Abstract
The M protein is the primary vaccine candidate to prevent group A streptococcal (GAS) infection and the subsequent development of rheumatic fever (RF). However, the large number of serotypes have made it difficult to design a vaccine against all strains. We have taken an approach of identifying amino-terminal M protein epitopes from GAS isolates that are highly prevalent in GAS-endemic populations within the Northern Territory (NT) of Australia. Australian Aboriginals in the NT experience the highest incidence of RF worldwide. To develop a vaccine for this population, 39 peptides were synthesized, representing the amino-terminal region of the M protein from endemic GAS. Mice immunized with these peptides covalently linked to tetanus toxoid and emulsified in complete Freund's adjuvant raised high-titer antibodies. Over half of these sera reduced bacterial colony counts by >80% against the homologous isolate of GAS. Seven of the peptide antisera also cross-reacted with at least three other heterologous peptides by enzyme-linked immunosorbent assay. Antiserum to one peptide, BSA101–28, could recognize six other peptides, and five of these peptides could inhibit opsonization mediated by BSA101–28 antiserum. Cross-opsonization studies showed that six of these sera could opsonize at least one heterologous isolate of GAS. These data reveal vaccine candidates specific to a GAS-endemic area and show the potential of some to cross-opsonize multiple isolates of GAS. This information will be critical when considering which epitopes may be useful in a multiepitope vaccine to prevent GAS infection.
Group A streptococci (GAS) are a major human pathogen responsible for suppurative and nonsuppurative pathology. The former group of conditions range from pharyngitis to the far more serious toxic shock-like syndrome and necrotizing fasciitis, whereas the latter include poststreptococcal glomerulonephritis and rheumatic fever (RF). RF and rheumatic heart disease (RHD) are responsible for 25 to 50% of cardiac conditions in children in developing countries (23). Australia's Aboriginal population experiences the highest rate of RF and RHD in the world. Aboriginal communities in the Northern Territory of Australia have RF incidence rates as high as 650 per 100,000, and the prevalence of RHD approaches 30 per 1,000 in some communities, compared to only 0.14 per 1,000 non-Aboriginals living in the same region (6). The median age for acquiring RF among Aboriginals is 11 years, and the life expectancy of Aboriginals who die of the disease is less than 35 years. Since RF and RHD only follow an infection with GAS, a strategy to prevent these conditions is to prevent GAS infections. The current approach of administering penicillin by injection has had limited success, as compliance with this regimen is low (approximately 50%), resulting in many recurrences in developing countries (6, 23). The best prospects for controlling RF rest with developing a vaccine to prevent streptococcal infection.
Immunity to GAS is mediated by antibodies to the M protein, which exists as a coiled-coil protein on the surface of the bacteria. The amino acid sequence of the amino terminus of the M protein is responsible for the serotype of the organism, with at least 80 distinct serotypes having been defined (19). Antibodies directed to the M protein opsonize streptococci in the presence of neutrophils; however, these antibodies are serotype specific and generally only opsonize the homologous GAS isolate (1, 2, 10, 13, 14, 21).
Potential problems exist when immunizing with subunits of the M protein, as accumulating evidence suggests that RF is likely to be an autoimmune disease, although the pathogenesis is not clear. It may be that an immune response directed to GAS proteins can also react with host tissues, including the heart (12, 16, 17, 24, 29). Therefore, care must he taken when designing vaccines for GAS. The safest approach is to use sequences which do not evoke host cross-reactive antibody or T-cell responses. Previous studies have focused on defining opsonic epitopes from the amino-terminal region, and most of these epitopes do not evoke cross-reactive antibody to human host tissues (1, 9, 10, 12, 13, 14).
Epidemiological studies of endemic GAS isolates suggest that some serotypes are much more common than others within a population, and these vary between distinct geographic locations (15, 22, 25, 27, 28, 30). Although amino-terminal M protein epitopes from common reference strains of GAS have been identified, protective epitopes from GAS strains prevalent within a GAS-endemic region have yet to be investigated. In the current study, we synthesized peptides to the amino-terminal region of the M protein from GAS isolated from Northern Territory Aboriginal communities. We show that mice immunized with these peptides covalently linked to tetanus toxoid (TT) and emulsified in complete Freund's adjuvant (CFA) raise high-titer antibodies to the immunizing peptide. We define numerous opsonic epitopes from the M proteins of endemic GAS isolates and determine the degree to which these induce antibodies that cross-opsonize heterologous GAS strains. This study lays the foundation for rationally designing a polyvalent immunogen which will prevent GAS infection in targeted communities.
MATERIALS AND METHODS
Syntheses of peptides and conjugation to TT.
Synthetic peptides were produced as described and were purified by high-pressure liquid chromatography (18). All peptides were conjugated via the C-terminal cysteine residue to TT using 6′-maleimido-caproyl n-hydroxy succinimide (MCS) coupling (7). Peptide sequences are given in Table 1.
TABLE 1.
