Abstract
In common with many bacterial virulence genes, the fimbrillin (fimA) gene of Porphyromonas gingivalis is modulated in response to environmental fluctuation. The trans-acting components that comprise the regulatory system for transcriptional activity of the fimA gene in P. gingivalis were investigated. Three major proteins were found to bind to the upstream region of the fimA promoter. One of these proteins was fimbrillin itself, and the other two were a major arginine protease (Rgp) and lysine protease (Kgp). Production of these proteins was necessary for maximal fimA transcription. An exogenous fimA promoter-lacZ reporter was inactive when introduced into a strain of P. gingivalis carrying a mutation in the indigenous fimA gene. Furthermore, fimA mRNA levels were significantly decreased in rgp and kgp mutant strains. These data indicate that P. gingivalis has evolved multiple levels of control of fimbrial gene expression to enhance its survival in hostile environments.
Periodontal diseases are a group of chronic inflammatory infections that affect millions of people worldwide and cause destruction of the periodontal tissues, eventually leading to exfoliation of the teeth. Periodontal diseases ensue following the establishment of a mixed microbial subgingival biofilm, and foremost among these pathogenic agents is the gram-negative anaerobe Porphyromonas gingivalis (25). A number of virulence factors contribute to the pathogenicity of P. gingivalis. These include proteolytic enzymes that degrade host tissue and inactivate immune effector molecules; hemagglutinins that target the cells towards hemin, a requisite iron source; and fimbriae that are required for attachment to oral surfaces such as epithelial cells and to antecedent plaque bacteria and for invasion of epithelial cells (4, 9, 11, 28). As the oral cavity is a continuously changing environment, successful colonizers such as P. gingivalis have the ability to sense and respond to environmental conditions. Regulation of proteases, hemagglutinins, superoxide dismutase, and fimbriae has been documented, primarily at the transcriptional level, although it has also been reported at the posttranscriptional level (2, 9, 11, 26). However, although regulation of virulence genes is known to occur, the nature of the regulatory proteins and pathways in P. gingivalis has not been defined.
To gain an understanding of the regulatory networks in P. gingivalis, we are studying the expression of fimbrillin, the monomeric subunit of the P. gingivalis major fimbriae. Previous work has shown that expression of the gene encoding fimbrillin (fimA) and subsequent fimbria-dependent phenotypic activity are tightly regulated by environmental cues (30). Expression of the fimA gene is maximal at a relatively lower temperature and higher hemin concentration compared to normal growth conditions. In contrast, serum and salivary molecules inhibit fimA promoter activity. More-detailed site-specific mutagenesis experiments suggested that the fimA upstream region has a ς70-recognized RNA polymerase binding site along with an upstream element and a site determining positive regulation (31). Thus, we have proposed that trans-acting element(s) are involved in positive regulation of expression of the fimA gene in P. gingivalis by interacting with the fimA promoter region. The objective of this study was, therefore, to identify the proteins involved in regulation of the fimA gene. The results indicate that fimA expression is regulated by fimbrillin itself and by two major proteases of P. gingivalis.
MATERIALS AND METHODS
Bacterial stains, vectors, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. P. gingivalis strains were grown in Trypticase soy broth (Becton Dickinson) or on 1.5% Trypticase soy broth agar plates, supplemented with yeast extract (1 mg/ml; Difco), hemin (5 μg/ml), and menadione (1 μg/ml), at 37°C in an anaerobic chamber (85% N2, 10% H2, 5% CO2). Antibiotics—erythromycin (20 μg/ml) and gentamicin (100 μg/ml)—were used where appropriate. Escherichia coli DH5α was used as the host strain for recombinant plasmids and grown in L broth with antibiotics—ampicillin (100 μg/ml), kanamycin (50 μg/ml), trimethoprim (200 μg/ml), and tetracycline (10 μg/ml) when necessary.
TABLE 1.
