Abstract
A 65-kDa mannoprotein (CaMp65) has long been studied as a major, immunodominant antigen of the human opportunistic pathogen Candida albicans. An expression library of C. albicans was screened with serum from mice immunized with ScMp65 (ScW10), a Saccharomyces cerevisiae recombinant protein of about 48 kDa. This serum recognized the CaMp65 from a cell wall extract of C. albicans. After cloning and sequencing of the relevant C. albicans cDNA, an open reading frame encoding a protein of 379 amino acids was identified. Its deduced amino acid sequence showed regions of identity with all previously characterized tryptic fragments of CaMp65, as well as with the corresponding regions of ScMp65. A prepeptide of 32 amino acids with signal peptidase and Kex2 cleavage sites as well as a high number of potential O-glycosylation sites but no N-glycosylation sites or GPI anchor were observed in sequence studies of CaMp65. A putative adhesin RGD sequence was also present in the C-terminal region of the molecule. This triplet was absent in the ScMp65. The relevant gene (designated CaMP65) was localized to chromosome R of C. albicans as determined by pulse-field gel electrophoresis. Northern blot analysis demonstrated that gene transcription was heat inducible and associated with germ-tube formation by the fungus. A recombinant, His6-tagged protein (rCaMp65) was expressed in Escherichia coli under an inducible promoter. After purification by nickel-chelate affinity chromatography, the recombinant product was detected as a 47-kDa protein band in immunoblots with the anti-ScMp65 serum, as well as with CaMp65-specific monoclonal antibodies. Both ScMp65 and CaMp65 were assayed for antigenic stimulation in cultures of peripheral blood mononuclear cells (PBMC) from 10 unselected human donors. While ScMp65 was substantially nonstimulatory, both rCaMp65 and the native CaMp65 were equally able to induce lymphoproliferation of the PBMC from all the donors. In addition, a number of CD4+ T-cell clones were generated using a C. albicans mannoprotein fraction as an antigenic stimulant. Several of these clones specifically responded to both the native and the recombinant C. albicans Mp65 but not to ScMp65. Thus, the recombinant Mp65 of C. albicans retains antigenicity and, as such, could be a valid, standardized reagent for serodiagnostic and immunological studies.
Despite the recognized importance of cell-mediated immunity (CMI) in the protective response against the human opportunistic fungus Candida albicans (33, 34), few antigenic targets of this response have so far been characterized. They include heat shock proteins, enolase, and a number of as-yet-uncharacterized mannoproteins, some with adhesive function (1–3, 6, 7, 9, 11, 15–18, 26, 36). The identification of these antigens and an understanding of the mechanisms whereby they elicit and regulate CMI is an obvious prerequisite for generation of molecules with potential immunoprophylactic or immunotherapeutic activity, or even for use as immunodiagnostic reagents.
We have studied a 65-kDa mannoprotein (here designated CaMp65) of C. albicans, structural and secreted, component of the fungus. It is particularly observed in extracellular fractions of hyphal cells (4, 10, 20, 37–39). Our major interest in this mannoprotein resides in its recognition by peripheral blood T cells of practically all healthy subjects tested (9, 10, 38, 39). In experimental models of disseminated murine candidiasis, animals vaccinated with a low-virulence Candida strain generated a strong and protective CMI which was promptly revealed by CaMp65 stimulation of splenocytes in vitro, as well as by a delayed-type hypersensitivity response to this antigen in vivo (10, 27). Moreover, a moderate yet significant level of protection was conferred upon mice by immunization with a mannoprotein fraction containing CaMp65 as a major antigenic, CMI-inductive component (27–28). This protection was clearly enhanced by the concomitant administration of interleukin-12 (IL-12) as an adjuvant (10).
Because of these interesting and potentially useful properties, we have recently addressed the biochemical characterization of CaMp65. A strong homology at the protein level was found between this protein and the glucanase or transglycosidase family of cell wall proteins of Saccharomyces cerevisiae (8, 20, 21). Interestingly, the least similarity between CaMp65 and the yeast proteins was found in the most antigenic peptides of the N-terminus regions of CaMp65 (21). The availability of the amino acid sequences of several tryptic and chymotryptic fragments of CaMp65 and the established sequence homology with S. cerevisiae cell wall proteins have allowed us to clone the relevant genes of both C. albicans and S. cerevisiae and to express them in Escherichia coli. Here we characterize the cloned products and show that the recombinant protein of C. albicans induces in vitro an intense CMI response by peripheral blood mononuclear cells (PBMC) of healthy subjects and derived CD4+ T-cell clones, with a magnitude comparable to that previously shown by the native antigen (10, 20, 37). These data indicate that CaMp65 provides a suitable reagent for studies of Candida-specific CMI generation and its role in the anti-Candida defense.
MATERIALS AND METHODS
Microrganisms, growth conditions, and mannoprotein extract.
C. albicans strain BP (3, 4) was used throughout this study. It was grown in YPD (2% glucose, 1% yeast extract, 2% Bacto Peptone; Difco, Detroit, Mich.), Winge (0.3% yeast extract, 0.2% glucose; Difco), or modified Lee (4, 31) media, as specified in single experiments. E. coli XL1-Blue cells [endA1 hsdR17 supE44 thi1 recA1 gyrA96 relA1 Δlac (F′ probAB lacIqZΔM15, Tn10) and M15 (Nals Strs Rifs Δlac− ara− gal− mtl F′ recA+ uvr+)(pUHA1)] were used as host strains for recombinant plasmids, while E. coli HL1-Blue MRF′ (ΔmcrA183 ΔmcrCB-hsdSMR-mrr173 endA1 supE44 thi1 recA1 gyrA96 relA1 Δlac [F′ proAB lacIqZΔM15 Tn10]) and SORL (e14− mrcA ΔmcrCB-hsdSMR-mrr171 sbsC recB recJ umuC::Tn5 uvrC endA1 Δsu thi1 gyr96 relA1 Δlac [F′ proAB lacIqZΔM15]) cells were the host strains for bacteriophage λZAPII. E. coli cells were usually grown in L broth (1% tryptone, 0.5% yeast extract, 0.5 NaCl; pH 7.0), Luria-Bertani (LB) plates (1% tryptone, 0.5% yeast extract, 0.5 NaCl, 1.5% agar; pH 7.0) or top agarose (1% tryptone, 0.8% NaCl, 0.6% agarose; Boehringer, Mannheim, Germany) supplemented when necessary with ampicillin (100 μg/ml), kanamycin (50 μg/ml), or tetracycline (12.5 μg/ml) (Boehringer).
The mannoprotein fraction MPF2 was obtained by ethanol precipitation followed by gel filtration of a crude extract of autoclaved cells, as reported elsewhere (37).
