Abstract
The genus Galerita Fabricius, 1801 belongs to the tribe Galeritini of the family Carabidae. In this study, the complete mitochondrial genome (GenBank: ON920164.1) of G. orientalis is newly sequenced, annotated, characterized, and composed of 37 typical genes, and one control region. Mitogenome is a circular DNA molecule of 16,137 bp with a 78.79% AT content. All 13 protein-coding genes are initiated using a typical ATN (Met) as the start codon, except for nad1, which has a TTG as the start codon, and are terminated using a typical TAN stop codon. Twenty-two tRNAs could fold into a typical cloverleaf structure, including trnS1-GCU, which lacks the DHU stem observed in other mitogenomes of the subfamily Harpalinae. Both rrnS and rrnL contain many helices. A conserved poly-T stretch (19 bp) and seven tandem repeats are observed in the control region, and a phylogenetic analysis indicated that the genus Galerita is an independent lineage. The complete mitogenome of G. orientalis will contribute to further studies on the molecular basis of the classification and phylogeny of Harpalinae, and even Carabidae.
Keywords: Galerita orientalis, mitochondrial genome, phylogenetic analysis
1. Introduction
Carabidae is the largest family in the order Coleoptera, comprising more than 32,000 species worldwide [1,2], and is known as carabid beetles or ground beetles [1,3]. Harpalinae is the largest group of Carabidae and contains more than 19,000 species [1,4]. These beetles live in diverse habitats, have diverse morphological forms, and exhibit a variety of unusual lifestyles [5]. Therefore, the study of phylogeny is critical for understanding the evolution and diversity of tribes, genera, and species within the Harpalinae [1,5]. The monophyly of Harpalinae is based on morphological characteristics, chemical defensive secretions, chromosome number in males, and molecular sequence data obtained from the 18S rDNA gene [1,5]. However, the boundaries of these character states do not exactly match Harpalinae [1,5]. The nominotypical subgenus of the pantropical genus Galerita Fabricius, 1801 is represented in all zoogeographic regions, except Australian [6,7,8,9], and includes more than 110 species [10], which belong to the tribe Galeritini of the subfamily Harpalinae. The taxonomy of the tribe Galeritini has been developed based on adult and larvae features [6,7,8,9,10,11]. Based on the DNA-sequence datasets obtained from nuclear genes (the 28S rDNA and wingless nuclear protein-coding genes), higher-level phylogenetic relationships within Harpalinae are investigated [1,5,12,13], including the tribe Galeritini. Galerita is a sister group to ctenodactylines in the Zuphiitae clade, based on the 28S rDNA gene and wingless data [13].
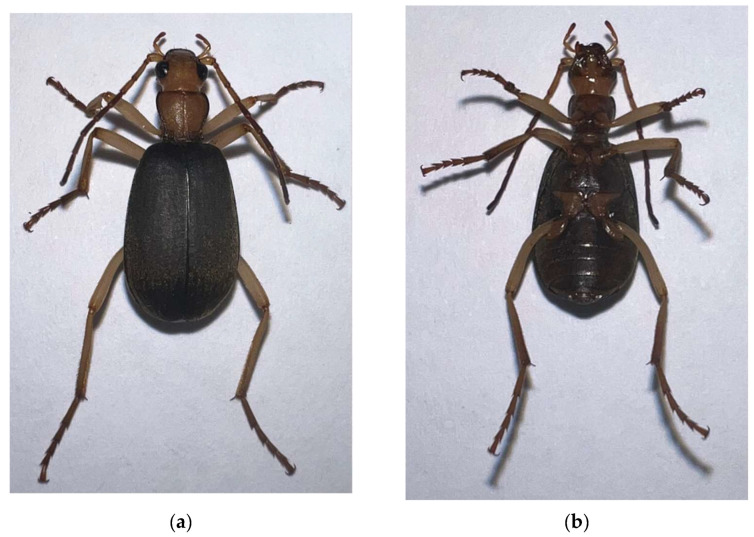
Galerita orientalis Schmidt-Göbel, 1846 (Figure 1) is widely distributed in continental Asia, Japan, and the Greater and Lesser Sunda Islands [9], and was revised by Reichardt (1965) [8,9]. However, our knowledge remains incomplete, with limited genetic and mitogenomic information from G. orientalis. As of August 2022, only three partial coding sequences of cox1 from G. orientalis had been published in GenBank. Insect mitochondrial genomes (mitogenomes) are closed, circular, double-stranded DNA molecules with lengths ranging from 15 to 19 kb that contain 37 typical genes, including 13 protein-coding genes (PCGs), 2 rRNAs, 22 tRNAs, and a control region (CR). Mitogenomes play an important role in the molecular phylogeny of insects [14]. Next-generation sequencing (NGS) is an important and effective strategy for mitogenome assembly and the phylogenetic analysis of Harpalinae [3,15,16,17].
Figure 1.
Species reference image of Galerita orientalis Schmidt-Göbel, 1846. (a) The dorsal view of G. orientalis; (b) the ventral view of G. orientalis.
In the present study, we sequence and characterize the G. orientalis mitogenome using NGS. Furthermore, we construct phylogenetic trees based on the mitogenomes of 30 species of the family Carabidae and two outgroup species, which will contribute to the research on its phylogenetic position in the family Harpalinae. These results will be useful to reconstruct the phylogenetic relationships within Carabidae in the future.
