Abstract
Extracellular proteins made by group A Streptococcus (GAS) play critical roles in the pathogenesis of human infections caused by this bacterium. Although many extracellular GAS proteins have been identified and characterized, there has been no systematic analysis of culture supernatant proteins. Proteins present in the culture supernatant of strains of serotype M1 (MGAS 5005) and M3 (MGAS 315) mutants lacking production of the major extracellular cysteine protease were separated by two-dimensional gel electrophoresis and identified by amino-terminal amino acid sequencing and interrogation of available databases, including a serotype M1 genome sequence. In the aggregate, amino-terminal amino acid sequence data for 66 protein spots were generated, 53 unique sequences were obtained, and 44 distinct proteins were identified. Sixteen of the 44 proteins had apparent secretion signal sequences and 27 proteins did not. Eight of the 16 proteins with apparent secretion signal sequences have not been previously described for GAS. Antibodies against most of the apparently secreted proteins were present in sera from mice infected subcutaneously with MGAS 5005 or MGAS 315. Humans with documented GAS infections (pharyngitis, acute rheumatic fever, and severe invasive disease) also had serum antibodies reacting with many of the apparently secreted proteins, indicating that they were synthesized in the course of GAS-human interaction. The genes encoding four of the eight previously undescribed and apparently secreted culture supernatant proteins were cloned, and the proteins were overexpressed in Escherichia coli. Western blot analysis with these recombinant proteins and sera from GAS-infected mice and humans confirmed the immunogenicity of these proteins. Taken together, the data provide new information about the molecular aspects of GAS-host interactions.
The human bacterial pathogen group A Streptococcus (GAS) causes a wide variety of diseases globally (35). GAS is the primary cause of pharyngitis, an infection that causes substantial morbidity and economic loss and can lead to acute rheumatic fever and rheumatic heart disease, the main cause of preventable pediatric heart disease globally. Although largely controlled in the United States and other western countries, the disease persists in certain regions, for example, Salt Lake City, Utah (49). GAS can also cause serious invasive infections such as septicemia, streptococcal toxic shock syndrome, and necrotizing fasciitis. The fatality rate for patients with invasive infections has exceeded 50% in some case series. The organism can cause other postinfection sequelae such as glomerulonephritis, a disease that is also a major public health problem in developing countries. There is currently no licensed vaccine to prevent human GAS infection.
Research conducted over many decades has shown that much of the host pathology caused by GAS is mediated by extracellular proteins (8, 12, 35). Three general categories of extracellular proteins have been recognized (12). First, proteins that are actively secreted into the extracellular environment and are largely or solely found free in the culture supernatant have been described. These proteins have secretion signal sequences and are presumed to be actively exported. Examples of these proteins include potent superantigens such as streptococcal pyrogenic exotoxins A and C. A second category of GAS extracellular proteins includes molecules with a secretion signal sequence and a conserved hexapeptide sequence (Leu-Pro-X-Thr-Gly [LPXTG]) located at the carboxy terminus that anchors the protein to the bacterial cell membrane (12). Some of these proteins or fragments thereof can also be present free in the culture supernatant, especially in the stationary phase of growth. These cell surface-anchored proteins include known virulence factors such as M protein and C5a peptidase. In recent years, evidence has been presented that a third category of GAS extracellular proteins exists (37–39, 44, 51). These proteins lack apparent secretion signal sequences and the LPXTG membrane anchor motif. Although their presence in culture supernatants may be related in part to passive release from the intracellular compartment due to cell wall autolysis, the observation that some of these proteins are found in relatively high concentrations in the logarithmic phase of growth suggests that an unknown transport mechanism participates. Interestingly, two proteins that are assigned to this class (glyceraldehyde-3-phosphate dehydrogenase and α-enolase) are enzymes involved in the glycolytic pathway of metabolism (37–39, 51).
Despite the reasonably detailed understanding of GAS extracellular proteins, there has been no systematic analysis of culture supernatant proteins with contemporary investigative methods. Importantly, this general line of investigation has provided critical new information about extracellular proteins made by diverse pathogens, such as Mycobacterium tuberculosis, Haemophilus influenzae, Listeria monocytogenes, and Helicobacter pylori (5, 6, 19–21, 25, 41, 54). For example, new vaccine candidates and drug and diagnostic targets have been identified.
