Abstract
Bartonella quintana, the agent of trench fever and a cause of endocarditis and bacillary angiomatosis in humans, has the highest reported in vitro hemin requirement for any bacterium. We determined that eight membrane-associated proteins from B. quintana bind hemin and that a ∼25-kDa protein (HbpA) was the dominant hemin-binding protein. Like many outer membrane proteins, HbpA partitions to the detergent phase of a Triton X-114 extract of the cell and is heat modifiable, displaying an apparent molecular mass shift from approximately 25 to 30 kDa when solubilized at 100°C. Immunoblots of purified outer and inner membranes and immunoelectron microscopy with whole cells show that HbpA is strictly located in the outer membrane and surface exposed, respectively. The N-terminal sequence of mature HbpA was determined and used to clone the HbpA-encoding gene (hbpA) from a lambda genomic library. The hbpA gene is 816 bp in length, encoding a predicted immature protein of approximately 29.3 kDa and a mature protein of 27.1 kDa. A Fur box homolog with 53% identity to the Escherichia coli Fur consensus is located upstream of hbpA and may be involved in regulating expression. BLAST searches indicate that the closest homologs to HbpA include the Bartonella henselae phage-associated membrane protein, Pap31 (58.4% identity), and the OMP31 porin from Brucella melitensis (31.7% identity). High-stringency Southern blots indicate that all five pathogenic Bartonella spp. possess hbpA homologs. Recombinant HbpA can bind hemin in vitro; however, it does not confer a hemin-binding phenotype upon E. coli. Intact B. quintana treated with purified anti-HbpA Fab fragments show a significant (P < 0.004) dose-dependent decrease in hemin binding relative to controls, suggesting that HbpA plays an active role in hemin acquisition and therefore pathogenesis. HbpA is the first potential virulence determinant characterized from B. quintana.
Trench fever is an arthropod-borne disease caused by Bartonella quintana and occurs about 10 days following the bite of an infected body louse (Pediculus humanus) (14). The morbidity impact of trench fever was second only to influenza in terms of lost man-hours during World War I, and thousands of troops were debilitated by the disease (56). Following a period of quiescence, trench fever reappeared during World War II (32) and appeared sporadically for the next four decades. Although the symptoms of trench fever can vary, the disease usually presents with mild to moderately severe fever, chills, malaise, myalgia, and bone pain that is prominent in the tibia (hence the nickname “shinbone fever”) (65). Occasionally, patients develop splenomegaly and a maculopapular rash resembling the rose spots of typhoid fever (56). Trench fever generally lasts about 1 week, but some cases can persist for up to 12 weeks with recurrent febrile episodes and protracted bacteremia (59).
B. quintana is currently reemerging as an etiologic agent primarily afflicting homeless, alcoholic males that live within the inner cities of the United States and Europe (28). Although many cases of “urban trench fever” present with symptoms of the classical disease (27), the pathogen can also cause potentially fatal bacillary angiomatosis (31), endocarditis (53), lymphadenopathy (31), infections of the central nervous system (45), and lytic bone lesions (30, 31). In addition, B. quintana infections have been reported in both immunocompromised individuals as well as in immunocompetent patients (54). However, in spite of B. quintana's emerging status, little is known about the pathogen's current reservoir and vector of transmission, although exposure to lice appears to correlate with incidence of disease (30).
To date, no virulence determinant has been characterized from B. quintana. Pili (6) and specialized extensions of the outer membrane (13) are thought to serve as host cell adhesins. Recent work has shown that B. quintana binds to membrane ruffles and can subsequently invade cultured vascular endothelial cells within 1 minute of coincubation. Tissue biopsies from endocarditis vegetations were also found to contain intracellular B. quintana (13). Internalized bacteria divide and form vacuoles that resemble morulae observed during ehrlichiosis. The morulae contain numerous bacteria and blebs, suggesting that membranes are sloughed as the pathogen grows. Blebs are apparently more common in infected, cultured host cells than in endocarditis tissues (13).
Bartonella species are the only bacterial pathogens for humans that engage in the practice of hemotrophy, i.e., erythrocyte parasitism. Because of this unusual parasitic strategy all Bartonella species require erythrocytes or hemin supplements in order to grow in vitro. In fact, B. quintana has the greatest known hemin requirement (20 to 40 μg/ml of medium) for a bacterium (41). This study was undertaken to elucidate the molecular mechanism whereby hemin is acquired by B. quintana in order to better understand the reasons for this pathogen's extraordinary hemin requirement.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli was grown overnight at 37°C in Luria-Bertani (LB) medium using standard antibiotic supplements when required. Bartonella cultures were grown on heart infusion agar supplemented with 4% sheep erythrocytes and 2% sheep serum. With the exception of B. bacilliformis, Bartonella cultures were incubated at 37°C in 5% CO2 and 100% relative humidity and were harvested at 3 days postinoculation (approximately mid-log phase [69]). B. bacilliformis cultures were grown and harvested as before (7). For gene expression in E. coli, cells were grown to mid-log phase in LB medium containing appropriate antibiotics, treated with IPTG (isopropyl-β-D-thiogalactopyranoside; 5 mM, final concentration), and grown for an additional 2 h prior to harvest. Bartonella and E. coli strains used or generated in this study are summarized in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| Bartonella spp. | ||
| B. bacilliformis KC583 | Type strain | 12 |
| B. clarridgeiae | Type strain | 34 |
| B. doshiae R18 | Type strain | 8 |
| B. elizabethae | Type strain | 19 |
| B. grahamii V2 | Type strain | 8 |
| B. henselae Houston R1302 | Type strain | 47 |
| B. quintana OK 90-268 | Human isolate | CDCa |
| B. vinsonii Baker | Vole agent | 69 |
| E. coli | ||
| DH5α | Host strain for cloning | Gibco-BRL |
| XL1-Blue MRF′ | Host strain for λ phage | Stratagene |
| XLOLR | Host strain for pBK-CMV and its derivatives | Stratagene |
| Plasmids | ||
| pBK-CMV | Excised vector from λ Zap | Stratagene |
| pHBP-CMV | pBK-CMV plus 3.5-kbp Sau3AI insert with hbpA | This study |
CDC, Centers for Disease Control and Prevention, Atlanta, Ga.
Membrane fractionation and purification.