Bacterial strains and plasmids used and constructed in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| P. gingivalis 33277 | Type strain from ATCC | This laboratory |
| P. gingivalis UPF | Derivative of 33277, fimAp::lacZ gene, Emr | 31 |
| P. gingivalis MPF35A | Derivative of P. gingivalis 33277, mpf35::fimA, fimAp::lacZ gene, Emr | This study |
| P. gingivalis MP150A | Derivative of P. gingivalis 33277, mp150::fimA, fimAp::lacZ gene, Emr | This study |
| P. gingivalis MPS10A | Derivative of P. gingivalis 33277, mps10::fimA, fimAp::lacZ gene, Emr | This study |
| P. gingivalis YPF1 | FimA-deficient mutant of 33277, Emr | 13 |
| P. gingivalis YPP1 | RgpA-deficient mutant of 33277, Emr | 23 |
| P. gingivalis YPP2 | Kgp-deficient mutant of 33277, Emr | 23 |
| E. coli DH5α | endA1 hsdR17 supE44 thi-1 recA gyrA96 relA1 Δ(lacZYA-argF) U169λ-φ80 dlacZ ΔM15, recipient for recombinant plasmids | Gibco-BRL |
| Plasmids | ||
| pJRD215 | Wide-host-range cosmid vector, Kmr Smr Mob+, unable to replicate in P. gingivalis | 22 |
| Tn4351 | Contains a 3.8-kb EcoRI fragment from pBF4, Tcr expressed in E. coli, Emr expressed in P. gingivalis | 24 |
| R751 | IncP plasmid used to mobilize vectors from E. coli to Bacteroides recipient Tpr Tra+ | 22 |
| pMP58 | fimAp::lacZ gene in pUC19 with a 16-bp deletion from −23 to −8 (mp58::lacZ), Amr | 31 |
| pMPF35 | mpf35::lacZ gene in pJRD215, three-base change from TTG to CCA at position −46 to −44, Kmr | 31 |
| pMP150 | mp150::lacZ gene in pJRD215, 150-bp deletion from −240 to −90, Kmr | 31 |
| pMPS10 | mps10::lacZ gene in pJRD215, three-base change from TAA to GCC at position −11 to −9, Kmr | 31 |
| pMP591 | mp150::lacZ gene in pJRD215, 15-bp deletion from −85 to −71 and 17-bp deletion from −64 to −48, Kmr | 31 |
Abbreviations: Kmr, Smr, Tcr, Emr, and Tpr, resistance to kanamycin, streptomycin, tetracycline, erythromycin, and trimethoprim, respectively; Mob+, can be mobilized; Tra+, capable of self transfer; fimAp, fimA promoter.
Purification of sequence-specific DNA binding proteins.
To purify P. gingivalis proteins binding to the regulatory sequences upstream from the fimA promoter, we utilized biomagnetic beads (Dynal, Oslo, Norway) according to the manufacturer's instructions. Briefly, the P. gingivalis 33277 fimA promoter region with a deletion of the RNA polymerase −35 binding site (31) was amplified by PCR. The template was plasmid MP58, which carries the mutated fimA promoter (31). The primers used were 5′-GGAATTCCGACGCTATATGCAAGACAA-3′ and 5′-TGTAACGGGTTCTGCCTCGT-3′, which was biotinylated at the 5′ end to facilitate binding to streptavidin-coated magnetic (M-280) beads. PCR mixtures contained 10 pmol of template DNA, 30 pmol of each primer, 1.5 mM of MgCl2, a 10 mM concentration of each deoxynucleoside triphosphate (dNTP) and 5 U of Taq DNA polymerase (Gibco-BRL). The amplification was performed in a thermal cycler (Techne) at 94°C for 45 s, 42°C for 1 min, and 72°C for 1 min for a total 30 cycles, followed by 10 min of elongation at 72°C. The PCR product was 166 bp (designated F166) and was immobilized on magnetic beads through a streptavidin-biotin interaction. A cell extract of P. gingivalis was prepared from the cells grown at 37°C to late log phase and disrupted by sonication. The extract was partially purified on a DEAE-cellulose column (Pharmacia Biotech) prior to reaction with F166-coated beads. The reaction mixture was separated in a magnetic field and washed, and specific DNA binding proteins were eluted with 1 M NaCl. Finally, the DNA binding proteins were resuspended in 10 mM Tris buffer by passing through CentriSpin-20 (Princeton Separations, Inc), and visualized by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS-PAGE) on 10% gels (7) stained with Coomassie blue. As a control, beads were coated with fimA promoter region from which the regulatory sequences were deleted. Template plasmid pMP591 was amplified by PCR with primers MP150 (31), 5′-GCTATGGTGTTGTTGGGTTGCATATTCA-3′, and biotin- 5′-TGTAACGGGTTCTGCCTCGT-3′. The PCR product was 103 bp (designated F103) and contains deletions from position −240 to −90, position −85 to −71, and position −64 to −48.
Recombinant protein, antisera, and immunoblotting.
Recombinant fimbrillin (rFim) was produced by PCR amplification of the fimA coding sequences from P. gingivalis 33277 chromosomal DNA (6) and cloning into the pET30 expression system (Novagen). Following induction in E. coli, rFim was purified by chromatography over a Ni2+ metal chelation resin and elution with imidazole. Upstream vector-derived sequences were then removed by cleavage with enterokinase. rFim is full-length mature fimbrillin without the leader amino acid sequence. Monospecific rabbit antibodies to rFim were produced by Covance Inc., Princeton, N.J. For blotting, proteins were separated by SDS-PAGE and transferred onto a nitrocellulose membrane. Antibodies to rFim or P. gingivalis cells (1:10,000) were used as the probe with peroxidase-conjugated secondary antibodies (1:3,000). Antigen-antibody binding was developed with 0.05% diaminobenzidine tetrahydrochloride.