2. Materials and Methods
2.1. Animal Materials, DNA Extraction, and Illumina Sequencing
Samples of adult G. orientalis were collected from Jingziguan Town (E 111.026°, N 33.244°), Xichuan County, Nanyang City, Henan Province, China, on 9 June 2022, the genomic DNA (gDNA) of which was extracted using the Qiagen DNeasy Blood and Tissue Extraction kit (Qiagen, Germantown, MD, USA). The purity and concentration of the obtained gDNA were tested using a NanoPhotometer® spectrophotometer (Implen, Calabasas, CA, USA) and a Qubit® 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA), respectively [18,19]. Sequencing libraries for the quality-checked gDNA were generated using a TrueLib DNA Library Rapid Prep Kit for Illumina sequencing (Illumina, Inc., San Diego, CA, USA) [18,19]. The libraries were subjected to size-distribution analysis using an Agilent 2100 bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA), followed by a real-time PCR quantitative test [18,19]. The successfully generated libraries were sequenced using an Illumina NovaSeq 6000 platform (Illumina, Inc., San Diego, CA, USA) [18,19], and 150 bp paired-end reads with a 300 bp insert library were generated.
2.2. DNA Data Cleaning and Mitogenome Assembly
The obtained raw reads were filtered to obtain clean reads using fastp version 0.23.2 (https://github.com/OpenGene/fastp (accessed on 23 October 2022)) [20]. The quality control (QC) standards of the reads obtained from the DNA were as follows:
-
(1)
Trimming adapter sequences with >6 bases;
-
(2)
Removing reads with >0 unidentified nucleotides (Ns);
-
(3)
Removing reads with >20% bases with Phred quality < Q30;
-
(4)
Removing reads with <150 bases.
The mitogenome of G. orientalis was assembled de novo from high-quality cleaned reads using NOVOPlasty v4.3.1 (https://github.com/ndierckx/NOVOPlasty (accessed on 23 October 2022)) [21] with default parameters and the G. orientalis voucher NSMK-IN-160200486 cox1 gene (GenBank: OL663076.1) [16] as a seed sequence.
2.3. Mitogenome Annotation and Analysis
The AT-skew [(A − T)/(A + T)] and GC-skew [(G − C)/(G + C)] of the sequence were estimated to investigate the nucleotide composition bias, using Perna and Kocher’s formula [22]. The G. orientalis mitogenome was initially annotated using GeSeq version 2.03 (https://chlorobox.mpimp-golm.mpg.de/geseq.html (accessed on 23 October 2022)) [23], using the third-party software tRNAscan-SE v2.0.7 [24], ARWEN v1.2.3 [25], BLAT v36×7 [26], with the mitogenome of Mastax latefasciata (ON674050.1) as a reference. Using the mitogenomes of M. latefasciata as references, the start and stop codons of PCGs were manually corrected. The order and orientation of the genes were determined and drawn using the CGView Web server (https://proksee.ca/ (accessed on 23 October 2022)) [27]. Relative synonymous codon usage (RSCU) was analyzed using MEGA v11.0.13 [28]. The secondary structures of tRNAs were analyzed and visualized using forna [29,30]. The secondary structures of rRNAs and CR were inferred and visualized using RNAfold Web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi (accessed on 23 October 2022)) [31,32], with minimum free energy (MFE) and partition functions [33]. Tandem repeats in the control region (CR) were predicted using the Tandem Repeats Finder program version 4.09 [34] with parameters = 2 7 7 80 10 50 500 -f -d m.
2.4. Phylogenetic Analysis
In this study, the phylogenetic analysis included sequences from mitogenomes of 30 Carabidae species and 2 outgroup species (Lepisma saccharina and Corydidarum magnifica) (Table 1). The concatenated sequences of 13 protein-coding genes from mitogenomes were used to reconstruct the phylogenetic relationships of Carabidae using PhyloSuite version 1.2.2 [35] with MAFFT version 7 [36], MACSE version 2.03 [37], Gblocks 0.91b [38], ModelFinder [39], MrBayes version 3.2.7 [40], and IQ-TREE 1.6.12 [41]. Nucleotide sequences of 13 PCGs were aligned using MAFFT version 7 with the default parameters, and MACSE version 2.03 with default parameters. Ambiguously aligned fragments of the alignments from the 13 PCGs were removed using Gblocks 0.91b [38] with default parameters. The nucleotide sequences were used to construct phylogenetic trees using two methods: Bayesian inference (BI) using MrBayes 3.2.0,7 and maximum likelihood (ML) using IQ-TREE 1.6.12. According to the Bayesian information criterion (BIC) scores, a GTR + F [state frequencies, fixed (empirical)] + I [proportion of invariable sites, uniformly distributed on the interval (0.00, 1.00)] + G4 (γ-distributed rate variation, four categories) model was selected as the best-fit partition model (edge-unlinked) for the BI of nucleotide sequences using PhyloSuite version 1.2.2 with ModelFinder. For the IQ-tree, owing to the selection of the partition mode, the best-fit partition model was automatically calculated before the phylogenetic trees were constructed. In the BI analysis, 2 runs of 2,000,000 generations were conducted for each matrix, and the initial 25% was discarded as burn-in, which had the same topology with an average standard deviation of split frequencies of 0.008349 (<0.01). In the ML analysis, node support values were assessed using 5000 bootstrap resampling replicates. The resulting phylogenetic trees were visualized using an interactive tree of life (iTOL) (https://itol.embl.de/ (accessed on 23 October 2022)) [42].
Table 1.
