Abstract
Directed mutagenesis of a gene coding for a membrane protein of the periodontopathogen Actinobacillus actinomycetemcomitans was achieved by conjugation. The gene was disrupted by insertion of an antibiotic cassette into a unique endonuclease restriction sequence engineered by inverse PCR. The disrupted gene was cloned into a conjugative plasmid and transferred from Escherichia coli to A. actinomycetemcomitans. The allelic replacement mutation resulted in the loss of a 22-kDa inner membrane protein. The loss of this protein (ImpA) resulted in changes in the outer membrane protein composition of the bacterium. Concurrent with the mutation in impA was a change in the pattern of growth of the mutant bacteria in broth cultures. The progenitor bacteria grew as a homogeneous suspension of cells compared to a granular, autoaggregating adherent cell population described for the mutant bacteria. These data suggest that ImpA may play a regulatory role or be directly involved in protein(s) that are exported and associated with colony variations in A. actinomycetemcomitans.
Actinobacillus actinomycetemcomitans is widely recognized as a major pathogen in the etiology of localized juvenile periodontal disease and cases of refractory adult periodontal disease (32, 33, 41). The bacteria initiate a cascade of events that involve both a cellular and a humoral immune response which results in chronic inflammation (14, 40). The contribution of both bacterial and host factors may lead to the loss of tissue-matrix components and ultimately the loss of teeth. Multiple virulence determinants have been described for this organism which may participate in the infection of the host and protection from the host's immune system (8). To define the mechanisms of bacterial factors in the infection process, molecular strategies need to be developed.
Recently, genes coding for exported proteins in A. actinomycetemcomitans have been identified using translational fusions to alkaline phosphatase (22). Based on the deduced amino acid sequence, several of these sequences were found to be homologous to bacterial membrane proteins but with no known associated function(s). A powerful method to deduce protein function is the generation of defined isogenic mutants. Therefore, one of the above gene sequences that has homology to a hypothetical 22-kDa transmembrane protein of Haemophilus influenzae was used as the prototypic gene to demonstrate the utility of conjugation for directed mutagenesis in A. actinomycetemcomitans.
Fresh clinical isolates of A. actinomycetemcomitans express a rough colony phenotype which grows in broth as granular, autoaggregate adherent cells that leave a clear broth (7, 11, 13). Upon successive rounds of in vitro subculturing on solid media, the colonies convert to a smooth phenotype that grows as a homogeneous suspension with no adherent cells (7, 11, 13). Allelic replacement mutagenesis of the gene coding for the 22-kDa protein of a smooth phenotype strain of A. actinomycetemcomitans resulted in a reversion of the growth, in broth, from a homogeneous suspension to an aggregated growth morphology. We describe here a method for directed mutagenesis in A. actinomycetemcomitans by conjugation and characterization of a strain mutant for the 22-kDa protein.
MATERIALS AND METHODS
Bacterial strains and plasmids.
A. actinomycetemcomitans strains in this study were grown statically in Trypticase soy broth supplemented with 0.6% yeast extract (TSBYE) in a humidified, 10% CO2 atmosphere at 37°C. E. coli strains DH5α(λpir) and SM10(λpir) and the conjugative plasmid pGP704 were obtained from the laboratory of Murry Stein (University of Vermont, Burlington). The E. coli strains were grown in Luria-Bertani (LB) medium at 37°C with aeration. The plasmid was propagated and purified from E. coli strain CC118la grown in LB medium containing 50 μg of ampicillin per ml. Mu phage was obtained from the laboratory of Howard A. Shuman (Columbia University, New York, N.Y.). Phage DNA was purified as described previously (22).
A nalidixic acid-rifampin-resistant strain of A. actinomycetemcomitans was derived from strain SUNY 465 (smooth phenotype). A rifampin-resistant spontaneous mutant was selected by plating 108 logarithmically growing bacteria on TSBYE agar plates containing 60 μg of rifampin per ml followed by incubation for 3 days in a humidified, 10% CO2 atmosphere at 37°C. The Rifr colonies were replated on fresh TSBYE agar plates containing antibiotic. A single colony was propagated and frozen in 10% dimethyl sulfoxide. This strain was shown to be resistant to rifampin at 200 μg/ml. The Rifr Nalr strain of A. actinomycetemcomitans was generated by plating 3 × 108 Rifr cells (grown in rifampin-containing medium) on TSBYE agar plates containing 0.1 to 10 μg of nalidixic acid per ml and incubated as described above. The colonies that grew on the highest concentration were replated on plates containing incremental increasing concentrations of nalidixic acid. The Rifr Nalr strain (VT1169) was routinely passaged using 100 and 50 μg of rifampin and nalidixic acid per ml, respectively.
Plasmid construction for allelic replacement mutagenesis.