Mobility shift DNA-binding assay.
Mobility shift DNA-binding assays were conducted by using the Bandshift kit (Pharmacia Biotech), as previously described (31). The F166 PCR product containing the fimA upstream region was used as probe. Briefly, the DNA fragment was labeled with [α-32P]dATP (3,000 μCi/mmol; NEN) using the Klenow fragment. For the protein-DNA reaction, 1 μg of 32P-labeled DNA and 1 μg of protein were mixed and incubated at room temperature for 20 min. The mixture was then loaded onto a 5% nondenaturing polyacrylamide gel and electrophoresed in 0.5 Tris-borate-EDTA buffer at 10 V/cm. Finally, the gel was dried and exposed to X-ray film at −70°C.
Construction of fimA mutant strains.
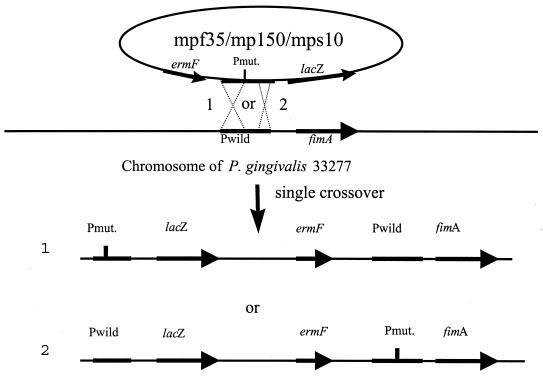
Site-specific mutations in the fimA upstream region were generated by a unique-site elimination mutagenesis kit (Pharmacia Biotech) as described in detail previously (31). Three mutant alleles of the fimA promoter were created and fused with a lacZ gene. One mutation (MPF35) was located in the −35 RNA polymerase binding site (−46 to −44), with a 3-bp substitution (TTG to CAA), and the second (MP150) carried a deletion from position −240 to −90, a region in which we have detected positive regulatory sequences (31). The third (MPS10) was a 3-bp substitution (TAA to GCC) at position −11 to −9, a region that is not involved in promotion of fimA transcription. The mutant alleles were then conjugated into P. gingivalis 33277 and integrated to the chromosomal DNA by a single crossover (Fig. 1). The authenticity of the mutated sequence was verified by DNA sequencing and PCR analysis with specific pairs of primers for each mutation (31).
FIG. 1.
Introduction of the mutated fimA promoter into P. gingivalis chromosomal DNA. Two genomic configurations can occur after a single crossover between plasmid MPF35 MPF150, or MPS10 and chromosomal DNA. In configuration 1, the crossover occurs upstream of the mutation. In configuration 2, the recombination occurs downstream of the mutation. Pmut. and Pwild represent the mutated and wild-type fimA promoter, respectively; ermF is an erythromycin resistance gene. Configuration 2 was selected by examination by PCR with primer pairs designed for Pmut. and fimA and for Pwild and lacZ.
RT-PCR.
The level of fimA mRNA expression was tested in the rgpA and kgp null mutants YPP1 and YPP2 (23). The oligonucleotides for the fimA gene were as follows: fimA1, 5′-AATCGTGCTTTTGGAGTTGG-3′; fimA2, 5′-ACCAACGAGAACCCACTCAG-3′. Reverse transcription was performed in the presence of 2 μg of total RNA, 50 ng of reverse primer, 50 U of reverse transcriptase (RT) (Ambion), 13 U of RNase inhibitor, a 10 mM concentration of each dNTP, and 1× RT buffer. The reaction was carried out at 72°C for 2 min and then at 48°C for 1 h. Controls without RT were included in all experiments. The resulting cDNA was amplified, with each 100 μl of PCR mixture containing 1× PCR buffer, 3 μl of cDNA, 1.5 mM MgCl2, a 10 mM concentration of each dNTP, 100 ng of each primer, and 2.5 U of Taq DNA polymerase. The amplification conditions were denaturation at 96°C for 30 s, annealing at 45°C for 30 s, and elongation at 72°C for 2 min. The expected size of the fimA PCR product was 1 kbp. As a control, an unrelated gene (an open reading frame demonstrating homology to the luxS gene) was also amplified.
Protein sequencing.