The mitogenomic sequences used for phylogenetic analysis in this study.
| Family | Subfamily | Species | Whole Length | GenBank Accession Number | Reference |
|---|---|---|---|---|---|
| Carabidae | Brachininae | M. latefasciata | 16,735 bp | ON674050.1 | Unpublished |
| Cicindelinae | Cicindela anchoralis | 16,388 bp | MG253029.1 | [43] | |
| Cicindela puritana | 15,676 bp | MW442537.1 | Unpublished | ||
| Manticora tibialis | 16,439 bp | MF497821.1 | [44] | ||
| Carabinae | Carabus changeonleei | 16,831 bp | MG253028.1 | [45] | |
| Carabus granulatus | 16,918 bp | MN122870.1 | Unpublished | ||
| Carabus lafossei | 16,793 bp | KY992943.1 | [46] | ||
| Calosoma sp. BYU-CO241 | 16,462 bp | GU176340.1 | [47] | ||
| Damaster mirabilissimusmirabilissimus | 16,823 bp | GQ344500.1 | [48] | ||
| Harpalinae | Abax parallelepipedus | 17,701 bp | KT876877.1 | [15] | |
| Amara aulica | 16,646 bp | MN335930.1 | [3] | ||
| Amara communis | 15,745 bp | KX035135.1 | Unpublished | ||
| Craspedophorus nobilis | 15,063 bp | JX412738.1 | [49] | ||
| Galerita orientalis | 16,137 bp | ON920164.1 | This study | ||
| Harpalus anxius | 16,429 bp | ON929899.1 | Unpublished | ||
| Harpalus discrepans | 16,027 bp | OP161482.1 | Unpublished | ||
| Harpalus griseus | 16,972 bp | OP133272.1 | Unpublished | ||
| Harpalus pensylvanicus | 16,434 bp | MN245975.1 | [17] | ||
| Harpalus sinicus | 16,521 bp | MN310888.1 | [16] | ||
| Hexagonia terminalis | 12,639 bp | JX412768.1 | [49] | ||
| Orthomus sp. BMNH 1042407 | 16,368 bp | MK692555.1 | Unpublished | ||
| Pterostichus madidus | 18,324 bp | KT876910.1 | [15] | ||
| Pterostichus niger | 17,160 bp | KT876909.1 | [15] | ||
| Stomis pumicatus | 17,265 bp | KT876914.1 | [15] | ||
| Nebriinae | Nebria brevicollis | 21,161 bp | KT876906.1 | [15] | |
| Notiophilus quadripunctatus | 15,312 bp | MW800883.1 | [50] | ||
| Omophroninae | Omophron limbatum | 15,438 bp | MW800882.1 | [50] | |
| Rhysodinae | Rhysodes sp. BMNH-844233 | 16,315 bp | KX035156.1 | Unpublished | |
| Scaritinae | Scarites subterraneus | 16,163 bp | OL872182.1 | [51] | |
| Trechinae | Tachyta nana | 16,165 bp | KX035142.1 | [15] | |
| Lepismatidae | L. saccharina | 15,244 bp | MT108230.1 | [52] | |
| Blaberidae | Perisphaerinae | Co. magnifica | 16,627 bp | MW630139.1 | [53] |
3. Results and Discussion
3.1. Sequencing, Quality Control, and Mitogenome Organization and Base Composition of G. orientalis
Approximately 43.38 Gb of high-quality, clean reads were obtained using the fastp software [6] from approximately 51.89 Gb of raw reads of a 300 bp insert library, using the Illumina NovaSeq 6000 platform for the G. orientalis mitogenome assembly. The Q20, Q30, and GC contents of the clean reads were 98.37%, 93.88%, and 35.88%, respectively (Table 2).
Table 2.
Reads statistics for G. orientalis.
| Raw Reads Base (bp) | Raw Reads num | Q20 (%) | Q30 (%) | Clean Reads Base (bp) | Clean Reads num | Q20 (%) | Q30 (%) | GC (%) |
|---|---|---|---|---|---|---|---|---|
| 51,892,104,000 | 345,947,360 | 96.90 | 91.37 | 43,375,436,400 | 289,169,576 | 98.37 | 93.88 | 35.88 |
Abbreviations: Q20, percentage of bases with quality value ≥20; Q30, percentage of bases with quality value ≥30; GC, GC content.
These high-quality clean short reads (0.39% reads from the mitogenome) defined the mitogenome of G. orientalis (ON920164.1) with 100% coverage at a high-average-reads depth (10,425 times), which consisted of a typical, single, circular DNA molecule 16,137 bp in length. The length of this mitogenome was between 16,027 bp for Ha. discrepans and 17,701 bp for Ab. parallelepipedus in the subfamily Harpalinae (Table 1).
The mitogenomes of Cr. nobilis and He. terminalis were sequenced using the Roche/454 sequencing platform [49]. The mitogenomes of Ab. parallelepipedus, S. pumicatus, P. niger, P. madidus, Ha. sinicus, Ha. pensylvanicus, and Am. aulica were sequenced on an Illumina sequencer [3,15,16,17]. Therefore, the complete Harpalinae mitochondrial genome could be assembled using NGS alone.
The mitogenome of G. orientalis contained 40.85% A, 37.94% T, 8.46% G, and 12.75% C, which showed an obvious AT bias with 78.79% AT content. The AT content of the G. orientalis mitogenome was slightly lower than those of Ha. sinicus [16], Am. communis [17], S. pumicatus [17], and Ha. pensylvanicus [17]. The AT- and GC-skews of the major strand of the G. orientalis mitogenome were 0.037 and −0.202, respectively, indicating a major strand compositional bias characterized by a slight excess of A over T nucleotides, and a strong excess of C over G nucleotides. Bias is generally observed in the mitogenomes of members of the subfamily Harpalinae, including Am. aulica [3] and Ha. pensylvanicus [17].
