ABSTRACT
Data currently generated in the field of nutrition are becoming increasingly complex and high-dimensional, bringing with them new methods of data analysis. The characteristics of machine learning (ML) make it suitable for such analysis and thus lend itself as an alternative tool to deal with data of this nature. ML has already been applied in important problem areas in nutrition, such as obesity, metabolic health, and malnutrition. Despite this, experts in nutrition are often without an understanding of ML, which limits its application and therefore potential to solve currently open questions. The current article aims to bridge this knowledge gap by supplying nutrition researchers with a resource to facilitate the use of ML in their research. ML is first explained and distinguished from existing solutions, with key examples of applications in the nutrition literature provided. Two case studies of domains in which ML is particularly applicable, precision nutrition and metabolomics, are then presented. Finally, a framework is outlined to guide interested researchers in integrating ML into their work. By acting as a resource to which researchers can refer, we hope to support the integration of ML in the field of nutrition to facilitate modern research.
Keywords: machine learning, personalized nutrition, omics, obesity, diabetes, cardiovascular disease, models, random forest, XGBoost
Statement of Significance: Many problems in nutrition are complex, multifactorial, and unlikely to be solved with data analysis methods that have been used traditionally; however, the capabilities of machine learning may be able to. For nutrition researchers to fully capitalize on the types of data that will be generated in the coming years, we provide a guide to machine learning in nutrition for nutrition researchers.
Introduction
There is a high prevalence of nutritionally mediated chronic diseases that have multifaceted origins and require complex and diverse data to be solved. Traditional research applications have approached these questions with focused and mechanistic techniques that may not fully capture the complexity of the interaction between nutrition and disease. Technological and computational advances have recently allowed investigators to utilize high-dimensional data approaches to better understand these diseases and other complex questions. Topical themes, such as obesity (1, 2), omics (3, 4), and the microbiome (5–7), as well as older subjects such as epidemiology, are deriving benefits from these developments (8–10). Due to the increasing complexity of the data generated, new trends in nutrition research, such as precision nutrition (PN) (11) and data-driven disease modeling (12, 13), require an increasing complexity in algorithms to make sense of these data; artificial intelligence (AI) and its subdivision, machine learning (ML), have been important for this.
The terms AI and ML are used interchangeably in some of the literature (13), exposing some of the conceptual confusion that surrounds these topics. The overall goal of AI is to simulate human-like intelligence in a computer system (14). ML is the overarching term used for a subset of algorithms that help achieve this goal. These algorithms are self-learning from the data with which they are presented and can identify complex underlying patterns in data; they are also capable of processing unstructured types of data that traditional statistical techniques are incapable of doing, such as free text, images, video, and audio. Making such unstructured data available for use by ML algorithms increases the amount and potentially the quality of information available, which can lead to better predictive capacity.
By using ML algorithms, the work in the field of AI so far has been able to build systems that are performing well in a specific task. However, outside the scope of that task, most of these systems perform poorly, meaning that true intelligence has yet to be achieved (14). For nutritionists, the application of ML algorithms to their data is not to approximate human intelligence but rather to process complex data to generate results relevant to health and disease or to process large volumes of complex data—in other words, to apply these algorithms to a very specific scope of the task. Within the field of nutrition, specific tasks that have already benefited from ML algorithms are related to finding causes and potential solutions for many nutrition-related noncommunicable diseases, such as obesity, diabetes, cancer, and CVD, all of which have a complex and multifactorial etiology (3, 4, 14–19). These works have shown promise to the application of ML in solving the biggest challenges in nutrition and to the opportunities before us.
The direction of research in the discipline of nutrition science is going increasingly toward one that would benefit from the use of advanced tools, from data generation through to explanation and prediction. ML has the potential to supplement existing techniques to generate and analyze complex data, but to do so, it must be applied appropriately. Although the use of AI and ML does not require extensive background knowledge in computer science or mathematics, the application of ML without appropriate understanding can lead to biased models and results that do not represent real-world representative. For a nutritionist without prior ML experience, approaching the subject area can be overwhelming, which subsequently hampers adoption of its use by interested researchers.
This article aims to deal with this by providing a resource to which nutrition researchers can refer to guide their efforts. First, ML itself and its distinction from traditional techniques are explained. Next, an overview of ML is provided, and key concepts are elucidated. This covers core ideas in ML, such as ML types, tasks, data types, common algorithms, explainable AI (xAI), and ML performance evaluation. Throughout the aforementioned sections, application examples from the literature are provided. Related terminology can be found in Supplemental Table 1. Following this, a short review of nutrition-orientated literature utilizing ML is elaborated in the form of case studies of areas in nutrition science that are currently employing ML. Finally, practical application is supported by providing a framework for implementing ML in nutrition research. By providing a key reference, we enable the continuation of groundbreaking research by circumventing the problem that ML-naive researchers encounter when dealing with complex problems.
Machine Learning Capabilities
ML is a subdivision of AI that employs algorithms to complete a task by learning from patterns in the data, rather than being explicitly programmed to do so. This is achieved by defining an objective (e.g., predicting a numerical value), evaluating performance, and then performing experiments recursively to optimize the model. Whereas AI falls short of replicating the complexity of human thinking, it excels in certain aspects of learning, making it faster, able to deal with high-dimensional data, and able to learn abstract patterns (15, 16). These aspects of ML make it more suitable for tasks than traditional statistical techniques and certain domain-specific techniques and so gift ML with practical advantages that make it attractive, as discussed throughout.
Whereas the current section focuses on situations in which ML may be advantageous over traditionally used methods, researchers should be encouraged to consider each option as another tool in the box rather than using one or the other. Understanding the advantages and disadvantages of various methods and learning when and how to apply each one can lead to synergy and more fruitful results by allowing methods to complement one another. Researchers should be encouraged to think deeply about their problem and the research questions that they would like to answer and to select the appropriate techniques that best do this. Whenever possible, researchers should also consider experimenting with multiple options and selecting those most suitable. The idea of selecting the pool of solutions to suit the problem at hand is discussed in detail in the Framework for Applying ML in Nutrition Science section.
Machine Learning and Traditional Statistical Methods
When the goal is inference; interpretability is paramount; and the features are well established, simple, known a priori, and low-dimensional, traditional statistical techniques such as regression methods may suffice (1). However, researchers often choose these approaches due to familiarity, despite that ML techniques can be more suitable and efficacious in certain circumstances. ML is suited for high-dimensional data and when the goal is predictive performance (17). The capability of ML to learn from patterns in the data means that precise and premeditated variable selection is not a necessity; instead, many variables can be trialed, and repeated experimentation can quickly identify those of most relevance. That is, ML can be applied exploratively (17), which may lead to the discovery of novel predictive features, sometimes serendipitously (18–24).
By perceiving data to have been generated by a stochastic model, traditional statistics is limited by the assumptions that it makes: assumptions that are increasingly unjust as data are increasingly complex in domains relevant to nutrition such as health (25). When predicting mortality with epidemiologic data sets, Song et al. (26) noted that the nonlinear capabilities of more sophisticated ML techniques explain their consistently superior performance. Stolfi and Castiglione (27) integrated metabolic, nutritional, and lifestyle data in an emulator for a handheld device application in the context of precision medicine that predicts processes in the development of type 2 diabetes (T2D). They noted that such a dynamic and high-dimensional system is too computationally demanding for statistical methods traditionally used for emulation. Compared with classifiers or regressors that assume linearity, ensemble predictors (see Supplemental Figure S1, for example) applied to medical data consistently perform better (1, 21, 25, 28). Whereas ensemble predictors are considered uninterpretable, Breiman (25) made the interesting case that although the mechanism by which outputs are generated is not entirely transparent, this is counterbalanced with the advantage of making a more accurate prediction and in this way better represents the true process of data generation than do white box models. Other authors noted the inadequacy of statistical techniques for dealing with complex data derived from such subject areas as obesity (1), omics (29), and the microbiota (5). Ultimately, as data become more complex, the advantage of presenting a simplified representation of the data comes at a cost, a characteristic of classic statistical techniques from which ML suffers less.
Machine Learning and Traditional Statistical Methods
ML can be used to supplement domain-specific data analysis techniques. Gou et al. (20) linked 2 domains suitable for ML application—diabetes and the microbiome—by using ML to generate risk scores for T2D development based on microbiome composition. After pointing out that analysis of microbiota data is beyond the capabilities of classical statistical tools, they proceeded with ML to predict T2D better than traditional, domain-specific diabetes risk factors while identifying 11 novel microbial taxa predictive for T2D risk. Tap et al. (22) showed that conventional ecologic approaches did not find differences in microbiota signatures between patients with irritable bowel syndrome (IBS) and controls, whereas their ML approach (Lasso) was able to link intestinal microbiota signatures with IBS symptom severity.
Activity tracking utilizes unstructured data of movement generated from wearable devices to predict activity types and calorie expenditure, making it suitable for ML applications. Compared with domain-specific cut points, classification via ML techniques reduces misclassification rate, increases generalizability, allows grading of movement quality, and simplifies experimental design (30–32). Energy expenditure estimation traditionally uses methods that are expensive (e.g., doubly labeled water), impractical (e.g., indirect calorimetry with breathing masks), or non–free-living (e.g., direct calorimetry). Systems that analyze accelerometer data, with or without other physiologic data, can be adequate alternatives for the prediction of energy expenditure in a free-living, practical, and cost-effective manner (33, 34). In CVD research, CVD risk scores may be generated by using various biomarkers and are deployed in clinical practice; here too, ML techniques outperform traditional risk scores, making use of and identifying novel biomarkers in the process (19, 23, 35–38). ML is also a promising alternative for domain-specific techniques that are expensive, invasive, or both, as with nonalcoholic fatty liver disease (NAFLD) (39–41) and cancer (42–47).
Machine Learning: Practical Advantages
There are practical advantages of ML. Since the computer learns itself to complete a task, time and effort need not be invested in instructing the computer on what to do. This not only saves time and effort that would otherwise be spent on programming but also increases adaptability to solve various problems. That is, the same algorithm can be retrained on various data sets and problems. On a similar note, ML accepts various data types as input, including structured (e.g., tabular data) and unstructured (e.g., image based). In some cases, the same ML algorithms can be applied to different problems, perform different tasks, and take as input different data types; neural networks and k-nearest neighbors (kNN) are such examples.
By predicting an outcome based on existing data, ML algorithms can save on the time and cost of having to verify such outcomes experimentally. For example, Sorino et al. (48) concluded that incorporating ML algorithms into the analysis of noninvasive and comparatively cheaper variables could avoid 81.9% of unnecessary ultrasound scans in NAFLD, which are expensive and have long waiting times for results.
The tools required for performing ML experiments are minimal; other than a computer and a virtual environment to work in, all that is needed is a data set. To this end, data are being increasingly generated, and recent pushes for data sharing are meaning that more and more data sets are publicly available, suggesting that researchers across the world are able to run experiments and derive meaningful results in their field without the need for grants, research equipment, or generation of the data themselves. This is an extremely empowering aspect of ML, and if more researchers were better able to mine their own and others’ data, more scientific progress could be expected.
