Abstract
Cytochrome P450 is an enzyme involved in the metabolism of phase 1 xenobiotics, toxins, endogenous hormones, and drugs, including those used in COVID-19 treatment. Cytochrome p450 genes are linked to the pathogenesis of some multifactorial traits and diseases, such as cancer, particularly prostate cancer, colorectal cancer, breast cancer, and cervical cancer. Genotyping was performed on 540 supposedly healthy individuals of 5 Finno-Permic populations from the territories of the European part of the Russian Federation. There was a statistically significant difference between Veps and most of the studied populations in the rs4986774 locus of the CYP2D6 gene; data on the rs3892097 locus of the CYP2D6 gene shows that Izhemsky Komis are different from the Mordovian and Udmurt populations.
Keywords: CYP1A1, CYP2D6, Finno-Permic populations, pharmacogenetic markers
1. Introduction
The cross-disciplinary integration of the latest research results is particularly important in modern science in order to ensure its effective practical application and reliability. Thus, population genetics studies may contribute to developing new pharmacogenetic approaches, such as tailored treatments targeting representatives of specific ethnic groups with drugs that are more effective for the population in question. The study of polymorphic allelic variants of the cytochrome P450 (CYP450s) genes is of a particular interest to us as its products are responsible for the Phase I biotransformation of drugs of some of the most important categories. CYP450s are heme-containing enzymes that are critical to many cellular processes including eicosanoid metabolism, cholesterol, and bile acid biosynthesis, synthesis, and metabolism of steroids and vitamin D3, biogenic amines formation, and the breakdown and hydroxylation of retinoic acid. There are many other functions of CYP450s including some still not fully understood. Evidence shows that polymorphic CYP450s genes variants are associated with predisposition to such serious diseases as malignant neoplasms (MNs), for example rs1048943 in the CYP1A1 gene [1].
A study of 100 patients with prostate cancer and 150 healthy people reported an association of the AA rs1048943 genotype with the risk of developing prostate cancer in Iraqi residents [2]. Similar data were obtained regarding the risk of developing colorectal cancer in a study of 200 patients and 200 healthy Iraqis [3]. This is confirmed by meta-analysis of 20 independent original studies involving 8665 patients and 9953 healthy individuals that revealed an association of rs1048943 A > G with an increased risk of developing colorectal cancer [4].
Another meta-analysis caried out in 2016 included 748 patients with laryngeal cancer and a control group of 1558 individuals; it concluded that the G allele and G/G rs1048943 homozygotes in the CYP1A1 gene were associated with the risk of developing this malignant neoplasm in the Asian population. At the same time, these associated risks were not true for the Caucasian population [1]. This indicates that pharmacogenetic analyses should take ethnicity into account, since such differences, in addition to predisposition to specific neoplasms, may also affect the biotransformation of xenobiotics. In 2018, a meta-analysis of 29 genetic studies showed an increased risk of developing cervical cancer in carriers of the G rs1048943 allele among women from India [5]. In 2021, a meta-analysis using data from 39 studies (7630 patients and 8169 healthy people) reported an association of rs1048943 (AG + GG) in Indians [6].
In 2017, a meta-analysis of 15 studies of breast cancer in Asia (total of 1794 individuals) revealed an association of the rs1065852 *10/*10 (TT) polymorphism in the CYP2D6 gene with worse disease-free survival and relapse in women receiving adjuvant treatment with tamoxifen dosage of 20 mg/day [7].
In addition, the role of CYP450s isoforms in the metabolism of neurotransmitters, neurosteroids, and cholesterol is currently being studied, as well as their possible effect on behavior including stress, depression, schizophrenia, cognitive processes, learning, and memory. Haduch and Daniel (2019) emphasize the significance of the CYP-mediated alternative pathways for the synthesis of dopamine and serotonin, as they are critical in the local production of these neurotransmitters specifically in the areas of the brain severely affected by depression and schizophrenia, where these neurotransmitter systems are impaired [8]. Most antipsychotics and antidepressants are also metabolized by polymorphic CYP2D6 enzymes and their capacity is genetically determined, which undoubtedly increases the importance of this study [9,10].
The role of cytochrome P450 genes in the pathogenesis of infectious diseases, COVID-19 in particular, is currently being studied, as well as its effect on drug therapy. It was shown that the expression of CYP2D6 decreases in mice infected with hepatitis C virus (HCV) [11], while the expression of CYP2A5 and CYP3A increases in transgenic mice infected with hepatitis B virus (HBV) [12]. Most drugs used to treat COVID-19 are metabolized by cytochrome P450 (CYP) enzymes, primarily CYP2D6 [13]. It was demonstrated that the CYP2D6 variant is associated with the hydroxychloroquine metabolic ratio, which was recently used in COVID-19 treatment [14]. The CYP2D6 and CYP2C19 genes were responsible for most treatment modifications, and the medications most often affected were ondansetron, oxycodone, and clopidogrel, commonly given to patients with COVID-19 [15].
The Finno-Ugric ethno-linguistic community is currently one of the largest language groups in Europe with total of more than 25 million people. The Finno-Ugric branch of languages is divided into two large sub-branches: Finno-Permic and Ugric. Most modern Finno-Ugric languages belong to the Finno-Permic sub-branch while accounting for less than half of the population [16,17].
It is assumed that the ancestral pre-Finno-Ugric population belonged to a single anthropological group originated in ancient Ural. However, modern Finno-Ugric peoples are extremely diverse. Thus, modern Karelians and Vepsians can be described as Caucasians of White Sea-Baltic type, while most of the Mordovians-Erzi are of the Atlanto-Baltic Sura type, and the Mordovians-Mokshas are of the Subural type. The Udmurts belong to the Vyatka-Kama sublaponoid anthropological type, which is characterized by the predominance of Caucasian features over the also present Mongoloid ones.
Population genetics studies of the Finno-Ugric peoples have shown that the modern speakers of these languages and their geographical neighbors are alike in terms of genetic composition of these biological populations. However, when studying connections between geographically distant populations, it is revealed that most of the speakers of these languages and some of their neighbors share a common genetic component, possibly of Siberian origin. In addition, it has been shown that the number of identical IBD segments is much higher among most Uralic-speaking peoples compared to their closest geographic neighbors belonging to other language families [18].
The objective of this research is to study the main pharmacogenetic markers among the Finno-Permic peoples populating the European part of Russian Federation.