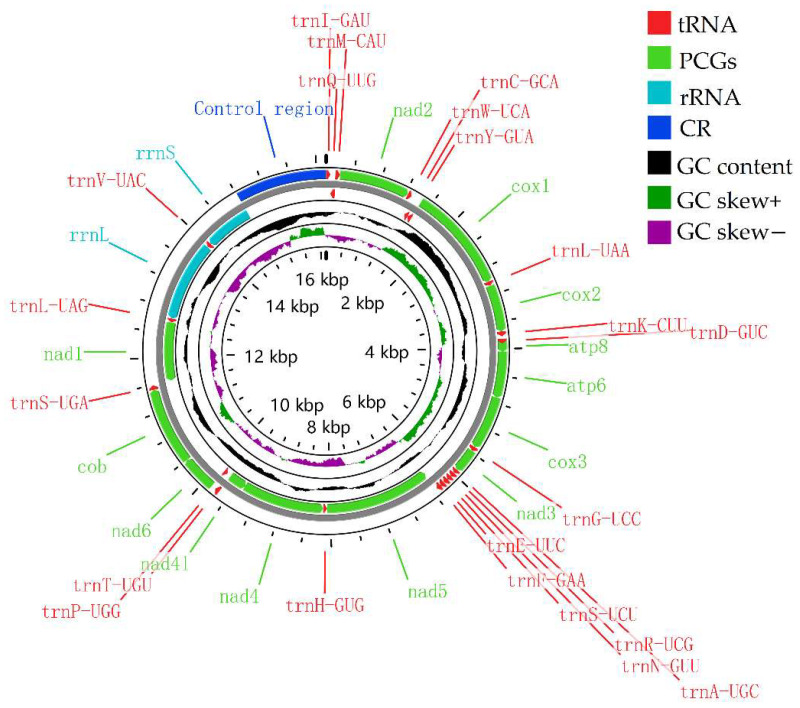
The G. orientalis mitogenome comprises 13 PCGs, 22 tRNA, 2 rRNA, and 1 CR (Figure 2), of which the order and orientation of the genes are the same as those in the mitogenomes of the subfamily Harpalinae [3,15,16,17,49]: 23 genes (9 PCGs and 14 tRNAs) on the majority strand (J-strand), and 4 PCGs, 8 tRNAs, and 2 rRNAs on the minority strand (N-strand) (Figure 2 and Table 3). Specifically, the mitogenome contains 12 overlapping genes (a total of 42 bp), ranging from 1 to 8 bp, with the longest region located between trnY-GUA and cox1 in the Ha. sinicus mitogenome [16] (Table 3). Most of the gene-overlap regions appeared between tRNA and PCGs. Intergenic spacers have 10 regions (a total of 45 bp), ranging from 1 to 16 bp, with the longest region located between 20 trnS-UGA and nad1 in the Ha. sinicus mitogenome [16] (Table 3).
Figure 2.
Mitogenome-pattern map of G. orientalis. The first circle shows the gene map (PCGs, rRNAs, tRNAs, and control region). Arrows in the second and third circles indicate the direction of gene transcription. The fourth circle shows the GC content and the fifth shows the GC-skew values. GC content and GC skew are plotted as the deviation from the average value of the entire sequence.
Table 3.
Organization of G. orientalis mitogenome.
| Gene | Strand | Location | Size (bp) | Anticodon | Start Codon | Stop Codon | Intergenic Nucleotides |
|---|---|---|---|---|---|---|---|
| trnI | J | 1–65 | 65 | GAT | |||
| trnQ | N | 66–140 | 75 | TTG | 0 | ||
| trnM | J | 136–206 | 71 | CAT | −5 | ||
| nad2 | J | 206–1234 | 1029 | ATA | TAA | −1 | |
| trnW | J | 1232–1306 | 75 | TCA | −3 | ||
| trnC | N | 1333–1396 | 64 | GCA | 2 | ||
| trnY | N | 1398–1465 | 68 | GTA | 1 | ||
| cox1 | J | 1458–2997 | 1540 | ATT | T | −8 | |
| trnL2 | J | 2998–3062 | 65 | TAA | 0 | ||
| cox2 | J | 3065–3749 | 685 | ATG | T | 2 | |
| trnK | J | 3750–3820 | 71 | CTT | 0 | ||
| trnD | J | 3821–3886 | 66 | GTC | 0 | ||
| atp8 | J | 3887–4048 | 162 | ATC | TAA | 0 | |
| atp6 | J | 4042–4719 | 678 | ATG | TAA | −7 | |
| cox3 | J | 4719–5507 | 789 | ATG | TAA | −1 | |
| trnG | J | 5509–5576 | 68 | TCC | 1 | ||
| nad3 | J | 5576–5929 | 354 | ATT | TAG | −1 | |
| trnA | J | 5930–5990 | 61 | TGC | 0 | ||
| trnR | J | 5993–6058 | 66 | TCG | 2 | ||
| trnN | J | 6059–6124 | 66 | GTT | 0 | ||
| trnS1 | J | 6125–6191 | 67 | GCT | 0 | ||
| trnE | J | 6193–6261 | 69 | TTC | 1 | ||
| trnF | N | 6257–6325 | 69 | GAA | −5 | ||
| nad5 | N | 6324–8052 | 1729 | ATT | T | −2 | |
| trnH | N | 8053–8117 | 65 | GTG | 0 | ||
| nad4 | N | 8118–9456 | 1339 | ATG | T | 0 | |
| nad4l | N | 9450–9743 | 294 | ATT | TAA | −7 | |
| trnT | J | 9744–9812 | 69 | TGT | 0 | ||
| trnP | N | 9811–9875 | 65 | TGG | −2 | ||
| nad6 | J | 9877–10,401 | 525 | ATT | TAA | 1 | |
| cob | J | 10,401–11,537 | 1137 | ATG | TAG | −1 | |
| trnS2 | J | 11,536–11,603 | 68 | TGA | −2 | ||
| nad1 | N | 11,620–12,570 | 951 | TTG | TAG | 16 | |
| trnL1 | N | 12,571–12,636 | 66 | TAG | 0 | ||
| rrnL | N | 12,637–13,948 | 1312 | 0 | |||
| trnV | N | 13,954–14,024 | 71 | TAC | 5 | ||
| rrnS | N | 14,025–14,810 | 786 | 0 | |||
| CR | J | 14,811–16,137 |
Abbreviations: J, J-strand (the majority strand); N, N-strand (the minority strand).