The DNA binding proteins eluted from magnetic beads were separated by SDS–10% PAGE. Following visualization with Coomassie blue, proteins were excised from the gel. Tryptic digestion followed by high-pressure liquid chromatography separation and Edman degradation were performed at the Biotechnology Center at the Fred Hutchinson Cancer Research Center, Seattle, Wash.
β-Galactosidase assays.
Expression of the lacZ gene under control of the fimA promoter was measured by the standard spectrophotometric β-galactosidase assay with o-nitrophenyl-β-d-galactopyranoside as the substrate, as described by Miller (17). P. gingivalis strains were recovered from late log phase and tested at an optical density at 600 nm of 0.4 to 0.6.
RESULTS
Interaction between P. gingivalis proteins and the fimA promoter.
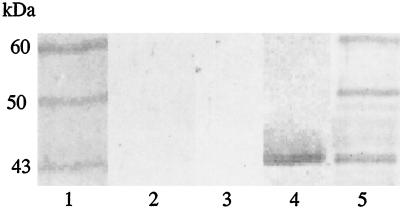
It is well documented that regulatory proteins can bind to specific sites in the upstream regions of genes, and this, in turn, influences the function of RNA polymerase (3). To search for regulatory proteins involved in fimA expression, we used a magnetic bead-bound DNA fragment of the fimA upstream region (F166) as bait for potential DNA-binding proteins from a cell extract of P. gingivalis 33277. After elution from the fimA promoter and visualization by SDS-PAGE, three major fimA binding proteins were observed, with molecular masses of approximately 60, 50, and 43 kDa (Fig. 2, lane 1). None of these proteins were recovered after incubation of the cell extract with magnetic beads conjugated with F103, the fimA upstream region from which the regulatory sequences were deleted (Fig. 2, lane 2). Furthermore, extract from the fimA null mutant, YPF1, did not bind to F166 (Fig. 2, lane 3). Western blot analysis with anti-FimA antibody indicated that the 43-kDa band was fimbrillin (Fig. 2, lane 4). Antibodies to P. gingivalis cells revealed additional fainter bands in the 45- to 49-kDa region (Fig. 2, lane 5).
FIG. 2.
Proteins binding to the fimA upstream region. Proteins were eluted from magnetic beads conjugated with F166 (lanes 1, 3, 4, and 5) or with F103 (lane 2) and separated by SDS-PAGE. Coomassie-stained proteins from strains 33277 are shown in lanes 1 and 2. Lane 3 contains a Coomassie stain of strain YPF1 (FimA deficient). Lanes 4 and 5 are immunoblots of 33277 eluted proteins, developed with antifimbrillin or anti-P. gingivalis antibodies, respectively. Sizes are indicated.
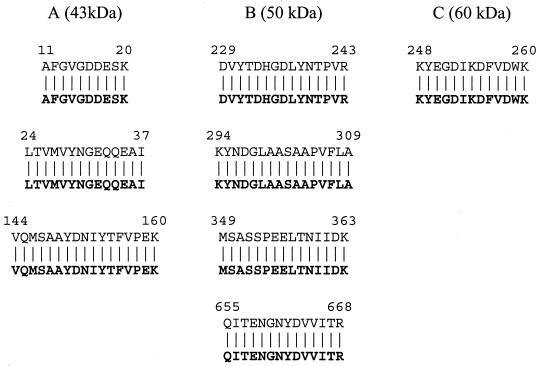
The identity of the major binding proteins was further investigated by amino acid sequencing. Three trypsin-digested peptide fragments from the 43-kDa band matched the fimbrillin amino acid sequence (Fig. 3A). Furthermore, a peptide derived from the 60-kDa peptide demonstrated 100% homology to the arginine-specific cysteine protease, Rgp (Fig. 3C). Thus, this molecule could be either RgpB or a posttranslational processed fragment of RgpA (4). Four of the peptides from the 50-kDa protein were 100% homologous to the lysine-specific cysteine protease, Kgp (Fig. 3B).
FIG. 3.
(A) Sequences of the digested peptides from the 43-kDa protein in Fig. 2. The fimbrillin (FimA) sequence is given in boldface letters. (B) Sequences of the peptides from the 50-kDa protein in Fig. 2. The lysine-specific cysteine proteinase (Kgp) sequence is given in boldface letters. (C) Sequence of the peptide from the 60-kDa protein in Fig. 2. The Gingipain R2 (RgpB) sequence is given in boldface letters. Numbers represent amino acid residues from sequences deposited in GenBank (accession numbers: FimA, BAA86887; Kgp, AAC26523; RgpB, A55426).
Mobility shift.