Ten plates of B. quintana were harvested into 1 ml of ice-cold Dulbecco phosphate-buffered saline (PBS; pH 7.4) and then homogenized for 3 min using 0.1-mm glass beads and a Mini Beadbeater-8 (BioSpec products, Bartlesville, Okla.). The resulting lysate was centrifuged for 5 s at 3,000 × g, and the supernatant was retained. To obtain a crude membrane preparation (CMP), the mixture was clarified twice by centrifuging for 15 min at 1,000 × g (4°C) and then retaining the supernatant. To obtain total membranes, CMP was centrifuged for 2 h at 100,000 × g (4°C) in an SW60 rotor (Beckman Instruments, Fullerton, Calif.), and the pellet was retained. Outer and inner membranes were prepared by centrifuging CMP on a 35 to 55% sucrose step gradient for 40 h at 222,000 × g in an SW41 rotor at 4°C (Beckman). Tea-colored inner membranes were collected near the top of the gradient, while fluffy white outer membranes were collected near the bottom of the tube (44). Membranes were dialyzed overnight at 4°C against 10-fold diluted PBS (pH 7.4) and then lyophilized, suspended in 0.5 ml sterile distilled water, and stored at −20°C until needed. Triton X-114 extracts of whole-cell lysates were prepared (9), followed by phase separation of the extract to obtain integral membrane proteins (18).
SDS-PAGE and hemin-binding blots.
Protein concentrations were determined with a bicinchoninic acid protein kit (Sigma Chemical, St. Louis, Mo.). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was done according to the general methods of Laemmli (33). Approximately 20 μg of protein were solubilized in Laemmli sample buffer (LSB) and separated by SDS-PAGE (12.5% [wt/vol] acrylamide), and the resulting gel was fixed and stained with Coomassie brilliant blue R (5). For hemin-binding blots, unfixed gels were transferred to nitrocellulose by the general methods of Towbin et al. (63). The resulting blots were rinsed with Tris-buffered saline (TBS; 10 mM Tris-HCl [pH 8.0] containing 150 mM NaCl) and Tween 20 (0.1% [vol/vol]) and subsequently probed for 1.5 h with TBS containing hemin (10 μg/ml). Blots were subsequently washed three times for 30 min with TBS-Tween 20 (0.1% [vol/vol]) and developed using enhanced chemiluminescence (ECL) reagents (Amersham Pharmacia, Piscataway, N.J.). Hemin-binding protein (HBP) bands were visualized by exposing the blot to autoradiographic film (Labscientific, Livingston, N.J.).
Preparation of polyclonal antibody and Fab fragments.
Precipitates from Triton X-114 extracts of the B. quintana cell, as prepared above, were solubilized in LSB and separated by SDS-PAGE. The resulting gel was briefly rinsed in water and stained with Coomassie brilliant blue (0.05%). HbpA bands (ca. 100 μg total protein) were excised and used to generate rabbit anti-HbpA antiserum as before (50). Immunoglobulin G (IgG) was purified from the antiserum using an Nab chromatography kit (Pierce, Rockford, Ill.). Fab fragments were prepared from IgG with an ImmunoPure Fab kit (Pierce).
N-terminal sequencing.
Triton X-114 extracts of B. quintana were precipitated with methanol-chloroform as described above, and the precipitate was solubilized into LSB and separated by SDS-PAGE. The resulting gel was blotted to polyvinylidene difluoride (37), and the excised HbpA band was subjected to Edman degradation using an ABI 431A automated peptide sequencer. Sequencing was performed on two separate samples.
Immunoblots and immunoelectron microscopy.
Immunoblots, immunogold analysis and transmission electron microscopy were done as previously described (50). Unless otherwise indicated, rabbit anti-HbpA antiserum was used at a 1:10,000 dilution.
Hemin binding and inhibition assays.
Four plates of B. quintana were harvested into Tris-HCl buffer (0.1 M, pH 8.0) and centrifuged for 5 min at 4,620 × g. The pellet was washed twice by resuspending it in 3 ml of Tris buffer and recentrifuging it. The final pellet was resuspended in Tris buffer to a final optical density at 600 nm of 1. For inhibition experiments, 1 ml of cells was preincubated for 1 h at 24°C with 40 or 80 μl (0.2 or 0.4 mg/ml, respectively) of anti-HbpA Fab fragments. Cells treated with equal volumes of PBS served as controls. Hemin binding was subsequently measured using a standard liquid binding assay (20, 22, 26, 43) and a Spectronic Genesys 2 spectrophotometer (Milton Roy, Rochester, N.Y.).
Preparation and manipulation of DNA.
Plasmids used or generated in this study are given in Table 1. Chromosomal DNA from Bartonella spp. was prepared using a hexadecyltrimethyl ammonium bromide technique (5). Plasmids were propagated in E. coli (DH5α or XLOLR) and isolated with a Perfectprep plasmid kit (Eppendorf Scientific, Westbury, N.Y.) or a Qiagen Midi Prep kit (Qiagen, Valencia, Calif.). When required, DNA fragments or amplicons were purified from agarose gels using a GeneClean II kit (Bio 101, La Jolla, Calif.). A genomic library of B. quintana was generated by partial digestion of B. quintana chromosomal DNA with Sau3AI and ligation of the resulting fragments into BamHI-cut lambda Zap Express arms according to the manufacturer's instructions (Stratagene, La Jolla, Calif.). Ligations were done using standard procedure (5), and transformations were performed by the methods of Chung et al. (16). High-stringency DNA hybridizations (Southern blots and lambda library screening) and PCR were conducted as previously described (7).
Nucleotide sequencing and analysis.
Both DNA strands of hbpA were sequenced using a BigDye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems) and an automated DNA sequencer (ABI model 377). Sequence data were compiled and analyzed by PC/GENE 6.8 software (Intelligenetics, Mountain View, Calif.). BLAST 2.0 (1) was used for database searches, while sequence alignments were done using FASTA 2.0 (46), CLUSTAL W 1.6 (61), and BOXSHADE 3.21 (K. Hofmann, and M. D. Baron, http://www.ch.embnet.org/software/BOX_form.html, 1998).
Statistical analysis.
Statistical analyses (Student's t test, standard error of the mean [SEM]) and graphs were done using SigmaPlot 3.0 software (Jandel Scientific, San Rafael, Calif.). A value of P < 0.05 was considered statistically significant.
Nucleotide sequence accession number.
The GenBank accession number for the B. quintana hbpA sequence is AF266281.
RESULTS
Identification of B. quintana HBPs.