3.2. Protein-Coding Genes
A total of 9 of the 13 PCGs were encoded on the majority strand (cox1, cox2, cox3, atp6, atp8, nad2, nad3, nad6, and cob), and 4 on the minority strand (nad1, nad4, nad4l, and nad5) (Figure 2 and Table 3). All 13 PCGs had a typical ATN (Met) start codon, except for nad1 (TTG as start codon): only 1 PCG (nad2) initiated with an ATA start codon, 5 PCGs (cox1, nad3, nad5, nad4l, and nad6) initiated with an ATT start codon, 5 PCGs (cox2, atp6, cox3, nad4, and cob) initiated with an ATG start codon, and only 1 PCG (atp8) initiated with an ATC start codon. The start codons in the G. orientalis mitogenome were consistent with those in the Ha. sinicus mitogenome. In the mitogenomes of the subfamily Harpalinae, CGA and TAT are used as start codons [3,17]. All 13 PCGs contained a typical TAN stop codon, 3 PCGs (nad3, cob, and nad1) terminated with a TAG stop codon, 6 PCGs (nad2, atp8, atp6, cox3, nad4l, and nad6) ended with a TAA stop codon, and 4 PCGs (cox1, cox2, nad5, and nad4) terminated with an incomplete stop codon (T), consisting of a codon that was completed by the addition of A nucleotides at the 3′ end of the encoded mRNA. Most of the stop codons of the genes in the G. orientalis mitogenome were identical to those in the Ha. sinicus and Am. aulica mitogenomes. Other types of stop codons are present in the mitogenomes of the subfamily Harpalinae, such as incomplete stop codons A, CTA, and TTA [16,17]. The diversity of the start and stop codons reflects the evolutionary diversity of species and makes it difficult to determine the start and stop positions of PCGs.
The results of the relative synonymous codon usage (RSCU) analysis for the 13 PCGs, comprising 3714 codons excluding the start and stop codons, showed codon usage bias in the G. orientalis mitogenome (Figure 3 and Supplementary Table S1). Among the amino acids (Supplementary Table S1), Leu was the predominant type (576), followed by Ile (381) and Phe (363). Among the codon usage counts, UUA (434) for Leu was dominant, followed by AUU (356) for Ile and UUU (323) for Phe. Thirteen PCGs had the biased usage of the A and T nucleotides (Supplementary Table S2).
Figure 3.
Relative synonymous codon usage (RSCU) of G. orientalis mitogenome.
3.3. Transfer and Ribosomal RNA Genes
The traditional 22 tRNA genes were interspersed among the PCGs. Fourteen of the 22 tRNAs were in the majority strand, and eight were in the minority strand (Figure 2 and Table 3). The lengths of the 22 tRNAs ranged from 61 bp (trnA-UGC) to 75 bp (trnQ-UUG and trnW-UCA) (Table 3 and Supplementary Table S2), which had a typical cloverleaf secondary structure (Figure 4). Most of the secondary structures of tRNAs in the G. orientalis mitogenome were consistent with those in the mitogenomes of Ha. sinicus and Ha. pensylvanicus. In the mitogenomes of the subfamily Harpalinae, trnS1-GCU lacked a DHU stem [3,16,17], which was replaced by a simple loop [3,16,17]. However, the trnS1-GCU of the G. orientalis mitogenome had a 2 bp DHU (Figure 4). The diversity of the secondary structures of tRNAs reflects the evolutionary diversity of the species. The length of the anticodon stems of the tRNAs ranged from 3 bp (trnT-UGU) to 8 bp (trnC-GCA) (Figure 4). The length of the DHU stem ranged from 2 bp (trnY-GUA and trnS1-GCU) to 5 bp (trnM-CAU and trnK-CUU) (Figure 4), most of which were 3–4 bp long. The length of the TΨC stem ranged from 3 bp (trnC-GCA and trnV-UAC) to 6 bp (trnL1-UAG) (Figure 4), most of which were 4–5 bp long. There are three types of mismatched base pairs for tRNA: U–U base pairs, A–G base pairs, and non-canonical G–U base pairs (Figure 4). The amino acid accepter stem of trnC-GCA has U–U base pairs (Figure 4), the amino acid accepter stem of trnW-UCA has A–G base pairs (Figure 4), and the anticodon stems of trnW-UCA and trnD-GUC and the TΨC stem of trnS1-GCU have G–U base pairs (Figure 4).
Figure 4.
Secondary structure of 22 tRNAs of G. orientalis mitogenome. The mismatched base pairs are presented in yellow; the matched base pairs are presented in green; the bases in the loop are presented in blue; and the nucleotide outlines of the anticodon are presented in red.
The rrnL and rrnS were 1312 and 786 bp long, respectively. The AU- and GC-skews’ values of rrnL and rrnS were −0.051, 0.305, −0.060, and 0.365, respectively (Supplementary Table S2). The secondary structures of both rrnL and rrnS (Supplementary Figures S1 and S2) contained many helices, similar to those of the Ha. sinicus and Am. aulica mitogenomes [3,16].
3.4. Control Region
The CR, also called the AT-rich region, is 1327 bp in length with an AT content of 88.62% and is located between the rrnS and trnI-GAU genes (Figure 2 and Supplementary Table S2). In the mitogenomes of the subfamily Harpalinae, the AT content of the CR of G. orientalis was slightly higher than that of Ha. sinicus, and lower than that of Ha. pensylvanicus. The AT- and GC-skews of CR were 0.005 and −0.125 on the majority strand, respectively, indicating a major-strand compositional bias characterized by a slight excess of A over T nucleotides, and a strong excess of C over G nucleotides. Bias is generally observed in the mitogenomes of members of the subfamily Harpalinae, such as Ha. pensylvanicus [17]. The secondary structure of the CR was inferred (Supplementary Figures S3) to contain more than 20 stem-loop structures. A conserved poly-T stretch (19 bp) and seven tandem repeats (TRs) were observed in the CR of the G. orientalis mitogenome (Figure 5 and Table S3). The total length of the TRs was 238 bp, contributing to 17.94% of the CR size. For TR3-TR6, these four TRs overlapped with each other (Figure 5 and Table S3). The region where TR3-TR6 was located, was the most enriched region of A + T, starting from 15,383 to 15,471, with a total length of 89 bp which was entirely composed of A and T bases; TR2 was a varied and typical microsatellite-like element (TA)24, with a left-flanking conserved polyT stretch (19 bp) (Figure 5).