2. Materials and Methods
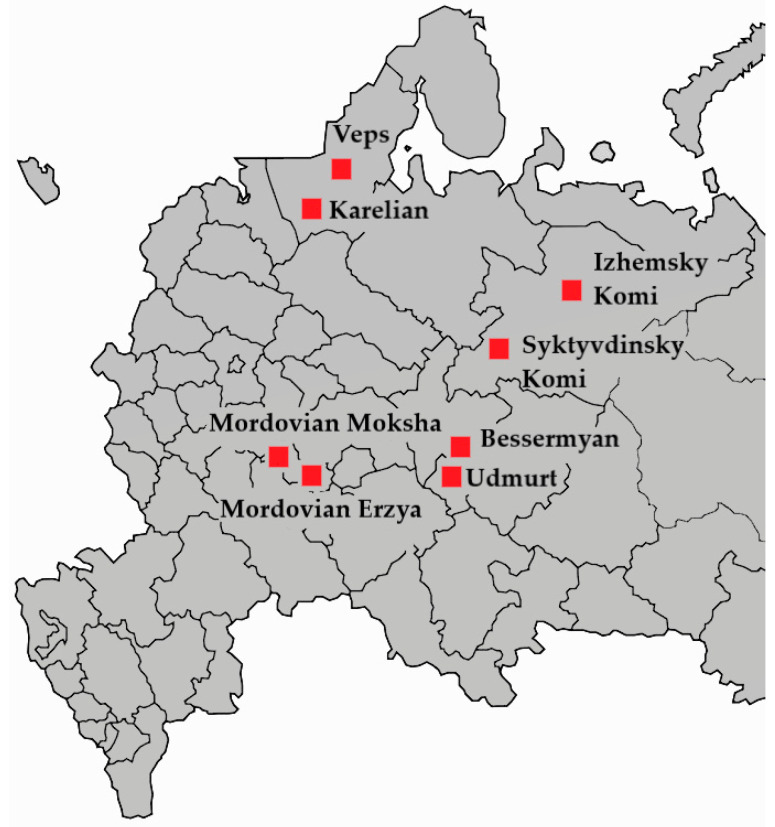
The sample used in this study included 540 presumably healthy individuals of 5 Finno-Permic populations from the territories in the European part of the Russian Federation (Figure 1). The sample was divided into 8 groups accounting for the ethnoterritorial distinctions within the populations (Table 1). Sampling was carried out in accordance with the ethical standards of the Bioethics Committee, developed by the WMA Declaration of Helsinki: “Ethical Principles for the Conduct of Medical Research Involving Human Subjects”. All subjects filled out a questionnaire taking into account nationality (up to three generations) and year of birth. All respondents signed an informed voluntary consent to participate in the study. The work was approved by the Local Ethics Committee of the Institute of Biochemistry and Genetics of the USC RAS (protocol No. 14 of 15 September 2016).
Figure 1.
Map of the European part of Russia indicating the places of biomaterial collection.
Table 1.
Linguistic picture of the Finno-Permic population of Russia, studied in the work, as well as the amount of material used.
| Sub-Branch of Finno-Permic Languages | Language Group | Language Sub-Group | Populations Included in the Study | N |
|---|---|---|---|---|
| Finno-Permic sub-branch | Permic | - | Udmurt | 96 |
| Besermyan | 94 | |||
| Komi | 94 | |||
| Finno-Volga | Mordovian | Erzya | 78 | |
| Moksha | 35 | |||
| Baltic-Finnish | Karelian | 60 | ||
| Veps | 60 |
The DNA was extracted from peripheral blood samples using phenol-chloroform [19]. Vacutainer® tubes were used to collect, transport, and store the blood samples using 0.5 M EDTA solution as a preservative. After drawing the sample, each tube was shaken and stored at 4 °C.
TaqMan real-time PCR technology was used for the genotyping of polymorphic loci. The incidence of allele variants in given populations were calculated based on observed genotype frequencies. The correspondence of the genotype frequencies to the Hardy–Weinberg equilibrium was assessed using Pearson’s χ2 test (at p > 0.05). The significance of differences in allele frequencies in the sample was calculated by the χ2 test using the Yates correction for continuity. Surfer 24.1.181 was used to produce allele distribution maps.
3. Results
In this paper we studied 4 polymorphic loci located in two genes of the cytochrome P450 system (rs1048943, rs1065852, rs3892097, and rs4986774). The distribution of genotype frequencies corresponded to the Hardy–Weinberg equilibrium in most Finno-Permic populations studied by us. Among the few exceptions were the polymorphisms rs1048943, rs1065852, rs3892097 in Mordovian population with p < 0.05, as well as the distribution of genotypes of the rs1065852 polymorphic variant of the CYP2D6 gene in the Udmurt population. Based on this data alone, it is impossible to determine the cause of such a deviation, however, the effect of natural selection on these loci seems to be the most reasonable explanation for this phenomenon.
Paired comparison tests were run for allele frequencies and all selected markers in the studied ethnic groups. In addition, the analysis included data on some other world populations previously published in the academic literature, as well as the data obtained inthe 1000 genomes project [20].
The ethnogenesis of modern Finno-Ugric peoples is an extremely complex topic. The collapse of the ancient Ural community, according to researchers [21,22], occurred in the forth and third millennium BC, when, as R.G. Kuzeev suggests, the tribes of the Keltiminar culture, who came from the south, broke up into two separate groups. The ancestors of the Samoyeds moved to the Yenisei, while the Western groups remained in their former territories. In the third millennium, the latter moved to the west, and mixed with the newcomers formed the Late Neolithic Finno-Ugric community on the territory of the Volga-Kama, Urals, and Trans-Urals [23]. The third and second millennia BC were marked by the continued migration of smaller groups of Finno-Ugric peoples to the north, to the White Sea. The geographical factors and the distance between the tribes led to the final breakup of Finno-Ugric community into Ugric and Finno-Perm groups. Thus, each ethnic group considered in this study developed unique features that are different from related populations due to the timescale and the complexity of its ethnogenesis, the sheer vastness of the populated territories, as well as the relatively large number of neighbors that are different in language and anthropology, which increases the value of this study.
In our study, it was found that the frequency of the genotype AA rs1048943 of the CYP1A1 gene is evenly distributed among all the studied populations and the minimum value is observed in the Udmurt population (88.54%) while the maximum value is in the Erzya subpopulation (96.15%). The GG genotype was found in the Mordovian and Komi populations with frequencies of 1.47% and 1.06%, respectively (Table 2). Table 3 shows the frequency of the minor allele rs1048943 of the CYP1A1 gene in the studied samples of Finno-Permic peoples, as well as in some world populations. Based on the cross-comparison of the populations (p-value), it can be concluded that there are no statistically significant differences between allele frequencies in all studied populations. At the same time, the statistically significant differences between the Besermyan and Erzya and the populations of Tatars and Bashkirs, as well as Komi and Bashkirs, are of interest. Even though both Tatars and Bashkirs live in the Volga-Ural region, may be explained by the greater effect of East Eurasian component on the gene pool of the indicated Turkic-speaking populations.
Table 2.