Machine Learning Overview
Types
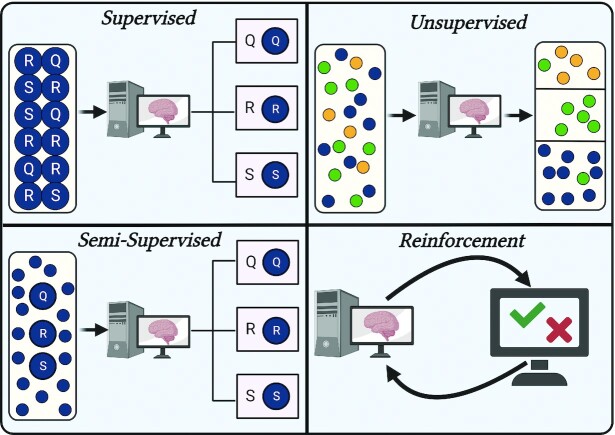
Four types of ML exist, each differing in the way that it learns, the algorithms that it employs, and its uses. A graphical depiction of each learning type is provided in Figure 1.
FIGURE 1.
Four types of machine learning. In supervised learning, labels are provided in the data for objective evaluation of algorithm performance, whereas in unsupervised learning, the algorithm partitions the data based on similarity. In semi-supervised, only a portion of the data comes with labels, although all data are eventually classified. Reinforcement learning makes use of penalties and rewards in a dynamic environment to train the algorithm.
Supervised learning
In supervised learning, the data come with labels in which the value or class being predicted by the algorithm is known, meaning that performance can be objectively verified. This is commonly observed in predictive models utilizing data sets with health variables and a disease outcome, such as CVD, T2D, and plasma nutrient prediction (49, 50). Since the labels of the data are required, human intervention plays a larger role than in other ML types, which can increase costs and time (51).
Unsupervised learning
Unsupervised learning occurs without labels; instead, the algorithms seek to find patterns in the data and partition them based on similarity. This reduces human intervention, saving time on feature engineering and labeling. The most common use of unsupervised learning is clustering, dimensionality reduction and anomaly detection are also unsupervised. Unsupervised learning has been applied extensively in phenotyping, such as grouping individuals for PN (11). Unsupervised learning can also be used as a processing step before a supervised task to homogenize the data, as evidenced by Ramyaa et al. (52), who predicted BMI in women more accurately after phenotyping than when using the data as a whole. Another attractive use of unsupervised learning is hypothesis generation; because this type of learning works on the detection of patterns, this may lead to the formation of previously unidentified groups in the data.
Semi-supervised learning
Semi-supervised is somewhat in between the 2 previously defined learning types in that labels are partially present but usually mostly absent. Providing labels on a subset of the data has the advantages of improving accuracy and generalizability while sparing the time and financial costs of labeling an entire data set (53). Consequently, semi-supervised learning has been used to study the influence of genes on disease outcomes when the known genes (i.e., the labeled data) are few (54).
Another example of semi-supervised learning is constrained clustering, which expects that certain criteria are satisfied during cluster formation, such as given data points being necessarily in the same or different clusters (55). This can be a way to circumvent potential issues that can arise when dealing with biological or health data in unsupervised learning, such as the grouping or separation of data points that violate plausibility—for example, the clustering of data of one biological sex with another in a system where this is not possible. However, it should be kept in mind that such findings may also provide interesting information about the data and adding such constraints may mask this.
Reinforcement learning
In reinforcement learning, the algorithm exists in a dynamic environment and is penalized or rewarded for the decisions that it makes within the environment. The algorithm then updates its behavior to maximize reward, minimize penalization, or both. This allows the algorithm to become proficient in a task without being explicitly programmed to behave in a certain way. A famous example of reinforcement learning is Alpha Go Zero, which was able to achieve superhuman performance playing Go (Weiqi) with only a few hours of training by playing against itself (56). The complex nature of reinforcement learning limits application in simple classification or regression tasks and is instead used where the integration of complex and varied data is concerned, such as recommender systems (57) or mobile-based fitness apps (58).
Tasks
ML algorithms are employed to complete tasks, which are distinguished into various categories. To complete these tasks, algorithms are used. Various of these are mentioned throughout the current subsection; for a more detailed description of each algorithm, see the Supplemental Material.
Regression
Regression involves the prediction of a continuous variable based on one or more input variables. In the case of linear regression, a linear relationship between the input variables and the dependent variable is assumed, whereas in nonlinear regression, relationships can be more complex. As well as basic linear regression and its variants (e.g., Ridge, Lasso), more complex algorithms can be used, including some that are more often associated with classification, such as random forest (RF) and support vector machines (SVMs) (59, 60).
Classification
Classification tasks aim to predict the class labels of data based on their independent variables. Data of the same class will likely have similar characteristics, at least for variables that contribute most to the classification decision; this forms the basis for how an algorithm learns to assign classes to a data point. In binary classification, there are only 2 labels, whereas with multiclass classification, there can be many.
Sample uses of classification include predicting adherence to exercise regimes (61) and image-based food recognition for dietary intake monitoring (11). Classification has significant overlap with regression in that oftentimes a regression problem can be converted to a classification task with only slight modifications and vice versa. This is reflected in the algorithms that can do both, such as SVM, RF, decision trees, and kNN.
Clustering
Similar to classification, algorithms in clustering split the data based on similar characteristics, but clustering differs in that it is unsupervised, meaning that there is no ground truth or class labels to which the data points should belong. Thus, the goal is to obtain clusters that are more homogeneous than the data as a whole. Because it is typically unsupervised, clustering can be performed with or without expectations, leading to new discoveries and hypothesis generation. A well-known and popular application of clustering in nutrition and health research is that of phenotyping individuals based on shared characteristics, such as microbiome profiles (62) or identifying activity patterns (63). The most common clustering algorithm is k-means (for numerical data), with adaptations including k-modes (for categorical data) and k-prototypes (for mixed data). Other examples include density-based spatial clustering and mean-shift clustering (51).
Recommendation
Recommender systems use data to generate a recommendation on a decision to be taken and have been used in nutrition to suggest meals to help manage chronic diseases (64–67). Recommender systems can be further classified into subtypes such as collaborative filtering, content based, and popularity based, as well as hybridizations of each. Recommender systems can be complex and may require the integration of multiple components, each of which may involve different ML tasks and algorithms. For example, Baek et al. (68) described a recommender method that clusters individuals based on chronic disease status, suggests suitable foods for each cluster, and considers the preferences of the individual and on the universal level. Because recommendation systems can involve lots of data, deep neural networks are often utilized.
Dimensionality reduction
When working with data sets with many features, dimensionality can be problematic; it slows computation, may reduce accuracy, and can cause overfitting (69). This is particularly relevant in the modern age where high-dimensional data are being generated (70). Dimensionality reduction techniques aim to reduce dimensionality while maintaining the most important characteristics of the data or the variance. Whereas at times this may cause a small reduction in predictive capability, it may be preferred in exchange for data with drastically fewer (irrelevant) features, which can enhance computational efficiency and interpretability. Conversely, by reducing noise and simplifying learning for the model, dimensionality reduction may sometimes even improve performance.
Modern techniques such as microarrays can generate high-dimensional data with few samples and thus benefit from dimensionality reduction techniques (69, 71). Principal component analysis (PCA) and t-distributed stochastic neighborhood embedding are linear and nonlinear dimensionality reduction techniques, respectively. Because of its capacity to eliminate redundant features, Lasso regression can also be used as a dimensionality reduction technique.
Explainable AI
The concept of xAI is concerned with not only generating an output but also how it was generated. Technological developments have enabled the creation of sophisticated algorithms such as ensemble methods and deep neural networks that, although usually superior, are not interpretable. If predictive ability is most relevant to the problem being solved, then this may not be an issue; however, in some practical applications of ML, it is important to know how the output was generated. This is understandable in medical situations where the predicting output can have serious consequences for, say, patient lifestyle or treatment avenues. The results of xAI can be informative in that they reveal which features contributed most to the algorithm output (72).
In certain situations, interpretable algorithms are preferred over ensemble methods, despite better performance in the latter, as is the case with nutrition care by Maduri et al. (73) and in the prediction of nutrient content in infant milk by Wong et al. (74). However, methods exist that facilitate interpretability without sacrificing performance. xAI techniques such as Shapley additive explanations (SHAP) and Shapley values (75), partial dependence plots (76), and local interpretable model-agnostic explanations (LIME) (77) exist to make transparent black box models. Choi et al. (28) emphasized the importance of being able to capture complex nonlinear interactions when predicting refeeding hypophosphatemia and thus chose to opt for XGBoost in place of linear models, especially since classification was much better in the former. Instead, they used SHAP values to elucidate the features most influential for classification decision by XGBoost. Zeevi et al. (78) used partial dependence plots to extract the relative contribution of their features for the prediction of postprandial glucose response (PPGR). This enabled the use of a gradient-boosting RF that, although black box, was able to capture the nonlinear relationships inherent to their complex feature set. Davagdorj et al. (79) made use of LIME to explain predictions of artificial neural networks and XGBoost, the best performers, when predicting hypertension in the Korean population. They emphasized the importance not only of prediction quality but also explainability for decision making in public health.
In conclusion, not only is the eventual output of ML relevant but so is the means by which it was produced. After the proof-of-principle stages of model development, researchers can further their fields by incorporating xAI into their work, therein providing transparency and encouraging public understanding.
Evaluating Performance
Metrics for evaluating ML algorithms are broad and can be task, type, or model specific (Supplemental Table 2). For example, evaluation approaches of supervised algorithms are often not suitable for unsupervised techniques since the data might be without labels. In clustering, metrics are instead used that focus on the purity of the data partitioning or similarity of the data after grouping (80). For PCA for dimensionality reduction, cumulative variance with a predetermined, arbitrary cutoff point is used (e.g., 95%).
Additionally, although some evaluation metrics reflect model performance similarly, the specification of evaluation metrics should not be made arbitrarily. For example, where higher accuracy at the cost of specificity might be less problematic in applications categorizing food for dietary intake purposes, the same trade-off can have serious consequences in disease prediction, as in the incorrect assignment of a serious disease (false positives) such as cancer (81). Accuracy is the most common classification metric but too often it is presented or interpreted at face value, whilst other metrics are neglected. The consequences of this can be easily witnessed in data sets with class imbalances in the target variable, as is common in health data. For example, a classifier that predicts negative for all data points on a given NHANES data set where the target variable is undiagnosed T2D would have an accuracy of approximately 97% (82), without actually having real predictive power. For these reasons, multiple metrics or meta-metrics, such as F1 score or area under the receiver operating characteristic curve (AUROC) (83), should be considered.
Evaluation metrics should be specific to the problem at hand. For example, the coefficient of determination R2 measures how well a continuous target variable is estimated by a set of predictors and is thus generalizable across problems, models, and data sets. However, at times it may be more relevant to know how well the model performs for a specific problem, such as mean error in prediction of plasma cholesterol or cost of meeting a healthy diet; in such cases, a metric such as mean absolute error would be preferred. Likewise, in circumstances where the consequences of the model output are less severe and the treatment response is risk-free, the emphasis would be on accuracy rather than specificity. An example of such a case could be the prediction of risk for overweight, with the treatment response being free admission to an education healthy eating course. These examples demonstrate the influence that the problem has on metric selection.
In sum, many methods exist for evaluating the performance of ML models. Metrics should be chosen thoughtfully, keeping in mind the task, the model being used, and the specific problem trying to be solved.