Figure 5.
Organization of control region in G. orientalis mitogenome. The orange block is the tandem repeat region; the green block indicates the poly-T stretch.
3.5. Phylogenetic Analysis
This study’s phylogenetic analyses were based on the nucleotide sequences of the 13 PCGs obtained from 32 mitogenomes (Figure 6 and Figure 7). A total of 11,136 alignment positions were obtained using Gblock [38], from 11,556 alignment positions of the 13 PCGs (Supplementary Table S4). The BI tree provided significantly higher support values than the ML tree for the same dataset, particularly for branches that involved the subfamily Harpalinae relationships of the 13 PCG nucleotide sequences. In the ML tree, significantly low ML bootstrap support values (21 and 36) were observed for the subfamily Harpalinae, which is consistent with the result of a previous phylogenetic study [3]. In the BI and ML trees, nine genera of the subfamily Harpalinae were clustered as (((((((Pterostichus + Stomis) + Orthomus) + Amara) + Harpalus) + (Abax + Craspedophorus)) + Hexagonia) + Galerita) (Figure 6 and Figure 7), which was similar to the results of previous phylogenetic studies [1,3,5,12,16]. At the genus level, Galerita is an independent lineage in the topology of the BI and ML trees (Figure 6 and Figure 7). The genus Galerita is closely related to the genus Trichognathus in the tribe Galeritini [10,12]. The tribe Galeritini and Dryptini have sister relationships [10]. However, the mitogenomes are unknown. An accurate phylogenetic position within the genus Galerita requires additional mitogenome sequences. At the subfamily level of the BI and ML trees, the positions of Brachininae and Omophroninae were unstable. Nevertheless, this study provided a molecular basis for the classification and phylogeny of the family Carabidae, especially the subfamily Harpalinae.
Figure 6.
Bayesian-inferred phylogenetic tree of the nucleotide sequences of 13 protein-coding genes (PCGs) of 32 mitogenomes using MrBayes under the GTR + F + I + G4 model. Bayesian inference (BI) poster probability values are indicated near the nodes. The newly determined Galerita orientalis is shown in red. The branches of the family Carabidae are shown in red. L. saccharina and Co. magnifica are outgroups, and the corresponding branches are shown in green or purple.
Figure 7.
Maximum likelihood phylogenetic tree of nucleotide sequences of 13 protein-coding genes (PCGs) of 32 mitogenomes using IQ tree under the GTR + F + I + G4 model. Maximum likelihood (ML) bootstrap support values are indicated near the nodes. The newly determined Galerita orientalis is shown in red. The branches of the family Carabidae are shown in red. L. saccharina and Co. magnifica are outgroups, and the corresponding branches are shown in green or purple.
4. Conclusions
The genus Galerita contains more than 110 species [10], which is relatively disorganized due to simplistic morphological characteristics [9] and the absence of mitogenome molecular phylogenetic evidence. Therefore, we newly assembled the G. orientalis mitogenome in the present study. Compared to other previously reported mitogenomes of subfamily Harpalinae, all of them presented similar structural characters and nucleotide compositions, which contained 13 PCGs, 22 tRNA, 2 rRNA, and a control region. All 13 protein-coding genes were initiated using a typical ATN (Met) as the start codon, except for nad1, which has a TTG as the start codon, and were terminated using a typical TAN stop codon. The 22 tRNA could fold into a typical cloverleaf structure, including trnS1-NCU, which lacks the DHU arm in other mitogenomes of subfamily Harpalinae. Both rrnS and rrnL contained a lot of helices. A conserved poly-T stretch (19 bp) and seven TRs were observed in the CR. The phylogenetic analysis indicated that the genus Galerita was an independent lineage. Considering the diversity of the family Carabidae and the limitations of the current mitogenome information, the accurate phylogeny within the genus Galerita will require additional mitogenomes.
Supplementary Materials
The following supporting information can be downloaded at: www.mdpi.com/article/10.3390/genes13122199/s1, Table S1: Codon number and RSCU of PCGs of G. orientalis mitogenome; Table S2: Composition and skewness of genes and CR of G. orientalis mitogenome; Table S3: Tandem repeats of CR of G. orientalis mitogenome; Table S4: Alignment results using Gblocks; Figure S1: Predicted secondary structure of rrnL of G. orientalis mitogenome; Figure S2: Predicted secondary structure of rrnS of G. orientalis mitogenome; Figure S3: Predicted secondary structure of control region of G. orientalis mitogenome.
Author Contributions
Conceptualization, Y.B. and X.G.; methodology, Y.B. and X.G.; software, Y.B.; validation, Y.B.; formal analysis, Y.B., L.Y. and K.Y.; investigation, Y.B. and K.Y.; resources, X.G.; data curation, Y.B. and X.G.; writing—original draft preparation, Y.B. and X.G.; writing—review and editing, Y.B. and X.G.; visualization, Y.B. and L.Y.; supervision, Y.B.; project administration, X.G.; funding acquisition, Y.B. and X.G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable. This research does not involve ethical research. Insects are invertebrates, and there are no ethics involved in using them in experiments.
