Abstract
Yersinia enterocolitica is one of three pathogenic Yersinia species that share a tropism for lymphoid tissues. However, infection of an immunocompromised host is likely to result in a systemic infection, which is often fatal. Little is known about the bacterial proteins needed to establish such an infection. The genes that encode these virulence factors are likely to be active only during systemic infection. A library of random cat fusions was used to inoculate BALB/c mice. Fusions expressed during a systemic infection were enriched by the administration of chloramphenicol-succinate. Y. enterocolitica isolates recovered from the mice were tested for chloramphenicol resistance in vitro. Fusions that were inactive in vitro were analyzed further and found to represent 31 allelic groups. Each was given a sif (for systemic infection factor) designation. Based on homology to known proteins, the sif genes are likely to encode proteins important for general physiology, transcription regulation, and other functions. During systemic infections, 13 of the sif-cat fusions were able to outcompete the wild type in the presence of chloramphenicol-succinate, confirming that the fusions were active. The in vitro expression of several sif genes was determined, showing modest changes in response to various growth conditions. A mutation in sif15, which encodes a putative outer membrane protein, caused attenuation during systemic infection but not during colonization of the Peyer's patches. Comparisons between the Y. enterocolitica sif genes and the previously identified hre genes imply that very different groups of genes are active during a systemic infection and during colonization of the Peyer's patches.
Three species of the genus Yersinia are able to infect humans, in whom they exhibit a tropism for lymphoid tissue (12). Yersinia pestis infections are characterized by colonization of the lymph nodes proximal to the site of infection and progression to a systemic disease, the bubonic plague. The enteric pathogens Yersinia pseudotuberculosis and Yersinia enterocolitica infect after being ingested in contaminated food or water. Y. pseudotuberulosis is primarily an animal pathogen, whereas Y. enterocolitica readily infects humans. In humans, Y. enterocolitica initially colonizes the Peyer's patches of the small intestine, after which the bacteria colonize the mesenteric lymph nodes. The infection results in gastroenteritis characterized by diarrhea, fever, and abdominal pain (3, 16). In humans who are immunocompromised due to medical conditions such as age, disease, or chemotherapy, the bacteria are not cleared from the lymphoid tissues. From those tissues, the bacteria gain access to the blood, leading to a systemic infection (12). While systemic infections are rare, they are associated with a striking 50% mortality rate (4). A BALB/c mouse model for Y. enterocolitica infection has been developed in which infections closely resemble human infections in both the progression of the infection and the immune response against the infection (9, 10). One important distinction is that BALB/c mice infected with Y. enterocolitica usually develop systemic infections, as determined by colonization of the spleen, liver, and lungs. Therefore, the mouse model affords the opportunity to study both the acute infection of the lymphoid tissue and the development of systemic infections.
Several virulence factors have been identified in the three pathogenic Yersinia species. All three species contain a highly conserved virulence plasmid, pYV, which encodes a type III secretion apparatus (composed of Ysc proteins) and the Yop effector proteins that it secretes (14, 15). While the virulence plasmid is necessary for infection, it is not sufficient, indicating that essential virulence factors are encoded on the chromosome. Previous studies have identified several chromosomally encoded virulence factors. Functional complementation of Escherichia coli K-l2 led to discovery of the inv and ail genes, whose expression in E. coli allows that bacterium to attach to and invade host cells in vitro (27, 37). A transcriptional regulator of inv, RovA, appears to regulate other genes that are important for virulence; a rovA mutant is less virulent than an inv mutant in vivo (41). A pathogenicity island encoding a high-affinity iron siderophore and its transport system was identified in the more virulent serotypes of Y. enterocolitica (7, 8, 19, 22). The genes necessary for O-antigen biosynthesis were found to be essential for full virulence of Y. enterocolitica (55). Mutants lacking functional phospholipase, encoded by yplA, show a defect in colonization of the Peyer's patches and mesenteric lymph nodes (43). The exotoxin, Yst, produced by Y. enterocolitica appears to be responsible for diarrhea in the young-rabbit model (13). Recently, signature-tagged mutagenesis was used to identify eight chromosomal loci that encode novel Y. enterocolitica virulence factors (17). One of these loci contains a homolog to the psp operon of E. coli. Transposon insertion into or deletion of the pspC gene renders Y. enterocolitica completely unable to infect BALB/c mice. The E. coli pspC gene is a member of the phage shock operon and has no known biochemical function, so the role that PspC plays in virulence remains unclear.
One approach used to identify novel virulence factors is to determine genes that are expressed during infection but not under laboratory conditions (in vivo expression technology [IVET]). This technique and subsequent adaptations have been used successfully for a number of pathogenic organisms, such as Salmonella enterica serovar Typhimurium, Pseudomonas aeruginosa, Vibrio cholerae, Y. enterocolitica, Staphylococcus aureus, and Actinobacillus pleuropneumoniae, resulting in the description of several novel virulence factors (6, 18, 23, 30–32, 50, 51, 54). Application of IVET to Y. enterocolitica led to the discovery of novel virulence factors necessary for the initial colonization of the Peyer's patches (54). For that study, a library of random chromosomal fusions to a promoterless cat gene was generated and integrated into the chromosome of wild-type Y. enterocolitica. The expression of genes that are transcriptionally fused to the cat gene can be detected by chloramphenicol (Cam) resistance of the bacterium. Pools of the cat fusion library were used to orally infect BALB/c mice. Administration of chloramphenicol-succinate allowed the in vivo enrichment of bacteria in which the cat gene was expressed. This enrichment was performed very early during the course of infection, and bacteria were recovered from the initial site of Y. enterocolitica infection, the Peyer's patches. The genes that were enriched in the Peyer's patches, the hre (host responsive elements) genes, are likely to encode putative virulence factors. This collection of hre genes encoded proteins involved in stress response, iron starvation response, cell envelope maintenance, transcription regulation, and other unknown functions. Seventy-five percent of the hre genes tested are important for virulence, since gene disruption led to an attenuated phenotype in vivo.