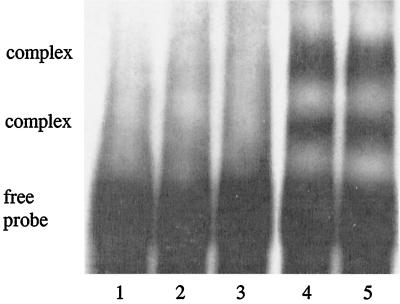
To test the ability of FimA to bind directly to the fimA promoter region, a bandshift experiment was performed (Fig. 4). Proteins eluted from the magnetic beads interacted with the fimA promoter, resulting in the formation of DNA-protein complex that halted movement of the DNA fragment (F166) (Fig. 4, lane 4). Mobility shift occurred in the presence of unrelated DNA (Fig. 4, lane 5), but was competitively inhibited by excess unlabeled F166 (Fig. 4, lane 3). The bandshift profile indicated more than one mobility change, which was probably due to instability of the multiple protein-DNA complex during the experimental process. Purified fimbrillin alone failed to associate with the fimA promoter (Fig. 4, lane 2), suggesting that fimbrillin may not bind to its own promoter directly but may associate with the promoter through other regulatory proteins. Proteases could function in this capacity; however, this appears unlikely, as in the absence of FimA the proteases could not be recovered from F166 (Fig. 2). Minor binding proteins visualized by Western blotting with whole P. gingivalis antibodies (Fig. 2) are also potential candidates, and work is under way to resolve this issue.
FIG. 4.
Mobility shift DNA binding assay. Lane 1, fimA upstream fragment (F166) only; lane 2, fimA promoter fragment and 1 μg of purified fimbrillin; lane 3, fimA promoter, the 33277 proteins eluted from magnetic beads, and 250 pmol of unlabeled F166; lane 4, fimA promoter and the 33277 eluted proteins from magnetic beads; lane 5, fimA promoter, the 33277 eluted proteins from magnetic beads, and 3 μg of calf thymus DNA.
Effect of fimA mutations on fimA promoter activity.
Based on the evidence provided by the binding assays, we predicted that fimbrillin serves as a regulatory protein in its own expression. To further test this hypothesis, we generated P. gingivalis strains containing a fimA promoter-lacZ reporter chromosomal fusion along with a mutation in the indigenous fimA promoter. Thus, these strains contain two fimA promoters, with one driving the fimA structural gene and the other promoting transcription of the lacZ gene. Insertion of the fimA::lacZ construct does not alter expression of fimA mRNA or levels of FimA protein (31). The resulting strains, MPF35A and MP150A, contain mutations at the −35 region (3-bp substitution) and the upstream regulatory sequence (150-bp deletion), respectively. These strains allow us to examine the influence of loss of production of FimA protein on fimA promoter activity (as reported by lacZ activity). RT-PCR confirmed both that fimA mRNA is either not produced (MPF35A) or significantly reduced (MP150A) and that there was no transcriptional read-through from the ermF gene (not shown). As shown in Table 2, lacZ activity decreased by around 40% in strain MP150A and to background levels in MPF35A. As an additional control, a mutation was introduced into a region upstream of the fimA structural gene and distal to the −10 site (MPS10A). This mutation does not affect promoter activity (31), and the resulting strain possessed wild-type levels of fimA mRNA and did not have reduced β-galactosidase activity (Table 2).
TABLE 2.
Effect of fimA promoter mutations (fimAMP) on the expression of fimA::lacZ fusion genes
| P. gingivalis strain | Promotera | Fusion | β-Galactosidase levelb |
|---|---|---|---|
| UPF | fimAWT | fimAWT::lacZ | 260 ± 9 |
| MP150A | fimAMP150 | fimAWT::lacZ | 158 ± 10 |
| MPF35A | fimAMPF35 | fimAWT::lacZ | 17 ± 1 |
| MPS10A | fimAMPS10 | fimAWT::lacZ | 300 ± 16 |
fimAWT represents wild-type promoter; fimAMP150 represents the fimA promoter with a 150-bp deletion of positive regulatory sequence spanning position −240 to −90 of the fimA gene; fimAMPF35 represents the fimA promoter with a substitution mutation, from TTG to CCA, at position −46 to −44, affecting the −35 RNA polymerase binding site; fimAMPS10 represents the fimA promoter with a substitution mutation, from TAA to GCC, at position −11 to −9, which does not affect promoter activity.
Miller units obtained from P. gingivalis grown at 37°C. Means ± standard deviations are indicated (n = 3).
Transcription of fimA in rgp and kgp mutants.
