Abstract
When Borrelia burgdorferi is transmitted from the tick vector to the mammalian host, the bacterium experiences alterations in its environment, such as changes in temperature and pH. Previously, we observed numerous alterations in the membrane protein profile when B. burgdorferi B31 was grown at pH 7.0 compared to pH 8.0. Here we identify 11 genes localizing to linear plasmids that are up-regulated at pH 7.0 relative to pH 8.0 in vitro. Seven genes (bba03, bba24, bba64, bba66, bbe31, bbj41/bbi39 [encoding products that are 99% identical], and bbk01) were indirectly identified by proteomic analysis of membrane proteins. Another gene, bba36, was identified by screening a B. burgdorferi B31 genomic library with cross-adsorbed hyperimmune rabbit serum. Two additional genes, bba65 and bba73, were identified by Northern blot analysis. Genes bba64, bba65, bba66, bbj41/bbi39, and bba73 are members of paralogous gene family 54, and bbe31 is a member of the closely related paralogous gene family 60. Gene bba24 is part of a bicistronic operon with bba25 that encodes the well-characterized decorin binding proteins A and B. All 11 genes were transcriptionally regulated, yet the degree of pH regulation varied, with some genes more tightly regulated than others. The regions upstream of these pH-regulated genes appeared to be unrelated, yet many contained dyad repeats ranging from 12 to 25 nucleotides in length that may be involved in the regulation of these genes.
Borrelia burgdorferi, the causative agent of Lyme disease, is transmitted to a mammalian host by a tick vector of the Ixodes ricinus complex. During transmission the spirochete encounters fluctuations in growth parameters such as temperature, pH, and available nutrients. In response to its environment, B. burgdorferi is able to regulate several genes and the synthesis of numerous proteins (5, 7, 8, 12, 22, 29, 31, 33, 34, 36, 38). These changes in gene expression are likely to play an important role in adaptation to its environment. The ability of B. burgdorferi to establish an infection in a potential host may rely on its ability to sense and adapt to these changing conditions.
Recently we reported over 37 alterations in the membrane protein profile when cells were grown at different pHs (5). The most striking changes were observed between cultures grown at pH 7.0 and 8.0. This is similar to the pH change encountered by the spirochetes during transmission from mammal to tick vector, respectively. Interestingly, one well-characterized protein, OspC, was observed to dramatically decrease in amount as the pH of the medium was raised to 8.0 (5). OspC synthesis is also influenced by temperature, where the amount of OspC produced is decreased at 23°C relative to 34°C (31); this suggests that ospC is under the coordinate regulation of pH and temperature. These observations correlate well with in vivo studies in which OspC is undetectable on spirochetes in the midguts of unfed ticks (alkaline pH, 23°C) (13, 31) but can be detected in the midguts of fed ticks (31) and within the skin of mammals infected by tick bite (pH 7.0 to 7.4, 34°C) (20, 24, 31, 32).
Using matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometry on proteins separated by two-dimensional nonequilibrinm pH gradient gel electrophoresis (2D-NEPHGE) in concert with immunoblotting, Northern analysis, and the screening of a B. burgdorferi B31 genomic library with cross-adsorbed serum, we have determined the identities of 11 genes that are regulated by the environmental pH. Some of these pH-regulated genes and the proteins they encode have been previously identified and partially characterized, whereas others of the genes appear to encode hypothetical proteins (14). Here we demonstrate that genes bba03, dbpAB (bba24 and bba25), bba36, bba64, bba65, bba66, bba73, bbe31, bbj41/bbi39 (encoding proteins that are 99% identical [see Results]), and bbk01 of B. burgdorferi are regulated in vitro by the environmental pH. Further analysis of the DNA sequences upstream of these genes revealed putative operator-promoter regions consisting of features indicative of regulator binding sites in other organisms. These regions may be involved in the regulation of genes in response to the environmental pH in B. burgdorferi.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Low-passage (<6 passages), infectious B. burgdorferi B31 (4) was grown to mid-log phase (5 × 107 cells per ml) under an atmosphere of 5% CO2 at 35°C in BSK-H medium (Sigma Chemical Co., Saint Louis, Mo.). For pH studies the cells were then concentrated by centrifugation (8,000 × g; 10 min; 24°C) and resuspension in BSK-H. The spirochetes were then inoculated at a final concentration of 107 per ml into BSK-H buffered with 25 mM HEPES and adjusted to pH 7.0 or 8.0 with the addition of either HCl or NaOH. Cells were incubated for 2 to 4 days and were harvested by centrifugation (8,000 × g; 10 min; 4°C) (5). Virulent strains were previously tested in Syrian hamsters as described elsewhere (23). Escherichia coli XL1-Blue MRF′ and XLOLR were obtained from Stratagene (La Jolla, Calif.). E. coli TOP10 was obtained from Invitrogen Carlsbad, Calif.). Transformation- competent E. coli DH5α was purchased from Life Technologies (Grand Island, N.Y.). All E. coli strains were grown in Luria broth (LB) supplemented with the appropriate antibiotic for selection according to the the instructions of the suppliers.
B. burgdorferi B31 genomic library construction and screening.
Genomic DNA from low-passage B. burgdorferi B31 was isolated by pheno-chloroform extraction as described by Marmur (26). A library was constructed by partial digestion of B. burgdorferi genomic DNA with Sau3AI (Promega, Madison, Wis.) and ligated into lambda Zap Express (Stratagene) digested with BamHI. The recombinant DNA was packaged, its titer was determined, and it was rescued as phagemid vector pBK-CMV (Stratagene). Plasmid DNA from recombinant clones was isolated using the Qiagen (Chatsworth, Calif.) plasmid minikit. The average insert size was determined by restriction endonuclease analysis of 20 random recombinant plasmids. DNA fragments were separated by agarose gel electrophoresis (0.8% agarose; 1× Tris-acetate-EDTA buffer; 80 V).
Antiborrelia rabbit serum raised against low-passage (<5 passages) B. burgdorferi B31 was cross-adsorbed with cell lysate from E. coli and virulent B. burgdorferi B31 grown at pH 8.0 using a previously described method (6). This yielded an antiserum that primarily recognized proteins synthesized at pH 7.0 but decreased in synthesis at pH 8.0. The recombinant phage genomic library was diluted and plated on LB per the manufacturer's instructions. The resulting plaques were transferred to nitrocellulose discs (Schleicher & Schuell, Keene, N.H.) and probed with the cross-adsorbed serum (1 h; 37°C). The filter discs were washed in Tris-buffered saline (150 mM NaCl in 10 mM Tris-HCl, pH 8.0) with the addition of 0.1% Tween 20 (TBS-T20), probed with goat anti-rabbit serum conjugated with horseradish peroxidase (Sigma Chemical Co.) at a dilution of 1:5,000 in TBS-T20 (45 min; 37°C), and washed in TBS-T20. Immunoreactive recombinant plaques were visualized using an enhanced chemiluminescence kit (Amersham Life Sciences, Inc., Arlington Heights, Ill). DNA inserts from positive clones were sequenced using standard T7 and T3 primers (Geneseek Inc., Lincoln, Nebr.), and positive clones were stored at −80°C in LB containing 25% glycerol.