Validation
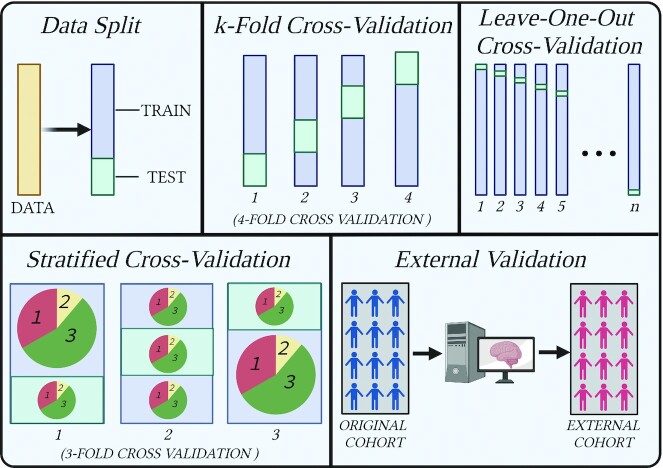
ML models will most likely perform better on the training data on which they were trained than on unseen data. Whilst this is to be expected, it can give a deceptive reflection of how well the algorithm has learned a task. If performance drops substantially when going from the training to unseen data, overfitting has occurred, and the model is therefore not generalizable or useful in real applications. Overfitting is a widespread problem and often unaccounted for in the literature. Whereas virtually all supervised techniques suffer from overfitting, the degree to which they do so can be lessened with validation techniques such as data splitting, cross-validation (CV), external validation, and combinations of these, as discussed later and shown in Figure 2. It is imperative that robust methods of validation are used to preserve generalizable model conclusions.
FIGURE 2.
Various validation techniques. Data split simply consists of excluding a portion of the data for testing after training. In k-fold cross-validation, the data are split into k number of folds, and each fold is used once for training and k – 1 times for training. Leave-one-out cross-validation uses the same concept except that k is equal to the number of data samples, so each individual sample is used once for testing and n – 1 times for training. Stratified cross-validation ensures that the proportions of classes remain the same in each split (training and test) and each fold. Finally, external validation consists of using data different from those on which the algorithm was trained.
Data split
Also known as the holdout method, the data split method splits the data into a training set and a test set, where the model is first trained on the training data and then applied to the test set to gauge generalizability. In this way, the test data act as the “unseen” data, although, since the data set came from the same source and was processed in the same way, it is not truly unseen. Splitting the data like this can be problematic in small data sets by reducing the instances from which the model has to learn. It can also increase vulnerability to outliers. Finally, unless data are sufficiently large, there can be large variations in results between different splits of the data. In most circumstances, a simple data split is not a sufficiently powerful tool for assessing the generalizable performance of ML models.
Cross-validation
CV and its variations run the model multiple times with different splits in the data so that every split is used for training and once for testing. This is most often achieved with k-fold CV, where k is an arbitrarily selected value by which to split the data, with a minimum value of 2 and a maximum value of n – 1. In the case of the latter, this represents another CV variation known as leave-one-out CV, where all instances but 1 are used to train the model, with the remaining data point representing the test data. The aforementioned instances use random sampling to split the data; in another variation, stratified CV, the data are split in a way that maintains the proportions of classes of the original data. This is useful in preventing issues arising when certain classes are particularly low since otherwise the model might be left with too few instances from which to learn. Eventually, results from each fold after performing CV should be averaged to give a balanced CV score, although examining the scores of each fold can also be informative; if they differ wildly, this can be indicative of problems such as outliers or class imbalances. CV provides a more robust way to validate a model and should be selected over a simple data split whenever possible.
Despite these advantages, biased models can still occur when the same CV scheme is used for hyperparameter tuning and model evaluation. Hyperparameter optimization schemes such as those discussed in the Supplemental Material (Hyperparameter Optimization section) often use CV, and although overfitting is reduced, the model is still yet to be tested on a pristine test sample that was not involved in either model training or hyperparameter tuning (84). Nested CV aims to overcome this by splitting the data into an outer loop, which itself is split into training and testing, and an inner loop, which is composed of the training folds of the outer loop. CV is used on the inner folds to select model hyperparameters; then, the outer loop is run by the optimal model identified in the inner loop. This is repeated k times, where k represents the number of folds of the outer loop. This prevents that all of the data are used for model selection, evaluation, and feature selection and maintains the ability to evaluate cross-validated performance on unseen data. Although this increases computational demands substantially, it provides a much more honest representation of model performance.
External validation
For an even truer representation of model generalizability, a different data set from that used to train and test the model can be used. For example, a model predicting glycemic response trained on an Israeli cohort was tested in an American cohort, meaning that different dietary elements, societal influences, and genetics are introduced (85). Capturing the effect of intercohort variation in this way informs the degree to which the model can perform its task in different populations. This is a common approach in health studies deploying ML that use cohorts (22, 36, 37, 40, 42, 78, 86, 87).
Case Studies: Applications of Machine Learning in Nutrition Domains
The current section briefly presents two case studies within the discipline of nutrition that are suitable for the application of ML techniques. It is hoped that readers will derive ideas and inspiration from the case studies, which they can then apply to their own research domains.
Precision Nutrition
PN concerns the use of personal information to generate nutritional advice that, in theory, leads to superior health outcomes than generic advice (11, 88). It rests on the basis that differences in a myriad of factors among individuals ultimately necessitate specific nutritional requirements that population-level guidelines cannot capture. The diversity, complexity, and, at times, high dimensionality of the data that represent these factors have created expectations for ML in PN. Such expectation is reflected in the commitment of the National Institutes of Health to supply $170 million in funding algorithm development in PN over the next 5 years (89). A detailed systematic review of ML in PN is provided by Kirk et al. (11); here, a short overview and recent developments are provided.
A model example of the application of ML in PN was in the high-impact study of Zeevi et al. (78), which made use of a gradient-boosting RF model to integrate plasma, microbiome, anthropometric, personal, and dietary data to predict PPGR to the challenge meal, with accuracy comfortably exceeding established methods. A striking finding was the remarkable interindividual variation in PPGR seen in response to the same foods, substantiating the claims of PN for improving health. Berry et al. (90) used a similar design to predict not only postprandial glucose but also triglyceride and C-peptide with an RF regressor. In both studies, the features most relevant to the decision outcomes were estimated, and the contribution of modifiable factors was shown to be large. These studies show not only that the influence of specific foods on metabolic parameters in an individual can be known but also the main factors that can be modified to change this. Such information is of great value to those attempting to manage metabolic health.
Obesity and overweight constitute another important subject area in nutrition, thus attracting attention in PN. Ramyaa et al. (52) found homogeneous phenotypes within a population of women and then proceeded to predict their body weight. The clusters associated with different dietary and physical activity variables suggested that the women responded differently to macronutrients and exercise in their propensity to gain weight and thus that personalized diets and exercise regimes would be effective. Zellerbach and Ruiz (91) aimed to predict instances of overeating based on macronutrient composition of an individual's diet. Although they were unsuccessful, the concept may have merit with the inclusion of other relevant variables (e.g., stress, sleep, and alcohol or drug use) and with higher-quality diet data than the self-recorded publicly available food logs used in their study, where data were collected outside a scientifically controlled setting and liable to bias. That Wang et al. (5) could predict obesity using gut flora data is interesting to PN because of the known relationship between diet and microbiota composition. Because the microbiota is involved in various health conditions (92), its targeting by PN interventions could be fruitful in subsets of individuals.
Malnutrition has been targeted by PN in various ways. Current screening tools for malnutrition in inpatients, for example, suffer a lack of agreement and poor adherence from hospital staff, suggesting that automatized approaches may be appreciated (93). The decision system of Yin et al. (94) sought to realize this by applying k-means to hospital record data to separate patients based on nutritional status. Well-nourished and mild, moderate, and severely malnourished clusters were identified, the characteristics of which formed the basis for a logistic regression classifier to assign unseen data points to 1 of the 4 clusters with perfect performance (AUROC: 1). Subramanian et al. (95) characterized a “healthy” microbiota index in children, from which RF could predict chronological age (AUROC: 0.73). Severe acute malnutrition could subsequently be predicted by deviation from the index for a given age, since malnourished children have a relatively immature microbiota when compared with healthy children of the same age. This approach allows targeted intervention in children at risk of malnutrition-induced growth stunting. Malnutrition can also be predicted with ML from demographic data in developing countries, which is attractive since such data are routinely collected and available to health organizations (96–98).
ML is helpful not only for generating PN outcomes but also for collecting PN data. Current dietary assessment methods have serious limitations such that estimated intakes can vary wildly from true intakes (99, 100). However, ML-assisted dietary intake monitoring could make more convenient and accurate the process of collecting intake data, the benefits of which would extend beyond PN to the broader nutrition domain. Indeed, examples in which ML has been used in dietary assessment include image-, smart watch–, piezoelectric-, and audio-based methods [see Table 4 in Kirk et al. (11)]. Although some of these instances are relatively primitive and often confined to controlled settings, it can be imagined that their successors will be refined and convenient in real-world settings. Natural language processing (NLP) can also be valuable in dietary assessment. NLP is a specific field in computer and linguistic sciences that has the goal to interpret written and spoken text in such a way that its meaning is understandable by a computer. In PN, this can automate the processing of food diaries (101) and consolidate multiple data sets (data integration) and food tables (102). Eventually, NLP can communicate messages to users of health-tracking apps to offer personalized advice and provide support, omitting the need for such advice to come exclusively from health professionals, which is expensive and time-consuming. Activity tracking is relevant to PN outcomes by providing information on the exercise and sedentarism of individuals. ML has been used to classify activity patterns (103–106) and estimate energy expenditure (33, 34, 107) based on accelerometer data, which can improve the quality of activity data acquisition, thus increasing its value as a feature in PN approaches.
Omics is a discipline that derives its name from the suffix of components from which it is composed: genomics, transcriptomics, epigenomics, proteomics, metabolomics, and occasionally others (microbiome, lipidomics, etc.) (108). Figure 3 shows the main omics components and their proximity to the genotype and phenotype. Data captured on any of these levels can be valuable in PN, and integrating their data (i.e., multi-omics) can provide a systems-level view capable of providing more information than the constituent parts independently (109). Genetic information has been used in tandem with ML for predicting obesity (110–112) and diabetes (113, 114), although, despite the wealth of information within the human genome, genetic information often explains little of the variance in complex health outcomes (115). Gene expression, transcripts, and the proteins that they encode can all be modified by environmental factors such as food, which can make the genetic sequence encoding them effectively redundant; hence, epigenomics, transcriptomics, and proteomics, respectively, exist in response to this. Further still, the microbiome is increasingly recognized as a key player in health and disease, at times being responsible for a significant portion of the variance in predictive health models (5, 20, 87, 90, 116, 117). The microbiome is of particular interest to PN because it can act as an input variable and a target variable to be modified in PN.
FIGURE 3.

The major components of the omics field and their proximity to the phenotype.
It should be emphasized that studies need not be designed with PN in mind to realize how responses can differ among individuals within a study. A prime example of this is seen in the weight loss study of Gardener et al. (118), where participants followed either a low-carbohydrate or low-fat diet for 12 months. Whereas the group means suggested that weight loss was similar, analyzing the data on an individual basis within the groups told a different story: although some lost a great deal of weight, others failed to lose or even gained weight. This highlights a pitfall of research in health sciences in that, although comparing groups means is convenient, it can mask individual differences that can be much more informative.
