Abstract
Purpose:
The Cancer Immune Monitoring and Analysis Centers – Cancer Immunologic Data Commons (CIMAC-CIDC) network supported by the NCI Cancer Moonshot initiative was established to provide correlative analyses for clinical trials in cancer immunotherapy, using state-of-the-art technology. Fundamental to this initiative is implementation of multiplex IHC assays to define the composition and distribution of immune infiltrates within tumors in the context of their potential role as biomarkers. A critical unanswered question involves the relative fidelity of such assays to reliably quantify tumor-associated immune cells across different platforms.
Experimental Design:
Three CIMAC sites compared across their laboratories: (i) image analysis algorithms, (ii) image acquisition platforms, (iii) multiplex staining protocols. Two distinct high-dimensional approaches were employed: multiplexed IHC consecutive staining on single slide (MICSSS) and multiplexed immunofluorescence (mIF). To eliminate variables potentially impacting assay performance, we completed a multistep harmonization process, first comparing assay performance using independent protocols followed by the integration of laboratory-specific protocols and finally, validating this harmonized approach in an independent set of tissues.
Results:
Data generated at the final validation step showed an intersite Spearman correlation coefficient (r) of ≥0.85 for each marker within and across tissue types, with an overall low average coefficient of variation ≤0.1.
Conclusions:
Our results support interchangeability of protocols and platforms to deliver robust, and comparable data using similar tissue specimens and confirm that CIMAC-CIDC analyses may therefore be used with confidence for statistical associations with clinical outcomes largely independent of site, antibody selection, protocol, and platform across different sites.
Translational Relevance.
The Cancer Immune Monitoring and Analysis Centers – Cancer Immunology Data Commons (CIMAC-CIDC) network was established to provide comprehensive correlative analyses for clinical trials in cancer immunotherapy. Central to this initiative is multiplex IHC assays to define the composition and distribution of tumor-associated immune infiltrates to determine their potential role as predictive biomarkers. To ensure fidelity of such assays to reliably quantify immune cells across different platforms, three CIMAC sites harmonized multiplex staining protocols, image acquisition platforms, and image analysis algorithms, using both multiplexed IHC consecutive staining on single slide (MICSSS) and multiplexed immunofluorescence (mIF). This work provides a template for aligning methodologies across multiple institutions, without the need to enforce identical protocols and instrumentation across sites, but rather by ensuring concordant quantification and interpretation of data. The resulting data yield comparable results, which is critical when performing cross-trial analyses enabled by cooperative clinical research.
Introduction
The landscape of cancer treatment has been transformed with the introduction of immunotherapy that destroys cancer cells through activation of the host immune system and has resulted in unprecedented clinical and pathologic responses. The successes and failures of immunotherapy have prompted the interrogation of the tumor-immune contexture to identify biomarkers predictive of response and/or resistance to these interventions to distinguish responders from nonresponders and ultimately inform and refine the next generation of therapeutic interventions. Central to this approach has been the development of novel imaging platforms to characterize tumor tissues. In addition to standard singleplex IHC, an array of different multiplex platforms have evolved to simultaneously identify, quantify, and spatially resolve the tumor-associated immune infiltrate (1–5). Such multiplexed methodologies provide several critical advantages over conventional singleplex IHC. Multiple markers can be quantified simultaneously, and the spatial relationships of those markers, including the relative proximity of immune cell populations to one another and to tumor cells as well as marker coexpression, can be simultaneously explored (6). Also, these techniques can be performed on a single tissue section, thus maximally preserving a valuable clinical resource (7). Studies using these technologies have unveiled important biomarkers predictive of response or resistance to immunotherapy (8, 9). Despite the advantages afforded by multiplexed technologies, there are important critical unanswered questions. The variety of different methodologies introduce important variables, including unique antibody clones, different staining platforms and protocols, and novel image acquisition and analysis platforms. Consequently, the inherent complexity of these assays and data reporting show variability in assay performance across different laboratories. Taken together, the extent to which these methodologic differences impact data integrity and whether different approaches provide comparable data independent of the laboratory site, platform, and reagents utilized, remains a critical unanswered question, particularly as these biomarkers enter into the clinical testing environment. To address these challenges, we compared the quantification of immune infiltrates and tumor cells across three institutions using two distinct multiplexed image technologies [multiplex immunofluorescence (mIF)-based tyramide signal amplification (TSA) system and chromogenic-based multiplexed immunohistochemical consecutive staining on single slide (MICSSS)] within the Cancer Immune Monitoring and Analysis Centers – Cancer Immunologic Data Commons (CIMAC-CIDC) Network. The goal of this study was to harmonize interpretation and quantification of data following multiplex image staining, acquisition, and cell segmentation procedures across distinct platforms of different CIMACs. Harmonization based on integration of adjusted laboratory-specific protocols will enable objective interpretation and comparison of correlative biomarker studies across laboratories agnostic of platforms, reagents and procedures for clinical trials and will provide sufficient sample size for biomarker development. Our data also provide a framework for future efforts seeking to compare biomarker studies across institutions and/or platforms.
Materials and Methods
Study design
A stepwise harmonization process was implemented to facilitate comparability of the data across three CIMAC laboratories:(i) Harmonization of singleplex IHC image analysis algorithms, (ii) harmonization of mIF/MICSSS image analysis algorithms, (iii) harmonization of mIF/MICSSS staining and image analysis algorithms on two head and neck (H&N) tumors, and (iv) harmonization of mIF/MICSSS staining and image analysis algorithms on a tissue microarray (TMA) for scoring immune-cell densities across 3 CIMACs.
All assays used in this harmonization study by each participating institution were internally optimized and validated using reference material and controls prior to initiation of this work, and they continue to be routinely used for translational research studies on clinical trial specimens at each respective institution. The resulting individual validation documentation and SOPs for site-specific assays can be openly accessed from the CIMAC-CIDC website (https://cimac-network.org/assays). Before performing data image analysis during the harmonization, 3-way staining comparisons were completed by pathologists qualitatively, and there was strong intersite agreement observed. The quantitative part of the harmonization was performed on image analysis level by adjusting primary aspects of imaging algorithms including nuclear segmentation and positive cell thresholding. The primary data generated for comparison across CIMAC sites was density (number of positive cells per mm2), and the threshold for positivity of each marker was adjudicated across the CIMAC sites by pathologists from each site as a gold standard, and final density data from each CIMAC was compared with each other after adjudication was completed.
Materials
IHC/IF harmonization was performed at 3 CIMAC sites, including the Icahn School of Medicine at Mount Sinai (ISMMS), MD Anderson Cancer Center (MDACC), and Dana-Farber Cancer Institute (DFCI). DFCI and MDACC utilized identical technologies, instruments, and pipelines for both singleplex and multiplex IHC/IF-based assays including staining, image acquisition, and digital image quantification, whereas ISMMS utilized a chromogenic-based multiplex staining technology and related instruments as described below.