Gene bba36 with its promoter region was PCR amplified using primers A36.3 and A36.4 (Table 1). The resultant amplicon was TA cloned into pCR2.1-TOPO and transformed into TOP10 One Shot chemically competent E. coli per the instructions of the manufacturer (Invitrogen). This gave rise to construct pCR2.1-A36, which was subsequently transformed into E. coli DH5α.
TABLE 1.
Oligonucleotides (5′ to 3′) used in this study
| Oligonucleotide | Sequence | Application |
|---|---|---|
| A03.1 | CCTAGTTGATGAAGATAGAATTG | bba03 gene-specific probe |
| A03.2 | GTAAGTATTTAGCTCTTCTGG | bba03 gene-specific probe |
| A24.1 | CATGTGGACTAACAGGAGC | bba24 gene-specific probe |
| A24.2 | GTCATCTCTTGTATTCCC | bba24 gene-specific probe |
| A36.1 | CGATGTTAAATCGCTTACAG | bba36 gene-specific probe |
| A36.2 | GTTGCTCAGTGGGGCGTCG | bba36 gene-specific probe |
| A36.3 | GACCCTATTTGTAGTTTTAAAG | Cloning of bba36 with promoter |
| A36.4 | CAAGTTCTCACTAGAACTGC | Cloning of bba36 with promoter |
| A64.1 | CCCAACGCTAATGCCAAC | bba64 gene-specific probe |
| A64.2 | CTGTATATTCGTGGCTGTC | bba64 gene-specific probe |
| A65.1 | GGGATAACGACATTTTTCTCTTG | bba65 gene-specific probe |
| A65.2 | CTTGTTGGATTCGTATACCAC | bba65 gene-specific probe |
| A66.1 | CAAGCAGCACCAAGCCCAC | bba66 gene-specific probe |
| A66.2 | GCTGTCTTGTTGGTTGAC | bba66 gene-specific probe |
| A73.1 | CTAAAGACGAGCAAAAGCGCC | bba73 gene-specific probe |
| A73.2 | GTTCATGCTCGTATCCAACCC | bba73 gene-specific probe |
| E31.1 | GGGTAGCATCATATGAAACG | bbe31 gene-specific probe |
| E31.2 | GGATATAAGTGATTAATCG | bbe31 gene-specific probe |
| J41.1 | GGCAATGTTAATCCAAACG | bbj41 gene-specific probe |
| J41.2 | GAGCTTCCAGGTCTTTACC | bbj41 gene-specific probe |
| K01.5 | GGATAGTGTTCAAGAAGATGGTC | bbk01 gene-specific probe |
| K01.6 | CTCTATTGTTCTTAGGATTCTC | bbk01 gene-specific probe |
| Osp8 | AATTTGGTGCCATTTGAGTC | ospAB gene-specific probe |
| Osp13 | AAGTACGATCTAATTGCAAC | ospAB gene-specific probe |
Isolation and quantitation of protein samples.
Once the spirochetes grew to the desired density (5 × 107 cells per ml), they were harvested by centrifugation (8,000 × g; 10 min; 4°C). The cell pellets were gently rinsed with cold 50 mM NaCl in 20 mM HEPES (pH 7.6) (HEPES buffer), centrifuged a second time, and suspended in HEPES buffer. The cell suspensions were lysed by two passes through a French pressure cell (16,000 lb/in2) (SLM-Aminco, Rochester, N.Y.), and cell debris was removed by centrifugation (10,000 × g; 10 min; 4°C). Total membranes (TM) were separated from the soluble protein fraction by ultracentrifugation (100,000 × g; 1 h; 4°C). The membranes were rinsed once in HEPES buffer to remove residual soluble proteins, pelleted again by ultracentrifugation, and resuspended with the aid of a glass tissue homogenizer (Kontes Glass Co., Vineland, N.J.) in 250 μl of HEPES buffer. Aliquots of cell lysates and rinsed membranes were stored at −20°C. Protein concentrations were determined by a modified Lowry protein assay (25) with bovine serum albumin as a standard.
Sera used for immunoblots.
Hyperimmune rabbit antiserum raised against live, low-passage B. burgdorferi B31 (hyperimmune serum) was produced as previously described (6). Polyclonal serum to P35 was kindly donated by Robert D. Gilmore of the Centers for Disease Control and Prevention in Fort Collins, Colo. Polyclonal sera to DbpA and -B were kindly donated by Mark Hanson of MedImmune, Inc.
Electrophoresis and immunoblotting.
Proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (SDS-PAGE) with an SE600 gel apparatus. (Hoefer Scientific, San Francisco, Calif.). Twenty-five to 35 μg of protein was applied to each lane. 2D-NEPHGE, using a Hoefer SE600 gel apparatus, was performed as previously described (5). Proteins were visualized by staining with the Silver Stain Plus kit (Bio-Rad Laboratories, Hercules, Calif.) or prepared for immunoblotting. Molecular weight standards were purchased from Bio-Rad Laboratories.
For immunoblotting, the proteins were electrophoretically transferred to nitrocellulose (0.45-μm-pore-size Trans-Blot Transfer Medium; Bio-Rad Laboratories) as described by Towbin et al. (37) with a Bio-Rad Trans Blot Cell (30 V; 12 h; 4°C). After transfer, the proteins were visualized with Ponceau red (0.1% Ponceau red dye in 1.0% acetic acid), and the standards were marked. The nitrocellulose membranes were blocked with 5% nonfat dry milk in TBS-T20 (3 h; 24°C), and immune serum diluted either 1:500, 1:1,000, or 1:10,000 in TBS-T20 (primary antibody) was applied to the blot (1 h; 24°C). The blot was washed twice in 100 to 200 ml of TBS-T20 for 10 min to remove residual primary antibody. Secondary antibody (horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse antibody) (Sigma Chemical Co.) was diluted 1:5,000 in TBS-T20 and applied to the blot (45 min; 24°C), followed by three washes with 100 to 200 ml of TBS-T20. Reactive bands were visualized with the enhanced chemiluminescence kit (Amersham Life Sciences, Inc.) in accordance with the manufacturer's specifications. The relative molecular masses of protein bands or spots were estimated by a two-variable statistic linear regression with molecular mass standards purchased from Bio-Rad Laboratories.
