Abstract
Diabetes represents one of the most significant, and rapidly escalating, global healthcare crises we face today. Diabetes already affects one-tenth of the world's adults—more than 537 million people, numbers that have tripled since 2000 and are estimated to reach 643 million by 2030. Type 2 diabetes (T2D), the most prevalent form, is a complex disease with numerous contributing factors, including genetics, epigenetics, diet, lifestyle, medication use, and socioeconomic factors. In addition, the gut microbiome has emerged as a significant potential contributing factor in T2D development and progression. Gut microbes and their metabolites strongly influence host metabolism and immune function, and are now known to contribute to vitamin biosynthesis, gut hormone production, satiety, maintenance of gut barrier integrity, and protection against pathogens, as well as digestion and nutrient absorption. In turn, gut microbes are influenced by diet and lifestyle factors such as alcohol and medication use, including antibiotic use and the consumption of probiotics and prebiotics. Here we review current evidence regarding changes in microbial populations in T2D and the mechanisms by which gut microbes influence glucose metabolism and insulin resistance, including inflammation, gut permeability, and bile acid production. We also explore the interrelationships between gut microbes and different T2D medications and other interventions, including prebiotics, probiotics, and bariatric surgery. Lastly, we explore the particular role of the small bowel in digestion and metabolism and the importance of studying small bowel microbes directly in our search to find metabolically relevant biomarkers and therapeutic targets for T2D.
Keywords: type 2 diabetes, insulin resistance, gut microbiome, small intestine, inflammation, gut permeability
Diabetes—Prevalence and Significance
The global prevalence of diabetes has reached crisis levels, affecting more than half a billion adults worldwide (537 million), or 1 in 10 of the adult population, in 2021 [1] (Fig. 1). These numbers have more than tripled since 2000 and, unless checked, are projected to rise to 643 million by 2030 and 783 million by 2045 [1]. Significantly, it is also estimated that almost 1 in 2 adults with diabetes (240 million) are undiagnosed and hence unaware of their condition and strategies for its management, and that a further 541 million adults have impaired glucose tolerance and thus have an increased risk for developing diabetes, specifically type 2 diabetes (T2D) [1]. In the United States, 37.3 million people are estimated to have diabetes (11.3% of the population), of whom 8.5 million are undiagnosed [2].
Figure 1.
Estimated age-adjusted prevalence of diabetes in adults aged 20 to 79 years in 2021. Reproduced with permission from: IDF Diabetes Atlas, 10th ed. Brussels, Belgium: International Diabetes Federation, 2021. http://www.diabetesatlas.org.
Pathophysiology of T2D
T2D accounts for approximately 90% of diabetes cases globally [1]. It can be precipitated by deficiencies in the production and release of insulin by pancreatic beta cells and in the response of muscle, adipose tissue, and/or liver cells to insulin [3–5]. A full analysis of T2D pathophysiology is beyond the scope of this review, but, as discussed in a recent review by Galicia-Garcia et al [6], the mechanisms underlying insulin production and release, and its detection and uptake by target tissues, are all tightly regulated, and defects in any of these may lead to metabolic imbalances and ultimately to the development of T2D [3, 6]. For example, impairments in any of the pathways involved in insulin binding can lead to reduced glucose uptake, resulting in higher circulating glucose levels (hyperglycemia) and placing increasing demands on insulin-producing pancreatic beta cells [3, 7] and, as noted above, impaired insulin detection and/or uptake by target tissues such as skeletal muscle, adipose tissue, and the liver can result in insulin resistance [3, 7]. Of note, this also leads to inflammation in the target tissues, with increases in circulating levels of proinflammatory cytokines [3, 7].
Risk Factors for T2D
While T2D is inextricably linked to obesity [which constitutes another escalating global crisis (8)], multiple other factors are also involved, including smoking, sleep quality, family history, hypertension, dyslipidema, and others [recently reviewed by Ismail et al (9)]. Tellingly, in the 2022 National Diabetes Statistics Report [2], the Centers for Disease Control and Prevention reports that the prevalence of diagnosed diabetes in US adults is highest in families with incomes below the federal poverty level (14.1%) and is also higher in American Indian and Alaska Native (14.5%), non-Hispanic Black (12.1%), Hispanic (11.8%), and Asian (9.5%) people when compared to non-Hispanic White people (7.4%) [2]. Rates are also higher in individuals with lower education levels, at 13.4% for those with less than a high school level vs 9.2% for high school and 7.1% for more than high school level educations [2]. In addition to these important societal and other (often interrelated) contributing factors such as diet, lifestyle, and medication use, data now indicate that changes in gut microbial populations may also contribute the development of T2D.
Alterations in the Salivary Microbiome in T2D
Most studies of the influences of the gut microbiome on T2D discussed in this review were performed using stool samples, as are the majority of gut microbiome studies. More recently, the salivary microbiome has also been shown to altered in T2D, including decreased abundances of Bifidobacterium and increased Streptococcus and Lactobacillus [10], decreased abundance of the genera Actinomyces and Atopobium (both phylum Actinobacteria [11]), and increased abundance of Blautia wexlerae, Lactobacillus fermentum, Nocardia coeliaca, and Selenomonas artemidis in treatment-naïve T2D subjects [12]. These findings indicate that T2D has effects on the whole host, including the entire alimentary tract, and that tracking changes in the salivary microbiome may provide another useful means to monitor the development and progression of T2D.
Alterations in the Gut Microbiome in T2D—Findings From Stool Studies
The human host relies on the approximately 3.8 × 1013 microorganisms that colonize various body sites and perform numerous essential functions in maintaining host health. The gut microbiome plays roles in vitamin and amino acid biosynthesis [13, 14], bile acid deconjugation and other transformations [15], immune responses and protection against pathogens [16], and many others [17]. This is made possible by the collective genomes of the bacteria, Eukarya [18], Archaea (primarily methanogens) [19], and viruses [20] that inhabit the gut, which together harbor over 150 times more genes than the human genome [21]. Although bacteria predominate, contributing an estimated 99.1% of genes in the gut, Archaea contribute around 0.8%, and the remaining 0.1% are contributed by Eukaryotes and viruses [22].
Gut microbes strongly influence host metabolism [23], breaking down otherwise indigestible complex carbohydrates via glycoside hydrolases and polysaccharide lyases that are not encoded in the human genome [24, 25], and regulating lipid accumulation, lipopolysaccharide content, and the production of short chain fatty acids (SCFAs), which influence many aspects of metabolism [26] including insulin signaling [27]. An early study of the gut microbiome in subjects with T2D found significant reductions in phylum Firmicutes, and specifically class Clostridia, when compared to controls, as well as increases in class Betaproteobacteria and genus Lactobacillus that correlated with increased plasma glucose levels [28]. A subsequent study by Karlsson et al found increased relative abundances of Lactobacillus gasseri and Streptococcus mutans, and decreased Roseburia intestinalis and Faecalibacterium prausnitzii in T2D subjects, as well as increased expression of microbial oxidative stress genes [29], which has been linked to insulin resistance, β-cell dysfunction, impaired glucose tolerance, and T2D [27, 30]. Moreover, there were similarities between these studies, performed using adult Danish males [28] and elderly Swedish females, [29] respectively, and a metagenome-wide association study of Chinese subjects, including increases in Lactobacillus species and increased microbial oxidative stress functions [31]. Since then, numerous studies have examined gut microbiota in human subjects with T2D. In a systematic review of the associated data, Gurung et al found that in addition to Roseburia and Faecalibacterium, the genera Bifidobacterium, Bacteroides, and Akkermansia were consistently found to be decreased in the gut microbiome in T2D subjects, whereas the genera Ruminococcus, Fusobacterium, and Blautia were consistently increased [32]. In addition, positive associations have been found between the genera Clostridium and Phascolarctobacterium and insulin sensitivity, and negative associations with genus Dialister [33], whereas fasting insulin correlated negatively with Phascolarctobacterium and positively with Dialister [33]. Taken together, these clinical data suggest that gut bacteria may have direct effects on the mechanisms underlying the development of T2D.
Role of Microbial Metabolites in T2D—Importance of Short Chain Fatty Acids
As noted earlier, a common alteration of the gut microbiome in T2D found in many studies is decreased abundance of R. intestinalis, F. prausnitzii, and other species that produce the SCFA butyrate [29, 31]. Butyrate and the other principal SCFAs acetate and propionate are important microbial metabolites produced via fermentation of dietary fibers, and together they have numerous effects on the host [34]. Acetate and propionate are substrates for lipogenesis and gluconeogenesis in the liver and peripheral tissues, whereas butyrate is an important energy substrate for the colonic epithelium [34] and has been shown in mouse studies to activate gluconeogenesis in the small bowel and colon [35]. Studies in mouse models have also revealed that SCFAs can modulate the secretion of key peptide hormones including glucagon-like peptide-1 (GLP-1), gastric inhibitory peptide (GIP), GLP-2, and peptide YY (PYY) in the gut, which can increase insulin sensitivity, decrease inflammation, and increase satiety [36–38]. Decreased abundance of butyrate producers has been linked to increased metabolic risks, whereas increased ingestion of dietary fibers has been specifically linked to enrichment of butyrate-producing bacteria in human T2D subjects [39] and has also been shown to improve glycemic control, decrease hyperinsulinemia, and lower plasma lipid concentrations [40]. Taken together, these data illustrate the importance of SCFAs, particularly butyrate, in the development of T2D.