Oligonucleotides and PCR.
Ca33, Ca34, Ca64, and Ca65 oligonucleotides were purchased from Pharmacia. Their sequence and specificity are shown in Table 1. PCR reactions with S. cerevisiae or C. albicans purified DNA were performed as previously described (26) by using primer pairs Ca33-Ca34 and Ca64-Ca65, respectively. Briefly, the reactions were carried out on a Gene AMP PCR System 9600 Apparatus (Perkin-Elmer/Cetus Corp., Norwalk, Conn.) in a volume of 100 μl containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2 mM MgCl2, 200 μM concentrations of each deoxynucleotide, 50 pmol of each primer, 1 U of Taq polymerase (Perkin-Elmer), and 100 ng of genomic DNA template. The PCRs were done in three steps of 60 s at 94°C, 60 s at 60°C, and 120 s at 72°C (25 cycles).
TABLE 1.
Sequence and localization of Ca oligonucleotides used for molecular cloning of CaMP65 and Scw10
| Oligonucleotide | Sequencea | Gene | Positionsb |
|---|---|---|---|
| Ca33 | CCCGGATCCATGCGTTTTTCAAATTTCC | Scw10 | 1–19 |
| Ca34 | CCCAAGCTTAATCACTTGATAGAATACC | Scw10 | 1168–1150 |
| Ca64 | AACGGATCCATGTTATTCAAGTCTTTC | CaMP65 | 1–18 |
| Ca65 | GGGCTGCAGGTGCTTAGTTAGAGTAA | CaMP65 | 1144–1128 |
The underlined sequences are flanking noncomplementary regions containing the restriction site (in boldface) used to clone the gene in the expression vector.
For both primers, position 1 is the first base of the start codon and position 1168 and 1140 are the third base of the stop codons of S. cerevisiae Scw10 and C. albicans MP65, respectively.
Generation of a mouse antiserum against recombinant S. cerevisiae MP65.
PCR amplification of S. cerevisiae genomic DNA was accomplished by using the Ca33 and Ca34 primers (Table 1). The PCR product, after digestion with BamHI and HindIII, was cloned into the BamHI/HindIII polylinker sites of the expression vector pDS56/RBSII6xhis/E− to give pRLV126 (26, 32), which was used to transform E. coli M15 carrying the lack repressor-producing pUHA1 plasmid (22). To obtain a recombinant His6-ScMp65 protein, induction was performed in LB medium containing kanamycin and ampicillin, by adding isopropyl-β-d-thiogalactopyranoside (IPTG; Boehringer) at a final concentration of 1 mM to a culture with an optical density at 600 nm (OD600) of 0.6, followed by an additional 5-h incubation at 37°C. The recombinant His6-ScMp65 protein was purified as previously described (26, 35). A hyperimmune murine serum against the purified ScMp65 was obtained by immunizing CD2F1 mice (18 to 21 g) with four intraperitoneal injections at weekly intervals of 10 μg of the recombinant product in complete (the first two) and incomplete (the last two) Freund adjuvant. The serum obtained had a titer of >1,280, as determined by an assay (performed as described in reference 3) with 1 μg of the recombinant protein used as coating antigen.
cDNA synthesis.
Poly(A)+ RNA was isolated from total RNA of C. albicans (grown as hyphae for 24 h in Lee medium [4]) by an Invitrogen Micro-Fast Track mRNA Isolation Kit (Leek, The Netherlands) (26), according to the manufacturer's instructions. Reverse transcription was performed using the Stratagene ZAP-cDNA synthesis kit (La Jolla, Calif.) as described by the manufacturer.
C. albicans ZAPII cDNA library.
Purified cDNA of C. albicans (see above) was ligated with dephosphorylated EcoRI-XhoI λZAPII vector arms and incubated with in vitro packaging extracts (Stratagene), according to the manufacturer's instructions. Recombinant phage particles were amplified by preparing plate lysates with E. coli strain XL1-Blue MRF′, yielding 3.5 × 105 plaques. The amplified library (initial density, 40,000 plaques/13-cm plate) was screened with a murine antiserum generated against ScMP65 recombinant protein (see above).
Cloning and sequencing of CaMP65 gene.
CaMP65 was subcloned from the λZAPII recombinant phage library into pBluescript by infecting E. coli SORL cells (Stratagene) as described by the manufacturer. Double-stranded dideoxy sequencing of recombinant plasmids was performed with the Sequenase Kit (USB, Cleveland, Ohio) using primers flanking the polylinker region of the vector and various internal CaMP65 primers. The cDNA sequence was compared to sequences in the Saccharomyces Genome Database (Stanford University) by using the BLAST search. The EMBL database accession no. was AJ010064.
Southern blot analysis.
Genomic DNA of C. albicans was restricted and hybridized with the full-length CaMP65 probe essentially as previously described (19, 26, 32). Briefly, the DNA was digested with the restriction endonucleases EcoRI, BamHI, HindIII, BglII, and PstI (Boehringer), separated by agarose gel electrophoresis, and transferred onto nitrocellulose transblot membranes (Bio-Rad, Hercules, Calif.). The blotted material was hybridized with 32P-labeled randomly primed (Boehringer) CaMP65 full-length cDNA insert. Hybridization and initial washing steps were carried out as described previously (26). Filters were exposed on 3M (St. Paul, Minn.) XDA Plus film, with 3M Trimax screens, at 80°C.
Northern blot analysis of CaMP65 transcription.
Total RNA from C. albicans cells grown under the conditions specified later in the text was isolated by the proteinase K method as previously described (26, 32). Approximately 5 μg of RNA per lane was run on denaturing 1.5% formaldehyde-agarose gel, transblotted to nitrocellulose filters, and hybridized with randomly prime labeled full-length CaMP65 or Act1 (25, 26) probes. Hybridization and washing were done as described elsewhere (26), with the final stringent washing step carried out in 0.1% SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) at 70°C for 30 min.
Generation of a mouse antiserum against the purified recombinant CaMP65.
CaMP65 was generated by PCR amplification of the C. albicans genomic DNA using the Ca64 and Ca65 primers (Table 1). The PCR product, after digestion with BamHI and PstI, was cloned into BamHI/PstI sites of the expression vector pDS56/RBSII6xhis/E− to give pRLV130. Expression and purification of the recombinant CaMp65 (rCaMp65), as well as generation of a mouse antiserum against the purified C. albicans protein, were performed as described above for rScMP65.
Chromosome separation and hybridization.