MALDI-TOF analysis.
MALDI-TOF mass spectrometry was performed at PerSeptive Biosystems (Framingham, Mass.) using a Voyager DE STR MALDI-TOF S/N 4113 Biospectrometry Workstation (PerSeptive Biosystems) on samples prepared as follows. 2D-NEPHGE was performed on TM protein samples isolated from B31 exposed to medium at pH 7.0 and 8.0 as previously described (5). Gels were stained with silver, and the protein spots were excised with a clean razor blade and shipped to PE Biosystems in 5% acetic acid. At PE Biosystems gel slices were prepared for analysis by destaining with a ferricyanide-thiosulfate solution and washed in 50% acetonitrile containing 25 mM ammonium bicarbonate (pH 8.0) (three times; 15 min each; 24°C). Gel slices were dehydrated in 100% acetonitrile for 10 min, the acetonitrile was removed, and the gel slices were dried under vacuum for 30 min. Samples were rehydrated with sequencing-grade trypsin solution (10 μg/ml in 25 mM ammonium bicarbonate, pH 8.0) and incubated overnight at 37°C. Peptides were extracted with 50% acetonitrile–5% trifluoroacetic acid in distilled H2O and concentrated with a Speed-Vac. Samples were mixed with the matrix α-cyano-4-hydroxycinnamic acid and analyzed by MALDI-TOF. Mass spectrometry profiles were searched against the National Center for Biotechnology Information database using Protein Prospector from University of California at San Francisco.
Northern analysis.
Total RNA was extracted from B. burgdorferi B31 cultures incubated at pH 7.0 and 8.0 using the Ultraspec-II RNA isolation system (Biotecx, Houston, Tex.). RNA was quantitated by absorbance at 260 nm and stored in 50-μl aliquots at −80°C. RNA was denatured with glyoxal and dimethyl sulfoxide for 1 h at 50°C, and 10 μg of total RNA per lane was separated on a 1% (wt/vol) agarose gel in 10 mM NaH2PO4 (pH 7.0) (80 V; 3 h). Separated RNA was transferred to a Hybond N+ nylon membrane using a vacuum blotter system (6,000 Pa, 1 h; 20× SSC) [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], air dried, auto-cross-linked, and stained with methylene blue (0.03% methylene blue in 1.0% acetic acid). Millennium RNA markers (Ambion, Inc., Austin, Tex.) were marked, and 23S and 16S rRNAs were noted as additional internal standards. RNA blots were stored dry in the dark at 24°C until probed.
Radioactive probes for Northern blots were generated using the RadPrime labeling kit (Life Technologies) according to the manufacturer's instructions with [α-32P]dATP (3,000 Ci/mmol) (NEN Life Science Products, Inc., Boston, Mass.) and primers specific for the target gene of interest (Table 1). Target sequences to be used as probes were amplified by PCR using the GeneAmp kit (Perkin-Elmer, Branchburg, N.J.) with genomic DNA as a template. First-round amplicons were purified by the process of agarose gel electrophoresis, excision from the gel, and extraction using the GenElute agarose spin column (Sigma Chemical Co.). PCR was performed a second time with the eluted amplicons as templates. Second-round amplicons were cleaned with the Quick Step PCR purification kit (Edge BioSystems, Gaithersburg, Md.) and stored at −20°C until labeled. RNA blots were placed in a hybridization oven and prehybridized and hybridized with the radiolabeled probes at 55°C in 1% (wt/vol) bovine serum albumin–7% (wt/vol) SDS in 0.5 M sodium phosphate, pH 7.0. Membranes were washed twice with 0.1% SDS in 2.0× SSC (55°C; 10 min.) and then washed again with 0.1% SDS in 0.2× SSC (two times; 55°C; 10 min). The rinsed membranes were then placed on autoradiography film at −70°C. Since ospA expression appears to be unaffected by pH (5), a Northern probe specific for ospA was used as a control to ensure that equivalent amounts of RNA were loaded per lane. Northern blots that had been probed previously were allowed to decay until no signal was detected and were then probed with the radiolabeled ospA fragment generated and labeled as described above. Membranes were treated as described above and placed on film. mRNA intensities and integrated density values were measured using an AlphaImager 2000 digital imaging system (Alpha Innotech Corporation, San Leandro, Calif.). All Northern blotting was performed independently at least twice.
Computer analysis of 5′ flanking regions of the B. burgdorferi B31 pH-regulated loci.
Sequences 200 nucleotides (nt) 5′ of bba03, bba25, bba36, bba64, bba65, bba66, bba73, bbe31, bbj41, and bbk01 were retrieved from the B. burgdorferi B31 genome sequence from The Institute of Genomic Research website (www.tigr.org) (14). The regions upstream of the pH-regulated genes were analyzed for percent identity, dyad repeats, direct repeats, and indirect repeats using Lasergene software (DNASTAR Inc., Madison, Wis.). Sequences were aligned by the Clustal method using Megalign (DNASTAR).
RESULTS
MALDI-TOF mass spectrometry analysis of proteins separated by 2D-NEPHGE.