The goal of the present study was to identify and characterize proteins found in relative abundance in culture supernatants of GAS strains grown in vitro. A comprehensive understanding of GAS pathogenesis requires molecular dissection of the pathogen proteome, especially proteins found in culture supernatants. As a first step toward this end, we analyzed culture supernatant proteins made by strains of serotype M1 and M3 GAS by two-dimensional electrophoresis and amino-terminal amino acid sequencing. Isolates expressing these M proteins are the two more abundant types causing human invasive infections such as septicemia and necrotizing fasciitis (33). To identify supernatant proteins expressed in the course of host-pathogen interactions, we used two-dimensional Western blot analyses with sera obtained from mice with experimentally induced soft tissue infection and from humans with pharyngitis, acute rheumatic fever, and severe invasive disease. The genes encoding four of eight apparently secreted and previously undescribed culture supernatant proteins were cloned, and recombinant proteins were overexpressed in Escherichia coli. Western blot analysis with these recombinant proteins and sera from GAS-infected mice and humans confirmed the immunogenicity of the four proteins.
MATERIALS AND METHODS
Bacterial strains and routine growth.
Strains MGAS 5005 (serotype M1) and MGAS 315 (serotype M3) have been described previously and characterized extensively (16, 17, 28–30, 33). Briefly, MGAS 5005 contains the gene (speA) encoding streptococcal pyrogenic exotoxin A, expresses the most common variant of M1 and streptococcal inhibitor of complement (1, 17), and is representative of the most abundant serotype M1 strains recovered from humans with invasive disease globally. Similarly, MGAS 315 has the speA gene and all genetic features typical of serotype M3 strains commonly cultured from patients with invasive disease.
E. coli XL-1 Blue (Stratagene, La Jolla, Calif.) or E. coli NovaBlue and BL21(DE3) (Novagen, Madison, Wis.) were used for gene cloning and protein expression. GAS strains were grown routinely in Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) supplemented with 0.2% yeast extract (THY). Brain heart infusion (BHI) agar (Difco Laboratories) or tryptose agar with 5% sheep blood (Becton Dickinson, Cockeysville, Md.) was used as a solid medium. For mutant selection, BHI agar supplemented with spectinomycin (150 μg/ml) was used. The GAS strains were grown at 37°C in a 5% CO2–20% O2 atmosphere.
Construction of speB-inactivated isogenic mutant strains.
A promoterless nonpolar spectinomycin resistance gene (aad gene) cassette was initially used to construct the speB-negative isogenic mutant (26, 28). However, the desired mutants of MGAS 5005 and MGAS 315 could not be obtained, presumably due to the failure of the mutant to survive antibiotic selection. The speB gene is expressed in the stationary phase of growth, well after antibiotic selective pressure has been supplied. Hence, when the speB gene is regulated by its native promoter, it is unlikely that transcription of the gene and the contiguous antibiotic resistance-conferring gene are expressed early during selection. To overcome this problem, a new nonpolar cassette with the spectinomycin resistance gene linked to a constitutive promoter was constructed. The aad gene and its promoter (26) were amplified using plasmid pEU904 (obtained from J. R. Scott, Emory University) DNA as the template and primers 5′-GGACCCGGGAATACATGTTATAATAACTATAAC-3′ and 5′-CCTCCCGGGCATGTGATTTTCCTCCTTTTTATAATTTTTTTAATCTGTTA-3′. The PCR product was digested with SmaI and inserted into pLEX5B (10) to generate recombinant plasmid pSL81. Plasmid pSEB1719 harboring the speB gene (36) was digested with StyI and blunt ended with Klenow fragment DNA polymerase I. The aad-containing SmaI fragment of pSL81 was inserted between the StyI sites to replace the majority of the speB gene. The suicide plasmid obtained (pSL82) was introduced into MGAS 5005 and MGAS 315 by electroporation. The recombinants were selected on medium with 150 μg of spectinomycin per ml. Presumed mutants were identified and confirmed by PCR, Southern blotting, and DNA sequencing. The speB mutants lacked proteolytic activity, as assessed by the casein plate assay (36).
Isolation of culture supernatant proteins.