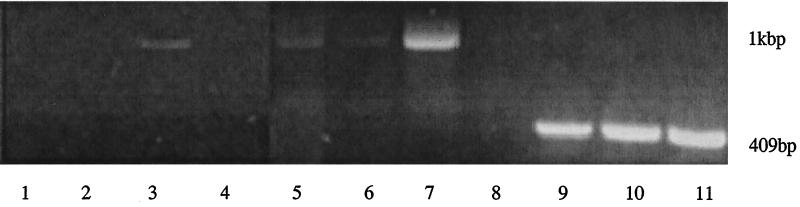
As the data indicated that the major cysteine proteases of P. gingivalis are also involved in regulation of fimA expression, loss of protease expression should cause downregulation of fimA. Thus, we examined fimA mRNA levels in P. gingivalis strains YPP1 (which contains an insertional inactivation of the rgpA gene) and YPP2 (which contains an insertional inactivation of the kgp gene) (23) by RT-PCR (Fig. 5). Expression of fimA mRNA was significantly reduced in these mutants compared to wild type. In contrast there were no differences in expression of a luxS-like gene that was identified by a homology search of the P. gingivalis genome database (http://www.tigr.org).
FIG. 5.
RT-PCR of fimA mRNA in YPP1 and YPP2 protease-deficient mutants. Lanes 1, 5, and 9, YPP1; lanes 2, 6, and 10, YPP2; lanes 3, 7, and 11, 33277; lanes 4 and 8, negative controls without RT. Lanes 1 to 4, cDNA amplified for 30 cycles with fimA primers; lanes 5 to 8, amplified for 40 cycles with fimA primers; lanes 9 to 11, amplified for 35 cycles with control primers for a luxS-like gene.
DISCUSSION
It is becoming increasingly apparent that fine control over the expression of bacterial virulence determinants is required for successful bacterial colonization and disease progression. Such control can be the outcome of complex and regulatory networks that sense the prevailing environmental conditions and transduce information to modulate gene expression (16). P. gingivalis is well adapted to life in the subgingival environment and regulates gene expression in response to orally relevant parameters (2, 9, 11, 26). However, the signal transduction pathways and regulatory mechanisms in this strict anaerobe are not well understood. Evidence provided here suggests that several well-known proteins of P. gingivalis are members of a fimA regulatory system. These include fimbrillin itself, along with an arginine-specific cysteine protease (Rgp) and a lysine-specific cysteine protease (Kgp). These proteins could bind to, and cause a mobility shift of, the fimA upstream region. Site-specific mutagenesis also showed that the wild-type fimA promoter was maximally activated only when the fimA structural gene was fully expressed. Indeed, a 3-bp replacement in the −35 region of fimA alone was sufficient to repress transcription from the intact fimA promoter in a reconstituted lacZ gene fusion system. Furthermore, transcriptional activity of fimA was significantly reduced in mutants lacking expression of either RgpA or Kgp. Thus, the presence of all three of these proteins appears to be necessary for maximal expression of fimA. Whether these proteins assemble prior to or after DNA association remains to be determined, and it is possible that the observed participation of the proteolytic enzymes is a consequence of their preexisting association with prefimbrillin in a processing capacity. In either event a role for additional direct DNA-binding proteins is suggested. Work is currently under way to identify these molecules.
The phenomenon of autoregulation is well established for several bacterial genes. Liberek and Georgopoulos (12) reported that in E. coli, the heat shock genes, dnak, dnaJ, and grpE, were negatively autoregulated, and mutations in any one of these genes could lead to their own constitutive expression. In some studies the mechanisms of gene autoregulation were revealed. The simplest autoregulation pathway involves the gene products serving as activators or repressors by binding to the promoter region of the same gene (5). An alternative pathway involves autoregulation at the translational level, whereby the protein can bind to its own mRNA (15). Autoregulation at a posttranscriptional level is unlikely in P. gingivalis, since an operon fusion was used in our experimental system. Autoregulation affords bacteria an additional level of genetic control and thus facilitates fine-tuning of protein expression. In the case of P. gingivalis fimbrillin, this may be important in optimizing protein levels during the transition from colonization to initiation of disease. Fimbriae are essential mediators of adherence and invasion and are thus necessary for colonization of the organism (1, 8, 9, 10, 29). However, fimbriae are also potent inducers of immune cell function (19, 20) and hence may be detrimental to the organism when it engages in a more intimate interaction with periodontal tissues. The ability to amplify down- and upregulation signals may thus provide P. gingivalis with a selective advantage in mixed microbial oral biofilms.
