Abstract
Vibrio cholerae is a facultative intestinal pathogen that lives in aquatic environments, often in association with planktonic species. In the suckling mouse, oral inoculation with V. cholerae leads to intestinal colonization and symptoms of diarrheal disease. Results reported here indicate a role for the alternative sigma factor, RpoS, in intestinal colonization in this model of cholera. We constructed within rpoS multiple independent mutations which consistently resulted in a fivefold decrease in colonization ability as assessed by competition assays. These mutations had no detectable effect on the in vitro growth of V. cholerae in a rich medium. The occurrence of spontaneous suppressor mutations potentially required for viability of rpoS strains was ruled out by determination of the frequency of insertional inactivation of rpoS in comparison to two other nonessential loci. Finally, both the in vitro and in vivo mutant phenotypes of rpoS strains were fully complemented by providing rpoS in trans or by allelic reversion, indicating that the observed decrease in colonization fitness was indeed due to the loss of functional RpoS.
A common theme emerges when one considers the diverse and sometimes harsh parameters encountered by bacteria during the course of their life cycles: microbes are experts at adaptation and survival within often tumultuous environments. Additionally, when one considers that a single bacterial species may encounter a broad spectrum of environments during the course of its life cycle, it is not surprising that bacteria have developed complex regulatory systems and stress adaptation mechanisms to best capitalize on each environment encountered. One particularly drastic change in environment is experienced by facultative pathogenic bacteria as they transition from their natural reservoir to the host organism. For instance, Vibrio cholerae is a gram-negative bacterium that naturally exists within an aquatic reservoir and infects human beings. Within the aquatic environment, V. cholerae is found in close association with planktonic species (13). This pathogen enters its human host through contaminated food and water, passes through the gastric acid barrier, and colonizes the relatively sterile environment of the small intestine, where it produces cholera toxin (reviewed in reference 20). Recently, a system involved in adaptation to low pH that likely plays a role in survival within the host gastrointestinal tract was described (17).
The ability to adapt to changing environments and to mount a general stress response has been the focus of intense study in many bacterial species. The ability to alter gene expression patterns via alternative sigma factors often plays an important role in the adaptation process (12). Specifically, ςS, which is encoded by rpoS, has been shown to play a key role in the adaptive processes of a diverse group of bacterial species (2, 10, 11, 12, 14, 21). RpoS has been best characterized in Escherichia coli, where it has been shown to control a regulon consisting of at least 30 genes. These genes are expressed upon entry into stationary phase and are also involved in the general stress response, which is required for survival upon exposure to starvation conditions, low pH, and oxidative stress (reviewed in reference 12).
In pathogenic bacterial species, the role of RpoS in the general stress response and virulence is varied. For example, it was recently shown that Legionella pneumophila requires RpoS for growth within an amoeboid species that serves as its natural reservoir. However, unlike in E. coli, RpoS seems to play no role in the growth phase-dependent stress responses of L. pneumophila (10). RpoS has been shown to be critical for stress response in Yersinia enterocolitica in a temperature-dependent manner (2). RpoS is a critical component of low-pH survival of Shigella flexneri (21) as well as Salmonella enterica serovar Typhimurium (7). The role that RpoS plays in virulence of each of these pathogenic organisms is also varied. For example, in animal models, an rpoS mutant of Y. enterocolitica was not attenuated in virulence, whereas rpoS mutants of S. enterica serovar Typhimurium showed significant attenuation (2, 7). In fact, the 50% lethal dose of an rpoS Salmonella strain is 1,000-fold higher than that of the wild type (7).
The rpoS orthologue of V. cholerae was recently identified and shown to play a crucial role in survival under a variety of stressful situations, including exposure to hydrogen peroxide, hyperosmolarity, and nutrient deprivation (22). In contrast to Salmonella, however, the V. cholerae RpoS was not required for colonization of the murine intestine. A separate study tested the role of RpoS in the ability of V. cholerae to mount an adaptive stress response, known as the acid tolerance response, upon exposure to acidic conditions (17). Once again, in contrast to the results with Salmonella, RpoS was not found to play a role in survival of V. cholerae under acidic conditions. In the present study, competition assays were used to analyze the ability of the rpoS strain to colonize the suckling mouse small intestine. Here we show that in contrast to previous reports, an rpoS null strain of V. cholerae is attenuated in its ability to colonize the small intestine.