The complete impA sequence is presented in Fig. 1 and can be obtained from GenBank (accession no. AF04561). impA was amplified by PCR using primers corresponding to sequences starting 67 bp 5′ of the start of the signal sequence (5′-ACA TAG CGA ACA AGT GGT GG-3′, residues 1 to 20; Fig. 1) and 55 bp 3′ of the stop codon (5′-CAT AGT AAG CCT TGA AGC G-3′, residues 729 to 741; Fig. 1) and cloned into pT7-Blue (Promega, Inc., Madison, Wis.). A unique StuI restriction site was engineered by inverse PCR (25) using primers corresponding to nucleotides 384 to 402 (5′-AAA GAC TTC CCA TTG GCA GAA-3′; Fig. 1) and 356 to 335 (5′-AGC AAG GCT AAG ACG GCA TAG-3′; Fig. 1). The gene was disrupted by insertion of the spectinomycin gene isolated from plasmid pDL269 (20). The aad9 was released from pDL269 by incubation with NdeI/HindIII and gel purified. The DNA fragment was incubated with Klenow (29) for blunt-end ligation with the inverse PCR product restricted with StuI. The ligation mixture was transformed into E. coli JM109 cells by electroporation and plated onto LB agar plates containing 50 μg of spectinomycin per ml. Spcr colonies were selected, and the plasmids were isolated using a rapid plasmid purification scheme (1). The constructs were confirmed by restriction mapping and PCR. The disrupted gene was released from the plasmid by digestion with HindIII/EcoRI and treated with Klenow. The fragment was ligated with pGP704 previously cleaved with EcoRV. Electrocompetent DH5α(λpir) cells were transformed with the ligation mixture, and transformants were selected on LB agar containing 50 μg of spectinomycin per ml. Plasmids were isolated and the construct was confirmed by PCR. Plasmid containing the disrupted gene was purified using Qiagen spin columns (Qiagen, Inc., Valencia, Calif.) and transformed by electroporation into E. coli SM10(λpir) cells for conjugation.
FIG. 1.
Nucleotide sequence of impA and flanking sequence from A. actinomycetemcomitans SUNY 465. Potential prokaryotic −35 and −10 promoter sequences are underlined. The possible Shine-Dalgarno sequence is in underlined boldface. The start (ATG) and stop (TAG) translational sequences are in boldface. Sequences that are in boldface and have arrows correspond to the sequences used for inverse PCR. The underlined sequences at the 3′ end of the sequence is the inverted repeat found adjacent to the translational stop signal.
Allelic replacement mutagenesis.
Mobilization of the plasmid containing the disrupted gene from E. coli SM10(λpir) to A. actinomycetemcomitans strain VT1169 (Rifr Nalr) was accomplished by conjugation as described by Goncharoff et al. (10) with modifications. Donor and recipient cells were mixed in a 1:10 ratio, respectively. E. coli SM10(λpir) cells containing the plasmid (donor cells) were grown to stationary phase in LB medium containing 50 μg of spectinomycin per ml, centrifuged, and resuspended in TSBYE. A. actinomycetemcomitans cells were harvested during the mid-log phase of growth following culture in nalidixic acid-rifampin-containing medium, centrifuged, and resuspended in the appropriate volume of TSBYE. Then, 100 μl of the recipient strain was spotted onto a TSBYE plate and overlaid with 100 μl of donor cells. The bacteria were incubated for 5 to 10 min at room temperature and transferred to a humidified atmosphere of 10% CO2 at 37°C for 5 h. Following the incubation period, 1 ml of TSBYE was added to the plate, and the cells were removed from the plate by scraping the cells with a sterile glass slide. Bacteria were plated on TSBYE agar containing nalidixic acid, rifampin, and spectinomycin (50, 100, and 50 μg/ml, respectively) and incubated as described above for 3 to 4 days. Spectinomycin-resistant colonies were replica plated on TSBYE plates containing 100 μg of ampicillin per ml. Bacteria that were Spcr Amps were grown in TSBYE containing 50 μg of spectinomycin per ml, and the DNA was isolated using Puregene DNA extraction Kit (Gentra Systems, Minneapolis, Minn.). Chromosomal DNA of the transconjugants was analyzed by PCR.
Southern analysis.
Chromosomal DNA was restricted with EcoRI, and the fragments were separated on a 0.7% agarose gel in TAE buffer. The DNA fragments were transferred to Hybond nylon membranes (Amersham Life Sciences, Buckinghamshire, England), and the membranes were treated according to the method of Sambrook et al. (29). The membranes were hybridized with DNA probes conjugated with horseradish peroxidase using the conditions suggested by the manufacturer (Amersham Life Sciences, Buckinghamshire, England). Hybridizing fragments were visualized using the enhanced chemiluminescence detection system (Amersham Life Sciences, Buckinghamshire, England) and exposure to photographic film (XAR-5; Eastman Kodak, Rochester, NY).
Northern analysis.
Total RNA from A. actinomycetemcomitans strain VT1169 was isolated using the Purescript RNA isolation kit according to the manufacturer's protocol (Gentra Systems, Minneapolis, Minn.). Total RNA was separated by electrophoresis and transferred to a nitrocellulose membrane as described previously (38). The immobilized RNAs were hybridized with impA labeled with [α-32P]dCTP by nick translation according to the manufacturer's protocol (Gibco-BRL, Grand Island, N.Y.). After hybridization overnight at 42°C in 50% formamide, the filter was washed by successive incubations (twice each) with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min and 0.1× SSC–0.1% sodium dodecyl sulfate (SDS) for 30 min at 65°C. The filter was air dried and exposed to radiographic film at −70°C for 7 days.
Isolation of inner and outer membrane proteins.