Using the murine model, both acute infections of the lymphoid tissue and systemic infections with Y. enterocolitica can be examined. Thus, a comparison of the virulence factors required for each can be made in this system. Additionally, the IVET technique could be modified to enrich for genes from the same fusion pool(s) that are active at different stages of infection. Therefore, these tools present the unique opportunity to identify virulence factors needed during one stage of infection and compare them to those needed at a different stage. This study is the result of using the available tools to identify putative virulence factors that are expressed during the development of a systemic infection and comparing them to those previously found to be active during an acute infection of the Peyer's patches.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains used in this study are shown in Table 1. Unless otherwise noted, E. coli strains were grown at 37°C and Y. enterocolitica strains were grown at 26°C. All strains were routinely grown in Luria-Bertani (LB) broth with agitation to ensure full aeration or on LB agar plates. Growth under nutrient-limiting conditions was assessed on M63 agar plates containing 0.2% glucose and 1 mM MgSO4. When indicated, LB was buffered with 20 or 80 mM 3-(N-morpholino)propanesulfonic acid (MOPS) to maintain either pH 7.0 or pH 7.5 and with 20 or 80 mM 2-(N-morpholino) ethanesulfonic acid (MES) to maintain pH 5.5. For log-phase cultures, 20 mM buffer was sufficient to maintain the pH, whereas stationary-phase cultures required 80 mM buffer to maintain the pH. A 1% (wt/vol) tryptone medium and supplemented M63 medium (0.2% glucose, 1 mM MgSO4, 0.25% casamino acids, and 0.8% thiamine) were used to study gene expression and growth phenotypes. Antibiotics were used in the following concentrations: nalidixic acid (Nal), 20 μg/ml; chloramphenicol (Cam), 10 μg/ml for Y. enterocolitica and 25 μg/ml for E. coli; rifampin (Rif), 100 μg/ml; streptomycin (Str), 50 μg/ml; spectinomycin (Spc), 50 μg/ml; and erythromycin (Erm), 50 μg/ml for Y. enterocolitica and 100 μg/ml for E. coli.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant genotypea | Source or reference |
|---|---|---|
| Y. enterocolitica | ||
| JB580v | Serogroup O:8 Nalr ΔyenR (R− M+) | 29 |
| JB41v | JB580v wild-type inv; inv::phoA Nalr Camr | 1 |
| ASG21 | As JB580v with a TnMax2 insertion in sif15, Nalr Ermr | This study |
| E. coli | ||
| CC118λpir | Δ(ara-leu) araD ΔlacX74 galE phoA20 thi-1 rpsE rpoB argE(Am) recAl λ pir lysogen | 24 |
| S17-1λpir | Tpr StrrrecA thi pro hsdR hsdM+ RP4::2-Tc::Mu::Km Tn7 λ pir lysogen | 38 |
| Plasmids | ||
| pEP185.2 | MobRP4 oriR6K Camr | 29 |
| pFUSE | MobRP4 oriR6K Camr, polylinker upstream of promoterless lacZYA | 2 |
| pGY2 | MobRP4 oriR6K Apr Spcr Strr, unique BglII site upstream of promoterless cat | 54 |
| pHG329 | lacZα Apr, medium-copy-number cloning vector | 47 |
| pRK2013 | Tra+ Kmr | 40 |
Tp, trimethoprim; Str, streptomycin; Ap, ampicillin; Km, kanamycin.
Generation of cat fusion library. The library of random cat fusions used was generated for a previous study (54). Briefly, chromosomal DNA was isolated from JB580c, a variant of wild-type Y. enterocolitica that lacks the pYV virulence plasmid, and partially digested with Sau3AI. The DNA was then cloned into a unique BglII site in pGY2 that is directly upstream of a promoterless cat gene. The DNA fused to the cat gene consisted exclusively of DNA from the chromosome of Y. enterocolitica. Clones were recovered and mated into JB580v (containing the virulence plasmid). pGY2 contains an R6K origin of replication and cannot replicate autonomously in JB580v. Selection with a vector-derived antibiotic marker ensured that the growing bacteria had the fusion plasmid integrated into the chromosome. Integrants were isolated on minimal medium to ensure that the resulting merodiploid strains were not auxotrophs. It is important to note that the alleles fused to the promoterless cat gene are also likely to be present in an unaltered form on the chromosome of the integrants (i.e., a merodiploid is formed). Therefore, the integration of the cat fusion should not alter the physiology of the bacterium. The library consists of 32 pools (each containing 103 to 104 fusions) that were created independently. Therefore, the library represents approximately 300,000 independent cat fusions and is considered comprehensive.
Enrichment for active cat fusions in vivo. Each pool of Y. enterocolitica strains containing cat fusions was grown overnight in LB medium and washed and resuspended in phosphate-buffered saline (PBS) to an estimated density of 108 CFU/ml. Since each pool contains an estimated 104 fusions, each fusion should be represented approximately 10,000 times at this culture density. Three female, 6- to 8-week-old, virus-free BALB/c mice were inoculated intraperitoneally with 100 μl of each pool (approximately 107 bacteria). At 16 and 24 h postinfection, the mice were given subcutaneous injections of chloramphenicol-succinate (150 μl of a 20-mg/ml solution). At 40 h postinfection, the mice were sacrificed and the spleens were harvested. Homogenates were diluted 1:50 and used to inoculate LB containing Nal to select for Y. enterocolitica JB580v derivatives. This culture of recovered Y. enterocolitica was used to inoculate a second group of three mice, using the protocol described above. Tissue homogenates were serially diluted in PBS and plated on LB agar containing Nal, Spc, and Str. The Y. enterocolitica organisms isolated from the second group of mice were enumerated and further analyzed in vitro.
Fusion classification based on in vitro growth. To initially classify the fusions based on growth phenotypes, bacteria recovered from the spleenic tissue were tested for resistance to Cam in vitro on both rich (LB) and minimal (M63) agar plates. The fusion-containing isolates were replica plated to LB and M63 plates containing Cam (10 μg/ml) (just above the MIC for Y. enterocolitica) and incubated at 26°C. Bacteria that appeared to be Cam sensitive were patched from the original LB plate containing Nal onto LB agar containing Nal, LB agar containing Cam, and M63 agar containing Cam and allowed to grow at 26°C. Again, those bacteria that were unable to grow in the presence of Cam were streaked onto the three media for isolated colonies. This procedure allowed reliable classification of the fusions by growth phenotype.
In vitro growth rates of fusion-containing strains and wild-type JB580v were also measured in either LB or supplemented minimal medium. Overnight cultures grown at 26°C were diluted at least 1:50 into fresh medium and incubated at either 26 or 37°C. Growth was monitored by measuring the optical density at 600 nm (OD600) of the cultures with time.
Classification of cat fusions into allelic groups. Chromosomal DNA was isolated from cat fusion-containing strains and digested with EcoRV, which has one site within the vector portion of the integrated fusion plasmid. A Southern blot was performed by probing the digested chromosomal DNA with a 2.5-kb BamHI fragment of the pGY2 vector that contains the single EcoRV site. The probe DNA was labeled, and its hybridization was detected using the ECL direct nucleic acid labeling and detection system (Amersham Life Science). Two bands of varying size were visible from each integrant using this probe. The Southern blot patterns of each were compared, allowing the identification of fusions that are likely to belong to the same allelic group.
Cloning and sequencing of cat fusions. Triparental matings were used to clone fusions of interest as previously described (40). Briefly, cultures of the Y. enterocolitica fusion-containing strain, an E. coli π+ recipient, and an E. coli strain harboring a helper plasmid (pRK2013) that encodes tra were mixed in equal amounts, pelleted, plated on an LB agar plate, and incubated at 26°C. At some frequency, the integrated cat fusion plasmid will recircularize but be unable to replicate in Y. enterocolitica. However, the Tra+ helper plasmid enables conjugation of the cat fusion plasmid to the π+ recipient, in which it can replicate autonomously. The bacteria were collected from the LB plate, plated on medium that selected for both the antibiotic resistance marker on the plasmid (Spc and Str resistance) and that of the π+ recipient (Rif resistance), and incubated at 37°C. Plasmid DNA was isolated using Wizard Plus SV miniprep kits (Promega).
DNA sequence data were obtained for the DNA directly upstream of the promoterless cat gene using a primer that hybridizes within the cat gene (5′ - CAA CGG TGG TAT ATC CAG TG- 3′; Gibco Life Technologies, Inc.). DNA sequencing was performed with a BigDye terminator cycle sequencing kit (Applied Biosystems) and analyzed at the Nucleic Acid Chemistry Laboratory, Washington University. Sequences were compared to those in the databases using the BLAST program available through the National Institutes of Health NCBI database (www.ncbi.nlm.nih.gov). Sequences were also analyzed using the program MacVector (Oxford Molecular).