Singleplex imaging harmonization
Anonymized normal tonsil (n = 3), malignant melanoma (MM; n = 3), colorectal adenocarcinoma (CRC; n = 3), and H&N squamous cell carcinoma (HNSCC; n = 3) archival formalin-fixed, paraffin-embedded (FFPE) specimens were sectioned and stained by DFCI with a panel of CD3- (n = 6), CD68- (n = 6), and Ki-67- (n = 6) -specific antibodies for the singleplex imaging harmonization (Supplementary Table S1). All stained sections were scanned with a whole slide scanner (Aperio AT2, Leica) and generated SVS-formatted whole slide images (WSI) were distributed to MDACC and ISMMS for independent digital quantification (Supplementary Table S2). Identical regions of interest (ROI) were selected with visual interpretation on each WSI by each CIMAC center, where independent image analysis and quantification was performed for a given marker to quantify the number of positive cells per mm². DFCI and MDACC used ImageScope (version 12.3.2, Leica) for quantifying the density of positive cells stained by the corresponding markers, whereas ISMMS used QuPath (version 0.2.0, open source; ref. 10) image analysis platform. Initial results were then compared across the 3 CIMAC sites, and adjustments to each site's quantification algorithms were made to for threshold and segmentation to achieve agreement on quantification. A Gaussian smoothing algorithm was applied for CD68 as an extra step due to intensity irregularities and large size of stained CD68-positive macrophages (11). It is a widely used methodology in imaging field, typically to reduce image noise and reduce detail.
Multiplex imaging harmonization
Two anonymized HNSCC archival FFPE blocks were sectioned, stained with mIF methodology at DFCI, and a slide scanner with spectral unmixing technology (Vectra) was used for low- and high-resolution image acquisition by DFCI (Supplementary Table S3). Low resolution images were distributed by DFCI to the other 2 CIMACs for ROI selection. A total of five identical ROIs were selected from each HNSCC images by consensus agreement of all 3 CIMACs after several teleconference meetings to agree upon the precise ROI location. Selected ROI coordinates were scanned at high resolution and tiff formatted ROI images were generated and distributed to the CIMACs for independent digital quantification. For the second step in harmonization, the same anonymized HNSCC FFPE blocks were subsequently consecutively sectioned by DFCI and distributed to the MDACC and ISMMS for multiplex staining, imaging, and quantification harmonization (DFCI used the same data they had already generated in step 1). MDACC and ISMMS additionally stained, scanned, and digitally quantified the matching regions in the newly stained tissue sections based on originally selected ROIs from samples previously stained, scanned, and quantified at DFCI. In the final step of harmonization, whole multiplex imaging pipeline harmonization was repeated on selected ROIs of a TMA consisting of various malignant tumor and normal tissue cores which was designed and constructed by DFCI (Supplementary Table S4). In this final step, staining, imaging, and quantification steps were performed independently at each CIMAC from consecutive sections distributed at the same time, which facilitated equivalent ROI selection and minimized geographic variability across sections in the block.
Multiplex staining and imaging platforms
MICSSS
MICSSS pipeline was used by ISMMS as a multiplex IHC platform that is based on iterative cycles of staining and scanning of a single slide with a panel of Abs that can include up to 10 markers. The complete protocol has been previously optimized, validated, and published (Supplementary Materials and Methods, Multiplex Immunostaining Methodologies; refs. 12, 13). Each staining cycle of MICSSS staining is identical to singleplex IHC staining as they both generate brightfield chromogenic IHC staining of 4-μm FFPE sections using an automated immunostainer (Leica Bond RX, Leica Biosystems; Supplementary Table S2). Slides were scanned by a slide scanner (NanoZoomer S60) after each staining cycle and chosen ROIs (669 × 500 μm size) on the WSIs generated by the MICSSS staining protocol were analyzed at full resolution (0.4414μm/pixel at 20×) by the QuPath image analysis platform (10).
mIF
For mIF staining, MDACC and DFCI followed similar protocols previously optimized, validated, and published (14–17). Each Ab was applied in an automated stainer (Leica Bond RX, Leica Biosystems) in sequential order using the respective Opal Polaris 7-Color Automation IHC Detection Kit's (Catalog no. #:NEL821001KT, Akoya Bioscienses//PerkinElmer) reagents prepared according to the manufacturer's instructions (Supplementary Materials and Methods, Multiplex Immunostaining Methodologies) and following the requirements described previously for each Ab during the optimization and validation process (ref. 17; Supplementary Table S2). For each round of staining, negative (for autofluorescence) and positive controls were run in parallel. Slides for negative control were run with the primary and secondary Abs omitting the fluorophore tyramide. Slides from mIF staining were scanned using the Vectra 3.0.3, multispectral imaging system (Akoya Biosciences/PerkinElmer), at low magnification 10× (1.0 μm/pixel). Five ROIs on each slide of the two HNSCC cases and a single ROI from each core from the TMA consisting of 28 cores were considered for analysis. High magnification using the Phenochart Software image viewer 1.0.9 (669 × 500 μm size at resolution 20×, 0.5 μm/pixel) to capture various elements of tissue heterogeneity was applied. To establish reproducibility across three CIMACs, images of low tissue magnification were shared in each step across sites to capture the same location of the ROI to quantify the cell phenotypes (counts/mm2) in the sequential slides stained at each institution. For image analysis, MDACC and DFCI used the image analysis software (InForm™ 2.6.2, Akoya Biosciences/PerkinElmer) following previously published guidelines (17).
Harmonization methodology
All assays used in this harmonization study by the corresponding institutions were optimized and validated by using proper controls, and they are routinely used for translational research studies on clinical trial specimens at our respective institutions. Each of the CIMAC institutions participating in this harmonization study also has its own validation documentations and SOPs for their assays, and all SOPs are open access on CIMAC-CIDC website. In addition, 3-way staining comparison was completed by pathologist agreement qualitatively before performing data image analysis, and there was strong intersite agreement observed (data not shown). The quantitative part of the harmonization was performed on image analysis level by adjusting primary aspects of imaging algorithms including nuclear segmentation and positive cell thresholding. The main data generated for comparison across CIMAC sites was density (number of positive cells per mm2), and the threshold for positivity and segmentation of one cell from another for each marker was adjudicated across the CIMAC sites by pathologists at each site as the ultimate gold standard, and final density data from each CIMAC was compared with each other afterwards.