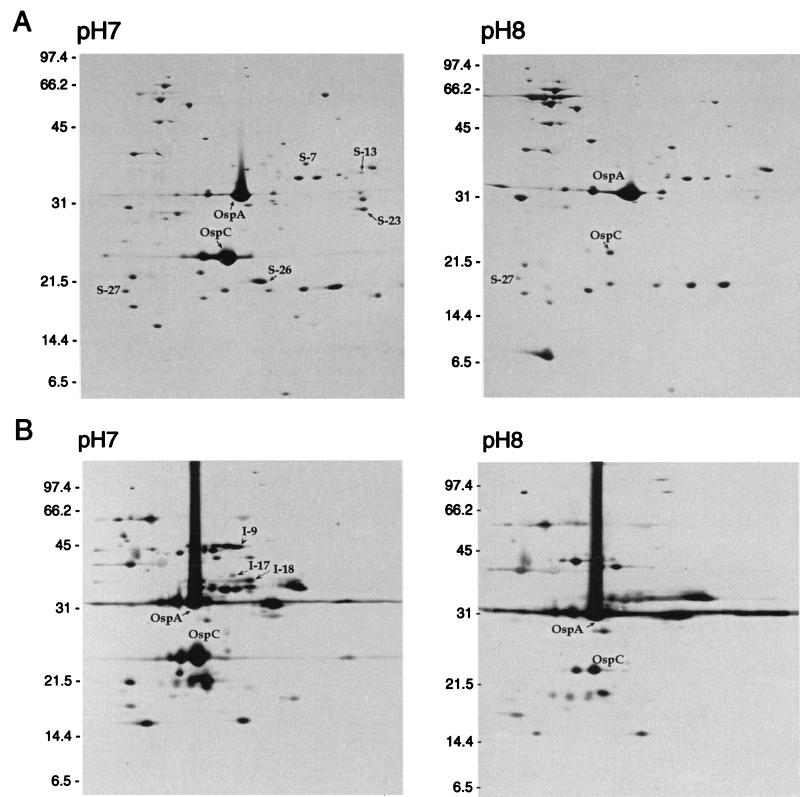
MALDI-TOF analysis was performed on several proteins separated by 2D-NEPHGE that were detected in greater amounts at pH 7.0 than at pH 8.0 (Fig. 1) (5). The report obtained from PE Biosystems indicated with high probability that seven of the peptide mass spectrometry profiles of proteins (spots I-9, I-18, S-7, S-13, S-23, S-26, and S-27 in Fig. 1) matched with gene products localizing to linear plasmid 25 (lp25), lp36, lp38, lp28-4, and lp54 (Table 2). Four of these genes (bba64, bba66, bbi39, and bbj41) were members of paralogous gene family 54, and one gene (bbe31) was a member of paralogous gene family 60. The gene product of bbj41 has 99% identity to that of bbi39 (a member of the same gene family but found on lp28-4). Since distinguishing between these two proteins would be difficult, we refer to the genes as bbj41/bbi39 throughout this paper.
FIG. 1.
Comparison of 2D-NEPHGE membrane protein profiles from B. burgdorferi B31 grown at pH 7.0 and 8.0 by silver staining (A) and by immunoblotting and probing with hyperimmune serum (B). 2D-NEPHGE profiles were reproduced from reference 5. The acidic ends are to the left. OspA and OspC are indicated as reference marks. Molecular mass standards in kilodaltons are indicated on the left of each panel.
TABLE 2.
MALDI-TOF identification of proteins separated by 2D-NEPHGE
| Spot designationa | Protein identity | Molecular mass/pIb | Molecular wt search scorec | Plasmid location |
|---|---|---|---|---|
| S-7 (I-17) | BBK01 | 34.1/9.06 | 3.19 × 104 | lp 36 |
| S-13 | BBJ41/BBI39 | 33.0/9.67 | 1.77 × 103 | lp 38/lp 28-4 |
| S-23 | BBE31 | 28.2/9.47 | 1.22 × 107 | lp 25 |
| S-26 | BBA24 (DbpA) | 21.2/9.24 | 5.42 × 105 | lp 54 |
| S-27 | BBA03 | 19.2/5.27 | 4.72 × 104 | lp 54 |
| I-9 | BBA66 | 45.9/9.37 | 9.29 × 103 | lp 54 |
| I-18 | BBA64 (P35) | 34.4/9.31 | 2.24 × 103 | lp 54 |
Sequence analysis of paralogous gene families 54 and 60 suggested that these two families are closely related, and phylogenetic analysis of the genes or the proteins they encode indicated that there was some overlap between the two putative families (data not shown). Gene bba64 encodes the well-characterized P35 (3, 22, 27, 29), a protein that is produced early during infection (10, 15) and whose expression was reported to be dependent on cell density (22). Genes bba66, bbe31, and bbj41/bbi39 encode lipoproteins with approximate molecular masses of 46, 28, and 33 kDa, respectively, and were identified by sequence analysis of the genome (14).
In addition to genes of families 54 and 60, we identified bba03, bba24, and bbk01 by MALDI-TOF analysis (Table 2). Gene bba03 encodes a putative lipoprotein of unknown function with a molecular mass of 19 kDa. Gene bba24 encodes decorin binding protein A (DbpA), where the gene is cotranscribed with bba25 (DbpB). Both are lipoproteins that have been shown by Western blot analysis to bind decorin (16, 17). Gene bbk01 is a member of gene family 12, which consists of four additional genes. One of these genes, bbg01, is found on lp28-2 and shares >94% sequence identity at the nucleotide level with bbk01. Even though the proteins encoded by bbk01 and bbg01 share 87.9% identity, they are easily distinguished from each other by MALDI-TOF analysis.
Comparison of B. burgdorferi TM samples from pH 7.0 and 8.0 probed with polyclonal sera to P35, DbpA, and DbpB.
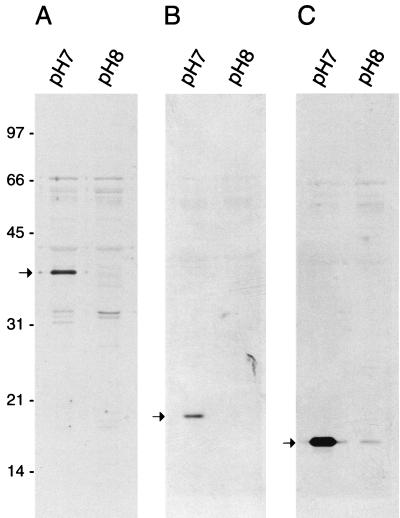
In order to verify the validity of the MALDI-TOF analysis, TM proteins from cultures incubated at pH 7.0 and 8.0 were separated by SDS-polyacrylamide gel electrophoresis and probed with polyclonal antibodies to P35, DbpA, and DbpB (Fig. 2). The anti-P35 polyclonal serum reacted strongly to a band with an approximate molecular mass of 35 kDa (Fig. 2A) that was present in the pH 7.0 lane but undetectable in the pH 8.0 lane. Similarly, anti-DbpA and anti-DbpB strongly recognized bands of the appropriate molecular masses that were again present in greater amounts at pH 7.0 than at pH 8.0 (Fig. 2B and C). This indicated that MALDI-TOF analysis could reliably identify proteins differentially expressed as a result of the altered pH.
FIG. 2.
Immunoblots of equivalent amounts (30 μg per lane) of total membrane proteins from B. burgdorferi grown at pH 7.0 or 8.0 and probed with polyclonal sera to P35 (A), to DbpA (B), and to DbpB (C). The arrow in each panel indicates the presence of an immunoreactive band of the appropriate molecular mass that was detected in cells grown at pH 7.0 but not in cells grown at pH 8.0. Molecular mass standards in kilodaltons are indicated on the left.