In vivo expression of cat fusions. The in vivo expression of several individual cat fusions was tested by competition infections with wild-type Y. enterocolitica in the presence of Cam. Cultures of the fusion-containing isolate or a cat+ control strain (JB41v) and JB580v were grown overnight in LB medium, washed, and resuspended in PBS at a density of approximately 108 CFU/ml. Equal amounts of fusion-containing or JB41v and wild-type bacteria were mixed, and dilutions were plated on LB with Nal only to determine the total number of bacteria, LB with Nal, Spc, and Str to determine the number of fusion-containing bacteria, or LB with Nal and Cam to determine the number of JB41v bacteria. From this, the exact input ratio was determined. A 100-μl sample of the mixture was used to inoculate two female, 6- to 8-week-old, virus-free BALB/c mice intraperitoneally. At 16 and 24 h postinfection, the mice were given subcutaneous injections of chloramphenicol-succinate (150 μl of a 20-mg/ml solution). Forty hours postinfection, the mice were sacrificed and the spleen tissue was harvested. The spleens from both mice were pooled, and homogenates were plated again on selective media to determine the ratio of fusion-containing or JB41v to wild-type bacteria present. The fold enrichment was calculated by dividing the ratio of fusion-containing bacteria to wild-type bacteria present in the murine tissue (CFU per gram) by the ratio of the starting inoculum (CFU per milliliter).
In vitro expression of selected sif alleles. The in vitro expression of nine sif alleles (sif1, sif15, sif16, sif18, sif20, sif21, sif23, sif25, and sif29) was determined by Western blot analysis. The Y. enterocolitica strains containing integrated sif-cat fusions were grown under a number of conditions. To achieve stationary phase, cultures were grown for approximately 16 h. Log-phase cultures were obtained after the dilution of 10 μl of 26°C overnight cultures into 4 ml of fresh medium and subsequent incubation for 4 h. The culture OD600 was measured. A volume of culture equivalent to 0.150 OD600 unit was harvested, and the cells were resuspended in sample buffer. Cellular proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and proteins were transferred to nitrocellulose using a Transblot SD (Bio-Rad). Cat was visualized using a polyclonal anti-Cat antibody as a primary antibody, anti-rabbit immunoglobulin G (IgG) linked to horseradish peroxidase as a secondary antibody, and the ECL Western detection kit (Amersham Pharmacia).
The in vitro expression of three sif alleles (sif2, sif6, and sif15) was assessed using transcriptional fusions to the reporter gene lacZ and measuring β-galactosidase activity. For each, an internal region of the sif gene was amplified by PCR and cloned upstream of a promoterless lacZYA in the plasmid pFUSE (2). These constructs were then integrated onto the chromosome of JB580v after selection for the antibiotic resistance encoded on pFUSE. The integration of each φ(sif-lacZ) construct was confirmed by Southern blot (data not shown). The expression of each sif gene was then determined by measuring the β-galactosidase activity of three independent cultures after growth under a number of conditions. Expression in stationary-phase cultures was determined after approximately 16 h of growth at either 26 or 37°C. To determine sif expression in the exponential phase, 10 μl of the 26°C stationary-phase culture was used to inoculate 3 ml of fresh medium, which was then incubated for 4 h under the conditions indicated. Cells used for β-galactosidase assays were washed in 10 mM Tris (pH 7.0) prior to lysis. Assays for β-galactosidase activity were performed as described previously (36). The values shown are the averages of the three independent cultures, with the error bars indicating the range of Miller units measured.
Construction of a sif15 mutant and in vivo characterization. A cosmid library of Y. enterocolitica chromosomal DNA (B. Young and V. Miller, unpublished results) was probed for sif15 using a 500-bp PCR probe. A. 4.5-kb KpnI fragment of one cosmid which contained sif15 was cloned into pHG329 (47) and confirmed by PCR analysis. This clone was mutagenized with the Erm transposon TnMax2 (20) to facilitate sequence analysis. Sequence data suggested that sif15 was not part of an operon, and therefore an insertional mutation could be used to test the role of sif15 in virulence. One of the TnMax2 insertions was determined to reside within sif15. This mutated allele was cloned into pEP185.2 (29), which is a mobilizable suicide vector. The resultant clone was mated into JB580v from S17-1λpir, and the presence of the transposon insertion in Y. enterocolitica was selected on LB with Nal and Erm. Isolates likely to contain insertions in sif15 were identified by screening the Nal Erm isolates for sensitivity to Cam, which indicated loss of the vector DNA. The disruption of sif15 was confirmed by Southern blot (data not shown). The in vitro growth rate of the sif15 mutant was determined as described above for the fusion-containing strains.
To test the role of Sif15 in virulence, competition infections with wild-type Y. enterocolitica were preformed. Cultures of the sif15 mutant and JB580v were grown overnight in LB medium at 26°C, washed, and resuspended in PBS. Equal numbers of sif15 mutant and JB580v bacteria were mixed, and dilutions were plated on LB with Nal to determine the total number of bacteria and LB with Nal and Erm to determine the number of sif15 bacteria. From this, an exact input ratio was determined. Four BALB/c mice were given 5 × 106 CFU intraperitoneally, and five were given 1 × 108 CFU intragastrically. The mice inoculated intraperitoneally were sacrificed 40 h postinfection, and the spleens were recovered. The mice inoculated intragastrically were sacrificed 48 h postinfection, and the Peyer's patches were recovered. Tissues were homogenized in PBS and plated on selective medium to determine the ratio of sif15 mutant to JB580v bacteria present. The competitive index was calculated by dividing the ratio of fusion-containing bacteria to wild-type bacteria (CFU per gram) present in the murine tissue by the ratio of the starting inoculum (CFU per milliliter).
RESULTS
Enrichment of active cat fusions during systemic infections in vivo.
To determine if a successful in vivo enrichment for Cam-resistant bacteria was possible during a systemic infection, a control was done to see if Cam-resistant bacteria would colonize the spleen better than wild-type (Cam-sensitive) bacteria in mice treated with chloramphenicol-succinate. Four BALB/c mice were infected intraperitoneally with approximately 107 bacteria, consisting of wild-type (JB580v) and 3% Cam-resistant (JB41v) bacteria. Intraperitoneal inoculations bypass the gastric barrier and deliver the bacteria directly to the peritoneal cavity and possibly to the bloodstream. After intraperitoneal inoculation, mice develop a systemic infection, and bacteria are recovered from the spleen within the first 24 h of infection (A. J. Darwin and V. L. Miller, unpublished results). At 16 and 24 h postinfection, the mice were given subcutaneous injections of chloramphenicol-succinate (150 μl of a 20-mg/ml solution). Forty hours postinfection, the mice were sacrificed and the spleens were harvested. The pooled tissues were homogenized, and the fraction of Cam-resistant bacteria was determined. Of the bacteria recovered from the first set of mice, 90% were Cam resistant. The enrichment for Cam-resistant bacteria was further improved by using the recovered bacteria to inoculate a second group of mice that were also given chloramphenicol-succinate subcutaneously. The second passage through mice increased the Cam-resistant fraction to more than 95%. Therefore, this protocol provides a strong enrichment for Cam-resistant bacteria during systemic infection. Interestingly, if the first chloramphenicol-succinate dose was given 8 h postinfection, the spleen was less efficiently colonized (data not shown).
In order to identify genes induced during infection, 32 independent pools of random cat fusions were used to infect BALB/c mice, and Cam-resistant bacteria were enriched as described above and in Materials and Methods. Following this protocol, the spleenic tissue was typically colonized with 106 to 108 bacteria/g of tissue at the time of harvest. A fusion-containing isolate is most likely recovered from the spleen because the cat fusion it contains is active in vivo and renders the bacterium resistant to chloramphenicol-succinate. However, because Cam is bacteriostatic rather than bacteriocidal, bacteria with inactive fusions that were present in the spleen prior to the treatment with chloramphenicol-succinate could be isolated from that tissue. This is possible even though the bacterium containing the inactive cat fusion would not be able to replicate in the presence of chloramphenicol-succinate. For each pool of cat fusions, more than 300 isolates recovered from the second infection were analyzed further.