Mechanisms by Which Gut Microbes Influence Glucose Metabolism and Insulin Resistance
Inflammation
Inflammation has emerged as a significant microbial mechanism underlying insulin resistance and the development of obesity and T2D [reviewed in (41, 42)]. The chronic low-grade inflammation associated with both obesity and T2D is driven by increased production of inflammatory cytokines including tumor necrosis factor (TNF)-α, the interleukins IL-1 and IL-6, and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) [42]. Alterations in gut microbial populations, often referred to as dysbiosis, lead to increasing bacterial encroachment on the gut epithelium, triggering an inflammatory response [43]. Bacterial components lipopolysaccharide (LPS) and flagellin are recognized by and bind to the toll-like receptors TLR4 and TLR5, respectively [44]. TLR4/5 binding results in expression of TNF-α, which has been linked to impaired insulin receptor signaling and insulin resistance [45]. Of note, the effects of TLR4 may be cell-type specific, as both TLR4 and its coreceptor CD14 are required to induce adipose tissue insulin resistance in mice [46–48], but knocking out TLR4 in intestinal epithelial cells impairs glucose metabolism and leads to the development of metabolic syndrome [49]. TNF-α in turn leads to NF-κB activation [50] [via c-Jun N-terminal kinases and IκB kinase (42)] and translocation to the nucleus, which propagates further inflammatory responses. IL-1 inhibits β-cell function, and IL-6 translocation to the liver results in the production of the inflammatory biomarker C-reactive protein, which is strongly associated with T2D and cardiometabolic health [51]. Of note, increases in LPS have been linked to decreases in Bifidobacterium spp, among others [52], whereas supplementing diets with specific probiotics including Lactobacillus, Bifidobacterium, Clostridium, and Akkermansia have been shown to reduce inflammatory responses and increase insulin sensitivity [53, 54], reinforcing the central role, and potential therapeutic implications, of gut microbes in T2D.
Increased Gut Permeability
An important factor contributing to the inflammation described here is increased gut permeability, which has been linked to consumption of a high-fat diet (HFD) [55]. The mucus lining the gut epithelium is essential for the barrier function of the gut (ie, preventing bacterial pathogens, viruses, and toxins crossing from the lumen in to the epithelium and potentially entering the circulatory or lymphatic systems) [56]. When this barrier fails, detection of LPS or flagellin on encroaching bacteria triggers these inflammatory responses. Mucin produced by goblet cells [57], antimicrobial peptides such as defensins and regenerating islet-derived 3-gamma produced by Paneth cells [58], GLP-2 produced by L-cells, as well as tight junctions between epithelial cells (composed of the proteins zonula occludens 1, occludin, and claudin) [59] are all important components of this defense system. Bacterial LPS directly increases tight junction permeability, in a TLR-dependent manner [59], whereas microbial SCFAs strengthen the intestinal epithelial barrier [26, 60]. Butyrate in particular has been shown to upregulate the expression of claudin-1 [61] and to alter the distribution of occludin and zonula occludens 1 [62, 63] in cell studies. SCFAs also increase mucin production by goblet cells [64], which is important as mucin turnover is critical for intestinal barrier integrity, and loss of the mucin-degrading bacteria Akkermansia muciniphila is consistently noted in T2D subjects. Conversely, restoring A. muciniphila levels normalized insulin resistance in obese mice [65]. Lastly, both the consumption of specific probiotic strains [66] and treatment with the prebiotic oligofructose (OFS) [67] appear to reduce gut permeability, underscoring that this is an important mechanism whereby gut microbiota and their metabolites influence T2D.
Bile Acids
Bile acids play key roles in regulating glucose homeostasis, and gut microbes are involved in modifying primary bile acids synthesized in the liver (cholic acid and chenodeoxycholic acid) into secondary bile acids (deoxycholate and lithocholate) [68]. Specifically, conjugated bile acids (bile salts) can be deconjugated by ileal microbes, particularly the genera Lactobacillus, Bifidobacterium, Enterobacter, Bacteroides, and Clostridium, which allows them to avoid being recycled back to the liver and secreted into the biliary system (enterohepatic circulation) and instead be further metabolized by colonic microbes into secondary bile acids [34]. Bile acids strongly influence glucose homeostasis—for example, cholic acid decreases triglyceride levels by reducing expression of the transcription factor sterol response element binding protein 1c, which is an important regulator of lipogenic gene expression [69], and feeding mice a HFD augmented with cholic acid increased energy expenditure and prevented insulin resistance [70]. The effects of primary bile acids are mediated through farnesoid X receptor, whereas the effects on secondary bile acids are mediated through Takeda G-protein-coupled receptor [34, 71, 72]. Knocking out the farnesoid X receptor gene in mice with genetic and diet-induced obesity decreased hyperglycemia and hyperinsulinemia and improved glucose tolerance [73], whereas Takeda G-protein-coupled receptor promotes GLP-1 secretion and thus glycemic regulation [74]. Interestingly, bile acid sequestrants have been tested as potential therapeutics and showed benefits in human trials [75–78]. For example, colesevelam improved glycemic control and reduced low-density lipoprotein (LDL) cholesterol in adult patients with poorly controlled T2D on sulfonylurea monotherapy or sulfonylurea in combination with additional oral antidiabetic agents [77], likely by increasing GLP-1 secretion [79], and colestimide treatment resulted in significantly decreased LDL cholesterol and increased A1C levels in Japanese subjects with T2D [75], again likely due to effects on GLP-1 [80]. Lasty, the beneficial effects of Roux-en-Y gastric bypass surgery have been suggested to be mediated via post-surgical increases in A. muciniphila and its effects on bile acids [81], and the negative effects of the persistent organic pollutants such as 2,3,7,8-tetrachlorodibenzofuran on hepatic lipogenesis, gluconeogenesis, and glycogenolysis are also associated with altered microbial bile acid metabolism [82].
T2D Medications and the Gut Microbiome
Biguanides
An important factor to consider in discussing the roles of the gut microbiome in T2D is the potential confounding effects of diabetes medications. The biguanide metformin suppresses hepatic gluconeogenesis via activation of adenosine monophosphate-activated protein kinase [83] and also enhances GLP-1 secretion [84]. It is the most commonly prescribed oral medication for T2D [85], and several human [86, 87] and mouse [65, 88] studies have shown that the beneficial effects of metformin are due, at least in part, to effects on gut microbes. Specifically, metformin increases the abundance of mucin-degrading A. muciniphila and SCFA-producing genera such as Megasphaera and Blautia [86, 87], resulting in increases in the beneficial effects of mucin and SCFAs on intestinal barrier integrity, bile acid metabolism, and glucose homeostasis described earlier. Metformin also increases the abundance of Bifidobacterium bifidum [87] and genus Lactobacillus [89] in human subjects, and it has been suggested that the increased abundance of Lactobacillus species previously identified in many studies of T2D subjects may in fact be due to confounding effects of metformin [90]. A variety of Bifidobacterium and Lactobacillus species are found in probiotic supplements [including yogurts (91)] with demonstrated antidiabetic effects [92, 93], and increases in these species may contribute to the improvements in glycemic profiles seen following metformin treatment [93–95]. Lastly, fecal transfers from metformin-treated human subjects to germ-free mice resulted in improved glucose tolerance [86]. These findings underscore both that gut microbiota play a key role in mediating metformin effects, and that this needs to be taken into consideration when assessing the effects of other potential therapeutics in T2D subjects taking metformin.
GLP-1 Receptor Agonists
While metformin is the preferred front-line therapy of the American Diabetes Association for treating T2D [96], GLP-1 receptor agonists (RAs), such as liraglutide, semaglutide, and dulaglutide, have been used for some time in the treatment of individuals with T2D [97], and more recently in particular in those with cardiovascular disease or risk factors [96]. GLP-1 RAs have also been used in the treatment of obesity (liraglutide, semaglutide) [98] and short bowel syndrome (exenatide) [99]. In addition to promoting insulin secretion, GLP-1 RAs inhibit pancreatic glucagon release, suppress appetite, and delay gastric emptying and also significantly affect gut microbial composition in both rodent [100–102] and human [103] studies. For example, liraglutide treatment significantly increased SCFA-producing taxa in a diabetic rat model [102], and enriched for genera including Allobaculum, Blautia, Desulfovibrio and Lactobacillus while reducing the abundance of orders Clostridiales (phylum Firmicutes) and Bacteroidales (phylum Bacteroidetes) in mice with induced hyperglycaemia [100]. Increased Akkermansia abundance has been demonstrated in human T2D subjects treated with liraglutide [103]. In an interesting study, Tsai et al [104] recently identified gut microbiome signatures that distinguished responders and nonresponders to GLP-1 RA treatment in T2D subjects. Specifically, responders had higher abundances of Bacteroides dorei and Lachnoclostridium species that correlated with glycemic reduction, whereas nonresponders had higher abundances of Prevotella copri (which can induce insulin resistance) and Mitsuokella multacida that negatively correlated with glycemic reduction [104].