Data Availability Statement
The following information was supplied regarding the deposition of DNA sequences: The raw data can be obtained from the Sequence Read Archive at NCBI under accession number SRR20727398. The associated BioProject and Bio-Sample numbers are PRJNA864596 and SAMN30075025, respectively.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This research was funded by the Discipline and Master’s Site Construction Project of Guiyang University by Guiyang City Financial Support Guiyang University under Grant No. KJY-2020; the Foundational Research Fund of Guangxi Academy of Agricultural Sciences under Grant No. 2021YT067 and GuiNongKe2022YM04; the fund of the Guangxi Key Laboratory of Biology for Crop Diseases and Insect Pests under Grant No. 2020-KF-03; Guangxi Natural Science Foundation under Grant No. 2020GXNSFBA297162, and the Guizhou Fundamental Research Program (Natural Science Project) under Grant No. QianKeHeJiChu-ZK[2022]YiBan006.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ober K.A. Phylogenetic relationships of the carabid subfamily Harpalinae (Coleoptera) based on molecular sequence data. Mol. Phylogenet. Evol. 2002;24:228–248. doi: 10.1016/S1055-7903(02)00251-8. [DOI] [PubMed] [Google Scholar]
- 2.Bousquet Y., Larochelle A. Catalogue of the Geadephaga (Coleoptera: Trachypachidae, Rhysodidae, Carabidae including Cicindelini) of America north of Mexico. Mem. Entomol. Soc. Can. 1993;167:1–397. doi: 10.4039/entm125167fv. [DOI] [Google Scholar]
- 3.Li Z., Li X., Song N., Tang H., Yin X. The Mitochondrial Genome of Amara aulica (Coleoptera, Carabidae, Harpalinae) and Insights into the Phylogeny of Ground Beetles. Genes. 2020;11:181. doi: 10.3390/genes11020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lorenz W. Systematic List of Extant Ground Beetles of the World. W. Lorenz; Tutzing, Germany: 2005. [Google Scholar]
- 5.Ober K.A., Maddison D.R. Phylogenetic relationships of tribes within Harpalinae (Coleoptera: Carabidae) as inferred from 28S ribosomal DNA and the wingless gene. J. Insect Sci. 2008;8:63. doi: 10.1673/031.008.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hovorka O. A new Galerita species from Bolivia (Coleoptera: Carabidae: Galeritini) Stud. Rep. Taxon. Ser. 2012;8:131–134. [Google Scholar]
- 7.Hovorka O. New species of Galerita Fabricius, 1801 from Panama (Coleoptera: Carabidae: Galeritini) Stud. Rep. Taxon. Ser. 2016;12:89–92. [Google Scholar]
- 8.Hovorka O. Three new Galerita Fabricius, 1801 species (Coleoptera: Carabidae: Galeritini) Stud. Rep. Taxon. Ser. 2017;13:323–334. [Google Scholar]
- 9.Hovorka O. Five new species of Galerita from Asia and new distributional records (Coleoptera: Carabidae: Galeritini) Folia Heyrovskyana Ser. A. 2019;27:26–41. [Google Scholar]
- 10.Makarov K.V., Matalin A.V. The preimaginal stages of Galerita ruficollis Dejean, 1825 and the position of the tribe Galeritini in the classification of ground beetles (Coleoptera, Carabidae) Zookeys. 2021;1044:527–561. doi: 10.3897/zookeys.1044.63085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hunting W. Female reproductive system of the tribe Galeritini (Coleoptera: Carabidae): Structural features and evolution. Ann. Carnegie Mus. 2008;77:229–242, 214. doi: 10.2992/0097-4463-77.1.229. [DOI] [Google Scholar]
- 12.Ober K.A., Heider T.N. Phylogenetic diversification patterns and divergence times in ground beetles (Coleoptera: Carabidae: Harpalinae) BMC Evol. Biol. 2010;10:262. doi: 10.1186/1471-2148-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ober K.A. The Evolution of Arboreal Carabid Beetles. The University of Arizona; Tucson, AZ, USA: 2001. [Google Scholar]
- 14.Liu Y.-Y., Zhou Z.-C., Chen X.-S. Characterization of the Complete Mitochondrial Genome of Epicauta impressicornis (Coleoptera: Meloidae) and Its Phylogenetic Implications for the Infraorder Cucujiformia. J. Insect Sci. 2020;20:16. doi: 10.1093/jisesa/ieaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linard B., Arribas P., Andújar C., Crampton-Platt A., Vogler A.P. Lessons from genome skimming of arthropod-preserving ethanol. Mol. Ecol. Resour. 2016;16:1365–1377. doi: 10.1111/1755-0998.12539. [DOI] [PubMed] [Google Scholar]
- 16.Yu X., Tan W., Zhang H., Jiang W., Gao H., Wang W., Liu Y., Wang Y., Tian X. Characterization of the Complete Mitochondrial Genome of Harpalus sinicus and Its Implications for Phylogenetic Analyses. Genes. 2019;10:724. doi: 10.3390/genes10090724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieran T.J. Mitochondrial, metagenomic, and phylogenetic analysis of the ground beetle Harpalus pensylvanicus (Coleoptera: Carabidae) Gene. 2020;740:144540. doi: 10.1016/j.gene.2020.144540. [DOI] [PubMed] [Google Scholar]
- 18.Bai Y., Ye L., Yang K., Wang H. Genome Survey and SSR Analysis of Camellia nitidissima Chi (Theaceae) Genet. Res. 2022;2022:5417970. doi: 10.1155/2022/5417970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai Y., Gao X., Wang H., Ye L., Zhang X., Huang W., Long X., Yang K., Li G., Luo J., et al. Comparative mitogenome analysis reveals mitochondrial genome characteristics in eight strains of Beauveria. PeerJ. 2022;10:e14067. doi: 10.7717/peerj.14067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S., Zhou Y., Chen Y., Gu J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dierckxsens N., Mardulyn P., Smits G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016;45:e18. doi: 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perna N.T., Kocher T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995;41:353–358. doi: 10.1007/BF01215182. [DOI] [PubMed] [Google Scholar]