Bacterial membranes were isolated by disruption of the bacteria using a French pressure cell at 2,000 lb in−2, followed by differential centrifugation (21). The membranes were resuspended in 10 mM HEPES (pH 7.4). Sodium N-lauroylsarcosinate was added to a final concentration of 1% and incubated at room temperature for 30 min (3). The mixture was centrifuged at 15,600 × g for 30 min. The supernatant was removed and stored on ice. The pellet was resuspended in 10 mM HEPES (pH 7.4) and centrifuged as described above. The final pellet was resuspended in 10 mM HEPES (pH 7.4). Based on bacterial membrane protein solubility in sarcosinate (6), inner membrane proteins are defined as proteins soluble in the detergent and outer membrane proteins are insoluble in the detergent. Protein concentrations were determined by absorbance at 280 nm. The insoluble proteins were solubilized in 1% SDS before determination of the protein concentration. Equal concentration of proteins were boiled for 10 min in sample buffer, before application to 5 to 15% polyacrylamide-SDS gels, and electrophoresis was performed according to the method of Laemmli (18).
Hydrophobicity assay.
Relative surface hydrophobicity was determined using a rapid method (organic phase partitioning) adapted from Rosenberg et al. (28).
RESULTS
Inactivation of the gene coding for ImpA of A. actinomycetemcomitans.
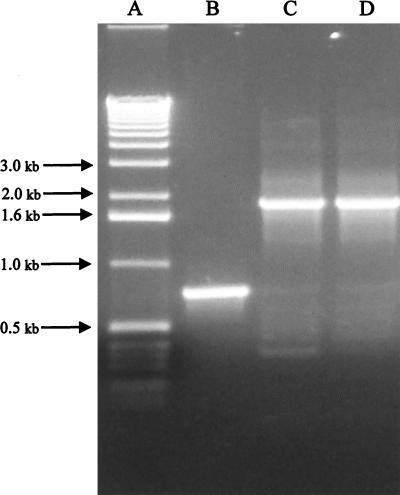
A PCR product (737 bp) comprised of impA including 67 bp 5′ of the start of the signal sequence and 55 bp 3′ of the stop codon was cloned into pT7Blue (Fig. 2, lane B). A unique StuI site was engineered into impA by inverse PCR, and the gene was disrupted by ligation of a spectinomycin gene (1.1 kb) by blunt-end ligation (Fig. 2, lane C). The primers selected for inverse PCR resulted in a 49-bp deletion in impA. The final construct was composed of ∼350 bp of impA (flanking and coding sequence) juxtaposed next to the spectinomycin gene, which was contiguous with the remaining ∼350 bp of the gene and flanking sequence.
FIG. 2.
Identification of a chromosomal mutation by PCR. Chromosomal DNA was isolated from transconjugants or wild-type cells and was used as a template for PCR with primers corresponding to the 5′ and 3′ ends of impA. Products were analyzed by agarose gel electrophoresis. Lane A, 1-kb DNA ladder; lane B, wild-type chromosomal DNA; lane C, cloned impA disrupted with a spectinomycin cassette; lane D, impA mutant chromosomal DNA.
Allelic replacement mutagenesis in A. actinomycetemcomitans.
Transformation of A. actinomycetemcomitans by electroporation (35) with the above construct resulted in few transformants, all of which contained the entire plasmid integrated into the chromosome (data not shown). However, the frequency of recombinatorial events was increased using bacterial mating or conjugation. Successful gene transfer was achieved following conjugation of E. coli SM10(λpir) containing the disrupted impA on the conjugative plasmid pGP704 with a spontaneous mutant of A. actinomycetemcomitans SUNY 465 resistant to nalidixic acid and rifampin. Transconjugants were recovered from this mating experiment after counterselection on TSBYE agar plates containing spectinomycin. Transconjugants containing an allelic replacement of impA were selected by replica plating on TSBYE agar plates containing ampicillin (plasmid marker) or spectinomycin (gene marker). Transconjugants that grew on the spectinomycin plates but not on the plates containing ampicillin indicated that the disrupted gene sequence but not the vector sequence integrated into the genome of these transconjugants.
Confirmation of the integration event was determined by PCR using primers that hybridized to the flanking sequences of impA. The antibiotic resistance profile of the transconjugants indicated that a double crossover event had occurred. However, PCR of the chromosomal DNA indicated that two products were generated for most of the transconjugants: one product corresponding to the intact gene and the other corresponding to the disrupted gene (data not shown). However, one of the transconjugants was observed to have a single PCR product. The gel mobility of this product (Fig. 2, lane D) corresponded to the mobility of the disrupted gene contained on the conjugative plasmid (Fig. 2, lane C). Both of these products migrated more slowly than the product generated using the parent or wild-type chromosomal DNA as the template. This difference in mobility corresponded to the size of the spectinomycin gene.
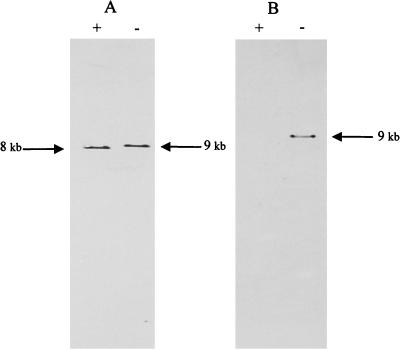
The allelic replacement was confirmed by Southern analysis (Fig. 3). DNA extracted from wild-type and impA mutant strains and probed with labeled impA hybridized to fragments which differed in size that corresponded to the size of the spectinomycin gene (Fig. 3A). The presence of the spectinomycin gene in this fragment was demonstrated by hybridization of a labeled spectinomycin gene exclusively with the ∼9-kb DNA fragment (Fig. 3B). Southern analysis using vector sequence as the probe did not hybridize with chromosomal DNA extracted from the impA mutant strain (data not shown). These data indicate that the disrupted gene replaced the wild-type gene in a site-directed and specific manner into the A. actinomycetemcomitans genome.