List of synthetic peptides, corresponding GAS isolates, M and ST type, OF, and murine serum antibody titer to immunizing peptidea
| Peptide | Sequence | Gas isolate | Originb | M/STc | OF | Titer to immunizing peptided |
|---|---|---|---|---|---|---|
| ML2 family | ||||||
| 20321–19 | NSKNPVPVKKEAKLSEAELC | 2032 | T | M2 | + | 204,800 |
| 203246–59 | KVEEEHKKDHEKLEC | 2032 | T | M2 | + | 204,800 |
| BSA101–28 | NSKTPAPAPAVPVKKEATKSKLSEAELHC | BSA10 | NTS | STBSA10 | + | 204,800 |
| BSA101–19 | NSKTPAPAPAVPVKKEATKC | BSA10 | NTS | STBSA10 | + | 204,800 |
| NS141–19 | DNPSSVPVKKAAELYDKIKC | NS14 | NTS | STNS14 | + | 6,400 |
| ML60 family | ||||||
| 20801–20 | ESSTVKAESSTVKAESSTISC | 2080 | T | M60 | + | 3,200 |
| 20241–20 | AEIKKPQADSAWNWPKEYNAC | 2034 | T | M4 | + | 12,800 |
| 203440–59 | DDKEKDPQYRALMGENQDLRC | 2034 | T | M4 | + | 102,400 |
| 25511–20 | ESSNNAESSNISQESKLINTC | 2551 | T | M22 | + | 12,800 |
| Y530S1–20 | EAPKSHSISNNEQLINELNDC | Y530S | NTS | M8 | + | 25,600 |
| M5 family | ||||||
| 20351–20 | AVTRGTINDPQRAKEALDKYC | 2035 | T | M5 | − | 12,800 |
| 20361–20 | RVFPRGTVENPDKARELLNKC | 2036 | T | M6 | − | 12,800 |
| 20451–20 | PLTRATADNKDELIKRANDYC | 2045 | T | M18 | − | 12,800 |
| 20541–20 | TIITRDIARDPDKLRKIAENC | 2054 | T | M30 | − | 12,800 |
| NS11–19 | RVTTRSQAQDAAGLKEKADC | NS1 | NTB | STNS1 | − | 25,600 |
| TIN family | ||||||
| Y504S1–20 | TEVKAAGQSAPKGTNVSADLC | Y504S | NTS | M11 | + | 204,800 |
| Y504S45–64 | EETKNKELDKKNKELDSRVTC | Y504S | NTS | M11 | + | 204,800 |
| 90/851–20 | DGERVPKNNRLSKKYSELSEC | 90/85 | NTS | ST90/85 | − | 204,800 |
| 88/5441–20 | DGPQKSVSQGGVSLDLYSKLC | 89/544 | NTS | ST89/544 | − | 204,800 |
| 20411–20 | ADPQKQNSVSNDSVTINVYNC | 2041 | T | M13 | + | 12,800 |
| M52/53/80 family | ||||||
| 411–19 | EGNARLAQAQEEALRDVLNC | 41 | T | M41 | − | 51,200 |
| 20721–20 | DQPVDHHRYTEANDAVLQGRC | 2072 | T | M52 | − | 3,200 |
| NS271–19 | ADDHPGAVAARNDVLSGFSC | NS27 | NTS | STNS27 | − | 25,600 |
| NS51–19 | ADHPSYTAAKDEVLSHFSVC | NS5 | NTB | STNS5 | − | 25,600 |
| 1561–19 | DNPRYTDAHNAVTQGRTVPC | 87/156 | NZ | ST156 | − | 25,600 |
| 88/251–32 | DRYTDAHNAVTQGRTVPLRNLLLEMDKNSKLRC | 88/25 | NTS | ST88/25 | − | 12,800 |
| 88/2533–53 | SENELLQAGLQEKERELEDLKC | 88/25 | NTS | ST88/25 | − | 12,800 |
| 23171–19 | HQLADAARREVLKGETVPAC | 2317 | T | M80 | − | 2,631,680 |
| M12/M55-related sequence | ||||||
| 20331–20 | DARSVNGEFPRHVKLKNEIEC | 2033 | T | M3 | − | 51,200 |
| 20401–12 | DHSDLVAEKQRLC | 2040 | T | M12 | − | 25,600 |
| 204050–69 | LKMLNRDLEQAYNELSGEAHC | 2040 | T | M12 | − | 102,400 |
| 2040239–258 | AALGHQHAHNEYQAKLAEKDC | 2040 | T | M12 | − | 102,400 |
| 20751–19 | NQTEPSQTHNRLYQERQRLC | 2075 | T | M55 | − | 12,800 |
| Unrelated sequences | ||||||
| 20691–20 | AEKKVEAKVEVAENNVSSVAC | 2069 | T | M49 | + | 12,800 |
| 20771–19 | NDDITSMTPILSGVGPGNAC | 2077 | T | M57 | − | 12,800 |
| P735541–20 | EEERTFTELPYEARYKAWKSC | P73554 | NZ | M75 | + | 51,200 |
| 88/301–20 | DNGKAIYERARERALQELGPC | 88/30 | NTS | ST88/30 | − | 12,800 |
| STBSB241–20 | DEGPKDITDSLPAPMWRDKAC | BSB24 | NTS | STBSB24 | + | 204,800 |
| PL11–20 | DNPSSVPKKAAELYDKIKC | PL1 | NTS | M54 | − | 2,621,450 |
Sequences are grouped into M families based on dendrogram analysis of amino-terminal nucleotide and amino acid sequence relatedness (25).
T, reference type strain, NZ, isolate from New Zealand derived from blood culture; NTS, Northern Territory isolate derived from skin swab; NTB, Northern Territory isolate derived from blood culture.
M type determined by serotyping; sequence type (ST) determined through sequence analysis of the amino terminus of the M proteins of reference strains and M-nontypeable proteins (25).
Antibody titer of pooled murine sera to immunizing peptide following primary immunization and three booster injections of the conjugate peptide.
Immunization of animals.