Similar to GLP-1 RAs, GLP-2 RAs (eg, teduglutide) also reduce insulin sensitivity in HFD-fed mice [105] but are not typically used to treat human subjects with T2D. Interestingly, GLP-1/GLP-2 coagonists have been shown to have greater glycemic effects in a diabetic mouse model when compared to either liraglutide or teduglutide alone [106], and treatment with one of these, the longer-acting GUB09-145, has also been shown to affect the gut microbiome, with Lachnospiraceae and Clostridiales species specifically correlating with improvements in total cholesterol and glucose tolerance [107].
GLP-1R/GIPR dual agonists (“twincretins”) showed benefits over individual GLP-1 and GIP analogs in rodent models, nonhuman primates, and human subjects [108], and following several successful phase 3 clinical trials [reviewed in (109)], tirzepatide was approved by the Food and Drug Administration for the treatment of adults with T2D in May 2022 [110]. However, the effects of tirzepatide on the gut microbiome remain to be determined.
Sodium-Glucose Cotransporter-2 (SGLT2) Inhibitors
SGLT2 inhibitors (eg, canagliflozin, dapagliflozin, and empagliflozin) have also been approved by the Food and Drug Administration for the treatment of adults with T2D. These drugs block SGLT2 cotransporters in the renal tubes of the kidneys, thus inhibiting glucose reabsorption and reducing glycemia [111]. Few studies have been performed exploring the relationship between SGLT2 inhibitors and the microbiome. A study in diabetic mice showed that dapagliflozin treatment improved vascular function and reduced hyperglycemia, with some subtle concomitant changes in the gut microbiome including a reduced Firmicutes:Bacteroidetes ratio and a trend toward increased A. muciniphila, although the authors acknowledged that these changes were not definitively associated with the improved vascular function [112]. A human study in which subjects with T2D were treated with dapagliflozin or gliclazide for 12 weeks also found no significant effects of treatment on the gut microbiome [113]. In contrast, a study of diabetic rats treated with dapagliflozin found higher abundance of class Deltaproteobacteria and of family Desulfovibrionaceae, as well as a negative association between Desulfovibrionaceae and blood glucose levels [114]. Taken together, these data suggest that while SGLT2 inhibitors may have some effects on the gut microbiome, this may not be a significant mechanism of action for this class of medications.
Other T2D Medications
Other oral antidiabetic medications include sulfonylureas and meglitinides, both of which stimulate the release of insulin from pancreatic beta cells, and α-glucosidase inhibitors. No studies exploring the effects of meglitinides on the gut microbiome were found. The sulfonylureas Glipizide and Glebclamide appear to have minimal effects on the gut microbiome in T2D subjects [115] and a streptozotocin-induced diabetic rat model [116], respectively, but conclusive statements cannot be made without larger studies. α-glucosidase inhibitors, which include acarbose, voglibose, and miglitol, inhibit upper gastrointestinal enzymes, slowing the absorption of dietary carbohydrate and reducing post-prandial blood glucose concentrations [117]. α-glucosidase inhibitors also have anti-arthritic effects, and studies in a mouse model of collagen-induced arthritis found that both acarose and miglitol treatments affected the gut microbiome, lessening the reduction of Firmicutes that occurred after onset of arthritis, reducing the abundances of genera Lactobacillus, Anaeroplasma, Adlercreutzia, and RF39 and improving overall bacterial diversity and richness [118].
Bariatric Surgery
Bariatric surgeries were originally intended to induce weight loss by limiting the amount of food the stomach can hold, thus reducing calorie intake [119] via restriction and malabsorption. Bariatric surgeries, particularly Roux-en-Y gastric bypass and sleeve gastrectomy, have also shown benefits for subjects for subjects with T2D, including improved glucose metabolism [120, 121]. It has been suggested that these improvements are, in part, linked to post-surgical changes in the gut microbiome [122] and resulting changes in SCFA levels [119, 123]. Different studies have shown varying results regarding which microbial taxa are altered, which may be due to differences in the type of surgery performed (eg, Roux-en-Y gastric bypass vs sleeve gastrectomy) [124] and/or resulting changes in stomach pH [125]. However, consistent findings include increases in the abundance of Sutterella, which has been linked to glycemic control [120, 121], and of mucin-degrading A. muciniphila [81, 120, 121, 126]. As noted earlier, the beneficial effects of A. muciniphila are thought to result from its effects on bile acids [127, 128]. Consistent with this, a recent study showed that A. muciniphila abundance is decreased in T2D, including lean subjects with newly diagnosed T2D, and that its abundance was positively correlated with insulin secretion and levels of fibroblast growth factor 15/19 but inversely correlated with serum 3β-chenodeoxycholic acid levels [129]. Further, mice supplemented with A. muciniphila were protected against sucrose-induced impairment of glucose tolerance, and this protection was linked to increased insulin secretion, increased fibroblast growth factor 15/19 levels, and decreased 3β-chenodeoxycholic acid levels [129]. These findings underscore the extent of the influences of gut microbes on host metabolism and physiology, as well as the importance of understanding these influences when studying T2D.
Microbiome-Based Interventions and T2D
Prebiotics
In addition to the medications described previously, a variety of prebiotic and probiotic supplements have been used to treat T2D. Prebiotics promote the growth or action of beneficial microbes [130] and include fructooligosaccharides or galactooligosaccharides, lactulose, and nondigestible carbohydrates (eg, inulin, cellulose, and pectin) [131]. Both inulin and the inulin-like fructan OFS have shown promise for ameliorating T2D phenotypes. Dietary supplementation with OFS promotes satiety [132] and has been associated with weight loss and improved glucose regulation in obese and overweight subjects [133], and also prevents weight gain and fat mass accumulation in HFD-fed animals [134]. These effects appear to be mediated by decreases in ghrelin levels and increases in PYY [133] and GLP-1 [134] and have also been linked to reductions in gut permeability [67], decreases in proinflammatory cytokines, and increases in Bifidobacterium [52]. Similarly, inulin supplementation protects against HFD-induced obesity, with decreases in food intake, adiposity, liver triglycerides, and leptin levels and also improves glucose tolerance [135], effects that were associated with increased L-cell density and PYY levels [135]. Inulin supplementation can also reduce insulin resistance in diabetic mice, as can dietary supplementation with SCFAs [136] Lastly, HFDs are associated with increased microbial encroachment on the gut epithelium, due to loss of enterocyte proliferation. Inulin supplementation prevents microbial encroachment and restores epithelial health [137], due to increased production of IL-22, which pays a key role in promoting enterocyte proliferation. It is important to note that inulin may have significant gastrointestinal side effects that may mitigate use in some patients.
Probiotics
Probiotics are live microorganisms, predominantly commensal gut bacteria such as Lactobacillus and Bifidobacterium, which maintain gut homeostasis and regulate the metabolic activities of other gut microbes [138]. Numerous studies have now evaluated the efficacy of a variety of probiotic strains or strain combinations in lowering glucose in T2D, many of which were compared in a recent systematic review and meta-analysis by Rittiphairoj et al [139]. This meta-analysis included 26 trials across the globe that investigated different combinations of Lactobacillus species and strains, Bifidobacterium species and strains, and Streptococcus thermophilus, delivered in a wide range of formats including yogurt, kimchi, fermented milk, bread, honey, powders, capsules, and tablets, at doses ranging from 106 to 1019 CFU daily. Despite this variability, the authors found that, overall, probiotics successfully reduced fasting blood glucose and serum cholesterol in T2D subjects and, where reported, also appeared to reduce HbA1c [139]. These effects were most pronounced in subjects who were not already on insulin therapy [139], findings that echo those of previous reviews [140, 141]. In a study performed in diabetic db/db mice, administration of a probiotic containing 10 different Lactobacillus strains and 4 yeast strains derived from camel's milk resulted in decreased fasting blood glucose, glucose area under the curve and HbA1c, decreased total cholesterol, triglycerides and LDL concentrations, and increased secretion of GLP-1 and insulin, due to upregulation of G protein-coupled receptor 43/41 [142], again suggesting that probiotics can yield significant benefits in improving T2D phenotypes. Mechanistically, the consumption of specific probiotic strains has been shown to help regulate gut membrane integrity and permeability, and thus prevent endotoxemia and the associated inflammation [66] and has independently been shown to decrease levels of pro-inflammatory cytokines while increasing anti-inflammatory cytokines [143]. Consistent with this, mucin production and intestinal barrier function were also improved in the previously described study of db/db mice administered the camel's milk probiotic, which the authors attributed to increases in the levels of the SCFAs propionate and butyrate, which in turn were likely due to increased abundances of the SCFA-producing genera Lactobacillus and Bifidobacterium, as well as C. leptum and Roseburia [142]. Lastly, Levels of claudin-1 and mucin-2 were also upregulated in these mice, and circulating levels of bacterial LPS were decreased, further supporting a role for these probiotics in promoting gut membrane integrity and preventing bacterial encroachment on the gut epithelium [142]. Of note, while these findings may be promising, the authors caution against the use of probiotics as a surrogate for standard care and note that the desiccation and shelf-handling of many brands may diminish their usefulness.