Distribution of the rs1048943 genotypes of the CYP1A1 gene in the Finno-Permic populations of the Russian Federation.
| Population | N | AA | AG | GG | Minor Allele Frequency (95% CI) |
χ2 | Deviations from HWE, p | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed (N) | Expected (N) | % | Observed (N) | Expected (N) | % | Observed (N) | Expected (N) | % | |||||
| Veps | 60 | 55 | 55.1 | 91.67 | 5 | 4.8 | 8.33 | 0 | 0.1 | 0 | 4.17% (1.36–9.45) | 0.113 | 0.736 |
| Karelian | 60 | 55 | 55.1 | 91.67 | 5 | 4.8 | 8.33 | 0 | 0.1 | 0 | 4.17% (1.36–9.45) | 0.113 | 0.736 |
| Udmurt | 96 | 85 | 85.3 | 88.54 | 11 | 10.4 | 11.46 | 0 | 0.3 | 0 | 5.73% (2.89–10.02) | 0.354 | 0.551 |
| Bessermyan | 94 | 90 | 90 | 95.74 | 4 | 3.9 | 4.26 | 0 | 0 | 0 | 2.13% (0.58–5.36) | 0.044 | 0.833 |
| Mordovian (total) | 136 | 128 | 126.2 | 94.12 | 6 | 9.6 | 4.41 | 2 | 0.2 | 1.47 | 3.68% (1.78–6.66) | 19.340 | 0.00001 |
| Erzya | 78 | 75 | 75 | 96.15 | 3 | 2.9 | 3.85 | 0 | 0 | 0 | 1.92% (0.40–5.52) | 0.03 | 0.862 |
| Moksha | 35 | 32 | 31.1 | 91.43 | 2 | 3.8 | 5.71 | 1 | 0.1 | 2.86 | 5.71% (1.58–13.99) | 7.721 | 0.005 |
| Komi (total) |
94 | 89 | 88.1 | 94.68 | 4 | 5.8 | 4.26 | 1 | 0.1 | 1.06 | 3.19% (1.18–6.82) | 9.113 | 0.002 |
| Izhemsk | 47 | 44 | 44 | 93.62 | 3 | 2.9 | 6.38 | 0 | 0 | 0 | 3.19% (0.66–9.04) | 0.051 | 0.821 |
| Syktyvdinsk | 47 | 45 | 44 | 95.74 | 1 | 2.9 | 2.13 | 1 | 0 | 2.13 | 3.19% (0.66–9.04) | 20.20 | 0.000007 |
Table 3.
Frequencies of the minor allele rs1048943 of the CYP1A1 gene in the studied samples of Finno-Permic peoples, as well as in some populations of the world.
| Population | N | Minor allele Frequency% | Veps | Karelian | Mordovian (Total) |
Erzya | Moksha | Udmurt | Komi (Total) |
Izhemsk | Syktyvdinsk | Bessermyan |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Veps | 60 | 4.17% | 0.747 | 0.958 | 0.459 | 0.896 | 0.730 | 0.893 | 0.992 | 0.992 | 0.491 | |
| Karelian | 60 | 4.17% | 0.747 | 0.958 | 0.459 | 0.896 | 0.730 | 0.893 | 0.992 | 0.992 | 0.491 | |
| Mordovian (total) | 136 | 3.68% | 0.958 | 0.958 | 0.412 | 0.984 | 0.917 | 0.917 | 0.500 | |||
| Erzya | 78 | 1.92% | 0.460 | 0.460 | 0.269 | 0.128 | 0.693 | 0.835 | 0.835 | 0.803 | ||
| Moksha | 35 | 5.71% | 0.896 | 0.896 | 0.269 | 0.767 | 0.568 | 0.689 | 0.689 | 0.283 | ||
| Udmurt | 96 | 5.73% | 0.730 | 0.730 | 0.418 | 0.128 | 0.767 | 0.343 | 0.520 | 0.520 | 0.124 | |
| Komi (total) | 94 | 3.19% | 0.893 | 0.893 | 0.984 | 0.693 | 0.568 | 0.343 | 0.749 | |||
| Izhensk | 47 | 3.19% | 0.992 | 0.992 | 0.917 | 0.835 | 0.689 | 0.520 | 0.678 | 0.892 | ||
| Syktyvdinsk | 47 | 3.19% | 0.992 | 0.992 | 0.917 | 0.835 | 0.689 | 0.520 | 0.678 | 0.892 | ||
| Bessermyan | 94 | 2.13% | 0.491 | 0.491 | 0.500 | 0.803 | 0.283 | 0.124 | 0.749 | 0.892 | 0.892 | |
| Finnish [20] | 99 | 5.1% | 0.930 | 0.930 | 0.619 | 0.204 | 0.923 | 0.942 | 0.509 | 0.677 | 0.677 | 0.207 |
| Russian [24] | 314 | 4.8% | 0.957 | 0.957 | 0.576 | 0.172 | 0.958 | 0.733 | 0.468 | 0.673 | 0.673 | 0.165 |
| Tatar, Russia [24] | 243 | 6.4% | 0.482 | 0.482 | 0.158 | 0.050 | 0.961 | 0.889 | 0.150 | 0.335 | 0.335 | 0.041 |
| Bashkir, Russia [24] | 134 | 10.5% | 0.063 | 0.063 | 0.004 | 0.002 | 0.329 | 0.105 | 0.006 | 0.051 | 0.051 | 0.001 |
| England and Scotland [20] | 91 | 3.3% | 0.935 | 0.935 | 0.964 | 0.658 | 0.603 | 0.379 | 0.813 | 0.756 | 0.756 | 0.709 |
| Spain [20] | 107 | 1.9% | 0.372 | 0.372 | 0.363 | 0.727 | 0.203 | 0.073 | 0.597 | 0.763 | 0.763 | 0.863 |
| Toscana, Italy [20] | 107 | 3.3% | 0.908 | 0.908 | 0.994 | 0.642 | 0.573 | 0.337 | 0.812 | 0.754 | 0.754 | 0.693 |
| Balkar [25] | 104 | 8.6% | 0.191 | 0.191 | 0.035 | 0.012 | 0.595 | 0.350 | 0.039 | 0.138 | 0.138 | 0.009 |
| Karachay [25] | 73 | 7.5% | 0.373 | 0.373 | 0.137 | 0.040 | 0.837 | 0.657 | 0.125 | 0.263 | 0.263 | 0.035 |
| Europe [20] | 503 | 3.5% | 0.901 | 0.901 | 0.977 | 0.438 | 0.524 | 0.199 | 0..984 | 0.881 | 0.881 | 0.463 |
| S. Asia [20] | 489 | 12.7% | 0.006 | 0.006 | 0.00002 | 0.00008 | 0.126 | 0.008 | 0.0001 | 0.011 | 0.011 | 0.00002 |
Bold indicates statistically significant differences (p < 0.05).