MATERIALS AND METHODS
Strain and plasmid construction.
All strains and plasmids used in this study are listed in Table 1. pDSM747 was constructed by PCR amplification of a 2,611-bp fragment from the chromosome of C6709-1, using Taq polymerase and primers RpoS1 (5′-GCGAGATCTGTTGAACCTGTCGGTAA-3′) and RpoS-R-mut (5′-GGTGTTGATGGATGAGAT-3′). The PCR product was ligated directly into pGEMT (Promega) and then liberated by digestion with SalI/SphI. The resulting fragment was subcloned into similarly digested pCVD442 (6). All plasmid integration mutations were made using pGP704 (18), while deletions were made using pCVD442. All plasmids used for construction of insertion mutations were mobilized into V. cholerae from E. coli SM10λpir as previously described (17), and all integration mutations were subsequently verified by Southern blot analysis.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Relevant phenotype(s) | Source or reference |
|---|---|---|
| E. coli strains | ||
| SM10λpir | thi recA thr leu tonA lacY supE | Laboratory strain |
| RP4-2-Tc::Mu λ::pir strain | ||
| AC-E184 | SM10λpir(pAC160) | 5 |
| AC-E189 | SM10λpir(pAC214) | 5 |
| DSM-E713 | SM10λpir(pDSM375) | 17 |
| V. cholerae strains | ||
| C6709-1 | El Tor biotype, Smr | Roberts et al.a |
| AC-V168 | C6709-1 lacZ::pGP704, LacZ− | 5 |
| DSM-V382 | C6709-1 rpoS::pGP704 | 17 |
| DSM-V490 | C6709-1 rpoS::pCVD442 | This work |
| DSM-V491 | C6709-1 ΔrpoS | 17 |
| DSM-V506 | C6709-1 ΔrpoS (pFY7) | This work |
| DSM-V583 | C6709-1(pMMB67EH), LacZ− | This work |
| DSM-V714 | C6709-1(pFY7) | This work |
| DSM-V717 | DSM-V506 cured of pFY7 | This work |
| DSM-V719 | DSM-V491 reverted to wild type | This work |
| DSM-V720 | DSM-V717 reverted to wild type | This work |
| Plasmids | ||
| pAC160 | pGP704::′iviVI′ | 5 |
| pAC214 | pGP704::′orf3′ | 5 |
| pFY7 | pMMB67EH::′nlpD-rpoS-mutS′ | 22 |
| pDSM375 | pGP704::′rpoS′ | 17 |
| pDSM747 | pCVD442::′nlpD-rpoS-mutS′ | This work |
| pMMB67EH | IncQ broad-host-range cloning vector | 22 |
A. Roberts, G. D. Pearson, and J. J. Mekalanos, Proc. 28th Joint Conf., U.S.-Japan Cooperative Med. Sci. Prog. Cholera Related Diarrheal Dis., p. 43–47, 1992.
Growth conditions.
All strains were maintained at −80°C in Luria-Bertani (LB) broth containing 30% glycerol. All strains were grown at 37°C in LB broth with the following exception: pFY7, encoding ampicillin resistance, was cured from DSM-V506 by growth at 42°C. After growth at 42°C, DSM-V506 was plated on LB agar (LB), and colonies were replica plated to LB supplemented with ampicillin. Ampicillin-sensitive colonies were colony purified on LB and retested for ampicillin sensitivity. Ampicillin and streptomycin were used at concentrations of 100 μg ml−1. Counterselection of pCVD442 was accomplished by plating on LB lacking NaCl but supplemented with 10% sucrose followed by growth at 30°C. All growth curves, whether single or competitive, were done by diluting overnight cultures into LB broth. Single-strain growth assays were done with a 200-fold dilution, while competitive assays used a 1,000-fold dilution of each of the appropriate strains. CFU were determined by serial dilution and plating on LB supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 40 μg ml−1) and/or the appropriate antibiotic.
Frequency of plasmid insertion.