Relevance of the Small Bowel in T2D
While studies of both the stool and salivary microbiomes have yielded significant insights and are important particularly in the identification of potential microbial markers of disease development and progression, the small bowel is central to digestion, nutrient absorption, and endocrine regulation [144], and our group has shown that its microbial populations differ significantly from those in stool [145]. Typically 3 to 5 meters in length in an adult and with an absorptive surface area of around 30 m2 [146], the small intestine is divided into the duodenum, jejunum, and ileum [147]. The duodenum, which is the meeting point for chyme from the stomach, enzymes from the pancreas, and bile from the liver, is where most digestion occurs and absorption begins [147], after which most absorption occurs in the jejunum, and residual nutrients, vitamin B12, and bile acids are absorbed in the ileum [147]. The surface area available for absorption is increased by the presence of numerous villi that project into the gut lumen, with invaginated crypts at the base of the villi [148–150] (see Fig. 2). Although comprised of a single layer of intestinal epithelial cells, differentiated cell types within villi and crypts perform specialized functions [148–150]. Columnar enterocytes (which cover the villus tips) absorb nutrients and secrete immunoglobulins, goblet cells secrete the mucus which lines and protects the gut epithelium [57], tuft cells are thought to sense luminal contents [148], and Paneth cells (located primarily at crypt bases) secrete antimicrobial peptides such as defensins [58] (see Fig. 2). In addition, a variety of different enteroendocrine cells secrete hormones and peptides [57], including K cells that secrete the incretin GIP and L cells that secrete the incretin GLP-1 [151]; the peptide hormone GLP-2, which induces gut epithelial cell growth and proliferation and maintains mucosal integrity and gut barrier function [152, 153]; and PYY, which regulates satiety [57], among many others (see Fig. 2). Incretins stimulate insulin release, and GLP-1 receptor agonists are widely used in T2D treatment [151, 154]. Of note, SCFAs (produced by gut microbes) modulate the secretion of GLP-1, GLP-2, and PYY by L-cells, as well as the secretion of GIP by K-cells [36, 37, 155]. Taken together, these findings underscore the central importance of the small intestine in metabolism and the need to study its microbial populations directly in developing novel, targeted therapies and interventions for T2D (discussed further next).
Figure 2.
Schematic showing specialized cell types and their products in the small bowel epithelium.
The Importance of Studying the Small Bowel Microbiome
Despite the importance of the small intestine in metabolism and the likely relevance of its microbial populations to metabolic conditions, including T2D, the majority of gut microbiome studies are still performed using stool samples. As recently noted in a review by Kastl et al, few studies have analyzed the small intestinal microbiome, which is less accessible and requires more invasive sampling procedures [156]. Studies that have been performed in human subjects have utilized techniques such as esophagoduodenogastroscopy and nasoduodenal catheters or have obtained ileal samples during procedures such as colonoscopy, intestinal resection, or small bowel transplantation or from ileostomy effluents [156–161]. While these have provided tremendously valuable insights, as Kastl et al correctly noted [156], EGD and nasoduodenal sampling are prone to sample contamination with microbes from the upper gastrointestinal tract, and colonoscopy and ileostomy samples are prone to contamination with microbes from the lower gastrointestinal tract and skin, respectively. Moreover, while these approaches have been used to study small intestinal microbes in subjects with inflammatory bowel disease [162, 163], small intestinal bacterial overgrowth, short bowel syndrome [164], and irritable bowel syndrome, and to characterize small intestinal microbial populations and their metabolites [165, 166], to date, small bowel microbial populations have not been studied in subjects with metabolic conditions such as T2D.
To explore and characterize the microbial populations of the human small bowel and their roles in health and disease, we created the REIMAGINE (Revealing the Entire Intestinal Microbiota and its Associations with the Genetic, Immunologic, and Neuroendocrine Ecosystem) study [167]. We developed a new catheter design to overcome cross-contamination with microbes from the mouth and stomach, which plagued traditional open aspiration catheters [168, 169], and as small intestinal samples have high viscosity, low microbial biomass, and small sample volumes, we also developed improved techniques for sample processing, DNA recovery and library preparation prior to sequencing, using duodenal samples [167]. The results indicated that the number of microbes in the small bowel was significantly higher than previously thought and also revealed expanded microbiome-related pathways [167], suggesting a greater range of microbial functions than previously thought.
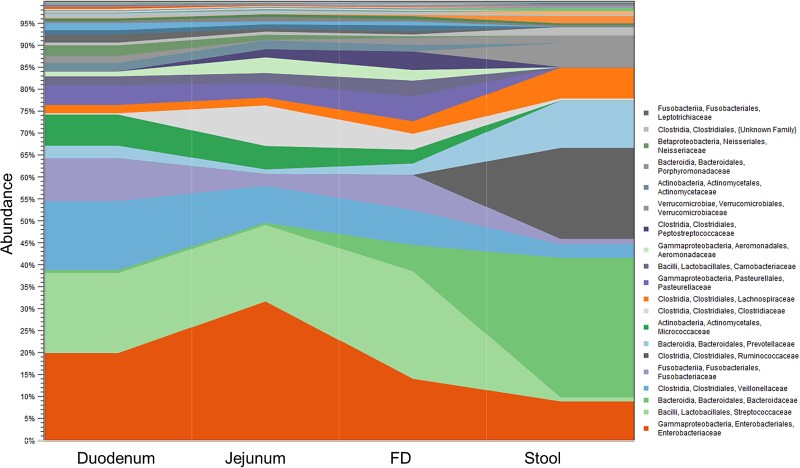
Using these validated techniques, we then explored the microbial populations of the small bowel (duodenum) and compared them to stool. We found significant differences between their respective microbiomes, even at the phylum level. The stool microbiome is characterized by high relative abundance of Bacteroidetes and Firmicutes, but in the small bowel Firmicutes and Proteobacteria are the most abundant, and Bacteroidetes is far less abundant [145]. Moreover, in the duodenum, phylum Firmicutes was primarily comprised of lactic acid bacteria, particularly families Streptococcaceae, Lactobacillaceae and Carnobacteriaceae, and family Veillonellaceae, whereas in stool, Firmicutes was dominated by families Ruminococcaceae, Lachnospiraceae, and Christensenellaceae (which is completely absent from the duodenum) [145] (see Fig. 3). There were also differences in Proteobacteria composition, with families Neisseriaceae, Pasteurellaceae, and Enterobacteriaceae predominating in the duodenum and class Deltaproteobacteria predominating in stool [145] (see Fig. 3). Clearly, the Firmicutes:Bacteroidetes ratio that is much quoted as being relevant to obesity and T2D has little relevance for the small bowel, illustrating the importance of studying the small bowel directly to find metabolically relevant biomarkers and therapeutic targets.
Figure 3.
Relative abundance of the major families in the duodenum, jejunum, furthest distance reached using endoscopy (FD), and stool, in the same subjects.
Since these first studies, we have utilized these validated techniques to explore the roles and contributions of changes in small bowel microbial populations in a variety of conditions, including small intestinal bacterial overgrowth [170], use of proton pump inhibitors [171], age and the ageing process [172], menopause and the use of hormone therapy [173], and smoking [174]. These studies have consistently demonstrated that overabundance of Proteobacterial taxa, particularly family Enterobacteriaceae and the disruptor genera [175] Escherichia-Shigella and Klebsiella, are associated with loss of microbial diversity and negative effects on the duodenal microbiome, as are overgrowth of family Lactobacillaceae and the genus Lactobacillus, which is another disruptor in the small bowel [175]. In contrast, families Prevotellaceae, Neisseriaceae, and Porphyromonadaceae are consistently associated with increased duodenal microbial diversity [173, 174], suggesting that they have positive effects in the small bowel. Our next steps will include applying these novel techniques to explore the roles of small bowel microbes and their metabolites in metabolic conditions such as obesity and T2D, which will hopefully allow us to identify novel, and successful, microbiome-based therapeutic targets.