The general procedure described by Vollrath and Davis (40), as summarized in previous reports (13, 26), was used to prepare DNA samples for pulsed-field electrophoresis and karyotype determination by contour-clamped homogenous electrophoresis field (CHEF) analysis. The electrophoretic analysis was performed with CHEF-DRII apparatus (Bio-Rad). Each gel (14.5 by 20.5 cm; 1-cm thick; 1% agarose) containing the agarose inserts of DNA was immersed in running buffer (50 mM Tris-HCl, 50 mM boric acid, 1.5 mM EDTA; pH 8.2) and run for 54 h at 150 V and 14°C with 90- to 325-s switches and a rotation angle of 120°. Gels were stained with ethidium bromide (0.5 mg/ml; 30 min), destained, and photographed under UV light. Hybridization was performed as described for Southern blot analysis, except that the labeled 5′ CaMP65 fragment of about 700 bp obtained by digestion of the pRLV213 with BamHI and HindIII endonucleases was used as a probe.
Immunoblotting.
Recombinant proteins from IPTG-induced and noninduced M15 (pUHA1 and pRLV130) or M15 (pUHA1 and pRLV126) cells, their purified counterparts, and cell wall extracts or secretory antigenic mannoprotein (SAM) from hyphal cells of C. albicans (4; see also below) were resuspended in sample buffer, at approximately 1 mg of protein/ml, boiled for 10 min, and subjected to 5 to 15% gradient-polyacrylamide gel eletrophoresis (PAGE). The electrophoresed materials were blotted onto nitrocellulose filters in a buffer containing 25 mM Tris, 192 mM glycine, 0.1% SDS, and 20% methanol. Filters were incubated with antibodies as described in specific experiments. In all cases, nonspecific binding of antibodies to nitrocellulose was prevented by blocking the filters with 1% bovine serum albumin in phosphate-buffered saline (PBS) for 2 h at room temperature (5). After an extensive washing with PBS, bound antibodies were detected by alkaline phosphatase-conjugated second antibodies, as described in specific experiments.
Whole-cell extracts of C. albicans grown at 22°C in Lee medium and after a shift to 37°C for 3 h were obtained by cell breakage with 0.1-mm glass beads and adsorption of mannan on a concanavalin A resin as described elsewhere (9, 26). A secreted mannoprotein-rich preparation of hyphal cells of C. albicans was obtained as described by Bromuro et al. (4).
Lymphocyte proliferation assay.
The procedure previously described by Ausiello et al. (1, 2) and by Torosantucci et al. (37–39) was used. Briefly, PBMC obtained from heparinated, venous, peripheral blood samples of healthy adult blood donors were washed twice and resuspended in RPMI medium (GIBCO) supplemented with 5% pooled AB serum and antibiotics (penicillin, 100 IU/ml; streptomycin, 0.1 mg/ml; GIBCO), hereafter referred to as complete medium. PBMC proliferation was measured by incubating 2 × 105 PBMC cells/well in 0.2 ml of complete medium in 96 flat-bottom microwell trays (3072 Falcon; Becton Dickinson) in triplicate in the presence of the relevant stimulants. The plates were incubated at 37°C in 5% CO2 and harvested after 7 days. Then, 0.5 μCi of [methyl-3H]thymidine (Amersham; specific activity, 2.5 Ci/mmol) was added to the culture 18 h before cell harvesting, and DNA synthesis was evaluated by measuring [3H]thymidine incorporation (3). The magnitude of lymphoproliferation was estimated by reference to the PBMC culture incubated in the absence of the antigen by calculation of the stimulation index (SI). Donors were arbitrarily classified as high, moderate, and low responders to the Candida antigen (MP65) if their SI, at an antigen concentration of 1 μg/ml, was >100, between 10 and 50, and <10, respectively. In addition, the high responders to the Candida recombinant mannoprotein were characterized by an appreciable lymphoproliferative response at as low a dose of antigen as 10 ng/ml. All reagents were of standard immunological laboratory grade and did not contain lipopolysaccharide, as verified by Limulus amebocyte assay. No lymphocyte culture or clone (see below) proliferated in the absence of mitogen or antigen.
Human CaMp65-reactive T-lymphocyte clones.
PBMC from a Candida antigen high responder were cultured in complete medium in the presence of a previously characterized mannoprotein fraction (MPF2) (9, 10, 27). After 5 days, 10 U of rIL-2 per well was added to the cultures. After 4 additional days, cells were cloned by limiting dilution at 3, 1, and 0.3 cells per well, as previously described (30). After 10 to 15 days, growing cultures were expanded and finally tested for MPF2 specificity in a proliferation assay using irradiated autologous PBMC as antigen-presenting cells (APC) with or without antigen at 10 μg/ml. MPF2-specific clones were expanded and maintained in culture with 25- to 30-day cycles of restimulation with phytohemagglutinin (PHA) and irradiated PBMC. Their phenotype was determined by fluorescence-activated cell sorting cytometry as previously described (30).
RESULTS
Molecular cloning, karyotype assignment, and expression of C. albicans Mp65 gene.
The amino acid sequences of tryptic and chymotryptic fragments of native CaMp65 were highly homologous to the deduced Scw10 and Scw4 proteins encoded by the S. cerevisiae open reading frames (ORFs), designated YMR305C and YGR279, respectively (Saccharomyces cerevisiae Genome Database) (8). On this basis, the YMR305C ORF was cloned by PCR using the Ca33 and Ca34 oligonucleotide primers containing the initiation and stop codons of the gene (Table 1). Molecular analysis of the amplified product (1,168 bp) confirmed the sequence of YMR305C DNA of S. cerevisiae. This fragment was cloned into an inducible E. coli expression vector to produce a recombinant His6-tagged protein (designated rScMp65). A mouse serum raised against this recombinant product was able to recognize, besides the immunogen itself, the native MP65 of C. albicans in a cell wall extract of the fungus (Fig. 1). The antiserum against rScMp65 was therefore used to screen a λZAPII expression library of C. albicans, as described in Materials and Methods. Six independent clones were isolated and purified. Molecular analysis of these clones displayed sequence identity. The clone (clone CaRLV213, 1,415 bp) containing both start and top codons of an ORF of 1,140 bp (designated CaMP65) was selected for further studies. By using the BLAST search, the nucleotide sequence of this clone was found to be 98 and 96% identical to contigs 5-2183 and 5-2970, respectively, of the Stanford Candida Genome Database.