The frequency distribution of the rs1065852 genotypes of the CYP2D6 gene in the studied Finno-Permic populations shows that TT genotype is absent, while the frequency of the minor T allele ranges from 6.38% (95% CI 2.38–13.38) in the Komi population from the Izhma region, and up to 18.23% (95% CI 13.04–24.43) in the Udmurt population (Table 4). The population comparison (p-value) revealed complex interrelations within the studied populations (Table 5). Thus, there are statistical differences between the Komi and Mordovian populations, the Moksha subpopulation, and the Udmurts. Komi from the Izhma region, in addition to all the listed populations, have a marked difference from the Erzya population. There is also a significant difference between Veps and Mordovians-Moksha.
Table 4.
Distribution of the rs1065852 genotypes of the CYP2D6 gene in the Finno-Permic populations of the Russian Federation.
| Population | N | CC | CT | TT | Minor Allele Frequency (95% CI) |
χ2 | Deviations from HWE, p | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed (N) | Expected (N) | % | Observed (N) | Expected (N) | % | Observed (N) | Expected (N) | % | |||||
| Veps | 60 | 47 | 47.7 | 78.33 | 13 | 11.6 | 21.67 | 0 | 0.7 | 0 | 10.83% (5.90–17.81) | 0.886 | 0.347 |
| Karelian | 60 | 45 | 45.9 | 75 | 15 | 13.1 | 25 | 0 | 0.9 | 0 | 12.50% (7.17–19.78) | 1.224 | 0.268 |
| Udmurt | 96 | 61 | 64.2 | 63.54 | 35 | 28.6 | 36.46 | 0 | 3.2 | 0 | 18.23% (13.04–24.43) | 4.771 | 0.029 |
| Bessermyan | 94 | 64 | 66.4 | 68.09 | 30 | 25.2 | 31.91 | 0 | 2.4 | 0 | 15.96% (11.03–21.99) | 3.389 | 0.066 |
| Mordovian (total) | 136 | 87 | 91.4 | 63.97 | 49 | 40.2 | 36.03 | 0 | 4.4 | 0 | 18.01% (13.63–23.11) | 6.566 | 0.01 |
| Erzya | 78 | 52 | 54.2 | 66.67 | 26 | 21.7 | 33.33 | 0 | 2.2 | 0 | 16.67% (11.19–23.46) | 3.12 | 0.077 |
| Moksha | 35 | 19 | 20.8 | 54.29 | 16 | 12.3 | 45.71 | 0 | 1.8 | 0 | 22.86% (13.67–34.45) | 3.073 | 0.08 |
| Komi (total) |
94 | 75 | 76 | 79.79 | 19 | 17.1 | 20.21 | 0 | 1 | 0 | 10.11% (6.20–15.33) | 1.188 | 0.276 |
| Izhemsk | 47 | 41 | 41.2 | 87.23 | 6 | 5.6 | 12.77 | 0 | 0.2 | 0 | 6.38% (2.38–13.38) | 0.218 | 0.640 |
| Syktyvdinsk | 47 | 34 | 34.9 | 72.34 | 13 | 11.2 | 27.66 | 0 | 0.9 | 0 | 13.83% (7.57–22.49) | 1.211 | 0.271 |
Table 5.
Frequencies of the minor allele rs1065852 of the CYP2D6 gene in the studied samples of Finno-Permic peoples, as well as in some populations of the world.
| Population | N | Minor allele Frequency% | Veps | Karelian | Mordovian (Total) |
Erzya | Moksha | Udmurt | Komi (Total) |
Izhemsk | Syktyvdinsk | Bessermyan |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Veps | 60 | 10.83% | 0.841 | 0.100 | 0.228 | 0.044 | 0.109 | 0.990 | 0.371 | 0.691 | 0.273 | |
| Karelian | 60 | 12.50% | 0.841 | 0.225 | 0.427 | 0.097 | 0.237 | 0.640 | 0.207 | 0.982 | 0.501 | |
| Mordovian (total) | 136 | 18.01% | 0.100 | 0.225 | 0.950 | 0.027 | 0.011 | 0.396 | 0.653 | |||
| Erzya | 78 | 16.67% | 0.228 | 0.427 | 0.357 | 0.811 | 0.102 | 0.031 | 0.626 | 0.975 | ||
| Moksha | 35 | 22.86% | 0.044 | 0.097 | 0.357 | 0.509 | 0.014 | 0.005 | 0.176 | 0.269 | ||
| Udmurt | 96 | 18.23% | 0.109 | 0.237 | 0.950 | 0.811 | 0.509 | 0.034 | 0.012 | 0.402 | 0.651 | |
| Komi (total) | 94 | 10.11% | 0.990 | 0.640 | 0.027 | 0.102 | 0.014 | 0.034 | 0.125 | |||
| Izhemsk | 47 | 6.38% | 0.371 | 0.207 | 0.011 | 0.031 | 0.005 | 0.012 | 0.161 | |||
| Syktyvdinsk | 47 | 13.83% | 0.691 | 0.982 | 0.396 | 0.626 | 0.176 | 0.402 | 0.161 | 0.717 | ||
| Bessermyan | 94 | 15.96% | 0.273 | 0.501 | 0.653 | 0.975 | 0.269 | 0.651 | 0.125 | 0.037 | 0.717 | |
| Finnish [20] | 99 | 14.6% | 0.422 | 0.712 | 0.399 | 0.709 | 0.163 | 0.413 | 0.231 | 0.066 | 0.940 | 0.829 |
| England and Scotland [20] | 91 | 24.7% | 0.004 | 0.014 | 0.107 | 0.093 | 0.884 | 0.160 | 0.0003 | 0.0004 | 0.043 | 0.049 |
| Spain [20] | 107 | 17.3% | 0.154 | 0.317 | 0.930 | 0.986 | 0.389 | 0.907 | 0.053 | 0.018 | 0.508 | 0.823 |
| Toscana, Italy [20] | 107 | 20.6% | 0.034 | 0.088 | 0.554 | 0.418 | 0.810 | 0.640 | 0.006 | 0.003 | 0.188 | 0.289 |
| Europe [20] | 503 | 20.2% | 0.014 | 0.044 | 0.426 | 0.305 | 0.700 | 0.535 | 0.001 | 0.002 | 0.152 | 0.180 |
| S. Asia [20] | 489 | 16.5% | 0.111 | 0.264 | 0.545 | 0.949 | 0.225 | 0.549 | 0.027 | 0.015 | 0.551 | 0.864 |
Bold indicates statistically significant differences (p < 0.05).
The AA homozygotes are also absent in the frequency distribution of the rs3892097 genotypes of the CYP2D6 gene, while the distribution of GA heterozygotes varies considerably from the lowest value of 12.77% in the Komi population of the Izhma region to the maximum in the Mordvin-Moksha population at 37.14% (Table 6). The minor allele A frequency observed in the Komi of the Izhma region is the least and is 6.38% (95% CI 2.38–13.38), while the maximum frequency is 18.57% (95% CI 10.28–29.66) found in the Mordovian-Moksha population. When calculating the significance level (p-value), a statistically significant difference was revealed between the Komi population from the Izhemsky district of the Komi Republic and three other populations: the Udmurt populations, the total sample of Mordovians, and with the Mordovian-Moksha subpopulation (Table 7).