Clinical Perspectives and Future Directions
The impact of the gut microbiome on human disease is undeniable. Current research is exploring the specific mechanisms this microcosm has on the human host in a variety of conditions. Diabetes is a complex and multifactorial disease. The role of the microbiome is likely variable depending on other host conditions and genetics. Having stated this, there is most probably a subgroup of patients with diabetes (and prediabetes) who have a more defined relationship with metabolic derangements and the gut microbiome. Future areas of research should involve identifying this particular subgroup of individuals and diving deep into the specifics of their microbial fingerprints. Perhaps a personalized approach to treating the microbiome will result. In addition, it is vital to recognize that these organisms live in a community and depend on each other and the collective environment. These communities and environments within the gut are also distinct and need to be treated as such. For example, the microbiome of the small bowel is distinct from that of stool, and using stool as a surrogate for analysis may lead to issues with interpretation. Future research needs to be able to manipulate, with specificity, the organisms in question, without perturbations in the environment as a whole. The understanding of the microbiome and its influence on human health is really in its early stages, and, at present, one must be cautious about applying interventions such as probiotics, which claim to influence disease and outcomes. Finally, it is vital to recognize the human as a whole and understand that manipulations of any factor in one direction can have implications in another. We need to learn more before advocating for microbial-based disease interventions.
Contributor Information
Gillian M Barlow, Medically Associated Science and Technology (MAST) Program, Cedars-Sinai, Los Angeles, CA, USA.
Ruchi Mathur, Email: ruchi.mathur@cshs.org, Medically Associated Science and Technology (MAST) Program, Cedars-Sinai, Los Angeles, CA, USA; Division of Endocrinology, Diabetes, and Metabolism, Department of Medicine, Cedars-Sinai, Los Angeles, CA, USA.
Conflicts of Interest
The authors declare that they have no relevant conflicts of interest.
Data Availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
- 1. IDF Diabetes Atlas, 10th ed. International Diabetes Federation; 2021. [Google Scholar]
- 2. National Diabetes Statistics Report, 2022. Centers for Disease Control and Prevention, US Department of Health and Human Services; 2022. [Google Scholar]
- 3. American Diabetes Association . 2. Classification and diagnosis of diabetes: standards of medical care in diabetes-2020. Diabetes Care. 2020;43(Suppl 1):S14–S31. [DOI] [PubMed] [Google Scholar]
- 4. Abdul-Ghani MA, DeFronzo RA. Pathogenesis of insulin resistance in skeletal muscle. J Biomed Biotechnol. 2010;2010:476279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Petersen KF, Shulman GI. Pathogenesis of skeletal muscle insulin resistance in type 2 diabetes mellitus. Am J Cardiol. 2002;90(5):11G–18G. [DOI] [PubMed] [Google Scholar]
- 6. Galicia-Garcia U, Benito-Vicente A, Jebari S, et al. Pathophysiology of type 2 diabetes mellitus. Int J Mol Sci. 2020;21(17):6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanches JM, Zhao LN, Salehi A, Wollheim CB, Kaldis P. Pathophysiology of type 2 diabetes and the impact of altered metabolic interorgan crosstalk. FEBS J. 2021. Published online November 30, 2021. [DOI] [PubMed] [Google Scholar]
- 8. Fact sheet—obesity and overweight. World Health Organization; 2020. [Google Scholar]
- 9. Ismail L, Materwala H, Al Kaabi J. Association of risk factors with type 2 diabetes: a systematic review. Comput Struct Biotechnol J. 2021;19:1759–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kampoo K, Teanpaisan R, Ledder RG, McBain AJ. Oral bacterial communities in individuals with type 2 diabetes who live in southern Thailand. Appl Environ Microbiol. 2014;80(2):662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Long J, Cai Q, Steinwandel M, et al. Association of oral microbiome with type 2 diabetes risk. J Periodontal Res. 2017;52(3):636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang Y, Liu S, Wang Y, et al. Changes of saliva microbiota in the onset and after the treatment of diabetes in patients with periodontitis. Aging (Albany NY). 2020;12(13):13090–13114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uebanso T, Shimohata T, Mawatari K, Takahashi A. Functional roles of B-vitamins in the gut and gut microbiome. Mol Nutr Food Res. 2020;64(18):e2000426. [DOI] [PubMed] [Google Scholar]
- 14. Waterhouse M, Hope B, Krause L, et al. Vitamin D and the gut microbiome: a systematic review of in vivo studies. Eur J Nutr. 2019;58(7):2895–2910. [DOI] [PubMed] [Google Scholar]
- 15. Poland JC, Flynn CR. Bile acids, their receptors, and the gut microbiota. Physiology (Bethesda). 2021;36(4):235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wiertsema SP, van Bergenhenegouwen J, Garssen J, Knippels LMJ. The interplay between the gut microbiome and the immune system in the context of infectious diseases throughout life and the role of nutrition in optimizing treatment strategies. Nutrients. 2021;13(3):886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Singer-Englar T, Barlow G, Mathur R. Obesity, diabetes, and the gut microbiome: an updated review. Expert Rev Gastroenterol Hepatol. 2019;13(1):3–15. [DOI] [PubMed] [Google Scholar]
- 18. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gaci N, Borrel G, Tottey W, O'Toole PW, Brugere JF. Archaea and the human gut: new beginning of an old story. World J Gastroenterol. 2014;20(43):16062–16078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shkoporov AN, Clooney AG, Sutton Thomas DS, et al. The human gut virome is highly diverse, stable, and individual specific. Cell Host Microbe. 2019;26(4):527–541.e525. [DOI] [PubMed] [Google Scholar]
- 21. Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fan Y, Pedersen O. Gut microbiota in human metabolic health and disease. Nat Rev Microbiol. 2021;19(1):55–71. [DOI] [PubMed] [Google Scholar]
- 24. Han H, Li Y, Fang J, et al. Gut microbiota and type 1 diabetes. Int J Mol Sci. 2018;19(4):995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ley RE, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blaak EE, Canfora EE, Theis S, et al. Short chain fatty acids in human gut and metabolic health. Benef Microbes. 2020;11(5):411–455. [DOI] [PubMed] [Google Scholar]
- 27. Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut. 2014;63(9):1513–1521. [DOI] [PubMed] [Google Scholar]
- 28. Larsen N, Vogensen FK, van den Berg Frans WJ, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5(2):e9085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karlsson FH, Tremaroli V, Nookaew I, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. [DOI] [PubMed] [Google Scholar]
- 30. Wright E, Jr., Scism-Bacon JL, Glass LC. Oxidative stress in type 2 diabetes: the role of fasting and postprandial glycaemia. Int J Clin Pract. 2006;60(3):308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. [DOI] [PubMed] [Google Scholar]
- 32. Gurung M, Li Z, You H, et al. Role of gut microbiota in type 2 diabetes pathophysiology. EBioMedicine. 2020;51:102590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Naderpoor N, Mousa A, Gomez-Arango L, et al. Faecal microbiota are related to insulin sensitivity and secretion in overweight or obese adults. J Clin Med. 2019;8(4):452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489(7415):242–249. [DOI] [PubMed] [Google Scholar]
- 35. De Vadder F, Kovatcheva-Datchary P, Goncalves D, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156(1-2):84–96. [DOI] [PubMed] [Google Scholar]
- 36. Tolhurst G, Heffron H, Lam YS, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61(2):364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin HV, Frassetto A, Kowalik EJ, et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One. 2012;7(4):e35240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Samuel BS, Shaito A, Motoike T, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A. 2008;105(43):16767–16772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao L, Zhang F, Ding X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359(6380):1151–1156. [DOI] [PubMed] [Google Scholar]
- 40. Chandalia M, Garg A, Lutjohann D, et al. Beneficial effects of high dietary fiber intake in patients with type 2 diabetes mellitus. N Engl J Med. 2000;342(19):1392–1398. [DOI] [PubMed] [Google Scholar]
- 41. Scheithauer TPM, Rampanelli E, Nieuwdorp M, et al. Gut microbiota as a trigger for metabolic inflammation in obesity and type 2 diabetes. Front Immunol. 2020;11:571731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sikalidis AK, Maykish A. The gut microbiome and type 2 diabetes mellitus: discussing a complex relationship. Biomedicines. 2020;8(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chassaing B, Raja SM, Lewis JD, Srinivasan S, Gewirtz AT. Colonic microbiota encroachment correlates with dysglycemia in humans. Cell Mol Gastroenterol Hepatol. 2017;4(2):205–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ringel Y. The gut microbiome in irritable bowel syndrome and other functional bowel disorders. Gastroenterol Clin North Am. 2017;46(1):91–101. [DOI] [PubMed] [Google Scholar]