FIG. 3.
Southern blot analysis of chromosomal DNA. Chromosomal DNA was isolated from wild-type and impA mutant strains and incubated with EcoRI. Products were separated by agarose gel electrophoresis and transferred to nylon membranes. Duplicate membranes were probed with labeled impA (A) or spc (B). Lanes: +, wild-type DNA; −, mutant DNA.
E. coli SM10(λpir) has an integrated form of a conjugative plasmid that is used to mobilize the suicide vector (31). This plasmid has functional copies of bacteriophage Mu, a temperate phage that integrates by random transposition. The RP-4-based system used to transfer plasmids has been successfully used in a number of bacteria (4, 15, 36). However, this system has proven to be problematic in Legionella pneumophila (37). In the L. pneumophila system, mutants resulting from matings with an RP-4-based plasmid containing TnphoA were found to be introduced by random insertions of Mu into the L. pneumophila genome. To determine the presence or absence of random integration of Mu into the impA mutant chromosome, DNA was restricted with EcoRI and transferred to nylon membrane for Southern blot analysis. The blot was probed a 1.1-kb HindIII fragment of Mu phage DNA conjugated with horseradish peroxidase. The blot did not reveal any detectable signal in the lane corresponding to the impA mutant DNA (data not shown). These data indicate that Mu sequence is not present in the genome of the impA mutant.
Characterization of the impA mutant strain of A. actinomycetemcomitans.
Broth cultures of primary derived impA mutant grew as aggregates on the side and bottom of the tube leaving a clear broth as compared to a turbid, homogeneous suspension with no aggregates, as observed with the parent strain (Fig. 4). These aggregates could be partially dissociated by sonication. When grown on agar for the same period of time, the impA mutant strain colonies were larger in size than the wild-type colonies and demonstrated an alteration in the colony morphology. Both strains were observed to be circular and did not show any differences in surface elevation. However, the edges of the colonies were observed to be slightly different. The edge of the wild-type colonies displayed an entire morphology, whereas the edges of impA mutant strain colonies had a more erose morphology (data not shown).
FIG. 4.
Comparison of broth culture phenotypes of wild-type impA and mutant impA of A. actinomycetemcomitans. A single bacterial colony was inoculated into 8 ml of TSBYE (impA+) or TSBYE containing 50 μg of spectinomycin per ml (impA mutant) and grown statically overnight at 37°C in 10% CO2. The left tube represents overnight broth culture of impA wild type, and the right tube represents overnight broth culture of impA mutant.
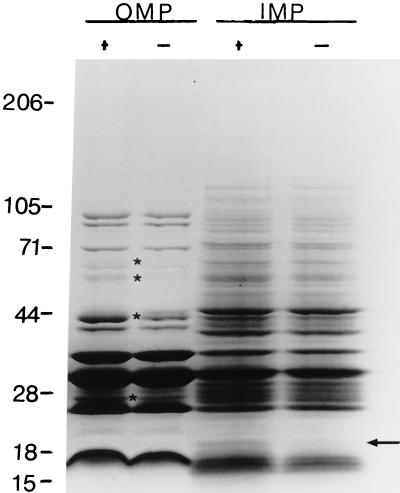
The differences in growth characteristics and colony morphology suggested that there may be differences in the expression of membrane proteins. These possible differences were investigated by analysis of the membrane proteins by SDS-polyacrylamide gel electrophoresis (PAGE). Inner and outer membrane proteins were separated by differential solubilization in the detergent sodium lauryl sarcosinate. Comparison of the outer membrane proteins of impA mutant and wild-type strains revealed a number of differences in the protein profiles (Fig. 5). Two proteins (∼67 and ∼60 kDa) were observed to be absent in the impA mutant outer membrane as compared to the parent strain. Conversely, an additional ∼44-kDa protein was present in the impA mutant profile compared to the parent strain. In addition, two proteins (∼42 and ∼26 kDa) stained with less intensity in the mutant strain compared to the wild-type strain following staining with Coomassie brilliant blue.
FIG. 5.
SDS-PAGE analysis of inner and outer membrane proteins from impA+ and impA mutant strains of A. actinomycetemcomitans. Membrane preparations were obtained as described in Materials and Methods. Membranes were solubilized in 1% sodium N-lauroyl sarcosinate. Outer membrane proteins (OMP) were recovered in the pellet following centrifugation, whereas inner membrane proteins (IMP) remained soluble in the supernatant following centrifugation. The pellets were washed twice with buffer before resuspension of the proteins. Equal protein concentrations were applied to the gel. +, impA+; −, impA (mutant). The arrow indicates the absence of a 22-kDa protein in the impA mutant strain. The asterisks denote proteins that are absent or reduced in staining intensity in the mutant strain compared with the parent strain. The numbers represent the molecular masses of protein standards in kildaltons.