Table 6.
Distribution of the rs3892097 genotypes of the CYP2D6 gene in the Finno-Permic populations of the Russian Federation.
| Population | N | GG | GA | AA | Minor Allele Frequency (95% CI) |
χ2 | Deviations from HWE, p | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed (N) | Expected (N) | % | Observed (N) | Expected (N) | % | Observed (N) | Expected (N) | % | |||||
| Veps | 60 | 48 | 48.6 | 80 | 12 | 10.8 | 20 | 0 | 0.6 | 0 | 10% (5.27–16.82) | 0.741 | 0.389 |
| Karelian | 60 | 43 | 44.2 | 71.67 | 17 | 14.6 | 28.33 | 0 | 1.2 | 0 | 14.17% (8.47–21.71) | 1.634 | 0.201 |
| Udmurt | 96 | 65 | 67.5 | 67.71 | 31 | 26 | 32.29 | 0 | 2.5 | 0 | 16.15% (11.24–22.13) | 3.559 | 0.059 |
| Bessermyan | 94 | 68 | 69.8 | 72.34 | 26 | 22.4 | 27.66 | 0 | 1.8 | 0 | 13.83% (9.24–19.60) | 2.421 | 0.119 |
| Mordovian (total) | 136 | 94 | 97.2 | 69.12 | 42 | 35.5 | 30.88 | 0 | 3.2 | 0 | 15.44% (11.36–20.29) | 4.535 | 0.033 |
| Erzya | 78 | 55 | 56.7 | 70.51 | 23 | 19.6 | 29.49 | 0 | 1.7 | 0 | 14.74% (9.58–21.30) | 2.333 | 0.127 |
| Moksha | 35 | 22 | 23.2 | 62.86 | 13 | 10.6 | 37.14 | 0 | 1.2 | 0 | 18.57% (10.28–29.66) | 1.821 | 0.177 |
| Komi (total) |
94 | 74 | 75.1 | 78.72 | 20 | 17.9 | 21.28 | 0 | 1.1 | 0 | 10.64% (6.62–15.95) | 1.332 | 0.248 |
| Izhemsk | 47 | 41 | 41.2 | 87.23 | 6 | 5.6 | 12.77 | 0 | 0.2 | 0 | 6.38% (2.38–13.38) | 0.218 | 0.640 |
| Syktyvdinsk | 47 | 33 | 34 | 70.21 | 14 | 11.9 | 29.79 | 0 | 1 | 0 | 14.89% (8.39–23.72) | 1.439 | 0.230 |
Table 7.
Frequencies of the minor allele rs3892097 of the CYP2D6 gene in the studied samples of Finno-Permic peoples, as well as in some populations of the world.
| Population | N | Minor allele Frequency% | Veps | Karelian | Mordovian (Total) |
Erzya | Moksha | Udmurt | Komi (Total) |
Izhemsk | Syktyvdinsk | Bessermyan |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Veps | 60 | 10% | 0.428 | 0.200 | 0.321 | 0.143 | 0.173 | 0.990 | 0.485 | 0.380 | 0.413 | |
| Karelian | 60 | 14.17% | 0.428 | 0.863 | 0.970 | 0.550 | 0.756 | 0.454 | 0.109 | 0.963 | 0.932 | |
| Mordovian (total) | 136 | 15.44% | 0.200 | 0.863 | 0.939 | 0.179 | 0.038 | 0.969 | 0.730 | |||
| Erzya | 78 | 14.74% | 0.321 | 0.970 | 0.596 | 0.833 | 0.326 | 0.072 | 0.880 | 0.931 | ||
| Moksha | 35 | 18.57% | 0.143 | 0.550 | 0.596 | 0.781 | 0.137 | 0.030 | 0.678 | 0.453 | ||
| Udmurt | 96 | 16.15% | 0.173 | 0.756 | 0.939 | 0.833 | 0.781 | 0.154 | 0.033 | 0.920 | 0.625 | |
| Komi (total) | 94 | 10.64% | 0.990 | 0.454 | 0.179 | 0.326 | 0.137 | 0.154 | 0.431 | |||
| Izhemsk | 47 | 6.38% | 0.485 | 0.109 | 0.038 | 0.072 | 0.030 | 0.033 | 0.098 | 0.097 | ||
| Syktyvdinsk | 47 | 14.89% | 0.380 | 0.963 | 0.969 | 0.880 | 0.678 | 0.920 | 0.098 | 0.952 | ||
| Bessermyan | 94 | 13.83% | 0.413 | 0.932 | 0.730 | 0.931 | 0.453 | 0.625 | 0.431 | 0.097 | 0.952 | |
| Finnish [20] | 99 | 13.6% | 0.434 | 0.972 | 0.679 | 0.886 | 0.423 | 0.580 | 0.456 | 0.103 | 0.913 | 0.926 |
| England and Scotland [20] | 91 | 24.2% | 0.003 | 0.048 | 0.027 | 0.042 | 0.433 | 0.070 | 0.0009 | 0.0005 | 0.101 | 0.016 |
| Spain [20] | 107 | 14.5% | 0.315 | 0.934 | 0.869 | 0.936 | 0.529 | 0.744 | 0.314 | 0.068 | 0.935 | 0.964 |
| Toscana, Italy [20] | 107 | 18.7% | 0.051 | 0.366 | 0.408 | 0.391 | 0.877 | 0.587 | 0.033 | 0.009 | 0.519 | 0.239 |
| Europe [20] | 503 | 18.6% | 0.019 | 0.234 | 0.230 | 0.245 | 0.877 | 0.421 | 0.008 | 0.005 | 0.455 | 0.118 |
| S. Asia [20] | 489 | 10.9% | 0.754 | 0.292 | 0.043 | 0.166 | 0.081 | 0.040 | 0.903 | 0.231 | 0.324 | 0.310 |
Bold indicates statistically significant differences (p < 0.05).
It was found that the frequency distributions of alleles and genotypes rs4986774 of the CYP2D6 gene in the studied populations differ significantly (Table 8). In the populations of the Udmurts, Besermens, and two ethnoterritorial groups of the Komi, there was no diversity in the frequencies of genotypes and alleles at all, and the AA genotype is detected in 100% of the samples. In the Karelian population and in the subpopulations of the Mordovians, the frequency of the minor allele (delA) was under 2%, while the Veps population is fundamentally different, and the minor allele frequency is 6.67% (95% CI 2.92–12.71). This difference in allele frequencies may be explained by drift and/or the founder effect. When calculating the p-value, a statistically significant difference was revealed between the Veps population and all the other populations in the sample, except for Karelians and Moksha-Mordovians. (Table 9). Due to the lack of rs4986774 of the CYP2D6 gene in the databases, only the samples analyzed in this study were compared in Table 9.