Analysis of recombinant clones screened with cross-adsorbed serum.
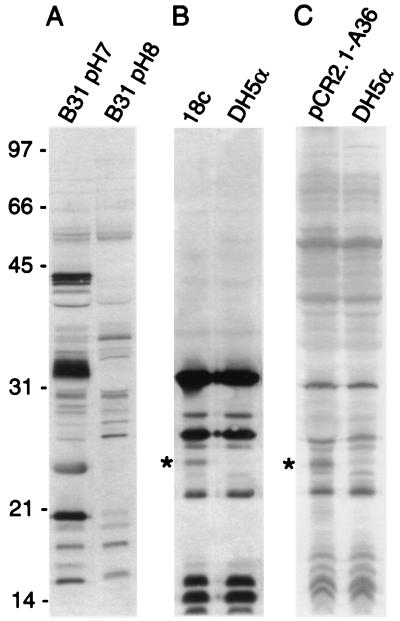
In addition to MALDI-TOF analysis of differentially expressed proteins, serum cross-adsorbed with cell lysates from spirochetes grown at pH 8.0 (Fig. 3A) was used to screen a B. burgdorferi B31 genomic library. Screening of 20,000 plaques of a genomic library yielded only 10 immunoreactive plaques. One recombinant clone, designated 18c, expressed a 25-kDa protein that reacted strongly with the cross-adsorbed serum when the cell lysate was probed by immunoblotting (Fig. 3B). Sequence analysis of the DNA insert and comparison to the B31 genome sequence (14) indicated that recombinant clone 18c contained a 4.6-kb portion of lp54. This 4.6-kb segment included six open reading frames (bba34 to bba39), five of which encode hypothetical proteins and one (bba36) that encodes a putative lipoprotein of approximately 24.2 kDa in molecular mass.
FIG. 3.
Immunoblots of cell lysates from spirochetes grown at pH 7.0 and 8.0 (A), E. coli DH5α recombinant clone 18c with control (B), and E. coil DH5α transformed with pCR2.1-A36 with control (C) probed with hyperimmune serum cross-adsorbed with cell lysates from E. coli and B. burgdorferi B31 grown at pH 8.0. The asterisk indicates the 25-kDa immunoreactive protein identified as the gene product of bba36. Molecular mass standards in kilodaltons are indicated on the left.
We focused on bba36 for two reasons: (i) by MALDI-TOF analysis we had identified several pH-regulated lipoproteins, and (ii) the apparent molecular mass of the bba36 product (24.2 kDa) was extremely similar to that of the heterologous protein produced by clone 18c (25 kDa). Gene bba36 along with 280 nt upstream of the first codon was amplified by PCR from clone 18c and TA cloned into pCR2.1-TOPO. The resultant construct expressed a heterologous protein that was immunoreactive with cross-adsorbed serum and was identical in molecular mass to the protein observed in clone 18c (Fig. 3B and C). This confirmed that gene bba36 encoded the immunoreactive 25-kDa protein observed with the cross-adsorbed serum by immunoblotting.
Northern analysis of pH-regulated genes.
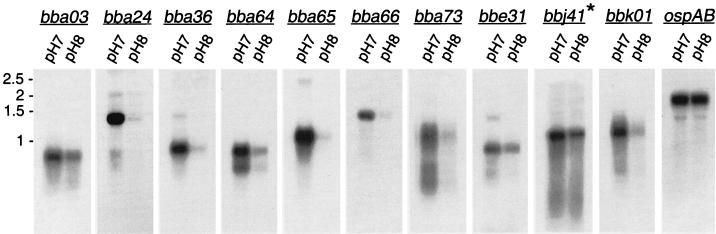
Probes specific for pH-regulated genes indirectly identified by MALDI-TOF analysis were hybridized with total RNA extracted from B. burgdorferi B31 grown in medium at pH 7.0 or pH 8.0 (Fig. 4). Transcripts of all genes appeared to be increased in abundance in RNA isolated from cultures at pH 7.0 compared to pH 8.0. Probes specific for bba66, bba24, and bbk01 strongly hybridized with mRNA bands of the expected size, and by densitometry they were found to be expressed >16-fold higher at pH 7.0 than at pH 8.0 (Fig. 4 and Table 3). Similar results were obtained when mRNA was hybridized with a probe specific for bba25 (data not shown), indicating bba24 and bba25 were cotranscribed in our strain. Probes specific for bba03, bba64, bbe31, and bbj41 also hybridized with mRNA bands of the appropriate size, but they showed only a two- to fivefold decrease in expression at 8.0 relative to pH 7.0 (Fig. 4 and Table 3).
FIG. 4.
Northern analysis of pH-regulated genes. Equivalent amounts of total RNA from B. burgdorferi B31 grown at pH 7.0 or 8.0 were separated, transferred to membranes, and hybridized with radiolabeled DNA probes specific for pH-regulated genes. The asterisk indicates that this probe will also hybridize to bbi39. Hybridization with an ospAB Northern probe served to ensure that equal RNA amounts were loaded per lane. RNA size markers in kilobases are on the left.
TABLE 3.
Integrated density values from the Northern blots in Fig. 4
| Gene | IDVa at pH:
|
|
|---|---|---|
| 7.0 | 8.0 | |
| bba03 | 118,404 | 47,619 |
| bba24 | 44,080 | 464 |
| bba36 | 98,600 | 10,846 |
| bba64 | 137,268 | 35,712 |
| bba65 | 262,200 | 11,400 |
| bba66 | 42,000 | 2,250 |
| bba73 | 168,795 | 13,640 |
| bbe31 | 39,600 | 8,580 |
| bbj41 | 101,640 | 55,440 |
| bbk01 | 116,348 | 6,902 |
| ospAB | 119,340 | 112,320 |
Integrated density value (IDV) = Σ(each pixel value − background).
The expression of bba36, which was identified by cross-adsorbed serum, was also observed to be transcriptionally regulated and by densitometry was expressed approximately ninefold higher at pH 7.0 than at pH 8.0 (Fig. 4 and Table 3). Because four of the six pH-regulated genes identified by MALDI-TOF analysis were members of paralogous gene family 54 or 60, the pH regulation of other members of these families was assessed by Northern blot analysis. Two additional pH-regulated loci (bba65 and bba73) were identified by this method. Genes bba65 and bba73, both members of paralogous gene family 54, were expressed at >23- and >12-fold higher levels, respectively, at pH 7.0 than at pH 8.0 (Fig. 4 and Table 3). A probe specific for ospAB, which is not regulated by pH, displayed no significant difference in expression at pH 7.0 and 8.0 (Fig. 4 and Table 3) and served as a control to ensure that equivalent RNA amounts were loaded.