The change in the outer membrane protein profile may be correlated with the absence of the 22-kDa inner membrane protein of the impA mutant strain (Fig. 5). This protein molecular mass corresponds to the molecular mass deduced from the impA sequence. Only minor differences were observed in the electrophoretic mobilities and staining intensities of the remaining inner membrane proteins isolated from impA wild-type and mutant strains of A. actinomycetemcomitans.
Associated with the changes in the outer membrane protein profile of the mutant, an increase in the hydrophobicity of the organism was also observed. Hydrophobicity, as determined by the percentage of organisms that partition into the organic phase (hexadecane) was increased 13% for the mutant compared to the parent strain (58.9 ± 5.5 versus 45.8 ± 5.2, respectively; P < 0.05).
Northern analysis and reverse transcription-PCR (RT-PCR) of impA.
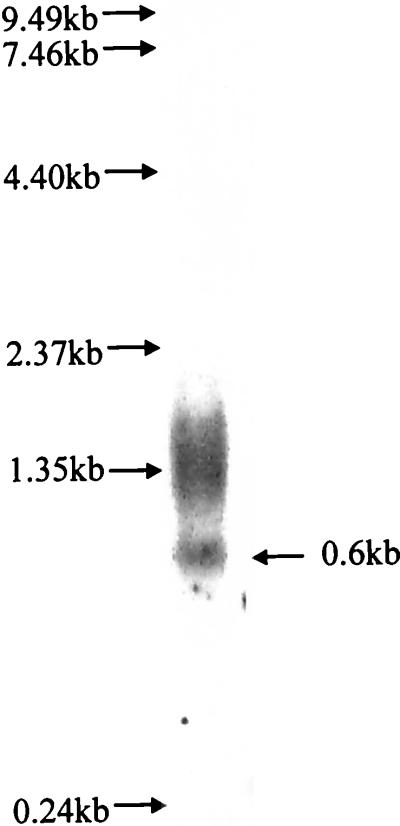
The inactivation of impA leads to a change in the protein profile of the outer membrane fraction (Fig. 5). These data suggest that impA is part of an operon and that the disruption of this gene may lead to polar effects of the downstream genes. To address this possibility, total RNA was isolated from the parent strain (VT1169) and used in Northern analysis with impA as the probe (Fig. 6). A 0.6-kb transcript was detected that corresponds to the size of the coding region of impA (612 bp). The hybridization signal in the region of the 1.35-kb marker corresponds to the ribosomal subunits of A. actinomycetemcomitans (D. Galli, personal communication).
FIG. 6.
Northern blot analysis of A. actinomycetemcomitans total RNA. Total RNA was isolated from A. actinomycetemcomitans VT1169, run on a 0.8% agarose gel with 8% formaldehyde, and transferred to nitrocellulose. The blot was probed with [α-32P]dCTP-labeled impA. The arrow indicates the 0.6-kb transcript.
Based on information obtained from the A. actinomycetemcomitans genome sequencing project being conducted at the University of Oklahoma, primers were designed for RT-PCR. A putative open reading frame transcribed in the same orientation as impA was indicated 500 bp downstream of the translational stop codon of impA. This protein shares homology with a putative protein of H. influenzae (HI0149). This primer was used as the RNA primer for the RT reaction using the SUPERSCRIPT One-Step RT-PCR System according to the manufacturer's instruction (Gibco-BRL). The resulting cDNA was used as the template in PCR with the above primer and a primer corresponding to the 5′ end of impA (nucleotides 1 to 20; Fig. 1). Analysis of this reaction did not result in any product (data not shown). The RT reaction was verified using the same RNA with the 3′ primer of impA as the RNA primer (nucleotides 729 to 741; Fig. 1). Analysis of the PCR product of the resulting cDNA with the 5′ and 3′ primers of impA resulted in the expected PCR product. Only one other open reading frame was found that was approximately 1 kb downstream of the translational stop codon of impA. However, this gene is transcribed in the opposite orientation of impA. This protein shares homology with DnaA, a replication initiator protein of E. coli (39). The primers used in the RT-PCR experiments were tested with A. actinomycetemcomitans DNA as the template and generated the expected PCR products (data not shown).
In addition, analysis of the impA sequence suggests the presence of a 7-bp inverted repeat (ΔG = −10.9) 7 bp from the stop codon TAG (Fig. 1). Collectively, the data indicate that impA is transcribed as a monocistronic mRNA. These data further suggest that the disruption of impA should not interfere with downstream gene expression and the phenotypes observed were associated with disruption of impA.
DISCUSSION
The molecular analysis of virulence factors of A. actinomycetemcomitans has been enhanced by the development of an efficient electroporation system and shuttle plasmids (2, 9, 19, 23, 24, 34, 35). This system is sufficient for the transformation of A. actinomycetemcomitans by replicating plasmids, but we found that the frequency for integration events mediated by homologous recombination is greatly reduced. An alternative approach to electrotransformation are conjugative methods which have been used to transfer broad-host-range group P and Q plasmids from E. coli to A. actinomycetemcomitans (10) and to transfer a Tn5 minitransposon into the Y4 strain of A. actinomycetemcomitans (16). However, conjugative transfer methods have not been used for the development of defined mutants by homologous recombination in A. actinomycetemcomitans.
