Abstract
Neolamarckia cadamba (N. cadamba) is a fast-growing tree species with tremendous economic and ecological value; the study of the key genes regulating photosynthesis and sugar accumulation is very important for the breeding of N. cadamba. Fructose 1,6-bisphosptase (FBP) gene has been found to play a key role in plant photosynthesis, sugar accumulation and other growth processes. However, no systemic analysis of FBPs has been reported in N. cadamba. A total of six FBP genes were identifed and cloned based on the N. cadamba genome, and these FBP genes were sorted into four groups. The characteristics of the NcFBP gene family were analyzed such as phylogenetic relationships, gene structures, conserved motifs, and expression patterns. A cis-acting element related to circadian control was first found in the promoter region of FBP gene. Phylogenetic and quantitative real-time PCR analyses showed that NcFBP5 and NcFBP6 may be chloroplast type 1 FBP and cytoplasmic FBP, respectively. FBP proteins from N. cadamba and 22 other plant species were used for phylogenetic analyses, indicating that FBP family may have expanded during the evolution of algae to mosses and differentiated cpFBPase1 proteins in mosses. This work analyzes the internal relationship between the evolution and expression of the six NcFBPs, providing a scientific basis for the evolutionary pattern of plant FBPs, and promoting the functional studies of FBP genes.
Keywords: phylogenetic analysis; fructose 1,6-bisphosptase; Neolamarckia cadamba; bioinformatics analysis
1. Introduction
N. cadamba (Roxb.) Bosser (Rubiaceae) is mainly distributed in Burma, India, Vietnam, Sri Lanka and other places in Southern Asia, as well as Papua New Guinea, Australia and other countries in the South Pacific [1]. At the World Forestry Congress in 1972, N. cadamba was described as ‘a miraculous tree’ due to its fast-growing. Under normal conditions, it can attain a height of 17.67 m and a trunk diameter of 25.3 cm at breast height within nine years [2]. As early as 1933, Indonesia began to establish the plantation of N. cadamba, and it has been introduced to African and Central American countries as a major industrial timber species [3]. N. cadamba is not only used for pulp and paper production, but also in furniture making [4]. Its flowers, fruit, leaves and bark are widely used in modern and traditional medicine [5]. Therefore, it has potential, in suitable regions, to meet the increasing demand for wood products.
Fructose 1,6-bisphosptase (FBPase, EC 3.1.3.11), first discovered by Gomori in 1943 [6], is widely found in animals and plants. It can catalyze the irreversible hydrolysis of fructose-1,6-bisphosphate to fructose-6-phosphate and inorganic phosphate. Two FBPase isozymes have been identified in eukaryotic cells of photosynthesis, with one in the cytoplasm (cyFBPase), the key enzyme of gluconeogenesis, and the other one in the chloroplast (cpFBPase), the rate-limiting enzyme involved in the Calvin cycle [7]. CpFBPase isozymes are widely distributed in photosynthetic organisms, such as bacteria [8], cyanobacteria [9], green algae [10], lichens [11] and higher plants [12].
FBPase in most higher plants exists in the form of monomer, dimer and tetramer, among which tetramer FBPase has catalytic activity [13]. Both chloroplast and cytoplasmic fructose-1,6-diphosphatase can be highly regulated, but the regulatory mechanism is different between them. CyFBPase plays a regulatory role in gluconeogenesis and is strongly inhibited by adenosine monophosphate (AMP) and fructose-1,6-diphosphate; on the contrary, cpFBPase plays a regulatory role in photosynthesis and is not sensitive to AMP and fructose-1,6-diphosphate [14].
Considerable effort has been made to investigate the roles of FBPase on plant growth, biotic stress and abiotic stress. Miyagawa et al. [15] transferred cyanobacteria chloroplast FBPase into tobacco, which increased the photosynthesis and sugar accumulation of transgenic tobacco and accelerated its plant growth. Thorbjornsen et al. [16] successfully isolated potato cpFBPase gene and analyzed its transformation. Compared with wild potato tubers, the transformed plants contained higher chloroplast FBPase activity and accumulated more starch. In Arabidopsis thaliana, it was found that the lack of cFBP1 resulted in lower soluble sugar content, less starch accumulation and higher superoxide dismutase (SOD) activity, in addition to smaller rosette size and lower photosynthetic rate [12]. The mutant also experienced some developmental changes, including stomatal opening defects and an increase in the number of root vascular layers. In addition, cpFBPase also plays an important role under high temperature or drought in Pyropia haitanensis [17]. FBPase also plays an important role in biotic stress. The expression of PmFBP gene in insect-resistant varieties was significantly higher than that of the control, speculating that PmFBP gene played a key role in the defense process of insect resistance of Pinus massoniana [18].
As FBPase plays a key role in plant photosynthesis, sugar accumulation and other growth processes, the study of FBPase is important for N. cadamba breeding. In the study, the NcFBP gene family members were cloned in the N. cadamba genome, followed by comprehensive analyses, including sequence alignment, gene structures, phylogenetic analyses, conserved motifs, and conserved domains. Therefore, the expression patterns of FBP genes in different tissues in N. cadamba and their phylogenetic analysis were investigated.
2. Materials and Methods
2.1. Identification and Cloning of FBP Genes in N. cadamba and Identification in 22 Other Species
The predicted NcFBP genes were identified as follows: first, the N. cadamba genomic sequences were downloaded from the National Center for Biotechnology Information (NCBI) BioProject database [19] and BioEdit biological sequence alignment editor software was used to create a local database file. Next, a hidden Markov model (HMM) program was used for gene prediction against the local database through the BlastP method (E value < 10−10). NcFBP gene sequences for potential protein-coding segments were searched by ‘ORF prediction’ in TBtools [20], and all candidates were examined for the FBP domain in the Pfam databases. Reverse transcription polymerase chain reaction (RT-PCR) was used to amplify full-length ORFs of these NcFBP genes. Total RNA from each sample was isolated using CTAB plus the OMEGA Plant RNA isolation kit, as described previously [21]. Total RNA (0.5 µg) was reverse transcribed into the first strand cDNA following the PrimeScript II First Strand cDNA Synthesis Kit (TaKaRa Bio, Tokyo, Japan) instructions. Candidate genes were amplified using KOD FX polymerase (Toyobo Co., Osaka, Japan) with the following thermocycling conditions: 94 °C for 2 min, 35 cycles (94 °C for 30 s, 53 °C for 30 s, and 72 °C for 40 s), with a final extension at 72 °C for 2 min. The amplified products were sequenced by Beijing Genomics Institute (China) and the sequences were used for further investigation.
To investigate variation of FBP gene family members, we used the HMM software package to identify putative FBP proteins in 22 other species′ completely sequenced genomes, including dicot and monocot plants, ferns, moss, and algae, followed by querying the candidates against the Phytozome database (http://www.phytozome.net/, accessed on 27 May 2021) to confirm the presence of the FBP domain. In total, 151 FBPs were identified in these 23 genomes (Table 1). The amino acid sequences were analyzed using the web-based Multiple Expectation–Maximization for Motif Elicitation (MEME) program, version 4.11.4, to identify and analyze conserved motifs [22]. The maximum number of motifs was set to 10 and the optimum motif width was set to ≥6 and ≤50. Finally, conserved domains were identified with the web-based Pfam search program [23] (http://pfam.xfam.org/, accessed on 27 May 2021). The intron–exon structures were drawn by the TBtools [20].
Table 1.