In vitro classification and analysis of enriched cat fusions.
Previous studies in which in vivo-induced genes were enriched found that the majority of the enriched genes were constitutively active. As we are interested in identifying genes whose products play a role in pathogenesis, it is critical to separate the constitutively active cat fusions from those that are induced only during infection. More than 10,000 isolates that were recovered from the spleen tissue were tested for the ability to grow in vitro in the presence of Cam at 26°C. The in vitro growth rate of Y. enterocolitica is higher at 26°C than at 37°C because Yop induction at 37°C has a negative effect on growth. Both rich (LB) and minimal (M63) media containing Cam were used to classify the in vitro growth phenotypes of each isolate. Isolates were classified into four separate groups. The vast majority of isolates were Cam resistant during growth on both media and were designated class I isolates. These isolates were thought to contain fusions to constitutively active genes. A smaller number of isolates, 256, were found to be Cam sensitive during growth on both media and were given a class II designation. These cat fusions seemed to be preferentially expressed in vivo. Class III isolates were Cam resistant only during growth on rich medium, and class IV isolates were Cam resistant only during growth on minimal medium. The regulation of these fusions is more complex, but expression was not limited to in vivo conditions. Because our primary interest is the genes that are active only under in vivo conditions, isolates belonging to class II were studied further.
A Southern blot was performed for each of the 256 class II isolates to determine the number of distinct alleles represented. After comparing the Southern blot patterns for each, 31 allelic groups were identified. The number of isolates in each allelic group varied from 1 to 30, and isolates of one allelic group were often enriched from more than one mouse (data not shown). Each was given a sif (for systemic infection factor) designation. These sif genes are likely to encode proteins important for virulence, because each gene is preferentially expressed during infection.
At least one representative of each allelic group was cloned from the fusion-containing Y. enterocolitica strain by triparental mating (see Materials and Methods). Sequence data were derived using a primer that hybridized within the promoterless cat gene and read into the chromosomal DNA to which it was fused. For each, 500 to 800 bases of DNA sequence was obtained and used to search the databases for protein homologs. Table 2 shows the function and/or properties of the sif products predicted by amino acid similarity of partial protein sequences to protein sequences in the databases. Two types of sif-cat fusions were represented. The majority were fusions to sif alleles (sif1 to sif11 and sif13 to sif17) that appear to encode known proteins (Table 2). For the remaining sif-cat fusions (sif12 and sif18 through sif31), we were unable to identify the open reading frame responsible for cat expression, although potential open reading frames were identified for each allele. The majority of these fusions were to regions of the Y. enterocolitica chromosome that do not have significant homology to sequences in the databases. A subset of fusions were determined to be inactive in vivo (see below) and were not included in Table 2 (data not shown).
TABLE 2.
sif alleles
| sif allele | Gene homologa | Predicted function or property | % amino acid identity/ source of homologb | In vivo fold enrichmentc |
|---|---|---|---|---|
| sif1 | sitC | 29-kDa ABC transporter component | 37/S. epidermidis | 3.5* |
| sif2 | lepA | Membrane-bound GTPase | 80/E. coli | 3.9* |
| sif3 | rffG | LPS biosynthesis | 76/Erwinia carotovora | 22* |
| sif4 | frdB | Fumarate reductase subunit | 84/Proteus vulgaris | ND |
| sif5 | metL | Apartokinase/homoserine reductase | 80/E. coli | ND |
| sif6 | yohI | Putative transcriptional regulator | 91/E. coli | 28* |
| sif8 | bioH | Biotin synthesis | 82/E. coli | ND |
| sif9 | fyuA | Iron siderophore precursor | 100/Y. enterocolitica | ND |
| sif10 | manB | O-antigen biosynthesis | 100/Y. enterocolitica | ND |
| sif11 | yjcD | 45.7-kDa protein in soxR-acs region | 78/E. coli | 35 |
| sif13 | nifJ | Pyruvate-flavodoxin oxidoreductase | 70/B. subtilis | ND |
| sif14 | rhlB | RNA helicase | 68/E. coli | ND |
| sif15 | HP0694 | Putative outer membrane protein | 69/Salmonella | 30* |
| sif16 | gacA | Transcriptional regulator | 32/P. syringae | 6.0* |
| sif12 | Unknown ORF | No homology | ND | |
| sif18 | Unknown ORF | Near yjgF homolog | 4.5 | |
| sif20 | Unknown ORF | Near cbiO homolog | 35* | |
| sif21 | Unknown ORF | No homology | 21 | |
| sif23 | Unknown ORF | No homology | 4.2 | |
| sif25 | Unknown ORF | No homology | 31* | |
| sif29 | Unknown ORF | No homology | 46* | |
| sif31 | Unknown ORF | near polI homolog | ND |
Unknown ORF, the open reading frame (ORF) responsible for cat expression was not identified from the sequence data obtained.
The percent amino acid identity was determined for partial protein sequences only.
ND, not determined. Values marked with an asterisk were determined in at least two separate experiments with similar results. The data shown are one representative data set.
In vivo expression.
To confirm that the sif genes are active in vivo, we decided to test the ability of the individual cat fusions to compete against wild-type Y. enterocolitica in the presence of Cam in vivo. We anticipated that fusions that are expressed in vivo would be able to multiply in the presence of Cam and should therefore colonize better than the wild type, which is Cam sensitive. However, if the cat fusion is not active in vivo, neither the fusion-containing strain nor the wild type should be able to multiply in the presence of Cam. Thus, the recovered bacterial population should contain the fusion strain and wild type in nearly the same ratio as found in the inoculum.
For each of the fusions tested, two BALB/c mice were inoculated with a 1:1 ratio of the fusion-containing isolate to the wild type. The mice were treated as described previously for the enrichment of active fusions. The fold enrichment shown in Table 2 was determined as described in Materials and Methods. The fold enrichment of the fusions tested ranged from 0.15 to 46 (data not shown and Table 2). The positive control strain, JB41v, which contains an intact cat gene, was enriched 11-fold in this assay. We considered a fold enrichment value of greater than 3 to indicate that the cat fusion was at least transiently active in vivo. Of the 22 fusion-containing isolates tested, 13 did outcompete the wild type in this assay. Seven fusion-containing strains that were unable to outcompete the wild type in these experiments contained fusions for which we were unable to identify the chromosomal gene fused to cat.
It is possible for an active fusion to fail to outcompete wild-type cells if integration of the sif-cat fusion resulted in a growth defect in vivo. Each of the fusions tested for in vivo expression was found to have in vitro growth rates similar to those for JB580v in LB medium at 37°C (data not shown). It seemed possible that growth in a rich medium may not adequately mimic in vivo growth conditions, making it more likely to detect a growth defect under nutrient-limiting growth conditions. A subset of fusion-containing strains were also tested for growth defects in supplemented minimal medium at 37°C and found to have wild-type growth rates under those conditions (data not shown). Therefore, while we cannot rule out the possibility of an in vivo growth defect, our in vitro data do not support that conclusion. It is possible that these fusions were never active in vivo but were recovered from the enrichment as background.
Characterization of in vitro sif expression.
One caveat to the approach used for in vitro classification of cat fusions is that fusions induced at 37°C would be included in class II, even though the fused genes may not be relevant for pathogenesis. To address this, a representative of each allelic group was tested for its ability to grow on LB agar containing Cam (10 μg/ml) at 37°C. None of the class II strains were able to form isolated colonies under these conditions (data not shown). In addition, the MIC of Cam was determined for 10 sif-cat fusion strains in LB broth, and no difference in Cam resistance was detected between 26 and 37°C (data not shown). These data suggest that while temperature may be an important factor for the regulation of sif genes, it is not the only signal for induction of sif gene expression.