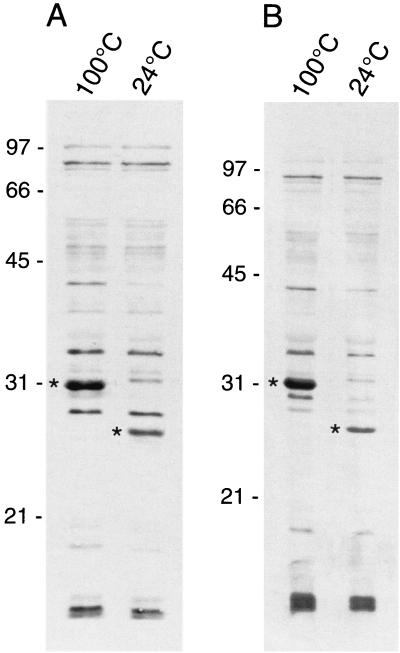
Eight HBPs of 11, 12, 19, 25 or 30 (depending upon solubilization temperature), 29, 36, 42, and 87 kDa were identified on hemin blots containing total cell lysates of B. quintana (Fig. 1A). HBPs with identical molecular masses were also observed in hemin blots of total membranes purified from B. quintana (Fig. 1B). Of the eight HBPs, a prominent HBP band of 25 kDa (termed HbpA) was observed to shift from approximately 25 to 30 kDa when heated, thus displaying a common characteristic of outer membrane proteins (see Discussion). The HbpA band also demonstrated the highest affinity for hemin relative to the seven other HBPs, based upon its unique ability to retain bound hemin after a 24-h wash with TBS (data not shown). In addition, the HbpA band was the only protein that was visibly brown on blots probed with hemin, prior to ECL detection (data not shown). Taken together, these data formed the basis of our hypothesis that HbpA was an HBP located in the outer membrane of B. quintana.
FIG. 1.
Identification of hemin-binding proteins of B. quintana using hemin blots. (A) Total cell lysates. (B) Total membranes. Samples were solubilized in LSB at 24 or 100°C as indicated and then separated by SDS-PAGE, transferred to nitrocellulose, probed with hemin, and developed using ECL reagents (Amersham Pharmacia) as described in Materials and Methods. The position of the two forms of HbpA are indicated by asterisks. Molecular mass standards are given to the left in kilodaltons.
Anti-HbpA antibodies and localization of HbpA in the B. quintana cell.
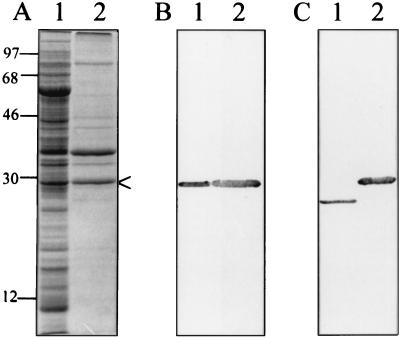
To characterize HbpA, whole-cell lysates were subjected to phase partitioning with Triton X-114. HbpA was observed predominantly in the detergent phase, which is typical of integral membrane proteins (9, 18). This phase was found to contain two dominant proteins, HbpA and a 36-kDa polypeptide, plus minor proteins of 33, 42, 46, and 87 kDa (Fig. 2A, lane 2). The Triton X-114 detergent fraction was further separated by SDS-PAGE and HbpA bands were excised from unfixed, Coomassie blue-stained gels to generate rabbit polyclonal antibody. Specificity of the anti-HbpA antibody for HbpA was verified using immunoblots (Fig. 2B). The antibody was able to recognize both the 25- and the 30-kDa forms of the molecule (Fig. 2C).
FIG. 2.
Monospecificity of the anti-HbpA antibody and reactivity against both molecular mass forms of HbpA. (A) Coomassie blue-stained SDS-PAGE gel containing a B. quintana cell lysate (lane 1) and a phase-separated Triton X-114 cell extract of the bacterium (lane 2). Both samples were solubilized at 100°C in LSB. HbpA is arrowed. (B) Immunoblot corresponding to panel A but developed with rabbit anti-HbpA antiserum showing monospecificity of the anti-HbpA antibody. (C) Immunoblot showing anti-HbpA antibody reactivity to phase-separated Triton X-114 cell extracts of B. quintana solubilized at 25°C (lane 1) and 100°C (lane 2). Note that the antibody recognizes both molecular mass forms of HbpA in panel C. Molecular mass standards are given to the left in kilodaltons.
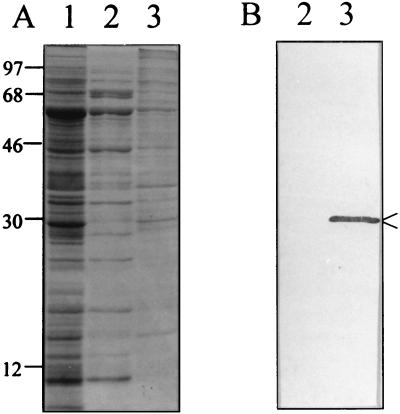
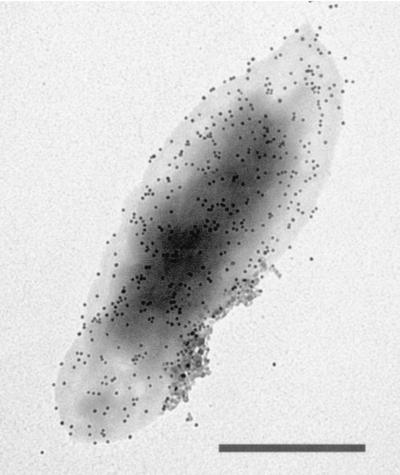
To verify HbpA's suspected outer membrane location in the cell, inner and outer membranes were isolated from B. quintana in a manner similar to that described for B. bacilliformis (39) using sucrose density gradient centrifugation (44). The average buoyant density values (ρ) were determined from three membrane preparations and were calculated as 1.08 for the inner membrane and 1.2 for the outer membrane. These density values are very close to those reported for E. coli (42) and Salmonella spp. (44). Likewise, the characteristic tea-color of the B. quintana inner membrane, due to cytochromes, and white floculence of the outer membrane was in keeping with characteristics of the respective membranes from E. coli. The protein profiles of the outer and inner membranes were distinct on SDS-PAGE (Fig. 3A). Immunoblot analysis using anti-HbpA antiserum detected HbpA in the outer membrane but not in the inner membrane (Fig. 3B). To corroborate these data, immunogold analyses were done using anti-HbpA and intact bacteria to determine if HbpA is surface exposed. The data clearly show that HbpA is both exposed and abundant on the surface of the B. quintana cell (Fig. 4). Immunogold controls prepared with an equal volume of PBS or preimmune rabbit serum showed insignificant protein A-gold binding (not shown).
FIG. 3.
Localization of HbpA in the outer membrane of B. quintana. Lanes: 1, cell lysate; 2, purified inner membrane; 3, outer membrane. (A) Coomassie blue-stained SDS-PAGE gel. (B) Corresponding immunoblot developed with anti-HbpA showing HbpA in the outer membrane (arrowed). All samples were solubilized at 100°C in LSB. Molecular mass standards are given to the left in kilodaltons.