Peptides were administered subcutaneously at the tail base to B10.BR (H-2k) mice (Animal Resources Centre WA). Each of three mice received 30 μg of peptide emulsified in CFA (Difco Laboratories, Detroit, Mich.) in a total volume of 50 μl. Mice were boosted after 21 days with 30 μg of peptide in phosphate-buffered saline (PBS), and two subsequent booster injections were given at 10-day intervals. Mice were bled via the tail artery at 2, 4, 6, and 8 weeks following primary immunization. If, after three booster injections, individual mice had titers of <12,800, mice were boosted with a further 30 μg of peptide in PBS prior to using sera in the bactericidal assay.
Detection of antibodies.
An enzyme-linked immunosorbent-assay (ELISA) was used to measure murine serum antibodies to the peptides as described (17, 26). Titer is defined as the lowest dilution that gave an optical density (OD) reading of more than 3 standard deviations above the mean OD of the control wells containing normal mouse serum. Western blot analysis of antipeptide cross-reactivity against cardiac myosin (Sigma), tropomyosin (Sigma), keratin (Sigma), and murine whole heart extract (26) was performed as described (26).
GAS reference strains and isolates.
GAS isolates and reference strains were obtained from the Menzies School of Health Research, Darwin, Northern Territory, Australia, and Diana Martin, Institute of Environmental Science and Research, Wellington, New Zealand. Details for each strain are given in Table 1. Full designations of reference strains are as follows (reference name in parentheses): 2032 (M2B), 2033 (B930/24 PHLS), 2034 (SS241), 2035 (T5/B/PS PHLS), 2036 (S43/75/8/W14), 2040 (R53/1077 PHLS), 2041 (SS31), 2045 (PHL J17C), 2054 (D24/46 PHLS), 2069 (B737/71/1), 2072 (10870), 2075 (Trinidad A-75), 2077 (3890-V-Ramkisson), 2080 (D335 Lfd), 2317 (R75/268), and 2551 (68/3116). Isolates from the Northern Territory were derived from blood culture or skin swabs from Aboriginal patients. All isolates were subjected to serotyping (M, T, and opacity factor [OF]) using standard methods (Table 1) (27, 28).
Bactericidal assay.
Murine antipeptide sera were assayed for their ability to opsonize GAS essentially as described (21). Briefly, the bacteria were grown overnight at 37°C in 5 ml of Todd-Hewitt broth (THB), and then 200 μl of overnight culture was subinoculated into 5 ml of warm (37°C) THB and allowed to grow for 2 h at 37°C. GAS were then serially diluted to 10−4 in saline. Fifty microliters of the bacteria dilution was mixed with 50 μl of fresh heat-inactivated serum and 400 μl of nonopsonic heparinized donor blood, as a source of neutrophil and complement. All donor blood was screened prior to assay to ensure that it could support the growth of the GAS strain to at least 32 times the inoculum level in a 3-h incubation at 37°C (4, 5). The mixture was incubated end over end at 37°C for 3 h, 50 μl from each tube was plated out, in duplicate, on THB blood agar pour plates and incubated overnight, and the number of colonies on each plate was counted. Opsonic activity of the antipeptide sera (percent kill) was calculated as [(1 − mean CFU in the presence of antipeptide sera)/(mean CFU in the presence of normal mouse sera)] × 100. Peptide inhibition opsonization assays were performed as described (4, 5) using log-phase bacteria.
RESULTS
Synthetic peptides from the N terminus of M proteins of endemic GAS.
The amino-terminal peptide sequences and sub-amino-terminal peptide sequences from the A, B, and C repeat regions (not containing the amino terminus of the protein) were derived from DNA sequence analysis of streptococcal reference strains and clinical isolates collected from the Northern Territory of Australia and New Zealand (15, 27, 28). Table 1 shows the amino acid sequence of each of the 39 synthetic peptide sequences with their corresponding GAS strain, M type, and OF type. Amino-terminal peptide sequences were derived from sequence typing (ST), a PCR-based sequencing method, of M proteins and M-like (ML) proteins (highly homologous to M protein) of reference strains and Australian and New Zealand GAS isolates (27, 28). Based on dendogram analysis, sequences have been grouped into five ML protein families according to their nucleotide and protein sequence relatedness; ML2, ML60, M5, TIN, M52/53/80, an M12/55-related sequence group, and an unrelated sequence group (Table 1) (27). Within each ML family, sub-amino-terminal sequences have higher homology between family isolates than amino-terminal sequences (Table 2). Therefore, we also synthesized six sub-amino-terminal peptides to determine whether opsonic epitopes exist within these common domains.
TABLE 2.
Comparison and homology of sub-amino-terminal sequences within ML family isolates
| Peptidea | Sequenceb | % Similarity | Location in M protein |
|---|---|---|---|
| ML2 family | |||
| 203246–59 | KVEEEHKKDHEKLE | B repeat | |
| BSA10(54–67) | -------------- | 100 | |
| NS14(45–58) | --K------K---- | 93 | |
| ML60 family | |||
| 203440–59 | DDKEKDPQYRALMGENQDLR | Nonrepetitive region | |
| 2080(56–75) | -NIR---------------M | 85 | |
| Y530S(41–60) | -NTR---------------M | 85 | |
| TIN family | |||
| Y504S45–64 | EETKNKELDKKNKELDSRVT | Follows A repeat | |
| 90/85(39–58) | LLD-QE--E-E-EK---Q-A | 45 | |
| 88/544(41–60) | LTE--E-------K---Q-D | 65 | |
| M52/53/80 family | |||
| 88/2533–53 | SENEELQAGLQEKERELEDL | A1 repeat | |
| 2072(39–58) | ------K-D--K--Q--KN- | 70 | |
| NS27(37–56) | --K---ETE-----Q--KN- | 65 | |
| NS5(36–55) | ----G-K-------Q--KN- | 75 | |
| 87/156(35–54) | ------K-D--K--Q--KN- | 70 | |
| 2317(34–53) | -A----ETT--K--Q--KG- | 60 | |
| M41(35–54) | RR-N--EKII--------G- | 60 | |
| M12/M55 family | |||
| 204050–69 | LKMLNRDLEQAYNELSGEAH | B1 repeat | |
| 2075(58–77) | --E--DEF---------DGV | 85 | |
| 2033(56–75) | --G--DWA-RLLQ--N--DV | 45 | |
| 2040239–258 | AALGHQHAHNEYQAKLAEKD | ||
| 2033(268–287) | -------------------- | 100 |
Subscripts in parentheses are the amino acid positions of each sequence corresponding to the peptide. Adapted from Relf (27).
