Abstract
The hydrolysis of urea by ureases of oral bacteria in dental plaque can cause a considerable increase in plaque pH, which can inhibit the development of dental caries. There is also indirect evidence that urea metabolism may promote the formation of calculus and that ammonia release from urea could exacerbate periodontal diseases. Actinomyces naeslundii, an early colonizer of the oral cavity and a numerically significant plaque constituent, demonstrates comparatively low levels of urease activity on isolation, so this organism has not been considered a major contributor to total oral urease activity. In this study it was observed that urease activity and urease-specific mRNA levels in A. naeslundii WVU45 can increase up to 50-fold during growth under nitrogen-limiting conditions. Using primer extension analysis, a putative, proximal, nitrogen-regulated promoter of the A. naeslundii urease gene cluster was identified. The functionality and nitrogen responsiveness of this promoter were confirmed using reporter gene fusions and 5′ deletion analysis. The data indicated that regulation of urease expression by nitrogen availability in A. naeslundii may require a positive transcriptional activator. Plaque bacteria may experience nitrogen limitation when carbohydrates are present in excess. Therefore, based on the results of this study and in contrast to previous beliefs, strains of A. naeslundii may have the potential to be significant contributors to total plaque ureolysis, particularly during periods when there is an increased risk for caries development.
Urease is a multisubunit enzyme that catalyzes the hydrolysis of urea to ammonia and carbonic acid, causing a net increase in the environmental pH. The three structural subunits of urease, α, β, and γ, are encoded by the ureC, ureB, and ureA genes, respectively (30). Four additional proteins, encoded by ureD, -E, -F, and -G, catalyze the incorporation of Ni2+ into the active site of urease, a process that is required for the production of a catalytically active urease (30). In bacteria, the enzyme subunit and accessory genes are usually arranged in operons. Some bacterial urease clusters contain additional genes, such as ureR, which is involved in the regulation of urease expression in members of the family Enterobacteriaceae (15, 32); ureHI, which are believed to encode a nickel transporter in a thermophilic Bacillus sp. (25); and ureI of Helicobacter pylori and Streptococcus salivarius, which may be involved in urea transport at low environmental pH (46).
In prokaryotes, ureolysis can provide a source of assimilable nitrogen (13), it can protect the organisms against lethal acidification (41), and in some bacteria, it can support ATP synthesis driven by the gradients established by ammonia generation (43). Accordingly, urease expression in bacteria is often regulated in response to environmental parameters such as nitrogen availability and pH (30). Control of urease expression by nitrogen availability occurs primarily at the level of transcription and can involve the action of either positive (NAC) (11) or negative (G1nR) regulatory molecules (50). The low-pH-dependent expression of urease in S. salivarius is negatively regulated, but the responsible trans-acting factor(s) has not yet been identified (7).
Bacterial ureases are implicated in the pathogenesis of a number of human clinical conditions, including gastritis and peptic ulcer formation, pyelonephritis, and urolithiasis (29). In the oral cavity, ammonia produced via the enzymatic hydrolysis of salivary urea appears to an important factor in inhibition of the development of dental caries (24). There is some suggestive evidence that ureolyis may be a factor in promoting calculus formation (17) and may contribute to periodontal disease progression by enhancing inflammatory processes and impairing key host cell functions (19). However, a role for urea breakdown in calculus formation or periodontal disease has not been conclusively demonstrated. In spite of the presumed importance of ureolysis in oral health and disease, the organisms that are responsible for the bulk of urea hydrolysis in natural human dental plaque have not been unequivocally established, and molecular aspects of oral ureolysis have only recently begun to be established (8).
Strains of Actinomyces naeslundii genospecies 1 are predominant members of supragingival and subgingival dental plaque (4, 45), and they routinely demonstrate low levels of urease activity on isolation (36, 47). The urease gene cluster of A. naeslundii WVU45 (ATCC 12104) comprises seven contiguously arranged open reading frames (ORFs), which demonstrate significant nucleotide and deduced amino acid sequence homologies to the ureA-G genes from other bacteria (31). A putative proximal promoter for the A. naeslundii urease gene cluster was identified by primer extension at 66 bp 5′ to ureA (31). The nucleotide sequence of this promoter region does not conform to established standards for bacterial promoters, supporting the notion that Actinomyces species may possess distinct types of promoter sequences, perhaps due to their unusually high (68%) G+C DNA content (52).
Comparison of the physiologic properties of the wild-type organism with those of an otherwise isogenic, urease-deficient A. naeslundii strain has demonstrated that a primary function of urease in this organism can be to provide ammonia, which is efficiently utilized as a nitrogen source (31). This knowledge led to the hypothesis that urease expression in A. naeslundii may be induced under conditions of limited nitrogen availability. This hypothesis was directly tested in this study by comparing the levels of urease activity and urease-specific mRNA present in A. naeslundii cells growing under nitrogen-limiting conditions to those in cells growing in the presence of a variety of nitrogen sources. Once the validity of the hypothesis was established, the nitrogen-regulated promoter of the urease genes was identified and partially characterized. The significance of regulation of urease expression by nitrogen availability in A. naeslundii is discussed in the context of the oral environment and of oral health.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and reagents.