Genome-wide identification of FBP in the 23 completely sequenced plant genomes.
| Classification | Species Name | Common Name | Genome Size (Mb) | Org Code | FBP (PF00316) |
|---|---|---|---|---|---|
| Algae | Chlamydomonas reinhardtii | Green algae | 111.1 | Cre | 3 |
| Mosses | Physcomitrella patens | Moss | 473.0 | Ppa | 12 |
| Ferns | Selaginella moellendorffii | Spikemoss | 212.5 | Smo | 5 |
| Monocots | Setaria italica | Foxtail millet | 405.7 | Sit | 5 |
| Monocots | Oryza sativa | Rice | 372.0 | Osa | 5 |
| Monocots | Zea mays PH207 | Maize | 2450.0 | Zma | 5 |
| Monocots | Brachypodium hybridum | 509.0 | Bhy | 10 | |
| Monocots | Miscanthus sinensis | Chinese silver grass | 2000.0 | Msi | 9 |
| Monocots | Sorghum bicolor | Cereal grass | 732.2 | Sbi | 5 |
| Dicots | Citrus sinensis | Sweet orange | 319.0 | Csi | 6 |
| Dicots | A. thaliana | Thale cress | 135.0 | Ath | 4 |
| Dicots | Populus trichocarpa | Western poplar | 422.9 | Ptr | 11 |
| Dicots | Amborella trichopoda | Amborella | 706.0 | Atr | 4 |
| Dicots | Eucalyptus grandis | Eucalyptus | 691.0 | Egr | 5 |
| Dicots | Aquilegia coerulea | Colorado blue columbine | 306.5 | Aco | 5 |
| Dicots | Solanum tuberosum | Potato | 723.0 | Stu | 7 |
| Dicots | Olea europaea | Olive | 1140.0 | Oeu | 10 |
| Dicots | Coffea arabica | Coffee | 1192.6 | Car | 10 |
| Dicots | Vitis vinifera | Grape vine | 487.0 | Vvi | 5 |
| Dicots | Solanum lycopersicum | Tomato | 900.0 | Sly | 7 |
| Dicots | Gossypium hirsutum | Upland cotton | 2305.2 | Ghi | 14 |
| Dicots | Cinnamomum kanehirae | Stout camphor tree | 730.4 | Cka | 5 |
| Dicots | N. cadamba | Kadan | 724.5 | Nca | 6 |
2.2. Analysis of Cis-Acting Elements of Promoter
The promoter sequences of NcFBP gene family members (2.0 kb upstream of transcription initiation site) were obtained from the N. cadamba genome, and the cis-acting elements of NcFBP gene family promoters were predicted by PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 30 May 2021).
2.3. Multiple Sequence Alignment and Phylogenetic Analyses of FBP Family
For phylogenetic analyses among the NcFBPs, the full length protein sequences were multiply aligned by the ClustalX2.1 program with default parameters, followed by constructing a phylogenetic tree with the maximum likelihood (ML) method using the MEGA X program [24]. The bootstrap values were calculated with 1000 replications. These alignments among 23 species were used to generate a phylogenetic tree with the neighbor-joining method using the Poisson correction in 1000 bootstrap test replicates [25].
2.4. Preparation of Plant Materials
The sampled N. cadamba plants were planted at 120 m northwest of the College of Forestry and Landscape Architecture, South China Agricultural University (113.36 E, 23.16 N) in 2011. The roots, cambium, phloem tissue, bark, young leaves, old leaves, buds and fruit were collected in July 2020, and all samples were immediately frozen in liquid nitrogen and stored at −80 °C in a refrigerator for further study. Each tissue was collected from three individual plants representing three biological replicates.
2.5. Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
The RNA isolation, cDNA synthesis, qRT-PCR and the selection of reference genes followed the methods described previously [21,26]. qRT-PCR amplification was performed on a LC480 instrument (Roche Diagnostics, Basel, Switzerland) with three technical replicates under the conditions as follows: 95 °C for 30 s, followed by 40 cycles (95 °C for 5 s, annealing at 60 °C for 30 s, and 72 °C for 30 s). The expression level of the roots was used as a control, and the relative expression levels were calculated using the 2−ΔΔCT method [27].
3. Results
3.1. Identification and Physicochemical Properties of NcFBP Family in N. cadamba
The whole genome amino acid sequence of N. cadamba was compared with A. thaliana FBP amino acid sequence by TBtools, and six genes encoding FBP in N. cadamba were screened, named as NcFBP1~NcFBP6, respectively. The nucleotide arrangement information of NcFBPs gene is shown in Table A1. In order to further understand the function of NcFBPs in the growth and development of N. cadamba, the physical and chemical properties of the protein sequences of the six FBP genes were analyzed (Table 2). The results showed that the length of NcFBP protein sequence ranged from 340 amino acid residues (aa) (NcFBP4) to 409 aa (NcFBP2), with an average length of 386 aa, which was conserved in evolution. The isoelectric points (pIs) of the FBP proteins ranged from 5.25 (NcFBP5) to 6.27 (NcFBP3), and all pIs were less than 7, indicating that all NcFBP were negatively charged and acidic. The molecular weights (MWs) ranged from 37.1957 kDa (NcFBP6) to 44.3803 kDa (NcFBP4). The instability index of NcFBP protein ranges from 35.88 (NcFBP1) to 44.59 (NcFBP3). Among them, the instability index of NcFBP3 and NcFBP4 were more than 40, indicating that they are less stable than the other members. The aliphatic index of NcFBP protein ranges from 84.56 (NcFBP6) to 94.04 (NcFBP2). The higher the aliphatic index, the better the stability of the protein, which is beneficial for the protein to play a role in different environments. The grand average hydrophobicity of all NcFBP proteins was negative (between −0.223 and −0.092), indicating that all members are hydrophilic proteins.
Table 2.
Physicochemical properties of FBP protein in N. cadamba.
| Gene Symbol | Gene ID | pI | MW (kDa) |
aa-num | Instability Index | Aliphatic Index | Grand Average of Hydropathicity |
|---|---|---|---|---|---|---|---|
| NcFBP1 | evm.model.Contig96.207 | 5.76 | 42.5726 | 393 | 35.88 | 86.11 | −0.092 |
| NcFBP2 | evm.model.Contig54.150 | 5.77 | 40.4778 | 374 | 37.52 | 94.04 | −0.108 |
| NcFBP3 | evm.model.Contig462.135 | 6.27 | 43.0109 | 392 | 44.59 | 89.29 | −0.223 |
| NcFBP4 | evm.model.Contig462.133_evm.modle.Contig462.134 | 5.45 | 44.3803 | 407 | 42.64 | 88.38 | −0.156 |
| NcFBP5 | evm.model.Contig296.294 | 5.25 | 44.3753 | 409 | 37.70 | 91.27 | −0.120 |
| NcFBP6 | evm.model.Contig172.7 | 5.87 | 37.1957 | 340 | 36.53 | 84.56 | −0.168 |
3.2. Bioinformatics Analysis of FBP Family in N. cadamba
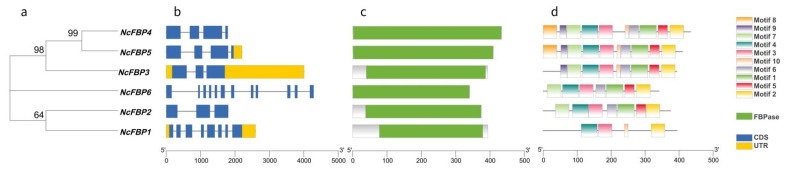
In order to explore the evolutionary relationship among members of NcFBP protein family, a phylogenetic tree was constructed by ML maximum likelihood method. The result is shown in Figure 1a. The NcFBPs can be divided into four subfamilies: NcFBP1 as the member of subfamily I, NcFBP2 as the member of subfamily II, NcFBP6 as the member of subfamily III, NcFBP3, and NcFBP4 and NcFBP5 as the members of subfamily IV.
Figure 1.
Phylogenetic relationships, gene structure, domains and motifs of FBP family in N. cadamba. (a) Protein maximum likelihood (ML) tree. (b) Gene structure, introns were shown as lines, exons and UTR were shown as blue and yellow rectangle, respectively. (c) The conserved domains, FBPase domains (PF00316) were highlighted by green. (d) The conserved motifs, all motifs were identified by MEME database.
The gene structure analysis of NcFBPs family in N. cadamba is shown in Figure 1b. The three genes of subfamily IV were similar in structure, while the three genes were also different. NcFBP4 had no untranslated regions (UTR), NcFBP3 had a small UTR at the 5′-end and a long UTR at the 3′-end, while NcFBP5 only had a small UTR at the 3′-end. There was only one gene in subfamily I, subfamily II or subfamily III, and the gene structures were quite different among them. In terms of the number of exons, there were 12 exons in subfamily III, significantly more than that in other three subfamilies containing 3–10 exons, indicating that there were great differences in gene structure among NcFBPs subfamilies.
In order to explore the evolutionary diversity of NcFBP proteins, the conserved domains of six NcFBP protein sequences were analyzed by Pfam (Figure 1c). The result showed that all NcFBP proteins have FBPase (PF00316) domain, indicating that the NcFBP proteins were highly conserved. The NcFBP proteins were analyzed by MEME online analysis software, and 10 conserved motifs were obtained as shown in Figure 1d, named motif1~motif10, respectively. All of the NcFBP protein sequences contained motif2, motif3 and motif4, NcFBP1 had the least number of motifs (four motifs), while NcFBP4 and NcFBP5 contained all 10 motifs.