Statistical approaches and acceptance criteria for the experiments
Analyses for correlations of positive cell counts/mm2 (cell density) for mIF/MICSSS harmonization among CIMACs were performed using Spearman correlation (Rs) based on the assumption that the data is not Gaussian-distributed across all samples and that these were paired comparisons. Logarithm transformations are applied when needed. Multiple individual ROIs selected within the same tissue were considered as independent counts, because the geographic morphologic variations in the ROIs within a tissue were chosen purposefully for the greatest variety to test the range of the quantification, and therefore may be considered as unique areas, rather than as a repeated sampling of a same specimen. The data generated from the multiplex imaging harmonization phase performed on 2 H&N tumors were shared with a panel of biostatisticians and overall intersite correlation coefficients and coefficient of variations (CV) were used as measures of agreement/error across the 3 CIMACs participating in the multiplex harmonization effort for properly powering the analyses and harmonization. Although there is no uniformly accepted criterion of an “acceptable” agreement, a correlation of greater than 0.7, 0.8, and 0.9 can be considered as having adequate, good, and excellent correlation, respectively. Similarly, there is no uniformly accepted criterion regarding the magnitude of an “acceptable” CV. However, a CV of less than 0.3, 0.2, and 0.1 can be considered as having adequate, good, and excellent precision between the measurements, respectively.
Results
Harmonization of singleplex IHC image quantification
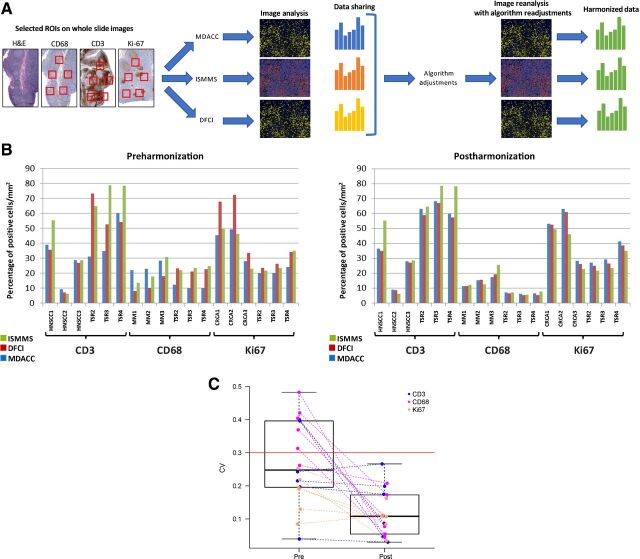
Harmonization of the singleplex imaging analysis was performed on tissue sections that had been centrally stained, scanned, and distributed by DFCI CIMAC using antibodies specific for CD3, CD68, and Ki67 as representatives of membranous, cytoplasmic, and nuclear signals location. To begin to address the challenges encountered with computer-assisted quantification of tissues that might contain cells of varying sizes and morphologies (i.e., the distinction of positive from negative signals and the separation of positive cells from one another agnostic of cellular cytomorphology), a wide variety of tissue morphologies were stained, including normal tonsil, MM, CRC, and HNSCC (Fig. 1A; Supplementary Table S1). A total of 18 stained tissue sections were stained, scanned, and generated WSIs were distributed across CIMACs for independent digital quantification. Identical ROIs were selected on each WSI by each CIMAC center, where independent image analysis and quantification was performed for a given marker to quantify the number of positive cells per mm2. Initial analyses revealed significant disparities in immune-cell density quantification across sites using independent protocols for data collection (Fig. 1B, left panel). Ki67 (MIB1), a nuclear marker of cellular proliferation; CD3, a pan T-cell marker; and CD68, a pan macrophage marker were quantified using nuclear and total cellular algorithms. Adjudication of differences in relative quantification between sites was done under the supervision of pathologists from each site to define a best performing consensus and it relied on adjustments to nuclear segmentation (how the software resolves and delineates the border of individual nuclei and generates shape- and intensity-related data for individual segmented cells inside this zone) and intensity-based thresholding (whether a cell is identified as positive or negative by the software; Supplementary Fig. 2A). The selected ROIs were reanalyzed using the harmonized algorithms based on integration of laboratory-specific protocols, resulting in greatly reduced variability of CD3- and Ki67-positive cell density data across the 3 CIMAC centers (Fig. 1B, right panel). The mean (SD) of the CV for the pre and postharmonization are 0.27 (0.12) and 0.12 (0.07), respectively. Each CV was calculated by taking the SD and dividing by the mean of the readings across the three institutions. When we compared the 18 preharmonization CVs and 18 postharmonization CVs using the Wilcoxon signed-rank test, the results were statistically significant (P = 0.0004; Fig. 1C). In contrast, the cytoplasmic staining pattern of pan-histiocytic marker CD68 is often of uneven intensity and distribution, which presented additional challenges to adjudicating segmentation and threshold settings. Therefore, a Gaussian noise-filtering strategy was applied to smooth the shape and intensity irregularities of CD68-stained macrophages (Supplementary Fig. 2B; ref. 11).
Figure 1.
Singleplex IHC image quantification harmonization. A, Representative illustration of singleplex IHC harmonization. Images shown are for illustrative purposes to demonstrate workflow only and are not intended as primary data figures. B, Percentage of cells positive for either CD3, CD68, or Ki67 per mm2 is plotted upon enumeration by each site (MDACC, DFCI, ISMMS) using the preharmonized and postharmonized algorithm settings. C, Wilcoxon rank-sum test showing the statistically significant reduction of CV (P = 0.0001). TSR, tonsil.