A. naeslundii WVU45 (ATCC 12104), a genospecies 1 strain, was the focus of this study. A. naeslundii strains were maintained on brain heart infusion (BHI) agar (Difco Laboratories, Detroit, Mich.), supplemented with antibiotics, if necessary, and organisms on solid medium were grown anaerobically in a Gas-Pak System (Becton Dickinson, Cockeysville, Md.) for 2 to 3 days at 37°C. Batch cultures of A. naeslundii were grown in a semidefined medium (Actinomyces defined medium [ADM]) (3) modified to contain either 0.05% (ADM/4) or 0.025% (ADM/8) casamino acids as the principal nitrogen source at 37°C in a 5% CO2 atmosphere. Escherichia coli strains were grown in Luria broth (37) with aeration. The antibiotics used in this study were ampicillin (100 μg/ml), kanamycin (Km, 50 μg/ml), streptomycin (Str, 50 μg/ml), and chloramphenicol (3 μg/ml). A. naeslundii strains carrying chloramphenicol acetyltransferase (CAT) gene fusions (AN70CAT1, AN150CAT8, AN220CAT11, ANC220CAT18, and ANCAT25) were grown to in ADM/8 or ADM/8 supplemented with 2% tryptone (Difco Laboratories) and also containing Km or Str. All chemical reagents and antibiotics were obtained from Sigma Chemical Co. (St. Louis, Mo.).
DNA manipulations.
Electrotransformation of E. coli cells was conducted as described by Sambrook et al. (37) using a CellPorator E. coli pulser (Gibco-BRL) at the medium setting. Transformants were identified on selective agar and screened using a rapid boiling method as described by Sambrook et al. (37). Plasmid DNA to be used for subcloning or nucleotide sequence analysis was extracted from E. coli using the QIAprep Spin Plasmid kit (Qiagen, Inc., Chatsworth, Calif.). Restriction and DNA-modifying enzymes were obtained from Life Technologies Inc. (LTI; Bethesda, Md.), U.S. Biochemicals (Cleveland, Ohio), or New England Biolabls (Beverly, Mass.). Transfer of exogenous DNA into A. naeslundii was performed as described by Yeung and Kozelsky (53). Rapid screening of the transformants was performed by plasmid isolation from 1.5-ml cultures in BHI broth, using the protocol of Anderson and McKay (1).
Amplification of the promoter region of the ure cluster (GenBank accession no. AF056321) for subsequent fusion to a reporter gene was achieved by PCRs (100 μl) containing 0.6 μg of chromosomal DNA, 50 pmol of each primer, 200 μM deoxynucleoside triphosphates, 2 mM MgSO4, 1 μl of Perfect Match Enhancer (Stratagene, La Jolla, Calif.), and 2 U of Vent polymerase (New England Biolabs). The reaction was carried out at 94°C for 5 min, then five cycles of 94°C for 1 min, 45°C for 30 s, and 72°C for 30 s, followed by 25 cycles of 94°C for 1 min, 55°C for 30 s, and 72°C for 30 s, and finally 72°C for 5 min. The following oligonucleotides were used as primers for the amplification: P+66 (5′-GGTGAGGTGCATAGATCTTCTCCCAG-3′), P-70 (5′-GGAAACCGATGGATCCGAAGCACC-3′), P-150 (5′-CCTTTCCTGGATCCCGACCCCG-3), and P-232 (5′-CCGTCTCAGGATCCGTCAGGGC-3′). Restriction sites (underlined) were introduced into these primers to facilitate subsequent manipulations. Nucleotide sequence analysis was performed using the Ladderman dideoxy sequencing method (Takara Shuzo Co., Otsu, Japan) or the Sequenase version 2.0 kit (U.S. Biochemicals). Primers used were oligonucleotides (18 to 22 bases) complementary to sequences of the A. naeslundii urease locus and of the E. coli cat gene obtained from LTI. Sequencing reactions were labeled with [α-35S]dATP (New England Nuclear, Boston, Mass.).
RNA isolation and analysis.
Total RNA was extracted from exponentially growing A. naeslundii WVU45 cells by the protocol of Putzer et al. (33) as modified by Chen et al. (8). Primer extensions were performed as described previously (31) using the 20-mer PE1 (5′-GCGGCGACGACGATGAGTAG-3′), which is complementary to the nucleotide sequence between positions 31 and 50 of the ureA structural gene. Slot blot analysis of total RNA was performed as described by Sambrook et al. (37) using a slot blot manifold (LTI). The membranes were prehybridized for 4 h and hybridized overnight in 7% sodium dodecyl sulfate (SDS)–1 mM EDTA–1% bovine serum albumin (BSA)–0.1 mg of fish sperm DNA per ml–250 mM potassium phosphate (pH 7.5) at 60°C. DNA probes were labeled with [α-32P] dCTP (New England Nuclear) using the Random Primers kit (LTI). After hybridization, the membranes were washed once in Church buffer (9) containing 5% SDS, 1 mM EDTA, 0.5% BSA, and 40 mM potassium phosphate buffer (pH 7.5) at 60°C and then washed four times in 1% SDS–1 mM EDTA–40 mM potassium phosphate (pH 7.5) at 60°C. Densitometric analysis was performed using an IS1000 digital imaging system (Alpha Innotech, San Leandro, Calif.).
Urease enzyme assays.
To measure urease activity, cells were collected by centrifugation at 2,900 × g and washed once in 10 mM sodium phosphate buffer (pH 7.0). Cells were then resuspended in 1 mM sodium phosphate buffer (pH 7.0) and incubated at 37°C in a reaction mixture containing 50 mM potassium phosphate buffer (pH 6.0) and 50 mM urea. The amount of ammonia released was quantitated with the Sigma ammonia color reagent using ammonium sulfate as the standard. Urease specific activity was expressed as nanomoles of ammonia produced per minute per milligram of cell dry weight.
Analysis of cat gene fusion strains.