Despite its potential, ML in PN must still prove itself able to reduce disease burden when applied in real-world situations. Most of the aforementioned studies are descriptive, and indeed more experimental studies are required to prove that PN is more effective than generic healthy eating recommendations when both are adhered to. Even if experimental studies can prove a theoretical role for PN in improving health, PN approaches must be practical. If the suggested dietary alterations are restrictive or infeasible, it is unlikely that they will be adhered to in the long term. For example, although the large-scale study Food4Me found that personalized advice favorably altered dietary habits in participants (119), there is insufficient evidence that personalized approaches will lead to sufficient adherence to reap the potential benefits of PN. This was demonstrated in a recent Korean study where only those in the highest adherence group saw improvements in the markers of health that were measured, and in fact, the same markers deteriorated in the group of lowest adherence (120). Whereas ML has much potential to help with data generation and analysis in PN, these approaches must be able to demonstrate practical application and ultimately a reduction in clinical burden, both of which require many more studies for their verification.
Metabolomics
One area of nutrition that has received much attention in recent times is metabolomics. Metabolomics is closely related to PN, as a predictor of health outcomes and for data collection (11), though it also has functions that do not necessarily relate to PN. Modern technologies are now enabling the profiling of many metabolites all at once, within one or a few samples, followed by analysis of their interactions (121). The profiling of thousands of metabolites makes for noisy raw data, which requires preprocessing and analysis, two tasks for which ML is highly suited. Some examples of metabolomics research using ML in nutrition are presented in turn.
A popular application of metabolomics is phenotyping, which overlaps significantly with PN. Metabolites are generally considered to give a much more representative picture of a phenotype than other omics varieties since they more closely reflect the reactions that actually occur in a system (122). The simultaneous assessment of many metabolites in an individual enables a form of phenotyping specific to shared metabolite characteristics known as metabotyping (115). A randomized controlled trial found no effect of 15 μg of vitamin D supplementation on markers of metabolic syndrome (123). However, after metabotyping via k-means clustering, a vitamin D–responsive cluster was found where, in contrast to the population as a whole, vitamin D supplementation did improve markers of metabolic syndrome. Also utilizing k-means, O'Donovan et al. (124, 125) used a range of metabolites to identify healthy and unhealthy clusters in 2 cohorts, and on both occasions targeted advice was given based on the defining characteristics of the clusters. For example, a cluster composed of individuals with elevated cholesterol was administered personalized advice oriented toward lowering cholesterol (125). Given the diversity of metabolic alterations that people can experience despite having similar demographic or anthropometric characteristics, tailoring nutritional recommendations to the individual is a logical approach.
The concept of “metabolically healthy obese” and whether it actually exists is another example of phenotyping. It is indeed curious that approximately 1 in 3 obese individuals does not show metabolic alterations on commonly investigated clinical parameters (126) and that the exact metabolic consequences of the remaining two-thirds vary greatly among individuals (127). The drivers of this divergence remain unknown; hence, investigating metabolomic signature differences between the phenotypes may be revealing. Studies using ML techniques have found key differences between the metabolite profiles of healthy and unhealthy obese subjects (128–130). In a systematic review, BCAAs, aromatic amino acids, lipids, and acylcarnitines were all found to be elevated in the unhealthy obese phenotype as compared with the healthy (131). Due to the high dimensionality of the data sets in metabolomics, PCA is typically opted for. PCA-reduced feature sets can then be used to identify differences in metabolites, amino acids, and lipid patterns (132, 133). Using ML to understand how and which metabolic aberrations could develop among obese phenotypes can inform targeted treatment to minimize obesity-imposed harm.
ML has been applied in metabolomics when studying the microbiota. Microbiota-derived metabolomics data are complex and high-dimensional, which has motivated researchers to consider applying ML. Some notable examples include distinguishing healthy and unhealthy metabolite signatures following a red wine intervention (134), distinguishing women with and without food addiction from fecal samples alone (135), identifying pediatric IBS (136), and comparing metabolic activity of the microbiota between vegans and omnivores (137). The last of these studies is particularly relevant to the metabolomics field because, although differences in microbiota composition were minimal, metabolic activity differed significantly, and an RF classifier could distinguish the groups with 91.7% accuracy. This shows the importance of not only the composition but also the metabolic activity of the microbiota.
Metabolomics is interesting to the nutrition community as a free-living, objective dietary assessment tool (138, 139). Biomarkers of intake have been found for various foods, such as bread (140), coffee (141), citrus consumption (142), and meat and fish (143), as well as dietary components such as polyphenols (144, 145) and fermented foods (146). Such information can be used not only for simply monitoring food intake but also for associations with health and disease outcomes (141, 142, 147). This function of metabolomics permits estimating adherence to dietary patterns. Acar et al. (139) used metabolite profiles to identify participants potentially noncompliant to a particular dietary pattern through partial least squares discriminant analysis with a reduced feature set. Aside from identifying regular noncompliers, they observed that at any given time approximately 10% of the participants may have been deviating from their prescribed diets, which, if generalizable, has clear implications for nutrition intervention studies. The capabilities of ML make it suitable for finding associations in metabolomics, as well as identifying new phenotypes or markers of intake in untargeted approaches. Many processing steps are required to transform raw metabolomics data into a form from which information can be derived. This, however, is essentially feature selection and pattern recognition. To this end, the competency of deep artificial neural networks, such as convolutional neural networks in feature selection, could be particularly useful, especially given the complexity of data used in metabolomics (148). Research investigating this in nutrition, though, is lacking but would be valuable.
In sum, ML can be a useful tool in the data preprocessing stages of metabolomics and in generating predictive models on the prepared data. Through clustering and classification, ML can analyze processed metabolomics data for applications such as disease prediction and understanding disease mechanisms, phenotyping, characterizing the metabolic environment, identifying biomarkers, and dietary assessment.
Framework for Applying Machine Learning in Nutrition Science
After understanding the advantages of using ML in research, researchers should be able to know when and how ML can be applied to a problem. The present section aims to support decision making in this process by providing a framework to guide researchers interested in using ML in their work. The framework takes inspiration from the concept of method engineering (149), though is adapted with nutrition research in mind.
Understanding the problem and the data
Whether ML can be used to solve a problem depends primarily on the problem itself and the data involved. If the problem is one concerned with predicting an outcome based on a given data set, ML can be considered. Data sets with many features and complex, nonlinear interactions suggest themselves suitable for application with ML because of its ability to identify patterns among the input variables that can then map the output variable, thus producing results that would otherwise go amiss. This is exemplified in cases where data are clustered without expectation yet new findings are discovered (52, 123).
ML could be considered in data preprocessing. This function of ML was evidenced on various occasions in the present article, such as the use of PCA for dimensionality reduction, deep learning for feature extraction, and ML approaches for data collection in PN and for processing noisy and complicated metabolomics data. “Missing data” is the often colloquial term denoting a data set in which not all the data entries that should have values are filled, regardless of the reason why. In nutrition, due to the practical challenges of longitudinal data gathering, missing data are a common issue. It is up to the researcher to understand the impact of values that are missing in the data set and how they are to be dealt with. Techniques exist for their imputation, and sometimes it is appropriate to remove entire variables or data entries (150). Since each of these approaches has advantages and disadvantages, the decision ultimately taken by the researcher should be done so after deliberation. Resources describing the missing data problem and its solutions exist (150–154). ML techniques can also be considered for imputation (155–158). The extent of the missing data can influence the modeling approach. Certain approaches in traditional statistics and ML can handle missing data well, including linear mixed models, decision trees, kNN, and XGBoost, sometimes even when missing data are as high as 20% (159, 160). Regardless of how this is done, the method of handling missing data should be explained and reported (161). Understanding the data will provide insight into how exactly ML can be applied.
Background research and existing solutions
An understanding of the existing solutions to the problem at hand is crucial to knowing exactly how ML can be applied. If existing solutions are already suitable, additional benefits from ML may be marginal. ML is also incapable of replacing the human aspect involved in managing health and nutrition. For example, although an algorithm may make accurate personalized nutritional recommendations, it cannot deliver the same information in a way that a trained professional would, and this aspect may be important for inducing behavior change.
ML, instead, is better applied when existing methods are insufficient. This is observed in data with complex relationships where classical statistical methods are incapable or in situations where domain-specific techniques are inadequate. Examples of the latter include the prediction of PPGR (78, 90), for which existing methods have low accuracy. It may be that current solutions, although accurate, have other limitations, such as invasiveness, as is the case for NAFLD detection (39–41). Further still, existing solutions that have a higher human element naturally suffer from human limitations such as fatigue or calculation mistakes. This was exemplified by Kondrup et al. (162), who found that a major reason why patients in clinical care were not screened for nutritional assessment was that nurses “just forgot.” ML in these cases can increase predictive capacity, improve efficiency, reduce patient risk, and mitigate human error.
Possible solutions
Of the options within the domain of ML, the eventual candidate solutions should be tailored to the needs of the problem, the data, and the ultimate goal of the project. Primarily, one must think about the task required, as this will naturally limit the options available since certain algorithms are capable of only certain tasks. Also important is the algorithm in relation to the data. For example, naive Bayes would be an unsuitable choice in data sets with classes that contain certain values of very low frequency due to the zero-frequency problem (see Supplemental Material) (163). Likewise, applying estimators that assume linearity to a data set that has predictors with nonlinear relationships with the dependent variable would lead to suboptimal performance. In this case, performing transformations on the data or choosing a model with nonlinear capabilities, such as RF or SVM with a nonlinear kernel, would be preferred.
Another possible solution might be to use dimensionality reduction techniques. Feature selection and engineering are valuable methods in ML and can be specifically helpful to nutrition data sets where collinearity is often present. Such techniques must be chosen with care as their improper use can affect performance, but due to the breadth of their possible applications, it is not possible to state which should be used; instead, they should be tailored to the problem. On a general level, if dimensionality reduction is applied with collinearity in mind, then the method should be chosen and applied in a way that preserves or increases the score on the test set. When dimensionality is applied to reduce computational strain, one must have preconceptions about the degree to which error is allowed to increase to enable decreases in computational time.
The end goal must be kept in mind, which too will dictate the pool of candidate solutions. For example, if predictive performance was the primary goal in the detection of cancer based on medical images, high-performance convolutional neural networks could be used. Alternatively, when interpretability is needed, algorithms can be selected that provide coefficients (e.g., regression) or can be easily understood (e.g., decision trees). Black box models (e.g., RF, XGBoost, artificial neural networks) can be understood through xAI techniques (e.g., SHAP, LIME, partial dependence plots), but such techniques have their pros and cons and should be considered in the context of the entire problem scope. When ML approaches are to be deployed or incorporated into an application, such as using ML for food tracking on a mobile device, pragmatic factors such as computational time are relevant. For example, whereas RF would usually outperform naive Bayes on a classification test, naive Bayes is much faster. These practicalities must be kept in mind.
Testing the available solutions
One of the advantages of using ML in research is the capacity to test multiple options in multiple configurations and converge on an optimal one. This capacity should be exercised by trying various, if not all, candidate models. The interpretation of the test data results should be given more importance than the training data to reduce overfitting. If possible, hyperparameters of each algorithm should be optimized with techniques such as grid or random search (see Hyperparameter Optimization section in Supplemental Material), and nested CV should always be considered. Although this can be time-consuming, it will give a fairer representation of the quality of the possible solutions since some models perform better in their default parameters than others.