Identical residues are shown as dashes.
Definition of opsonic amino-terminal epitopes.
Mice (three per group) were immunized with the peptide-TT conjugates listed in Table 1. An ELISA was used to detect the level of antibody in the pooled murine sera to the homologous peptide (unconjugated) on the plate. Pooled murine sera had high-titer antibodies (>3,200) against homologous peptides following one primary immunization and three boosts (Materials and Methods) (Table 1). In all cases, immunoglobulin G2b (IgG2b) was the predominant antibody isotype produced. As negative controls, sera from mice immunized with PBS or TT emulsified in CFA were tested by ELISA against each of the peptides. There was no significant recognition (titer of <100) by these sera of any of the amino-terminal peptides listed in Table 1.
Each of the pooled antipeptide sera with a titer greater than >12,800 to the immunizing peptide was examined by Western blot to determine whether the antibodies cross-reacted with cardiac myosin, tropomyosin, keratin, or whole murine heart extract. None of the sera investigated recognized any of the proteins investigated (data not shown).
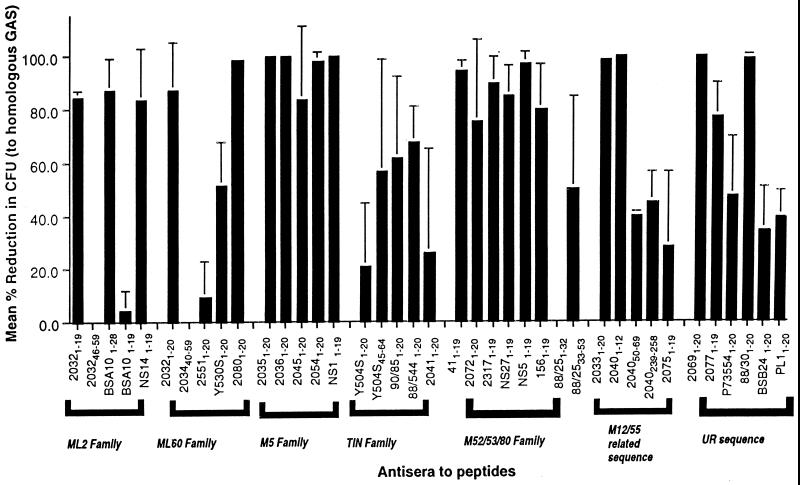
Sera with high-titer antibodies raised to each of the peptides were then tested for their ability to opsonize the representative GAS reference strains and endemic isolates (listed in Table 1) in an in vitro bactericidal assay (Fig. 1). For the 33 individual amino-terminal peptides, 19 (58%) peptide-specific antisera demonstrated greater than 80% mean reduction in colony counts against the homologous GAS isolate. An opsonic activity of 60 to 80% was observed with four (12%) peptide antisera; two (6%) had between 40 and 60% opsonic activity; and eight (24%) had less than 40% opsonic activity against the homologous GAS isolate. Antisera to all six sub-N-terminal peptides were also tested for their ability to opsonize the corresponding GAS isolate: four antisera (Y504S45–64, 88/2533–53, 204050–69, and 2040239–258) had an opsonic activity of between 40 and 60%, and two peptide antisera (203242–59 and 203440–49) had no opsonic activity at all against the homologous GAS isolate (Fig. 1).
FIG. 1.
Opsonization of GAS by antisera to amino-terminal peptides derived from the M protein of reference and clinical isolates. Peptide antisera are grouped by their sequence type into M families. Results are given as the mean percent kill (of at least two separate experiments) against the homologous GAS isolate. UR, unrelated sequences.
Epitope specificity.
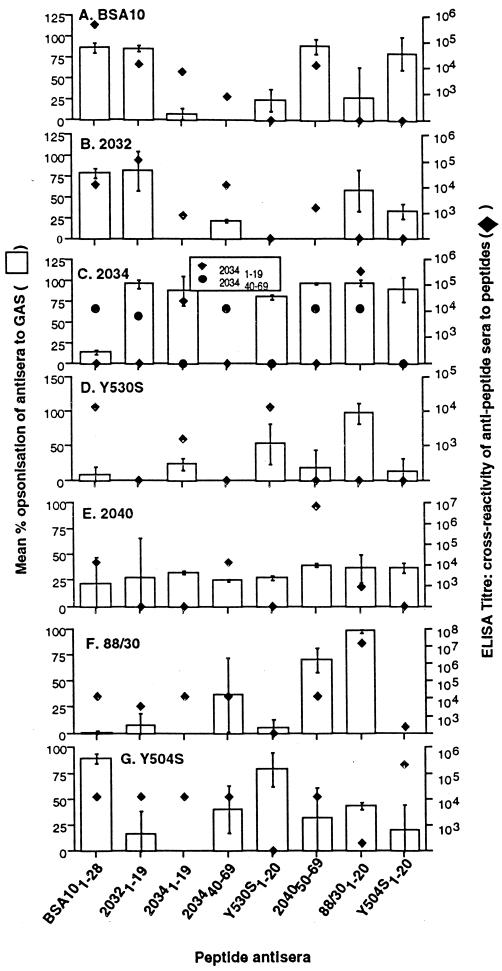
To determine the specificity of the murine amino-terminal and sub-amino-terminal peptide antisera, pooled antipeptide sera with a titer of >12,800 to the immunizing peptide were tested in an ELISA for binding to homologous and heterologous peptides. The majority of the antisera bound only the homologous peptide. However, seven antisera to amino-terminal and sub-amino-terminal peptides, BSA101–28, 20321–19, 20341–19, 203440–69, Y504S45–60, 204050–69, and 88/301–20, cross-reacted (defined as a titer of ≥1,600) with at least three other heterologous peptides in an ELISA (Fig. 2). Antisera to BSA101–28 and 20321–19 cross-reacted with six and four heterologous peptides, respectively (Fig. 2). Peptide antisera to 203246–59, NS141–19, NS11–19, 90/951–20, 88/5441–20, 20401–12, and 20331–19 cross-reacted with at least one other heterologous peptide (data not shown).