Cells from an early-exponential-phase (optical density at 600 nm [OD600] ≅ 0.2 to 0.3) culture were washed with 10 mM Tris-HCl (pH 7.8) and resuspended in 0.5 ml of the same buffer on ice. The concentrated cell suspensions were mixed with an equal volume of glass beads (0.1-mm diameter) and subjected to mechanical disruption by homogenization in a Bead Beater for two 30-s pulses at 4°C, with a 2-min incubation on ice between the pulses. The CAT specific activity of the cell-free extracts was determined using the spectrophotometric method of Shaw (40). One unit of CAT activity was defined as the amount of enzyme required to acetylate 1 nanomole of chloramphenicol per minute. Total protein content of the cell lysates was measured using the Bio-Rad protein assay (Bio-Rad, Hercules, Calif.) with BSA as the standard.
RESULTS
Identification of possible nitrogen sources for A. naeslundii WVU45.
We have previously demonstrated that buffered ADM supplemented with ammonia, urea, or an amino acid mixture (casamino acids) could comparably support the growth of A. naeslundii WVU45 cells (31). In order to identify additional nitrogen sources for this organism, A. naeslundii WVU45 cells were cultured in ADM containing 0.05% casamino acids (ADM/4) and supplemented with amino acids that have been shown to be preferred nitrogen sources for microorganisms (Table 1). The medium was buffered to neutrality by the addition of 35 mM potassium phosphate (pH 7.5). As shown in Table 1, cultures supplemented with glutamine or asparagine achieved final turbidities comparable to those of cultures that were supplemented with ammonia, urea, or casamino acids, indicating that glutamine and asparagine can be utilized by A. naeslundii as nitrogen sources. ADM supplemented with arginine or histidine appeared to support the growth of A. naeslundii but only to a small extent, while aspartic and glutamic acids and lysine were not utilized as nitrogen sources by this organism (Table 1).
TABLE 1.
Growth of A. naeslundii WVU45 on ADM/4 supplemented with various nitrogen sourcesa
| Supplementary nitrogen source | Final OD600 at supplement concn (mM):
|
Final pH | ||||
|---|---|---|---|---|---|---|
| 0 | 1.25 | 2.5 | 5 | 10 | ||
| Arginine | 0.43 | 0.5 | 0.55 | 0.6 | 0.64 | 7.0 |
| Asparagine | 0.43 | 1.15 | 1.27 | 1.29 | 1.29 | 5.0–5.5 |
| Aspartate | 0.43 | 0.46 | 0.48 | 0.49 | 0.46 | 6.5–7.0 |
| Glutamine | 0.43 | 0.98 | 1.17 | 1.13 | 1.17 | 5.0–6.0 |
| Glutamate | 0.43 | 0.46 | 0.48 | 0.47 | 0.46 | 6.5–7.0 |
| Histidine | 0.43 | 0.56 | 0.56 | 0.55 | 0.61 | 6.5 |
| Lysine | 0.43 | 0.46 | 0.47 | 0.46 | 0.46 | 7.0 |
| NH4Cl | 0.43 | 0.95 | 1.16 | 1.25 | 1.34 | 5.0–6.0 |
| Urea | 0.43 | 1.17 | 1.36 | 1.35 | 1.49 | 5.5 |
Cells were grown in ADM/4 supplemented with the indicated concentrations of amino acids, ammonium chloride, or urea to stationary phase as detailed in Materials and Methods. The pH of the culture fluid was measured when the cells were harvested.
Effects of nitrogen availability on urease levels in A. naeslundii.
A. naeslundii WVU45 was grown in ADM containing 0.025% casamino acids (ADM/8) in the presence or absence of additional nitrogen sources (Table 2). The concentration of glucose was 0.5% so cells would not become limited for carbohydrate. Exponentially growing cultures (OD600 ≅ 0.2 to 0.3 for cultures without a supplementary nitrogen source and OD600 ≅ 0.2 to 0.4 for nitrogen-supplemented cultures) were harvested and assayed for urease activity as described in Materials and Methods. Although buffers were not included in the culture medium, the pH of all cultures at this specific growth stage did not differ significantly and was greater than 6.0 in all cases. As shown in Table 2, urease activity of A. naeslundii cells grown in the presence of any added nitrogen source was lower than the activity of cells grown under nitrogen-limiting conditions. Repression of urease appeared to be more significant in the presence of casamino acids or tryptone. However, tryptone was able to repress urease activity almost completely, i.e., to the levels usually observed in rich medium (0.5 to 1.5 nmol/min/mg dry cell weight), such as BHI or tryptone (3%)–yeast extract (0.5%) medium. The data demonstrated that urease activity in A. naeslundii is regulated in response to the availability of readily utilized nitrogen sources.
TABLE 2.
Urease activity in A. naeslundii WVU45 cells grown on various nitrogen sourcesa
| Medium | Urease sp act (nmol/mm/mg) | SD |
|---|---|---|
| ADM/8 (0.025% Casamino Acids) | 61.1 | 11.95 |
| ADM/4 (0.05% Casamino Acids) | 31.7 | 15.3 |
| ADM/8 + 25 mM ammonium chloride | 25.2 | 9.37 |
| ADM/8 + 50 mM ammonium chloride | 18.1 | 1.1 |
| ADM/8 + 25 mM asparagine | 32.0 | 7.15 |
| ADM/8 + 50 mM asparagine | 29.56 | 4.86 |
| ADM/8 + 25 mM glutamine | 23.49 | 10.58 |
| ADM/8 + 50 mM glutamine | 21.4 | 2.6 |
| ADM/8 + 25 mM urea | 40.6 | 10.48 |
| ADM/8 + 2% Casamino Acids | 11.3* | 5.99 |
| ADM/8 + 2% tryptone | ND† |
ND, none detected [<1.5 nmol of urea hydrolyzed min−1 (mg of cell dry weight)−1]. ∗, value differs significantly from that for cells cultivated in 0.025% Casamino Acids; † , value differs significantly from that for cells cultivated in 0.05% Casamino Acids.
Regulation of urease activity by nitrogen availability in A. naeslundii occurs at the level of transcription.