The frequency of plasmid insertion was determined at the rpoS, iviVI, and orf3 loci in the following manner. E. coli SM10λpir (donor) was used for mobilization of each suicide construct into V. cholerae C6709-1 (recipient). Overnight cultures of donor and recipient were mixed together at a 1:1 (vol/vol) ratio, and 50 μl was spotted onto dry LB plates and incubated at 37°C for 4 h. The titer of each donor and recipient strain in the overnight cultures was concurrently determined by serial dilution and plating on LB. After 4 h, the mating mixture was excised from the agar plate, resuspended in 2 ml of LB, and then plated on LB supplemented with ampicillin and streptomycin to select for transconjugants. The frequency of plasmid integration at each locus was calculated by division of the total number of transconjugants by the total number of recipient cells per mating.
Reversion of the ΔrpoS mutation.
A DNA fragment that includes the entire 1,008-bp rpoS coding sequence, as well as 1,009 bp of upstream and 594 bp of downstream sequence, was amplified from C6709-1 and cloned into pCVD442 (6), resulting in the allelic exchange vector pCVD442::′nlpD-rpoS-mutS′ (named pDSM-747). pDSM-747 was mobilized into DSM-V491 and DSM-V717 as previously described (17), and subsequent double-crossover products were isolated as previously described (6). Replacement of ΔrpoS by the wild-type rpoS allele in the sucrose-resistant and ampicillin-sensitive strains was confirmed by Southern blot analysis.
In vitro and in vivo competition assays and peroxide exposure assays.
Hydrogen peroxide killing assays (22) and competition assays (17) were conducted as previously described. The output ratio from each in vivo and in vitro competition was corrected for any deviations in the inoculum ratio from a value of 1:1.
RESULTS
An rpoS null derivative of V. cholerae C6709-1 is defective for intestinal colonization.
Strain DSM-V382, which contains a plasmid insertion within the V. cholerae rpoS coding sequence, was previously constructed to assess the phenotype of an rpoS mutant in response to acid exposure (17). DSM-V382 exhibited enhanced sensitivity to hydrogen peroxide exposure, as expected for an rpoS strain (22), but was fully competent for survival upon exposure to low pH (17). Nevertheless, we hypothesized that RpoS might play an important role in the establishment and persistence of V. cholerae infections since it has been shown to be essential for full virulence of S. enterica serovar Typhimurium, which also infects its host via an oral route. To address this hypothesis, DSM-V382 was coinoculated with the fully virulent, isogenic, LacZ− strain AC-V168 into 5-day-old suckling CD1 mice. In this competition assay, DSM-V382 was attenuated in its ability to successfully colonize the suckling mouse small intestine, as indicated by its in vivo competition index of 0.2 (Table 2). Strains with an equivalent ability to colonize would be expected to show a competition index of approximately 1.0. Thus, the V. cholerae rpoS strain was fivefold attenuated in colonizing the suckling mouse intestine. A second rpoS strain with a different plasmid insertion mutation, DSM-V490, was found to have the same defect in intestinal colonization (Table 2).
TABLE 2.
In vitro and in vivo competition assays
| Competing straina | Relevant genotype | Competition indexb
|
nc | |
|---|---|---|---|---|
| In vitro | In vivo | |||
| DSM-V382 | rpoS::pGP704 | 2.0 | 0.20 | 4 |
| DSM-V490 | rpoS::pCVD442 | 1.0 | 0.25 | 8 |
| DSM-V491 | ΔrpoS | 1.2 | 0.21 | 8 |
| DSM-V506 | ΔrpoS (pFY7) | 32 | 1.2 | 8 |
| DSM-V506d | ΔrpoS (pFY7) | 2.0 | 1.7 | 5 |
| DSM-V717 | DSM-V506 cured of pFY7 | 4.2 | 0.12 | 5 |
| DSM-V719 | DSM-V491 reverted | 1.1 | 1.1 | 8 |
| DSM-V720 | DSM-V717 reverted | 2.9 | 0.88 | 5 |
All competition assays were between the indicated strain and the isogenic wild-type strain C6709-1 (LacZ+) or AC-V168 (LacZ−) unless otherwise indicated.
Calculated by dividing mutant output number by wild-type output number and correcting the quotient for input deviations from 1:1.
n, number of animals used per competition experiment. All competitions were conducted in duplicate. Results from a single representative experiment are shown.