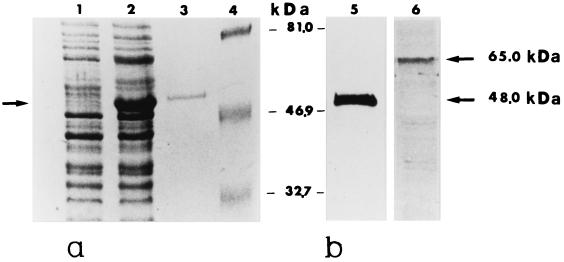
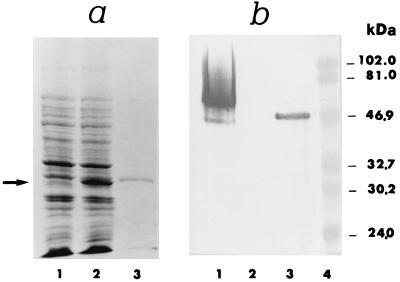
FIG. 1.
(a) Expression, purification, and immunoblots of the recombinant ScMp65 protein. Cells of E. coli M15 (pUHA1 and pRLV126) either noninduced (lane 1) or IPTG induced (lane 2) were extracted and subjected to SDS-PAGE, together with the recombinant ScMp65 (nickel-nitrilotriacetic acid [Ni-NTA] affinity chromatography purified) (lane 3). The gels were stained with Coomassie blue. Lane 4 shows the molecular mass standards (in kilodaltons). (b) Immunoblots of ScMp65 (lane 5) or of a cell wall extract of C. albicans (lane 6) with a mouse antiserum raised against the purified ScMp65. A total of 1 μg of the purified protein or 10 μg of cell wall extract was run, electrotransferred to nitrocellulose membranes, and immunoblotted with the anti-ScMp65 serum (dilution, 1:800). The reaction was developed with alkaline phosphatase-conjugated anti-mouse (1:5,000) immunoglobulin G. The arrows to the right side of lane 6 point to the apparent molecular mass of the ScMp65 in lane 5 and its correspondent of the natural Mp65 of C. albicans in lane 6.
The amino acid sequence of the deduced CaMp65 showed regions identical or similar to all previously characterized tryptic fragments of the native MP65 protein (21) and, of course, with the corresponding regions of the Scw10 protein (Fig. 2). Both contain a prepeptide (32 and 29 amino acids [aa] in C. albicans and S. cerevisiae, respectively) with a signal sequence (aa 17 in CaMp65 and aa 21 in Scw10), as well as a Kex2 peptidase site (KR) where the protein is cleaved for secretion. Both proteins are also characterized by a high number of potential O-glycosylation sites, in particular a long serine-rich stretch corresponding to aa 96 to 120 in CaMp65. One potential N-glycosylation site (aa 279) was present in the Scw10 but not in CaMp65, thus confirming the sequence analysis of trypsin and chymotrypsin digests of the native mannoprotein (21). No GPI anchor sequence was present in CaMp65, which contained an RGD site (aa 323 to 325) which was absent in the Scw10 protein.
FIG. 2.
Alignment of the predicted amino acid sequences of CaMp65 (upper) and Scw10 (lower). Since the two proteins had different lengths, it was necessary to introduce gaps (dashes) for sequence alignment. The amino acid is given in the single-letter code and the symbols (∗, ., :) denote the identity or decreasing order of similarity between the corresponding pairs of amino acids. The arrows point to the experimentally determined N-terminal sequence of the two mature proteins (8, 20).
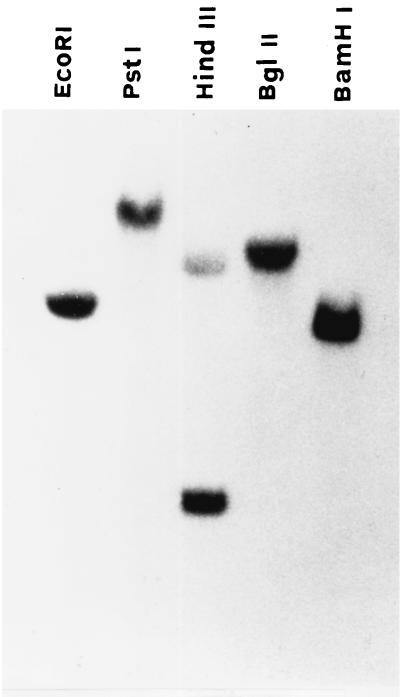
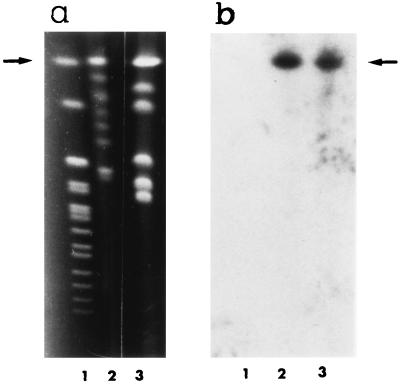
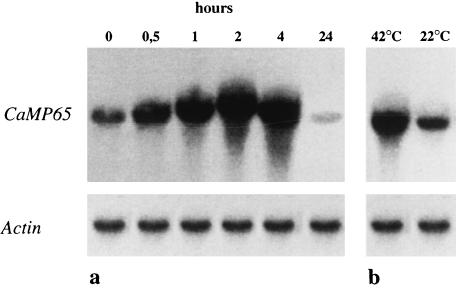
Southern blotting of C. albicans chromosomal DNA digested with various restriction endonucleases and hybridized with the full-length CaMP65 cDNA insert, under high-stringency conditions, is shown in Fig. 3. The presence of two bands in the HindIII lane would appear to be justified by the presence of two sites (nucleotides 701 and 725 in the codifying sequence) for this endonuclease in CaMP65. With CHEF analysis, the ethidium bromide-stained chromosome-sized bands of C. albicans (strain BP and those of a recent clinical isolate, SA-247) showed a different pattern, mostly in the area encompassing the chromosomal bands lower than 2.5 Mb (Fig. 4a). Nonetheless, in both strains, the molecular probing in Southern blots with the CaMP65 cDNA gave a hybridization signal on the 3.8-Mb R1 chromosome-sized band (Fig. 4b). Northern blotting of mRNA from yeast cells incubated at 37°C in Lee medium (Fig. 5a) demonstrated induction of CaMP65 gene transcription, with a maximum after 2 h of incubation corresponding to full germ tube formation. Moreover, as shown in Fig. 5b, transcription was higher at 42°C than at 22°C when the cells were grown, under yeast form, in YPD medium for 30 min.
FIG. 3.
Southern blot analysis of genomic DNA of C. albicans digested with the indicated enzymes and hybridized with the CaMp65 full-length probe. For technical details, see Materials and Methods.
FIG. 4.
Chromosome mapping of the CaMP65 locus. Chromosome-sized bands of C. albicans DNA were separated by CHEF analysis as described in Materials and Methods. (a) CHEF karyotype of C. albicans strain BP (lane 2) or of a fresh clinical isolate (SA-247, laboratory code) (lane 3). Lane 1, S. cerevisiae chromosomal bands. (b) Autradiographs of the blots probed with 5′ 700-bp fragment of CaMP65 cDNA under high-stringency conditions. For details, see Materials and Methods.