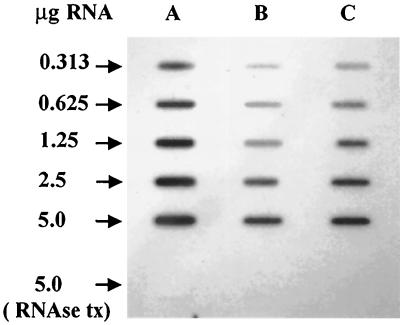
Total RNA was isolated from A. naeslundii cells grown in ADM/8, ADM/8 supplemented with 2% tryptone, and ADM/8 supplemented with 2% casamino acids. All cultures were harvested during exponential growth (OD600 ≅ 0.25) and had comparable pH values (6.2 to 6.6). Serial dilutions of RNA were subjected to slot blot analysis using a probe composed of portions of ureA and ureC and the entire ureB gene. As demonstrated in Fig. 1, significantly less ure mRNA was present in cells grown in the presence of either casamino acids or tryptone compared to cells grown under nitrogen-limiting conditions (unsupplemented ADM/8). In accordance with the observed urease activity levels (Table 2), the lowest amounts of ure-specific RNAs were present in cells grown in tryptone. These data demonstrated that regulation of urease expression by nitrogen availability in A. naeslundii occurs in large part at the level of transcription.
FIG. 1.
Slot blot analysis of total RNA from A. naeslundii WVU45 cells grown under (A) nitrogen limitation in ADM/8 or under nitrogen excess in (B) ADM/8 supplemented with 2% tryptone or (C) ADM/8 supplemented with 2% casamino acids. The blot was probed with a 0.7-kbp DNA fragment containing the 3′ region of ureA, all of ureB, and the 5′ region of ureC. RNase tx, samples were treated with RNase before being applied to the membrane.
Identification of a nitrogen-regulated promoter of the urease gene cluster of A. naeslundii.
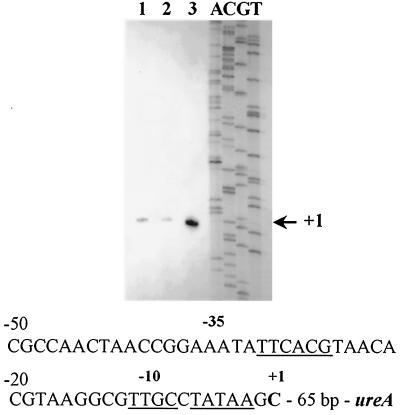
A transcription initiation site for the urease gene cluster located 66 bases 5′ to ureA was previously mapped by primer extension, using RNA from cells growing in BHI medium (31). In order to determine whether this putative promoter was nitrogen regulated or if alternative transcription initiation sites were utilized under nitrogen-limiting conditions, primer extension analysis was performed using RNA from cells grown in either plain ADM/8 or ADM/8 supplemented with tryptone or casamino acids, as described above. A single transcription initiation site was observed in all three reactions, corresponding to the same cytosine residue 66 bp 5′ to ureA that was identified previously in cells grown in BHI (Fig. 2). The primer extension products shown in Fig. 2 were generated from equal amounts of total RNA and radiolabeled primer PE1 under identical reaction conditions. However, significantly lower amounts of the product were present in cells grown under conditions of nitrogen excess compared to those grown under nitrogen-limiting conditions. The relative amounts of mRNA under each condition correlated well with the data obtained from urease activity assays and RNA slot blot analysis.
FIG. 2.
Primer extension analysis of total RNA from A. naeslundii WVU45 cells grown under conditions of nitrogen excess (lane 1, ADM/8 supplemented with 2% casamino acids; lane 2, ADM/8 supplemented with 2% tryptone) or nitrogen limitation (lane 3, ADM/8). The nucleotide sequence of the promoter region immediately 5′ to the transcription initiation site is indicated. DNA sequences that demonstrate significant homologies to consensus recognition sequences for RNA polymerase associated with ς70 or ς54 are underlined.
Functional characterization of the nitrogen-regulated promoter of the A. naeslundii urease gene cluster.
To demonstrate the functionality and nitrogen responsiveness of this promoter region, transcriptional fusions to a promoterless cat gene were constructed on plasmid pJRD215, a low-copy-number, broad-host-range plasmid (14) known to replicate in Actinomyces spp. (53). The constructs were introduced into A. naeslundii WVU45, and the levels of CAT activity expressed in the transformed strains under conditions of nitrogen limitation and nitrogen excess were measured. To generate the reporter gene constructs, a 297-bp region extending from −232 to +65 bases relative to the transcripition initiation site (up to the start codon for ureA) was amplified by PCR as described in Materials and Methods. Two deletions of this region were also amplified by the same method: one extending from ureA (+65) to −150, (215 bp) and one from ureA (+65) to −70 (135 bp). Each of the amplified fragments included the putative ribosome-binding site for ureA and contained a BamHI restriction site on the 5′ end and a BglII site on the 3′ end. The fragments were cloned in the appropriate orientation into the BamHI site of pCW24 (8), which carries a cat gene lacking a promoter and ribosome-binding site. Subsequently, BamHI-HindIII fragments containing the urease promoter-cat fusions were cloned into the corresponding sites of pJRD215, and the resulting recombinant plasmids were introduced into A. naeslundii WVU45 as described in Materials and Methods. Transformants were selected on BHI agar containing Km (50 μg/ml) and Str (50 μg/ml). Plasmid DNA was reisolated from the transformants, and restriction mapping confirmed that the fusions were intact.