FIG. 2.
Antibody and bactericidal cross-reactivity of murine antisera to amino-terminal peptides. Columns represent mean percent reduction in CFU from at least two separate experiments (left vertical axis). Symbols represent mean antibody titer from pooled murine sera to amino-terminal peptides (right vertical axis). (A) GAS isolate BSA10, peptide BSA101–28 (peptide on ELISA plate). (B) 2032 GAS, peptide 20321–19. (C) 2034 GAS, peptides 20341–19 and 203440–69. (D) Y530S GAS, peptide Y530S1–20. (E) 2040 GAS, peptide 204050–59. (F) 88/30 GAS, peptide 88/301–20. (G) Y504S GAS, peptide Y504S1–20.
The epitope specificity of antisera to BSA101–28 and 20321–19 was further evaluated by comparing the ability of cross-reactive and non-cross-reactive peptides to inhibit opsonization against GAS isolates BSA10 and 2032, respectively. Antisera to BSA101–28 and 20321–19 showed bactericidal activity against their respective GAS isolates. Peptides BSA101–28, 20321–19, 20341–19, 88/301–20, and Y504S1–20, which cross-react to BSA101–28 antisera by ELISA, were added to BSA101–28 antiserum prior to addition of nonopsonic donor blood and BSA10 GAS. Bactericidal activity of the BSA10 antiserum was inhibited significantly by peptides BSA101–28, 20321–19, 88/301–20, and Y504S1–20 by between and 78 and 84% and by 43% for peptide 20341–19 (Table 3). Antiserum to peptide 20321–19 also shows ELISA cross-reactivity to peptides BSA101–28, 20321–19, 20341–19, 88/301–19, and Y504S1–20 but not peptide Y530S1–20. Following addition of these peptides to 20321–19 antiserum in a peptide inhibition opsonization assay against GAS strain 2032, all cross-reactive peptides could inhibit bactericidal activity, whereas in the presence of the non-cross-reactive peptide Y530S1–20, the 20321–19 antiserum could still significantly opsonize 2032 GAS (Table 3).
TABLE 3.
Inhibition of opsonic activity by cross-reactive peptides
| Antiserum | GAS isolate (inoculum size) | Inhibiting peptide | Mean CFUa | % Inhibitionb | ELISA cross-reactivity to peptidec |
|---|---|---|---|---|---|
| BSA101–28 | BSA10 (87) | None | 530 | ||
| BSA101–28 | 3,100 | 83 | + | ||
| 20321–19 | 3,300 | 84 | + | ||
| 20341–19 | 970 | 45 | + | ||
| 88/301–20 | 2,460 | 78 | + | ||
| Y5041–20 | 2,640 | 80 | + | ||
| 20321–19 | 2032 (101) | None | 280 | ||
| BSA101–28 | 5,300 | 95 | + | ||
| 20321–19 | 5,000 | 94 | + | ||
| 20341–19 | 4,000 | 93 | + | ||
| Y530S1–20 | 180 | NI | − | ||
| 88/301–20 | 2,000 | 86 | + | ||
| Y504S1–20 | 5,600 | 95 | + |
Mean colony count (CFU) from two plates times the dilution factor.
Percent inhibition was calculated as described in the text. NI, no inhibition. In controls containing blood and bacteria (no antiserum, no peptide), the BSA10 titer was 2,800 and the 2032 titer was 3,650.
+, ELISA cross-reactive titer to inhibiting peptide >1,600; −, titer 800. Data derived from Fig. 2.
Bactericidal cross-reactivity of the amino-terminal peptide antisera.
We have shown that antiserum raised to ML2 family peptide BSA101–28 reacted with itself and six other heterologous peptides, and antisera raised to three of the six peptides that cross-react with BSA101–28 antiserum (20321–19, 20341–19, and 204050–69) also recognized peptide BSA101–28. To determine whether these antisera could also cross-opsonize the representative heterologous GAS isolates, opsonization assays were performed using antisera to peptides BSA101–28, 20321–19, 20341–19, 203440–69, Y530S1–20, 204050–69, 88/301–20, and Y504S1–20 against the homologous (amino-terminal representative GAS) and heterologous isolates represented by the peptides listed above.