P. gingivalis is an asaccharolytic organism and utilizes a number of proteolytic enzymes to provide peptides for growth. These proteinases also have a variety of effects with relevance to pathogenicity, including tissue destruction and inactivation of effector molecules of the immune system (4, 28). Previous studies have also reported a role for proteases in the fimbriation of P. gingivalis. Mutants that are defective in Rgp production possess very few fimbriae on their cell surfaces (18). These observations have been explained on the basis of posttranslational processing, since proteases can cleave the N-terminal amino acid (leader peptide) of FimA protein in vitro (21). However, reports vary on whether fimbriation is affected in single rgpA or kgp mutants, or whether a double rgpAB mutation is required to reduce fimbrillin production. For example, Nakayama et al. (18) showed that an rgpA-rgpB double mutant possessed very few fimbriae on the cell surface, whereas fimbriation was normal in an rgpA single mutant. In contrast, Tokuda et al. (27) demonstrated that an rgpA single mutant did not express detectable levels of the FimA protein as determined by Western blotting, nor were fimbriae visible following electron microscopy of the cells. Moreover, the rgpA single mutant in the latter study had reduced expression of fimA mRNA. Our observations are more consistent with those of Tokuda et al. (27) and demonstrate that individual proteases can play a direct role in fimbrillin production at the transcriptional level. Linkage of fimbrillin expression to proteolytic activity may allow coordination of activity of a number of the molecules that drive the pathophysiology of the organism.
The fimbriae of P. gingivalis contribute significantly to virulence. Indeed, fimbria-deficient mutant cells are less pathogenic in animal models (14) and are unable to form biofilms (Y. Park, J. W. Costerton, G. S. Cook, D. R. Demuth, and R. J. Lamont, Abstr. 77th Meet. Int. Assoc. Dent. Res., abstr. 2514, 1999). P. gingivalis fimbriae do not exhibit homology to other gram-negative fimbriae, either in terms of sequence or chromosomal arrangement, and may represent a unique class of fimbrial structure (6). An understanding of the mechanism of transcriptional control of fimbria production should provide important insights into the basis of the pathogenicity of this important periodontal pathogen.
ACKNOWLEDGMENT
The support of the NIDCR (DE00401, DE11111, and DE12505) is gratefully acknowledged.
REFERENCES
- 1.Amano A, Sharma A, Lee J-Y, Sojar H T, Raj P A, Genco R J. Structural domains of Porphyromonas gingivalisrecombinant fimbrillin that mediate binding to salivary proline-rich protein and statherin. Infect Immun. 1996;64:1631–1637. doi: 10.1128/iai.64.5.1631-1637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amano A, Sharma A, Sojar H T, Kuramitsu H K, Genco R J. Effects of temperature stress on expression of fimbriae and superoxide dismutase by Porphyromonas gingivalis. Infect Immun. 1994;62:4682–4685. doi: 10.1128/iai.62.10.4682-4685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busby S, Ebright R H. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 4.Curtis M A, Kuramitsu H K, Lantz M, Macrina F L, Nakayama K, Potempa J, Reynolds E C, Aduse-Opoku J. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J Periodontal Res. 1999;34:464–472. doi: 10.1111/j.1600-0765.1999.tb02282.x. [DOI] [PubMed] [Google Scholar]
- 5.Davis T L, Helinski D R, Roberts R C. Transcription and autoregulation of the stabilizing functions of broad-host-range plasmid RK2 in Escherichia coli, Agrobacterium tumefaciens and Pseudomonas aeruginosa. Mol Microbiol. 1992;6:1981–1994. doi: 10.1111/j.1365-2958.1992.tb01371.x. [DOI] [PubMed] [Google Scholar]
- 6.Dickinson D P, Kubiniec M A, Yoshimura F, Genco R J. Molecular cloning and sequencing of the gene encoding the fimbrial subunit protein of Bacteroides gingivalis. J Bacteriol. 1988;170:1658–1665. doi: 10.1128/jb.170.4.1658-1665.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 8.Lamont R J, Bevan C A, Gil S, Persson R E, Rosan B. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol Immunol. 1993;8:272–276. doi: 10.1111/j.1399-302x.1993.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 9.Lamont R J, Jenkinson H F. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J Y, Sojar H T, Bedi G S, Genco R J. Synthetic peptides analogous to the fimbrillin sequence inhibit adherence of Porphyromonas gingivalis. Infect Immun. 1992;60:1662–1670. doi: 10.1128/iai.60.4.1662-1670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lepine G, Progulske-Fox A. Duplication and differential expression of hemagglutinin genes in Porphyromonas gingivalis. Oral Microbiol Immunol. 1996;11:65–78. doi: 10.1111/j.1399-302x.1996.tb00339.x. [DOI] [PubMed] [Google Scholar]
