fever and Rift Valley Fever at the human and livestock interface in northern Côte d’Ivoire. Acta Trop. 2017;165:66–75. doi: 10.1016/j.actatropica.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 144.Larson P.S., Espira L., Grabow C., Wang C.A., Muloi D., Browne A.S., Deem S.L., Fèvre E.M., Foufopoulos J., Hardin R., et al. The sero-epidemiology of Coxiella burnetii (Q fever) across livestock species and herding contexts in Laikipia County, Kenya. Zoonoses Public Health. 2019;66:316–324. doi: 10.1111/zph.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Espí A., Del Cerro A., Oleaga Á., Rodríguez-Pérez M., López C.M., Hurtado A., Rodríguez-Martínez L.D., Barandika J.F., García-Pérez A.L. One Health Approach: An Overview of Q Fever in Livestock, Wildlife and Humans in Asturias (Northwestern Spain) Animals. 2021;11:1395. doi: 10.3390/ani11051395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Burns R.J.L., Douangngeun B., Theppangna W., Khounsy S., Mukaka M., Selleck P.W., Hansson E., Wegner M.D., Windsor P.A., Blacksell S.D. Serosurveillance of Coxiellosis (Q-fever) and Brucellosis in goats in selected provinces of Lao People’s Democratic Republic. PLoS Negl. Trop. Dis. 2018;12:e0006411. doi: 10.1371/journal.pntd.0006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schack M., Sachse S., Rödel J., Frangoulidis D., Pletz M.W., Rohde G.U., Straube E., Boden K. Coxiella burnetii (Q fever) as a cause of community-acquired pneumonia during the warm season in Germany. Epidemiol. Infect. 2014;142:1905–1910. doi: 10.1017/S0950268813002914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.European Centre for Disease Prevention and Control . ECDC. Annual Epidemiological Report for 2019. ECDC; Stockholm, Sweden: 2021. Q fever. [Google Scholar]
- 149.European Centre for Disease Prevention and Control . Annual Epidemiological Report 2011, Reporting on 2009 Surveillance Data and 2010 Epidemic Intelligence Data. ECDC; Stockholm, Sweden: 2011. p. 224. [Google Scholar]
- 150.European Centre for Disease Prevention and Control . Annual Epidemiological Report 2014, Emerging and Vector-Borne Diseases. ECDC; Stockholm, Sweden: 2014. p. 52. [Google Scholar]
- 151.Cutler S.J., Bouzid M., Cutler R.R. Q fever. J. Infect. 2007;54:313–318. doi: 10.1016/j.jinf.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 152.Szymanska-Czerwinska M., Galinska E.M., Niemczuk K., Knap J.P. Prevalence of Coxiella burnetii Infection in Humans Occupationally Exposed to Animals in Poland. Vector Borne Zoonotic Dis. 2015;15:261–267. doi: 10.1089/vbz.2014.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Valencia M.C.S., Rodriguez C.O., Puñet O.G., Giral I.B. Q fever seroprevalence and associated risk factors among students from the Veterinary School of Zaragoza, Spain. Eur. J. Epidemiol. 2000;16:469–476. doi: 10.1023/A:1007605414042. [DOI] [PubMed] [Google Scholar]
- 154.Dorko E., Rimárová K., Kecerová A., Pilipčinec E., Dudríková E., Lovayová V., Petrovičová J., Boroš E. Potential association between Coxiella burnetii seroprevalence and selected risk factors among veterinary students in Slovakia. Ann. Agric. Environ. Med. 2011;18:47–53. [PubMed] [Google Scholar]
- 155.de Rooij M.M., Schimmer B., Versteeg B., Schneeberger P., Berends B.R., Heederik D., van der Hoek W., Wouters I.M. Risk factors of Coxiella burnetii (Q fever) seropositivity in veterinary medicine students. PLoS ONE. 2012;7:e32108. doi: 10.1371/journal.pone.0032108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Njeru J., Henning K., Pletz M.W., Heller R., Forstner C., Kariuki S., Fèvre E.M., Neubauer H. Febrile patients admitted to remote hospitals in Northeastern Kenya: Seroprevalence, risk factors and a clinical prediction tool for Q-Fever. BMC Infect. Dis. 2016;16:244. doi: 10.1186/s12879-016-1569-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Schimmer B., Lenferink A., Schneeberger P., Aangenend H., Vellema P., Hautvast J., van Duynhoven Y. Seroprevalence and Risk Factors for Coxiella burnetii (Q Fever) Seropositivity in Dairy Goat Farmers’ Households in