Computer analysis of the 5′ flanking sequences upstream of pH-regulated loci.
With the exception of bba24 and bba25, the pH-regulated genes identified in this study were all monocistronic as determined by Northern analysis (Fig. 4). We analyzed the regions 200 nt upstream of the first codon of each gene using Lasergene software in order to gain a better understanding of how these particular genes may be regulated. With multiple-sequence alignments of these upstream regions analyzed by the Clustal method, we observed no significant homologies among the 11 pH-regulated loci. Even the sequences upstream of the pH-regulated genes of paralogous family 54 that localize to lp54 (bba64, bba65, bba66, and bba73) displayed little similarity to one another. For example, the 200 nt 5′ of bba64 and bba65 share the greatest identity, at 44.3%, where a comparison of the same region upstream of bba64 and bba73 revealed the lowest sequence identity, at only 29.6%. Interestingly, many of the upstream sequences contained regions of dyad symmetry ranging from 12 to 25 nt in length (Table 4). We could not discern a suitable regulator consensus sequence by comparison of the dyads but presumed that these regions could be involved in the positive or negative control of these genes in response to the environmental pH.
TABLE 4.
Regions of dyad symmetry upstream of some pH-regulated loci
| Gene | Upstream dyada | Location (nt) upstream of the first codon |
|---|---|---|
| bba25 | TTTATTTTATTTTATTT | 55 |
| →← | ||
| bba25 | AATTTAAATTTAA | 25 |
| →← | ||
| bba64 | TTAATAATAATT | 151 |
| →← | ||
| bba64 | ATTAATGGGGTAATTA | 52 (overlapping − 10 region) |
| →← | ||
| bba65 | ACTAAATTTAAATCA | 73 |
| →← | ||
| bba66 | TAATTAATTTTAAATTTTAATTAAT | 69 |
| →← | ||
| bba73 | AATTTTTTTTAA | 22 |
| →← |
Arrows indicate the axis of symmetry.
DISCUSSION
We reported in an earlier study that there are numerous alterations in the membrane protein profile of B. burgdorferi B31 when grown in BSK-H medium at pH 7.0 (the pH of mammals) compared to pH 8.0 (the pH of ticks) (5). Thus far we have identified 11 genes located on linear plasmids in B. burgdorferi B31 that are transcribed in larger amounts by spirochetes grown at pH 7.0 than by those grown at pH 8.0. Seven of the 11 genes (bba03, bba24, bba64, bba66, bbe31, bbj41/bbi39, and bbk01) were identified by MALDI-TOF analysis of membrane proteins separated by 2D-NEPHGE (Fig. 1 and Table 2), while immunoblotting confirmed that DbpB (encoded by bba25) was regulated by pH as well. A ninth gene, bba36, was identified by screening a B. burgdorferi B31 genomic library with cross-adsorbed serum that had been enriched to recognize immunogens expressed at pH 7.0 (Fig. 3). Lastly, two genes (bba65 and bba73) were identified by Northern blot analysis alone (Fig. 4).
MALDI-TOF analysis and Northern blotting suggested that five members of paralogous gene family 54 (bba64, bba65, bba66, bba73, and bbj41/bbi39) were differentially expressed as the pH of the medium was altered from 7.0 to 8.0. Paralogous gene family 54 is composed of one pseudogene and 13 paralogs, most of which are defined as hypothetical proteins with unknown function (14). One gene in particular, bba64, encodes the well-characterized lipoprotein P35 (3, 15, 22). P35, an immunogen often associated with early Lyme disease (10, 15), was previously observed to be synthesized in larger amounts as the culture cell density increased, and the control of its expression has been attributed to the growth phase of the organism (21, 22). We have found that spirochetes inoculated to the same cell density from a single starter culture into medium at either pH 7.0 or 8.0 displayed no apparent change in growth rate, but when cells from both culture conditions were harvested during the same phase of growth (mid-log phase), P35 was detectable in cultures grown at pH 7.0 but not in cultures grown at pH 8.0 (Fig. 2). These experiments strongly suggest that the increase in P35 in stationary-phase cultures (22) may actually be a response to the acidification of the medium, which occurs in standard BSK-H medium as cells enter late log and stationary phases. We have eliminated this pH change due to growth by adding 25 mM HEPES to the medium to allow for increased buffering capacity as the cell density increases.
Northern blots indicated that the seven genes identified by MALDI-TOF analysis (bba03, bba24, bba64, bba66, bbe31, bbj41/bbi39, and bbk01), four of which are members of gene families 54 and 60, were most likely regulated at the level of transcription. Transcripts of these genes were observed in larger amounts at pH 7.0 than at pH 8.0 (Fig. 4). This led to subsequent analysis of other members of paralogous gene families 54 and 60 by Northern blotting. Hence, we identified two additional members of paralogous gene family 54, bba65 and bba73, that were regulated by the in vitro environmental pH (Fig. 4). Interestingly, bba64, bba65, and bba66 were recently observed by Anguita et al. to be up-regulated in mice infected with a clonal isolate of B. burgdorferi N40 (cN40) (1). More importantly, a high-passage isolate of cN40 which was found to have lost the ability to up-regulate these genes during infection also lacked the ability to cause disease in the C3H/HeN mouse model (1). We determined that bba64, bba65, and bba66 were up-regulated at pH 7.0 (consistent with expression in the mammalian environment) yet down-regulated at pH 8.0 (similar to the tick vector environment), suggesting that pH-regulated genes may play a role in adaptation and/or in the manifestation of disease within the mammalian host.
Similar to some members of paralogous gene family 54, DbpA and -B are expressed during mammalian infection and elicit high antibody titers with low-dose inocula of cultured spirochetes (7, 11, 18, 19), but researchers have shown that immunization with DbpA, but not DbpB, confers protection in mice challenged by a needle inoculm (11, 19). By MALDI-TOF, immunoblot, and Northern blot analyses we were able to determine that DbpA and -B (products of bba24 and bba25, respectively) were up-regulated at pH 7.0 compared to pH 8.0. DbpA and -B appear to localize to the outer surface of the cell (16, 18) and are in a bicistronic operon encoded on lp48 in B. burgdorferi N40 (11) but on lp56 in B. burgdorferi B31 (14). Recent evidence suggests that hyperimmune DbpA antiserum is bactericidal in vitro and is effective against a large number of diverse B. burgdorferi isolates. These data indicate a conserved protective epitope within DbpA and have made this immunogenic lipoprotein an attractive alternative vaccine candidate (30). Interestingly, DbpA and -B appear to be coordinately regulated by pH and temperature (7, 33). These observations highlight the importance of the effects of pH and temperature on differential expression in B. burgdorferi and adaptation to the mammalian host.