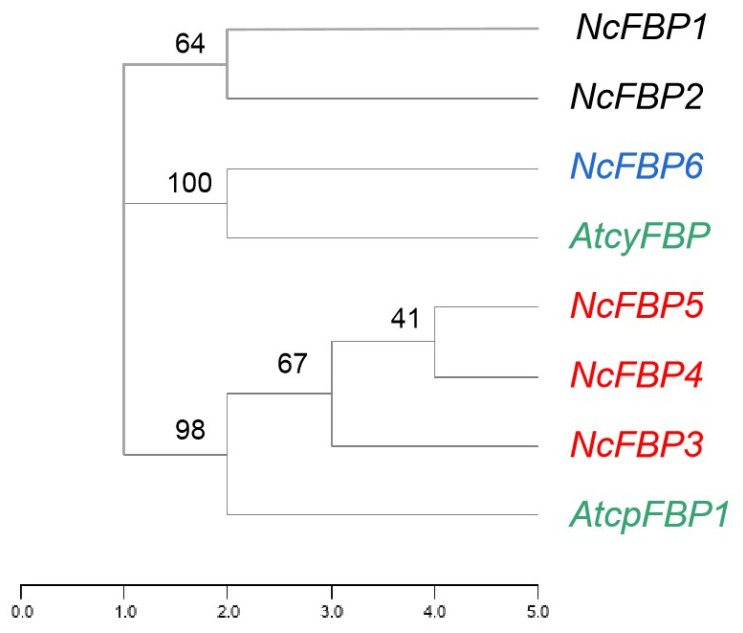
In order to further explore the functional characteristics of the FBP family, the phylogenetic analysis of AtFBPs and NcFBPs was carried out by using the “Find Best Homology” function of TBtools (Figure 2). The results showed that subfamily IV may be chloroplast type FBP (cpFBP) and subfamily III may be cytoplasmic FBP (cyFBP). Since another newly discovered chloroplast type FBP (FBP2) has not been confirmed in A. thaliana [28], it is speculated that subfamily I and subfamily II in the NcFBPs family belongs to FBP2.
Figure 2.
Evolutionary Analysis of FBP homologous genes between N. cadamba and A. thaliana. The AtcyFBP and AtcyFBP1 represent the cytoplasmic FBP and the chloroplast FBP1 of A. thaliana, respectively.
3.3. Analysis of Cis-Acting Elements of FBP Gene Family Promoter in N. cadamba
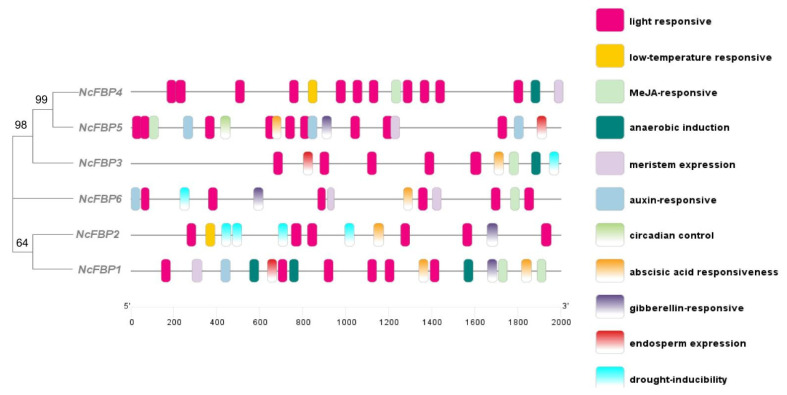
To assess the transcriptional regulation and potential functions of the NcFBP gene family, PlantCARE website was used to predict the cis-acting elements of the promoters of NcFBP gene family (Figure 3). It was found that the promoter sequences of six members of NcFBP gene family contained three kinds of stress-related cis-acting elements (drought-inducibility elements, low-temperature responsive elements and anaerobic induction elements), four kinds of hormone-related cis-acting elements (auxin-responsive elements, gibberellin-responsive elements, abscisic acid responsiveness elements, and MeJA-responsive elements), and four other kinds of cis-acting elements (circadian control elements, endosperm expression elements, light responsive elements and meristem expression elements).
Figure 3.
Prediction of cis-acting elements in the promoter regions of FBP gene family in N. cadamba.
The promoter region of each NcFBP gene contained light responsive cis-acting elements, which is essential for the photosynthesis. Except NcFBP5, other family members all contained stress-related cis-acting elements. These cis-acting elements would respond to changes in the external environment, thus regulating the expression of NcFBP genes, suggesting that NcFBP genes are closely related to the growth and development of N. cadamba, biotic and abiotic stress and other biological processes, and their transcriptional regulations are related to a variety of factors.
3.4. Analysis of Tissue Expression Pattern of NcFBPs Gene Family in N. cadamba
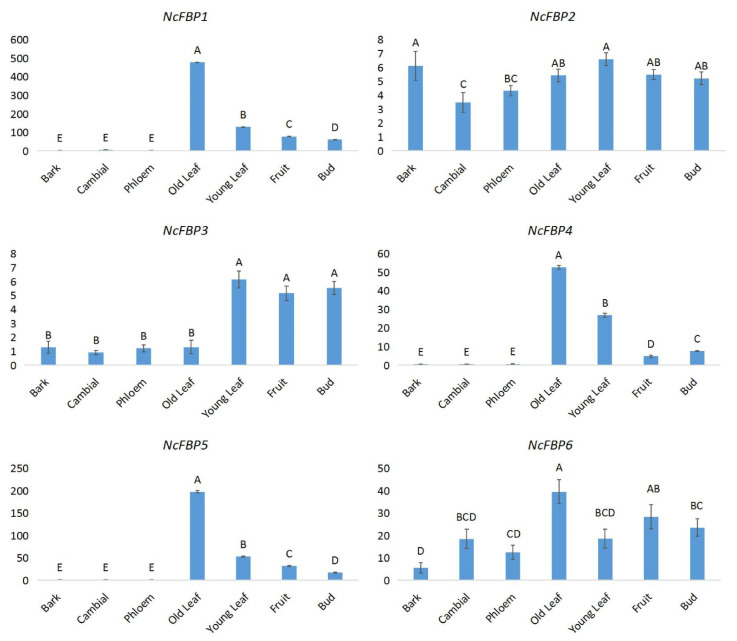
In order to understand the tissue expression characteristics of FBP genes in N. cadamba, the expression was analyzed by qRT-PCR (Figure 4). The results showed that the expression patterns of NcFBP1, NcFBP4 and NcFBP5 were similar, with high expression level in green tissues such as fruits, buds, old leaves and young leaves, while very low expression level in non-green tissues including cambium, phloem and bark. The difference of expression level between NcFBP1 and NcFBP4/NcFBP5 was that the NcFBP1 had a certain amount of expression in the cambium, while the expression of NcFBP4 and NcFBP5 in non-green tissues barely occurred. The expression patterns of NcFBP2 and NcFBP3 were similar, with a low expression level in all tissues. The expression of NcFBP6 was relatively stable in all tissues, however, it showed a higher expression level in tissues with relatively high metabolic rates, such as leaves, fruits, buds and cambium. Of all NcFBP members, the expression level of NcFBP1 was the highest in the green tissues such as fruits, buds, old leaves and tender leaves.
Figure 4.
The expression patterns of NcFBPs genes in tissues of N. cadamba. The same capital letters indicate groups that are not significantly different from each other according to Duncan’s multiple range test, p = 0.01.