Competed against DSM-V714 (wild-type carrying pFY7).
Since it was previously reported that rpoS mutants of V. cholerae did not exhibit defective colonization of the suckling mouse intestine (15, 22), we considered the possibility that certain plasmid integrations within the rpoS coding sequence might result in the production of truncated polypeptides that could have deleterious effects, resulting in the observed in vivo defect. To eliminate this possibility, DSM-V491, which contains an in-frame deletion of virtually all of the rpoS coding sequence, was constructed (17). The deletion was confirmed by Southern blot analysis, and the mutant was shown to be sensitive to hydrogen peroxide (Fig. 1). Strain DSM-V491 was also found to have a four- to fivefold defect in colonization of the suckling mouse intestine (Table 2). This result excludes the truncated polypeptide hypothesis and suggests that RpoS does play a role in establishment and/or persistence of V. cholerae infection in this animal model.
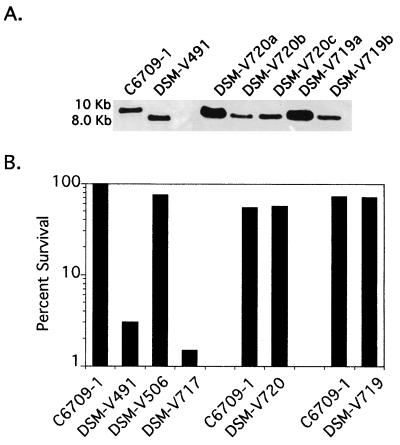
FIG. 1.
(A) Southern blot analysis of the rpoS region. Chromosomal DNA harvested from the indicated strains was digested with HindIII and hybridized with a probe specific for nlpD, which lies immediately upstream of the rpoS coding sequence. Lane C6709-1 shows the presence of the wild-type rpoS HindIII fragment, while lane DSM-V491 shows a smaller band due to an in-frame deletion of the rpoS coding sequence. Three and two independent isolates of reverted DSM-V506 and DSM-V491, respectively, are shown. In each case, the ΔrpoS band is absent and the wild-type rpoS band is regained. (B) Hydrogen peroxide sensitivity, shown as percent survival after 30 min of exposure to hydrogen peroxide as described in Materials and Methods. In each of the three data sets, a C6709-1 wild-type control is shown, as the three experiments were performed on different days.
rpoS strains are viable in the absence of second-site suppressor mutations.
Since our results differed from published reports of the role of rpoS in colonization of the suckling mouse, we considered the possibility that second-site suppressor mutations might be required for the viability of an rpoS strain and that such mutations might have effects on virulence. To address this possibility, quantitative mating assays were conducted to determine the frequency of plasmid disruption at the rpoS locus in comparison with two nonessential genes.
If rpoS is nonessential, rpoS insertion mutants should be recovered at a frequency similar to that obtained with other insertion vectors possessing similar-sized fragments of homology. If, however, a second-site suppressor mutation is required for the viability of an rpoS mutant strain, then the observed frequency of insertion should be significantly decreased. The vector pDSM-375, constructed by inserting a 287-bp internal fragment of rpoS into the suicide vector pGP704, was used previously to construct the rpoS strain DSM-V382 (17). Two other pGP704 derivatives were constructed with similar-sized inserts: pAC160, which contains a 249-bp internal fragment of iviVI, a nonessential gene coding for a putative ATP-binding cassette transporter (5); and pAC214, which contains a 295-bp internal fragment of orf3, a nonessential gene coding for a protein involved in chemotaxis (5). The frequency of plasmid insertion at each locus was determined by quantitative matings. As shown in Table 3, insertions within the rpoS locus occurred at a slightly higher frequency than at the other two loci. In addition, all transconjugants had colony sizes and morphologies similar to those of the parental strain (data not shown). These results strongly argue that a second-site suppressor mutation is not necessary for the viability of a V. cholerae rpoS strain.
TABLE 3.
Frequencies of replication-defective plasmid integration into rpoS and two other loci
| Site of insertion | Transferred plasmid (insert size [bp]) | Insertion frequency (transconjugants/recipient)a |
|---|---|---|
| rpoS | pDSM375 (287) | 1.4 × 10−6 ± 1.1 × 10−6 |
| iviVI | pAC160 (249) | 3.1 × 10−7 ± 1.0 × 10−7 |
| orf3 | pAC214 (295) | 9.5 × 10−7 ± 3.4 × 10−7 |
Average ± standard deviation of three separate matings.