Antisera to both BSA101–28 and 20321–19 could clearly opsonize both corresponding ML2 family GAS isolates (Fig. 2A and B). This result is not surprising, as the peptide sequence of 20321–19 has 90% sequence homology to BSA101–28 (Table 4). The only other isolate that was repeatedly opsonized by BSA101–28 antiserum was Y504S (Fig. 2G). Interestingly, antiserum to peptide Y504S1–20 could opsonize isolate BSA10 even though the cross-reactive ELISA titer to peptide BSA101–28 was only 100 (Fig. 2A), suggesting that other cross-reactive epitopes may exist on BSA10. In addition, antiserum to 204050–69 also cross-opsonized GAS isolate BSA10 (Fig. 2A).
TABLE 4.
Amino acid homology between ELISA-cross-reactive amino-terminal peptides
| Peptide | % similaritya with peptide:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| BSA101–28 | 20321–19 | 20341–19 | 203440–59 | Y530S1–20 | 204050–69 | 88/301–20 | Y504S1–20 | |
| BSA101–28 | 100 | |||||||
| 20321–19 | 90 | 100 | ||||||
| 20341–19 | 36 | 42.8 | 100 | |||||
| 203440–59 | 35 | 40 | 22 | 100 | ||||
| Y530S1–20 | 24 | 30 | 21 | 34 | 100 | |||
| 204050–69 | 38 | 38 | 47 | 50 | 42 | 100 | ||
| 88/301–20 | 33 | 45 | 58 | 40 | 32 | 44 | 100 | |
| Y504S1–20 | 18 | 40 | 75 | 41 | 34 | 43 | 43 | 100 |
Calculated using the GCG Pileup program.
Although antiserum to 204050–69 was only moderately opsonic (40%) against the representative isolate, 2040, it was able to cross-opsonize three other isolates, BSA10 (89.3%), 2034 (96.8%), and 88/30 (70%) (Fig. 2 A, C, and F, respectively), which correlated to ELISA cross-reactivity. Antiserum to 88/301–20 could also cross-opsonize two other isolates, 2034 (97.1%), which correlated with ELISA cross-reactivity, and Y530S (97.4%), which did not correlate with ELISA cross-reactivity (Fig. 2C and D, respectively). Similarly, antiserum to peptide Y530S1–20 reduced mean colony counts of two nonhomologous isolates of GAS, Y504S (79.2%) and 2034 (80%) (Fig. 2C and G, respectively), even though no ELISA cross-reaction was previously observed. Antisera to peptides 20341–19 and 203440–59 only poorly cross-opsonized heterologous isolates of GAS.
DISCUSSION
This study is the first to take the approach of investigating and defining opsonic epitopes from a specific region where GAS are endemic. We have synthesized amino-terminal and sub-amino-terminal peptides from M protein of clinical GAS isolates collected from Australian Aboriginal communities and RF outbreaks in the Northern Territory and reference strains. All 33 amino-terminal peptides and six sub-amino-terminal peptides were highly immunogenic in mice when conjugated to TT, and over half of the amino-terminus-specific antibodies were highly opsonic to the homologous GAS strain. Antisera to the sub-amino-terminal peptides did not significantly opsonize the homologous strains of GAS (less than 50% kill). These findings are in agreement with previous studies that show that antibodies to sub-amino-terminal domains generally do not effectively opsonize GAS (1, 13).
We also identified several antipeptide sera that could opsonize not only the homologous strain of GAS, but heterologous strains as well. Although other studies have identified antipeptide sera that cross-react with heterologous serotypes of M protein by ELISA, none of these sera cross-opsonized the related heterologous GAS (1). We were able to show that opsonization mediated by antisera to BSA101–28 and 20321–19 (against BSA10 and 2032 GAS, respectively) could be inhibited by those peptides that were cross-reactive by ELISA. Antisera to BSA101–28 and 20321–19 could each directly opsonize two GAS isolates represented by the ELISA-cross-reactive peptides. In addition, antisera to peptides Y530S1–20, 204050–69, 88/301–20, and Y504S1–20 could opsonize at least one nonhomologous isolate. In some cases where there was cross-opsonization between strains, no ELISA cross-reactivity was observed between amino-terminal peptide antisera and the nonhomologous amino-terminal peptide. It may be that other regions within the M protein, or indeed other surface antigens of GAS, cross-react and are opsonized by antibodies to these sequences.
With the exception of BSA101–28 and 20321–19, which have 90% similarity and are both in the ML2 family, very little sequence homology between the other amino-terminal peptides existed (Table 4). Cross-opsonization by the peptide antisera may in part reflect structural similarities of the peptides. The predicted secondary structures of peptides BSA101–28, 20321–19, 204050–69, 88/301–20, and 203440–59 are predominantly alpha-helical (Table 5). Investigation of the helical propensity of BSA101–28 using circular dichroism indicated that this peptide has high alpha-helix potential, with 62 to 81% alpha helix in 100% trifluoroethanol and 40 to 63% alpha helix in 50% trifluoroethanol (data not shown). Therefore, the cross-reactive antipeptide sera may recognize a common structure produced by the alpha-helical coiled folding of the primary sequence, either as a peptide on an ELISA plate or on the M or ML proteins of the streptococci. Several studies have in fact reported that antipeptide antibodies from the M protein can cross-react with several alpha-helical proteins, including laminin, myosin, and tropomyosin (8, 9, 31). It is thus conceivable that the cross-reactive antipeptide antibodies can recognize and cross-opsonize other nonhomologous alpha-helical M or ML proteins.
TABLE 5.