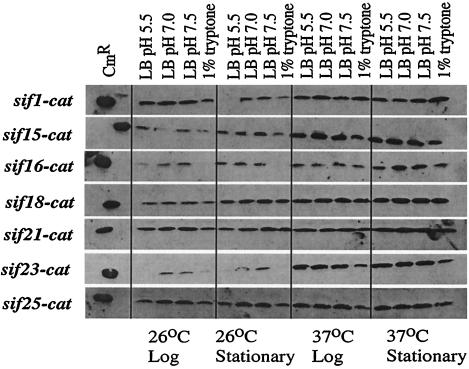
Studying the genes from the class II fusions presents a technical challenge, as these genes are not expressed enough in vitro to result in resistance to Cam. To facilitate further study, it is advantageous to identify in vitro conditions that allow expression of the sif genes. Qualitative measurements of sif expression were made using Western blot analysis. Y. enterocolitica strains containing integrated sif-cat fusions were grown in four different media (LB at pH 7.0, LB at pH 5.5, LB at pH 7.5, and 1% tryptone broth) to the exponential and stationary phases at either 26 or 37°C. These conditions were chosen because other Y. enterocolitica virulence factors respond to these conditions. For example, the gene encoding invasin shows high levels of expression after growth at 37°C and pH 5.5 (39), and the gene encoding phospholipase shows maximal expression after growth at 26°C in 1% tryptone (42). The relative Cat expression was determined from equivalent numbers of cells using a polyclonal anti-Cat antibody (Fig. 1). The expression of sif1, sif15, and sif23 was highest in cultures grown to stationary phase at 37°C, but each showed different expression in response to pH, nutrients, and growth phase at 26°C. The expression of sif18, sif21, and sif25 did not change significantly among the conditions tested. The expression of sif16 was uniform among the conditions tested except that it was highly repressed after growth in 1% tryptone medium at 26°C. The in vitro expression of two other sif-cat fusions (sif20 and sif29) was not detectable by this assay, implying that it was extremely low.
FIG. 1.
Western blot analysis of sif-cat fusions. Cultures for Western blot analysis were grown under the conditions indicated. The culture density was determined at 600 nm. The amount of culture loaded per lane was 0.150 OD600 unit, so that each lane contains an equal number of cells. The primary antibody used was a polyclonal anti-Cat antibody; the secondary antibody used was a horseradish peroxidase-conjugated anti-rabbit IgG antibody.
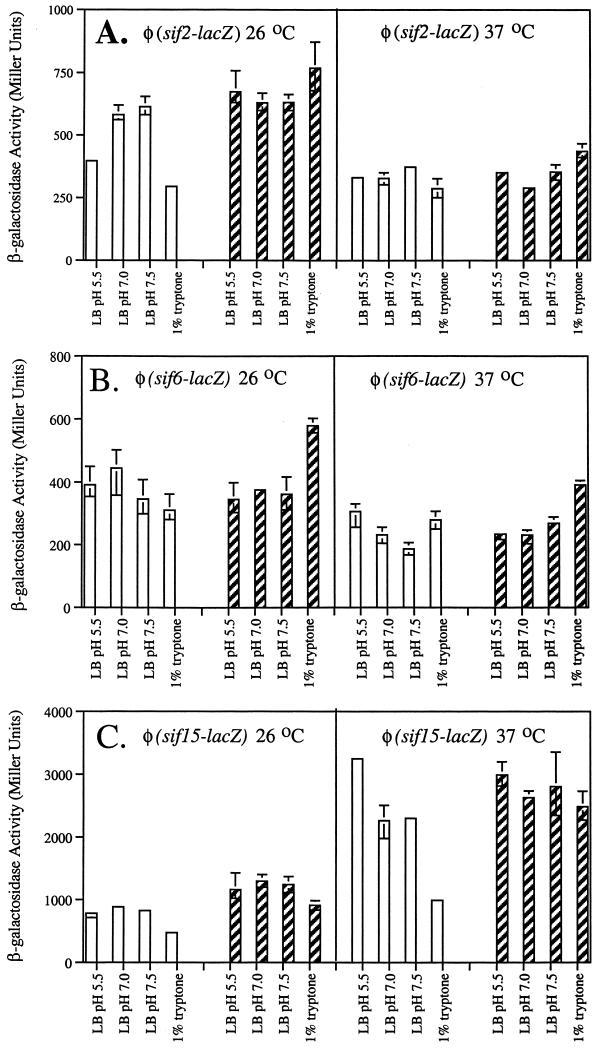
In addition, φ(sif-lacZ) transcriptional fusions were constructed for a subset of sif alleles to allow quantitative measurements of sif expression. The expression of each φ(sif-lacZ) fusion was measured after growth under the same conditions used for the Western blot analyses (Fig. 2). The expression of sif2 was slightly higher in stationary-phase cultures grown at 26°C, but did not seem to respond to pH or medium conditions. φ(sif6-lacZ) expression did not vary much among any of the conditions tested. The expression of sif15 was significantly higher in both the stationary- and exponential-phase LB cultures grown at 37°C. Importantly, the expression pattern of sif15-cat seen in the Western blot analysis strongly correlates with that obtained from the φ(sif15-lacZ) transcriptional fusion, further validating the use of Western blot analysis to determine in vitro expression.
FIG. 2.
φ(sif-lacZ) expression. φ(sif-lacZ) expression was measured by determining β-galactosidase activity in Miller units. (A) φ(sif2-lacZ); (B) φ(sif6-lacZ); (C) φ(sif15-lacZ). Open columns represent the activity in exponential phase, and solid columns represent the activity in stationary phase. Each column represents the average of three independent cultures grown under the conditions indicated. The error bars indicate the ranges of β-galactosidase activities measured.
Identification of a role for Sif15 during systemic infection.
A mutation in sif15, which encodes a putative outer membrane protein, was generated by disrupting the gene with TnMax2 (see Materials and Methods). This mutant was tested for growth defects in LB medium at both 26 and 37°C, but exhibited wild-type growth in both conditions (data not shown). The effect of a sif15 mutation on virulence was tested by competition infections with wild-type (JB580v) bacteria. When BALB/c mice were inoculated intraperitoneally the sif15 mutant was attenuated compared to JB580v, suggesting that Sif15 is necessary during systemic infections. The average competitive index of the sif15 mutant during such infections was 0.08. Interestingly, when BALB/c mice were inoculated intragastrically, the sif15 mutant was not significantly attenuated. The average competitive index of the sif15 mutant in the Peyer's patches after oral inoculation was 0.27. These results suggest that the sif15 gene product is not necessary during initial colonization of the Peyer's patches but plays an important role during systemic infection.
DISCUSSION
In this study we were able to identify a number of putative systemic virulence factors after in vivo enrichment for active Y. enterocolitica genes. The experiments were absolutely dependent on the ability to provide a strong enrichment for Cam-resistant bacteria during a systemic infection (i.e., colonization of the spleen). This was accomplished in the BALB/c mouse model for Y. enterocolitica after intraperitoneal inoculation and the administration of chloramphenicol-succinate. After two passages through mice, a 32-fold increase in the Cam-resistant Y. enterocolitica population was detected in the spleen. This protocol provided the enrichment necessary to identify cat fusions that were active in vivo from an existing fusion library.
Because genes that are expressed preferentially in vivo are more likely to encode virulence factors, it was necessary to classify the enriched fusion-containing isolates according to their in vitro expression. Of those tested, 3% (256 isolates) were unable to grow in the presence of Cam in vitro. Other applications of the IVET technique found that a similar fraction of the total isolates contained fusions that were only expressed in vivo (18, 23, 28, 31, 32, 54). The 256 isolates represented 31 separate allelic groups. For each, the sequence of the chromosomal DNA fused to cat was determined and analyzed to identify homologous sequences.