Multiplex imaging harmonization
Harmonization of image analysis algorithms for scoring immune-cell densities across 3 centers
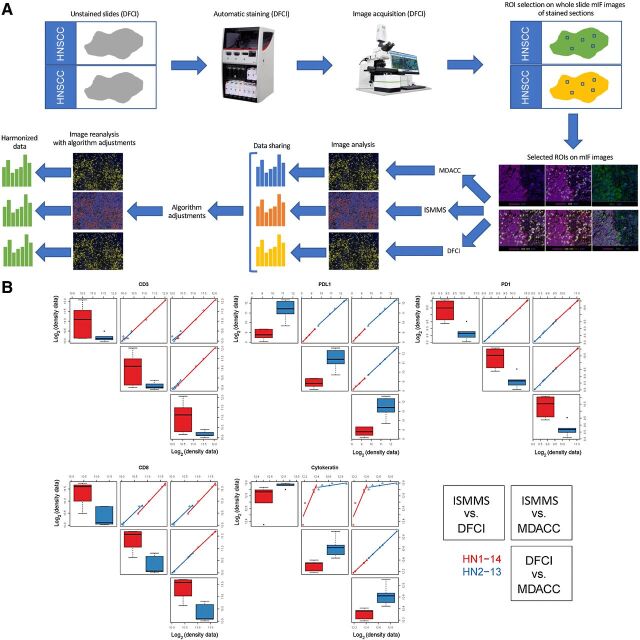
Multiplex imaging harmonization was performed in stepwise fashion. In the initial step, we utilized two HNSCCs stained with a panel for CD3, CD8, PD-L1, PD-1, and PanCK and scanned at a single CIMAC site (DFCI; Supplementary Table S3). WSIs were shared across the 3 institutions and quantification algorithms were harmonized using identical images. The differences in cellular quantification were adjudicated to harmonize threshold and segmentation as described above, and image analysis was repeated using the adjusted algorithms (Fig. 2A). All other sources of variability were eliminated in this step, enabling a singular focus on harmonizing quantification algorithms for each marker. Image quantification data of cell density expressing a specific marker showed excellent concordance (i.e., Rs = 1 for PD-1 and PD-L1) and strong agreement for all markers in the panel across all 3 CIMACs (all Rs > 0.8; Fig. 2B). We have also generated the Bland–Altman plots in which Spearman correlation coefficients were used to calculate the concordance and they showed excellent concordance between sites (Supplementary Fig. S1). Together, these findings argued that the differences in subsequent efforts would reflect differences in staining platform/protocol, image acquisition platform, and geographic differences in the tissue using deeper sections (even when the same ROI was selected) but would not reflect differences in quantification algorithms.
Figure 2.
MICSSS/mIF image analysis algorithm harmonization. A, Representative illustration of image analysis algorithm harmonization on prestained mIF images. Images shown are for illustrative purposes to demonstrate workflow only and are not intended as primary data figures. B, Concordance plot of CD3, CD8, PD-1, PD-L1, and cytokeratin log2 transformed cell density data between three centers within each tissue type. The distribution of each marker is shown separately. On the diagonals, box plots are shown to display the values for each sample (HN1-14 and HN2-13) measured at each site (ISMMS, DFCI, and MDACC), respectively. Off diagonals, pairwise scatter plots are shown to illustrate the concordance between two sites. The two samples are coded by red and blue, respectively. All Spearman correlation coefficients are greater than 0.7 except one, which is between ISMMS and MDACC for CD3. We observe that all median Spearman correlation coefficients are greater than or equal to 0.7.
Harmonization of staining and image analysis algorithms for scoring immune-cell densities from multiplex IF/MICSSS across 3 centers
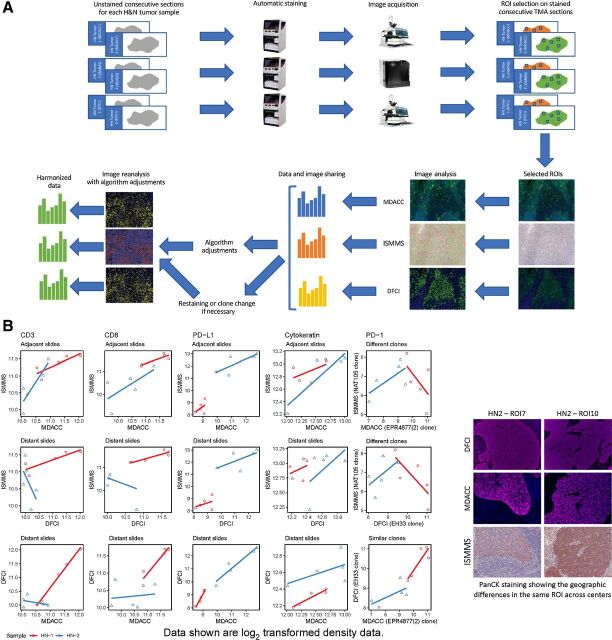
In step 3, we aimed to harmonize multiplex tissue staining and image acquisition protocols (Fig. 3A). Freshly-cut unstained consecutive sections from the 2 HNSCC cases previously processed and stained at DFCI (and used in step 1) were distributed to the other two CIMACs. Slides were independently stained at MDACC and ISSMS with a panel of antibodies (CD3, CD8, PD-1, PD-L1, and PanCK) to match the staining previously performed at DFCI, scanned, and digitally quantified using consensus ROI selection among all CIMACs, as in step 1. DFCI and MDACC performed mIF staining and scanning with spectral unmixing technology, while ISMMS performed MICSSS staining and brightfield whole-slide scanning. Notably, some of the consecutive sections utilized during this step of harmonization contained major geographic differences in the tissue due to deeper tissue levels being cut, which especially negatively impacted the pair-wise comparison between DFCI/MDACC versus MDACC/ISMMS (Fig. 3B). Therefore, ROIs were manually positioned based on best estimates of coordinates following consensus discussions among the 3 CIMACs. CD3+ T-cell densities (number of positive cells per mm2) showed stronger correlation between ISMMS and MDACC, due to better geographical match in ROI (adjacent slides) of these deeper sections (Rs = 0.94; Fig. 3B). In contrast, the pair-wise comparison between ISMMS and DFCI (Rs = 0.38), and MDACC and DFCI (Rs = 0.56) showed less concordance, emphasizing the lack of similarity in ROIs from the original section stained at DFCI in comparison with the newly-cut deeper sections stained at MDACC and ISMMS (distant slides; Supplementary Fig. S3). Results showing a similar trend were obtained for CD8, showing highest concordance between ISMMS and MDACC (Rs = 0.88) with slightly lower concordance values for ISMMS-DFCI (Rs = 0.78) and MDACC-DFCI (Rs = 0.79; Fig. 3B). In contrast, PD-1 quantification varied considerably between DFCI and MDACC compared with ISMMS, and this was attributed to site-specific differences in PD-1 clones. Singleplex IHC staining confirmed significant differences among the various anti–PD-1 antibody clones (data not shown). Specifically, the PD-1 EH33 clone (Cell Signaling used by DFCI) and the EPR4877 (2) clone (Abcam used by MDACC) both demonstrated a similar, discrete pattern of staining for PD-1+ cells. In contrast, NAT105 antibody (Abcam used by ISMMS), highlighted fewer PD-1+ cells with less intensity in comparison. Given these qualitative and quantitative differences (which were interpreted to explain the initial differences in the multiplex results), ISMMS replaced PD-1 NAT105 clone with PD-1 clone EPR4877 for the next phase of harmonization (2). PD-1 staining and image analysis harmonization was reserved for the TMA harmonization at ISMMS, to apply it on a wider range of tissue types.
Figure 3.