FIG. 4.
Immunoelectron microscopy showing surface localization of HbpA. Immunogold analysis and negative staining were done as previously described (50), using anti-HbpA and intact bacteria. Bar, 0.5 μm.
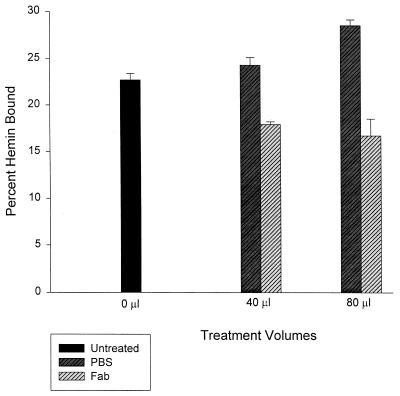
In vivo hemin binding by HbpA and inhibition with Fabs.
A standard liquid hemin-binding assay was done with freshly harvested B. quintana. Untreated B. quintana cells bound approximately 22% of exogenous hemin (1.4 μg of hemin/mg of B. quintana protein) during a 60-min assay; this percentage falls within the range of hemin binding exhibited by several human bacterial pathogens (20, 26, 43, 51). B. quintana pretreated with anti-HbpA Fab fragments showed a very significant (P < 0.004), dose-dependent decrease in hemin binding, relative to controls treated with an equal volume of PBS. For example, B. quintana treated with 0.2 mg of Fab per ml showed a 26% decrease in hemin binding relative to its respective PBS control, whereas B. quintana treated with 0.4 mg of Fab per ml exhibited a 41% decrease in hemin binding relative to PBS controls (Fig. 5). These data show a dose-dependent decrease in hemin binding by B. quintana when cells are treated with anti-HbpA Fabs prior to the liquid hemin-binding assay.
FIG. 5.
Anti-HbpA Fab fragments inhibit hemin binding by B. quintana. The percent hemin uptake as a function of treatment volumes is shown. B. quintana was pretreated with 0, 40, or 80 μl of PBS or anti-HbpA Fab (0.2 and 0.4 mg/ml, respectively) and then assayed by a standard liquid hemin-binding assay. The values are the average of three determinations ± the SEM.
Cloning the hbpA gene.
The N terminus of mature HbpA was determined from two separate samples and was found to be ADVIATHEAAPVITTPNF. BLASTp searches with this amino acid sequence showed a high probability hit (63% identity) with the Pap31 protein from B. henselae (10). This discovery prompted us to design PCR primers based upon the first 31 nucleotides and the last 25 nucleotides (inverse complement) of the pap31 open reading frame (ORF) (GenBank accession no. AF001274). The pap31 primers produced PCR products of identical size (∼840 bp) from both B. henselae and B. quintana DNA templates (data not shown). The B. quintana amplicon was subsequently used to screen a lambda ZAP Express (Stratagene) genomic library of B. quintana for hbpA. A set of positive plaques was identified, isolated, and tested by PCR to determine if a full-length copy of hbpA was present. A positive lambda clone that contained the full-length hbpA gene was excised in vivo to produce pHBP-CMV. This plasmid contains a Sau3AI insert of approximately 3.5 kbp.
Nucleotide sequence of hbpA.
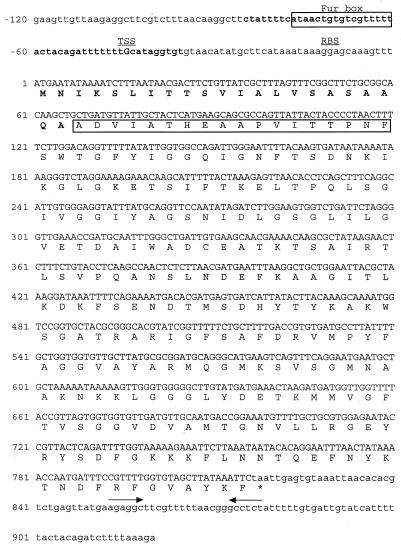
The sequence of hbpA was determined from both strands of pHBP-CMV (Fig. 6). The B. quintana hbpA gene is 816 bp long and has a 39.1 mol% G+C, a level in close agreement with the 38.5 mol% G+C for the B. quintana genome (64). The region upstream of the start codon of hbpA contains a consensus promoter sequence (0.92 score by prokaryotic promoter neural network prediction) and a potential ribosome-binding site with perfect identity to the E. coli consensus sequence, AGGA (23). A Fur box homolog is nested within the predicted promoter sequence of hbpA and has 53% identity to the E. coli Fur consensus (21). The hbpA ORF is followed 37 bp downstream by a 6-bp inverted repeat that may act as a rho-independent transcriptional terminator.
FIG. 6.
Sequence of hbpA, its encoded protein, and identification of a potential Fur box in the promoter region. Nucleotides within the hbpA ORF are given in uppercase letters. The promoter, as determined by prokaryotic promoter prediction by neural network, is shown in boldface lowercase letters, with a potential transcriptional start site (TSS) indicated by a boldface uppercase “G.” The fur box homolog within the promoter region is boxed. A potential ribosome-binding site (RBS) is indicated above the sequence. The deduced amino acid sequence of HbpA is shown below the second base of its respective triplet codon. Amino acids constituting the secretory signal sequence are shown in boldface. The N-terminal sequence experimentally determined from mature HbpA is boxed. The stop codon is denoted by an asterisk. Inverted repeats constituting a potential ρ-independent terminator of transcription are indicated by opposing arrows.
Computer analysis of the encoded HbpA protein predicts that it has 272 amino acids, a molecular mass of 29,270 Da, and a pI of approximately 9.5. The predicted protein also contains an internal 18-amino-acid stretch that is identical to the N terminus as determined from HbpA protein (Fig. 6). A predicted secretory signal cleavage site (67) was found between residues 21 and 22; one residue upstream of the sequence that was determined by Edman degradation (Fig. 6). Proteolytic cleavage at the actual peptidase cleavage site would yield a mature, secreted protein of 27,098 Da. Transmembrane computer predictions (TMPred program; ISREC server) identified three potential transmembrane helices in mature HbpA, encompassing amino acid residues 34 to 55, 74 to 101, and 218 to 236, that may serve to anchor HbpA to the outer membrane. Finally, the predicted HbpA sequence has a C-terminal phenylalanine, a characteristic of most integral outer membrane proteins (57).