FIG. 5.
CaMP65 gene expression in C. albicans cells grown for different time intervals at 37°C in Lee medium (a) or following a 30-min temperature shift from 22 to 42°C in YPD medium (b). The nitrocellulose filter containing the blotted RNA was hybridized under high-stringency conditions with the full-length CaMP65 probe or with the Act1 gene, as indicated. For further details, see Materials and Methods.
Generation of recombinant CaMp65 protein.
pRLV130-transformed E. coli cells were cultured in antibiotic LB broth, and total cell protein extracts, under induced or uninduced conditions, were subjected to SDS-PAGE, Coomassie blue staining, and Western blotting with various antibodies. As shown in Fig. 6a, a novel, prominent protein was detected in the extract of induced E. coli, as revealed by Coomassie blue staining. Figure 6b shows the positive reaction of this recombinant protein (with an apparent molecular mass of ca. 47 kDa) and of the MP65-rich secretion from the hyphal cells, but not of the ScMP65, with the anti-MP65 monoclonal antibody 4C8. As expected from previous data (20), most of the reactivity of the monoclonal antibody 4C8 with the mycelial secretion was detected in a molecular mass region roughly corresponding to 65 kDa, i.e., the native glycosylated Mp65 of C. albicans (Fig. 6b). The recombinant product was also reactive with another monoclonal antibody (7H8) raised against the native CaMp65 and was also recognized by the antiserum raised against the purified recombinant ScMp65 (data not shown).
FIG. 6.
Expression, purification, and immunoblot of the rCaMp65 protein. (a) Cells of E. coli M15 (pUHA1 and pRLV130) either noninduced (lane 1) or induced with IPTG (lane 2) were extracted and subjected to SDS-PAGE. The same was done with the His6-Mp65 (rCaMp65), Ni-NTA affinity chromatography-purified polypeptide (lane 3). The gels were stained with Coomassie blue. (b) Immunoblot of the secretory mannoprotein material (SAM; see also reference 4) from hyphal cells of C. albicans (lane 1), purified rScMP65 (lane 2), and purified rCaMP65 (lane 3). Each purified polypeptide (1 μg) or SAM (20 μg) was electrophoresed, electrotransferred to nitrocellulose membranes, and immunoblotted with the anti-CaMp65 monoclonal antibody 4C8 (dilution, 1:2,500). The reaction was developed by phosphatase-conjugated anti-mouse (1:7,500) immunoglobulin G. Lane 4 shows the molecular mass standards. The arrow in panel a points to the IPTG-induced rCaMP65.
Antigenicity of the recombinant CaMp65.
The recombinant CaMp65 and ScMp65, together with the native Mp65 of C. albicans and control antigen, were tested for reactivity with PBMC from healthy human subjects. Table 2 shows the lymphoproliferation data from three blood donors with different degrees of responsiveness to the antigenic stimulation out of the 10 examined with similar results. The recombinant protein of C. albicans was roughly as effective, at equal doses, as the native protein of the fungus. In particular, no subject tested was unresponsive to the recombinant, as well as to the native candidal protein, and the two antigens induced comparable responses in high (donor 1)-, intermediate (donor 2)-, and low (donor 3)-responder subjects (Table 2). Remarkably, in the high-responder donors, low doses of rCaMp65 (10 ng/ml) were still effective at inducing lymphoproliferation. The recombinant protein of S. cerevisiae was minimally effective in stimulating cell proliferation even in the high-grade responder shown in Table 2 (see also below).
TABLE 2.
PBMC proliferation induced by natural Mp65 of C. albicans and recombinant proteins of C. albicans and S. cerevisiae
| Antigenic or mitogenic stimulant | Concn (μg/ml) | PBMC proliferationa (mean cpm ± SD)
|
||
|---|---|---|---|---|
| Donor 1 (high responder) | Donor 2 (intermediate responder) | Donor 3 (low responder) | ||
| None | 1,000 ± 600 | 300 ± 70 | 1,400 ± 600 | |
| PHA | 3 | 35,600 ± 1,700 | 7,400 ± 3,400 | 26,400 ± 3,800 |
| SAMb | 1 | 116,100 ± 10,900 | 23,600 ± 1,400 | 41,400 ± 16,000 |
| 0.1 | 45,600 ± 26,200 | 15,000 ± 3,300 | 8,200 ± 1,000 | |
| Mp65 (native)c | 1 | 123,300 ± 400 | 13,500 ± 800 | 12,400 ± 2,800 |
| 0.1 | 103,400 ± 5,900 | 7,900 ± 800 | 3,300 ± 2,400 | |
| 0.01 | 9,100 ± 4,700 | 800 ± 300 | 1,800 ± 900 | |
| CaMp65d | 1 | 108,600 ± 6,600 | 28,500 ± 3,300 | 9,700 ± 4,900 |
| 0.1 | 51,300 ± 5,100 | 24,300 ± 2,000 | 3,900 ± 2,400 | |
| 0.01 | 5,500 ± 660 | 900 ± 100 | 1,100 ± 200 | |
| ScMp65d | 1 | 7,600 ± 4,300 | 8,500 ± 5,800 | 2,500 ± 900 |
| 0.1 | 4,900 ± 4,700 | 3,000 ± 1,400 | 600 ± 300 | |
| 0.01 | 1,200 ± 400 | 500 ± 200 | ||
Evaluated as [3H]thymidine incorporation and expressed as counts per minute. The PBMC donors were arbitrarily classified as high, moderate, and low responders as described in Materials and Methods.
SAM is the secretory mannoproteic material from hyphal cells (see reference 4), with concentration expressed as polysaccharide.
Affinity chromatography purified (20, 21), with concentration expressed as protein.
Recombinant proteins produced in E. coli and purified by Ni-NTA affinity chromatography.
Generation of CaMP65-specific human T-cell clones.