Table 8.
Distribution of the rs4986774 genotypes of the CYP2D6 gene in the Finno-Permic populations of the Russian Federation.
| Population | N | A/A | A/del | del/del | Minor Allele Frequency (95% CI) |
χ2 | Deviations from HWE, p | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Observed (N) | Expected (N) | % | Observed (N) | Expected (N) | % | Observed (N) | Expected (N) | % | |||||
| Veps | 60 | 52 | 52.3 | 86.67 | 8 | 7.5 | 13.33 | 0 | 0.3 | 0 | 6.67% (2.92–12.71) | 0.306 | 0.580 |
| Karelian | 60 | 58 | 58 | 96.67 | 2 | 2 | 3.33 | 0 | 0 | 0 | 1.67% (0.20–5.89) | 0.017 | 0.895 |
| Udmurt | 96 | 96 | 96 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Bessermyan | 94 | 94 | 94 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Mordovian (total) | 136 | 132 | 132 | 97.06 | 4 | 3.9 | 2.94 | 0 | 0 | 0 | 1.47% (0.40–3.72) | 0.03 | 0.861 |
| Erzya | 78 | 76 | 76 | 97.44 | 2 | 2 | 2.56 | 0 | 0 | 0 | 1.28% (0.16–4.55) | 0.013 | 0.908 |
| Moksha | 35 | 33 | 33 | 94.29 | 2 | 1.9 | 5.71 | 0 | 0 | 0 | 1.99% (0.35–9.94) | 0.03 | 0.862 |
| Komi (total) |
94 | 94 | 94 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Izhemsk | 47 | 47 | 47 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Syktyvdinsk | 47 | 47 | 47 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | |||
Table 9.
Frequencies of the minor allele rs4986774 of the CYP2D6 gene in the studied samples of Finno-Permic peoples.
| Population | N | Minor allele Frequency% | Veps | Karelian | Mordovian (Total) |
Erzya | Moksha | Udmurt | Komi (Total) |
Izhemsk | Syktyvdinsk | Bessermyan |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Veps | 60 | 6.67% | 0.106 | 0.015 | 0.040 | 0.425 | 0.0008 | 0.0009 | 0.030 | 0.030 | 0.0009 | |
| Karelian | 60 | 1.67% | 0.106 | 0.763 | 0.808 | 0.978 | 0.214 | 0.223 | 0.640 | 0.640 | 0.223 | |
| Mordovian (total) | 136 | 1.47% | 0.015 | 0.763 | 0.272 | 0.282 | 0.734 | 0.734 | 0.282 | |||
| Erzya | 78 | 1.28% | 0.040 | 0.808 | 0.776 | 0.346 | 0.358 | 0.842 | 0.842 | 0.358 | ||
| Moksha | 35 | 1.99% | 0.425 | 0.978 | 0.776 | 0.053 | 0.057 | 0.293 | 0.293 | 0.057 | ||
| Udmurt | 96 | 0% | 0.0008 | 0.214 | 0.272 | 0.346 | 0.053 | |||||
| Komi (total) | 94 | 0% | 0.0009 | 0.223 | 0.282 | 0.358 | 0.057 | |||||
| Izhemsk | 47 | 0% | 0.030 | 0.640 | 0.734 | 0.842 | 0.293 | |||||
| Syktyvdinsk | 47 | 0% | 0.030 | 0.640 | 0.734 | 0.842 | 0.293 | |||||
| Bessermyan | 94 | 0% | 0.0009 | 0.223 | 0.282 | 0.358 | 0.057 |
Bold indicates statistically significant differences (p < 0.05).
4. Discussion
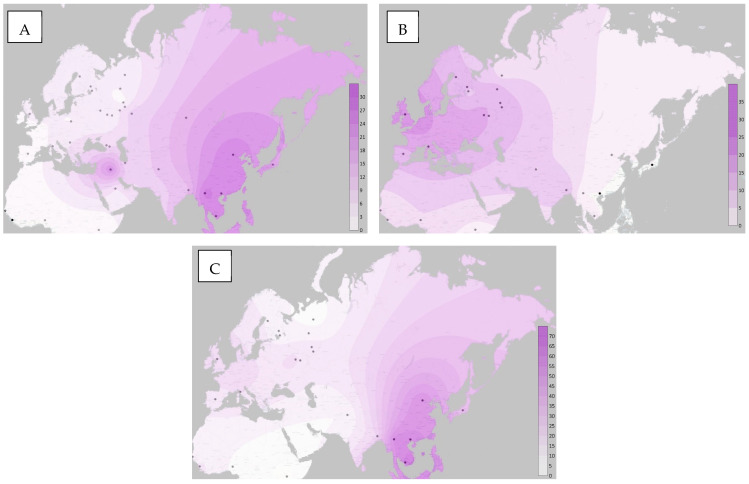
Based on the results of our study, as well as literature data, we built maps of the frequency distribution of minor alleles of the studied loci in the Surfer program (Figure 2).
Figure 2.
Distribution maps of minor alleles (A) rs1048943, (B) rs3892097, (C) rs1065852.
The frequency distribution of alleles and genotypes of the rs1048943 polymorphic variant in various populations of the world has already been described. Thus, the maximum values of the 462Val variant were found in the indigenous peoples of North and South America (over 70%), as well as in the populations of East Asia (over 30%) and Kazakhstan (28.4%) [26,27,28,29,30]. The lowest values are typical for the population of Africa, Europe, and some populations of Western Asia (0–3%, 2–7% and 5.8–9.5%, respectively) [2,20,24,31,32].
In this paper, the polymorphism of the Phase I gene of the CYP1A1 xenobiotic biotransformation system (A2455G, rs1048943) was studied for the first time in the Finno-Permic populations inhabiting the vast territories of the European part of the Russian Federation. The data obtained show no significant differences between the Finno-Permic populations included in the study, with the exception of Turkic-speaking populations of the Bashkirs and Tatars. In the studied populations, the CYP1A1 (2455G) allele occurs with frequencies typical for Western Asian and European populations. These new data on the xenobiotic biotransformation genes allele frequency distribution should be considered in further pharmacogenetic studies.
Cytochrome P450 2D6 is involved in the metabolism of many drugs including those used in treatment of cancer and cardiac diseases [33]. Our study shows that the frequencies of genotypes and alleles rs1065852 of the CYP2D6 gene obtained for our sample of the Komi, Komi of the Izhma region, Veps, and Karelians are statistically different from the typical pan-European frequency values. The populations with remarkably similar results are of a particular interest, especially the Karelians and the Finn population [20], which are closely related culturally, linguistically, and geographically.