Further analysis of the predicted full-length HbpA protein was done using BLAST 2.0. The highest-probability alignments resulting from this search included the Pap31 phage-associated membrane protein from B. henselae (10) with 58.4% identity, followed by the OMP31 porin protein of Brucella melitensis (66) with 31.7% identity. Alignments of these three protein sequences are shown in Fig. 7 and indicate several regions of homology. Inspection of the genes encoding these two homologs reveal that, like hbpA, each gene contains a potential, previously unrecognized Fur box in its respective promoter sequence.
FIG. 7.
Multiple sequence alignment of B. quintana HbpA with B. henselae Pap31 and B. melitensis Omp31. Identical amino acid residues are noted in black, conserved residues in gray and introduced gaps by hyphens. The GenBank accession numbers for the Pap31 and Omp31 homologs are AF001274 and U39453, respectively.
High-stringency Southern blots (7% DNA mismatch) showed that the four other Bartonella species known to be human pathogens (i.e., B. bacilliformis, B. clarridgeiae, B. elizabethae, and B. henselae) possess hbpA homologs. Likewise, B. doshiae DNA also produced a positive hybridization signal. However, B. grahamii and B. vinsonii did not possess homologs when analyzed at this level of stringency (data not shown).
Expression of hbpA in E. coli and characteristics of rHbpA.
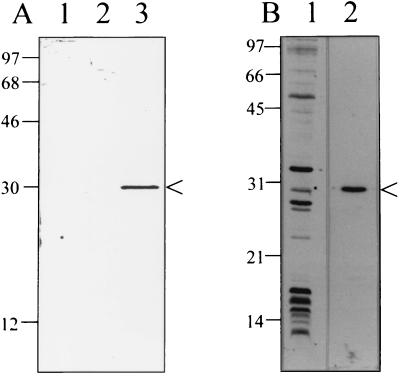
E. coli XLOLR containing pHBP-CMV were found to synthesize recombinant HbpA (rHbpA) (Fig. 8). Although the protein was not apparent on Coomassie blue-stained gels (data not shown), it was clearly detected on immunoblots (Fig. 8A). The cloned hbpA gene is apparently expressed in E. coli from its own promoter, as the gene is in opposite orientation to the lac promoter in the pBK-CMV vector. E. coli XLOLR or XLOLR containing the cloning vector pBK-CMV (Fig. 8A, lanes 1 and 2, respectively) did not produce rHbpA.
FIG. 8.
Expression of hbpA in E. coli and ability of recombinant HbpA to bind hemin in vitro. (A) Immunoblot developed with anti-HbpA antibody, showing lysates of E. coli XLOLR containing no plasmid (lane 1), pBK-CMV cloning vector (lane 2), or pHBP-CMV (lane 3). (B) Blots of an E. coli XLOLR(pHBP-CMV) lysate probed with hemin showing rHbpA binding hemin (indicated by an asterisk) (lane 1). The corresponding immunoblot developed with anti-HbpA antiserum and ECL reagents (Amersham), identifying the recombinant HbpA (arrowed) (lane 2), is also shown. All samples were solubilized at 100°C in LSB. Molecular mass standards are given to the left in kilodaltons.
Liquid hemin-binding assays using XLOLR, XLOLR(pBK-CMV), or XLOLR(pHBP-CMV) did not reveal significant differences in hemin binding, indicating that rHbpA does not confer a hemin-binding phenotype upon E. coli (data not shown). However, in vitro analyses with rHbpA clearly show that it can bind hemin on blots (Fig. 8B, lane 1), as observed with B. quintana HbpA (Fig. 1). Taken as a whole, these results suggest that rHbpA, although capable of binding hemin, cannot do so in E. coli, perhaps because of improper folding or lack of surface exposure.
DISCUSSION
A number of bacterial pathogens have evolved systems for accumulating hemin in order to satisfy their iron (43, 60), protoporphyrin ring (15, 17), or cytochrome cofactor (40) requirements. In addition, hemin has been shown to facilitate entry of certain bacterial pathogens into their respective host cells (20, 58). Early studies with B. quintana showed that the pathogen has the highest in vitro hemin requirement for any known bacterium: 20 to 40 μg/ml (41). The reasons for this extraordinary need and the mechanism(s) whereby hemin is acquired are unknown. The unusual strategy of erythrocyte parasitism (hemotrophy) practiced by all Bartonella species may have evolved to meet the bacterium's tremendous need for this molecule. Early studies with B. quintana indicated that iron, and not the protoporphyrin ring, is the critical component provided by hemin supplements (41). In either case the bacterium would require the synthesis of a hemin receptor on the outer surface to facilitate the acquisition of the iron needed for growth.
We have identified and characterized a gene, hbpA, in B. quintana that encodes a protein, designated HbpA, that retained the ability to bind hemin after SDS-PAGE and electrophoretic transfer to nitrocellulose. Initial analyses suggested HbpA was a membrane protein with an apparent molecular mass of 30 kDa. Triton X-114 phase partitioning and heat modification studies strongly suggested that the protein was an integral membrane protein that localized to the outer envelope of B. quintana. These results were confirmed by immunoblot, comparing inner and outer membrane preparations probed with polyclonal antibody to HbpA, and immunoelectron microscopy, which indicated that the protein was not only found in the outer membrane but was also surface exposed. The observed phenomenon of heat modification has been associated with numerous outer membrane proteins from other bacteria and may be a reflection of HbpA's interaction with lipopolysaccharide (2), interaction with peptidoglycan (4, 49), or tertiary structure (25). Interestingly, the increase in the apparent molecular mass of HbpA when heated (heated 30 kDa versus unheated 25 kDa) is the opposite of what has been observed in Porphyromonas gingivalis, where the reported HBPs decrease their apparent molecular masses from 30 to 24 kDa (29) and from 32 to 19 kDa (11) when heated.
Fab fragments purified from a polyclonal antibody specific for HbpA were able to significantly decrease the amount of hemin bound by B. quintana in a dose-dependent manner. With Fab concentrations of 0.2 and 0.4 mg/ml we observed average decreases of 26 and 41%, respectively, in the amount of hemin bound relative to controls. These results suggest that HbpA plays a role in the ability of the organism to acquire hemin from its surroundings and that antibody to HbpA may inhibit that interaction. The inhibition of hemin binding by antibodies specific for HbpA was similar to what has been demonstrated with other bacterial pathogens, where antibodies to components of the iron scavenging mechanism were shown to inhibit ligand-receptor interactions in vitro (24, 68). Hemin binding was not fully abolished in the presence of anti-HbpA Fab fragments, implying that B. quintana has multiple receptors that bind hemin, as is the case for P. gingivalis (52, 62) and Haemophilus influenzae (26, 48). This possibility is supported by the hemin blot (Fig. 1), which demonstrates that HbpA is only one of eight membrane-associated proteins in B. quintana that has an affinity for hemin. The exact localization of the additional hemin-binding membrane proteins and their involvement in iron acquisition are currently under investigation.