The speB mutants of MGAS 315 and MGAS 5005 were cultured in THY lacking proteins with a molecular mass greater than 10 kDa (protein-reduced THY [PR-THY]). To prepare PR-THY, THY was passed through a 0.22-μm-pore-size filter and the resulting material was filtered with a membrane cartridge with a 10-kDa molecular mass cutoff (S3Y10; Millipore Corporation, Bedford, Mass.) installed in a ProFlux M12 tangential-flow filtration system (Millipore Corporation). Proteins with a molecular mass greater than 10 kDa were retained by the cartridge and discarded. To isolate proteins produced in the exponential phase, MGAS 315 speB or MGAS 5005 speB was grown overnight on BHI agar and inoculated into 400 ml of PR-THY. The organisms were grown to exponential phase (optical density at 600 nm [OD600] of ∼0.4) in 5% CO2 at 37°C. Forty milliliters of the culture was added to 900 ml of PR-THY in each of eight 1-liter bottles. The bottles were incubated in 5% CO2 at 37°C and harvested at an OD600 of 0.4 to 0.5. To isolate proteins produced in the stationary phase, 1 ml of exponential-phase (OD600 ∼0.4) GAS was inoculated into 900 ml of PR-THY in each of eight 1-liter bottles. The bacteria were grown in 5% CO2 at 37°C for 16 to 18 h and pelleted by centrifugation at 12,000 × g for 10 min. For all growth conditions, the supernatant was concentrated to 350 ml by passage through a 0.22-μm-pore-size filter and the Millipore filtration system equipped with cartridge S3Y10. Proteins in the concentrated supernatant were precipitated by addition of (NH4)2SO4 to 90% saturation and recovered by centrifugation at 18,000 × g for 20 min. The pellet was dissolved in a small amount of water. The proteins were dialyzed against 3 liters of water in Slide-A-Lyzer dialysis cassettes with a 10-kDa molecular mass cutoff (Pierce, Rockford, Ill.) at 4°C for 20 h with one change of water. The sample was stored at −20°C in 1.5-ml aliquots.
Two-dimensional electrophoresis.
The first-dimension isoelectric focusing was performed with the IPGphor IEF system and Immobiline DryStrip gel strips (7 cm) with a linear immobilized pH gradient of 3 to 10 (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.). The sample was prepared by adding a saturating amount of solid urea to the starting protein solution and mixing this solution with an equal volume of rehydration stock solution {8 M urea, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 4% IPG buffer, 50 mM dithiothreitol}. The sample (130 μl), gel strip, and mineral oil were sequentially added to a 7-cm strip holder. The holder was loaded into the IPGphor IEF system. The strip was rehydrated with the sample solution for 11 h, and the proteins were focused in three steps consisting of 500 V for 1 h, 1,000 V for 2 h, and 8,000 V for 7 h at 20°C.
The second dimension (sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) was carried out with a Tris-HCl Ready Gel (Bio-Rad Laboratories, Hercules, Calif.). After isoelectric focusing, the gel strip was rocked gently in SDS equilibration buffer (50 mM Tris-Cl [pH 8.8], 6 M urea, 30% glycerol, 2% SDS, 10 mM mercaptoethanol, bromophenol blue) for 15 min. SDS-PAGE was conducted at 150 V for 1 h, and the gel was stained with GelCode Blue Stain Reagent (Pierce). Alternatively, the proteins were transferred to a membrane for amino-terminal amino acid sequencing or Western blotting.
Amino-terminal amino acid sequencing.
Proteins separated by two-dimensional electrophoresis were transferred to a polyvinylidene difluoride membrane (Bio-Rad Laboratories) with Towbin transfer buffer using a Trans-Blot SD semidry transfer cell (Bio-Rad Laboratories) at 15 V for 40 min. The membrane was washed three times with water, stained with 0.1% Coomassie blue in 40% methanol and 1% acetic acid, and destained with 50% methanol aqueous solution. Amino-terminal amino acid sequences of the stained spots in the polyvinylidene difluoride membrane were determined by Edman degradation with an ABI/Perkin-Elmer 494 cLC protein sequencer.
Mouse sera.
Mouse sera were obtained from adult (18- to 20-g) male outbred CD-1 Swiss mice (strain ICR; Harlan-Sprague-Dawley Inc., Houston, Tex.) inoculated subcutaneously with wild-type MGAS 5005 (serotype M1) and MGAS 315 (serotype M3). The animals were anesthetized by Metofane (Mallinckrodt Veterinary, Mundelein, Ill.) inhalation before all experimental procedures. Control sera were obtained prior to mouse inoculation. Inocula were prepared from GAS cultures grown to mid-logarithmic phase (OD600 of ∼0.5). The bacterial cells were harvested and washed once with sterile ice-cold and pyrogen-free phosphate-buffered saline (PBS). The cells were resuspended in an appropriate volume of PBS to give the required inoculum, which was verified by colony counts with tryptose agar plates containing 5% sheep blood.
Groups of ∼115 mice were injected subcutaneously in the right flank with wild-type MGAS 5005 (2.3 × 107 CFU) or MGAS 315 (1.7 × 107 CFU). Preliminary experiments found that about 70 to 80% of mice survive and develop prominent gross cutaneous lesions with these inocula. Cutaneous lesions caused by these inocula healed over time in the surviving mice. The animals were sacrificed 20 to 28 days after inoculation. Serum was obtained by cardiac puncture and stored at −20°C. Sera obtained from cohorts of animals inoculated with each strain were pooled and used for Western blot analyses.