3.5. Phylogenetic Analysis of FBP
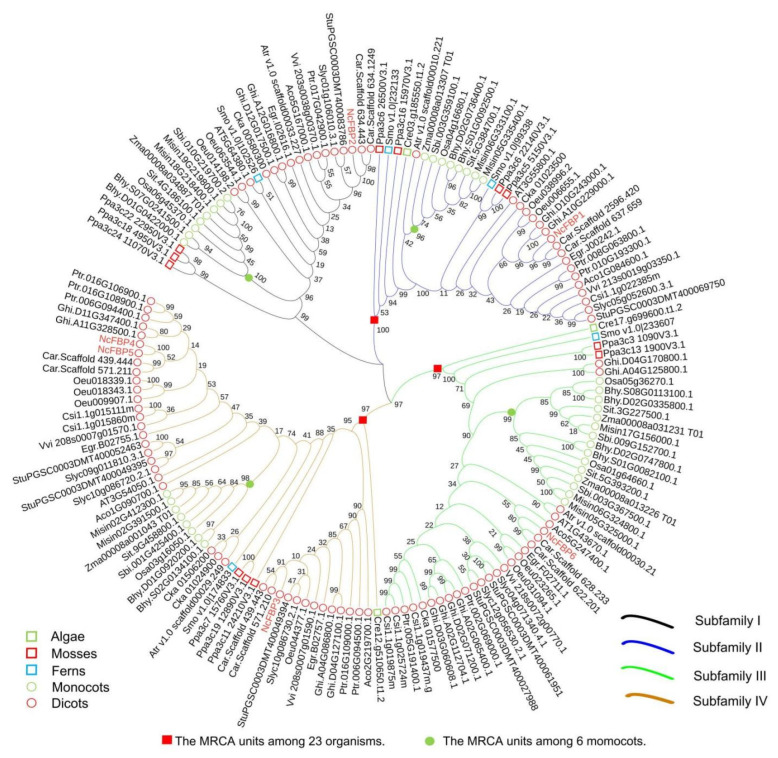
A total of 157 FBPs were identified from 23 species (Table 1) including algae (C. reinhardtii), moss (P. patens), ferns (Selaginella moellendorffii), monocotyledons (Setaria italica, Oryza sativa, Zea mays PH207, Brachypodium hybridum, Miscanthus sinensis, Sorghum bicolor) and dicotyledons (Citrus sinensis, A. thaliana, Populus trichocarpa, Amborella trichopoda, Eucalyptus grandis, Aquilegia coerulea, Solanum tuberosum, Olea europaea, Coffea arabica, Vitis vinifera, Solanum lycopersicum, Gossypium hirsutum, Cinnamomum kanehirae and N. cadamba). The results showed that all species have less than 15 members of FBP family, and only six species (P. patens, B. hybridum, P. trichocarpa, O. europaea, C. arabica, G. hirsutum) have more than or equal to 10 members of FBP family, while the vast majority of species (15 of 23) have 4~7 FBP family members. The member of the FBP family of C. reinhardtii was the least, with only three members, while G. hirsutum has the most members of the FBP family, as many as 14 members.
Figure 5 showed phylogenetic relationships and evolutionary history of FBP family in 23 species. The result shows that 157 FBPs were divided into four groups: subfamily I (28 FBPs), subfamily II (33 FBPs), subfamily III (45 FBPs) and subfamily IV (51 FBP). Except for C. reinhardtii, at least one member of each subfamily comes from 22 species. In each subfamily, the FBP of monocotyledons were basically clustered together, and dicotyledons had the similar pattern. In all subfamilies, the distance between N. cadamba and C. arabica was the closest, this was consistent with the results of plant taxonomy in which both N. cadamba and C. arabica were classified into Rubiaceae.
Figure 5.
Phylogenetic relationships and evolutionary history of FBP family in 23 species. Cre represents C. reinhardtii, Ppa represents P. patens, Smo represents S. moellendorffii, Sit represents S. italica, Osa represents O. sativa, Zma represents Z. mays, Bhy represents B. hybridum, Msi represents M. sinensis, Sbi represents S. bicolor, Csi represents C. sinensis, Ath represents A. thaliana, Ptr represents P. trichocarpa, Atr represents A. trichopoda, Egr represents E. grandis, Aco represents A. coerulea, Stu represents S. tuberosum, Oeu represents O. uropaea, Car represents C. arabica, Vvi represents V. vinifera, Sly represents S. lycopersicum, Ghi represents G. hirsutum, Cka represents C. kanehirae, Nc represents N. cadamba.
To evaluate the patterns of evolutionary and expansion history of these gene families, we broke down the phylogenetic tree into ancestral units and estimated the most recent common ancestor (MRCA) among the 23 species (Figure 5). The results showed that there were only three MRCA shared by the 23 species, and in each subfamily, monocotyledons had specific MRCA, while dicotyledons had no specific MRCA.
4. Discussion
The FBPase gene originated in bacteria in conjunction with the endosymbiotic event giving rise to mitochondria, and widely distributed in bacteria, fungi, higher plants and animals [29]. FBP genes have been identified and analyzed in many plants, such as potato [30,31], wheat [32,33], pea [34,35,36], spinach [37], tomato [38], A. thaliana [39], Porteresia coarctata [40], P. haitanensis [17], Euglena gracilis [41], P. massoniana [18] and others. To date, the genome-wide analysis of the FBP gene family has not been reported in forestry trees. In the present study, we systematically predicted and identified six FBP genes in the whole N. cadamba genome. The proteins coded by the six FBP gene members were negatively charged and contain FBP domain. Based on the analysis of motif, gene structure and phylogenetic tree, the six members can be divided into four subfamilies, which is consistent with the results of Li [28]. The gene structure of subfamily III was basically consistent with that of AtcpFBP1 in A. thaliana [12] and cpFBP1 in Gossypium species [42], and the gene structure of subfamily IV was consistent with that of AtcyFBP in A. thaliana [12] and cyFBP in Gossypium species [42], indicating that subfamily III was chloroplast type 1 FBP (cpFBP1), subfamily IV was cytoplasmic FBP (cyFBP), and both of cpFBP1 and cyFBP were highly conserved in plant species, respectively. In addition, all NcFBPs have three common FBP motifs (motifs 2, 3, 4). However, motif 8 and/or motif 9 only appear in subfamily IV, which was similar to that in cotton [42]. It is speculated that subfamily IV has a special function compared with other subfamilies. Moreover, within subfamily IV, not all members have motif 8, suggesting that these motifs may be involved in the functional differentiation of subfamily IV members.
FBP gene has been found to play a key role in plant photosynthesis, sugar accumulation, vascular bundle development and other growth processes in tobacco [15], potato [16], A. thaliana [12] and other plant species. Our study found that the most common cis-acting elements in the promoter region of NcFBPs were light responsive elements, which was similar to that in cotton [42], indicating that FBP gene mainly affects plant growth process by responding to light. Other studies have shown that FBP gene plays an important role in biotic stress [18] and abiotic stress [17,42]. This study also found that there were many cis-acting elements in the promoter region of NcFBPs related to biotic and abiotic stresses such as drought-inducibility elements, low-temperature responsive elements and anaerobic induction elements, which adds more evidence for the involvement of FBP gene in the process of biotic and abiotic stress. For breeders, it is necessary to further elucidate more gene functions involved in biotic and abiotic stresses, such as the ERECTA gene family [43] and DREB gene family [44]. Interestingly, a cis-acting element related to circadian control was found in the promoter region of NcFBP5, indicating that NcFBP5 may be involved in the regulation of circadian rhythm of N. cadamba. This is only a preliminary prediction, and thus more research is needed to verify it. So far, it has not been reported that FBP gene was involved in the regulation of circadian control in other plant species.
qRT-PCR was employed to analyze the tissue expression characteristics of FBP genes in N. cadamba. The expression level of each NcFBP gene were analyzed in different tissues, including barks, cambiums, phloems, young leaves, old leaves, fruits, and buds. The results showed that all members of NcFBP gene family were expressed at different levels in green tissues (young leaves, old leaves, fruits and buds), while NcFBP1/NcFBP2/NcFBP6 were expressed at a lower level in non-green tissues. This was similar with the expression pattern of PmFBPs in P. massoniana [18], while different from the expression pattern of HbcpFBPs in Hevea brasiliensis, with a higher expression level in bark than that in leaves [45]. The reason for the different tissue expression patterns of FBP gene may be caused by different plant species, as genomic differences are associated with differences in ecological adaptation [46]. The expression patterns of NcFBP4 and NcFBP5 were similar, except that the expression of NcFBP5 in green tissue was higher (Figure 4), indicating that NcFBP5 may be chloroplast FBP, which was consistent with the previous evolutionary analysis (Figure 5). The expression of NcFBP6 was relatively stable in all tissues, except it had a higher expression in tissues with relatively high metabolic rates (Figure 4), indicating that NcFBP6 may be a cytoplasmic FBP, which was consistent with the results of previous evolutionary analysis (Figure 5). In green tissues, the gene with the highest expression was NcFBP1, indicating that this member may play a very important role in the green tissues. In non-green tissues such as phloem, cambium and bark, the gene with the highest expression was NcFBP6; this may be due to the speculation that the NcFBP6 is a cytoplasmic FBP and participates in the pentosephosphate pathway, which is more important than the functions performed by other members of the FBP family in non-green organs. In future research, innovative research tools such as genome-environment associations (GEAs) [47,48,49] should be used to analyze the function and regulation mechanism of NcFBP genes, which can speed up the breeding process of N. cadamba.