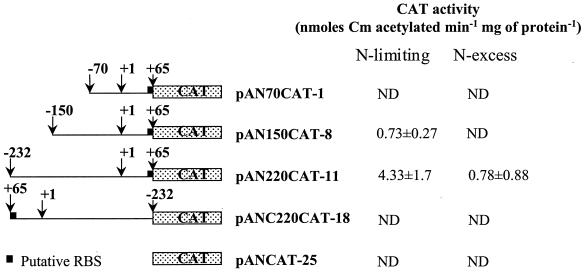
A. naeslundii WVU45 cells containing the recombinant plasmids were grown in ADM/8 or ADM/8 supplemented with 2% tryptone in the presence of Km (50 μg/ml). Cell-free extracts were prepared from exponentially growing cultures (OD600 = 0.2 to 0.3) and assayed for CAT activity as described in Materials and Methods. As shown in Fig. 3, the largest construct, pAN220CAT-11, which extends 232 bp 5′ from the transcriptional start site, expressed CAT activity under nitrogen-limiting conditions (ADM/8). However, CAT activity was reduced 4- to 10-fold in cells growing in the presence of tryptone. The two smallest constructs (pAN150CAT-8 and pAN70CAT-1), which extend to −150 and −70 from the transcriptional start site, respectively, expressed very low (pAN150CAT-8) or no (pAN70CAT-1) CAT activity under any conditions. These results confirm that the nitrogen-regulated promoter of the A. naeslundii urease is located in this region and that expression of urease from this particular promoter under nitrogen-limiting conditions may require the action of a positive regulatory protein.
FIG. 3.
Reporter gene constructs and corresponding CAT activity levels under conditions of nitrogen limitation (ADM/8) and nitrogen excess (ADM/8 plus 2% tryptone). ND, none detected (<0.25 nmol of chloramphenicol (Cm) acetylated min−1 mg of protein−1. RBS, ribosome-binding site.
DISCUSSION
Natural dental plaque demonstrates significant ureolytic activity (42), but the organisms that are primarily responsible for this activity and the extent to which particular species contribute to total plaque ureolysis have not been established. Strains of A. naeslundii genospecies 1 routinely demonstrate low levels of urease activity when assayed in vitro, and for that reason these organisms have not been considered major contributors to total plaque ureolysis. However, A. naeslundii is generally considered a fastidious organism, and it is routinely cultivated in rich medium. Urease expression is highly repressed under such conditions, as demonstrated in this study. Notably, A. naeslundii is one of the first organisms to colonize the oral cavity (16), and it constitutes a major proportion of supragingival and subgingival dental plaque (4, 45). Given its abundance in plaque and the demonstration that relatively high levels of urease can be expressed by this organism under particular environmental conditions, the contribution of A. naeslundii to total plaque ureolysis may have been substantially underestimated.
Most of the available evidence suggests that carbohydrates rather than nitrogen sources are the limiting nutrient for bacteria in dental plaque, although some controversy exists around this subject (12, 21, 44). However, during periods of dietary intake of carbohydrate, oral bacteria may experience nitrogen limitation. Those periods are also characterized by increased acid production from glycolysis and, thus, increased cariogenic activity. Induction of urease expression by A. naeslundii during conditions of limited nitrogen availability may be a mechanism that ensures the availability of adequate levels of urease to protect the cell from acidification and allow the bacteria to scavenge additional nitrogen sources (31). At the same time, the alkalinization of dental plaque by ureolysis would inhibit demineralization of dental enamel, which occurs in acidic environments. Therefore, the potential for increased production of urease by A. naeslundii during periods of carbohydrate excess could deter the development of dental caries.
The observation that A. naeslundii is able to use asparagine as a nitrogen source was not surprising. A number of investigators have reported that Actinomyces viscosus, which has since been reclassified as A. naeslundii genospecies 2, consumed all of the available asparagine when growing in a defined medium in continuous culture under glucose limitation (20, 35, 44). Since asparagine can be converted to succinate by A. viscosus with concomitant generation of ATP (5), it had been hypothesized that asparagine was utilized by A. viscosus for energy generation. Since the nitrogen source in these previous experiments consisted exclusively of individual amino acids, asparagine was probably used as the preferred nitrogen source and was therefore consumed first. In this study, asparagine could be utilized by A. naeslundii as a nitrogen source, consistent with previous findings. Previous work also indicated that A. viscosus probably could not utilize glutamine as a nitrogen source, at least not nearly as effectively as it uses asparagine (20). In contrast, the two amino acids appear to be good nitrogen sources for A. naeslundii and were used as efficiently as ammonia or urea. Assimilation of nitrogen from glutamine may represent another important biochemical difference between A. naeslundii genospecies 1 and 2.
Tryptone and casamino acids are both rich nitrogen sources, and both are excellent for supporting the growth of A. naeslundii. However, tryptone repressed urease expression more efficiently than did casamino acids. The explanation for this observation probably lies in the fact that tryptone consists mainly of small peptides, whereas casamino acids is a mixture of individual amino acids resulting from acid hydrolysis of caseins. It is likely that A. naeslundii possesses oligopeptide transport systems analogous to the ABC-type transporters identified in a number of organisms (22, 23) and therefore is able to transport peptides more efficiently than individual amino acids. More rapid uptake and hydrolysis of peptides than of amino acids would not necessarily affect the final growth yield in each medium, but it could result in greater intracellular amino acid pools being available in the tryptone-grown cells, leading to tighter repression of urease in cells growing on tryptone compared to those growing on casamino acids.