Human sera.
Inasmuch as GAS causes several very different types of infections, sera obtained from humans with three distinct diseases were used for Western blot analysis. Convalescent-phase serum was obtained from a child with uncomplicated GAS pharyngitis. The convalescent-phase serum was drawn 18 days after the pharyngitis diagnosis. The M protein type of the infecting strain was not known, and the organism was not available for analysis. Paired acute- and convalescent-phase sera obtained from a patient with an invasive GAS infection were also studied. The convalescent-phase serum was drawn 48 days after the invasive-disease diagnosis. The M type of the infecting strain (serotype M1) was inferred by sequence analysis of the region of the emm gene encoding the hypervariable amino terminus of this protein. We also used serum drawn from a patient in the acute phase of an initial rheumatic fever episode. The M protein type of the infecting strain was not known, and the organism was not available for analysis. However, serologic analysis indicated infection in an M1 organism.
Western blot analysis.
Proteins separated by two-dimensional electrophoresis or SDS-PAGE were transferred to nitrocellulose membranes (Immobilon-NC; Millipore Corporation) with Towbin transfer buffer using a Trans-Blot SD semidry transfer cell (Bio-Rad Laboratories) at 15 V for 40 min. The membrane was treated with 10 ml of block solution (1:2,000 Amersham Liquid Block in 150 mM NaCl and 100 mM Tris-HCl, pH 7.4) for 1 h and incubated for 1 h with primary antibodies added to the block solution. The membrane was then rinsed twice and washed three times for 15 min each with 0.1% Tween 20 in PBS. The membrane was incubated with goat anti-mouse or anti-human immunoglobulin G (heavy plus light chains) horseradish peroxidase-conjugated secondary antibodies (Sigma, St. Louis, Mo.) for 1 h and rinsed and washed as described above. Antigen-antibody reactivity was visualized by enhanced chemiluminescence.
Cloning and expression of the genes encoding SP13, SP22, SP24, and SP35.
The primers, vectors, and restriction enzymes used for PCR amplification and cloning of the sp13, sp22, and sp35 genes and two sp24 gene fragments are listed in Table 1. The primers were designed on the basis of gene sequences present in an available M1 GAS genome sequence (Streptococcus pyogenes Genome Sequencing Project, University of Oklahoma [http://genome.ou.edu/strep.html]). and some of the nucleotides were altered to create restriction enzyme sites for cloning. PCR products obtained with MGAS 5005 DNA were digested with the appropriate restriction enzymes and cloned into pET-21b or pET-21d to obtain recombinant plasmids (Table 1). The resulting cloned genes and gene fragments were sequenced to rule out introduction of spurious mutations.
TABLE 1.
Primers, vectors, cloning sites, and recombinant plasmids used in gene cloning
| Genea | Primers | Vector/cloning sites | Plasmid |
|---|---|---|---|
| sp13 | 5′-GTGGCTAGCCAAGATGTAATGCTTGAGACG-3′ | pET21b/NheI, BamHI | pSP13 |
| 5′-CGGATCCTTACTCTTGCCAATAACATATGTTAG-3′ | |||
| sp22 | 5′-GTGTTCATATGGATAGTTTTTCTGCTAATCAAG-3′ | pET21b/NdeI, BamHI | pSP22 |
| 5′-AGGATCCTTAATTGGTCTGATTCCAAC-3′ | |||
| sp24-f1 | 5′-GCGCAACATATGACCAATACAGAGTTGAGC-3′ | pET21b/NdeI, BamHI | pSP24-f1 |
| 5′-AGGATCCTAATGCATACCGTGTGACTCG-3′ | |||
| sp24-f2 | 5′-ACCATGGATCCTGAAACAAACCGATTC-3′ | pET21d/NcoI, EcoRI | pSP24-f2 |
| 5′-CGAATTCTAACCATCTCTTTTTGAAATGGT-3′ | |||
| sp35 | 5′-ACCATGGAGGATTTAAGTACTAAG ATTG-3′ | pET21d/NcoI, BamHI | pSP35 |
| 5′-CGGATCCTTAGTGTGGATAAATAAATGTAAC-3′ |
Fragment 1 (f1) represents amino acid residues 57 to 282. Fragment 2 (f2) represents amino acid residues 1029 to 1366.
To assess expression of the cloned genes or gene fragments, recombinant E. coli BL21 was grown at 37°C for 10 h in 3 ml of Luria-Bertani broth supplemented with 100 mg of ampicillin per liter. The OD600 of the cultures was measured. Cells were pelleted by centrifugation and resuspended in 1× SDS-PAGE loading buffer at a ratio of 100 μl of buffer per 1 OD600 unit per ml. The samples were boiled for 3 min before being loaded for protein separation with SDS-PAGE.