The FBPs of C. reinhardtii does not appear in subfamily III, indicating that FBP may not differentiate until the emergence of C. reinhardtii. In the previous literature, cpFBPase2 (subfamily I) was not a newly evolved enzyme limited in terrestrial plants, but in the early stages of the evolution of photosynthetic organisms [28], probably in the common ancestor of photosynthetic eukaryotes. CyFBPase (subfamily II) may first replicate to produce cpFBPase2, and subsequently replicate to produce cpFBPase1 (subfamily III). The universal coexistence of these two kinds of cpFBPase in chloroplasts was likely to be the result of adapting to different redox conditions of photosynthesis, especially caused by repeated changes in light conditions. There were 12 FBP members in P. patens, which was four-times that in C. reinhardtii, indicating that the FBP family may have expanded during the evolution of algae to mosses and differentiated cpFBPase1 proteins in mosses. The FBPs of dicotyledons and monocotyledons were clustered separately by themselves, while the FBPs of algae, mosses and ferns were clustered together, which was consistent with the evolutionary status of plants. The molecular evolution mechanism of tree species is very important for forest tree breeding, while the molecular evolution mechanism of tree species is closely related to climate change. In future research, innovative methods such as predictive breeding platform [50], which can capture and harness natural tree pre-adaptations to biotic stresses by merging tools from the ecology, phylo-geography and omnigenetics fields, and modern strategies such as genomic prediction and machine learning [51] are needed to assess and breed forest tree adaptation to the changing climate [52]. This may provide technical support for tree breeding projects to achieve the purpose of accelerating breeding.
5. Conclusions
In this study, a total of six FBP genes were identified and cloned based on the N. cadamba genome, and comprehensively analyzed in their molecular evolution including phylogenetic relationships, gene structures and conserved motifs. The six NcFBP genes were divided into four groups according to phylogenetic analysis. This study provide evidence that there are many cis-acting elements related to biotic and abiotic stresses in the promoter region of NcFBPs, and a cis-acting element related to circadian control first found in the promoter region of FBP gene. Furthermore, in combination with phylogenetic analysis, analyses of the expression profiles of six NcFBP genes based on qRT-PCR in various N. cadamba tissues provide evidence that NcFBP5 may be chloroplast type 1 FBP, and NcFBP6 may be cytoplasmic FBP. The phylogenetic relationships and evolutionary history of FBP family in 23 species showed that FBP family may have expanded during the evolution of algae to mosses and differentiated cpFBPase1 proteins in mosses. This study represents the first investigation of the FBP gene family in N. cadamba, providing a foundation for further study on the function of the FBP genes in this species.
Appendix A
Table A1.
Nucleotide arrangement information of NcFBPs.
| Gene Symbol | Complete CDS |
|---|---|
| NcFBP1 | ATGGAAACTGGGATTGCTTACTGTGCTGGCGGAGCACTGCTGCCAAATGTTTCTTCTCAGCATTCGACTGCATTAGTTTCTCCACGTTTAATTTCTCCTTCATTCAGCTCCAGAAGTCTAAAATCAAGCTCATTATTCGGTGAATCCTTGCGCATCATGCCTCCAAGTTCGTCACTTAAGTTTTCCAGGCCGAAGAACTCATCGCTCGTGACTAAATGTGAGATTGGTGATAGCCTGGAGGAGTTTCTTACAAAAGCAACCCCGGATAAGGGCCTGATTAGGCTGCTCATGTGCATGGGAGAAGCTTTGCGAACAATTTCTTTCAAGGTAAGAACAGCTTCTTGTGGAGGAACGGCTTGTGTCAACTCTTTTGGGGATGAGCAGCTTGCTGTTGATTTGCTTGCCAACAAACTTCTATTTGAGGCCTTGAAGTACTCTCACTTCTGCAAGTATGCTTGCTCTGAGGAAGTCCCTGAGCTCCAAGACATGGAAGGCCCTGTTGAAGGGGGATTCAGTGTTGCTTTCGACCCACTCGATGGTTCCAGTATTGTCGACACAAACTTTACTGTAGGCACCATTTTTGGAGTATGGCCTGGAGATAAGCTAACTGGGGTAACTGGAAGAGACCAAGTTGCTGCAGCCATGGGGATCTATGGACCTCGAACAACATACATTCTTTCCCTTAAGGACATTCCAGGAACCCATGAATTTCTACTCCTTGATGAAGGCAAATGGCAACACGTCAAAGATACAACTGAAATTGGAGAAGGAAAAATGTTCTCCCCTGGAAACTTGAGAGCCACATCTGACAACCCTGACTATGCCAAGCTGATTGACTTCTATGTCAGAGAGAAATACACCTTGCGATACACAGGAGGAATGGTGCCTGATGTCAACCAGATAATTGTGAAGGAAAAGGGCATATTTACAAATGTGGCATCCCCATCTGCCAAAGCCAAGCTGAGATTGCTATTTGAAGTAGCTCCACTGGGGTTCTTGATCGAGAAAGCTGGTGGGTACAGCAGTGATGGAACCAAGTCAGTGCTAGACAGGGTTATCAATAATCTTGATGACAGAACTCAAGTGGCTTATGGTTCTAAGAACGAGATCATTCGATTCGAGGAATTTCTATATGGCTCCTCTAGGCTCAAGGCTCCTGAACCAGTTGGAGCTGCTGCCTGA |
| NcFBP2 | ATGGCCTCTGTTTCATCTTTTCTTGGTGGATTGAACTTGAGGAAGGTCAGTGCAAAAGTGGGTGGTGATGCTTCTTCTTCTGTTAGTAGTTCTGATGATGATCAGAATAGTGGGTTTTGTACATTGATGGATTATATAGGTAAAGAGGGACTTGATGTTGGTGATGAATTGGTGGTTTTGTTTGGTCATTTGACCTACGCTTGCAAGAAAATTGCTGCTCTTGTGGCTTCCCCTTTCAACTCTAGCCTTGGCAAAAATGTCAGCAGCATTGCCAGTGGCTCTTACGCCTCTGATAGAGATAAGCCTAAGCCTCTTGATGTTGTCTCGAATGAGATTATTTTGTCTTCTCTGCGGAGTTCTGGTAAAGTTGCAGTTATGGCCTCAGAAGAAGATGATTTGCCAGTTTGGATAAATGATGATGGCCCGTATGTGGTGGTCACAGACCCTCTAGATGGATCCCGAAATATTGATGCCTGTATTCCCACCGGAACGATCTTTGGCATCTACAAACGTCTTGTGGAGCTTGATCATCTGCCAGTGGAGGAAAAGGCAATCCTGAATTCCCTGCAAAGTGGAACAAAGCTTGTTGCTGCTGGCTATGTTCTCTACTCATCAGCTACAATTCTCTGCACCAGCTTCGGTTCAGGAACACATGCATTCACTCTCGATCACTCAACAGGGGACTTCATTCTCACCCATCCAAATATTAAAATTCCCCCTAGAGGGCAAATTTATTCTGTCAATGATGCACGGTATTTTGACTGGCCTGAAGGTTTAAGGCAATATATTGACACTGTGAGACAAGGCAAGGGAAAATATCCTAAGAAATATTCAGCCCGTTATGTATGTTCTCTTGTTGCTGATTTTCATCGTACTATCTTATATGGGGGTGTGGCAATGAATCCCAGGGATCATCTCCGTCTTGTTTATGAAGCAAATCCATTAAGTTTTGTGGTGGAGCAGGCTGGAGGTAGAGGTAGCGATGGTAAAATTAGGATTTTGTCTCTGCAGCCAGTCAAGCTACACCAAAGACTTCCCTTGTTTTTGGGGAGTCCAGATGATATAGATGAATTAGTGAGTTATGGCAATGTTCAACAGAAAGTTAATCCTGGTTATGAAGTCTAA |