The demonstration that urease expression in A. naeslundii is controlled primarily by nitrogen availability does not preclude the possibility that additional factors may also play a role. For example, we have some preliminary evidence from batch-grown cells that growth rate or growth stage may affect urease expression in A. naeslundii. Upregulation of urease expression at high growth rates has also been observed in S. salivarius (7). The effects of growth rate and possibly pH on urease expression by A. naeslundii should ideally be studied in a continuous-culture system. Although we have made numerous attempts, A. naeslundii forms tenacious biofilms on the chemostat surfaces, so establishing reliable, homogeneous, steady-state cultures has not been possible. Other investigators (20) have reported similar difficulties, and at least in our experience, this problem cannot be overcome by the use of different types of growth media. Additionally, although our data indicate that regulation of urease expression by nitrogen availability in A. naeslundii occurs to a great extent at the level of transcription, we cannot exclude the possibility that additional factors, such as posttranscriptional or posttranslational modifications, may also be involved.
As was mentioned in the introduction, regulation of urease gene expression by nitrogen availability in gram-negative organisms is mediated through the ntr regulatory cascade and requires activation by NAC, which acts at a region immediately upstream to the −10 concensus sequence of ς70 promoters (11). The nitrogen-regulated urease promoter of A. naeslundii contains two sequences with significant homology to the −10 and −35 consensus binding sites for RNA polymerase associated with the vegetative sigma factors ς70 and ςA. However, it is highly improbable that these sequences function as recognition sites for A. naeslundii RNA polymerase due to their inappropriate spacing in relation to the transcription initiation site. In addition, the region immediately upstream of the TATAA sequence does not contain a consensus binding site for NAC (ATAN9TAT) (26). The actual −10 sequence of the A. naeslundii urease promoter is similar to the −12 consensus binding site for RNA polymerase associated with ς54, which is frequently involved in the transcription of nitrogen-regulated genes (27), but no homology is present to this class of promoters in the respective −24 region. Activation of transcription from nitrogen-regulated promoters by RNA polymerase associated with ς54 requires activation by the phosphorylated form of NtrC. No sequences with similarity to the NtrC consensus binding sequence (GCACN5TGGTGC) (34, 38) appear to be present in the A. naeslundii urease promoter region. Overall, it does not seem likely that regulation of the expression of the A. naeslundii urease by nitrogen availability involves a mechanism analogous to the ntr/NAC regulatory system, since, with the exception of an NtrC homologue in Bacillus subtilis (6, 18), we know of no other evidence of a classical Ntr pathway or of an alternative global regulatory system in gram-positive bacteria (28). In B. subtilis, urease transcription is activated by TnrA and repressed by GlnR in response to low and high nitrogen availability, respectively (50). GlnR also regulates transcription of the glnA gene, encoding glutamine synthetase, in response to nitrogen availability in B. subtilis (39) and in Streptomyces coelicolor (48, 49), which is more closely related to A. naeslundii. The putative nitrogen-regulated promoter of the A. naeslundii urease, however, does not contain sequences with significant homology to the consensus binding sequence of GlnR/TnrA (TGTNAN7TNACA) (51).
To our knowledge, this is the first study to utilize reporter genes in A. naeslundii, although we subsequently were able to use cat to study fructosyltransferase gene expression (2). The levels of CAT expression in A. naeslundii observed in this study were relatively low but repeatable and consistent with mRNA and urease activity levels under each set of conditions. Low levels of CAT expression may be attributable to codon utilization differences between A. naeslundii, which has a very high (68%) G+C DNA content, and the cat gene (45%; GenBank accession no. J01841). Since no recA-deficient A. naeslundii strain is available, transient integration of the plasmid carrying the gene fusions into the chromosome of A. naeslundii WVU45 could also potentially occur, resulting in alterations in the plasmid copy number and juxtaposition of deleted upstream regions. However, plasmid DNA could be consistently recovered from the recombinant strains, and complete loss of CAT activity was observed in the deletion derivatives, arguing that the potential transient interaction of the homologous DNA in the gene fusion constructs with the chromosome had no detectable impact on the outcome of the experiments.
The results of this study provide evidence that a functional promoter is included in the 150-bp region upstream of the transcription initiation site that was mapped by primer extension (31). In addition, this region appears to contain the cis elements involved in the regulation of this promoter by nitrogen availability. The fact that the smallest construct, which extends only to −70 from the transcription initiation site, expressed no CAT activity under any conditions suggests that expression from this promoter requires the action of a positive regulatory protein(s). The DNA-binding site for this molecule, according to these data, should then be located between −70 and −150. The involvement of a transcriptional activator in the expression of the A. naeslundii urease in response to nitrogen availability is in accord with the previously described examples of other nitrogen-regulated urease promoters (10). However, these data do not preclude the possibility that more than one transcription factor regulates the expression of the A. naeslundii urease.
This study has provided novel information about the regulation of a key enzymatic activity of an abundant oral microorganism. A. naeslundii is considered to play major roles in the homeostasis of sub- and supragingival oral biofilms, and ureolysis appears to be a major metabolic activity controlling biofilm stability and influencing oral health status. This work contributes to a small but growing body of knowledge about the molecular biology of Actinomyces, and it has established methodology for more detailed studies on gene regulation and nitrogen metabolism in these organisms. In addition, the finding that A. naeslundii may have a greater capacity to produce urease, and thus contribute to the pH-moderating activity of oral biofilms, suggests the potential for a greater involvement of A. naeslundii in prevention of caries.
ACKNOWLEDGMENTS
We acknowledge the late Maria Yeung (University of Texas Health Science Center at San Antonio) for plasmid pJRD215 as well as for advice and encouragement.
This study was supported by National Institute of Dental and Craniofacial Research grants RO1 DE10362 and T32 DE07165.