RESULTS
Two-dimensional electrophoresis profiles of culture supernatant proteins.
We chose to focus this analysis on serotype M1 and M3 GAS strains, 2 of greater than 100 distinct M types that have been described. Because genetic heterogeneity exists among strains expressing these serotypes (16, 17, 33, 34), we selected strains that have been extensively characterized genetically and phenotypically and are known to represent the abundantly occurring M1 and M3 subclones of each serotype infecting humans globally. These strains also are readily genetically manipulated and are virulent for mice. The major drawback of use of the chosen strains is that the genome sequences are not yet available for these organisms. Isogenic derivatives in which the gene encoding the major extracellular protease had been insertionally inactivated were used for three main reasons. First, we thought that use of the speB mutants would enhance the probability of successful identification of stationary-phase proteins, since degradation of culture supernatant proteins would be greatly reduced. Second, we thought that use of the speB mutants would enhance identification of immunogenic proteins, also due to decreased target protein degradation. Third, decreased proteolysis of culture supernatant proteins would also result in the need to identify fewer protein spots and would be more efficient.
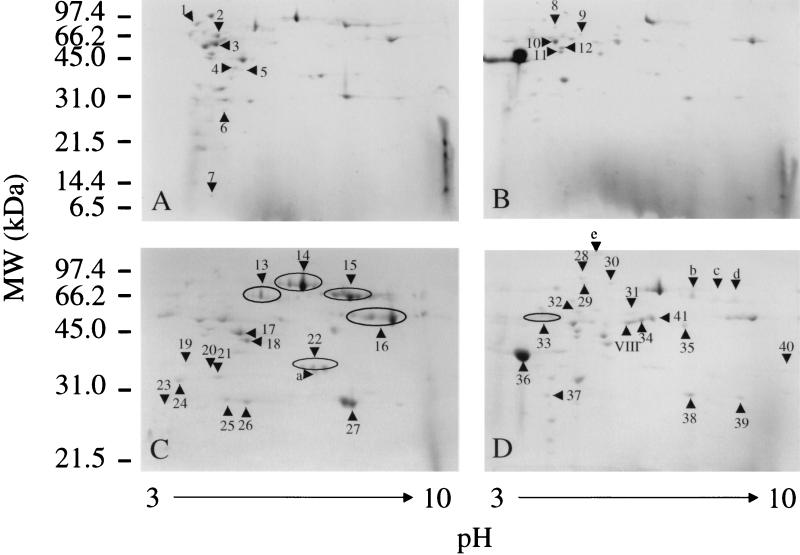
Two-dimensional electrophoresis of the culture supernatant proteins from the speB mutants of serotype M1 MGAS 5005 and serotype M3 MGAS 315 grown to exponential and stationary phases identified about 50 abundant spots in each sample (Fig. 1). (For purposes of description and uniformity, we will refer to these spots as protein spots throughout this paper.) The supernatant protein profiles were similar for the two strains in each growth phase, but some differences were detected. For example, protein spots a, 15, 25, and 27 in Fig. 1C (MGAS 315 speB) are absent in Fig. 1D (MGAS 5005 speB), and conversely, protein spots 31, 34, 36, and 41 in Fig. 1D are absent in Fig. 1C. In addition, certain protein spots were present uniquely or predominantly in culture supernatant proteins in a growth phase-dependent fashion. For example, protein spots 17, 27, and 34 were preferentially present at stationary phase (Fig. 1). Numerous protein spots had neighboring protein spots with very similar apparent molecular weights but distinct pIs.
FIG. 1.
Two-dimensional electrophoresis profiles of GAS culture supernatant proteins. Supernatant proteins (340 μg) were subject to isoelectrofocusing in a pH range of 3 to 10 and then resolved by SDS-PAGE with a 15% (A and B) or 12% (C and D) gel. The proteins were stained with GelCode Blue Stain Reagent (Pierce). The numbered and lettered triangles indicate the protein spots whose amino-terminal amino acid sequences were determined by Edman degradation. (A and B) Proteins obtained from culture supernatants of speB mutants of MGAS 315 (serotype M3) (A) and MGAS 5005 (serotype M1) (B) grown to exponential phase. (C and D) Proteins obtained from stationary-phase culture supernatants of speB mutants of MGAS 315 (C) and MGAS 5005 (D).
Protein spot identification.