Partially purified HbpA was subjected to N-terminal sequence analysis, where we were able to reproducibly determine the first 18 amino acids of the mature protein. A BLASTp search of the NCBI database indicated a close match (63% identity) to the N terminus of a phage-associated protein (Pap31) identified in B. henselae (10). Pap31 was previously reported to be a membrane protein of B. henselae that copurifies with bacteriophage during phage isolation and purification (3). Due to the similarity between the N terminus of mature Pap31 and HbpA, the deduced nucleotide sequence of pap31 was utilized as a template to develop primers in order to PCR amplify hbpA from B. quintana. A PCR amplicon was obtained, cloned, and used to probe a B. quintana genomic library, where the full-length gene and accompanying flanking sequences were cloned and analyzed.
Southern analysis showed that all pathogenic Bartonella spp. harbor hbpA homologs, implying a correlation between the presence of hbpA and pathogenicity. It was also evident that HbpA is synthesized as a preprotein with a 22-amino-acid signal sequence and a signal peptidase cleavage site. The HbpA signal sequence was strikingly similar (>90% identity, 100% similarity) to the signal sequence of Pap31 (10), suggesting that Pap31 may also localize to the outer membrane in B. henselae.
A BLASTp search of the NCBI database using the full-length HbpA indicated that it was related not only to Pap31 of B. henselae but also to Omp31 of Brucella melitensis, which encodes for a 31- to 34-kDa outer surface protein proposed to be a porin (66). Interestingly, neither Pap31 nor Omp31 have been implicated in hemin binding or iron acquisition. Yet, within the putative promoter regions of hbpA, pap31, and omp31 we identified an imperfect palindrome overlapping their putative −10 regions that closely resembles the ferric uptake regulator (Fur) consensus sequence found in E. coli (21). Similar Fur consensus sequences in Yersinia (55), Neisseria (36), and Shigella (38) spp. have been observed upstream of genes that encode proteins that are involved in, or associated with, iron acquisition. Due to the presence of a Fur consensus sequence (53% identity to the E. coli Fur consensus) and the recent identification of fur homologs in B. bacilliformis and B. henselae (L. Hendrix, Abstr. 15th Sesquiannu. Meet. Am. Soc. Rickettsiol. abstr. 10, 2000), hbpA (as well as pap31 and omp31) may be regulated by Fur in response to fluctuating cellular iron levels. We have yet to demonstrate any regulation of hbpA as a result of hemin or iron availability in vitro, but the abundance of HbpA on the cell surface of B. quintana grown on blood agar would suggest that cells grown under typical plating conditions (presumably iron replete) express hbpA.
The similarities between pap31 and hbpA suggest that they are homologs, and this brings into question the function of Pap31 in B. henselae. While this study does not directly address this question, we propose that both Pap31 of B. henselae and Omp31 of B. melitensis are involved in iron acquisition. Pap31's original designation as a phage-associated membrane protein in B. henselae (3, 10) could be explained by the hypothesis that the bacteriophage may use Pap31 as a receptor on the outer surface of the bacterium. Thus, the receptor (presumably Pap31) may copurify with the phage.
Recombinant HbpA did not confer a hemin-binding phenotype to E. coli but instead retained the ability to bind hemin after SDS-PAGE and electrophoretic transfer to blots. This observation suggests that rHbpA is either improperly folded or not localized to the outer surface in E. coli. The signal sequence preceding HbpA may not be recognized or properly translocated by the secretory machinery of E. coli. Similar results were reported for the cloning and expression of B. melitensis omp31, where recombinant Omp31 was not surface exposed but still maintained the ability to form SDS-resistant oligomers (characteristic of bacterial porins) in E. coli (66).
HbpA is the first potential virulence determinant characterized from B. quintana. This pathogen's high in vitro requirement for hemin makes it an ideal model to study iron acquisition and hemin binding in this genus. Obtaining iron needed for growth from hemin is typically a TonB-dependent process that entails the synthesis of a surface receptor to facilitate the binding of the ligand, a protein in the periplasmic space to ferry the hemin to the cytoplasmic membrane, and finally a permease to bring the molecule into the bacterial cell (35). We have cloned and characterized the first component, a gene encoding an HBP, in this choreographed chain of events. The remaining components must be identified and characterized in order to fully understand the role that hemin binding plays in the pathogenesis of Bartonella.
ACKNOWLEDGMENTS
We thank Elizabeth Fischer and Christian Eggers for their help with the electron microscopy; Russ Regnery for B. quintana; Amplicon Express, Inc., for sequencing service; and J. M. Battisti and D. S. Samuels for critical reviews of the manuscript.