In concert with MALDI-TOF analysis, a genomic library was screened with cross-adsorbed serum that reacted primarily with immunogens expressed at pH 7.0 and not at pH 8.0 (Fig. 3A). Using this method, we were able to identify an additional pH-regulated gene, bba36. By Northern blot analysis we observed that bba36 was expressed at a ninefold higher level at pH 7.0 than at pH 8.0, and we have evidence that this gene is under the coordinate regulation of temperature and pH (not shown), similar to the case for ospC (5). Gene bba36, like the other pH-regulated genes described in this study, is located on a linear plasmid. However, we have preliminary data that suggest that not all pH-regulated genes are found on linear plasmids and that some may map to circular plasmids (like ospC) and to the chromosome as well (not shown).
Ten of the 11 pH-regulated genes that we have identified either encode or are predicted to encode lipoproteins, yet bbk01 seems to be the exception. Gene bbk01 encodes a protein (BBK01) with an estimated molecular mass of 34 kDa and is one of five genes that make up paralogous gene family 12. BBK01 (spot S-7) was originally identified as a membrane protein (Fig. 1) (5), and a search of the predicted amino acid sequence using a dense alignment surface algorithm (9) identified a putative transmembrane segment at the N terminus, suggesting that BBK01 is an integral membrane protein. The proteins encoded by the remaining members of this gene family (bbg01, bbh37, bbj08, and bb0844) appear to have similar transmembrane motifs, but the extent of their regulation by pH has not been determined.
It was apparent by Northern blot analysis that the degrees of pH regulation observed among the genes identified in this study differed significantly (Fig. 4). Some were highly up-regulated (i.e. bba65), others displayed only moderate up-regulation (e.g., bbe31), and a few presented with a lower level of up-regulation (e.g., bbj41) at pH 7.0 compared to pH 8.0 (Fig. 4 and Table 3). We were curious if any features indicative of regulatory regions could be found upstream of these pH-regulated loci, and if so, how similar were these regions. Computer analysis of approximately 200 nt 5′ to these genes demonstrated that the regions upstream were quite dissimilar even among paralogous genes. This is different from what has been observed for the erp genes, where the regions 5′ to the erp genes share greater than 80% identity (35). We did determine that upstream of bba03, bba25, bba64, bba65, bba66, bba73, and bbe31 were regions of dyad symmetry that ranged from 12 to 25 nt in length. Furthermore, upstream of bba25 and bba64 were two dyads (Table 4).
Not all of the pH-regulated genes we observed had dyad repeats upstream. In other genes, like bbk01 and ospC, we noticed large inverted repeats, and upstream of bba36 we identified an overlapping direct repeat of 22 nt in length. We could not readily identify any similar features 5′ to bbj41, but we did find an inverted repeat 5′ to bbi39. Not surprisingly, bbj41/bbi39 displayed the least difference in the amount of transcript between pH 7.0 and 8.0 (Fig. 4). The role that these features may play in the pH regulation of these genes remains to be determined, but their location just upstream of these pH-regulated genes warrants further investigation.
We have identified in B. burgdorferi B31 11 genes located on linear plasmids that are regulated by the environmental pH, where transcript was more abundant at pH 7.0 than at pH 8.0. Understanding how these and other genes are regulated by environmental cues, such as pH and temperature, will aid in elucidating how this spirochete adapts to the changing environments of the tick vector and the mammalian host. These adaptations that occur as the bacteria are passed from vector to host are likely to be vital in allowing the organism to invade, infect, and cause disease in susceptible hosts. The role that these 11 pH-regulated loci play in the disease process remains to be determined, but considering the recent advancements in Borrelia genetics (2), the resolution of their function in pathogenesis is approaching.
ACKNOWLEDGMENTS
We thank P. Rosa, T. Schwan, P. Stewart, and J. Bono for comments on the manuscript; G. Hettrick and A. Mora for artwork and photography; and R. Gilmore for antibodies to P35.
REFERENCES
- 1.Anguita J, Samanta S, Revilla B, Suk K, Das S, Barthold S W, Fikrig E. Borrelia burgdorferi gene expression in vivo and spirochete pathogenicity. Infect Immun. 2000;68:1222–1230. doi: 10.1128/iai.68.3.1222-1230.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bono J L, Elias A F, Kupko III J J, Stevenson B, Tilly K, Rosa P. Efficient targeted mutagenesis in Borrelia burgdorferi. J Bacteriol. 2000;182:2445–2452. doi: 10.1128/jb.182.9.2445-2452.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunet L R, Sellitto C, Spielman A, Telford S R., III Antibody response of the mouse reservoir of Borrelia burgdorferi in nature. Infect Immun. 1995;63:3030–3036. doi: 10.1128/iai.63.8.3030-3036.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgdorfer W, Barbour A G, Hayes S F, Benach J L, Grunwaldt E, Davis J P. Lyme disease—a tick borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 5.Carroll J A, Garon C F, Schwan T G. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect Immun. 1999;67:3181–3187. doi: 10.1128/iai.67.7.3181-3187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll J A, Gherardini F C. Membrane protein variations associated with in vitro passage of Borrelia burgdorferi. Infect Immun. 1996;64:392–398. doi: 10.1128/iai.64.2.392-398.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassatt D R, Patel N K, Ulbrandt N D, Hanson M S. DbpA, but not OspA, is expressed by Borrelia burgdorferi during spirochetemia and is a target for protective antibodies. Infect Immun. 1998;66:5379–5387. doi: 10.1128/iai.66.11.5379-5387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Champion C I, Blanco D R, Skare J T, Haake D A, Giladi M, Foley D, Miller J N, Lovett M A. A 9.0-kilobase-pair circular plasmid of Borrelia burgdorferi encodes an exported protein: evidence for expression only during infection. Infect Immun. 1994;62:2653–2661. doi: 10.1128/iai.62.7.2653-2661.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 10.Engstrom S M, Shoop E, Johnson R C. Immunoblot interpretation criteria for serodiagnosis of early Lyme disease. J Clin Microbiol. 1995;33:419–427. doi: 10.1128/jcm.33.2.419-427.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng S, Hodzic E, Stevenson B, Barthold S W. Humoral immunity to Borrelia burgdorferi N40 decorin binding proteins during infection of laboratory mice. Infect Immun. 1998;66:2827–2835. doi: 10.1128/iai.66.6.2827-2835.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fikrig E, Feng W, Aversa J, Schoen R T, Flavell R A. Differential expression of Borrelia burgdorferi genes during erythema migrans and Lyme arthritis. J Infect Dis. 1998;178:1198–1201. doi: 10.1086/515684. [DOI] [PubMed] [Google Scholar]