The amino-terminal 10 or 12 amino acid residues for a total of 66 protein spots were determined by Edman degradation, and the data were used to interrogate available databases, including a GAS M1 genome sequence (http://genome.ou.edu/strep.html). Fifty-two of the 53 unique amino-terminal sequences obtained were unambiguous, and one had ambiguous identification of the first three residues (Table 2). Forty-six of the 53 amino-terminal amino acid sequences were 100% identical to the inferred sequence of an open reading frame (ORF) present in the available M1 genome sequence. Four of the 53 amino-terminal sequences had one or two mismatched amino acid residues and had a cognate ORF in the genome. It is unknown if the amino acid mismatches were due to naturally occurring polymorphisms or nucleotide sequencing errors. The genes encoding 47 protein spots were identified from an available GAS M1 genome sequence. The emm3 and speA genes, encoding protein spots 15 and 28, respectively, were obtained by analysis of the GenBank database. The identities of three MGAS 315 protein spots and one MGAS 5005 protein spot could not be discerned from available database information.
TABLE 2.
Proteins identified in the culture supernatants of the speB mutants of GAS serotype M1 MGAS 5005 and serotype M3 MGAS 315
| Spot no.a | Amino-terminal amino acid sequenceb | GenBank accession or ORF no. | Identical or homologous protein | Molecular mass (kDa)c | pIc |
|---|---|---|---|---|---|
| 1 | 2-TEMLKGIAAS | RST01348 | Phosphotransferase enzyme I | 63.1 | 4.48 |
| 2 | 1-MkKRVKIVATL | RST00804 | Pyruvate kinase | 54.5 | 4.86 |
| 3 | 1-TAQTPIHVYSE | RST01429 | Arginine deiminase | 46.6 | 4.93 |
| 4 | 2-TATKQHKKVIL | RST00350 | Lactate dehydrogenase | 35.3 | 5.07 |
| 5 | 1-MKRIAVLTSGG | RST00803 | 6-Phosphofructokinase | 35.7 | 5.36 |
| 6 | 2-VKLVFARHGES | RST01563 | Phosphoglyceromutase | 26.1 | 5.01 |
| 7 | 45-GGAEEAAKDSF | RST00830 | Ribosomal protein L7/L12 | 12.2 | 4.40 |
| 8 | 2-AKDIKFSADAR | RST00771 | GroEL | 57.1 | 4.65 |
| 9 | 2-AKQYKNLVNGE | RST01349 | NADP+-dependent glyceraldehyde-3-phosphate dehydrogenase | 50.3 | 4.99 |
| 10 | 2-SIITDVYAREV | RST00465 | Enolase | 47.3 | 4.64 |
| 11 | 2-AEITAKLVKEL | RST00510 | Elongation factor EF-Ts | 37.2 | 4.76 |
| 12 | 2-AKLTVKDVDLK | RST00513 | Phosphoglycerate kinase | 37.2 | 4.76 |
| 13 | 28-QDVMLETHKA | RST00809 | Immunogenic secreted protein homologue | 51.5 | 5.67 |
| 14 | 35-ESNKQNTASTE | RST00389 | Streptolysin O | 60.4 | 6.11 |
| 15d | 42-DARSVNGEFP | U40231 | M3 protein | 55.9 | 7.01 |
| 16 | 38-VSGKENKKSDV | RST00387 | NAD+-glycohydrolase | 46.5 | 7.92 |
| 17 | 2-TQVFQGRSFL | RST01431 | Ornithine carbamoyltransferase | 37.9 | 5.16 |
| 18 | 2-VVKVGINGFGR | RST00038 | Plasmin receptor/glyceraldehyde-3-phosphate dehydrogenase | 35.9 | 5.33 |
| 19 | 2-TKQKIVxALG | RST01436 | Carbamate kinase | 33.2 | 4.60 |
| 20 | 1-MLVYIAGSGAM | RST00882 | Dehydropantoate 2-reductase | 33.8 | 4.77 |
| 21 | 2-AIVSAEKFVQ | RST00186 | Aldolase | 31.2 | 4.81 |
| 22 | 30-DSFSANQEIR | RST01222 | Human Mac-1 | 34.9 | 6.41 |
| 23 | 38-ETIASSSTRHF | RST01220 | Autolysin | 21.6 | 4.23 |
| 24 | 35-DELSTMSEPT | RST00815 | Serine protease | 178 | 6.15 |
| 25d | [DSG][APV][RP]SVNGEFP | U40231 | M3 protein | ||
| 26d | 23-QQDPDPSQLHR | U40453 | Streptococcal pyrogenic exotoxin A | 25.8 | 5.45 |
| 27 | 28-HDNIDEKGKVH | RST01215 | Unknown function | 24.5 | 7.61 |