Trying many solutions is always advisable, even when the choice might seem obvious beforehand. For example, although ensemble methods consistently outperform logistic regression in classification, this is not always the case (164).
Indeed, one example of this was by Yin et al. (94), where logistic regression achieved perfect performance predicting malnutrition in a data set of 14, 000 patients with cancer, outperforming ensemble and deep learning techniques (94). Additionally, techniques such as stacking, which involve combining multiple learners into 1 meta-learner, should be explored. Packages such as SuperLearner (165) exist to facilitate this, but it can also be done manually. The process ensures a result at least as good as, if not better than, the best single learner alone. Stacking has been used in some nutrition research (166–168) but is typically underutilized.
Understanding and communicating the results
The evaluation process must be undertaken in the context of the solution (i.e., the algorithm) and the problem (see Evaluating Performance). Comparisons of the possible solutions should be made by the most suitable metrics to enable the optimal solution to be chosen. It is common to visualize results in various plots, such as AUROC plots for classification, R2 for regression, silhouette score and cluster plots for clustering, PCA score plots for PCA, and heat plots for correlations, among others. Common libraries for achieving this include ggplot in R and Matplotlib, Seaborn, and Yellowbrick in Python. If xAI techniques are used, these results too can be communicated. Feature importance plots, plots of the SHAP library (waterfall, force plots, bee swarm, etc.), and partial dependence plots, among others, can allow visualization of the features most relevant to a decision.
Limitations of Machine Learning in Nutrition Research
Although the present article has focused on the promise that ML is demonstrating, it does not come without limitations. There is an apparent overoptimism in ML research that exists due to nonrigorous methodologies. Although we described methods for detecting this, such as CV, it is not uncommon to see circumstances where these are not made use of. For example, unless a data set is sufficiently large, different training-test splits can lead to different results when using a simple data split for validation. This opens the possibility to the generation of interesting results based solely on how the data were split. Further still, it is rare to see nested CV used in nutrition literature, the consequences of which were discussed in the Validation section. Another consideration is that ML algorithms are generally evaluated on homogeneous data collected in affluent societies; the performance of these models in distinct populations and with different data generation techniques is not guaranteed. Both these considerations compromise generalizability, meaning that if such models cannot be applied outside the setting in which they were tested, ultimately their utility is greatly diminished. Another often overlooked issue is flawed feature selection and derivation of importance. For example, feature importances from algorithms such as RF and XGBoost are readily available and often reported in studies that utilize them. However, the mechanism by which such methods estimate importance means that correlated features, though scoring similarly, appear less important than they are. This, relatively speaking, also means that the importance of less important features are inflated. Such similar phenomena occur with other algorithms and xAI techniques but are often not checked, for example, by corresponding with other feature importance techniques for corroboration and, instead, are reported as is.
Finally, the application of ML in certain circumstances has practical drawbacks. There can be substantial costs for data collection, hardware, ML engineers, infrastructures (data storage, cloud computing), integration (pipeline development and documentation), and maintenance. In certain applications, data are generated from different sources by using different programming languages and arriving in different forms. Unifying this in a multimodal approach can be very challenging. Similarly, although the ability of ML to transform unstructured data into data suitable for use in models represents a stark advantage for ML over traditional methods, such techniques can be challenging even for those specialized in the area and thus may not be fruitful for researchers specialized in nutrition at the time of writing. In closing, ML is showing much potential in research in nutrition but still has much to prove to reduce the burden of nutrition-related ill-health in society.
Conclusion
In conclusion, there is much potential for ML to make progress in nutrition science. ML can capture the complex interactions that exist and are increasingly generated with modern technologies in nutrition and health data. The failure to be able to use techniques that can analyze complex data, such as ML, represents an unnecessary barrier to scientific progress. Although still relatively new, it is evident that AI approaches have much potential to supplant traditional and domain-specific methods in predictive capabilities, efficiency, costs, and convenience. ML can also be helpful in the data collection and data preprocessing stages in various fields of nutrition. To realize this potential, researchers must be familiar with ML concepts, be knowledgeable on when AI can be suitably applied to a problem and how to use it, and be willing to branch out of the techniques historically used in their disciplines. We hope that the intuitive explanation of ML and the examples of its application in nutrition science in the current article will facilitate this and be a useful reference guide to researchers of health and nutrition who would like to make use of ML in answering their research questions.
Supplementary Material
Acknowledgements
The authors’ responsibilities were as follows — DK: performed the literature review, wrote and designed the article, and made the figures; GC: spawned the idea for the article, providing machine learning and nutritional expertise, and was involved in writing the article and editing; EJMF: involved in editing and providing nutritional expertise; BT: involved in editing and providing machine learning expertise; EK: provided valuable feedback and machine learning expertise and edited text; MT: provided valuable feedback and was involved in editing; and all authors: involved in brainstorming and structuring the article and read and approved the final manuscript.
Notes
This project was partially funded by the 4TU–Pride and Prejudice program (4TU-UIT-346; 4 Dutch Technical Universities).
Author disclosures: The authors report no conflicts of interest.
Supplemental Tables 1 and 2, Supplemental Figure 1, and Supplemental Material are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/advances/.
Abbreviations used: AI, artificial intelligence; AUROC, area under the receiver operating characteristic curve; CV, cross-validation; IBS, irritable bowel syndrome; kNN, k-nearest neighbors; LIME, local interpretable model-agnostic explanations; ML, machine learning; NAFLD, nonalcoholic fatty liver disease; NLP, natural language processing; PCA, principal component analysis; PN, precision nutrition; PPGR, postprandial glucose response; RF, random forest; SHAP, Shapley additive explanations; SVM, support vector machine; T2D, type 2 diabetes; xAI, explainable AI.
Contributor Information
Daniel Kirk, Division of Human Nutrition and Health, Wageningen University and Research, Wageningen, The Netherlands.
Esther Kok, Division of Human Nutrition and Health, Wageningen University and Research, Wageningen, The Netherlands.
Michele Tufano, Division of Human Nutrition and Health, Wageningen University and Research, Wageningen, The Netherlands.
Bedir Tekinerdogan, Information Technology Group, Wageningen University and Research, Wageningen, The Netherlands.
Edith J M Feskens, Division of Human Nutrition and Health, Wageningen University and Research, Wageningen, The Netherlands.
Guido Camps, Division of Human Nutrition and Health, Wageningen University and Research, Wageningen, The Netherlands; OnePlanet Research Center, Wageningen, The Netherlands.
References
- 1. Colmenarejo G. Machine learning models to predict childhood and adolescent obesity: a review. Nutrients. 2020;12(8):2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lecroy MN, Kim RS, Stevens J, Hanna DB, Isasi CR. Identifying key determinants of childhood obesity: a narrative review of machine learning studies. Child Obes. 2021;17(3):153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reel PS, Reel S, Pearson E, Trucco E, Jefferson E. Using machine learning approaches for multi-omics data analysis: a review. Biotechnol Adv. 2021;49:107739. [DOI] [PubMed] [Google Scholar]
- 4. Li R, Li L, Xu Y, Yang J. Machine learning meets omics: applications and perspectives. Brief Bioinform. 2022;23(1):bbab460. [DOI] [PubMed] [Google Scholar]
- 5. Wang X, Liu J, Ma L. Identification of gut flora based on robust support vector machine. J Phys Conf Ser. 2022;2171(1):012066. [Google Scholar]
- 6. Namkung J. Machine learning methods for microbiome studies. J Microbiol. 2020;58(3):206–16. [DOI] [PubMed] [Google Scholar]
- 7. Cammarota G, Ianiro G, Ahern A, Carbone C, Temko A, Claesson MJet al. Gut microbiome, big data and machine learning to promote precision medicine for cancer. Nat Rev Gastroenterol Hepatol. 2020;17(10):635–48. [DOI] [PubMed] [Google Scholar]
- 8. Jorm LR. Commentary: towards machine learning-enabled epidemiology. Int J Epidemiol. 2021;49(6):1770–3. [DOI] [PubMed] [Google Scholar]
- 9. Wiemken TL, Kelley RR. Machine learning in epidemiology and health outcomes research. Annu Rev Public Health. 2020;41:21–36. [DOI] [PubMed] [Google Scholar]
- 10. Wiens J, Shenoy ES. Machine learning for healthcare: on the verge of a major shift in healthcare epidemiology. Clin Infect Dis. 2018;66(1):149–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kirk D, Catal C, Tekinerdogan B. Precision nutrition: a systematic literature review. Comput Biol Med. 2021;133:104365. [DOI] [PubMed] [Google Scholar]
- 12. Goecks J, Jalili V, Heiser LM, Gray JW. How machine learning will transform biomedicine. Cell. 2020;181(1):92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vilne B, Ķibilds J, Siksna I, Lazda I, Valciņa O, Krūmiņa A. Could artificial intelligence/machine learning and inclusion of diet-gut microbiome interactions improve disease risk prediction? Case study: coronary artery disease. Front Microbiol. 2022;13::627892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chollet F. On the measure of intelligence. [Internet]. 2019. Nov 5 [cited 2022 Jul 29]. Available from: https://arxiv.org/abs/1911.01547v2
- 15. Wang H, Ma C, Zhou L. A brief review of machine learning and its application. In: 2009 International Conference on Information Engineering and Computer Science. New York (NY): IEEE; 2009. doi:10.1109/ICIECS.2009.5362936 [Google Scholar]
- 16. Witten IH, Frank E, Hall MA, Pal CJ. Data mining: practical machine learning tools and techniques. Amsterdam (Netherlands): Elsevier; 2016. [Google Scholar]