MICSSS/mIF whole tissue imaging pipeline harmonization, including staining, image acquisition, and image analysis combined on the H&N tumor samples; A, Representative illustration of MICSSS/mIF whole tissue imaging pipeline harmonization on H&N tumor samples. Images shown are for illustrative purposes to demonstrate workflow only and are not intended as primary data figures. B, Concordance plot of CD3, CD8, PD-1, PD-L1, and cytokeratin log2 transformed cell density data between three centers within each tissue type. The distribution of each marker is shown separately. For CD3, CD8, PD-L1, and cytokeratin, similar ROIs were examined by ISMMS and MDACC (adjacent slides), while geographically different ROIs (distant slides) were analyzed by DFCI. For PD-1, the three sites used different Ab clones, two of which performed similarly (at DFCI and MDACC, similar clones) while one had weaker staining (at ISMMS, different clones). Scatter plots are shown to illustrate the concordance between two sites. The two samples are coded by red and blue, respectively.
Harmonization of TMA staining and algorithm for scoring immune-cell densities from multiplex IF/MICSSS across 3 centers
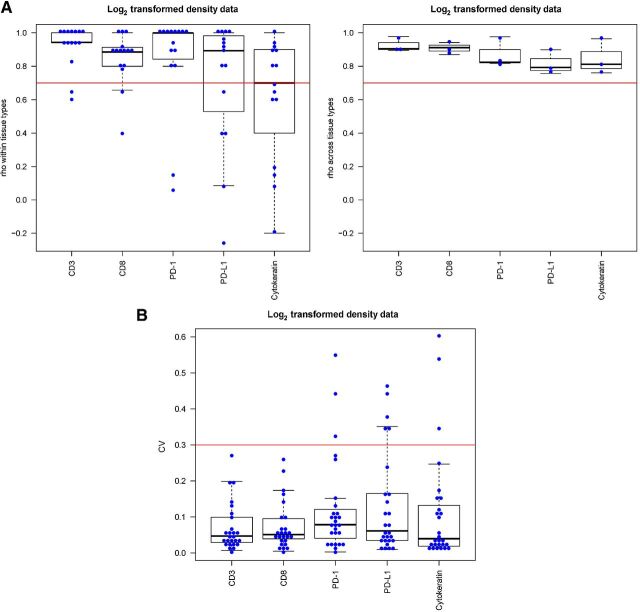
In step 4, the harmonization of staining protocols, image acquisition, and quantification described above was validated across a broad spectrum of cellular morphologies using a TMA constructed to include 27 individual patient tumors, including urothelial carcinoma (n = 4), HNSCC (n = 4), lung adenocarcinoma (n = 6), MM (n = 6), and clear cell renal cell carcinoma (ccRCC; n = 7; Fig. 4A; Supplementary Table S4). Pair-wise concordance plots comparing the quantification of CD3, CD8, PD-1, PD-L1, and PanCK from each of the 3 centers revealed overall strong correlation for each marker in each tissue core tested (Fig. 4B). Due to the relatively few numbers of cores for certain tissues and small variations in the quantification, some markers (PD-L1) showed greater fluctuation. Most importantly, the Spearman correlation coefficient for each marker across all of the ROIs tested exceeded 0.7 (Fig. 5A), while the majority of CV assessed for each ROI comparing 3 centers was less than 0.4, with median of CVs for each marker less than 0.1 (Fig. 5B). Each CV was calculated by taking the SD and dividing by the mean of the readings across the 3 institutions.
Figure 4.
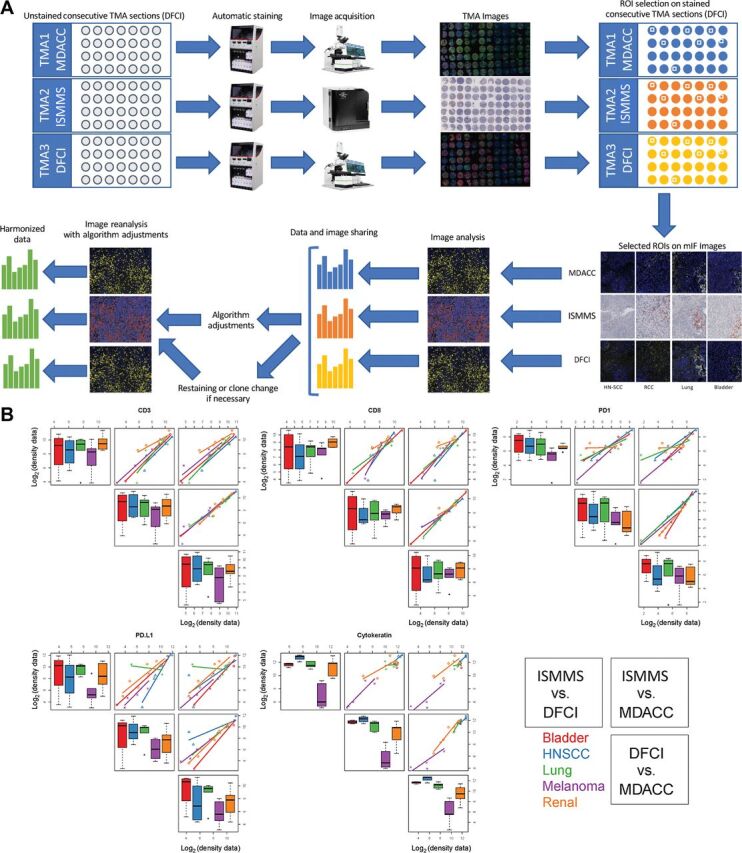
MICSSS/mIF whole tissue imaging pipeline harmonization, including staining, image acquisition, and image analysis combined on the TMA. A, Representative illustration of MICSSS/mIF whole tissue imaging pipeline harmonization on the TMA. Images shown are for illustrative purposes to demonstrate workflow only and are not intended as primary data figures. B, Concordance plot of CD3, CD8, PD-1, PDL1, and cytokeratin log2 transformed cell density data between three centers within each tissue type. The distribution of each marker is shown separately. On the diagonals, box plots are shown to display the values for each sample (bladder, H&N, lung, melanoma, and renal) measured at each site (ISMMS, DFCI, and MDACC), respectively. Off diagonals, pairwise scatter plots are shown to illustrate the concordance between two sites. The five samples are coded by different colors, respectively.
Figure 5.
Spearman rank correlation coefficient analysis on the data generated from the TMA. A, Spearman correlation analysis of indicated biomarkers within (left) and across (right) tissue types (horizontal red and green line indicate cutoff of 0.7 and 0.5, respectively). B, CV of log2 transformed cell density data for every sample in each marker. Horizontal red line indicates cutoff of 0.4 for all biomarkers. We observe that the medians of the CVs for all markers are below 0.1. We observe that all medians of the Spearman correlation coefficients across tissue types are greater than or equal to 0.7.