REFERENCES
- 1.Anderson P G, McKay L L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983;46:549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergeron L J, Morou-Bermudez E, Burne R A. Characterization of the fructosyl transferase gene of Actinomyces naeslundii WVU45. J Bacteriol. 2000;182:3649–3654. doi: 10.1128/jb.182.13.3649-3654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowden G H, Hardie J M, Fillery E D. Antigens from Actinomyces species and their value in identification. J Dent Res. 1976;55:A192–A204. doi: 10.1177/002203457605500112011. [DOI] [PubMed] [Google Scholar]
- 4.Bowden G H, Hardie J M, Slack G L. Microbial variations in approximal dental plaque. Caries Res. 1975;9:253–277. doi: 10.1159/000260162. [DOI] [PubMed] [Google Scholar]
- 5.Brown A T, Breeding L C. Carbon dioxide metabolism by Actinomyces viscosus: pathways for succinate and aspartate production. Infect Immun. 1980;28:82–91. doi: 10.1128/iai.28.1.82-91.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calogero S, Gardan R, Glaser P, Schweitzer J, Rapoport G, Debarbouille M. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J Bacteriol. 1994;176:1234–1241. doi: 10.1128/jb.176.5.1234-1241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y M, Burne R A. Analysis of Streptococcus salivarius urease expression using continuous chemostat culture. FEMS Microbiol Lett. 1996;135:223–229. doi: 10.1111/j.1574-6968.1996.tb07993.x. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y M, Weaver C A, Mendelson D R, Burne R A. Transcriptional regulation of the Streptococcus salivarius 57.I urease operon. J Bacteriol. 1998;180:5769–5775. doi: 10.1128/jb.180.21.5769-5775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins C M, D'Orazio S E. Bacterial ureases: structure, regulation of expression and role in pathogenesis. Mol Microbiol. 1993;9:907–913. doi: 10.1111/j.1365-2958.1993.tb01220.x. [DOI] [PubMed] [Google Scholar]
- 11.Collins C M, Gutman D M, Laman H. Identification of a nitrogen-regulated promoter controlling expression of Klebsiella pneumoniae urease genes. Mol Microbiol. 1993;8:187–198. doi: 10.1111/j.1365-2958.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- 12.Cowman R A, Fitzgerald R J, Perrella M M, Cornell A H. Human saliva as a nitrogen source for oral streptococci. Caries Res. 1977;11:1–8. doi: 10.1159/000260242. [DOI] [PubMed] [Google Scholar]
- 13.Cruz-Ramos H, Glaser P, Wray J L V, Fisher S H. The Bacillus subtilis ureABC operon. J Bacteriol. 1997;179:3371–3373. doi: 10.1128/jb.179.10.3371-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davison J, Heusterpreute M, Hevalier M, Vinh H-T, Brunel F. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene. 1987;51:275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- 15.D'Orazio S E, Collins C M. The plasmid-encoded urease gene cluster of the family Enterobacteriaceae is positively regulated by UreR, a member of the AraC family of transcriptional activators. J Bacteriol. 1993;175:3459–3467. doi: 10.1128/jb.175.11.3459-3467.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellen R P. Establishment and distribution of Actinomyces viscosus and Actinomyces naeslundii in the human oral cavity. Infect Immun. 1976;14:1119–1124. doi: 10.1128/iai.14.5.1119-1124.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Epstein S R, Mandel I, Scoop I W. Salivary composition and calculus formation in patients undergoing hemodialysis. J Periodontol. 1980;51:336–338. doi: 10.1902/jop.1980.51.6.336. [DOI] [PubMed] [Google Scholar]
- 18.Gardan R, Rapoport G, Debarbouille M. Expression of the rocDEF operon involved in arginine catabolism in Bacillus subtilis. J Mol Biol. 1995;249:843–856. doi: 10.1006/jmbi.1995.0342. [DOI] [PubMed] [Google Scholar]
- 19.Golub L, Borden S, Kleinberg I. Urea content of gingival crevice fluid and its relationship to periodontal disease in humans. J Periodont Res. 1971;6:243–251. [PubMed] [Google Scholar]
- 20.Hamilton I R, Ellwood D C. Carbohydrate metabolism by Actinomyces viscosus growing in continuous culture. Infect Immun. 1983;42:19–26. doi: 10.1128/iai.42.1.19-26.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hotz P, Guggenheim B, Schmid R. Carbohydrates in pooled dental plaque. Caries Res. 1972;6:103–121. doi: 10.1159/000259783. [DOI] [PubMed] [Google Scholar]
- 22.Jenkinson H F, Baker R A, Tannock G W. A binding-lipoprotein-dependent oligopeptide transport system in Streptococcus gordonii essential for uptake of hexa- and heptapeptides. J Bacteriol. 1996;178:68–77. doi: 10.1128/jb.178.1.68-77.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juillard V, Le Bars D, Kunji E R S, Konings W N, Gripon J C, Richard J. Oligopeptides are the main source of nitrogen for Lactococcus lactis during growth in milk. Appl Environ Microbiol. 1995;61:3024–3030. doi: 10.1128/aem.61.8.3024-3030.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kleinberg I. Effect of urea concentration on human plaque pH levels in situ. Arch Oral Biol. 1967;12:1475–1484. doi: 10.1016/0003-9969(67)90183-5. [DOI] [PubMed] [Google Scholar]
- 25.Maeda M, Hidaka M, Nakamura A, Masaki H, Uozumi T. Cloning, sequencing, and expression of thermophilic Bacillus sp. strain TB-90 urease gene complex in Escherichia coli. J Bacteriol. 1994;176:432–442. doi: 10.1128/jb.176.2.432-442.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magasanik B. Regulation of nitrogen utilization. In: Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1344–1356. [Google Scholar]
- 27.Merrick M J. In a class of its own—the RNA polymerase sigma factor ς54 (ςN) Mol Microbiol. 1993;10:903–909. doi: 10.1111/j.1365-2958.1993.tb00961.x. [DOI] [PubMed] [Google Scholar]