- 17. Bzdok D, Altman N, Krzywinski M. Points of significance: statistics versus machine learning. Nat Methods. 2018;15(4):233–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Silva K, Jönsson D, Demmer RT. A combined strategy of feature selection and machine learning to identify predictors of prediabetes. J Am Med Inform Assoc. 2020;27(3):396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Poss AM, Maschek JA, Cox JE, Hauner BJ, Hopkins PN, Hunt SCet al. Machine learning reveals serum sphingolipids as cholesterol-independent biomarkers of coronary artery disease. J Clin Invest. 2020;130(3):1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gou W, Ling CW, He Y, Jiang Z, Fu Y, Xu Fet al. Interpretable machine learning framework reveals robust gut microbiome features associated with type 2 diabetes. Diabetes Care. 2021;44(2):358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dinh A, Miertschin S, Young A, Mohanty SD. A data-driven approach to predicting diabetes and cardiovascular disease with machine learning. BMC Med Inform Decis Mak. 2019;19(1):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tap J, Derrien M, Törnblom H, Brazeilles R, Cools-Portier S, Doré Jet al. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology. 2017;152(1):111–23.e8. [DOI] [PubMed] [Google Scholar]
- 23. Ambale-Venkatesh B, Yang X, Wu CO, Liu K, Gregory Hundley W, McClelland Ret al. Cardiovascular event prediction by machine learning: the multi-ethnic study of atherosclerosis. Circ Res. 2017;121(9):1092–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. De Silva K, Lim S, Mousa A, Teede H, Forbes A, Demmer RTet al. Nutritional markers of undiagnosed type 2 diabetes in adults: findings of a machine learning analysis with external validation and benchmarking. PLoS One. 2021;16(5):e0250832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Breiman L. Statistical modeling: the two cultures (with comments and a rejoinder by the author). [Internet]. 2001;16(3):199–231.. Available from: https://projecteuclid.org/journals/statistical-science/volume-16/issue-3/Statistical-Modeling–The-Two-Cultures-with-comments-and-a/10.1214/ss/1009213726.full [Google Scholar]
- 26. Song X, Mitnitski A, Cox J, Rockwood K. Comparison of machine learning techniques with classical statistical models in predicting health outcomes. Stud Health Technol Inform. 2004;107:736–40. [PubMed] [Google Scholar]
- 27. Stolfi P, Castiglione F. Emulating complex simulations by machine learning methods. BMC Bioinformatics. 2021;22(Suppl 14):483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Choi TY, Chang MY, Heo S, Jang JY. Explainable machine learning model to predict refeeding hypophosphatemia. Clin Nutr ESPEN. 2021;45:213–9. [DOI] [PubMed] [Google Scholar]
- 29. Khorraminezhad L, Leclercq M, Droit A, Bilodeau JF, Rudkowska I. Statistical and machine-learning analyses in nutritional genomics studies. Nutrients. 2020;12(10):3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ahmadi MN, Brookes D, Chowdhury A, Pavey T, Trost SG. Free-living evaluation of laboratory-based activity classifiers in preschoolers. Med Sci Sports Exercise. 2020;52(5):1227–34. [DOI] [PubMed] [Google Scholar]
- 31. Chowdhury AK, Tjondronegoro D, Chandran V, Trost SG. Ensemble methods for classification of physical activities from wrist accelerometry. Med Sci Sports Exercise. 2017;49(9):1965–73. [DOI] [PubMed] [Google Scholar]
- 32. Pavey TG, Gilson ND, Gomersall SR, Clark B, Trost SG. Field evaluation of a random forest activity classifier for wrist-worn accelerometer data. J Sci Med Sport. 2017;20(1):75–80. [DOI] [PubMed] [Google Scholar]
- 33. Catal C, Akbulut A. Automatic energy expenditure measurement for health science. Comput Methods Programs Biomed. 2018;157:31–7. [DOI] [PubMed] [Google Scholar]
- 34. Ahmadi MN, Chowdhury A, Pavey T, Trost SG. Laboratory-based and free-living algorithms for energy expenditure estimation in preschool children: a free-living evaluation. PLoS One. 2020;15(5):e0233229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rigdon J, Basu S. Machine learning with sparse nutrition data to improve cardiovascular mortality risk prediction in the USA using nationally randomly sampled data. BMJ Open. 2019;9(11):e032703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sánchez-Cabo F, Rossello X, Fuster V, Benito F, Manzano JP, Silla JCet al. Machine learning improves cardiovascular risk definition for young, asymptomatic individuals. J Am Coll Cardiol. 2020;76(14):1674–85. [DOI] [PubMed] [Google Scholar]
- 37. Kakadiaris IA, Vrigkas M, Yen AA, Kuznetsova T, Budoff M, Naghavi M. Machine learning outperforms ACC/AHA CVD risk calculator in MESA. J Am Heart Assoc. 2018;7(22):e009476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alaa AM, Bolton T, Angelantonio E Di, Rudd JHF, van der Schaar M. Cardiovascular disease risk prediction using automated machine learning: a prospective study of 423,604 UK Biobank participants. PLoS One. 2019;14(5):e0213653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sorino P, Campanella A, Bonfiglio C, Mirizzi A, Franco I, Bianco Aet al. Development and validation of a neural network for NAFLD diagnosis. Sci Rep. 2021;11(1):20240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Canbay A, Kälsch J, Neumann U, Rau M, Hohenester S, Baba HAet al. Non-invasive assessment of NAFLD as systemic disease—a machine learning perspective. PLoS One. 2019;14(3):e0214436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Khusial RD, Cioffi CE, Caltharp SA, Krasinskas AM, Alazraki A, Knight-Scott Jet al. Development of a plasma screening panel for pediatric nonalcoholic fatty liver disease using metabolomics. Hepatol Commun. 2019;3(10):1311–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frantzi M, Gomez Gomez E, Blanca Pedregosa A, Valero Rosa J, Latosinska A, Culig Zet al. CE-MS-based urinary biomarkers to distinguish non-significant from significant prostate cancer. Br J Cancer. 2019;120(12):1120–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cao R, Mohammadian Bajgiran A, Afshari Mirak S, Shakeri S, Zhong X, Enzmann Det al. Joint prostate cancer detection and Gleason score prediction in mp-MRI via FocalNet. IEEE Trans Med Imaging. 2019;38(11):2496–506. [DOI] [PubMed] [Google Scholar]
- 44. Yala A, Lehman C, Schuster T, Portnoi T, Barzilay R. A deep learning mammography-based model for improved breast cancer risk prediction. Radiology. 2019;292(1):60–6. [DOI] [PubMed] [Google Scholar]
- 45. Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng Let al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25(6):954–61. [DOI] [PubMed] [Google Scholar]
- 46. Cruz JA, Wishart DS. Applications of machine learning in cancer prediction and prognosis. Cancer Inform. 2007;2:59–77. [PMC free article] [PubMed] [Google Scholar]
- 47. Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sorino P, Caruso MG, Misciagna G, Bonfiglio C, Campanella A, Mirizzi Aet al. Selecting the best machine learning algorithm to support the diagnosis of non-alcoholic fatty liver disease: a meta learner study. PLoS One. 2020;15(10):e0240867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zou Q, Qu K, Luo Y, Yin D, Ju Y, Tang H. Predicting diabetes mellitus with machine learning techniques. Front Genet. 2018;9:515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kirk D, Catal C, Tekinerdogan B. Predicting plasma vitamin C using machine learning. Applied Artificial Intelligence. 2022;36(1):2042924. [Google Scholar]
- 51. Sarker IH. Machine learning: algorithms, real-world applications and research directions. SN Comput Sci. 2021;2(3):160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ramyaa R, Hosseini O, Krishnan GP, Krishnan S. Phenotyping women based on dietary macronutrients, physical activity, and body weight using machine learning tools. Nutrients. 2019;11(7):1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Basu S. Semi-supervised learning. In: Liu L, Özsu MT, editors. Encyclopedia of database systems. [Internet]. Boston (MA): Springer; 2009; [cited 2022 Feb 28]. p. 2613–5.. Available from: https://link.springer.com/referenceworkentry/10.1007/978-0-387-39940-9_609 [Google Scholar]
- 54. Nguyen TP, Ho TB. Detecting disease genes based on semi-supervised learning and protein–protein interaction networks. Artif Intell Med. 2012;54(1):63–71. [DOI] [PubMed] [Google Scholar]
- 55. Davidson I. Clustering with constraints. In: Liu L, Özsu MT, editors. Encyclopedia of database systems. [Internet]. Boston, MA: Springer; 2009; [cited 2022 Feb 28]. p. 393–6.. Available from: https://link.springer.com/referenceworkentry/10.1007/978-0-387-39940-9_610 [Google Scholar]
- 56. Silver D, Hubert T, Schrittwieser J, Antonoglou I, Lai M, Guez Aet al. A general reinforcement learning algorithm that masters chess, shogi, and go through self-play. Science. 2018;362(6419):1140–4. [DOI] [PubMed] [Google Scholar]
- 57. Mulani J, Heda S, Tumdi K, Patel J, Chhinkaniwala H, Patel Jet al. Deep reinforcement learning based personalized health recommendations. In: Dash S, Acharya B, Mittal M, Abraham A, Kelemen A, editors. Deep learning techniques for biomedical and health informatics. Cham (Switzerland): Springer; 2020. p. 231–55. [Google Scholar]
- 58. Zhou M, Mintz Y, Fukuoka Y, Goldberg K, Flowers E, Kaminsky Pet al. Personalizing mobile fitness apps using reinforcement learning. [Internet]. CEUR Workshop Proc. 2018;2068. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7220419/ [PMC free article] [PubMed] [Google Scholar]
- 59. Breiman L. Random forests. Machine Learning. 2001;45(1):5–32. [Google Scholar]
- 60. Awad M, Khanna R, Awad M, Khanna R. Support vector regression. In: Efficient learning machines. Cham (Switzerland): Springer; 2015. p. 67–80. [Google Scholar]
- 61. Zhou M, Fukuoka Y, Goldberg K, Vittinghoff E, Aswani A. Applying machine learning to predict future adherence to physical activity programs. BMC Med Inform Decis Mak. 2019;19(1):169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu WK, Panyod S, Liu PY, Chen CC, Kao HL, Chuang HLet al. Characterization of TMAO productivity from carnitine challenge facilitates personalized nutrition and microbiome signatures discovery. Microbiome. 2020;8(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jones PJ, Catt M, Davies MJ, Edwardson CL, Mirkes EM, Khunti Ket al. Feature selection for unsupervised machine learning of accelerometer data physical activity clusters—a systematic review. Gait Posture. 2021;90:120–8. [DOI] [PubMed] [Google Scholar]
- 64. Kim J, Lin S, Ferrara G, Hua J, Seto E. Identifying people based on machine learning classification of foods consumed in order to offer tailored healthier food options. In: Advances in intelligent systems and computing. Cham (Switzerland): Springer; 2020. p. 190–4. [Google Scholar]
- 65. Sowah RA, Bampoe-Addo AA, Armoo SK, Saalia FK, Gatsi F, Sarkodie-Mensah B. Design and development of diabetes management system using machine learning. Int J Telemed Appl. 2020;2020:8870141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mitchell EG, Heitkemper EM, Burgermaster M, Levine ME, Miao Y, Hwang MLet al. From reflection to action: combining machine learning with expert knowledge for nutrition goal recommendations. Proc SIGCHI Conf Hum Factor Comput Syst. 2021;2021:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Metwally AA, Leong AK, Desai A, Nagarjuna A, Perelman D, Snyder M. Learning personal food preferences via food logs embedding. 2022;2281–6..Available from: 10.48550/arXiv.2110.15498. [DOI] [Google Scholar]