Discussion
The CIMAC-CIDC Network of 4 academic centers was established to perform biomarker analysis and provide correlative analyses for clinical trials in cancer immunotherapy, using state-of-the-art technologies. The network also includes a data coordination center supporting integrative analysis of biomarker and clinical data across different trials and sites. To allow such cross-trial and cross-site analysis and address variability associated with individual laboratory-specific protocols, multiplex IHC/IF assay performance and interpretation was harmonized across centers to ensure accuracy and reproducibility of any putative biomarkers identified. In the current study, the IHC Working Group of the CIMAC-CIDC performed validation of quantification of biomarkers in multiple human tissues using different singleplex and multiplex IF/IHC platforms for tissue staining, image acquisition, and quantification across 3 different CIMACs. They showed a correlation coefficient for each marker exceeding 0.7, with a median CV for each marker of less than 0.1. Our findings confirm our capability to reproducibly and accurately quantify tumor-associated immune cells independent of technique, platform, or CIMAC site.
Several important conclusions emerged in the course of the study. Through the process of comparing tissue staining and image acquisition/analysis protocols, we found that some markers are more amenable to harmonization than others. In particular, quantification of nuclear markers such as Ki67 and markers treated as ‘modified nuclear markers’, such as CD3/CD8, showed less variability and were ultimately robustly harmonized across different sites. Reproducible quantification of these types of antigens was largely dependent on the specificity and sensitivity of the Ab used to detect them. While Ki67, CD3, and CD8 were concordant after minimal adjustment of image quantification algorithms across centers, PD-1 was highly variable across centers, attributable to a less sensitive Ab clone (used initially at ISMMS), which was corrected in the course of the validation experiments by replacing this Ab clone with the clone used at MDACC.
In contrast, cytoplasmic/membranous markers, such as CD68, PD-L1, and cytokeratin, were more difficult to harmonize due to the large and often irregular staining patterns and cellular morphologies of the cells expressing these markers (including histiocytes and tumor cells). For example, the irregularity of shape and staining intensity of singleplex CD68+ histiocytes complicated their segmentation and resulted in lower initial concordance between CIMACs. However, implementation of a Gaussian smoothing filter reduced shape and intensity irregularities across the image and improved concordance. Similar approaches facilitated quantification of PD-L1 and cytokeratin in the multiplex harmonization, although these markers still showed greater variability across the centers. For cytokeratin, we emphasize, however, that this marker is typically not included for the purpose of quantification per se, but rather as a marker to spatially delineate tumor nests for their relationship to immune cells. Moreover, although PanCK is excellent at identification of cytokeratin-expressing tumor cells, the intensity of staining of different cytokeratins has been demonstrated to be highly variable within tumor cells (18). Variability identified in this study may represent intrapatient and/or intratumoral heterogeneity; thus, studies for which more precise measurement of tumor-cell density is important might consider using a nuclear marker (for example, p63, TTF-1, GATA-3) for more reproducible and accurate quantification. The multiplex harmonization panel of Abs included PD-L1 given its importance as a checkpoint molecule; however, its membranous staining pattern on tumor cells and variable staining intensity on different cells during the cell classification training made harmonization of PD-L1 especially difficult. Harmonization and interpretation of PD-L1 IHC have been investigated by many groups, and achieving concordance has been demonstrated, albeit with numerous challenges (19–23). Our harmonization efforts for PD-L1 quantification, which included both staining and image analysis, resulted in a high level of concordance (Rs = 0.898 for DFCI-MDACC, Rs = 0.793 for DFCI-ISMMS, Rs = 0.757 for ISMMS-MDACC) across different sites and platforms. Although IHC and IF test methods have improved over time, results are still affected by various preanalytical and analytical factors that affect assay reproducibility. While reproducibility concordance of a robust laboratory test is expected to be greater than 90% or a CV greater than 0.1, this is achieved by about 50% of the IHC clinical tests using a single Ab as per a review performed by the College American of Pathologists (CAP; ref. 24). No such of data are available for multiplex tissue assays as the mIF and ISMMS assays we studied. In addition, the outlier data (>0.1 CV) in our study were only for weakly stained ROIs or for ROIs with low density of the marker in question, where variation will appear larger based on low average counts (i.e., smaller denominator). We acknowledge that markers such as PD-1 and PD-L1 remain a critical challenge for digital quantification, due to lack of consensus for thresholding, challenges in segmentation, and generally low density, but we still achieved an average CVof more than 0.1 despite some outliers for unique ROI concordance.
Cell segmentation was an additionally challenging variable encountered throughout the multiplex harmonization process. This was largely attributed to the image differences between DFCI/MDACC and ISMMS. DFCI and MDACC employ a mIF platform in which multichannel fluorescence images are generated, and DAPI nuclear staining channel is utilized for cell segmentation, such that cell segmentation applied on the DAPI channel also applies to other channels. In contrast, ISMMS uses the MICSSS platform, where individual brightfield chromogenic images are generated and cell segmentation is performed on individual, consecutively stained images. This procedural difference in methodologies complicated the generation of a common cell segmentation algorithm and required adjudication of cell segmentation on a case-by-case basis to ensure fidelity of these decisions.
There were several limitations from current harmonization efforts that we did not directly address, but inferred from our data. First, it is important to control for preanalytical variables, including tissue procurement, fixation, and processing, all of which might contribute to potential differences in staining quality across different assays and institutions. Since CIMAC assays are to be deployed in multi-institutional clinical trials, rigorous preanalytic standardization may not be feasible unless the standardized protocols for specimen collection and processing developed for CIMACs are implemented in clinical trials. To guide CIMAC-CIDC investigators through sample distribution options among a variety of assays and provide standardized methods for specimen collection and handling, the Network and NCI developed a Specimen Collection “Umbrella” protocol that addresses various steps in the “sample flow” from tissue or blood sample acquisition at the trial sites, to immediate processing and storage at biorepositories, to subsequent processing and downstream distribution to the CIMAC laboratories (found at https://cimac-network.org/documents/).
We did not plan on addressing preanalytic differences in our harmonization study and purposedly used the same tissue blocks for sectioning, with the goal to compare the staining and quantification of markers tested using our respective mIHC/IF platforms. Nevertheless, we observed that geographic variability due to morphologic differences in the tissue created by nonconsecutive sectioning was still a major variable adversely impacting harmonization (Supplementary Fig. S3). This was particularly evident in the early efforts with mIF/MICSS harmonization in which we observed significant geographic differences in the relative content of tumor and tumor-associated lymphoid infiltrate, even when an ROI was placed in the “same area” of the tissue, due to unequal tissue trimming between stained sections, resulting in misaligned tissue sections. This variability produced poor correlation between DFCI and MDACC/ISMMS (Fig. 3B). This is an inherent limitation when using human tissues and can significantly impact interpretation and reproducibility. Notably, this produced significant differences between the ROIs stained and analyzed by each CIMAC for markers such as CD3. This is an important consideration for future biomarker analyses in clinical trial design, where differences in marker quantification are to be expected if using tissue sections from different areas or depth within a tissue block. Moreover, when the tumor/stroma architecture did not vary in an ROI, there was better agreement among the centers (particularly between MDACC and ISMMS in Fig. 3B), arguing that harmonization had in fact been achieved. This experience directly informed the TMA validation design, in which we ensured that each center stained immediately sequential sections. Reflective of this more careful design, concordance was uniformly higher for staining data from TMA generated independently for each marker at each site. Future biomarker studies in clinical trials should incorporate these critical lessons learned into how the process and allocate tissues for correlative studies. Finally, Ab clone differences among the centers can negatively impact data concordance levels, as shown by the experience with PD-1. Future efforts will require similar clone comparisons prior to application.