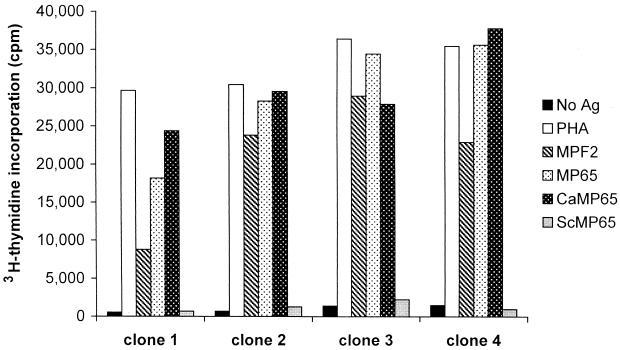
Forty-five T-cell clones were generated from a high-responder subject to a mannoprotein fraction (MPF2) of C. albicans. All of them were of the CD4+ T-cell phenotype. Several of these clones were specific for CaMp65. Figure 7 shows the proliferation of four representative clones following activation with different stimuli in the presence of irradiated autologous PBMC. All of the clones proliferated when stimulated with MPF2 and showed a remarkable increase in the [3H]thymidine incorporation following stimulation with CaMp65. The proliferation in the presence of rCaMp65 was of the same order of magnitude as that obtained with the native mannoprotein. With both the native and the recombinant Candida antigens, the values of [3H]thymidine incorporation were similar to those of PHA-stimulated clones. Importantly, none of the CaMp65-reactive clones responded to the recombinant ScMp65.
FIG. 7.
Proliferation of four representative human T-cell clones specific for MPF2. Each clone was incubated with autologous irradiated PBMC as APC alone or in the presence of PHA, MPF2, native CaMp65, recombinant CaMp65, or recombinant ScMp65. After 2 days of culture, clones were incubated with [3H]thymidine and harvested 18 h later. Results are expressed as the mean counts per minute of triplicates.
DISCUSSION
In this study, data on molecular cloning, expression, and characterization of recombinant Mp65, a protein which has long been identified as a major target of CMI response against the human opportunistic fungus C. albicans (10, 20, 21, 37–39), have been presented. Instrumental to this (and as an immunogenicity control), a recombinant protein of S. cerevisiae (rScmp65), corresponding to the Scw10 protein previously characterized by Cappellaro et al. (8), was also generated. A serum against this protein was raised in mice and used to screen a C. albicans library for homologous genes.
Besides sequence homology with previously characterized proteolytic fragments of natural Mp65 (21), the recombinant product of C. albicans (but not that of S. cerevisiae) reacted in Western blots with two monoclonal antibodies raised against the purified natural product, demonstrating that, as previously suggested (21), these antibodies recognize a peptide rather than a saccharide epitope of C. albicans protein. Moreover, a mouse serum from animals immunized with the S. cerevisiae recombinant product specifically reacted in immunoblots with the natural C. albicans Mp65. Altogether, these data leave little doubt that the recombinant product fully corresponds to a nonglycosylated antigenically functional form of C. albicans mannoprotein (see also below).
Previous biochemical characterization of the native CaMp65 indicated a substantial glycosylation of this protein, occurring as acid-labile O side chains with numerous serine and threonine residues, which together accounted for about 20% of the whole amino acid composition (20, 21). The deduced amino acid sequence of CaMP65 cDNA, as well as the determination of the molecular mass of the recombinant product, indicates a molecular mass of about 45 kDa; thus, glycosylation of the natural product would account for about one-third of its mass. Previous findings indirectly suggested that most of the glycosylation occurs at a long protein stretch where repeated serine-threonine sequence blocks were present (21). This was confirmed here by deducing the amino acid sequence of the cloned gene, a sequence which also confirmed the lowest homology between the two proteins in the immunodominant N-terminal region of CaMp65 (see also below). In contrast to the homologous protein of S. cerevisiae (8), no potential N-glycosylation site was present in the Mp65 deduced amino acid sequence. This confirms the previous determination (21) of the amino acid sequence of tryptic and chymotryptic fragments of the natural Mp65, inclusive of the region containing the potential N-glycosylation triplet of S. cerevisiae (NYS; aa 279). It is also in keeping with the resistance of natural Mp65 to digestion with endo-N and -F glycosidases (20, 38). Overall, the natural C. albicans Mp65 appear to be only O glycosylated, as in some other fungal mannoproteins (23). The presence of a prepeptide with a signal sequence for processing in the endoplasmic reticulum, as well as with the KR doublet of a Kex-like peptidase before the amino terminus start, clearly confirms the nature of a secretory mannoprotein which was already inferred from functional studies (3, 4).
Another interesting and perhaps functionally relevant difference between CaMp65 and Scw10-Scw4 proteins (8) is the presence of a RGD site in the C. albicans mannoprotein. This site has repeatedly been shown to characterize some integrin-like proteins of C. albicans, including a complement-like receptor of the fungus. In this context, RGD-containing molecules have been implicated in the increased adherence of hyphal cells of C. albicans to plastic and animal cells (6). One of these adhesins was previously characterized as a 60- to 65-kDa protein but its sequence was not determined (7). The data would suggest a role of “secretory adhesin” for CaMp65, and we are currently working on determining the functional features of this mannoprotein.
CaMp65 and the closest homologous Scw10 probably belong to a family of enzymes of glucan metabolism (glucanases or transglycosidases) (8). Since both rCaMp65 and rScMp65 have been here expressed under denaturing conditions, they could not be tested for their possible enzymatic activity. However, a number of observations suggest that these kinds of proteins are indeed involved in cell wall metabolism. The natural Mp65 could be preferentially purified from the hyphal secretory materials under tunicamycin inhibition of glycosylation (21), and much more immunogenic Mp65-containing, secretory mannoprotein material was obtained from hypha- than from yeast-growing cells (4, 37). The higher CaMP65 gene transcription at 37 to 42°C than at 22 to 28°C is more in keeping with the conditions favoring mycelial rather than yeast cell growth of C. albicans (31). However, more specific and quantitative expression studies are required before drawing a definitive conclusion regarding this.
rCaMp65 has been shown here to be highly antigenic and immunogenic. As with the natural mannoprotein, it was able to stimulate in vitro proliferation of lymphocytes from all of the normal subjects tested so far. Also, the magnitude of the proliferative response was comparable to that recorded in the same subjects under stimulation with the natural product. Moreover, the recombinant product was as efficient as the native mannoprotein in stimulating specific T-cell clones, indicating that the recombinant protein gains access to the endocytic compartment of the APC and that it is processed to generate the same peptides generated by the processing of the natural protein. Interestingly, the recombinant product of S. cerevisiae was much less effective at inducing, if not totally unable to induce, PBMC lymphoproliferation. Despite an overall rather large homology between the two proteins, there are several peptide regions of CaMp65 where this homology is rather low. Strong proliferation-inducing peptide fragments of the native CaMp65 were among those showing the least similarity with the corresponding peptide sequences of S. cerevisiae homologs Scw10 and Scw4 (21). These highly antigenic MP65 epitopes were found in the N-terminus region of CaMp65, suggesting that, as happens for other O-linked, saccharide-rich cell wall proteins of C. albicans (11), the N-terminal region of the mannoprotein is the one more likely to be exposed on the cell surface or accessible to immunocompetent cells. This idea is clearly in line both with the immunodominance of the molecule and with its potential adhesive role. In agreement with the data obtained with PBMC proliferation, none of the CaMp65-specific clones was stimulated by ScMp65. These data confirm that the immunogenic epitopes of CaMp65 are enriched with those regions of the protein sharing the least homology with the S. cerevisiae mannoprotein.