- 45. Moller DE. Potential role of TNF-alpha in the pathogenesis of insulin resistance and type 2 diabetes. Trends Endocrinol Metab. 2000;11(6):212–217. [DOI] [PubMed] [Google Scholar]
- 46. Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–1772. [DOI] [PubMed] [Google Scholar]
- 47. Saberi M, Woods N-B, de Luca C, et al. Hematopoietic cell-specific deletion of toll-like receptor 4 ameliorates hepatic and adipose tissue insulin resistance in high-fat-fed mice. Cell Metab. 2009;10(5):419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Poggi M, Bastelica D, Gual P, et al. C3h/HeJ mice carrying a toll-like receptor 4 mutation are protected against the development of insulin resistance in white adipose tissue in response to a high-fat diet. Diabetologia. 2007;50(6):1267–1276. [DOI] [PubMed] [Google Scholar]
- 49. Lu P, Sodhi CP, Yamaguchi Y, et al. Intestinal epithelial toll-like receptor 4 prevents metabolic syndrome by regulating interactions between microbes and intestinal epithelial cells in mice. Mucosal Immunol. 2018;11(3):727–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ding S, Chi MM, Scull BP, et al. High-fat diet: bacteria interactions promote intestinal inflammation which precedes and correlates with obesity and insulin resistance in mouse. PLoS One. 2010;5(8):e12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kreiner FF, Kraaijenhof JM, von Herrath M, Hovingh GKK, von Scholten BJ. Interleukin 6 in diabetes, chronic kidney disease, and cardiovascular disease: mechanisms and therapeutic perspectives. Expert Rev Clin Immunol. 2022;18(4):377–389. [DOI] [PubMed] [Google Scholar]
- 52. Cani PD, Neyrinck AM, Fava F, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50(11):2374–2383. [DOI] [PubMed] [Google Scholar]
- 53. Rad AH, Abbasalizadeh S, Vazifekhah S, et al. The future of diabetes management by healthy probiotic microorganisms. Curr Diabetes Rev. 2017;13(6):582–589. [DOI] [PubMed] [Google Scholar]
- 54. Salles BIM, Cioffi D, Ferreira SRG. Probiotics supplementation and insulin resistance: a systematic review. Diabetol Metab Syndr. 2020;12(1):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Brun P, Castagliuolo I, Di Le V, et al. Increased intestinal permeability in obese mice: new evidence in the pathogenesis of nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2007;292(2):G518–G525. [DOI] [PubMed] [Google Scholar]
- 56. Hartmann P, Chen P, Wang HJ, et al. Deficiency of intestinal mucin-2 ameliorates experimental alcoholic liver disease in mice. Hepatology. 2013;58(1):108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Noah TK, Donahue B, Shroyer NF. Intestinal development and differentiation. Exp Cell Res. 2011;317(19):2702–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lueschow SR, McElroy SJ. The Paneth cell: the curator and defender of the immature small intestine. Front Immunol. 2020;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guo S, Al-Sadi R, Said HM, Ma TY. Lipopolysaccharide causes an increase in intestinal tight junction permeability in vitro and in vivo by inducing enterocyte membrane expression and localization of TLR-4 and CD14. Am J Pathol. 2013;182(2):375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hansen TH, Gobel RJ, Hansen T, Pedersen O. The gut microbiome in cardio-metabolic health. Genome Med. 2015;7(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang HB, Wang PY, Wang X, Wan YL, Liu YC. Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein Claudin-1 transcription. Dig Dis Sci. 2012;57(12):3126–3135. [DOI] [PubMed] [Google Scholar]
- 62. Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139(9):1619–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Miao W, Wu X, Wang K, et al. Sodium butyrate promotes reassembly of tight junctions in Caco-2 monolayers involving inhibition of MLCK/MLC2 pathway and phosphorylation of PKCbeta2. Int J Mol Sci. 2016;17(10):1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Willemsen LE, Koetsier MA, van Deventer SJ, van Tol EA. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut. 2003;52(10):1442–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Everard A, Belzer C, Geurts L, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci. 2013;110(22):9066–9071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. de Oliveira GLV, Leite AZ, Higuchi BS, Gonzaga MI, Mariano VS. Intestinal dysbiosis and probiotic applications in autoimmune diseases. Immunology. 2017;152(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Muccioli GG, Naslain D, Bäckhed F, et al. The endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol. 2010;6(1):392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile acids and the gut microbiome. Curr Opin Gastroenterol. 2014;30(3):332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Watanabe M, Houten SM, Wang L, et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113(10):1408–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Watanabe M, Houten SM, Mataki C, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439(7075):484–489. [DOI] [PubMed] [Google Scholar]
- 71. Kawamata Y, Fujii R, Hosoya M, et al. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278(11):9435–9440. [DOI] [PubMed] [Google Scholar]
- 72. Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102(6):731–744. [DOI] [PubMed] [Google Scholar]
- 73. Prawitt J, Abdelkarim M, Stroeve JHM, et al. Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes. 2011;60(7):1861–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Thomas C, Gioiello A, Noriega L, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10(3):167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Suzuki T, Oba K, Igari Y, et al. Effects of bile-acid-binding resin (colestimide) on blood glucose and visceral fat in Japanese patients with type 2 diabetes mellitus and hypercholesterolemia: an open-label, randomized, case-control, crossover study. J Diabetes Complications. 2012;26(1):34–39. [DOI] [PubMed] [Google Scholar]
- 76. Yamakawa T, Takano T, Utsunomiya H, Kadonosono K, Okamura A. Effect of colestimide therapy for glycemic control in type 2 diabetes mellitus with hypercholesterolemia. Endocr J. 2007;54(1):53–58. [DOI] [PubMed] [Google Scholar]
- 77. Fonseca VA, Rosenstock J, Wang AC, Truitt KE, Jones MR. Colesevelam HCl improves glycemic control and reduces LDL cholesterol in patients with inadequately controlled type 2 diabetes on sulfonylurea-based therapy. Diabetes Care. 2008;31(8):1479–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rosenstock J, Truitt K, Baz-Hecht M, Ford D, Tao B, Chou H. Efficacy and safety of colesevelam in combination with pioglitazone in patients with type 2 diabetes mellitus. Horm Metab Res. 2014;46(13):943–949. [DOI] [PubMed] [Google Scholar]
- 79. Shang Q, Saumoy M, Holst JJ, Salen G, Xu G. Colesevelam improves insulin resistance in a diet-induced obesity (F-DIO) rat model by increasing the release of GLP-1. Am J Physiol Gastrointest Liver Physiol. 2010;298(3):G419–G424. [DOI] [PubMed] [Google Scholar]
- 80. Suzuki T, Oba K, Igari Y, et al. Colestimide lowers plasma glucose levels and increases plasma glucagon-like PEPTIDE-1 (7-36) levels in patients with type 2 diabetes mellitus complicated by hypercholesterolemia. J Nippon Med Sch. 2007;74(5):338–343. [DOI] [PubMed] [Google Scholar]
- 81. Palmisano S, Campisciano G, Silvestri M, et al. Changes in gut microbiota composition after bariatric surgery: a new balance to decode. J Gastrointest Surg. 2020;24(8):1736–1746. [DOI] [PubMed] [Google Scholar]
- 82. Zhang L, Nichols RG, Correll J, et al. Persistent organic pollutants modify gut microbiota-host metabolic homeostasis in mice through aryl hydrocarbon receptor activation. Environ Health Perspect. 2015;123(7):679–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. An H, He L. Current understanding of metformin effect on the control of hyperglycemia in diabetes. J Endocrinol. 2016;228(3):R97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Maida A, Lamont BJ, Cao X, Drucker DJ. Metformin regulates the incretin receptor axis via a pathway dependent on peroxisome proliferator-activated receptor-alpha in mice. Diabetologia. 2011;54(2):339–349. [DOI] [PubMed] [Google Scholar]
- 85. Wu T, Horowitz M, Rayner CK. New insights into the anti-diabetic actions of metformin: from the liver to the gut. Expert Rev Gastroenterol Hepatol. 2017;11(2):157–166. [DOI] [PubMed] [Google Scholar]
- 86. Wu H, Esteve E, Tremaroli V, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23(7):850–858. [DOI] [PubMed] [Google Scholar]
- 87. de la Cuesta-Zuluaga J, Mueller NT, Corrales-Agudelo V, et al. Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care. 2017;40(1):54–62. [DOI] [PubMed] [Google Scholar]