| NcFBP3 | ATGGCTGAAGCAATTCCAAGTCTGTATCGGAGCATCAGCTTCTCAAGCTCACCTTCGATCTCTCGCTTCTCCCCTTTCCATGTCCCCTCAACGCAAAGCAAAAGAAGGCATGTTATCAAAGGCAGTTTTGGTCCGAAAATCCGCTGTAAAGCAGTCGGATTAGAAGCAAACCCAGCTGCAGGAGAGGCCAGAAGAAAGAAAAACAGATACGAGATGGAAAATCTGACAACGTGGTTATTAAGGCAAGAACAGTCGGGTAAGATTGACGCTGAGCTTACCGTTGTTCTCTCAAGCATATCATTAGCTTGCAAACAGATCGCTTCGCTGCTTCAGAGATCAAACATTATCAACCTTACTGGAGGTCAGGGGACGATTAATGTCCAGGGGGAAGATCAGAAGAAACTTGATGTCATATCTAATGAGTTATTCTGTAACTGCTTAAGATCGAGCGGACGAACAGGCATTATTGCATCAGAAGAAGAAAGTACACCTATAGCTGTAGAAGAAACCAATTCTGGAAACTACATTGTTGTGTTTGACCCTATTGACGGATCTGCCAACATTGATACCTCTTTAACCACAGGATCAATCTTCGGGATATATGGGCCTGACGAACAATGCCTTGTAGACTTAGATGATGAATCGATGCTAGATGAAGCAAAACAGAAGTGCATAGTCAGCGTTTGCCAGCCGGGCAGCAATTTGCTTGCTGCAGGATACTGCCTGTATTCAAGTTCAGTGGTCTTTACAGTATCAGTTGGGAAGGGAGTTTTCGCATTCACACTTGACCCAGCATATGGAGAATTTGTTTTGACACATGAAAATATCAAGATACCAAAGTCTGGCAAGATCTACTCTTTCAATGAAGGAAACTATGATTTATGGGATGACGACCTGAAGCAATATCTTGACCACTTGAGGAAACCTGGTCCTAATGGTAAACCATATTCCGGCCGTTACATAGGCTGCCTTGTTGGTGAAATACACCGAATGTTGTTATATGGCGGCATTTATGGAAACCCCAAGAACAAGAACAACAAGAAAGGTAATTTAAGGCTTTTGTATGAATGTGCACCAATGAGCTATTTAGTAGAACAAGCTGGAGGAAAAGCAATAGATGGTTGCCAAAGAATACTTGAAATCATGCCTGAGCAGGTAAATCTTTCATTTATTCTTTAA |
| NcFBP4 | ATGGCCGCGGCTACAACAGCACAACATGTATTCTCCAGCTCTCACTCATTCTCTCGCCTCTCCCCCTTCCAACTCCGGGCTTTGGGCTCCAAGTCCTTCCTCTCATGTTCCAACAAACACTGCATCAGAAGGAGGCAAGGGCTTGGAGTACGGTGCATGGCTGTAGAAACTGCTTCTGAAGCCAAGACAAAGAAGGGCGGTTTCGAGATTCAGACCTTAACTGGCTGGCTGTTGAAGCAAGAACAAGCTGGAGTTATTGATGCTGAGCTTACCATTGTTATTTCCAGCATTTCCATGGCATGCAAGCAGATTGCTGCCCTGGTTCAGAGGGCTAGCATCTCCAATCTTACTGGAGTCCAGGGAGCTGTTAACGTTCAAGGAGAAGATCAGAAGAAACTAGATGTTGTCTCCAACGAGGTATTCTCTAATTGTTTGAGATCGAGCGGAAGAACAGGGATCATAGCATCAGAGGAGGAGGATGTGCCAGTGGCGGTGGAGGAGAGTTACTCTGGCAACTACATTGTGGTATTCGACCCTCTTGATGGATCATCCAACATTGATGCCGCTGTTTCCACTGGTTCCATTTTTGGGATATATAGCCCCAATGATGAGTGCCTTGCAGATGTTGAGGATGGTGATTCCACGGTAATTATTGCCAAACAATGTTTATATGCTACTACTCAATAGATGAACAATTCTTCTACGTGGTGCATTGGATTGCAGTTAGACCAAGTGAAACAGAGGTGTGTTGTGAACGTATGTCAGCCAGGAAATAATCTTCTTGCTGCAGGCTATTGCATGTACTCAAGCTCCGTGATCTTTGTGCTGACATTGGGAAATGGTGTCTTTGCCTTCACCTTAGATCCCACGTACGGAGAATTCGTACTTGCTCAAGAAAACATTCAAATACCAAAGGCAGGGAAGATTTATGCATTCAATGAAGGAAACTACCAGCTTTGGGATGATAAGTTGAAGAAGTACATTGATGATCTCAAGGACCCTGGTCCTAGCTTCAAGCCCTATTCTGCTCGTTACATTGGTAGTTTGGTTGGCGATTTCCACAGGACACTGTTGTATGGTGGCATTTACGGTTACCCCCGAGACAAGAAGAGCAAGAATGGCAAACTGAGGCTGCTGTATGAATGCGCGCCAATGAGCTTTATTGTGGAACAAGCTGGTGGTAAAGGATCAGATGGAAATCAGAGGATTCTTGATATCGAACCAACGGAGATTCATCAAAGAGTTCCACTTTACATTGGAAGTGTGGAGGAAGTAGAGAAATTAGAAAAGTATTTGTCATAG |
| NcFBP5 | ATGGCCACAGCAACAACAGCACACCACATCCTCTCCACTTCTCATTCGATCTCCCGTATATCTTCCTTTCAATTATGTGTTTTGGACCCCAAGTTGTTCCTCTCGTGTTCCAACACCAGGAGGAGGCAAGGGGTGGTGAATGGCGGCAGAGGAGGAGTACGGTGCATGGCTGTGGAGACTTCTTCAGAAGCCAAGACAAAGAAGGCTAGCTACGAGATTCAAACCTTAACTGGCTGGCTGTTGAAGCAAGAACAAGCTGGAGTTATTGATGCTGAGCTTACCATTGTGATTTCCAGTATTTCAATGGCATGCAAGCAGATTGCTGCCTTGGTTCAGAGGGCTAGCATTTCTAACCTTACTGGGGTCCAGGGTGCTGTTAACGTTCAAGGAGAAGATCAGAAGAAACTTGATGTTGTCTCCAACGAGGTTTTCTCTAATTGCTTGAAATCGAGCGGAAGGACAGGTATCATTGCATCAGAGGAGGAGGATGTGCCGGTGGCAGTGGAAGAGAGTTACTCTGGCAACTACATCGTGGTATTTGATCCTCTTGATGGATCATCCAACATTGACGCTGCGGTCTCTACTGGATCTATCTTCGGGATATACAGCCCCAATGATGAATGCCTAGCAGATATTGGTGATGATGCCACGCTAGACCAAGTGGAACAGAGGTGTGTTGTGAACGTATGCCAGCCAGGAAACAATCTCCTTGCTGCAGGCTATTGCATGTACTCGAGCTCAGTAATCTTCGTGCTAACCTTGGGAAGTGGCGTCTTTGCATTCACCTTGGATCCCATGTATGGAGAATTTGTACTCACTCAAGAAAAAATTCAAATACCAAAGGCAGGGAAGATTTATGCATTTAATGAAGGTAACTACCAGTTGTGGGATGATAAGTTGAAGAAGTACATTGATGATCTCAAGGACCCTGGCCCTAGTGGCAAGCCCTATTCTGCTCGATACATTGGTAGTTTGGTTGGTGATTTCCATAGGACTCTGTTGTATGGTGGCATTTACGGGTATCCCCGAGACAAGAAGAGCAAGAATGGCAAATTGAGGCTCTTGTATGAATGCGCGCCAATGAGCTTTATTGTGGAACAAGCTGGTGGTAAAGGATCAGATGGATCTGGGAGGATTCTTGATATCGAACCAACAGAGATACATCAACGAGTTCCGCTGTACATTGGAAGCTTGGAGGAAGTAGAGAAATTAGAAAAATATCTGTCTTAA |
| NcFBP6 | ATGGATCACAGCGCTGATGCACACCGGACGGATTTGATGACCATAACAAGGTATGTCTTGAACGAGCAGTCCAAGCATCCAGAATCTCGCGGGGATTTCACTATCTTGCTCAGTCACATTGTTTTGGGCTGCAAGTTCGTTTGCTCTGCTGTCAGCAAGGCAGGCTTGGCCAAGCTCATTGGGCTTGCGGGTGAGACAAATGTGCAGGGTGAAGAGCAAAAGAAACTAGATGTACTATCAAATGAAGTTTTCATCAAAGCTTTGGTCAGCAGTGGTCGAACTTGCATCCTTGTCTCTGAAGAAGATGAACAGGCAACTTTTGTTGAGCCATCCATGCAGGGAAGGTATTGTGTGGTGTTTGATCCCTTGGATGGATCCTCCAACATTGACTGTGGTGTCTCAATTGGAACTATCTTTGGGATTTACATGATCAAAGATGGTGGTGATCCGAAATTGGAAGATGTGTTGCAGCCTGGGAAAAACATGGTAGCTGCTGGCTACTGCATGTATGGTAGCTCTTGCATGCTTGTGTTGAGCACTGGAAATGGTGTTAACGGATTTACTCTTGATCCATCTCTTGGAGAGTTCATATTGACTCATCCAGATATTAAGATTCCTAAGAAAGGAAAGATTTACTCGGTCAATGAGGGGAATGCCAAGAATTGGGATGCTCCGACAGCTAAATATGTGGAAAAGTGCAAGTTCCCAAAAGATGGTTCATCAGCTAAATCATTACGATACATTGGAAGCATGGTAGCTGATGTCCACCGGACATTGCTTTATGGAGGTATCTTTATGTATCCAGCTGATAAGAAAAGCCCTAATGGGAAATTGCGAGTTCTATATGAGGTCTTCCCCATGTCATTCTTGATGGAACAAGCTGGAGGACAAGCATTCACCGGGAAGGAACGGGCACTTGACCTAGTTCCAAAGAAGATTCATGAACGATCACCAATATTTCTCGGTAGCTATGATGATGTTGAGGAGGTCAAAACACTTTATGCTGAAGAGCAGAAAGCATAA |
Author Contributions
Conceptualization, X.C., W.Z. and K.O.; methodology, Q.Q.; software, Q.Q.; validation, C.L., X.L. and H.S.; formal analysis, K.O. and R.P.; investigation, Q.Q., C.L., X.L. and H.S.; resources, W.Z. and K.O.; data curation, Q.Q. and C.L.; writing—original draft preparation, Q.Q.; writing—review and editing, P.L., R.P. and K.O.; visualization, Q.Q.; supervision, W.Z. and K.O.; project administration, P.L., R.P. and K.O.; funding acquisition, X.C., W.Z. and K.O. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by “Forestry Science and Technology Innovation Project in Guangdong Province [2023KJCX005, 2018KJCX001]” and “The National Key R & D Program of China during the 14th Five-Year Plan Period [2022YFD2200205]”.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Krisnawati H., Kallio M.H., Kanninen M. Anthocephalus cadamba Miq.: Ecology, Silviculture and Productivity. Center for International Forestry Research; Bogor Barat, Indonesia: 2011. [Google Scholar]
- 2.Mojiol A.R., Lintangah W., Maid M., Julius K. Neolamarckia cadamba (Roxb.) Bosser 1984. In: Roloff A., editor. Enzyklopädie der Holzgewächse: Handbuch und Atlas der Dendrologie. Wiley-VCH Verlag GmbH & Co. KGaA.; Weinheim, Germany: 2014. pp. 1–12. [Google Scholar]
- 3.Ismaill J., Jusoh M.Z., Sahri M.H. Anatomical variation in planted kelempayan (Neolamarckia cadamaba, Rubiaceae) IAWA J. 1995;16:277–287. doi: 10.1163/22941932-90001411. [DOI] [Google Scholar]
- 4.Pang S.L., Ho W.S., Mat-Isa M.N., Abdullah J. Gene discovery in the developing xylem tissue of a tropical timber tree species: Neolamarckia cadamba (Roxb.) Bosser (kelampayan) Tree Genet. Genomes. 2015;11:47. doi: 10.1007/s11295-015-0873-y. [DOI] [Google Scholar]