The prototypic gene used in this study was identified to code for an exported protein in A. actinomycetemcomitans using translational gene fusions to alkaline phosphatase (22). The gene codes for a 22-kDa inner membrane protein (ImpA). Due to the lack of a suitable endonuclease restriction site, an engineered site was generated by inverse PCR to disrupt the gene by insertion of a spectinomycin cassette. The disrupted gene was cloned into the suicide vector plasmid pGP704 contained in E. coli SM10(λpir) for conjugation with a strain of A. actinomycetemcomitans developed to be resistant to both nalidixic acid and rifampin. The use of a double antibiotic selection was used to reduce the number of spontaneous mutations of the donor cells based on the high numbers of bacteria used in plating the conjugation mixture.
Transconjugants that were sensitive to ampicillin and resistant to spectinomycin, indicating the absence of the vector marker, were screened for the double-crossover event by PCR. Interestingly, only one of the transconjugants displaying this antibiotic profile contained a single copy of impA disrupted with the spectinomycin cassette. The remaining transconjugants contained two genes corresponding to an intact and a disrupted impA as determined by PCR. The presence of these transconjugants suggests that either the vector inserted into the chromosome by nonhomologous or illegitimate recombination that leads to inactivation of bla or by homologous recombination of the suicide vector. Based on our studies, transconjugants were not obtained after transfer of the suicide plasmid, pGP704, into A. actinomycetemcomitans VT1169. This is in contrast to side-by side conjugation experiments using the identical plasmid containing homologous A. actinomycetemcomitans DNA sequence which resulted in thousands of transconjugants (data not shown). In addition, Southern analysis of wild-type chromosomal DNA using the conjugation vector as the probe under high-stringency conditions did not generate any signal (data not shown). These data indicate that the suicide plasmid alone does not integrate into the chromosome when introduced into A. actinomycetemcomitans by conjugation.
Prior to this study, there has been only one report of directed mutagenesis in A. actinomycetemcomitans (17). The successful mutation in the leukotoxin gene was generated by electroporation of a nonreplicating plasmid containing a construct in which the disrupted gene was flanked by 1 kb of homologous DNA sequence. In the majority of transformants, the plasmid integrated into the genome via a single crossover event. Only one transformant appeared to be generated by a reciprocal double crossover. In the present study, only 0.35 kb of homologous DNA flanking sequence was present for directed mutagenesis and one transconjugate was obtained that was generated by a reciprocal double crossover. Based on studies in Streptococcus parasanguis by electroporation using a nonreplicating plasmid, the number of reciprocal double crossover transformants increased with increasing size of homologous flanking sequence in the target DNA (5). Therefore, increasing the amount of homologous flanking sequence in our construct should increase the number of transconjugants and, therefore, the number of reciprocal double crossover events. These studies are currently under way in our laboratory.
Allelic replacement of the wild-type impA with a disrupted gene leads to the deletion of a 22-kDa protein localized to the inner membrane. Concurrent with the deletion of impA was a change in the phenotype of the cells when grown in broth culture and on agar. Fresh clinical isolates of A. actinomycetemcomitans display a rough colony phenotype characterized by a transparent, dull, circular colony with irregular borders when grown on agar that express fimbriae (13, 26, 27, 30). The bacteria change to a smooth colony phenotype that has reduced or lack of fimbriation (a more opaque, glistening, circular colony with regular borders) following extensive subculturing (13). An intermediate colonial variant of A. actinomycetemcomitans has also been characterized that grows as transparent colonies but lack the fimbriae of the rough phenotype (13). The morphology of the mutant colonies grown on agar was more closely related to the smooth phenotype than to the rough morphology. However, the outer edges of the mutant colonies did appear to be more irregular than the edges of the parent or smooth phenotype.
The rough and intermediate phenotype grow in aggregates in broth that adhere to the walls of the tube (13). This is the same pattern of growth that was demonstrated for the impA mutant strain compared with the parent strain that grows as a turbid, homogeneous solution. Based on these data, the impA mutant strain appeared to display an intermediate phenotype compared to the smooth phenotype of the parent strain when grown in broth cultures. To determine if the phenotype is directly correlated with the inactivation of impA, complementation studies should be performed. Currently, complementation in A. actinomycetemcomitans has not been demonstrated. However, studies in this laboratory are under way to develop a complementation system for A. actinomycetemcomitans. Further studies will be required to determine if these pleiotropic effects are primary or secondary events of the mutation.
The changes in colony morphology suggested a change in the surface composition of the individual bacteria within the colony. These morphological changes maybe related to changes in the outer membrane proteins of the impA mutant bacteria. Based on SDS-PAGE analysis, several outer membrane proteins were either absent or reduced in staining intensity of the mutant strain compared to the parent strain. In contrast, a protein with a molecular mass of ∼44 kDa was present in the impA mutant profile that was not observed in the parent strain. This protein may correspond to the 43-kDa protein that is found associated with the rough phenotype of A. actinomycetemcomitans but is absent following the transition to the smooth variant (11). The changes in outer membrane proteins due to a deletion of an inner membrane protein suggest that ImpA may play a regulatory role or is directly involved in protein export.
The differences in the protein composition may be related to the increase in hydrophobicity observed for the impA mutant strain. Differences in cell surface hydrophobicity are found between laboratory strains and fresh isolates (12). Fresh isolates are more hydrophobic than laboratory strains. Taken together, the data suggest that the impA mutant strain may represent a transitional stage in the rough-to-smooth-phenotype conversion.