The Netherlands, 2009–2010. PLoS ONE. 2012;7:e42364. doi: 10.1371/journal.pone.0042364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Glazunova O., Roux V., Freylikman O., Sekeyova Z., Fournous G., Tyczka J., Tokarevich N., Kovacova E., Marrie T.J., Raoult D. Coxiella burnetii Genotyping. Emerg. Infect. Dis. 2005;11:1211–1217. doi: 10.3201/eid1108.041354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Svraka S., Toman R., Skultety L., Slaba K., Homan W.L. Establishment of a genotyping scheme for Coxiella burnetii. FEMS Microbiol. Lett. 2006;254:268–274. doi: 10.1111/j.1574-6968.2005.00036.x. [DOI] [PubMed] [Google Scholar]
- 160.Astobiza I., Tilburg J.J., Piñero A., Hurtado A., García-Pérez A.L., Nabuurs-Franssen M.H., Klaassen C.H. Genotyping of Coxiella burnetii from domestic ruminants in northern Spain. BMC Vet. Res. 2012;8:241. doi: 10.1186/1746-6148-8-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Santos A.S., Tilburg J.J.H.C., Botelho A., Barahona M.J., Núncio M.S., Nabuurs-Franssen M.H., Klaassen C.H.W. Genotypic diversity of clinical Coxiella burnetii isolates from Portugal based on MST and MLVA typing. Int. J. Med. Microbiol. 2012;302:253–256. doi: 10.1016/j.ijmm.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 162.Ceglie L., Guerrini E., Rampazzo E., Barberio A., Tilburg J.J., Hagen F., Lucchese L., Zuliani F., Marangon S., Natale A. Molecular characterization by MLVA of Coxiella burnetii strains infecting dairy cows and goats of north-eastern Italy. Microbes Infect. 2015;17:776–781. doi: 10.1016/j.micinf.2015.09.029. [DOI] [PubMed] [Google Scholar]
- 163.Joulié A., Sidi-Boumedine K., Bailly X., Gasqui P., Barry S., Jaffrelo L., Poncet C., Abrial D., Yang E., Animal diagnostic laboratories consortium et al. Molecular epidemiology of Coxiella burnetii in French livestock reveals the existence of three main genotype clusters and suggests species-specific associations as well as regional stability. Infect. Genet Evol. 2017;48:142–149. doi: 10.1016/j.meegid.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 164.Tilburg J.J.H.C., Rossen J.W., van Hannen E.J., Melchers W.J., Hermans M.H., van de Bovenkamp J., Roest H.J., de Bruin A., Nabuurs-Franssen M.H., Horrevorts A.M., et al. Genotypic diversity of Coxiella burnetii in the 2007–2010 Q fever outbreak episodes in The Netherlands. J. Clin. Microbiol. 2012;50:1076–1078. doi: 10.1128/JCM.05497-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sulyok K.M., Kreizinger Z., Hornstra H.M., Pearson T., Szigeti A., Dán Á., Balla E., Keim P.S., Gyuranecz M. Genotyping of Coxiella burnetii from domestic ruminants and human in Hungary: Indication of various genotypes. BMC Vet. Res. 2014;10:107. doi: 10.1186/1746-6148-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rousset E., Sidi-Boumedine K., Kadra B., Kupcsullk B. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. 8th ed. World Organization for Animal Health; Paris, France: 2018. [(accessed on 30 September 2022)]. Chapter 3.1.17—Q fever. Available online: https://www.woah.org/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access/ [Google Scholar]
- 167.Database on Multi Spacers Typing of Coxiella burnetii. [(accessed on 25 September 2022)]. Available online: https://ifr48.timone.univ-mrs.fr/mst/coxiella_burnetii/
- 168.Hemsley C.M., Essex-Lopresti A., Norville I.H., Titball R.W. Correlating Genotyping Data of Coxiella burnetii with Genomic Groups. Pathogens. 2021;10:604. doi: 10.3390/pathogens10050604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pearson T., Hornstra H.M., Hilsabeck R., Gates L.T., Olivas S.M., Birdsell D.M., Hall C.M., German S., Cook J.M., Seymour M.L., et al. High prevalence and two dominant host-specific genotypes of Coxiella burnetii in U.S. milk. BMC Microbiol. 2014;14:41. doi: 10.1186/1471-2180-14-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: WAHIS Interface. Available online: https://wahis.woah.org/#/home and Database on Multi Spacers Typing of Coxiella burnetii. Available online: https://ifr48.timone.univ-mrs.fr/mst/coxiella_burnetii/.