| 28 | 2-TFDAIDQLAV | RST01509 | Transketolase | 71.3 | 4.81 |
| 29 | 2-AREFSLAKTRN | RST00093 | Elongation factor G | 76.5 | 4.73 |
| 30 | 31-ADNASVTNKADF | RST00768 | Cyclodextrin glucosyltransferase | 76.2 | 5.41 |
| 31 | 42-NGDGNPREVIE | RST01579 | M1 protein | 49.9 | 5.51 |
| 32 | 2-TTIDFKAEVDK | RST00833 | Dipeptidase | 51.3 | 4.77 |
| 33 | 38-VSGKENKKSDV | RST00387 | NAD+-glycohydrolase | 46.5 | 7.92 |
| 34 | 66-LRhENKDLKARL | RST01579 | M1 protein | 47.3 | 5.71 |
| 35 | 25-EDLSTKIAK | RST01497 | L. lactis secreted 45-kDa protein | 39.6 | 6.65 |
| 36 | 33-ETYTSXXXX | RST00460 | Streptococcal inhibitor of complement | 31.2 | 4.17 |
| 37 | 2-SRKPIIAGN | RST00099 | Triosephosphate isomerase | 26.6 | 4.44 |
| 38 | 44-QTQVSNDVVLN | RST01835 | Mitogenic factor (DNase) | 25.4 | 7.38 |
| 39 | 43-RQTQVSNDVVL | RST01835 | Mitogenic factor (DNase) | 25.5 | 8.25 |
| 40 | 217-SSEKEQLTIEK | RST01579 | M1 protein | 29.4 | 9.04 |
| 41 | 27-IAGYGWLPDRPP | RST00593 | Streptokinase | 47.1 | 6.21 |
| Id | SIRSLFGGLRE | —d | |||
| IId | KKIDRVALETL | —d | |||
| III | 2-STSFENKATNR | RST00259 | RopA | 47.1 | 4.29 |
| IVd | AIEASKLQAG | —d | |||
| V | 2-ASKDFHIVAET | RST01347 | Phosphocarrier protein | 9.0 | 4.63 |
| VI | 2-AeYEILYIIRP | RST01606 | Ribosome protein S6 | 11.1 | 4.89 |
| VII | 2-ANKQDLIAKV | RST00136 | Histone-like protein | 9.6 | 9.04 |
| VIIId | MHQPHQPLPP | —d | |||
| IX | 25-SGPKVPYTQE | RST01706 | Class B acid phosphatase | 24.5 | 9.10 |
| X | 2-SKIIGIDLGTT | RST00474 | DnaK protein | 64.9 | 4.49 |
| XI | 2-ANAIIETAKE | RST00322 | Ribosome recycling factor | 20.6 | 5.66 |
| XII | 2-SRIGNKVITM | RST00748 | Ribosomal protein L6 | 19.4 | 9.57 |
Spots I to XII are not in Fig. 1.
The number indicates the position of the first detected amino acid residue in the amino acid sequence inferred on the basis of the ORF. Lowercase letters indicate amino acids that do not match the residues in the amino acid sequences derived from the M1 genome sequence. X indicates amino acid residues that could not be determined.
Theoretical values calculated on the basis of the mature protein, presuming an intact carboxy terminus.
Protein whose gene was not found in the available M1 genome sequence.
Several distinct protein spots had amino-terminal amino acid sequences indicating that they were related proteins. For example, two protein spots (spots 38 and 39 [Fig. 1]) identified as mitogenic factor apparently arose by alternative proteolytic cleavage of the signal peptide. Protein spots 31, 34, and 40 had amino-terminal amino acid sequences corresponding to sequences beginning at amino acid residues 42, 66, and 217, respectively, of M1 protein. Protein spot 31 was the expected mature M1 protein whereas protein spots 34 and 40 apparently arose due to internal proteolysis by an enzyme other than the SpeB cysteine protease. Protein spots 16 and 33 each had the identical amino-terminal amino acid sequence appropriate for NAD+-glycohydrolase (3, 14, 24) (referred to as NADase in this report) but had different pIs (Fig. 1). The apparent molecular weight of protein spot 33 was slightly less than that of protein spot 16, and the spot may have arisen as a consequence of proteolytic cleavage toward the carboxy terminus of the protein. In the aggregate, 66 protein spots were analyzed, 53 unique amino-terminal amino acid sequences were obtained, and 44 unique proteins were identified (Table 2).
Proteins with apparent secretion signal sequences.