Thus, this C. albicans putative glucanase protein seems to have acquired rather unique antigenic properties among members of this family of cell wall proteins, in keeping with its cell wall expression, and human commensalism of the fungus. Interestingly, this marked immunogenicity seems to be preferentially addressed to the cell-mediated arm of host immune response, since normal human sera, which are usually rich in anti-Candida antibodies (31), show rather low reactivity with the natural or the recombinant Mp65 protein (unpublished data).
Several data have long suggested that peptide epitopes of the highly immunogenic mannoproteins of C. albicans were those inducing and revealing CMI (9, 10, 16, 17, 29, 38). The high frequency of responses by PBMC and T-cell clones to the whole recombinant protein detected in normal, healthy subjects further supports the above assumption. This is not to say, however, that the presumably numerous O-saccharide chains present on the natural Mp65 of C. albicans do not participate in CMI recognition, either enhancing or downmodulating the response. Recent evidence points to this possibility (14), as well as to the capacity of selected lymphocyte populations with gamma or delta T-cell receptors to start an adaptive cellular response against lipidated or nonlipidated polysaccharides. A comparison of the stimulating capacity of suitably designed synthetic peptides with the corresponding natural ones may help to distinguish between the two events. The generation of human T-cell clones against the recombinant protein will help to obtain further insight into the mechanisms of CaMp65 recognition and processing. The provision of a highly immunogenic, recombinant product of a common recall antigen may prove useful for all studies addressing the state of immune response to Candida and its modulation during disease (e.g., in AIDS or other immunodeficiencies), which greatly predispose to infection by this fungus (24, 34). In contrast to the natural Mp65, the recombinant product can be obtained in large quantities and can be standardized for purity, potency, and overall quality.
ACKNOWLEDGMENTS
We thank A. Botzios, C. Belotti, and F. Girolamo for help in manuscript preparation.
This work was in part supported by a grant from the National AIDS Program (contract 50 C/B).
REFERENCES
- 1.Ausiello C M, Spagnoli G C, Boccanera M, Casalinuovo I, Malavasi F, Casciani C U, Cassone A. Proliferation of human peripheral blood mononuclear cells induced by Candida albicans and its cell wall fractions. J Med Microbiol. 1986;22:195–202. doi: 10.1099/00222615-22-3-195. [DOI] [PubMed] [Google Scholar]
- 2.Ausiello M C, Urbani F, Gessani S, Spagnoli G C, Gomez M J, Cassone A. Cytokine gene expression in human peripheral blood mononuclear cells stimulated by mannoprotein constituents from Candida albicans. Infect Immun. 1993;61:4105–4111. doi: 10.1128/iai.61.10.4105-4111.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bromuro C, La Valle R, Sandini S, Urbani F, Ausiello C M, Morelli L, Fè d'Ostiani C, Romani L, Cassone A. A 70-kDa recombinant heat shock protein of C. albicans is highly immunogenic and enhances systemic murine candidiasis. Infect Immun. 1998;66:2154–2162. doi: 10.1128/iai.66.5.2154-2162.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bromuro C, Torosantucci A, Gomez M J, Urbani F, Cassone A. Differential release of an immunodominant 65-kDa mannoprotein antigen from yeast and mycelial forms of Candida albicans. J Med Vet Mycol. 1994;32:447–459. doi: 10.1080/02681219480000601. [DOI] [PubMed] [Google Scholar]
- 5.Burnette N. “Western blotting”: electrophoretic transfer of protein from dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- 6.Calderone R, Enache E, Eskandri T, Wadsworth E, Sturtevant J. Adherence of Candida albicans to mammalian cells in vitro: nutritional influences. In: Suzuki S, Suzuki M, editors. Fungal cells in biodefense mechanism. Tokyo, Japan: Saikon Publishing Co., Ltd.; 1997. pp. 73–83. [Google Scholar]
- 7.Calderone R A, Linehan L, Wadsworth E, Sandberg L. Identification of C3d receptors on Candida albicans. Infect Immun. 1988;56:252–258. doi: 10.1128/iai.56.1.252-258.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cappellaro C, Marsa V, Tanner W. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J Bacteriol. 1998;18:5030–5037. doi: 10.1128/jb.180.19.5030-5037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassone A, Torosantucci A. Immunological moieties of the cell wall. In: Prasad R, editor. Candida albicans. Berlin, Germany: Springer-Verlag; 1991. pp. 89–107. [Google Scholar]
- 10.Cassone A, De Bernardis F, Ausiello C M, Gomez M J, Boccanera M, La Valle R, Torosantucci A. Immunogenic and protective Candida albicans constituents. Res Immunol. 1998;149:289–298. doi: 10.1016/s0923-2494(98)80753-0. [DOI] [PubMed] [Google Scholar]
- 11.Chaffin W L, Lopez-Ribot J L, Casanova M, Gozalbo D, Martinez J M. Cell wall secreted proteins of Candida albicans: identification, function and expression. Microbiol Mol Biol Rev. 1988;62:130–180. doi: 10.1128/mmbr.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Bernardis F, Molinari A, Boccanera M, Stringaro A, Robert R, Senet J M, Arancia G, Cassone A. Modulation of cell surface-associated mannoprotein antigen expression in experimental candidal vaginitis. Infect Immun. 1994;62:509–519. doi: 10.1128/iai.62.2.509-519.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Bernardis F, Mondello F, San Millan R, Ponton J, Cassone A. Biotyping and virulence properties of skin isolates of Candida parapsilosis. J Clin Microbiol. 1999;37:3481–3486. doi: 10.1128/jcm.37.11.3481-3486.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deck M B, Sjölin P, Unanue E R, Kihlber J. MHC-restricted, glycopeptide-specific T cells shows specificity for both carbohydrate and peptide residues. J Immunol. 1999;162:4740–4744. [PubMed] [Google Scholar]
- 15.Deepe G S., Jr Prospects for the development of fungal vaccines. Clin Microbiol Rev. 1997;10:585–596. doi: 10.1128/cmr.10.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Domer J E. Candida cell wall mannan: a polysaccharide with diverse immunologic properties. Crit Rev Immunol. 1989;17:33–51. doi: 10.3109/10408418909105721. [DOI] [PubMed] [Google Scholar]