The rpoS phenotype can be complemented in trans.
To demonstrate that the rpoS mutation was responsible for the decreased competition index, we complemented the rpoS mutation by providing rpoS in trans using pFY7, a low-copy-number plasmid containing the entire rpoS promoter and coding region (22). This construct was previously shown to complement an rpoS mutation during in vitro growth under nutrient and oxidative stress test conditions (22). pFY7 was mobilized into DSM-V491 (ΔrpoS) to generate strain DSM-V506. DSM-V506 regained wild-type level hydrogen peroxide resistance (Fig. 1B), indicating that functional RpoS was being produced. In competition assays with C6709-1, DSM-V506 showed an in vivo competition index of 1.2 (Table 2), establishing that the low-copy-number plasmid containing rpoS complements the in vivo colonization defect.
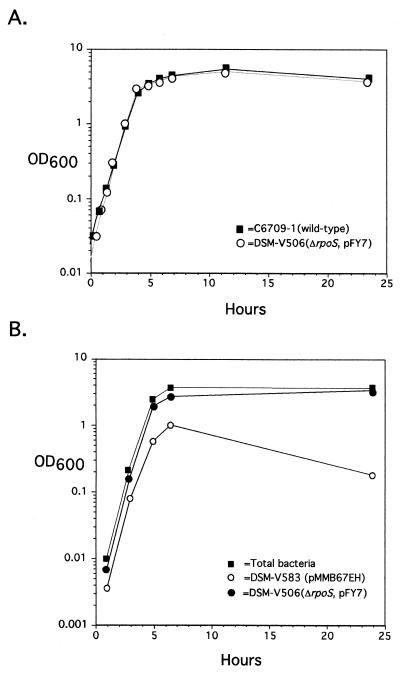
Interestingly, DSM-V506 showed an in vitro competition index of 32, indicating that DSM-V506 greatly outcompeted the wild-type strain during growth in LB. Since pFY7 has a copy number greater than one, we hypothesized that the increased in vitro competition index of DSM-V506 was due to the overproduction of RpoS. The ability of DSM-V506 to outcompete the wild-type strain in vitro could be a result of any number of altered growth phenotypes, including but not limited to (i) a decreased lag phase, (ii) a decreased doubling time, (iii) increased survival in stationary phase, and/or (iv) the ability to reach a higher cell density in stationary phase. To discriminate among these possibilities, we assessed the growth kinetics of the wild type, DSM-V491, and DSM-V506 in a standard growth curve assay. The presence of pFY7 did not cause a noticeable change in lag phase or doubling time of DSM-V506 (Fig. 2A). This strain also displayed no increase in cell density or viable CFU in stationary phase (Fig. 2A and data not shown). Thus, the presence of pFY7 had no detectable effects on the growth of V. cholerae in monoculture.
FIG. 2.
(A) Growth kinetics of DSM-V506 and C6709-1. For each strain, overnight cultures were diluted 1:200 into fresh LB and grown with aeration at 37°C. At the indicated times, the OD600 was read for each and plotted as a function of time. (B) In vitro competition growth kinetics of DSM-V506 and DSM-V583. A 1:1 mixture of overnight cultures of each strain was diluted 1:1,000 into fresh LB. The titer of each strain was determined at the indicated time points by plating on LB supplemented with X-Gal. In addition, the OD600 was read at each time point. The percentage of each colony type found at each time point was used to extrapolate the OD600 of each strain based on the total OD600.