For sixteen sif-cat fusions, a partial gene was fused to the cat gene that encodes a gene product with homology to a known protein. Based on partial protein homology, the sif genes encode proteins involved in general physiology, transcription regulation, and other unknown functions. The expression of seven known proteins (encoded by sif1, sif2, sif3, sif6, sif11, sif15, and sif16) during systemic infection was confirmed by competition assays. While IVET studies in other organisms have also identified in vivo-induced genes that encode known proteins, none encoded proteins homologous to those encoded by the sif alleles.
Some of the sif homologs were previously shown to be important for full virulence of Y. enterocolitica and other organisms. sif3 (rffG) and sif10 (manB) encode proteins important for lipopolysaccharide (LPS) and O-antigen biosynthesis, respectively. Both LPS and O-antigen are necessary for virulence of Y. enterocolitica as well as other enteric pathogens (44, 49, 55). Signature-tagged mutagenesis in Y. enterocolitica also identified manB as important for virulence (17). sif9 (fyuA), encoding an iron siderophore precursor protein, is found in the Y. enterocolitica pathogenicity island of the more virulent serotypes (48). This gene was previously found to be active during colonization of the Peyer's patches by Y. enterocolitica (54). sif16 is homologous to gacA, which encodes a transcriptional regulator of Pseudomonas syringae (26). For that organism, GacA is necessary for the formation of lesions on bean plants (52).
Sif1 shows homology to SitC of S. epidermidis. This protein was first identified as a 32-kDa cell envelope protein that was present in high amounts after growth of Staphylococcus epidermidis in human peritoneal dialysis fluid and not after growth under laboratory conditions (45). The gene encoding the 32-kDa protein was cloned and found to be part of an operon consisting of sitABC that encodes an ABC transporter (11). Based on homology to known proteins, SitA is an ATPase, SitB is a cytoplasmic protein, and SitC is a lipoprotein component of the ABC transporter that also has homology to adhesins. In S. epidermidis, the specificity of this transporter is not clear; both Mn2+ and Fe3+ seem to inhibit expression of the sitABC genes. The expression of this type of transporter in vivo is likely to benefit the bacterium, since the acquisition of cations, such as manganese, iron, and zinc, under such growth-limiting conditions is necessary.
Sif2 shows homology to LepA of E. coli, which is predicted to act as a membrane-bound GTPase (35). It is clear that LepA is found in the membrane and binds GTP; however, the physiological role for LepA remains unknown. LepA has been postulated to have a negative effect on translation, based on the observation that increasing LepA levels leads to a two- to threefold decrease in the cellular content of elongation factor G (53). Homologs of lepA are present in a number of bacterial species and the nematode Caenorhabditis elegans. In E. coli, lepA is the promoter-proximal gene in an operon with the gene encoding signal peptidase I (lepB) (34). However, that gene organization may not be conserved, since in Bacillus subtilis lepA is the promoter-proximal gene in an operon with hemN (25). The presence of lepA in a number of different organisms suggests that its product has an important cellular function. Eukaryotic GTPases are involved in a number of cellular processes: protein translocation, vesicle transport, hormonal responses, neurotransmission, nutrient sensing, cellular differentiation, and cell cycle regulation (33). Similar processes may be important for colonization in vivo.
Sif15 is a putative outer membrane protein, based on its homology to proteins in the databases. Sif15 shows 69% amino acid identity to proteins of unknown function encoded by several S. enterica serovars, including Typhi, Typhimurium, and Paratyphi. Sif15 shows a lower (20%) amino acid identity to a protein found in Helicobacter pylori whose function is also unknown. One very interesting feature of Sif15 is that no homolog is found in Y. pestis. Since we have found a sif15 mutant to be attenuated during systemic infections, the further characterization of this gene and its gene product is a priority. Preliminary results suggest that Sif15 contributes to the dissemination of Y. enterocolitica and the mortality seen in the BALB/c mouse model (A. S. Gort and V. L. Miller, unpublished results).
One common argument against the use of IVET to identify putative virulence factors is that a large fraction of the genes that are active in vivo may only be important for bacterial growth, so-called housekeeping genes. However, the identification of such genes allows determination of the metabolic and biosynthetic pathways that are necessary for bacterial growth during infection. For example, genes involved in methionine and biotin biosynthesis (sif5 and sif8) and anaerobic respiration (sif4) were identified in this enrichment for active genes. MetL is a bifunctional enzyme that acts at two steps during the conversion of oxaloacetate to homoserine, a precursor to threonine, lysine, and methionine. Therefore, the activity of this gene during a systemic infection may indicate a growth-limiting concentration of one or more of these amino acids or another need for homoserine. The activity of frdB in vivo suggests the bacteria are found in an oxygen-limiting environment, as FrdB is a component of fumarate reductase, an anaerobic respiratory enzyme. Information about the physiology of bacteria growing in vivo is particularly hard to obtain. Since very little is known about the conditions in vivo, it is very difficult to speculate which genes are needed for in vivo growth or to mimic in vivo conditions in the laboratory. The identification of metabolic and biosynthetic genes that are active during infection is therefore important for understanding bacterial physiology and may eventually lead to the identification of new targets for antibacterial therapeutics.
There were cases when the DNA sequence obtained from the sif-cat fusion did not reveal the open reading frame responsible for cat expression. In most of these cases, the cat gene was fused to a region of the Y. enterocolitica chromosome that did not exhibit significant homology to sequences in the databases. Four cat fusions to DNA with no known homology were found to be expressed in vivo (sif21, sif23, sif25, and sif29). These sif alleles are some of the most exciting putative virulence factors indentified, since the genes have not been described previously. None of these sif alleles were identified as putative virulence factors in a previous IVET study or in a signature-tagged transposon mutagenesis study, both done in Y. enterocolitica (17, 54). Many other IVET studies have also identified fusions to sequences that did not have any significant homology to those in the databases. It would be interesting to compare those sequences from the other published screens to see if any are represented in more than one study. Such an effort would require collaboration among several research groups but would allow the classification of each sequence as containing potential virulence factors that are important for a number of bacterial species or virulence factors that are unique to a given bacterium. A smaller number of cat fusions occurred in regions of DNA that did show some homology to the sequence databases; however, the open reading frame responsible for cat expression could not be identified. Two sif alleles of this type (sif18 and sif20) were found to be inactive in vivo. Similar fusions were described in one other IVET report (18), but the significance of these fusions is difficult to interpret.
The Western blot analysis of sif-cat fusions and the examination of lacZ transcriptional fusions revealed the in vitro gene expression patterns for several sif loci. For each tested, only modest differences in gene expression were found in response to pH, temperature, and nutrient availability. This may not be surprising, given that many genes encoding virulence factors are induced in response to numerous complex signals. Often the growth conditions in the laboratory do not sufficiently mimic those in vivo that are required for optimal gene expression. Similar results have been obtained for hre22, which encodes a serine protease, identified in a previous IVET enrichment and later found to be important for Y. enterocolitica virulence (54). Despite testing several growth conditions, in vitro induction of hre22 has not been detected (G. Heusipp and V. Miller, unpublished data). In addition, two of the sif alleles tested are thought to encode transcriptional regulators (sif6 and sif16) whose induced expression may be low and/or may not differ greatly from the uninduced level. Interestingly, cat fusions to sif20 and sif29 were expressed in vivo, as predicted by their ability to outcompete the wild type in the presence of Cam, but were expressed at such low levels in vitro that Western blot analysis was unable to detect any Cat (data not shown).