- 68. Baek JW, Kim JC, Chun J, Chung K. Hybrid clustering based health decision-making for improving dietary habits. Technol Health Care. 2019;27(5):459–72. [DOI] [PubMed] [Google Scholar]
- 69. Hua J, Tembe WD, Dougherty ER. Performance of feature-selection methods in the classification of high-dimension data. Pattern Recognit. 2009;42(3):409–24. [Google Scholar]
- 70. Waggoner PD. Modern dimension reduction. Cambridge (UK): Cambridge University Pres; 2021. doi:10.1017/9781108981767 [Google Scholar]
- 71. Gavai AK. Bayesian networks for omics data analysis. Wageningen (Netherlands); Wageningen University; 2009. [Google Scholar]
- 72. Barredo Arrieta A, Díaz-Rodríguez N, Del Ser J, Bennetot A, Tabik S, Barbado Aet al. Explainable artificial intelligence (XAI): concepts, taxonomies, opportunities and challenges toward responsible AI. Information Fusion. 2020;58:82–115. [Google Scholar]
- 73. Maduri C, Sabrina Hsueh, PY, Li Z, Chen CH, Papoutsakis C. Applying contemporary machine learning approaches to nutrition care real-world evidence: findings from the national quality improvement data set. J Acad Nutr Diet. 2021;121(12):2549–59.e1. [DOI] [PubMed] [Google Scholar]
- 74. Wong RK, Pitino MA, Mahmood R, Zhu IY, Stone D, O'Connor DLet al. Predicting protein and fat content in human donor milk using machine learning. J Nutr. 2021;151(7):2075–83. [DOI] [PubMed] [Google Scholar]
- 75. Lundberg SM, Lee SI. A unified approach to interpreting model predictions. [Internet]. Adv Neural Inf Process Syst. 2017;2017:4766–75.. Available from: https://arxiv.org/abs/1705.07874v2 [Google Scholar]
- 76. Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat. 2001;29(5):1189–232. [Google Scholar]
- 77. Ribeiro MT, Singh S, Guestrin C. Model-Agnostic interpretability of machine learning. [Internet]. 2016. Jun 16 [cited 2022 Sep 2]. Available from: https://arxiv.org/abs/1606.05386v1
- 78. Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger Aet al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163(5):1079–94. [DOI] [PubMed] [Google Scholar]
- 79. Davagdorj K, Li M, Ryu KH. Local interpretable model-agnostic explanations of predictive models for hypertension. In: Pan JS, Li J, Ryu KH, Meng Z, Klasnja-Milicevic A, editors. Advances in intelligent information hiding and multimedia signal processing. Singapore: Springer; 2021. p. 426–33. [Google Scholar]
- 80. Palacio-Niño JO, Berzal F. Evaluation metrics for unsupervised learning algorithms. [Internet]. 2019. May 14 [cited 2022 Feb 28]. Available from: https://arxiv.org/abs/1905.05667v2
- 81. Trevethan R. Sensitivity, specificity, and predictive values: foundations, pliabilities, and pitfalls in research and practice. Front Public Heal, 2017;5:307.doi: 10.3389/fpubh.2017.00307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Centers for Disease Control and Prevention. National diabetes statistics report 2020: Estimates of diabetes and its burden in the United States. Atlanta (GA): US Department of Health and Human Services; 2020.; [Google Scholar]
- 83. Linden A. Measuring diagnostic and predictive accuracy in disease management: an introduction to receiver operating characteristic (ROC) analysis. J Eval Clin Pract. 2006;12(2):132–9. [DOI] [PubMed] [Google Scholar]
- 84. Cawley GC, Talbot NLC. On over-fitting in model selection and subsequent selection bias in performance evaluation. Journal of Machine Learning Research. 2010;11:2079–107. [Google Scholar]
- 85. Mendes-Soares H, Raveh-Sadka T, Azulay S, Ben-Shlomo Y, Cohen Y, Ofek Tet al. Model of personalized postprandial glycemic response to food developed for an Israeli cohort predicts responses in midwestern american individuals. Am J Clin Nutr. 2019;110(1):63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Berry S, Valdes A, Davies R, Delahanty L, Drew D, Chan ATet al. Predicting personal metabolic responses to food using multi-omics machine learning in over 1000 twins and singletons from the UK and US: the PREDICT I study (OR31-01-19). Curr Dev Nutr. 2019;3(Suppl 1):nzz037. [Google Scholar]
- 87. Wu H, Tremaroli V, Schmidt C, Lundqvist A, Olsson LM, Krämer Met al. The gut microbiota in prediabetes and diabetes: a population-based cross-sectional study. Cell Metab. 2020;32(3):379–90.e3. [DOI] [PubMed] [Google Scholar]
- 88. Chatelan A, Bochud M, Frohlich KL. Precision nutrition: hype or hope for public health interventions to reduce obesity?. Int J Epidemiol. 2019;48(2):332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. National Institutes of Health. NIH awards $170 million for precision nutrition study. [Internet]. 2022; [cited 2022 Mar 21]. Available from: https://www.nih.gov/news-events/news-releases/nih-awards-170-million-precision-nutrition-study
- 90. Berry SE, Valdes AM, Drew DA, Asnicar F, Mazidi M, Wolf Jet al. Human postprandial responses to food and potential for precision nutrition. Nat Med. 2020;26(6):964–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zellerbach K, Ruiz C. Machine learning to predict overeating from macronutrient composition. In: Proceedings—2019 IEEE International Conference on Bioinformatics and Biomedicine, BIBM 2019. Piscataway (NJ): Institute of Electrical and Electronics Engineers Inc; 2019. p. 1268–70. [Google Scholar]
- 92. Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. 2018;361:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Raphaeli O, Singer P. Towards personalized nutritional treatment for malnutrition using machine learning-based screening tools. Clin Nutr. 2021;40(10):5249–51. [DOI] [PubMed] [Google Scholar]
- 94. Yin L, Song C, Cui J, Lin X, Li N, Fan Yet al. A fusion decision system to identify and grade malnutrition in cancer patients: machine learning reveals feasible workflow from representative real-world data. Clin Nutr. 2021;40(8):4958–70. [DOI] [PubMed] [Google Scholar]
- 95. Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MAet al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510(7505):417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Fenta HM, Zewotir T, Muluneh EK. A machine learning classifier approach for identifying the determinants of under-five child undernutrition in Ethiopian administrative zones. BMC Med Inform Decis Mak. 2021;21(1):291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Talukder A, Ahammed B. Machine learning algorithms for predicting malnutrition among under-five children in Bangladesh. Nutrition. 2020;78:110861. [DOI] [PubMed] [Google Scholar]
- 98. Islam MM, Rahman MJ, Islam MM, Roy DC, Ahmed N, Hussain Set al. Application of machine learning based algorithm for prediction of malnutrition among women in Bangladesh. International Journal of Cognitive Computing in Engineering. 2022;3:46–57. [Google Scholar]
- 99. Bingham SA. Limitations of the various methods for collecting dietary intake data. Ann Nutr Metab. 1991;35(3):117–27. [DOI] [PubMed] [Google Scholar]
- 100. Schoeller DA. Limitations in the assessment of dietary energy intake by self-report. Metabolism. 1995;44(Suppl 2):18–22. [DOI] [PubMed] [Google Scholar]
- 101. Mezgec S, Eftimov T, Bucher T, Koroušić Seljak B. Mixed deep learning and natural language processing method for fake-food image recognition and standardization to help automated dietary assessment. Public Health Nutr. 2019;22(7):1193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. van Erp M, Reynolds C, Maynard D, Starke A, Ibáñez Martín R, Andres Fet al. Using natural language processing and artificial intelligence to explore the nutrition and sustainability of recipes and food. Front Artif Intell. 2021;3:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ahmadi MN, Pavey TG, Trost SG. Machine learning models for classifying physical activity in free-living preschool children. Sensors (Basel). 2020;20(16):4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Fridolfsson J, Arvidsson D, Doerks F, Kreidler TJ, Grau S. Workplace activity classification from shoe-based movement sensors. BMC Biomed Eng. 2020;2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Fergus P, Hussain AJ, Hearty J, Fairclough S, Boddy L, Mackintosh Ket al. A machine learning approach to measure and monitor physical activity in children. Neurocomputing. 2017;228:220–30. [Google Scholar]
- 106. Kingsley MIC, Nawaratne R, O'Halloran PD, Montoye AHK, Alahakoon D, De Silva Det al. Wrist-specific accelerometry methods for estimating free-living physical activity. J Sci Med Sport. 2019;22(6):677–83. [DOI] [PubMed] [Google Scholar]
- 107. O'Driscoll R, Turicchi J, Hopkins M, Horgan GW, Finlayson G, Stubbs JR. Improving energy expenditure estimates from wearable devices: a machine learning approach. J Sports Sci. 2020;38(13):1496–505. [DOI] [PubMed] [Google Scholar]
- 108. Perakakis N, Yazdani A, Karniadakis GE, Mantzoros C. Omics, big data and machine learning as tools to propel understanding of biological mechanisms and to discover novel diagnostics and therapeutics. Metabolism. 2018;87:A1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Subramanian I, Verma S, Kumar S, Jere A, Anamika K. Multi-omics data integration, interpretation, and its application. Bioinf Biol Insights. 2020;14:117793221989905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Montanez CAC, Fergus P, Hussain A, Al-Jumeily D, Abdulaimma B, Hind Jet al. Machine learning approaches for the prediction of obesity using publicly available genetic profiles. In: Proceedings of the International Joint Conference on Neural Networks. Piscataway (NJ): Institute of Electrical and Electronics Engineers Inc; 2017. p. 2743–50. [Google Scholar]
- 111. Montañez CAC, Fergus P, Hussain A, Al-Jumeily D, Dorak MT, Abdullah R. Evaluation of phenotype classification methods for obesity using direct to consumer genetic data. In: Lecture notes in computer science. Berlin (Germany): Springer Verlag; 2017. p. 350–62. [Google Scholar]
- 112. Rodríguez-Pardo C, Segura A, Zamorano-León JJ, Martínez-Santos C, Martínez D, Collado-Yurrita Let al. Decision tree learning to predict overweight/obesity based on body mass index and gene polymporphisms. Gene. 2019;699:88–93. [DOI] [PubMed] [Google Scholar]
- 113. López B, Torrent-Fontbona F, Viñas R, Fernández-Real JM. Single nucleotide polymorphism relevance learning with random forests for type 2 diabetes risk prediction. Artif Intell Med. 2018;85:43–9. [DOI] [PubMed] [Google Scholar]
- 114. Wang Y, Zhang L, Niu M, Li R, Tu R, Liu Xet al. Genetic risk score increased discriminant efficiency of predictive models for type 2 diabetes mellitus using machine learning: cohort study. Front Public Heal. 2021;9::606711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Holmes E, Wilson ID, Nicholson JK. Metabolic phenotyping in health and disease. Cell. 2008;134(5):714–7. [DOI] [PubMed] [Google Scholar]
- 116. Korem T, Zeevi D, Zmora N, Weissbrod O, Bar N, Lotan-Pompan Met al. Bread affects clinical parameters and induces gut microbiome-associated personal glycemic responses. Cell Metab. 2017;25(6):1243–53.e5. [DOI] [PubMed] [Google Scholar]
- 117. Nielsen RL, Helenius M, Garcia SL, Roager HM, Aytan-Aktug D, Hansen LBSet al. Data integration for prediction of weight loss in randomized controlled dietary trials. Sci Rep. 2020;10(1):20103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Gardner CD, Trepanowski JF, Gobbo LCD, Hauser ME, Rigdon J, Ioannidis JPAet al. Effect of low-fat VS low-carbohydrate diet on 12-month weight loss in overweight adults and the association with genotype pattern or insulin secretion the DIETFITS randomized clinical trial. JAMA. 2018;319(7):667–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Celis-Morales C, Livingstone KM, Marsaux CFM, Macready AL, Fallaize R, O'Donovan CBet al. Effect of personalized nutrition on health-related behaviour change: evidence from the Food4Me European randomized controlled trial. Int J Epidemiol. 2017;46(2):578–88. [DOI] [PubMed] [Google Scholar]