In conclusion, our effort to harmonize the quantification of immune cell markers across three CIMAC-CIDC sites showed that: (i) harmonization should be performed in a step-wise manner, to assess each variable as independently as possible before combining them; (ii) reference tissues are recommended and useful not only to broadly validate a variety of immune markers but also to maintain harmonization thereafter (25); (iii) harmonization as opposed to more rigid standardization does not enforce identical protocols and reagents but still allows achievement of highly concordant results, provided that variables critical for assay performance are well controlled and compared; and (iv) harmonized data is achievable agnostic to platform used. Our harmonization of different multiplex immunostaining platforms may serve as a template for other similar efforts, where despite major differences in protocols and instruments, harmonized results were obtained without enforcing standardized elements. The CIMAC assay protocols for mIHC/mIF are accessible to the immuno-oncology community at https://cimac-network.org/assays/.
Remarkably, our data met the statistical criteria of harmonization with a Spearman correlation coefficient > 0.85 for most markers and > 0.7 for all markers, not only as a relative score but also for absolute quantification. Given the median CV < 0.1, it would be expected that any CIMAC lab would be able to quantify T-cell density within 10% of one another. Implementation of harmonized assays enhances the potential of biomarker development as it enables comparison and interpretation of data across multi-site clinical trials and overcomes the limitations of single-site analysis. However, for future clinical utilization of multiplex imaging methodologies in clinical laboratories under the Clinical Laboratory Improvement Amendments (CLIA), further validation and interlaboratory harmonization studies should be performed addressing the preanalytical variables and other parameters as established by the clinical assay validation recommendations established in the guidelines from the CAP (24, 26).
Authors’ Disclosures
A. Lako reports personal fees from Bristol Myers Squibb outside the submitted work. J.J. Lee reports grants from NCI during the conduct of the study. D. Neuberg reports grants from NCI 5 U24 CA224331 during the conduct of the study. B. Sanchez-Espiridion reports grants from NIH during the conduct of the study. C. Wu reports grants from NIH/NCI (grant 5 U24 CA224316-01) during the conduct of the study, as well as other support from BioNTech, Inc and Pharmacyclics outside the submitted work. I.I. Wistuba reports grants and personal fees from Genentech/Roche, Bayer, Bristol-Myers Squibb, Astra Zeneca/Medimmune, Pfizer, HTG Molecular, Merck, and Guardant Health; personal fees from GlaxoSmithKline, Daiichi Sankyo, Eli-Lilly, Oncocyte, and MSD; and grants from Adaptive, Adaptimmune, EMD Serono, Takeda, Amgen, Karus, Johnson & Johnson, Iovance, Flame, 4D, Novartis and Akoya outside the submitted work. S. Rodig reports grants from Bristol Myers Squibb and KITE/Gilead outside the submitted work; in addition, S. Rodig is a member of the scientific advisory board for Immunitas Therapeutics and has received equity in the company. S. Gnjatic reports personal fees from Merck and OncoMed and grants from Regeneron, Takeda, Genentech, Immune Design, Janssen R&D, and Bristol Myers Squibb outside the submitted work; in addition, S. Gnjatic has a patent for MICSSS licensed to Caprion. M.T. Tetzlaff reports personal fees from Myriad Genetics, Nanostring LLC, and Novartis LLC and personal fees from Merck outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Materials and Methods
Supplementary Tables
Acknowledgments
Scientific and financial support for the CIMAC-CIDC Network is provided through the NCI Cooperative Agreements U24CA224319 (to the Icahn School of Medicine at Mount Sinai CIMAC), U24CA224331 (to the Dana-Farber Cancer Institute CIMAC), U24CA224285 (to the MDACC CIMAC), U24CA224309 (to the Stanford University CIMAC), and U24CA224316 (to the CIDC at Dana-Farber Cancer Institute). Additional support is made possible through the NCI CTIMS Contract HHSN261201600002C (to the Emmes Company, LLC). MDACC received support from NIH Cancer Center Support Grant P30CA016672 and the University of Texas SPORE NCI P50CA70907. The Human Immune Monitoring Center at ISMMS received support from Cancer Center Support Grant CA196521. S. Gnjatic is additionally supported by grants U01 DK124165, P30 CA196521, and P01 CA190174. DFCI received funding from the Center for Immuno-Oncology at the Dana-Farber Cancer Institute. Scientific and financial support for the PACT project are made possible through funding support provided to the FNIH by AbbVie Inc., Amgen Inc., Boehringer-Ingelheim Pharma GmbH & Co. KG., Bristol-Myers Squibb, Celgene Corporation, Genentech Inc, Gilead, GlaxoSmithKline plc, Janssen Pharmaceutical Companies of Johnson & Johnson, Novartis Institutes for Biomedical Research, Pfizer Inc., and Sanofi.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
G. Akturk: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. E.R. Parra: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. E. Gjini: Conceptualization, data curation, formal analysis, supervision, validation, investigation, visualization, methodology. A. Lako: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J.J. Lee: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. D. Neuberg: Conceptualization, formal analysis, validation, writing–original draft, writing–review and editing. J. Zhang: Conceptualization, data curation, software, formal analysis, validation, investigation, methodology, writing–original draft, writing–review and editing. S. Yao: Conceptualization, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. I. Laface: Conceptualization, formal analysis, validation, investigation, methodology. A. Rogic: Data curation, formal analysis, validation, investigation, visualization, methodology. P.