To determine the basis for the in vitro growth advantage of DSM-V506 over the wild type, competition assays were repeated. Overnight cultures of DSM-V506 and DSM-V583 (C6709-1 carrying the empty pMMB67EH cloning vector) were mixed together at a ratio of approximately 1:1, inoculated into fresh LB, and grown with aeration. At various times, the ratio of LacZ+ (DSM-V506) to LacZ− (DSM-V583) colonies was determined and subsequently used to calculate the percentage of each strain in relationship to the total optical density at 600 nm (OD600). These values were then plotted as a function of time (Fig. 2B). The two competing strains had similar doubling times during the logarithmic phase of growth. However, during the late log and particularly stationary phases of growth, DSM-V506 outcompeted DSM-V583. After 24 h a final sample was plated, and the ratio was shown to be 19:1 (Fig. 2B). We are unsure of the basis for this competitive advantage of the pFY7-containing strain. As expected, this competitive advantage was abolished by mobilization of pFY7 into the competing wild-type strain (DSM-V714 [Table 2]), thus confirming that the survival advantage of DSM-V506 was due to the presence of plasmid pFY7 and presumably to the overproduction of RpoS.
The rpoS phenotype is revertible.
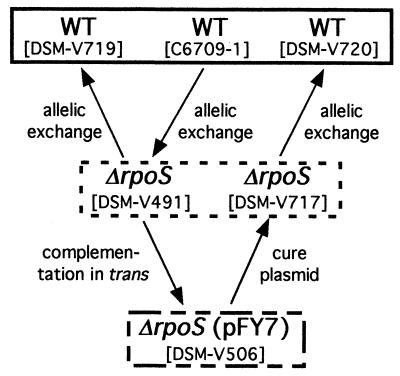
Since interpretation of the intestinal colonization experiments utilizing pFY7 plasmid complementation is confounded by the ability of strains containing this plasmid to outcompete wild-type strains in vitro, we decided to revert the rpoS deletion by allelic exchange. This was done in the original ΔrpoS strain, DSM-V491, and in the pFY7-containing strain DSM-V506, in order to confirm that phenotypes seen throughout the course of these experiments were not due to the presence of second-site mutations acquired spontaneously. The various strain constructions made in this study are depicted in Fig. 3.
FIG. 3.
Genetic manipulations of V. cholerae C6709-1 presented in this report. Strains of equivalent genotype and phenotype are in boxes. The box with a solid line represents the presence of a wild-type chromosomal copy of rpoS and wild-type growth and virulence; the box with a dashed line represents loss of rpoS function, sensitivity to H2O2 in vitro, and reduced colonization in vivo; the box with alternating short and long dashes represents the presence of a plasmid expressing rpoS in trans, resistance to H2O2, increased competitive fitness in vitro, and wild-type colonization.
DSM-V506 was cured of pFY7 by growth in LB in the absence of antibiotic selection. Initially, standard overnight cultures of DSM-V506 grown at 37°C were used to screen for colonies cured of pFY7. Screening of more than 2,500 colonies resulted in no such strain. To increase the level of plasmid loss, DSM-V506 was grown at the partially growth-restrictive temperature of 42°C. Cultures grown at this temperature yielded multiple cured strains from approximately 1,500 colonies screened. One of the strains was designated DSM-V717. Loss of plasmid pFY7 by DSM-V717 was additionally confirmed by the regained sensitivity to hydrogen peroxide (Fig. 1B).
Both DSM-V491 and DSM-V717 were subsequently restored to wild type by allelic exchange at the rpoS locus as described in Materials and Methods. Revertants of DSM-V491 and DSM-V717 were named DSM-V719 and DSM-V720, respectively (Fig. 3). Restoration of the rpoS locus in the reverted strains was verified by Southern blot analysis (Fig. 1A) and the regained resistance to hydrogen peroxide (Fig. 1B). To determine if DSM-V719 and DSM-V720 were also restored to full virulence, competition assays were done. In both cases, the reverted strains were able to colonize the suckling mouse at wild-type levels (Table 2). These data show that the originally observed decrease in in vivo competitive fitness of the rpoS strains DSM-V382 and DSM-V491 could be complemented by the restoration of the rpoS chromosomal locus.
DISCUSSION
The intestinal pathogen V. cholerae has a complicated life cycle that includes growth within an aquatic environment, oral ingestion by human hosts, passage through the low pH environment of the stomach, colonization within the small intestine, and subsequent dissemination in the cholera stool back into its aquatic niche. The study of environmentally induced gene regulation of V. cholerae has primarily focused on the production and regulation of virulence factors within the host environment (16, 20). Recently, however, the role of the stationary-phase sigma factor RpoS in regulation of genes required for surviving a variety of environmental stresses was demonstrated (22). In addition, the V. cholerae RpoS was found to positively regulate expression of at least 25 different genes upon entry into stationary phase (22). Similar but diverse roles for RpoS in response to environmental stresses have been demonstrated in a variety of bacterial species, including E. coli, S. flexneri, Y. enterocolitica, L. pneumophila, and S. enterica serovar Typhimurium (2, 3, 10, 21).