- 88. Derrien M, Van Baarlen P, Hooiveld G, Norin E, Müller M, de Vos WM, . Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol. 2011;2:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rodriguez J, Hiel S, Delzenne NM. Metformin: old friend, new ways of action-implication of the gut microbiome? Curr Opin Clin Nutr Metab Care. 2018;21(4):294–301. [DOI] [PubMed] [Google Scholar]
- 90. Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ejtahed HS, Mohtadi-Nia J, Homayouni-Rad A, et al. Probiotic yogurt improves antioxidant status in type 2 diabetic patients. Nutrition. 2012;28(5):539–543. [DOI] [PubMed] [Google Scholar]
- 92. Kesika P, Sivamaruthi BS, Chaiyasut C. Do probiotics improve the health status of individuals with diabetes mellitus? A review on outcomes of clinical trials. Biomed Res Int. 2019;2019:1531567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lee CB, Chae SU, Jo SJ, Jerng UM, Bae SK. The relationship between the gut microbiome and metformin as a key for treating type 2 diabetes mellitus. Int J Mol Sci. 2021;22(7):3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Shin NR, Lee J-C, Lee H-Y, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63(5):727–735. [DOI] [PubMed] [Google Scholar]
- 95. Vardarli I, Arndt E, Deacon CF, Holst JJ, Nauck MA. Effects of sitagliptin and metformin treatment on incretin hormone and insulin secretory responses to oral and “isoglycemic” intravenous glucose. Diabetes. 2014;63(2):663–674. [DOI] [PubMed] [Google Scholar]
- 96. American Diabetes Association . Pharmacologic approaches to glycemic treatment: standards of medical care in diabetes-2020. Diabetes Care. 2020;43(Suppl 1):S98–S110. [DOI] [PubMed] [Google Scholar]
- 97. Andersen A, Lund A, Knop FK, Vilsboll T. Glucagon-like peptide 1 in health and disease. Nat Rev Endocrinol. 2018;14(7):390–403. [DOI] [PubMed] [Google Scholar]
- 98. Singh G, Krauthamer M, Bjalme-Evans M. Wegovy (semaglutide): a new weight loss drug for chronic weight management. J Investig Med. 2022;70(1):5–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kunkel D, Basseri B, Low K, et al. Efficacy of the glucagon-like peptide-1 agonist exenatide in the treatment of short bowel syndrome. Neurogastroenterol Motil. 2011;23(8):739–e328. [DOI] [PubMed] [Google Scholar]
- 100. Wang L, Li P, Tang Z, Yan X, Feng B. Structural modulation of the gut microbiota and the relationship with body weight: compared evaluation of liraglutide and saxagliptin treatment. Sci Rep. 2016;6(1):33251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhao L, Chen Y, Xia F, et al. A glucagon-like peptide-1 receptor agonist lowers weight by modulating the structure of gut microbiota. Front Endocrinol (Lausanne). 2018;9:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhang Q, Xiao X, Zheng J, et al. Featured article: structure moderation of gut microbiota in liraglutide-treated diabetic male rats. Exp Biol Med (Maywood). 2018;243(1):34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wang Z, Saha S, Van Horn S, et al. Gut microbiome differences between metformin- and liraglutide-treated T2DM subjects. Endocrinol Diabetes Metab. 2018;1(1):e00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tsai CY, Lu H-C, Chou Y-H, et al. Gut microbial signatures for glycemic responses of GLP-1 receptor agonists in type 2 diabetic patients: a pilot study. Front Endocrinol (Lausanne). 2022;12:814770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Baldassano S, Rappa F, Amato A, Cappello F, Mule F. GLP-2 as beneficial factor in the glucose homeostasis in mice fed a high fat diet. J Cell Physiol. 2015;230(12):3029–3036. [DOI] [PubMed] [Google Scholar]
- 106. Wismann P, Pedersen SL, Hansen G, et al. Novel GLP-1/GLP-2 co-agonists display marked effects on gut volume and improves glycemic control in mice. Physiol Behav. 2018;192:72–81. [DOI] [PubMed] [Google Scholar]
- 107. Madsen MSA, Holm JB, Pallejà A, et al. Metabolic and gut microbiome changes following GLP-1 or dual GLP-1/GLP-2 receptor agonist treatment in diet-induced obese mice. Sci Rep. 2019;9(1):15582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Finan B, Ma T, Ottaway N, et al. Unimolecular dual incretins maximize metabolic benefits in rodents, monkeys, and humans. Sci Transl Med. 2013;5(209):209ra151. [DOI] [PubMed] [Google Scholar]
- 109. Chow E, Chan JCN. The emerging role of incretins and twincretins. Nat Rev Endocrinol. 2022;18(2):73–74. [DOI] [PubMed] [Google Scholar]
- 110. US Food and Drug Administration . FDA approves novel, dual-targeted treatment for type 2 diabetes. FDA News Release, May 13, 2022.
- 111. Fonseca-Correa JI, Correa-Rotter R. Sodium-glucose cotransporter 2 inhibitors mechanisms of action: a review. Front Med (Lausanne). 2021;8:777861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lee DM, Battson ML, Jarrell DK, et al. SGLT2 inhibition via dapagliflozin improves generalized vascular dysfunction and alters the gut microbiota in type 2 diabetic mice. Cardiovasc Diabetol. 2018;17(1):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. van Bommel EJM, Herrema H, Davids M, Kramer MHH, Nieuwdorp M, van Raalte DH. Effects of 12-week treatment with dapagliflozin and gliclazide on faecal microbiome: results of a double-blind randomized trial in patients with type 2 diabetes. Diabetes Metab. 2020;46(2):164–168. [DOI] [PubMed] [Google Scholar]
- 114. Yang M, Shi F-H, Liu W, et al. Dapagliflozin modulates the fecal microbiota in a type 2 diabetic rat model. Front Endocrinol (Lausanne). 2020;11:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Gu Y, Wang X, Li J, et al. Analyses of gut microbiota and plasma bile acids enable stratification of patients for antidiabetic treatment. Nat Commun. 2017;8(1):1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sheng Y, Zheng S, Ma T, et al. Mulberry leaf alleviates streptozotocin-induced diabetic rats by attenuating NEFA signaling and modulating intestinal microflora. Sci Rep. 2017;7(1):12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hedrington MS, Davis SN. Considerations when using alpha-glucosidase inhibitors in the treatment of type 2 diabetes. Expert Opin Pharmacother. 2019;20(18):2229–2235. [DOI] [PubMed] [Google Scholar]
- 118. Zhang L, Song P, Zhang X, et al. Alpha-glucosidase inhibitors alter gut microbiota and ameliorate collagen-induced arthritis. Front Pharmacol. 2020;10:1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Peat CM, Kleiman SC, Bulik CM, Carroll IM. The intestinal microbiome in bariatric surgery patients. Eur Eat Disord Rev. 2015;23(6):496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Cortez RV, Petry T, Caravatto P, et al. Shifts in intestinal microbiota after duodenal exclusion favor glycemic control and weight loss: a randomized controlled trial. Surg Obes Relat Dis. 2018;14(11):1748–1754. [DOI] [PubMed] [Google Scholar]
- 121. Wang C, Zhang H, Liu H, et al. The genus Sutterella is a potential contributor to glucose metabolism improvement after Roux-en-Y gastric bypass surgery in T2D. Diabetes Res Clin Pract. 2020;162:108116. [DOI] [PubMed] [Google Scholar]
- 122. Crommen S, Mattes A, Simon MC. Microbial adaptation due to gastric bypass surgery: the nutritional impact. Nutrients. 2020;12(4):1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Farup PG, Valeur J. Changes in faecal short-chain fatty acids after weight-loss interventions in subjects with morbid obesity. Nutrients. 2020;12(3):802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Davies N, O'Sullivan JM, Plank LD, Murphy R. Gut microbial predictors of type 2 diabetes remission following bariatric surgery. Obes Surg. 2020;30(9):3536–3548. [DOI] [PubMed] [Google Scholar]
- 125. Murphy R, Tsai P, Jüllig M, Liu A, Plank L, Booth M. Differential changes in gut microbiota after gastric bypass and sleeve gastrectomy bariatric surgery vary according to diabetes remission. Obes Surg. 2017;27(4):917–925. [DOI] [PubMed] [Google Scholar]
- 126. Zhang H, DiBaise JK, Zuccolo A, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A. 2009;106(7):2365–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Flynn CR, Albaugh VL, Abumrad NN. Metabolic effects of bile acids: potential role in bariatric surgery. Cell Mol Gastroenterol Hepatol. 2019;8(2):235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Albaugh VL, Banan B, Antoun J, et al. Role of bile acids and GLP-1 in mediating the metabolic improvements of bariatric surgery. Gastroenterology. 2019;156(4):1041–1051.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Zhang J, Ni Y, Qian L, et al. Decreased abundance of Akkermansia muciniphila leads to the impairment of insulin secretion and glucose homeostasis in lean type 2 diabetes. Adv Sci (Weinh). 2021;8(16):e2100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Shokryazdan P, Faseleh Jahromi M, Navidshad B, Liang JB. Effects of prebiotics on immune system and cytokine expression. Med Microbiol Immunol. 2017;206(1):1–9. [DOI] [PubMed] [Google Scholar]