- 13.Fingerle V, Hauser U, Liegl G, Petko B, Preac-Mursic V, Wilske B. Expression of outer surface proteins A and C of Borrelia burgdorferi in Ixodes ricinus. J Clin Microbiol. 1995;33:1867–1869. doi: 10.1128/jcm.33.7.1867-1869.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams M D, Gocayne J, Venter J C, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 15.Gilmore R D, Jr, Kappel K J, Johnson B J. Molecular characterization of a 35-kilodalton protein of Borrelia burgdorferi, an antigen of diagnostic importance in early Lyme disease. J Clin Microbiol. 1997;35:86–91. doi: 10.1128/jcm.35.1.86-91.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo B P, Brown E L, Dorward D W, Rosenberg L C, Hook M. Decorin-binding adhesins from Borrelia burgdorferi. Mol Microbiol. 1998;30:711–723. doi: 10.1046/j.1365-2958.1998.01103.x. [DOI] [PubMed] [Google Scholar]
- 17.Guo B P, Norris S J, Rosenberg L C, Hook M. Adherence of Borrelia burgdorferi to the proteoglycan decorin. Infect Immun. 1995;63:3467–3472. doi: 10.1128/iai.63.9.3467-3472.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagman K E, Lahdenne P, Popova T G, Porcella S F, Akins D R, Radolf J D, Norgard M V. Decorin-binding protein of Borrelia burgdorferi is encoded within a two-gene operon and is protective in the murine model of Lyme borreliosis. Infect Immun. 1998;66:2674–2683. doi: 10.1128/iai.66.6.2674-2683.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanson M S, Cassatt D R, Guo B P, Patel N K, McCarthy M P, Dorward D W, Hook M. Active and passive immunity against Borrelia burgdorferi decorin binding protein A (DbpA) protects against infection. Infect Immun. 1998;66:2143–2153. doi: 10.1128/iai.66.5.2143-2153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmeister E K, Childs J E. Analysis of Borrelia burgdorferi sequentially isolated from Peromyscus leucopus captured at a Lyme disease enzootic site. J Infect Dis. 1995;172:462–469. doi: 10.1093/infdis/172.2.462. [DOI] [PubMed] [Google Scholar]
- 21.Indest K J, Philipp M T. DNA-binding proteins possibly involved in regulation of the post-logarithmic-phase expression of lipoprotein P35 in Borrelia burgdorferi. J Bacteriol. 2000;182:522–525. doi: 10.1128/jb.182.2.522-525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Indest K J, Ramamoorthy R, Sole M, Gilmore R D, Johnson B J, Philipp M T. Cell-density-dependent expression of Borrelia burgdorferi lipoproteins in vitro. Infect Immun. 1997;65:1165–1171. doi: 10.1128/iai.65.4.1165-1171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson R C, Marek N, Kodner C. Infection of Syrian hamsters with Lyme disease spirochetes, J. Clin Microbiol. 1984;20:1099–1101. doi: 10.1128/jcm.20.6.1099-1101.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leuba-Garcia S, Martinez R, Gern L. Expression of outer surface proteins A and C of Borrelia afzelii in Ixodes ricinus ticks and in the skin of mice. Zentbl Bakteriol. 1998;287:475–484. doi: 10.1016/s0934-8840(98)80187-4. [DOI] [PubMed] [Google Scholar]
- 25.Markwell M A, Haas S M, Bieber L L, Tolbert N E. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal Biochem. 1978;87:206–210. doi: 10.1016/0003-2697(78)90586-9. [DOI] [PubMed] [Google Scholar]
- 26.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 27.Oschmann P, Wellensiek H J, Zhong W, Dorndorf W, Pflughaupt K W. Relationship between the Borrelia burgdorferi specific immune response and different stages and syndromes in neuroborreliosis. Infection. 1997;25:292–297. doi: 10.1007/BF01720399. [DOI] [PubMed] [Google Scholar]
- 28.Pappin D J C, Hojrup P, Bleasby A J. Rapid identification of proteins by peptide-mass fingerprinting. Curr Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 29.Ramamoorthy R, Philipp M T. Differential expression of Borrelia burgdorferi proteins during growth in vitro. Infect Immun. 1998;66:5119–5124. doi: 10.1128/iai.66.11.5119-5124.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts W C, Mullikin B A, Lathigra R, Hanson M S. Molecular analysis of sequence heterogeneity among genes encoding decorin binding proteins A and B of Borrelia burgdorferi sensu lato. Infect Immun. 1998;66:5275–5285. doi: 10.1128/iai.66.11.5275-5285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwan T G, Piesman J, Golde W T, Dolan M C, Rosa P A. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc Natl Acad Sci USA. 1995;92:2909–2913. doi: 10.1073/pnas.92.7.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seinost G, Dykhuizen D E, Dattwyler R J, Golde W T, Dunn J J, Wang I N, Wormser G P, Schriefer M E, Luft B J. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect Immun. 1999;67:3518–3524. doi: 10.1128/iai.67.7.3518-3524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skare J T, Foley D M, Hernandez S R, Moore D C, Blanco D R, Miller J N, Lovett M A. Cloning and molecular characterization of plasmid-encoded antigens of Borrelia burgdorferi. Infect Immun. 1999;67:4407–4417. doi: 10.1128/iai.67.9.4407-4417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevenson B, Schwan T G, Rosa P A. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect Immun. 1995;63:4535–4539. doi: 10.1128/iai.63.11.4535-4539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevenson B, Tilly K, Rosa P A. A family of genes located on four separate 32-kilobase circular plasmids in Borrelia burgdorferi B31. J Bacteriol. 1996;178:3508–3516. doi: 10.1128/jb.178.12.3508-3516.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suk K, Das S, Sun W, Jwang B, Barthold S W, Flavell R A, Fikrig E. Borrelia burgdorferi genes selectively expressed in the infected host. Proc Natl Acad Sci USA. 1995;92:4269–4273. doi: 10.1073/pnas.92.10.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallich R, Brenner C, Kramer M D, Simon M M. Molecular cloning and immunological characterization of a novel linear-plasmid-encoded gene, pG, of Borrelia burgdorferi expressed only in vivo. Infect Immun. 1995;63:3327–3335. doi: 10.1128/iai.63.9.3327-3335.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]