The inactivation of impA, presented here, is the first example of allelic replacement mutagenesis in A. actinomycetemcomitans achieved by conjugative transfer. The number of genes identified from A. actinomycetemcomitans will likely increase due to the sequencing of the entire genome. This genetic transfer system has significant potential for the elucidation of the function of gene products identified from the A. actinomycetemcomitans genome.
ACKNOWLEDGMENTS
We thank Hui Wu for his assistance in the Northern Blot analysis.
This work was supported by Public Health Service grant RO1-DE09760.
REFERENCES
- 1.Berghammer H, Auer B. “Easypreps”: Fast and easy plasmid minipreparation for the analysis of recombinant clones in E. coli. BioTechniques. 1993;14:527–528. [PubMed] [Google Scholar]
- 2.Brogan J M, Lally E T, Demuth D R. Construction of pYGK, an Actinobacillus actinomycetemcomitans-Escherichia coli shuttle vector. Gene. 1996;169:141–142. doi: 10.1016/0378-1119(95)00792-x. [DOI] [PubMed] [Google Scholar]
- 3.Carlone G M, Thomas M L, Rumschlag H S, Sottnek F O. Rapid microprocedure for isolating detergent-insoluble outer membrane proteins from Haemophilus species. J Clin Microbiol. 1986;24:330–332. doi: 10.1128/jcm.24.3.330-332.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fenno J C, Shaikh A, Fives-Taylor P. Characterization of allelic replacement in Streptococcus parasanguis: transformation and homologous recombination in a “nontransformable” streptococcus. Gene. 1993;130:81–90. doi: 10.1016/0378-1119(93)90349-8. [DOI] [PubMed] [Google Scholar]
- 6.Filip C, Fletcher G, Wulff J L, Earhart C F. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fine D H, Furgang D, Schreiner H C, Goncharoff P, Charlesworth J, Ghazwan G, Fitzgerald-Bocarsly P, Figurski D H. Phenotypic variation in Actinobacillus actinomycetemcomitans during laboratory growth: implications for virulence. Microbiology. 1999;145:1335–1347. doi: 10.1099/13500872-145-6-1335. [DOI] [PubMed] [Google Scholar]
- 8.Fives-Taylor P, Meyer D, Mintz K. Virulence factors of the periodontopathogen Actinobacillus actinomycetemcomitans. J Periodontol. 1996;67:291–297. doi: 10.1902/jop.1996.67.3s.291. [DOI] [PubMed] [Google Scholar]
- 9.Galli D M, Polan-Curtain J L, LeBlanc D J. Structural and segregational stability of various replicons in Actinobacillus actinomycetemcomitans. Plasmid. 1996;36:42–48. doi: 10.1006/plas.1996.0030. [DOI] [PubMed] [Google Scholar]
- 10.Goncharoff P, Yip J K, Wang H, Schreiner H C, Pai J A, Furgang D, Stevens R H, Figurski D H, Fine D H. Conjugal transfer of broad-host-range incompatibility group P and Q plasmids from Escherichia coli to Actinobacillus actinomycetemcomitans. Infect Immun. 1993;61:3544–3547. doi: 10.1128/iai.61.8.3544-3547.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haase E M, Zmuda J L, Scannapieco F E. Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect Immun. 1999;67:2901–2908. doi: 10.1128/iai.67.6.2901-2908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holm A, Kalfas S. Cell surface hydrophobicity and electrokinetic potential of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. Oral Microbiol Immunol. 1991;6:236–240. doi: 10.1111/j.1399-302x.1991.tb00483.x. [DOI] [PubMed] [Google Scholar]
- 13.Inouye T, Ohta H, Kokeguchi S, Fukui K, Kato K. Colonial variation and fimbriation of Actinobacillus actinomycetemcomitans. FEMS Microbiol Lett. 1990;57:13–17. doi: 10.1016/0378-1097(90)90405-f. [DOI] [PubMed] [Google Scholar]
- 14.Kinane D F, Mooney J, Ebersole J L. Humoral immune response to Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in periodontal disease. Periodontol 2000. 1999;20:289–340. doi: 10.1111/j.1600-0757.1999.tb00164.x. [DOI] [PubMed] [Google Scholar]
- 15.Knapp S, Mekalanos J J. Two trans-acting regulatory genes (vir and mod) control antigenic modulation in Bordetella pertussis. J Bacteriol. 1988;170:5059–5066. doi: 10.1128/jb.170.11.5059-5066.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolodrubetz D, Kraig E. Transposon Tn5 mutagenesis of Actinobacillus actinomycetemcomitans via conjugation. Oral Microbiol Immunol. 1994;9:290–296. doi: 10.1111/j.1399-302x.1994.tb00073.x. [DOI] [PubMed] [Google Scholar]
- 17.Kolodrubetz D, Phillips L H, Ezzo P J, Kraig E. Directed genomic integration in Actinobacillus actinomycetemcomitans: generation of defined leukotoxin-negative mutants. Infect Immun. 1995;63:2780–2784. doi: 10.1128/iai.63.7.2780-2784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.LeBlanc D J, Lee L N, Abu-Al-Jaibat A R, Sreenivasan P K, Fives-Taylor P M. Identification of plasmids in Actinobacillus actinomycetemcomitans and construction of intergeneric shuttle plasmids. Oral Microbiol Immunol. 1993;8:94–99. doi: 10.1111/j.1399-302x.1993.tb00552.x. [DOI] [PubMed] [Google Scholar]