-H. Chen: Validation, investigation, visualization, methodology. B. Sanchez-Espiridion: Resources, writing–original draft, project administration, writing–review and editing. D.M. Del Valle: Validation, investigation, visualization, methodology, project administration. R. Moravec: Investigation, methodology, writing–original draft, project administration, writing–review and editing. R. Kinders: Resources, validation, investigation, writing–original draft, project administration. C. Hudgens: Conceptualization, formal analysis, validation, investigation, visualization, methodology. C. Wu: resources, funding acquisition, writing–original draft, project administration, writing–review and editing. I.I. Wistuba: Conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. M. Thurin: Conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. S.M. Hewitt: Conceptualization, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. S. Rodig: Conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. S. Gnjatic: Conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. M.T. Tetzlaff: Conceptualization, resources, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Steiner C, Ducret A, Tille JC, Thomas M, McKee TA, Rubbia-Brandt L, et al. Applications of mass spectrometry for quantitative protein analysis in formalin-fixed paraffin-embedded tissues. Proteomics 2014;14:441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stauber J, MacAleese L, Franck J, Claude E, Snel M, Kaletas BK, et al. On-tissue protein identification and imaging by MALDI-ion mobility mass spectrometry. J Am Soc Mass Spectrom 2010;21:338–47. [DOI] [PubMed] [Google Scholar]
- 3. Sood A, Miller AM, Brogi E, Sui Y, Armenia J, McDonough E, et al. Multiplexed immunofluorescence delineates proteomic cancer cell states associated with metabolism. JCI Insight 2016;1:e87030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gorris MAJ, Halilovic A, Rabold K, van Duffelen A, Wickramasinghe IN, Verweij D, et al. Eight-color multiplex immunohistochemistry for simultaneous detection of multiple immune checkpoint molecules within the tumor microenvironment. J Immunol 2018;200:347–54. [DOI] [PubMed] [Google Scholar]
- 5. Rost S, Giltnane J, Bordeaux JM, Hitzman C, Koeppen H, Liu SD. Multiplexed ion beam imaging analysis for quantitation of protein expresssion in cancer tissue sections. Lab Invest 2017;97:992–1003. [DOI] [PubMed] [Google Scholar]
- 6. Carvajal-Hausdorf DE, Schalper KA, Neumeister VM, Rimm DL. Quantitative measurement of cancer tissue biomarkers in the lab and in the clinic. Lab Invest 2015;95:385–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dixon AR, Bathany C, Tsuei M, White J, Barald KF, Takayama S. Recent developments in multiplexing techniques for immunohistochemistry. Expert Rev Mol Diagn 2015;15:1171–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ascierto ML, McMiller TL, Berger AE, Danilova L, Anders RA, Netto GJ, et al. The intratumoral balance between metabolic and immunologic gene expression is associated with Anti-PD-1 response in patients with renal cell carcinoma. Cancer Immunol Res 2016;4:726–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bankhead P, Loughrey MB, Fernandez JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 2017;7:16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Iwabuchi S, Kakazu Y, Koh JY, Harata NC. Evaluation of the effectiveness of Gaussian filtering in distinguishing punctate synaptic signals from background noise during image analysis. J Neurosci Methods 2014;223:92–113. [DOI] [PubMed] [Google Scholar]
- 12. Remark R, Merghoub T, Grabe N, Litjens G, Damotte D, Wolchok JD, et al. In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci Immunol 2016;1:aaf6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Akturk G, Sweeney R, Remark R, Merad M, Gnjatic S. Multiplexed immunohistochemical consecutive staining on single slide (MICSSS): Multiplexed chromogenic IHC assay for high-dimensional tissue analysis. Methods Mol Biol 2020;2055:497–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carey CD, Gusenleitner D, Lipschitz M, Roemer MGM, Stack EC, Gjini E, et al. Topological analysis reveals a PD-L1-associated microenvironmental niche for Reed-Sternberg cells in Hodgkin lymphoma. Blood 2017;130:2420–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Francisco-Cruz A, Parra ER, Tetzlaff MT, Wistuba II. Multiplex immunofluorescence assays. Methods Mol Biol 2020;2055:467–95. [DOI] [PubMed] [Google Scholar]
- 16. Parra ER, Uraoka N, Jiang M, Cook P, Gibbons D, Forget MA, et al. Validation of multiplex immunofluorescence panels using multispectral microscopy for immune-profiling of formalin-fixed and paraffin-embedded human tumor tissues. Sci Rep 2017;7:13380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parra ER, Jiang M, Solis L, Mino B, Laberiano C, Hernandez S, et al. Procedural requirements and recommendations for multiplex immunofluorescence tyramide signal amplification assays to support translational oncology studies. Cancers (Basel) 2020;12:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chung JY, Braunschweig T, Hu N, Roth M, Traicoff JL, Wang QH, et al. A multiplex tissue immunoblotting assay for proteomic profiling: a pilot study of the normal to tumor transition of esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 2006;15:1403–8. [DOI] [PubMed] [Google Scholar]
- 19.Abdul Karim L, Wang P, Chahine J, Kallakury B. Harmonization of PD-L1 immunohistochemistry assays for lung cancer: A working progress. J Thorac Oncol 2017;12:e45. [DOI] [PubMed] [Google Scholar]
- 20. Adam J, Le Stang N, Rouquette I, Cazes A, Badoual C, Pinot-Roussel H, et al. Multicenter harmonization study for PD-L1 IHC testing in non-small-cell lung cancer. Ann Oncol 2018;29:953–8. [DOI] [PubMed] [Google Scholar]
- 21. Hernandez-Martinez JM, Zatarain-Barron ZL, Cardona AF, Arrieta O. The importance of PD-L1 diagnostic assay harmonization for the selection of lung cancer immunotherapy. J Thorac Dis 2018;10:S4096–S100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ionescu DN, Downes MR, Christofides A, Tsao MS. Harmonization of PD-L1 testing in oncology: a Canadian pathology perspective. Curr Oncol 2018;25:e209–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neuman T, London M, Kania-Almog J, Litvin A, Zohar Y, Fridel L, et al. A Harmonization Study for the Use of 22C3 PD-L1 Immunohistochemical Staining on Ventana's Platform. J Thorac Oncol 2016;11:1863–8. [DOI] [PubMed] [Google Scholar]
- 24. Fitzgibbons PL, Bradley LA, Fatheree LA, Alsabeh R, Fulton RS, Goldsmith JD, et al. Principles of analytic validation of immunohistochemical assays: Guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med 2014;138:1432–43. [DOI] [PubMed] [Google Scholar]
- 25. Tripodi SA, Rocca BJ, Hako L, Barbagli L, Bartolommei S, Ambrosio MR. Quality control by tissue microarray in immunohistochemistry. J Clin Pathol 2012;65:635–7. [DOI] [PubMed] [Google Scholar]
- 26. Goldstein NS, Hewitt SM, Taylor CR, Yaziji H, Hicks DG. Members of Ad-Hoc Committee On Immunohistochemistry Standardization. Recommendations for improved standardization of immunohistochemistry. Appl Immunohistochem Mol Morphol 2007;15:124–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Materials and Methods
Supplementary Tables