The role of RpoS in colonization and virulence is as diverse as the many pathogenic bacterial species in which it has been studied. A potential role for V. cholerae RpoS in the colonization and survival within a murine model of cholera was previously considered by two separate studies. In each case, it was concluded that RpoS plays no role in colonization within the suckling mouse model of cholera (15, 22). Data presented here suggest that RpoS is indeed important for intestinal colonization in this model by the El Tor biotype clinical isolate C6709-1. Specifically, we found that rpoS mutant derivatives of C6709-1 exhibit a four- to fivefold decrease in colonization compared to the wild type. This in vivo phenotype was fully complemented by a plasmid expressing rpoS in trans and by restoration of the rpoS allele on the V. cholerae genome.
We found that the provision of rpoS in trans from a low-copy-number plasmid resulted in the ability of the complemented strain to outcompete the wild-type strain in vitro. Other studies have noted that providing rpoS in trans from plasmids often results in aberrant complementation phenotypes, which likely result from overproduction or altered regulation of RpoS (10, 22). Curiously, growth curve analysis of the complemented strain showed the same growth kinetics as the wild type. We do not know the nature of the competitive advantage, but it is interesting to speculate that the complemented strain is able to acquire nutrients more efficiently than the wild-type strain. This phenomenon would be similar to the growth advantage in stationary phase (GASP) phenotype that has been demonstrated to arise spontaneously in E. coli (8, 9, 23). The GASP phenotype is often caused by mutations in rpoS, but there are no reported cases that are the result of increased rpoS expression. Rather, the GASP rpoS mutations usually result in decreased RpoS function (8). An alternative hypothesis is that the complemented strain has acquired the ability to produce and release a compound that is toxic to the wild-type bacteria. However, attempts to mimic this killing phenomenon using sterile culture supernatants of the complemented strain have been unsuccessful.
There are multiple explanations for the reported differences in the involvement of rpoS in V. cholerae colonization of suckling mice (15, 22), the most obvious and perhaps most likely being that of different strain usage by the various groups. While our mutation was constructed in the El Tor biotype strain C6709-1, an epidemic isolate from Peru in 1991, Klose and Mekalanos (15) constructed mutations in the Classical biotype strain O395. Multiple instances of differences in gene regulation and expression between the two biotypes of V. cholerae have been noted (19, 20), though none have addressed rpoS. The study by Yildiz and Schoolnik (22) was conducted using the El Tor biotype strain 92A1552, a clinical isolate from Latin America in 1992 (F. H. Yildiz, personal communication). It has previously been noted that different strains of E. coli show different phenotypes as a result of mutations or polymorphisms in rpoS (12). Other explanations include variations in the methodology of the competition assay and possible differences between the litters of mice in the different studies. Regardless, this work demonstrates a role for rpoS in V. cholerae C6709-1 colonization of the suckling mouse model.
What might be the role of the V. cholerae RpoS in colonization of the murine intestine? RpoS has been shown to function as both a positive and a negative regulator of expression of as many as 41 different proteins in V. cholerae (22). This RpoS-dependent regulation functions not only in the stationary phase but also in the exponential phase of growth (22), suggesting that RpoS is involved in expression of gene products which benefit the growing cell. Though the dynamics of colonization in suckling mice are not fully understood, after transit through the stomach and transit through the upper portion of the small intestine, V. cholerae encounters a niche that is permissive for colonization (1). Indeed, after an initial decline in bacterial number, V. cholerae undergoes a rapid growth phase that results in large numbers of progeny cells accompanied by fluid accumulation in the intestinal lumen (4). Therefore, RpoS may serve either of two functions: to aid in surviving environmental stresses encountered in vivo and/or to aid in optimizing the rapid growth phase that occurs subsequent to intestinal colonization.