5. Conclusions
A worldwide study of rs1048943 in the CYP1A1 gene revealed an association of the G allele with prostate cancer [2] and colorectal cancer in Iraqi population [3]. Meta-analyses have shown this allele to be associated with colorectal cancer regardless of the ethnic origin [4], with laryngeal cancer in Asians (but not in Caucasians) [1], cervical cancer [5], and lung cancer in Indians [6]. The rs1065852 (TT) in the CYP2D6 gene is reported to be associated with worse prognosis for breast cancer in Asian women [7]. Thus, rs1048943 allele variants in the CYP1A1 gene represent the risk of developing the most common malignancies, both regardless of population origin (colorectal cancer) and for certain regions and specific ethnic groups. In this regard, it is relevant to study the distribution of alleles in different populations, which can aid early diagnostics and more accurately predict risks related to specific cancers. It can be assumed that such a distribution of alleles affects not only the risk of developing cancer, but also the effectiveness of chemotherapy and, therefore, the survival of patients. With further comparative studies of the malignant neoplasms in these populations, it is possible to predict the development of tumors and select the best pharmacotherapy for specific groups of patients.
It is also important that a large number of antipsychotics and antidepressants, as well as drugs used for COVID-19 treatment, are metabolized by polymorphic CYP2D6 enzymes. Thus, understanding the distribution of CYP2D6 alleles in the Finno-Permic populations will allow us to develop new approaches to treatment targeting particular ethnic groups.
This study of the main pharmacogenetic markers shows statistically significant difference between Veps and most of the studied populations in the rs4986774 locus of the CYP2D6 gene; as for the rs3892097 locus of the CYP2D6 gene, here we can see that Izhemsky Komis are different from the Mordovian and Udmurt populations.
Acknowledgments
We thank all the individuals who participated in this research work.
Author Contributions
Research concept and design, analysis and interpretation of data, writing and editing M.D.; statistical analysis and interpretation of data, writing and editing N.E.; writing original drift, R.M.; genotyping end interpretation of data, L.G.; genotyping and interpretation of data, Y.G.; genotyping and interpretation of data, A.N.; genotyping and interpretation of data, Y.V.; interpretation of data, D.P.; research concept and design, writing and editing, E.K. A critical review of the manuscript was carried out for important intellectual content. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Sampling was carried out in accordance with the ethical standards of the Bioethics Committee, developed by the WMA Declaration of Helsinki, “Ethical Principles for the Conduct of Medical Research Involving Human Subjects”. All subjects filled out a questionnaire taking into account nationality up to three generations and year of birth. All respondents signed an informed voluntary consent to participate in the study. The work was approved by the Local Ethics Committee of the Institute of Biochemistry and Genetics of the USC RAS (protocol No. 14 of 15 September 2016).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
The work was supported by the State Assignment of the Ministry of Science and Higher Education of the Russian Federation No. 075-03-2021-193/5 and was funded by the Ministry of Science and Education of the Republic of Bashkortostan (agreement no. 1, 2 December 2022) in the part of genotyping; by the Ministry of Science and Higher Education of Russian Federation, grant number 075-15-2021-595, in the part of collection of biological materials. This work has been supported by the grants the Russian Science Foundation, RSF 21-74-00104, in the part of statistical analysis.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zeng W., Li Y., Lu E., Ma M. CYP 1A1 rs1048943 and rs4646903 polymorphisms associated with laryngeal cancer susceptibility among Asian populations: A meta-analysis. J. Cell. Mol. Med. 2016;20:287–293. doi: 10.1111/jcmm.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoidy W.H., Jaber F.A., Al-Askiry M.A. Association of CYP1A1 rs1048943 Polymorphism with Prostate Cancer in Iraqi Men Patients. Asian Pac. J. Cancer Prev. 2019;20:3839–3842. doi: 10.31557/APJCP.2019.20.12.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ibrahem S.Q., Al-Dalawi Z.T., Bahaaldin A.S. Sequence Polymorphism in Xenobiotic Metabolising Genes in Iraqi Colorectal Cancer Patients. Asian Pac. J. Cancer Prev. 2021;22:1203–1210. doi: 10.31557/APJCP.2021.22.4.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu X., Wang Z., He J., Wang W., Xue W., Wang Y., Zheng L., Zhu M.-L. Associations between CYP1A1 rs1048943 A > G and rs4646903 T > C genetic variations and colorectal cancer risk: Proof from 26 case-control studies. Oncotarget. 2016;7:51365–51374. doi: 10.18632/oncotarget.10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sengupta D., Guha U., Mitra S., Ghosh S., Bhattacharjee S., Sengupta M. Meta-Analysis of Polymorphic Variants Conferring Genetic Risk to Cervical Cancer in Indian Women Supports CYP1A1 as an Important Associated Locus. Asian Pac. J. Cancer Prev. 2018;19:2071–2081. doi: 10.22034/APJCP.2018.19.8.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sengupta D., Banerjee S., Mukhopadhyay P., Mitra R., Chaudhuri T., Sarkar A., Bhattacharjee G., Nath S., Roychoudhury S., Bhattacharjee S., et al. A comprehensive meta-analysis and a case–control study give insights into genetic susceptibility of lung cancer and subgroups. Sci. Rep. 2021;11:14572. doi: 10.1038/s41598-021-92275-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu J., Li H., Guo P., Shen R., Luo Y., Ge Q., Shi W., Li Y., Zhu W. The effect of CYP2D6* 10 polymorphism on adjuvant tamoxifen in Asian breast cancer patients: A meta-analysis. OncoTargets Ther. 2017;10:5429–5437. doi: 10.2147/OTT.S149197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haduch A., Daniel W.A. The engagement of brain cytochrome P450 in the metabolism of endogenous neuroactive substrates: A possible role in mental disorders. Drug Metab. Rev. 2018;50:415–429. doi: 10.1080/03602532.2018.1554674. [DOI] [PubMed] [Google Scholar]