Many adaptations of the original IVET technique, which enriched for active purA transcriptional fusions by complementation of a purA auxotroph in vivo (31), have been developed. Other gene fusions have been used to enrich for active genes in vivo by complementing riboflavin and cell wall auxotrophs (18, 21). However, for an active infection to occur, a constitutive low level of fusion expression must be present to allow bacterial growth in vivo. Therefore, genes that are tightly repressed under specific conditions may not be enriched in vivo even if the gene is active at another stage of infection. Fusions to a site-specific recombinase have also been used to detect in vivo gene expression through the heritable loss of a marker gene on the bacterial chromosome (5, 46). This allows the detection of transiently expressed fusions whose expression may not be sufficient to complement an auxotroph in vivo. The ability to enrich for active genes at different infection stages is unique to the cat fusion IVET technique (32). The positive enrichment for bacteria containing active cat fusions in vivo can be initiated at any time by the administration of chloramphenicol-succinate.
This study, along with the work of Young and Miller, represents the first description of using one IVET fusion library to enrich for active genes during two distinct stages of infection in a single model system (54). Early doses of chloramphenicol-succinate successfully enriched for active cat fusions in the Peyer's patches in a previous study, and here we have enriched for active genes during a systemic infection by delaying drug treatment. The fact that the sif15 mutant is not significantly attenuated during colonization of the Peyer's patches suggests that our enrichment for genes that are active during later infection stages was successful. If one compares the Y. enterocolitica hre genes which were enriched in the Peyer's patches to the sif genes identified in this study, only one allele was enriched in both, the fyuA gene. This is surprising, since the same fusion library was used in both studies. One potential explanation is that the hre genes were identified after oral inoculation, whereas the sif genes were identified after intraperitoneal inoculation. This difference in inoculation method is likely to alter the conditions encountered by the bacteria and may affect the enrichment. Although the saturation of the fusion library was not determined in either study, in both studies there were cases in which the same fusion was enriched from separate pools. This suggests that both screens were approaching saturation. Nevertheless, the small number of overlapping putative virulence factors implies that the colonization of lymphoid tissues by Y. enterocolitica requires different virulence factors than are needed to develop a systemic infection. Another implication of having two distinct groups of genes that were active in the Peyer's patches and during systemic infection is that the overall metabolism differs significantly between the two infection types. The distinct sets of in vivo-induced genes also hint at the very large number of virulence factors that must be involved in a multistage infection. Unfortunately, the model systems used to study many different bacterial pathogens are limited to one infection stage and thus may lead to simplification of the pathogen's life cycle. Analysis of the IVET studies performed in Y. enterocolitica suggests that each stage of infection, or even each tissue, may represent a unique environment to the bacterial pathogen, adaptation to which is likely to require careful transcriptional regulation of a large number of genes that are important for virulence.
ACKNOWLEDGMENTS
We thank Glenn Young, Andrew Darwin, and Paula Revell for their experimental and technical advice. Thanks to Andrew Darwin, Steve Gort, Kristin Nelson, and Joe Vogel for critical review of the manuscript prior to submission. We are grateful to Krisitin Nelson for her assistance in generating the lacZ fusion strains.
This work was supported by a National Institutes of Health Infectious Disease Training Grant (A.S.G.), an NSRA postdoctoral fellowship (A.S.G.), and research grant AI42736 (V.L.M.).
REFERENCES
- 1.Badger J L, Miller V L. Expression of invasin and motility are coordinately regulated in Yersinia enterocolitica. J Bacteriol. 1998;180:793–800. doi: 10.1128/jb.180.4.793-800.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumler A J, et al. Identification of a new iron regulated locus of Salmonella typhi. Gene. 1996;183:207–213. doi: 10.1016/s0378-1119(96)00560-4. [DOI] [PubMed] [Google Scholar]
- 3.Bottone E J. Yersinia enterocolitica: a panoramic view of a characteristic microorganism. Crit Rev Microbiol. 1977;5:211–241. doi: 10.3109/10408417709102312. [DOI] [PubMed] [Google Scholar]
- 4.Butler T. Plague and other Yersinia infections. New York, N.Y: Plenum Medical Book Company; 1983. [Google Scholar]
- 5.Camilli A, Beattie D T, Mekalanos J J. Use of genetic recombination as a reporter of gene expression. Proc Natl Acad Sci USA. 1994;91:2634–2638. doi: 10.1073/pnas.91.7.2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camilli A, Mekalanos J J. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carniel E, Guilvout I, Prentice M. Characterization of a large chromosomal “high pathogenicity island” in biotype 1B Yersinia enterocolitica. J Bacteriol. 1996;178:6743–6751. doi: 10.1128/jb.178.23.6743-6751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carniel E, Mazigh D, Mollaret H H. Expression of iron-regulated proteins in Yersinia species and their relation to virulence. Infect Immun. 1987;55:277–280. doi: 10.1128/iai.55.1.277-280.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter P B. Oral Yersinia enterocolitica infection of mice. Am J Pathol. 1975;81:703–705. [PMC free article] [PubMed] [Google Scholar]
- 10.Carter P B. Pathogenicity of Yersinia enterocolitica for mice. Infect Immun. 1975;11:164–170. doi: 10.1128/iai.11.1.164-170.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cockayne A, Hill P J, Powell N B L, Bishop K, Sims C, Williams P. Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. Infect Immun. 1998;66:3767–3774. doi: 10.1128/iai.66.8.3767-3774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cornelis G, Laroche Y, Balligand G, Sory M-P, Wauters G. Y. enterocolitica, a primary model for bacterial invasiveness. Rev Infect Dis. 1987;9:64–87. doi: 10.1093/clinids/9.1.64. [DOI] [PubMed] [Google Scholar]
- 13.Cornelis G R. Yersinia pathogenicity factors. Curr Top Microbiol Immunol. 1994;192:243–263. doi: 10.1007/978-3-642-78624-2_11. [DOI] [PubMed] [Google Scholar]
- 14.Cornelis G R, Boland A, Boyd A P, Geuigen C, Iriate M, Neyt C. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornelis G R, Wolf-Watz H. The Yersinia Yop virulon: a bacterial system for subverting eukaryotic cells. Mol Microbiol. 1997;23:861–867. doi: 10.1046/j.1365-2958.1997.2731623.x. [DOI] [PubMed] [Google Scholar]
- 16.Cover T L, Aber R C. Yersinia enterocolitica. N Engl J Med. 1989;321:16–24. doi: 10.1056/NEJM198907063210104. [DOI] [PubMed] [Google Scholar]
- 17.Darwin A J, Miller V L. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol Microbiol. 1999;32:51–62. doi: 10.1046/j.1365-2958.1999.01324.x. [DOI] [PubMed] [Google Scholar]
- 18.Fuller T E, Shea R J, Thacker B J, Mulks M H. Identification of in vivo induced genes in Actinobacillus pleuropneumoniae. Microbiol Pathol. 1999;27:311–327. doi: 10.1006/mpat.1999.0309. [DOI] [PubMed] [Google Scholar]