Secondary-structure characteristics of the ELISA-cross-reactive amino-terminal peptidesa
| Peptide | Peptide sequence and predicted secondary structure |
|---|---|
| BSA101–28 | NSKTPAPAPAVPVKKEATKSKLSEAELH |
| ttcccceehhhhhhhhhhhhhhhhhhhh | |
| 20321–19 | NSKNPVPVKKEAKLSEAEL |
| ttcccehhhhhhhhhhhhh | |
| 20341–19 | AEIKKPQADSAWNWPKEYNA |
| hhhhhhhhhcttcccttttt | |
| 20344–59 | DDKEKDPQYRALMGENQDLR |
| hhhhhhhhhheeehhhhhhh | |
| Y530S1–20 | EAPKSHSISNNEQLINELND |
| hhhhhccccccheeeehhtt | |
| 204050–69 | LKMLNRDLEQAYNELSGEAH |
| hhhhhhhhhhhhhhhhhhhh | |
| 88/301–20 | DNGKAIYERARERALQELGP |
| hhhhhhhhhhhhhhhhttct | |
| Y504S1–20 | TEVKAAGQSAPKGTNVSADL |
| hhhhhhhhecctteeeeeee |
Secondary-structure analysis was determined by Garnier secondary-structure prediction. h, helix; t, beta turn; c, random coil; e, extended strand.
The advantages of using a vaccine incorporating peptides representing amino-terminal sequences of the M protein is that they are more opsonic than C-terminal antisera (26) and less likely to evoke tissue-cross-reactive antibodies (10, 12, 14). None of the peptide-specific antibodies cross-reacted with host proteins, suggesting that a vaccine using these epitopes may not induce antibody-mediated pathology. In terms of vaccine development, we have now identified numerous opsonic epitopes specifically targeted to GAS-endemic region. In order to protect against the numerous strains of GAS, a vaccine would need to incorporate multiple protective epitopes. Several studies have shown the potential of constructing multivalent streptococcal vaccines capable of opsonizing multiple serotypes of GAS (1, 2, 10, 11, 14). Thus far, up to eight amino-terminal epitopes have been linked in tandem to produce a hybrid protein that can produce bactericidal antibodies in immunized rabbits (10, 11, 14). To develop a successful antistreptococcal vaccine, it is critical to define epitopes that would induce broad protective coverage. The frequency of GAS isolates collected from the Northern Territory of Australia, represented by the amino-terminal M protein peptides used in this study, has recently been investigated (15; unpublished data). Based on these studies, it may be possible to design a vaccine that will incorporate opsonic amino-terminal epitopes from GAS commonly found in this region in combination with cross-opsonic epitopes, including conserved-region epitopes (3, 4, 5, 24, 26), to produce coverage against the significant diversity of GAS strains found within the Aboriginal communities of the Northern Territory.
ACKNOWLEDGMENTS
This work was supported by NHMRC (Australia), National Heart Foundation of Australia, the Prince Charles Hospital Foundation, and Cooperative Research Centre for Vaccine Technology.
We thank Laszlo Otvos, the Wistar Institute, Philadelphia, Pa., for performing the circular dichroism analysis.
REFERENCES
- 1.Beachey E H, Seyer J M. Protective and non-protective epitopes of chemically synthesised peptides of the NH2-terminal region of type 6 streptococcal M protein. J Immunol. 1986;136:2287–2292. [PubMed] [Google Scholar]
- 2.Beachey E H, Gras-Masse H, Tarter A, Jolivet M, Audibert F, Chedid L, Seyer J M. Opsonic antibodies evoked by hybrid peptides copies of types 5 and 24 streptococcal M proteins synthesise in tandem. J Exp Med. 1986;163:1451–1458. doi: 10.1084/jem.163.6.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessen D, and Fischetti V A. Influence of intranasal immunization with synthetic peptides corresponding to conserved epitopes of M protein on mucosal colonization by group A streptococci. Infect Immun. 1988;56:2666–2672. doi: 10.1128/iai.56.10.2666-2672.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandt E R, Hayman W A, Currie B, Carapetis J, Wood Y, Jackson D, Cooper J, Melrose W D, Saul A J, Good M F. Opsonic human antibodies from an endemic population specific for a conserved epitope on the M protein of group A streptococci. Immunology. 1996;89:331–337. doi: 10.1046/j.1365-2567.1996.d01-754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandt E R, Hayman W A, Currie B, Pruksakorn S, Good M F. Human antibodies to the conserved region of the M protein: opsonisation of heterologous strains of group A streptococci. Vaccine. 1997;16:1805–1812. doi: 10.1016/s0264-410x(97)00178-3. [DOI] [PubMed] [Google Scholar]
- 6.Carapetis J R, Wolff D R, Currie B J. Acute rheumatic fever and rheumatic heart disease in the top end of Australia's Northern Territory. Med J Aust. 1996;164:146–149. doi: 10.5694/j.1326-5377.1996.tb122012.x. [DOI] [PubMed] [Google Scholar]
- 7.Coligan J E, Kruisbeek A M, Margulie D H, Shevach E M, Strober W. Current protocols in immunology. Vol. 2. New York, N.Y: John Wiley; 1992. [Google Scholar]
- 8.Cunningham M W, Antone S M, Smart M, Liu R, Kosanke S. Molecular analysis of human cardiac myosin-cross-reactive B- and T-cell epitopes of the group A streptococcal M protein. Infect Immun. 1997;65:3913–3923. doi: 10.1128/iai.65.9.3913-3923.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cunningham M W, McCormack J M, Fenderson P G, Ho M, Beachey E H, Dale J B. Human and murine antibodies cross-reactive with streptococcal M protein and myosin recognise the sequence GLN-LYS-SER-LYS-GLN in M protein. J Immunol. 1989;143:2677–2683. [PubMed] [Google Scholar]