M.F.M. was supported by Public Health Service grant AI45534 and American Heart Association Established Investigator Award 9940002N. J.A.C. was supported through an NIAID Intramural Research Training Award.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ames G F, Spudich E N, Nikaido H. Protein composition of the outer membrane of Salmonella typhimurium: effect of lipopolysaccharide mutations. J Bacteriol. 1974;117:406–416. doi: 10.1128/jb.117.2.406-416.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson B, Goldsmith C, Johnson A, Padmalayam I, Baumstark B. Bacteriophage-like particle from Rochalimaea henselae. Mol Microbiol. 1994;13:67–73. doi: 10.1111/j.1365-2958.1994.tb00402.x. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong S K, Parker C D. Heat-modifiable envelope proteins of Bordetella pertussis. Infect Immun. 1986;54:109–117. doi: 10.1128/iai.54.1.109-117.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1995. [Google Scholar]
- 6.Batterman H J, Peek J A, Loutit J S, Falkow S, Tompkins L S. Bartonella henselae and Bartonella quintana adherence to and entry into cultured human epithelial cells. Infect Immun. 1995;63:4553–4556. doi: 10.1128/iai.63.11.4553-4556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battisti J M, Minnick M F. Development of a system for genetic manipulation of Bartonella bacilliformis. Appl Environ Microbiol. 1999;65:3441–3448. doi: 10.1128/aem.65.8.3441-3448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birtles R J, Harrison T G, Saunders N A, Molyneux D H. Proposals to unify the genera Grahamella and Bartonella, with descriptions of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int J Syst Bacteriol. 1995;45:1–8. doi: 10.1099/00207713-45-1-1. [DOI] [PubMed] [Google Scholar]
- 9.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 10.Bowers T J, Sweger D, Jue D, Anderson B. Isolation, sequencing and expression of the gene encoding a major protein from the backeriophage associated with Bartonella henselae. Gene. 1998;206:49–52. doi: 10.1016/s0378-1119(97)00580-5. [DOI] [PubMed] [Google Scholar]
- 11.Bramanti T E, Holt S C. Hemin uptake in Porphyromonas gingivalis: Omp26 is a hemin-binding surface protein. J Bacteriol. 1993;175:7413–7420. doi: 10.1128/jb.175.22.7413-7420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brenner D J, O'Connor S P, Hollis D G, Weaver R E, Steigerwalt A G. Molecular characterization and proposal of a neotype strain for Bartonella bacilliformis. J Clin Microbiol. 1991;29:1299–1302. doi: 10.1128/jcm.29.7.1299-1302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brouqi P, Raoult D. Bartonella quintana invades and multiplies within endothelial cells in vitro and in vivo and forms intracellular blebs. Res Microbiol. 1996;147:719–731. doi: 10.1016/s0923-2508(97)85119-4. [DOI] [PubMed] [Google Scholar]
- 14.Byam W. Trench fever. In: Loyd L L, editor. Lice and their menace to man. Oxford, England: Oxford University Press; 1919. pp. 120–130. [Google Scholar]
- 15.Carman R J, Ramakrishnan M D, Harper F H. Hemin levels in culture medium of Porphyromonas (Bacteroides) gingivalis regulate both hemin binding and trypsinlike protease production. Infect Immun. 1990;58:4016–4019. doi: 10.1128/iai.58.12.4016-4019.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cope L D, Yogev R, Muller-Eberhard U, Hansen E J. A gene cluster involved in the utilization of both free heme and heme:hemopexin by Haemophilus influenzae type b. J Bacteriol. 1995;177:2644–2653. doi: 10.1128/jb.177.10.2644-2653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cunningham T M, Thomas D D, Thompson S D, Miller J N, Lovett M A. Identification of Borrelia burgdorferi surface components by Triton X-114 phase partitioning. Ann N Y Acad Sci. 1988;539:376–378. [Google Scholar]
- 19.Daly J S, Worthington M G, Brenner D J, Moss C W, Hollis D G, Weyant R S, Steigerwalt A G, Weaver R E, Daneshvar M I, O'Connor S P. Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–881. doi: 10.1128/jcm.31.4.872-881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daskaleros P A, Payne S M. Congo red binding phenotype is associated with hemin binding and increased infectivity of Shigella flexneri in the HeLa cell model. Infect Immun. 1987;55:1393–1398. doi: 10.1128/iai.55.6.1393-1398.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genco C A, Odusanya B M, Brown G. Binding and accumulation of hemin in Porphyromonas gingivalis are induced by hemin. Infect Immun. 1994;62:2885–2892. doi: 10.1128/iai.62.7.2885-2892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gold L, Pribnow D, Schneider T, Shinedling S, Singer B S, Stormo G. Translational initiation in prokaryotes. Annu Rev Microbiol. 1981;35:365–403. doi: 10.1146/annurev.mi.35.100181.002053. [DOI] [PubMed] [Google Scholar]
- 24.Gray-Owen S D, Schryvers A B. The interaction of primate transferrins with receptors on bacteria pathogenic to humans. Microb Pathog. 1993;14:389–398. doi: 10.1006/mpat.1993.1038. [DOI] [PubMed] [Google Scholar]
- 25.Hancock R E, Carey A M. Outer membrane of Pseudomonas aeruginosa: heat-2-mercaptoethanol-modifiable proteins. J Bacteriol. 1979;140:902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanson M S, Hansen E J. Molecular cloning, partial purification, and characterization of a haemin-binding lipoprotein from Haemophilus influenzae type b. Mol Microbiol. 1991;5:267–278. doi: 10.1111/j.1365-2958.1991.tb02107.x. [DOI] [PubMed] [Google Scholar]
- 27.Jackson L A, Spach D H. Emergence of Bartonella quintana infection among homeless persons. Emerg Infect Dis. 1996;2:141–144. doi: 10.3201/eid0202.960212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson L A, Spach D H, Kippen D A, Sugg N K, Regnery R L, Sayers M H, Stamm W E. Seroprevalence to Bartonella quintana among patients at a community clinic in downtown Seattle. J Infect Dis. 1996;173:1023–1026. doi: 10.1093/infdis/173.4.1023. [DOI] [PubMed] [Google Scholar]
- 29.Kim S J, Chu L, Holt S C. Isolation and characterization of a hemin-binding cell envelope protein from Porphyromonas gingivalis. Microb Pathog. 1996;21:65–70. doi: 10.1006/mpat.1996.0043. [DOI] [PubMed] [Google Scholar]
- 30.Koehler J E, Sanchez M A, Garrido C S, Whitfeld M J, Chen F M, Berger T G, Rodriguez-Barradas M C, LeBoit P E, Tappero J W. Molecular epidemiology of Bartonella infections in patients with bacillary angiomatosis-peliosis. N Engl J Med. 1997;337:1876–1883. doi: 10.1056/NEJM199712253372603. [DOI] [PubMed] [Google Scholar]
- 31.Koehler J E, Quinn F D, Berger T G, LeBoit P E, Tappero J W. Isolation of Rochalimaea species from cutaneous and osseous lesions of bacillary angiomatosis. N Engl J Med. 1992;327:1625–1631. doi: 10.1056/NEJM199212033272303. [DOI] [PubMed] [Google Scholar]
- 32.Kostrzewski J. The epidemiology of trench fever. Med Dosw Mikrobiol. 1950;11:233–263. [PubMed] [Google Scholar]
- 33.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Lawson P A, Collins M D. Description of Bartonella clarridgeiae sp. nov. isolated from the cat of a patient with Bartonella henselae septicemia. Med Microbiol Lett. 1996;5:64–73. [Google Scholar]