ACKNOWLEDGMENTS
This research was supported by NIH grants AI 40262 and AI 45746 to A.C. and the Center for Gastroenterology Research on Absorptive and Secretory Processes, NEMC (P30 DK34928).
We thank F. Yildiz and G. Schoolnik for providing the pFY7 complementation plasmid and A. Sonenshein for helpful discussion. In addition, we thank E. Joyce, M. Malamy, C. Kumamoto, and M. Waldor for critical readings of the manuscript.
REFERENCES
- 1.Angelichio M J, Spector J, Waldor M K, Camilli A. Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect Immun. 1999;67:3733–3739. doi: 10.1128/iai.67.8.3733-3739.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badger J L, Miller V L. Role of RpoS in survival of Yersinia enterocolitica to a variety of environmental stresses. J Bacteriol. 1995;177:5370–5373. doi: 10.1128/jb.177.18.5370-5373.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baik H S, Bearson S, Dunbar S, Foster J W. The acid tolerance response of Salmonella typhimurium provides protection against organic acids. Microbiology. 1996;142:3195–3200. doi: 10.1099/13500872-142-11-3195. [DOI] [PubMed] [Google Scholar]
- 4.Baselski V, Briggs R, Parker C. Intestinal fluid accumulation induced by oral challenge with Vibrio cholerae or cholera toxin in infant mice. Infect Immun. 1977;15:704–712. doi: 10.1128/iai.15.3.704-712.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camilli A, Mekalanos J J. Use of recombinase gene fusions to identify Vibrio cholerae genes induced during infection. Mol Microbiol. 1995;18:671–683. doi: 10.1111/j.1365-2958.1995.mmi_18040671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finkel S E, Zinser E R, Kolter R. Long-term survival and evolution in the stationary phase. In: Storz G, Hengge-Aronis R, editors. Bacterial stress response. Washington, D.C.: ASM Press; 2000. pp. 231–238. [Google Scholar]
- 9.Finkel S E, Kolter R. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci USA. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hales L M, Shuman H A. The Legionella pneumophila rpoS gene is required for growth within Acanthamoeba castellanii. J Bacteriol. 1999;181:4879–4889. doi: 10.1128/jb.181.16.4879-4889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hengge-Aronis R. Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 12.Hengge-Aronis R. The general stress response in Escherichia coli. In: Storz G, Hengge-Aronis R, editors. Bacterial stress response. Washington, D.C.: ASM Press; 2000. pp. 161–178. [Google Scholar]
- 13.Islam M S, Drasar B S, Sack R B. The aquatic flora and fauna as reservoirs of Vibrio cholerae: a review. J Diarrhoeal Dis Res. 1994;12:87–96. [PubMed] [Google Scholar]
- 14.Jorgensen F, Bally M, Chapon-Herve V, Michel G, Lazdunski A, Williams P, Stewart G S. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology. 1999;145:835–844. doi: 10.1099/13500872-145-4-835. [DOI] [PubMed] [Google Scholar]
- 15.Klose K E, Mekalanos J J. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol Microbiol. 1998;28:501–520. doi: 10.1046/j.1365-2958.1998.00809.x. [DOI] [PubMed] [Google Scholar]
- 16.Lee S H, Hava D L, Waldor M K, Camilli A. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell. 1999;99:625–634. doi: 10.1016/s0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- 17.Merrell D S, Camilli A. The cadA gene of Vibrio cholerae is induced during infection and plays a role in acid tolerance. Mol Microbiol. 1999;34:836–849. doi: 10.1046/j.1365-2958.1999.01650.x. [DOI] [PubMed] [Google Scholar]
- 18.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murley Y M, Carroll P A, Skorupski K, Taylor R K, Calderwood S B. Differential transcription of the tcpPH operon confers biotype-specific control of the Vibrio cholerae ToxR virulence regulon. Infect Immun. 1999;67:5117–5123. doi: 10.1128/iai.67.10.5117-5123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skorupski K, Taylor R K. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol. 1997;25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 21.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski J L. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yildiz F H, Schoolnik G K. Role of rpoS in stress survival and virulence of Vibrio cholerae. J Bacteriol. 1998;180:773–784. doi: 10.1128/jb.180.4.773-784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zambrano M M, Siegele D A, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]