- 12.Liberek K, Georgopoulos C. Autoregulation of the Escherichia coliheat shock response by the DnaK and DnaJ heat shock proteins. Proc Natl Acad Sci USA. 1993;90:11019–11023. doi: 10.1073/pnas.90.23.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Love R M, McMillan M D, Park Y, Jenkinson H F. Coinvasion of dentinal tubules by Porphyromonas gingivalis and Streptococcus gordoniidepends upon binding specificity of streptococcal antigen I/II adhesin. Infect Immun. 2000;68:1359–1365. doi: 10.1128/iai.68.3.1359-1365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malek R, Fisher J G, Caleca A, Stinson M, van Oss C J, Lee J Y, Cho M I, Genco R J, Evans R T, Dyer D W. Inactivation of Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J Bacteriol. 1994;176:1052–1059. doi: 10.1128/jb.176.4.1052-1059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer C, Kohrer C, Grobner P, Piendl W. MvaL 1 autoregulates the synthesis of the three ribosomal proteins encoded on the Mval 1 operon of the archaeon Methanococcus vannieliiby inhibiting its own translation before or at the formation of the first peptide bond. Mol Microbiol. 1998;27:455–468. doi: 10.1046/j.1365-2958.1998.00693.x. [DOI] [PubMed] [Google Scholar]
- 16.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 18.Nakayama K, Yoshimura F, Kadowaki T, Yamamoto K. Involvement of arginine-specific cysteine proteinase (Arg-gingipain) in fimbriation of Porphyromonas gingivalis. J Bacteriol. 1996;178:2818–2824. doi: 10.1128/jb.178.10.2818-2824.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ogawa T, Ogo H, Uchida H, Hamada S. Humoral and cellular immune responses to the fimbriae of Porphyromonas gingivalisand their synthetic peptides. J Med Microbiol. 1994;40:397–402. doi: 10.1099/00222615-40-6-397. [DOI] [PubMed] [Google Scholar]
- 20.Ogawa T, Uchida H, Hamada S. Porphyromonas gingivalisfimbriae and their synthetic peptides induce proinflammatory cytokines in human peripheral blood monocyte cultures. FEMS Microbiol Lett. 1994;116:237–242. doi: 10.1111/j.1574-6968.1994.tb06707.x. [DOI] [PubMed] [Google Scholar]
- 21.Onoe T, Hoover C I, Nakayama K, Ideka T, Nakamura H, Yoshimura F. Identification of Porphyromonas gingivalisprefimbrillin possessing a long leader peptide: possible involvement of trypsin-like protease in fimbrillin maturation. Microb Pathog. 1995;19:351–364. doi: 10.1016/s0882-4010(96)80006-4. [DOI] [PubMed] [Google Scholar]
- 22.Park Y, McBride B C. Characterization of the tpr gene product and isolation of a specific protease-deficient mutant of Porphyromonas gingivalisW83. Infect Immun. 1993;61:4139–4146. doi: 10.1128/iai.61.10.4139-4146.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park Y, Lamont R J. Contact-dependent protein secretion in Porphyromonas gingivalis. Infect Immun. 1998;66:4777–4782. doi: 10.1128/iai.66.10.4777-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoemaker N B, Getty C, Gardner J F, Salyers A A. Tn4351 transposes in Bacteroides spp. and mediates the integration of plasmid R751 into the Bacteroideschromosome. J Bacteriol. 1986;165:929–936. doi: 10.1128/jb.165.3.929-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Socransky S S, Haffajee A D. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–331. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- 26.Tokuda M, Chen W, Karunakaran T, Kuramitsu H K. Regulation of protease expression in Porphyromonas gingivalis. Infect Immun. 1998;66:5232–5237. doi: 10.1128/iai.66.11.5232-5237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokuda M, Karunakaran T, Duncan M, Hamada N, Kuramitsu H K. Role of Arg-gingipain A in virulence of Porphyromonas gingivalis. Infect Immun. 1998;66:1159–1166. doi: 10.1128/iai.66.3.1159-1166.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Travis J, Pike R, Imamura T, Potempa J. Porphyromonas gingivalisproteinases as virulence factors in the development of periodontitis. J Periodontal Res. 1997;32:120–125. doi: 10.1111/j.1600-0765.1997.tb01392.x. [DOI] [PubMed] [Google Scholar]
- 29.Weinberg A, Belton C M, Park Y, Lamont R J. Role of fimbriae in Porphyromonas gingivalisinvasion of gingival epithelial cells. Infect Immun. 1997;65:313–316. doi: 10.1128/iai.65.1.313-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie H, Cai S, Lamont R J. Environmental regulation of fimbrial gene expression in Porphyromonas gingivalis. Infect Immun. 1997;65:2265–2271. doi: 10.1128/iai.65.6.2265-2271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie H, Lamont R J. Promoter architecture of the Porphyromonas gingivalisfimbrillin gene. Infect Immun. 1999;67:3227–3235. doi: 10.1128/iai.67.7.3227-3235.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]