- 10.Dale J B, Chiang E Y, Lederer J W. Recombinant tetravalent group A streptococcal M protein vaccine. J Immunol. 1993;151:2188–2194. [PubMed] [Google Scholar]
- 11.Dale J B. Multivalent group A streptococcal vaccine designed to optimise the immunogenicity of six tandem M protein fragments. Vaccine. 1999;17:193–200. doi: 10.1016/s0264-410x(98)00150-9. [DOI] [PubMed] [Google Scholar]
- 12.Dale J B, Beachey E H. Epitopes of streptococcal M proteins shared with cardiac myosin. J Exp Med. 1985;162:583–591. doi: 10.1084/jem.162.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dale J B, Seyer J M, Beachey E H. Type specific immunogenicity of a chemically synthesised peptide fragment of type 5 streptococcal M protein. J Exp Med. 1983;158:1727–1732. doi: 10.1084/jem.158.5.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dale J B, Simmons M, Xhiang E C, Chiang E Y. Recombinant, octavalent group A streptococcal M protein vaccine. Vaccine. 1996;14:944–948. doi: 10.1016/0264-410x(96)00050-3. [DOI] [PubMed] [Google Scholar]
- 15.Gardiner D L, Goodfellow A M, Martin D R, Sriprakash K S. Group A streptococcal Vir types are M protein gene (emm) sequence-type specific. J Clin Microbiol. 1998;36:902–907. doi: 10.1128/jcm.36.4.902-907.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilherme L, Cunha-Neto E, Coelho V, Snitcowsky R, Pomerantzeff P M A, Assis R V, Pedra B S, Neumann J, Goldberg A, Pararroyo M E, Pileggi F, Kalil J. Human heart infiltrating T-cell clones from rheumatic heart disease patients recognise both streptococcal and cardiac proteins. Circulation. 1995;92:415–420. doi: 10.1161/01.cir.92.3.415. [DOI] [PubMed] [Google Scholar]
- 17.Hayman W A, Brandt E R, Relf W A, Cooper J, Saul A, Good M F. Mapping the minimal T and B cell epitome within a peptide vaccine candidate from the conserved region of the M protein of group A streptococci. Int Immunol. 1997;9:1723–1733. doi: 10.1093/intimm/9.11.1723. [DOI] [PubMed] [Google Scholar]
- 18.Houghten R A. General method for the rapid solid-phase synthesis of large numbers of peptides: specificity of antigen-antibody interaction at the level of individual amino acids. Proc Natl Acad Sci USA. 1985;82:5231–5235. doi: 10.1073/pnas.82.15.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kehoe M A. New aspects of Streptococcus pyogenes pathogenicity. Rev Med Microbiol. 1991;2:147–152. [Google Scholar]
- 20.Lancefield R C. Differentiation of group A streptococci with a common R antigen into three serological types, with special reference to the bactericidal test. J Exp Med. 1957;106:525–529. doi: 10.1084/jem.106.4.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malmanche S A, Martin D R. Protective immunity to the group A streptococcus may only be strain specific. Med Microbiol Immunol. 1994;183:299–306. doi: 10.1007/BF00196680. [DOI] [PubMed] [Google Scholar]
- 22.Martin D R, Voss L M, Walker SJ, Lennon D. Acute rheumatic fever in Auckland, New Zealand: spectrum of associated GAS different from expected. Pediatr Infect Dis J. 1994;13:264–269. doi: 10.1097/00006454-199404000-00004. [DOI] [PubMed] [Google Scholar]
- 23.Nordet P. WHO programme for prevention of rheumatic fever/rheumatic heart disease in 16 developing countries: report for phase I (1986–90) Bull WHO. 1992;70:213–218. [PMC free article] [PubMed] [Google Scholar]
- 24.Pruksakorn S, Currie B, Brandt E, Phornphutkul C, Hunsakunachi S, Manmontri A, Robinson J A, Kehoe M A, Galbraith A, Good M F. Identification of T-cell autoepitopes that cross-react with the carboxyterminal segment of the M protein of group A streptococci. Int Immunol. 1994;6:1235–1244. doi: 10.1093/intimm/6.8.1235. [DOI] [PubMed] [Google Scholar]
- 25.Pruksakorn S, Phornphutkul C, Boonchoo S. Prevalence of group A streptococci from school children in Chiang Mai, Thailand. Chiang Mai Med Bull. 1990;29:15–26. [Google Scholar]
- 26.Pruksakorn S, Galbraith A, Houghten R A, Good M F. Conserved T and B cell epitopes on the M protein of group A Streptococci: Induction of bactericidal antibodies. J Immunol. 1992;149:2729–2735. [PubMed] [Google Scholar]
- 27.Relf W A. Molecular diversity in the M protein of Streptococcus pyogenes. Ph.D. thesis. Sydney, Australia: University of Sydney; 1993. [Google Scholar]
- 28.Relf W A, Martin D R, Sriprakash K S. Identification of sequence types among the M-nontypeable group A streptococci. J Clin Microbiol. 1992;30:3190–3194. doi: 10.1128/jcm.30.12.3190-3194.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sargent S J, Beachey E H, Corbett C E, Dale J B. Sequence of protective epitopes of streptococcal M proteins shared with cardiac sarcolemmal membranes. J Immunol. 1987;139:1285–1290. [PubMed] [Google Scholar]
- 30.Single L A, Martin D R. Clonal differences within M types of the group A streptococcus revealed by pulsed field gel electrophoresis. FEMS Microbiol Lett. 1992;91:85–90. doi: 10.1016/0378-1097(92)90567-8. [DOI] [PubMed] [Google Scholar]
- 31.Vashishtha A, Fischetti V A. Surface exposed conserved region of the streptococcal M protein induces antibodies cross-reactive with denatured forms of myosin. J Immunol. 1993;150:4693–4701. [PubMed] [Google Scholar]