- 35.Lee B C. Quelling the red menace: haem capture by bacteria. Mol Microbiol. 1995;18:383–390. doi: 10.1111/j.1365-2958.1995.mmi_18030383.x. [DOI] [PubMed] [Google Scholar]
- 36.Legrain M, Mazarin V, Irwin S W, Bouchon B, Quentin-Millet M J, Jacobs E, Schryvers A B. Cloning and characterization of Neisseria meningitidis genes encoding the transferrin-binding proteins Tbp1 and Tbp2. Gene. 1993;130:73–80. doi: 10.1016/0378-1119(93)90348-7. [DOI] [PubMed] [Google Scholar]
- 37.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 38.Mills M, Payne S M. Identification of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect Immun. 1997;65:5358–5363. doi: 10.1128/iai.65.12.5358-5363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minnick M F. Identification of outer membrane proteins of Bartonella bacilliformis. Infect Immun. 1994;62:2644–2648. doi: 10.1128/iai.62.6.2644-2648.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monson E K, Weinstein M, Ditta G S, Helinski D R. The FixL protein of Rhizobium meliloti can be separated into a heme-binding oxygen sensing domain and a functional C-terminal kinase domain. Proc Natl Acad Sci USA. 1992;89:4280–4284. doi: 10.1073/pnas.89.10.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myers W F, Cutler L D, Wisseman C L. Role of erythrocytes and serum in the nutrition of Rickettsia quintana. J Bacteriol. 1969;97:663–666. doi: 10.1128/jb.97.2.663-666.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nikaido H. Outer membrane. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 29–47. [Google Scholar]
- 43.O'Connell W A, Hickey E K, Cianciotto N P. A Legionella pneumophila gene that promotes hemin binding. Infect Immun. 1996;64:842–848. doi: 10.1128/iai.64.3.842-848.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osborn M J, Gander J E, Parisi E, Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membranes. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- 45.Parrott J H, Dure L, Sullender W, Buraphacheep W, Frye T A, Galliani C A, Marston E, Jones D, Regnery R. Central nervous system infection associated with Bartonella quintana: a report of two cases. Pediatrics. 1997;100:403–408. doi: 10.1542/peds.100.3.403. [DOI] [PubMed] [Google Scholar]
- 46.Pearson W R. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 1990;183:63–98. doi: 10.1016/0076-6879(90)83007-v. [DOI] [PubMed] [Google Scholar]
- 47.Regnery R L, Anderson B E, Clarridge III J E, Rodriguez-Barradas M C, Jones D C, Carr J H. Characterization of a novel Rochalimaea species, R. henselae sp. nov., isolated from blood of a febrile, human immunodeficiency virus-positive patient. J Clin Microbiol. 1992;30:265–274. doi: 10.1128/jcm.30.2.265-274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reidl J, Mekalanos J J. Lipoprotein e(P4) is essential for hemin uptake by Haemophilus influenzae. J Exp Med. 1996;183:621–629. doi: 10.1084/jem.183.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reithmeier R A, Bragg P D. Cross-linking of the proteins in the outer membrane of Escherichia coli. Biochim Biophys Acta. 1977;466:245–256. doi: 10.1016/0005-2736(77)90222-x. [DOI] [PubMed] [Google Scholar]
- 50.Scherer D C, DeBuron-Connors I, Minnick M F. Characterization of Bartonella bacilliformis flagella and effect of antiflagellin antibodies on invasion of erythrocytes. Infect Immun. 1993;61:4962–4971. doi: 10.1128/iai.61.12.4962-4971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott D, Chan E C, Siboo R. Iron acquisition by oral hemolytic spirochetes: isolation of a hemin-binding protein and identification of iron reductase activity. Can J Microbiol. 1996;42:1072–1079. doi: 10.1139/m96-137. [DOI] [PubMed] [Google Scholar]
- 52.Smalley J W, Birss A J, McKee A S, Marsh P D. Hemin regulation of hemoglobin binding by Porphyromonas gingivalis. Curr Microbiol. 1998;36:102–106. doi: 10.1007/s002849900287. [DOI] [PubMed] [Google Scholar]
- 53.Spach D H, Callis K P, Paauw D S, Houze Y B, Schoenknecht F D, Welch D F, Rosen H, Brenner D J. Endocarditis caused by Rochalimaea quintana in a patient infected with human immunodeficiency virus. J Clin Microbiol. 1993;31:692–694. doi: 10.1128/jcm.31.3.692-694.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spach D H, Kanter A S, Dougherty M J, Larson A M, Coyle M B, Brenner D J, Swaminathan B, Matar G M, Welch D F, Root R K, Stamm W E. Bartonella (Rochalimaea) quintana bacteremia in inner-city patients with chronic alcoholism. N Engl J Med. 1995;332:424–428. doi: 10.1056/NEJM199502163320703. [DOI] [PubMed] [Google Scholar]
- 55.Staggs T M, Perry R D. Identification and cloning of a fur regulatory gene in Yersinia pestis. J Bacteriol. 1991;173:417–425. doi: 10.1128/jb.173.2.417-425.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strong R P. Trench fever. Report of commission: Medical Research Committee, American Red Cross. Oxford, England: Oxford University Press; 1918. pp. 40–60. [Google Scholar]
- 57.Struyve M, Moons M, Tommassen J. Carboxyl-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J Mol Biol. 1991;218:141–148. doi: 10.1016/0022-2836(91)90880-f. [DOI] [PubMed] [Google Scholar]
- 58.Stugard C E, Daskaleros P A, Payne S M. A 101-kilodalton heme-binding protein associated with Congo red binding and virulence of Shigella flexneri and enteroinvasive Escherichia coli strains. Infect Immun. 1989;57:3534–3539. doi: 10.1128/iai.57.11.3534-3539.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swift H F. Trench fever. Arch Intern Med. 1920;26:76–98. [Google Scholar]
- 60.Tai S S, Wang T R, Lee C-J. Characterization of hemin binding activity of Streptococcus pneumoniae. Infect Immun. 1997;65:1083–1087. doi: 10.1128/iai.65.3.1083-1087.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tompkins G R, Wood D P, Birchmeier K R. Detection and comparison of specific hemin binding by Porphyromonas gingivalis and Prevotella intermedia. J Bacteriol. 1997;179:620–626. doi: 10.1128/jb.179.3.620-626.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tyeryar F J, Weiss E, Millar D B, Bozeman F M, Ormsbee R A. DNA base composition of rickettsiae. Science. 1973;180:415–417. doi: 10.1126/science.180.4084.415. [DOI] [PubMed] [Google Scholar]
- 65.Varela G, Vinson J W, Molina-Pasquel C. Trench fever II. Propagation of Rickettsia quintana on cell-free medium from the blood of two patients. Am J Trop Med Hyg. 1969;18:708–712. [PubMed] [Google Scholar]
- 66.Vizcaino N, Cloeckaert A, Zygmunt M S, Dubray G. Cloning, nucleotide sequence, and expression of the Brucella melitensis omp31 gene coding for an immunogenic major outer membrane protein. Infect Immun. 1996;64:3744–3751. doi: 10.1128/iai.64.9.3744-3751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Webb D C, Cripps A W. Immunization with recombinant transferrin binding protein B enhances clearance of nontypeable Haemophilus influenzae from the rat lung. Infect Immun. 1999;67:2138–2144. doi: 10.1128/iai.67.5.2138-2144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiss E, Dasch G A. Differential characteristics of strains of Rochalimaea: Rochalimaea vinsonii sp. nov., the Canadian vole agent. Int J Syst Bacteriol. 1982;32:305–314. [Google Scholar]