- 131. Yoo JY, Kim SS. Probiotics and prebiotics: present status and future perspectives on metabolic disorders. Nutrients. 2016;8(3):173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Cani PD, Joly E, Horsmans Y, Delzenne NM. Oligofructose promotes satiety in healthy human: a pilot study. Eur J Clin Nutr. 2006;60(5):567–572. [DOI] [PubMed] [Google Scholar]
- 133. Parnell JA, Reimer RA. Weight loss during oligofructose supplementation is associated with decreased ghrelin and increased peptide YY in overweight and obese adults. Am J Clin Nutr. 2009;89(6):1751–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Cani PD, Neyrinck AM, Maton N, Delzenne NM. Oligofructose promotes satiety in rats fed a high-fat diet: involvement of glucagon-like peptide-1. Obes Res. 2005;13(6):1000–1007. [DOI] [PubMed] [Google Scholar]
- 135. Brooks L, Viardot A, Tsakmaki A, et al. Fermentable carbohydrate stimulates FFAR2-dependent colonic PYY cell expansion to increase satiety. Mol Metab. 2017;6(1):48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Weitkunat K, Stuhlmann C, Postel A, et al. Short-chain fatty acids and inulin, but not guar gum, prevent diet-induced obesity and insulin resistance through differential mechanisms in mice. Sci Rep. 2017;7(1):6109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Zou J, Chassaing B, Singh V, et al. Fiber-mediated nourishment of gut microbiota protects against diet-induced obesity by restoring il-22-mediated colonic health. Cell Host Microbe. 2018;23(1):41–53.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wilkins T, Sequoia J. Probiotics for gastrointestinal conditions: a summary of the evidence. Am Fam Physician. 2017;96(3):170–178. [PubMed] [Google Scholar]
- 139. Rittiphairoj T, Pongpirul K, Janchot K, Mueller NT, Li T. Probiotics contribute to glycemic control in patients with type 2 diabetes mellitus: a systematic review and meta-analysis. Adv Nutr. 2021;12(3):722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Kocsis T, Molnár B, Németh D, et al. Probiotics have beneficial metabolic effects in patients with type 2 diabetes mellitus: a meta-analysis of randomized clinical trials. Sci Rep. 2020;10(1):11787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Tiderencel KA, Hutcheon DA, Ziegler J. Probiotics for the treatment of type 2 diabetes: a review of randomized controlled trials. Diabetes Metab Res Rev. 2020;36(1):e3213. [DOI] [PubMed] [Google Scholar]
- 142. Wang Y, Dilidaxi D, Wu Y, et al. Composite probiotics alleviate type 2 diabetes by regulating intestinal microbiota and inducing GLP-1 secretion in db/db mice. Biomed Pharmacother. 2020;125:109914. [DOI] [PubMed] [Google Scholar]
- 143. Marino E, Richards JL, McLeod KH, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol. 2017;18(5):552–562. [DOI] [PubMed] [Google Scholar]
- 144. Leser TD, Molbak L. Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ Microbiol. 2009;11(9):2194–2206. [DOI] [PubMed] [Google Scholar]
- 145. Leite GGS, Weitsman S, Parodi G, et al. Mapping the segmental microbiomes in the human small bowel in comparison with stool: a REIMAGINE study. Dig Dis Sci. 2020;65(9):2595–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Helander HF, Fandriks L. Surface area of the digestive tract—revisited. Scand J Gastroenterol. 2014;49(6):681–689. [DOI] [PubMed] [Google Scholar]
- 147. Collins JT, Nguyen A, Badireddy M. Anatomy, Abdomen and Pelvis, Small Intestine. StatPearls; 2022. [PubMed] [Google Scholar]
- 148. Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154(2):274–284. [DOI] [PubMed] [Google Scholar]
- 149. Kiela PR, Ghishan FK. Physiology of intestinal absorption and secretion. Best Pract Res Clin Gastroenterol. 2016;30(2):145–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Kong S, Zhang YH, Zhang W. Regulation of intestinal epithelial cells properties and functions by amino acids. Biomed Res Int. 2018;2018:2819154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Holst JJ. From the incretin concept and the discovery of GLP-1 to today's diabetes therapy. Front Endocrinol (Lausanne). 2019;10:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Cani PD, Possemiers S, Van de Wiele T, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58(8):1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Sun W, Chen L-N, Zhou Q, et al. A unique hormonal recognition feature of the human glucagon-like peptide-2 receptor. Cell Res. 2020;30(12):1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Nauck MA, Quast DR, Wefers J, Meier JJ. GLP-1 receptor agonists in the treatment of type 2 diabetes—state-of-the-art. Mol Metab. 2021;46:101102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Gill PA, Zelm MC, Muir JG, Gibson PR. Review article: short chain fatty acids as potential therapeutic agents in human gastrointestinal and inflammatory disorders. Aliment Pharmacol Ther. 2018;48(1):15–34. [DOI] [PubMed] [Google Scholar]
- 156. Kastl AJ, Terry NA, Wu GD, Albenberg LG. The structure and function of the human small intestinal microbiota: current understanding and future directions. Cell Mol Gastroenterol Hepatol. 2020;9(1):33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Baumgart M, Dogan B, Rishniw M, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 2007;1(5):403–418. [DOI] [PubMed] [Google Scholar]
- 158. Wang M, Ahrne S, Jeppsson B, Molin G. Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol. 2005;54(2):219–231. [DOI] [PubMed] [Google Scholar]
- 159. Willing B, Halfvarson J, Dicksved J, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's Disease. Inflamm Bowel Dis. 2009;15(5):653–660. [DOI] [PubMed] [Google Scholar]
- 160. Hartman AL, Lough DM, Barupal DK, et al. Human gut microbiome adopts an alternative state following small bowel transplantation. Proc Natl Acad Sci U S A. 2009;106(40):17187–17192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Booijink CC, El-Aidy S, Rajilić-Stojanović M, et al. High temporal and inter-individual variation detected in the human ileal microbiota. Environ Microbiol. 2010;12(12):3213–3227. [DOI] [PubMed] [Google Scholar]
- 162. Ruigrok R,Collij V, Sureda P, et al. The composition and metabolic potential of the human small intestinal microbiota within the context of inflammatory bowel disease. J Crohns Colitis. 2021;15(8):1326–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Tuganbaev T, Mor U, Bashiardes S, et al. Diet diurnally regulates small intestinal microbiome-epithelial-immune homeostasis and enteritis. Cell. 2020;182(6):1441–1459.e21. [DOI] [PubMed] [Google Scholar]
- 164. Gutierrez IM, Kang KH, Calvert CE, et al. Risk factors for small bowel bacterial overgrowth and diagnostic yield of duodenal aspirates in children with intestinal failure: a retrospective review. J Pediatr Surg. 2012;47(6):1150–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Zoetendal EG, Raes J, van den Bogert B, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012;6(7):1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Seekatz AM,Schnizlein MK, Koenigsknecht MJ, et al. Spatial and temporal analysis of the stomach and small-intestinal microbiota in fasted healthy humans. mSphere. 2019;4(2):e00126-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Leite G, Morales W, Weitsman S, et al. Optimizing microbiome sequencing for small intestinal aspirates: validation of novel techniques through the REIMAGINE study. BMC Microbiol. 2019;19(1):239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Erdogan A, Weitsman S, Parodi G, et al. Small intestinal bacterial overgrowth: duodenal aspiration vs glucose breath test. Neurogastroenterol Motil. 2015;27(4):481–489. [DOI] [PubMed] [Google Scholar]
- 169. Jacobs C, Coss Adame E, Attaluri A, Valestin J, Rao SS. Dysmotility and proton pump inhibitor use are independent risk factors for small intestinal bacterial and/or fungal overgrowth. Aliment Pharmacol Ther. 201337;(11):1103–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Leite G, Morales W, Weitsman S, et al. The duodenal microbiome is altered in small intestinal bacterial overgrowth. PLoS One. 2020;15(7):e0234906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Weitsman S, Celly S, Leite G, et al. Effects of proton pump inhibitors on the small bowel and stool microbiomes. Dig Dis Sci. 2021;67(1):224–232. [DOI] [PubMed] [Google Scholar]
- 172. Leite G, Pimentel M, Barlow GM, et al. Age and the aging process significantly alter the small bowel microbiome. Cell Rep. 2021;36(13):109765. [DOI] [PubMed] [Google Scholar]
- 173. Leite G, Barlow GM, Parodi G, et al. Duodenal microbiome changes in postmenopausal women: effects of hormone therapy and implications for cardiovascular risk. Menopause. 2022;29(3):264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Leite G, Barlow GM, Hosseini A, et al. Smoking has disruptive effects on the small bowel luminal microbiome. Sci Rep. 2022;12(1):6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Barlow JT, Leite G, Romano AE, et al. Quantitative sequencing clarifies the role of disruptor taxa, oral microbiota, and strict anaerobes in the human small-intestine microbiome. Microbiome. 2021;9(1):214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.