- 5.Pandey A., Negi P.S. Traditional uses, phytochemistry and pharmacological properties of Neolamarckia cadamba: A review. J. Ethnopharmacol. 2016;181:118–135. doi: 10.1016/j.jep.2016.01.036. [DOI] [PubMed] [Google Scholar]
- 6.Gomori G. HEXOSEDIPHOSPHATASE. J. Biol. Chem. 1943;148:139–149. doi: 10.1016/S0021-9258(18)72326-0. [DOI] [Google Scholar]
- 7.Chueca A., Sahrawy M., Pagano E.A., Lopez G.J. Chloroplast fructose-1,6-bisphosphatase: Structure and function. Photosynth. Res. 2002;74:235–249. doi: 10.1023/A:1021243110495. [DOI] [PubMed] [Google Scholar]
- 8.Gibson J.L., Chen J.H., Tower P.A., Tabita F.R. The form II fructose 1,6-bisphosphatase and phosphoribulokinase genes form part of a large operon in Rhodobacter sphaeroides: Primary structure and insertional mutagenesis analysis. Biochemistry. 1990;29:8085–8093. doi: 10.1021/bi00487a014. [DOI] [PubMed] [Google Scholar]
- 9.Crawford N.A., Sutton C.W., Yee B.C., Johnson T.C., Carlson D.C., Buchanan B.B. Contrasting modes of photosynthetic enzyme regulation in oxygenic and anoxygenic prokaryotes. Arch. Microbiol. 1984;139:124–129. doi: 10.1007/BF00401986. [DOI] [PubMed] [Google Scholar]
- 10.Grotjohann N. Fructose 1,6-bisphosphatase in Chlorella kessleri grown in red or blue light. Z. Naturforsch. 1993;48:707–712. doi: 10.1515/znc-1993-9-1005. [DOI] [Google Scholar]
- 11.Brown D., Kershaw K.A. Seasonal Changes in the Kinetic Parameters of a Photosynthetic Fructose-1,6-Bisphosphatase Isolated from Peltigera rufescens. Plant Physiol. 1986;82:457–461. doi: 10.1104/pp.82.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rojas-González J.A., Soto-Súarez M., García-Díaz Á., Romero-Puertas M.C., Sandalio L.M., Mérida Á., Thormählen I., Geigenberger P., Serrato A.J., Sahrawy M. Disruption of both chloroplastic and cytosolic FBPase genes results in a dwarf phenotype and important starch and metabolite changes in Arabidopsis thaliana. J. Exp. Bot. 2015;66:2673–2689. doi: 10.1093/jxb/erv062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnarrenberger C., Martin W. The Calvin cycle—A historical perspective. Photosynthetica. 1997;33:331–345. [Google Scholar]
- 14.Ye B.-Y., Huang J.-C., Shao W.-H., Chen Y.-Q., Chen R.-K. Cloning and Sequence Analysis of Fructose-1,6-bisphosphatase cDNA from Sugarcane Stalk. Sugar Crops China. 2009;3:1–4. [Google Scholar]
- 15.Miyagawa Y., Tamoi M., Shigeoka S. Overexpression of a cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase in tobacco enhances photosynthesis and growth. Nat. Biotechnol. 2001;19:965–969. doi: 10.1038/nbt1001-965. [DOI] [PubMed] [Google Scholar]
- 16.Thorbjornsen T., Asp T., Jorgensen K., Nielsen T.H. Starch biosynthesis from triose-phosphate in transgenic potato tubers expressing plastidic fructose-1,6-bisphosphatase. Planta. 2002;214:616–624. doi: 10.1007/s004250100647. [DOI] [PubMed] [Google Scholar]
- 17.Xiao H., Chen C., Xu Y., Ji D., Xie C. Cloning and expression analysis of the chloroplast fructose-1,6-bisphosphatase gene from Pyropia haitanensis. Acta Oceanol. Sin. 2014;33:92–100. doi: 10.1007/s13131-014-0455-0. [DOI] [Google Scholar]
- 18.Chen H., Tan J.-H., Yan P.-D., Wu D.-S., Luo Q.-F., Yang Z.-Q. Cloning and Expression Analysis of PmFBP Gene in Pinus massoniana. Guangxi For. Sci. 2016;45:12–18. doi: 10.19692/j.cnki.gfs.2016.01.003. [DOI] [Google Scholar]
- 19.Zhao X., Hu X., Ouyang K., Yang J., Que Q., Long J., Zhang J., Zhang T., Wang X., Gao J., et al. Chromosome—Level assembly of the Neolamarckia cadamba genome provides insights into the evolution of cadambine biosynthesis. Plant J. 2022;109:891–908. doi: 10.1111/tpj.15600. [DOI] [PubMed] [Google Scholar]
- 20.Chen C., Chen H., Zhang Y., Thomas H.R., Frank M.H., He Y., Xia R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020;13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Ouyang K., Li J., Huang H., Que Q., Li P., Chen X. A simple method for RNA isolation from various tissues of the tree Neolamarckia cadamba. Biotechnol. Biotechnol. Eq. 2014;28:1008–1013. doi: 10.1080/13102818.2014.981086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L., Ren J., Li W.W., Noble W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn R.D., Coggill P., Eberhardt R.Y., Eddy S.R., Mistry J., Mitchell A.L., Potter S.C., Punta M., Qureshi M., Sangrador-Vegas A., et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., Battistuzzi F.U. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang D., Li J., Li B., Li C., Chen X., Ouyang K. Internal Reference Gene Selection under Different Hormone Stresses in Multipurpose Timber Yielding Tree Neolamarckia cadamba. Forests. 2020;11:1014. doi: 10.3390/f11091014. [DOI] [Google Scholar]
- 27.Livak K.J., Schmittgen T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Li Y., Ye Q., He D., Bai H., Wen J. The ubiquity and coexistence of two FBPases in chloroplasts of photosynthetic eukaryotes and its evolutionary and functional implications. Plant Divers. 2020;42:120–125. doi: 10.1016/j.pld.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutle D.D., Roret T., Muller S.J., Couturier J., Lemaire S.D., Hecker A., Dhalleine T., Buchanan B.B., Reski R., Einsle O., et al. Chloroplast FBPase and SBPase are thioredoxin-linked enzymes with similar architecture but different evolutionary histories. Proc. Natl. Acad. Sci. USA. 2016;113:6779–6784. doi: 10.1073/pnas.1606241113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kossmann J., Muller-Rober B., Dyer T.A., Raines C.A., Sonnewald U., Willmitzer L. Cloning and expression analysis of the plastidic fructose-1,6-bisphosphatase coding sequence from potato: Circumstantial evidence for the import of hexoses into chloroplasts. Planta. 1992;188:7–12. doi: 10.1007/BF01160706. [DOI] [PubMed] [Google Scholar]
- 31.Kobmann J., Sonnewald U., Willmitzer L. Reduction of the chloroplastic fructose-16-bisphosphatase in transgenic potato plants impairs photosynthesis and plant growth. Plant J. 1994;6:637–650. [Google Scholar]