- 20.LeBlanc D J, Lee L N, Inamine J M. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:1804–1810. doi: 10.1128/aac.35.9.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mintz K P, Fives-Taylor P M. Identification of an immunoglobulin Fc receptor of Actinobacillus actinomycetemcomitans. Infect Immun. 1994;62:4500–4505. doi: 10.1128/iai.62.10.4500-4505.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mintz K P, Fives-Taylor P M. Identification of genes coding for exported proteins of Actinobacillus actinomycetemcomitans. Infect Immun. 1999;67:6217–6220. doi: 10.1128/iai.67.11.6217-6220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakano Y, Yoshida Y, Yamashita Y, Koga T. Construction of a series of pACYC-derived plasmid vectors. Gene. 1995;162:157–158. doi: 10.1016/0378-1119(95)00320-6. [DOI] [PubMed] [Google Scholar]
- 24.Nakano Y, Yoshida Y, Yamashita Y, Koga T. New shuttle vectors for Actinobacillus actinomycetemcomitans and Escherichia coli. Gene. 1996;169:139–140. doi: 10.1016/0378-1119(95)00811-x. [DOI] [PubMed] [Google Scholar]
- 25.Ochman H, Ajoka J W, Garaza D, Hartl D L. Inverse polymerase chain reaction. In: Erlich H A, editor. PCR technology: principles and applications for DNA amplification. New York, N.Y: Stockton Press; 1989. pp. 105–111. [Google Scholar]
- 26.Preus H R, Namork E, Olsen I. Fimbriation of Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 1988;3:93–94. doi: 10.1111/j.1399-302x.1988.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 27.Rosan B, Slots J, Lamont R J, Listgarten M A, Nelson G M. Actinobacillus actinomycetemcomitans fimbriae. Oral Microbiol Immunol. 1988;3:58–63. doi: 10.1111/j.1399-302x.1988.tb00082.x. [DOI] [PubMed] [Google Scholar]
- 28.Rosenberg M, Gutnick D, Rosenberg E. Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiology Lett. 1980;9:29–33. [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Scannapieco F A, Millar S J, Reynolds H S, Zambon J J, Levine M J. Effect of anaerobiosis on the surface ultrastructure and surface proteins of Actinobacillus actinomycetemcomitans (Haemophilus actinomycetemcomitans) Infect Immun. 1987;55:2320–2323. doi: 10.1128/iai.55.9.2320-2323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 32.Slots J, Listgarten M A. Bacteroides gingivalis, Bacteroides intermedius and Actinobacillus actinomycetemcomitans in human periodontal diseases. J Clin Periodontol. 1988;15:85–93. doi: 10.1111/j.1600-051x.1988.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 33.Slots J, Reynolds H S, Genco R J. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect Immun. 1980;29:1013–1020. doi: 10.1128/iai.29.3.1013-1020.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sreenivasan P K, Fives-Taylor P. Isolation and characterization of deletion derivatives of pDL282, an Actinobacillus actinomycetemcomitans/Escherichia coli shuttle plasmid. Plasmid. 1994;31:207–214. doi: 10.1006/plas.1994.1022. [DOI] [PubMed] [Google Scholar]
- 35.Sreenivasan P K, LeBlanc D J, Lee L N, Fives-Taylor P. Transformation of Actinobacillus actinomycetemcomitans by electroporation, utilizing constructed shuttle plasmids. Infect Immun. 1991;59:4621–4627. doi: 10.1128/iai.59.12.4621-4627.1991. . (Erratum, 60:1728, 1992.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor R K, Manoil C, Mekalanos J J. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiater L A, Marra A, Shuman H A. Escherichia coli F plasmid transfers to and replicates within Legionella pneumophila: an alternative to using an RP4-based system for gene delivery. Plasmid. 1994;32:280–294. doi: 10.1006/plas.1994.1067. [DOI] [PubMed] [Google Scholar]
- 38.Wu H, Mintz K P, Ladha M, Fives-Taylor P M. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol Microbiol. 1998;28:487–500. doi: 10.1046/j.1365-2958.1998.00805.x. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto Y, Aiba H, Baba T, Hayashi K, Inada T, Isono K, Itoh T, Kimura S, Kitagawa M, Makino K, Miki T, Mitsuhashi N, Mizobuchi K, Mori H, Nakade S, Nakamura Y, Nashimoto H, Oshima T, Oyama S, Saito N, Sampei G, Satoh Y, Sivasundaram S, Tagami H, Horiuchi T, et al. Construction of a contiguous 874-kb sequence of the Escherichia coli K-12 genome corresponding to the 50.0–68.8 min on the linkage map and analysis of its sequence features. DNA Res. 1997;4(Suppl.):169–178. doi: 10.1093/dnares/4.2.169. [DOI] [PubMed] [Google Scholar]
- 40.Zadeh H H, Nichols F C, Miyasaki K T. The role of the cell-mediated immune response to Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in periodontitis. Periodontol 2000. 1999;20:239–288. doi: 10.1111/j.1600-0757.1999.tb00163.x. [DOI] [PubMed] [Google Scholar]
- 41.Zambon J J. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol. 1985;12:1–20. doi: 10.1111/j.1600-051x.1985.tb01348.x. [DOI] [PubMed] [Google Scholar]