- 9.Hiemke C., Bergemann N., Clement H.W., Conca A., Deckert J., Domschke K., Eckermann G., Egberts K., Gerlach M., Greiner C., et al. Consensus guidelines for therapeutic drug monitoring in neuropsycho-pharmacology: Update 2017. Pharmacopsychiatry. 2018;51:9–62. doi: 10.1055/s-0043-116492. [DOI] [PubMed] [Google Scholar]
- 10.Gaedigk A., Sangkuhl K., Whirl-Carrillo M., Klein T., Leeder J.S. Prediction of CYP2D6 phenotype from genotype across world populations. Anesthesia Analg. 2017;19:69–76. doi: 10.1038/gim.2016.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kikuchi R., McCown M., Olson P., Tateno C., Morikawa Y., Katoh Y., Bourdet D.L., Monshouwer M., Fretland A.J. Effect of Hepatitis C Virus Infection on the mRNA Expression of Drug Transporters and Cytochrome P450 Enzymes in Chimeric Mice with Humanized Liver. Drug Metab. Dispos. 2010;38:1954–1961. doi: 10.1124/dmd.109.031732. [DOI] [PubMed] [Google Scholar]
- 12.Kirby G.M., Chemin I., Montesano R., Chisari F.V., Lang M.A., Wild C.P. Induction of specific cytochrome P450s involved in aflatoxin B1 metabolism in hepatitis B virus transgenic mice. Mol. Carcinog. 1994;11:74–80. doi: 10.1002/mc.2940110204. [DOI] [PubMed] [Google Scholar]
- 13.Fakhouri E.W., Peterson S.J., Kothari J., Alex R., Shapiro J.I., Abraham N.G. Genetic Polymorphisms Complicate COVID-19 Therapy: Pivotal Role of HO-1 in Cytokine Storm. Antioxidants. 2020;9:636. doi: 10.3390/antiox9070636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J.Y., Vinayagamoorthy N., Han K., Kwok S.K., Ju J.H., Park K.S., Jung S.H., Park S.W., Chung Y.J., Park S.H., et al. Association of polymorphisms of cytochrome P450 2D6 with blood hy-droxychloroquine levels in patients with systemic lupus erythematosus. Arthritis Rheumatol. 2016;68:184–190. doi: 10.1002/art.39402. [DOI] [PubMed] [Google Scholar]
- 15.Stevenson J.M., Alexander G.C., Palamuttam N., Mehta H.B. Projected Utility of Pharmacogenomic Testing among Individuals Hospitalized with COVID-19: A Retrospective Multicenter Study in the United States. Clin. Transl. Sci. 2020;14:153–162. doi: 10.1111/cts.12919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klementiev E.I., Shlygina N.V. Baltic-Finnish Peoples of Russia. Nauka; Moscow, Russia: 2003. p. 670. [Google Scholar]
- 17.Mokshin N.F., Fedyanovich T.P., Khristolyubova L.S. In: The Peoples of the Volga and Ural Regions. Mokshin N.F., Fedyanovich T.P., Khristolyubova L.S., editors. Nauka; Moscow, Russia: 2000. p. 579. Komi-Zyrians. Komi-Permyaks. Mari. Mordva. Udmurts. [Google Scholar]
- 18.Tambets K., Yunusbayev B., Hudjashov G., Ilumäe A.-M., Rootsi S., Honkola T., Vesakoski O., Atkinson Q., Skoglund P., Kushniarevich A., et al. Genes reveal traces of common recent demographic history for most of the Uralic-speaking populations. Genome Biol. 2018;19:139. doi: 10.1186/s13059-018-1522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathew C.G.P. The Isolation of High Molecular Weight Eukaryotic DNA. Methods Mol. Biol. 1985;2:31–34. doi: 10.1385/0-89603-064-4:31. [DOI] [PubMed] [Google Scholar]
- 20.The 1000 Genomes Project Consortium An integrated map of genetic variation from 1092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Veresh P.K. On the Issue of the Finno-Ugric Ancestral Home and the Ethnogenesis of the Hungarians in the Light of the Latest Paleontological Data. Studia Hungarica VI MKFU; Syktyvkar, Russia: 1985. [Google Scholar]
- 22.Kuzeev R.G. Peoples of the Middle Volga and Southern Urals: An Ethnogenetic View of History. Nauka; Moscow, Russia: 1992. p. 347. [Google Scholar]
- 23.Kh Khalikov A. Ancient History of the Middle Volga Region. Science; Moscow, Russia: 1969. p. 396. [Google Scholar]
- 24.Korytina G., Kochetova O., Akhmadishina L., Viktorova E., Victorova T. Polymorphisms of cytochrome P450 genes in three ethnic groups from Russia. Balk. Med. J. 2012;2012:252–260. doi: 10.5152/balkanmedj.2012.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dzhaubermezov M.A., Mingazheva E.T., Axberg D.S., Ekomasova N.V., Khusnutdinova E.K. Polymorphism of the cytochrome P450 CYP1A1 (Ile462Val) gene in the populations of Balkars and Karachays. Ecol. Genet. 2021;19:273–279. doi: 10.17816/ecogen71456. [DOI] [Google Scholar]
- 26.Nakachi K., Imai K., Hayashi S., Kawajiri K. Polymorphisms of the CYP1A1 and glutathione S-transferase genes associated with susceptibility to lung cancer in relation to cigarette dose in a Japanese population. Cancer Res. 1993;53:2994–2999. [PubMed] [Google Scholar]
- 27.Chen J., Cheng M., Li Y., Jiang C. Relationship between CYP1A1 genetic polymorphisms and renal cancer in China. Asian Pac. J. Cancer Prev. 2011;12:2163–2166. [PubMed] [Google Scholar]
- 28.Tiis R.P., Osipova L.P., Churkina T., Tabikhanova L., Lichman D.V., Voronina E.N., Filipenko M.L. The ILE462VAL polymorphism of the cytochrome P450 CYP1A1 gene in the Tundra Nenets of the Yamalo-Nenets Autonomous Okrug, Nganasans of the Taimyr Peninsula, and Russians of Siberia. Russ. J. Genet. Appl. Res. 2016;6:864–870. doi: 10.1134/S2079059716070133. [DOI] [Google Scholar]
- 29.Tabikhanova L.E., Osipova L.P., Churkina T., Voronina E.N., Filipenko M.L. Genetic polymorphism of CYP1A1 and CYP2D6 in populations of Buryats, Teleuts and Russians of Eastern Siberia. Vavilov J. Genet. Breed. 2018;22:205–211. doi: 10.18699/VJ18.348. [DOI] [Google Scholar]
- 30.Massabayeva M., Chaizhunusova N., Aukenov N., Bulegenov T., Apsalikov B., Shapihanova A., Zhunussov Y. Association of radiation risk in the second and third generations with polymorphisms in the genes CYP1A1, CYP2E1, GSTP1 and changes in the thyroid. Mol. Med. 2019;25:48. doi: 10.1186/s10020-019-0117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elhawary N.A., Nassir A., Saada H., Dannoun A., Qoqandi O., Alsharif A., Tayeb M.T. Combined Genetic Biomarkers Confer Susceptibility to Risk of Urothelial Bladder Carcinoma in a Saudi Population. Dis. Markers. 2017;2017:1474560. doi: 10.1155/2017/1474560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sindi I.A., Babalghith A.O., Tayeb M.T., Mufti A.H., Naffadi H., Ekram S.N., Elhawary E.N., Alanezi M., Elhawary N.A. Risk of Colorectal Carcinoma May Predispose to the Genetic Variants of the GST, CYP450, and TP53 Genes among Nonsmokers in the Saudi Community. Int. J. Gen. Med. 2021;14:1311–1323. doi: 10.2147/IJGM.S294802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin J., Johnson J.A. Pharmacogenetics of beta-blockers. Pharmacotherapy. 2007;27:874–887. doi: 10.1592/phco.27.6.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.