- 120. An J, Yoon SR, Lee JH, Kim H, Kim OY. Importance of adherence to personalized diet intervention in obesity related metabolic improvement in overweight and obese Korean adults. Clin Nutr Res. 2019;8(3):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Letertre MPM, Giraudeau P, de Tullio P. Nuclear magnetic resonance spectroscopy in clinical metabolomics and personalized medicine: current challenges and perspectives. Front Mol Biosci. 2021;8:698337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Fiehn O. Metabolomics—the link between genotypes and phenotypes. Plant Mol Biol. 2002;48(1):155–71. [PubMed] [Google Scholar]
- 123. O'Sullivan A, Gibney MJ, Connor AO, Mion B, Kaluskar S, Cashman KDet al. Biochemical and metabolomic phenotyping in the identification of a vitamin D responsive metabotype for markers of the metabolic syndrome. Mol Nutr Food Res. 2011;55(5):679–90. [DOI] [PubMed] [Google Scholar]
- 124. O'Donovan CB, Walsh MC, Woolhead C, Forster H, Celis-Morales C, Fallaize Ret al. Metabotyping for the development of tailored dietary advice solutions in a European population: the Food4Me study. Br J Nutr. 2017;118(8):561–9. [DOI] [PubMed] [Google Scholar]
- 125. O'Donovan CB, Walsh MC, Nugent AP, McNulty B, Walton J, Flynn Aet al. Use of metabotyping for the delivery of personalised nutrition. Mol Nutr Food Res. 2015;59(3):377–85. [DOI] [PubMed] [Google Scholar]
- 126. Rochlani Y, Pothineni NV, Kovelamudi S, Mehta JL. Metabolic syndrome: pathophysiology, management, and modulation by natural compounds. Ther Adv Cardiovasc Dis. 2017;11(8):215–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Saklayen MG. The global epidemic of the metabolic syndrome. Curr Hypertens Reports 2018;20(2):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Korduner J, Nilsson PM, Melander O, Gerl MJ, Engström G, Bachus Eet al. Proteomic and metabolomic characterization of metabolically healthy obesity: a descriptive study from a Swedish cohort. J Obes. 2021;2021::6616983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Cirulli ET, Guo L, Leon Swisher C, Shah N, Huang L, Napier LAet al. Profound perturbation of the metabolome in obesity is associated with health risk. Cell Metab. 2019;29(2):488–500.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Chen HH, Tseng YJ, Wang SY, Tsai YS, Chang CS, Kuo TCet al. The metabolome profiling and pathway analysis in metabolic healthy and abnormal obesity. Int J Obes. 2015;39(8):1241–8. [DOI] [PubMed] [Google Scholar]
- 131. Cheng D, Zhao X, Yang S, Cui H, Wang G. Metabolomic signature between metabolically healthy overweight/obese and metabolically unhealthy overweight/obese: a systematic review. Diabetes Metab Syndr Obes. 2021;14:991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Bagheri M, Farzadfar F, Qi L, Yekaninejad MS, Chamari M, Zeleznik OAet al. Obesity-related metabolomic profiles and discrimination of metabolically unhealthy obesity. J Proteome Res. 2018;17(4):1452–62. [DOI] [PubMed] [Google Scholar]
- 133. Chashmniam S, Madani NH, Ghoochani B, Safari-Alighiarloo N, Khamseh ME. The metabolome profiling of obese and non-obese individuals: metabolically healthy obese and unhealthy non-obese paradox. Iran J Basic Med Sci. 2020;23(2):186–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Vázquez-Fresno R, Llorach R, Perera A, Mandal R, Feliz M, Tinahones FJet al. Clinical phenotype clustering in cardiovascular risk patients for the identification of responsive metabotypes after red wine polyphenol intake. J Nutr Biochem. 2016;28:114–20. [DOI] [PubMed] [Google Scholar]
- 135. Dong TS, Mayer EA, Osadchiy V, Chang C, Katzka W, Lagishetty Vet al. A distinct brain-gut-microbiome profile exists for females with obesity and food addiction. Obes. 2020;28(8):1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Hollister EB, Oezguen N, Chumpitazi BP, Luna RA, Weidler EM, Rubio-Gonzales Met al. Leveraging human microbiome features to diagnose and stratify children with irritable bowel syndrome. J Mol Diagn. 2019;21(3):449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Prochazkova M, Budinska E, Kuzma M, Pelantova H, Hradecky J, Heczkova Met al. Vegan diet is associated with favorable effects on the metabolic performance of intestinal microbiota: a cross-sectional multi-omics study. Front Nutr. 2022;8:783302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wang DD, Hu FB. Precision nutrition for prevention and management of type 2 diabetes. Lancet Diabetes Endocrinol. 2018;6:416–26. [DOI] [PubMed] [Google Scholar]
- 139. Acar E, Gürdeniz G, Khakimov B, Savorani F, Korndal SK, Larsen TMet al. Biomarkers of individual foods, and separation of diets using untargeted LC-MS-based plasma metabolomics in a randomized controlled trial. Mol Nutr Food Res. 2019;63(1):1800215. [DOI] [PubMed] [Google Scholar]
- 140. Garcia-Aloy M, Llorach R, Urpi-Sarda M, Tulipani S, Salas-Salvadó J, Martínez-González MAet al. Nutrimetabolomics fingerprinting to identify biomarkers of bread exposure in a free-living population from the PREDIMED study cohort. Metabolomics. 2015;11(1):155–65. [Google Scholar]
- 141. Cornelis MC, Erlund I, Michelotti GA, Herder C, Westerhuis JA, Tuomilehto J. Metabolomic response to coffee consumption: application to a three-stage clinical trial. J Intern Med. 2018;283(6):544–57. [DOI] [PubMed] [Google Scholar]
- 142. Heinzmann SS, Brown IJ, Chan Q, Bictash M, Dumas ME, Kochhar Set al. Metabolic profiling strategy for discovery of nutritional biomarkers: proline betaine as a marker of citrus consumption. Am J Clin Nutr. 2010;92(2):436–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Cheung W, Keski-Rahkonen P, Assi N, Ferrari P, Freisling H, Rinaldi Set al. A metabolomic study of biomarkers of meat and fish intake. Am J Clin Nutr. 2017;105(3):600–8. [DOI] [PubMed] [Google Scholar]
- 144. Cerdá B, Tomás-Barberán FA, Espín JC. Metabolism of antioxidant and chemopreventive ellagitannins from strawberries, raspberries, walnuts, and oak-aged wine in humans: identification of biomarkers and individual variability. J Agric Food Chem. 2005;53(2):227–35. [DOI] [PubMed] [Google Scholar]
- 145. Neveu V, Perez-Jiménez J, Vos F, Crespy V, du Chaffaut L, Mennen Let al. Phenol-Explorer: an online comprehensive database on polyphenol contents in foods. Database (Oxford). 2010;2010:bap024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Li KJ, Brouwer-Brolsma EM, Burton-Pimentel KJ, Vergères G, Feskens EJM. A systematic review to identify biomarkers of intake for fermented food products. Genes Nutr. 2021;16(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Hang D, Zeleznik OA, He X, Guasch-Ferre M, Jiang X, Li Jet al. Metabolomic signatures of long-term coffee consumption and risk of type 2 diabetes in women. Diabetes Care. 2020;43(10):2588–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Pomyen Y, Wanichthanarak K, Poungsombat P, Fahrmann J, Grapov D, Khoomrung S. Deep metabolome: applications of deep learning in metabolomics. Comput Struct Biotechnol J. 2020;18:2818–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Brinkkemper S. Method engineering: engineering of information systems development methods and tools. Inf Softw Technol. 1996;38(4):275–80. [Google Scholar]
- 150. Scheffer J. Dealing with missing data. [Internet]. Research Letters in the Information and Mathematical Sciences. 2002;3:153–60.. Available from: https://mro.massey.ac.nz/handle/10179/4355 [Google Scholar]
- 151. Kang H. The prevention and handling of the missing data. Korean J Anesthesiol. 2013;64(5):402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Jakobsen JC, Gluud C, Wetterslev J, Winkel P. When and how should multiple imputation be used for handling missing data in randomised clinical trials—a practical guide with flowcharts. BMC Med Res Method. 2017;17(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Harrell FE. Missing data. In: Regression modeling strategies. Cham (Switzerland); Springer Nature; 2015. p. 45–61. [Google Scholar]
- 154. Ware JH, Harrington D, Hunter DJ, D'Agostino RBS. Missing data. N Engl J Med. 2012;367(14):1353–4. [Google Scholar]
- 155. Jerez JM, Molina I, García-Laencina PJ, Alba E, Ribelles N, Martín Met al. Missing data imputation using statistical and machine learning methods in a real breast cancer problem. Artif Intell Med. 2010;50(2):105–15. [DOI] [PubMed] [Google Scholar]
- 156. Lakshminarayan K, Harp SA, Goldman R, Samad T. Imputation of missing data using machine learning techniques. [Internet]. 1996; [cited 2022 Jul 30]. Available from: http://www.aaai.org
- 157. Hong S, Lynn HS. Accuracy of random-forest-based imputation of missing data in the presence of non-normality, non-linearity, and interaction. BMC Med Res Method. 2020;20(1):199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Stekhoven DJ, Bühlmann P. MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics. 2012;28(1):112–8. [DOI] [PubMed] [Google Scholar]
- 159. Chen T, Guestrin C. XGBoost: a scalable tree boosting system. [Internet]. Available from: https://arxiv.org/abs/1603.02754v3
- 160. Krueger C, Tian L. A comparison of the general linear mixed model and repeated measures ANOVA using a dataset with multiple missing data points. Biol Res Nurs. 2004;6(2):151–7. [DOI] [PubMed] [Google Scholar]
- 161. Nijman SWJ, Leeuwenberg AM, Beekers I, Verkouter I, Jacobs JJL, Bots MLet al. Missing data is poorly handled and reported in prediction model studies using machine learning: a literature review. J Clin Epidemiol. 2022;142:218–29. [DOI] [PubMed] [Google Scholar]
- 162. Kondrup J, Johansen N, Plum LM, Bak L, Larsen HI, Martinsen Aet al. Incidence of nutritional risk and causes of inadequate nutritional care in hospitals. Clin Nutr. 2002;21(6):461–8. [DOI] [PubMed] [Google Scholar]
- 163. Wu J, Cai Z, Zhu X. Self-adaptive probability estimation for naive Bayes classification. In: The 2013 International Joint Conference on Neural Networks. New York (NY): IEEE; 2013. [Google Scholar]
- 164. Levy JJ, O'Malley AJ. Don't dismiss logistic regression: the case for sensible extraction of interactions in the era of machine learning. BMC Med Res Method. 2020;20(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. van der Laan MJ, Polley EC, Hubbard AE. Super learner. [Internet]. Statistical Applications in Genetics and Molecular Biolog. 2007;6(1). Available from: https://www.degruyter.com/document/doi/10.2202/1544-6115.1309/html [DOI] [PubMed] [Google Scholar]
- 166. Taha AA, Malebary SJ. A hybrid meta-classifier of fuzzy clustering and logistic regression for diabetes prediction. Computers, Materials and Continua. 2022;71(2):6089–105. [Google Scholar]
- 167. Naimi AI, Balzer LB. Stacked generalization: an introduction to super learning. Eur J Epidemiol. 2018;33(5):459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Bodnar LM, Cartus AR, Kirkpatrick SI, Himes KP, Kennedy EH, Simhan HNet al. Machine learning as a strategy to account for dietary synergy: an illustration based on dietary intake and adverse pregnancy outcomes. Am J Clin Nutr. 2020;111(6):1235–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.