- 19.Guilvout I, Mercereau-Puijalon O, Bonnefoy S, Pugsley A P, Carniel E. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to AngR of Vibrio anguillarium and belong to a family of proteins involved in nonribosomal peptide synthesis. J Bacteriol. 1993;175:5488–5504. doi: 10.1128/jb.175.17.5488-5504.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haas R, Kahrs A F, Facius D, Allmeier H, Schmitt R, Meyer T F. TnMax—a versatile mini-transposon for the analysis of cloned genes and shuttle mutagenesis. Gene. 1993;130:23–31. doi: 10.1016/0378-1119(93)90342-z. [DOI] [PubMed] [Google Scholar]
- 21.Handfield M, Schwiezer H P, Mahan M J, Sanschagrin F, Hoang T, Levesque R C. ASD-GFP vectors for in vivo expression technology in Pseudomonas aeruginosa and other gram-negative bacteria. BioTechniques. 1998;24:261–264. doi: 10.2144/98242st02. [DOI] [PubMed] [Google Scholar]
- 22.Heesemann J, Hanke K, Vocke T, Saken E, Rakin A, Stojilkovic I, Berner R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron-repressible outer membrane protein of 65,000 Da and pestisin sensitivity. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 23.Heithoff D M, Conner C P, Hanna P C, Julio S M, Hentschel U, Mahan M J. Bacterial infection as assessed by in vivo gene expression. Proc Natl Acad Sci USA. 1997;94:934–939. doi: 10.1073/pnas.94.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrero M. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Homuth G, Heinemann M, Zuber U, Shumann W. The genes lepA and hemN form a bicistronic operon in Bacillus subtilis. Microbiology. 1996;142:1642–1649. doi: 10.1099/13500872-142-7-1641. [DOI] [PubMed] [Google Scholar]
- 26.Hrabak E M, Willis D K. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isberg R R, Voorhis D L, Falkow S. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell. 1987;50:769–778. doi: 10.1016/0092-8674(87)90335-7. [DOI] [PubMed] [Google Scholar]
- 28.Kiliç A O, Herzberg M C, Meyer M W, Zhao X, Tao L. Streptococcal reporter gene-fusion vector for identification of in vivo expression genes. Plasmid. 1999;42:67–72. doi: 10.1006/plas.1999.1408. [DOI] [PubMed] [Google Scholar]
- 29.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R−M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 30.Lowe A M, Beattie D T, Deresiewicz R L. Identification of novel staphylococcal virulence genes by in vivo expression technology. Mol Microbiol. 1998;27:967–976. doi: 10.1046/j.1365-2958.1998.00741.x. [DOI] [PubMed] [Google Scholar]
- 31.Mahan M J, Slauch J M, Mekalanos J J. Selection of bacterial virulence genes that are specifically induced in host tissues. Science. 1993;259:686–688. doi: 10.1126/science.8430319. [DOI] [PubMed] [Google Scholar]
- 32.Mahan M J, Tobias J W, Slauch J M, Hanna P C, Collier R J, Mekalanos J J. Antibiotic-based selection for bacterial genes that are specifically induced during infection of a host. Proc Natl Acad Sci USA. 1995;92:669–673. doi: 10.1073/pnas.92.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.March P E. Membrane-associated GTPases in bacteria. Mol Microbiol. 1992;6:1253–1257. doi: 10.1111/j.1365-2958.1992.tb00845.x. [DOI] [PubMed] [Google Scholar]
- 34.March P E, Inouye M. Characterization of the lep operon of Escherichia coli. J Biol Chem. 1985;260:7206–7213. [PubMed] [Google Scholar]
- 35.March P E, Inouye M. GTP-binding membrane protein of Escherichia coli with sequence homology to initiation factor 2 and elongation factors Tu and G. Proc Natl Acad Sci USA. 1985;82:7500–7504. doi: 10.1073/pnas.82.22.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 37.Miller V L, Falkow S. Evidence for two genetic loci from Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulaton of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pepe J C, Badger J L, Miller V L. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol Microbiol. 1994;11:123–135. doi: 10.1111/j.1365-2958.1994.tb00295.x. [DOI] [PubMed] [Google Scholar]
- 40.Rainey P B. Singe step conjugative cloning of bacterial gene fusions involved in microbe-host interactions. Mol Gen Genet. 1997;256:2271–2276. doi: 10.1007/s004380050548. [DOI] [PubMed] [Google Scholar]
- 41.Revell P A, Miller V L. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol Microbiol. 2000;35:677–685. doi: 10.1046/j.1365-2958.2000.01740.x. [DOI] [PubMed] [Google Scholar]
- 42.Schmiel D H, Young G M, Miller V. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar operon. J Bacteriol. 2000;182:2314–2320. doi: 10.1128/jb.182.8.2314-2320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmiel D H, Wagar E, Karamanou L, Weeks D, Miller V L. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect Immun. 1998;66:3941–3951. doi: 10.1128/iai.66.8.3941-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skurnik M, Venho R, Bengoechea J, Moriyon I. The lipopolysaccharide outer core of Yersinia enterocolitica serotype O:3 is required for virulence and plays a role in outer membrane integrity. Mol Microbiol. 1999;31:1443–1462. doi: 10.1046/j.1365-2958.1999.01285.x. [DOI] [PubMed] [Google Scholar]
- 45.Smith D G E, Wilcox M H, Williams P, Finch R G, Denyer S P. Characterization of cell envelope proteins of Staphylococcus epidermidis cultured in human periotoneal dialysate. Infect Immun. 1991;59:617–624. doi: 10.1128/iai.59.2.617-624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Staib P, Kretschmar M, Nichterlein T, Kohler G, Michel S, Hof H, Hacker J, Morschhauser J. Host-induced, stage-specific virulence gene activation in Candida albicans during infection. Mol Microbiol. 1999;32:533–546. doi: 10.1046/j.1365-2958.1999.01367.x. [DOI] [PubMed] [Google Scholar]
- 47.Stewart G S A B, Lubinsky-Mink S, Jackson C G, Cassel A, Kuhn J. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid. 1986;15:172–181. doi: 10.1016/0147-619x(86)90035-1. [DOI] [PubMed] [Google Scholar]
- 48.Straley S C, Perry R D. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 1995;3:310–317. doi: 10.1016/s0966-842x(00)88960-x. [DOI] [PubMed] [Google Scholar]
- 49.Van Den Bosch L, Manning P A, Morona R. Regulation of O-antigen chain length is required for Shigella flexneri virulence. Mol Microbiol. 1997;23:765–775. doi: 10.1046/j.1365-2958.1997.2541625.x. [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Lory S, Ramphal R, Jin S. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol Microbiol. 1996;22:1005–1012. doi: 10.1046/j.1365-2958.1996.01533.x. [DOI] [PubMed] [Google Scholar]
- 51.Wang J, Mushegian A, Lory S, Jin S. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc Natl Acad Sci USA. 1996;93:10434–10439. doi: 10.1073/pnas.93.19.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willis D K, Hrabak E M, Rich J J, Barta T M, Lindow S E, Panopoulos N J. Isolation and characterization of a Pseudomonas syringae pv. syringae mutant deficient in lesion formation on bean. Mol Plant-Microbe Interact. 1990;3:149–156. [Google Scholar]
- 53.Yaskowiak E S, March P E. Small clusters of divergent amino acids surrounding the effector domain mediate the varied phenotypes of EF-G and LepA expression. Mol Microbiol. 1995;15:943–953. doi: 10.1111/j.1365-2958.1995.tb02363.x. [DOI] [PubMed] [Google Scholar]
- 54.Young G M, Miller V L. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol Microbiol. 1997;25:319–328. doi: 10.1046/j.1365-2958.1997.4661829.x. [DOI] [PubMed] [Google Scholar]
- 55.Zhang L, Radziejewshka-Lebrecht J, Krajewska-Pietrasik D, Toivanen P, Skurnik M. Molecular and chemical characterization of lipopolysaccharide O-antigen and its role in the virulence of Yersinia enterocolitica serotype O:8. Mol Microbiol. 1997;23:63–76. doi: 10.1046/j.1365-2958.1997.1871558.x. [DOI] [PubMed] [Google Scholar]