- 28.Merrick M J, Edwards R A. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mobley H L, Hausinger R P. Microbial ureases: significance, regulation, and molecular characterization. Microbiol Rev. 1989;53:85–108. doi: 10.1128/mr.53.1.85-108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mobley H L, Island M D, Hausinger R P. Molecular biology of microbial ureases. Microbiol Rev. 1995;59:451–480. doi: 10.1128/mr.59.3.451-480.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morou-Bermudez E, Burne R A. Genetic and physiologic characterization of urease of Actinomyces naeslundii WVU45. Infect Immun. 1999;67:504–512. doi: 10.1128/iai.67.2.504-512.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholson E B, Concaugh E A, Foxall P A, Island M D, Mobley H L. Proteus mirabilis urease: transcriptional regulation by UreR. J Bacteriol. 1993;175:465–473. doi: 10.1128/jb.175.2.465-473.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Putzer H, Gendron N, Grunberg-Manago M. Co-ordinate expression of two threonyl-tRNA synthetase genes in Bacillus subtilis: control by transcriptional antitermination involving a conserved regulatory sequence. EMBO J. 1992;11:3117–3127. doi: 10.1002/j.1460-2075.1992.tb05384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reitzer L J, Magasanik B. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine. In: Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington D.C.: American Society for Microbiology; 1987. p. 302-320. [Google Scholar]
- 35.Rogers A H, de Jong M H, Zilm P S, van der Hoeven J S. Estimation of growth parameters for some oral bacteria grown in continuous culture under glucose-limiting conditions. Infect Immun. 1986;52:897–901. doi: 10.1128/iai.52.3.897-901.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salako N O, Kleinberg I. Incidence of selected ureolytic bacteria in human dental plaque from sites with differing salivary access. Arch Oral Biol. 1989;34:787–791. doi: 10.1016/0003-9969(89)90029-0. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 38.Sasse-Dwight S, Gralla J D. Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc Natl Acad Sci USA. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schreier H J, Brown S W, Hirshci K D, Nomellini J F, Sonenshein A L. Regulation of Bacillus subtilis glutamine synthetase gene expression by the product of the glnR gene. J Mol Biol. 1989;210:51–63. doi: 10.1016/0022-2836(89)90290-8. [DOI] [PubMed] [Google Scholar]
- 40.Shaw W V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1979;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 41.Sissons C H, Hancock E M. Urease activity in Streptococcus salivarius at low pH. Arch Oral Biol. 1993;38:507–516. doi: 10.1016/0003-9969(93)90187-q. [DOI] [PubMed] [Google Scholar]
- 42.Sissons C H, Hancock E M, Cutress T W. The source of variation in ureolysis in artificial plaques cultured from human salivary bacteria. Arch Oral Biol. 1988;33:721–726. doi: 10.1016/0003-9969(88)90005-2. [DOI] [PubMed] [Google Scholar]
- 43.Smith D G, Russell W C, Ingledew W J, Thirkell D. Hydrolysis of urea by Ureaplasma ureolyticum generates a transmembrane potential with resultant ATP synthesis. J Bacteriol. 1993;175:3253–3258. doi: 10.1128/jb.175.11.3253-3258.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Hoeven J S, de Jong M H, Rogers A H, Camp P J M. A conceptual model for the co-existence of Streptococcus spp. and Actinomyces spp. in dental plaque. J Dent Res. 1984;63:389–392. doi: 10.1177/00220345840630030601. [DOI] [PubMed] [Google Scholar]
- 45.van Houte J, Lopman J, Kent R. The predominant cultivable flora of sound and carious human root surfaces. J Dent Res. 1994;73:1727–1734. doi: 10.1177/00220345940730110801. [DOI] [PubMed] [Google Scholar]
- 46.Weeks D L, Eskandari S, Scott D R, Sachs G. A H+-gated urea channel: the link between Helcobacter pylori urease and gastric colonization. Science. 2000;287:482–485. doi: 10.1126/science.287.5452.482. [DOI] [PubMed] [Google Scholar]
- 47.Wijeyeweera R L, Kleinherg I. Arginolytic and ureolytic activities of pure cultures of human oral bacteria and their effects on the pH response of salivary sediment and dental plaque in vitro. Arch Oral Biol. 1989;34:43–53. doi: 10.1016/0003-9969(89)90045-9. [DOI] [PubMed] [Google Scholar]
- 48.Wray L, Atkinson M, Fisher S. Identification and cloning of the glnR locus, which is required for transcription of the glnA gene in Streptomyces coelicolor A3(2) J Bacteriol. 1991;173:7351–7360. doi: 10.1128/jb.173.22.7351-7360.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wray L, Fisher S H. The Streptomyces coelicolor glnR gene encodes a protein similar to other bacterial response regulators. Gene. 1993;130:145–150. doi: 10.1016/0378-1119(93)90359-b. [DOI] [PubMed] [Google Scholar]
- 50.Wray L V, Jr, Ferson A E, Fisher S H. Expression of the Bacillus subtilis ureABC operon is controlled by multiple regulatory factors, inluding CodY, GlnR, TnrA, and Spo0H. J Bacteriol. 1997;179:5494–5501. doi: 10.1128/jb.179.17.5494-5501.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wray L V, Jr, Ferson A E, Rohrer K, Fisher S H. TnrA, a transcriptional factor required for global nitrogen regulation in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:8841–8845. doi: 10.1073/pnas.93.17.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeung M K. Complete nucleotide sequence of the Actinomyces viscosus T14V sialidase gene: presence of a conserved repeating sequence among strains of Actinomyces spp. Infect Immun. 1993;61:109–116. doi: 10.1128/iai.61.1.109-116.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yeung M K, Kozelsky C S. Transformation of Actinomyces spp. by a gram-negative broad-host-range plasmid. J Bacteriol. 1994;176:4173–4176. doi: 10.1128/jb.176.13.4173-4176.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]