- 32.Miles A.J., Potts S.C., Willingham N.M., Raines C.A., Lloyd J.C. A light- and developmentally-regulated DNA-binding interaction is common to the upstream sequences of the wheat Calvin cycle bisphosphatase genes. Plant Mol. Biol. 1993;22:507–516. doi: 10.1007/BF00015979. [DOI] [PubMed] [Google Scholar]
- 33.Ma W., Shi D., Wang Q., Wei L., Chen H. Exogenous expression of the wheat chloroplastic fructose-1,6-bisphosphatase gene enhances photosynthesis in the transgenic cyanobacterium, Anabaena PCC7120. J. Appl. Phycol. 2005;17:273–280. doi: 10.1007/s10811-005-4850-y. [DOI] [Google Scholar]
- 34.Dong S.M., Rhim J.H., Hahn T.R. Nucleotide sequence analysis of a cDNA encoding chloroplastic fructose-1,6-bisphosphatase from pea (Pisum sativum L.) Plant Physiol. 1995;107:313–314. doi: 10.1104/pp.107.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S.W., Hahn T.R. Two light-responsive elements of pea chloroplastic fructose-1,6-bisphosphatase gene involved in the red-light-specific gene expression in transgenic tobaccos. Biochim. Biophys. Acta. 2002;1579:8–17. doi: 10.1016/S0167-4781(02)00498-0. [DOI] [PubMed] [Google Scholar]
- 36.Serrato A.J., Romero-Puertas M.C., Lazaro-Payo A., Sahrawy M. Regulation by S-nitrosylation of the Calvin-Benson cycle fructose-1,6-bisphosphatase in Pisum sativum. Redox. Biol. 2018;14:409–416. doi: 10.1016/j.redox.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fonolla J., Hermoso R., Carrasco J.L., Chueca A., Lazaro J.J., Prado F.E., Lopez-Gorge J. Antigenic relationships between chloroplast and cytosolic fructose-1,6-bisphosphatases. Plant Physiol. 1994;104:381–386. doi: 10.1104/pp.104.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Obiadalla-Ali H., Fernie A.R., Lytovchenko A., Kossmann J., Lloyd J.R. Inhibition of chloroplastic fructose 1,6-bisphosphatase in tomato fruits leads to decreased fruit size, but only small changes in carbohydrate metabolism. Planta. 2004;219:533–540. doi: 10.1007/s00425-004-1257-y. [DOI] [PubMed] [Google Scholar]
- 39.Sahrawy M., Avila C., Chueca A., Canovas F.M., Lopez-Gorge J. Increased sucrose level and altered nitrogen metabolism in Arabidopsis thaliana transgenic plants expressing antisense chloroplastic fructose-1,6-bisphosphatase. J. Exp. Bot. 2004;55:2495–2503. doi: 10.1093/jxb/erh257. [DOI] [PubMed] [Google Scholar]
- 40.Chatterjee J., Patra B., Mukherjee R., Basak P., Mukherjee S., Ray S., Bhattacharyya S., Maitra S., GhoshDastidar K., Ghosh S., et al. Cloning, characterization and expression of a chloroplastic fructose-1,6-bisphosphatase from Porteresia coarctata conferring salt-tolerance in transgenic tobacco. Plant Cell Tiss. Org. 2013;114:395–409. doi: 10.1007/s11240-013-0334-y. [DOI] [Google Scholar]
- 41.Ogawa T., Kimura A., Sakuyama H., Tamoi M., Ishikawa T., Shigeoka S. Characterization and physiological role of two types of chloroplastic fructose-1,6-bisphosphatases in Euglena gracilis. Arch. Biochem. Biophys. 2015;575:61–68. doi: 10.1016/j.abb.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 42.Ge Q., Cui Y., Li J., Gong J., Lu Q., Li P., Shi Y., Shang H., Liu A., Deng X., et al. Disequilibrium evolution of the Fructose-1,6-bisphosphatase gene family leads to their functional biodiversity in Gossypium species. BMC Genom. 2020;21:379. doi: 10.1186/s12864-020-6773-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blair M.W., Cortes A.J., This D. Identification of an ERECTA gene and its drought adaptation associations with wild and cultivated common bean. Plant Sci. 2016;242:250–259. doi: 10.1016/j.plantsci.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Cortes A.J., This D., Chavarro C., Madrinan S., Blair M.W. Nucleotide diversity patterns at the drought-related DREB2 encoding genes in wild and cultivated common bean (Phaseolus vulgaris L.) Theor. Appl. Genet. 2012;125:1069–1085. doi: 10.1007/s00122-012-1896-5. [DOI] [PubMed] [Google Scholar]
- 45.He B., Zhu J., Long X., Qin Y., Tang C. Cloning and Expression Analysis of a Chloroplast Fructose-1,6-Bisphosphatase Gene in Hevea brasiliensis. Chin. J. Trop. Crops. 2015;36:448–455. [Google Scholar]
- 46.Wang X., Yu X., Zi-Han H., Ling-Ling L., Yan-Wen L., Xin-Sheng H. Evolutionary Divergence between Toona ciliata and Toona sinensis Assayed with Their Whole Genome Sequences. Genes. 2022;13:1799. doi: 10.3390/genes13101799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cortes A.J., Lopez-Hernandez F., Blair M.W. Genome-Environment Associations, an Innovative Tool for Studying Heritable Evolutionary Adaptation in Orphan Crops and Wild Relatives. Front. Genet. 2022;13:910386. doi: 10.3389/fgene.2022.910386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buitrago-Bitar M.A., Cortés A.J., López-Hernández F., Londoño-Caicedo J.M., Muñoz-Florez J.E., Muñoz L.C., Blair M.W. Allelic Diversity at Abiotic Stress Responsive Genes in Relationship to Ecological Drought Indices for Cultivated Tepary Bean, Phaseolus acutifolius A. Gray, and Its Wild Relatives. Genes. 2021;12:556. doi: 10.3390/genes12040556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez-Hernandez F., Cortes A.J. Last-Generation Genome-Environment Associations Reveal the Genetic Basis of Heat Tolerance in Common Bean (Phaseolus vulgaris L.) Front. Genet. 2019;10:954. doi: 10.3389/fgene.2019.00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guevara-Escudero M., Osorio A.N., Cortés A.J. Integrative Pre-Breeding for Biotic Resistance in Forest Trees. Plants. 2021;10:2022. doi: 10.3390/plants10102022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cortés A.J., López-Hernández F., Osorio-Rodriguez D. Predicting Thermal Adaptation by Looking into Populations’ Genomic Past. Front. Genet. 2020;11:564515. doi: 10.3389/fgene.2020.564515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cortes A.J., Restrepo-Montoya M., Bedoya-Canas L.E. Modern Strategies to Assess and Breed Forest Tree Adaptation to Changing Climate. Front. Plant Sci. 2020;11:583323. doi: 10.3389/fpls.2020.583323. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.