- 17.Domer J E, Li S, Wang Y, Lee S. Immunoregolatory studies with Candida albicans mannan. In: Suzuki S, Suzuki M, editors. Fungal cells in biodefense mechanism. Tokyo, Japan: Saikon Publishing Co., Ltd.; 1997. [Google Scholar]
- 18.Gilmore B J, Retsinas E M, Lorenz J S, Hostetter M K. An iC3b receptor on Candida albicans: structure, function and correlates for pathogenicity. J Infect Dis. 1988;157:38–41. doi: 10.1093/infdis/157.1.38. [DOI] [PubMed] [Google Scholar]
- 19.Glover D M. DNA cloning: a practical approach. Oxford, England: IRL Press; 1985. [Google Scholar]
- 20.Gomez M J, Torosantucci A, Arancia S, Maras B, Parisi L, Cassone A. Purification and biochemical characterization of a 65-kilodalton mannoprotein (MP65), a main target of anti-Candida cell-mediated immune responses in humans. Infect Immun. 1996;64:2577–2584. doi: 10.1128/iai.64.7.2577-2584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gomez M J, Maras B, Barca A, La Valle R, Barra D, Cassone A. Biochemical and immunological characterization of MP65, a major mannoprotein antigen of the opportunistic human pathogen Candida albicans. Infect Immun. 2000;68:694–701. doi: 10.1128/iai.68.2.694-701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hochuli E, Bannwarth W, Dobli H, Gentz R, Stuber D. Genetic approach to facilitate purification of recombinant proteins with a novel metal-chelate absorbant. Bio/Technology. 1988;6:1321–1325. [Google Scholar]
- 23.Kapteyn J C, Van Den Ende H, Klis F M. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim Biophys Acta. 1999;1426:373–383. doi: 10.1016/s0304-4165(98)00137-8. [DOI] [PubMed] [Google Scholar]
- 24.Klein R S, Harris C A, Small C B, Moll B, Lesser M, Friedland G H. Oral candidiasis in high-risk patients as the initial manifestation of the acquired immunodeficiency syndrome. N Engl J Med. 1984;311:354–358. doi: 10.1056/NEJM198408093110602. [DOI] [PubMed] [Google Scholar]
- 25.Lasker A A, Riggsby W S. Transcription of the single actin gene of Candida albicans during the yeast-to-mycelium conversion. Exp Mycol. 1992;161:155–162. [Google Scholar]
- 26.La Valle R, Bromuro C, Ranucci L, Muller H M, Crisanti A, Cassone A. Molecular cloning and expression of 70-kilodalton heat shock protein of Candida albicans. Infect Immun. 1995;63:4039–4045. doi: 10.1128/iai.63.10.4039-4045.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mencacci A, Torosantucci A, Spaccapelo R, Romani L, Bistoni F, Cassone A. A mannoprotein constituent of Candida albicans that elicits different levels of delayed-type hypersensitivity, cytokine production, and anticandidal protection in mice. Infect Immun. 1994;62:5353–5360. doi: 10.1128/iai.62.12.5353-5360.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mencacci A, Spaccapelo R, Del Sero G, Essle K H, Cassone A, Bistoni F, Romani L. CD4+ T-helper cell responses in mice with low-level Candida albicans infection. Infect Immun. 1996;64:4907–4914. doi: 10.1128/iai.64.12.4907-4914.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson R D, Shibata N, Podzorski P R, Herror M J. Candida mannan: chemistry, suppression of cell-mediated immunity, and possible mechanisms of action. Clin Microbiol Rev. 1991;4:1–19. doi: 10.1128/cmr.4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nisini R, Paroli M, Accapezzato D, Bonino F, Rosina F, Santantonio T, Sallusto F, Amoroso A, Houghton M, Barnaba V. Human CD4+ T-cell response to hepatitis delta virus: identification of multiple epitopes and characterization of T-helper cytokine profiles. J Virol. 1997;71:2241–2251. doi: 10.1128/jvi.71.3.2241-2251.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Odds F C. Candida and candidosis. 2nd ed. London, England: Baillière Tindall; 1988. [Google Scholar]
- 32.Perbal B. A practical guide to molecular cloning. New York, N.Y: Wiley-Interscience Publications; 1988. [Google Scholar]
- 33.Puccetti P, Romani L, Bistoni F. A TH-1-TH-2-like switch in candidiasis: new perspectives for therapy. Trends Microbiol. 1995;3:237–240. doi: 10.1016/s0966-842x(00)88931-3. [DOI] [PubMed] [Google Scholar]
- 34.Romani L, Howard D H. Mechanisms of resistance to fungal infection. Curr Opin Immunol. 1995;7:517–523. doi: 10.1016/0952-7915(95)80097-2. [DOI] [PubMed] [Google Scholar]
- 35.Stuber D, Matile H, Garotta G. System for high-level production in E. coli and rapid purification of recombinant proteins: application to epitope mapping preparation of antibodies and structure/function analysis. In: Lefkovitz I, Pernis B, editors. Immunological methods. Vol. 4. Orlando, Fla: Academic Press, Inc.; 1990. pp. 121–152. [Google Scholar]
- 36.Sundstrom P, Jensen J, Balish E. Humoral and cellular immune response to enolase after alimentary tract colonization or intravenous immunization with Candida albicans. J Infect Dis. 1994;170:390–395. doi: 10.1093/infdis/170.2.390. [DOI] [PubMed] [Google Scholar]
- 37.Torosantucci A, Palma C, Boccanera M, Ausiello C M, Spagnoli G C, Cassone A. Lymphoproliferation and cytotoxic responses of human peripheral blood mononuclear cells to mannoprotein constituents of Candida albicans. J Gen Microbiol. 1990;136:664–672. doi: 10.1099/00221287-136-11-2155. [DOI] [PubMed] [Google Scholar]
- 38.Torosantucci A, Gomez M J, Bromuro C, Casalinuovo J, Cassone A. Biochemical and antigenic characterization of mannoprotein constituents released from yeast and mycelial form of Candida albicans. J Med Vet Mycol. 1991;29:361–372. doi: 10.1080/02681219180000591. [DOI] [PubMed] [Google Scholar]
- 39.Torosantucci A, Bromuro C, Gomez M J, Ausiello C M, Urbani F, Cassone A. Identification of a 65-kDa mannoprotein as a main target of human cell-mediated immune response to Candida albicans. J Infect Dis. 1993;168:427–435. doi: 10.1093/infdis/168.2.427. [DOI] [PubMed] [Google Scholar]
- 40.Vollrath D, Davis R W. Resolution of greater than megabasepair DNA molecules by contour clamped homogeneous electric fields. Nucleic Acids Res. 1987;15:7865–7866. doi: 10.1093/nar/15.19.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]