- 23.Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E.S., Fischer A., Bock R., Greiner S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan P.P., Lowe T.M. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. In: Kollmar M., editor. Gene Prediction: Methods and Protocols. Springer; New York, NY, USA: 2019. pp. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laslett D., Canbäck B. ARWEN: A program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 2007;24:172–175. doi: 10.1093/bioinformatics/btm573. [DOI] [PubMed] [Google Scholar]
- 26.Kent W.J. BLAT—The BLAST-like Alignment Tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stothard P., Wishart D.S. Circular genome visualization and exploration using CGView. Bioinformatics. 2004;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 28.Tamura K., Stecher G., Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerpedjiev P., Hammer S., Hofacker I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics. 2015;31:3377–3379. doi: 10.1093/bioinformatics/btv372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gendron P., Lemieux S., Major F. Quantitative analysis of nucleic acid three-dimensional structures11Edited by I. Tinoco. J. Mol. Biol. 2001;308:919–936. doi: 10.1006/jmbi.2001.4626. [DOI] [PubMed] [Google Scholar]
- 31.Lorenz R., Bernhart S.H., Höner zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathews D.H., Disney M.D., Childs J.L., Schroeder S.J., Zuker M., Turner D.H. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang D., Gao F., Jakovlić I., Zou H., Zhang J., Li W.X., Wang G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 36.Katoh K., Standley D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ranwez V., Douzery E.J.P., Cambon C., Chantret N., Delsuc F. MACSE v2: Toolkit for the Alignment of Coding Sequences Accounting for Frameshifts and Stop Codons. Mol. Biol. Evol. 2018;35:2582–2584. doi: 10.1093/molbev/msy159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talavera G., Castresana J. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 39.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen L.-T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2014;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I., Bork P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang A.R., Kim M.J., Jeong S.Y., Kim I. Complete mitochondrial genome sequence of Cicindela anchoralis Chevrolat, 1845 (Coleoptera: Carabidae) Mitochondrial DNA Part B. 2018;3:282–283. doi: 10.1080/23802359.2018.1443040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.López-López A., Vogler A.P. The mitogenome phylogeny of Adephaga (Coleoptera) Mol. Phylogenet. Evol. 2017;114:166–174. doi: 10.1016/j.ympev.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 45.Wang A.R., Kim M.J., Hong E.J., Jeong J.-C., Kim S.S., Kim I. Complete mitochondrial genome sequence of Acoptolabrus changeonleei Ishikawa et Kim, 1983 (Coleoptera: Carabidae) Mitochondrial DNA Part B. 2019;4:1883–1885. doi: 10.1080/23802359.2019.1613184. [DOI] [Google Scholar]
- 46.Liu N., Wang S., Yang X., Song J., Wu J., Fang J. The complete mitochondrial genome of Carabus (Damaster) lafossei (Coleoptera: Carabidae) Conserv. Genet. Resour. 2018;10:157–160. doi: 10.1007/s12686-017-0787-0. [DOI] [Google Scholar]
- 47.Song H., Sheffield N.C., Cameron S.L., Miller K.B., Whiting M.F. When phylogenetic assumptions are violated: Base compositional heterogeneity and among-site rate variation in beetle mitochondrial phylogenomics. Syst. Entomol. 2010;35:429–448. doi: 10.1111/j.1365-3113.2009.00517.x. [DOI] [Google Scholar]
- 48.Wan X., Hong M.Y., Liao A., Kim M.I., Kim K.-G., Han Y.S., Kim I. Complete mitochondrial genome of a carabid beetle, Damaster mirabilissimus mirabilissim (Coleoptera: Carabidae) Entomol. Res. 2012;42:44–54. doi: 10.1111/j.1748-5967.2011.00355.x. [DOI] [Google Scholar]
- 49.Timmermans M.J.T.N., Barton C., Haran J., Ahrens D., Culverwell C.L., Ollikainen A., Dodsworth S., Foster P.G., Bocak L., Vogler A.P. Family-Level Sampling of Mitochondrial Genomes in Coleoptera: Compositional Heterogeneity and Phylogenetics. Genome Biol. Evol. 2015;8:161–175. doi: 10.1093/gbe/evv241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raupach M.J., Deister F., Villastrigo A., Balke M. The complete mitochondrial genomes of Notiophilus quadripunctatus Dejean, 1826 and Omophron limbatum (Fabricius, 1777): New insights into the mitogenome phylogeny of the Carabidae (Insecta, Coleoptera) Insect Syst. Evol. 2022;53:242–263. doi: 10.1163/1876312X-bja10027. [DOI] [Google Scholar]
- 51.Kyndt E.C., Kyndt J.A. Illumina Short-Read Sequencing of the Mitogenomes of Novel Scarites subterraneus Isolates Allows for Taxonomic Refinement of the Genus Scarites Fabricius 1775, within the Carabidae Family. Insects. 2022;13:190. doi: 10.3390/insects13020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bai Y., Chen J., Li G., Wang H., Luo J., Li C. Complete mitochondrial genome of the common silverfish Lepisma saccharina (Insecta: Zygentoma: Lepismatidae) Mitochondrial DNA Part B. 2020;5:1552–1553. doi: 10.1080/23802359.2020.1742598. [DOI] [Google Scholar]
- 53.Bai Y., Yang K., Ye L., Gao X. Complete mitochondrial genome of Pseudoglomeris magnifica (Shelford, 1907) (Insecta: Dictyoptera: Blaberidae) Mitochondrial DNA Part B. 2022;7:1672–1675. doi: 10.1080/23802359.2022.2119823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding the deposition of DNA sequences: The raw data can be obtained from the Sequence Read Archive at NCBI under accession number SRR20727398. The associated BioProject and Bio-Sample numbers are PRJNA864596 and SAMN30075025, respectively.