The amino-terminal sequences of 17 proteins did not start with the first or second inferred amino acid residue of the ORF and apparently were products of a proteolytic process. The inferred amino-terminal amino acids of 16 of the 17 proteins have characteristics typical of secretion signal sequences, including charged residues followed by a stretch of hydrophobic residues and cleavage after an alanine residue (Fig. 2). Ribosomal protein L7/L12 (protein spot 7) was excluded from further consideration because it lacks an apparent secretion signal sequence. Eight of these 16 extracellular proteins have been previously identified in GAS and characterized, including NADase, streptolysin O, M1 and M3 proteins, mitogenic factor, streptococcal pyrogenic exotoxin A, streptococcal inhibitor of complement, and streptokinase (8, 9, 23) (Table 3). Eight new proteins were identified, including homologues of class B acid phosphatase (spot IX [not shown in Fig. 1), serine protease (spot 24), and cyclomaltodextrin glucanotransferase (spot 30). Protein spots 13 and 35 are homologues of two proteins with unknown functions, i.e., GAS immunogenic secreted protein (31) and a putative secreted protein made by Streptococcus mutans (GenBank accession no. U78607), respectively. Protein spot 22 lacks a known bacterial homologue but has regions of identity with a human protein known as Mac-1 (2, 7, 53). Protein spot 23 is homologous to several peptidoglycan hydrolases (43). Protein spot 27 has no homologue in available protein databases and is of unknown function.
FIG. 2.
Putative secretion signal sequences. The first amino acids of the mature proteins (rightmost residue) are the residues after the italicized alanine residue. The arrow indicates the processing cleavage site. The amino acid sequences of the cleaved peptides for 13 of the 16 proteins were derived from the cognate ORFs in an available M1 genome sequence. The amino acid sequences of SP15, SP26, and SP36 were inferred from the nucleotide sequences deposited in GenBank under accession numbers X80186, U40453, and AF232306. The single-letter amino acid abbreviations are used.
TABLE 3.
Identified streptococcal proteins that contain secretion signal sequences
| Function and characterization status | Extracellular proteins |
|---|---|
| Previously characterized | NAD+-glycohydrolase (SP16), streptolysin O (SP14), M1 (SP31) and M3 (SP15) protein, mitogenic factor (DNase) (SP38), streptococcal pyrogenic exotoxin A (SP26), streptococcal inhibitor of complement (SP36), streptokinase (SP41) |
| Homologous to protein with known function | Class B acid phosphatase (SPIX), serine protease (SP24), cyclodextrin glucosyltransferase (SP30), autolysin (SP23), SP22 |
| Homologous to known protein but no known function | Immunogenic secreted protein homologue (SP13), L. lactis putative secreted protein (SP35) |
| No known homologue and no known function | SP27 |
Proteins that lack apparent secretion signal sequences.
Twenty-seven of the 43 identified proteins had no apparent secretion signal sequence. Among them, 2 proteins started with the first inferred amino acid residue (methionine), 24 proteins started with the second inferred amino acid residue, and, as noted above, one protein (ribosomal protein L7/L12) started with inferred amino acid 45. Consistent with the lack of a secretion signal sequence, these proteins are presumed to be intracellular proteins, including eight glycolytic pathway metabolic enzymes, three chaperonins, three proteins involved in translation, two sugar transport proteins, three urea cycle proteins, three ribosomal proteins, one DNA binding protein, and five other enzymes (Table 4). The other protein, referred to as antitumor glycoprotein or arginine deiminase (spot 3), may be an extracellular protein that lacks a secretion signal sequence (8, 9, 23).
TABLE 4.
Presumed or potential cytosolic proteins identified in the GAS culture supernatants
| Protein class | Protein(s) |
|---|---|
| Glycolysis | 6-Phosphofructokinase (SP5), aldolase (SP21), triosephosphate isomerase (SP37), phosphoglyceromutase (SP6), glyceraldehyde-3-phosphate dehydrogenase (SP18), phosphoglycerate kinase (SP12), enolase (SP10), pyruvate kinase (SP2) |
| Chaperonin | GroEL (SP8), DnaK (SPX), RopA (SPIII) |
| Ribosomal | L6 (SPXII), L7/12 (SP7), S6 (SPVI) |
| DNA binding | Histone-like protein (SPVII) |
| Protein synthesis | Elongation factor G (SP29), elongation factor EF-Ts (SP11), ribosome recycling factor (SPXI) |
| Urea cycle | Carbamate kinase (SP19), ornithine carbamoyltransferase (SP17), arginine deiminase (SP3) |
| Sugar transport | Phosphotransferase enzyme I (SP1), phosphocarrier protein (SPV) |
| Other enzymes | Lactate dehydrogenase (SP4), NADP+-dependent glyceraldehyde-3-phosphate dehydrogenase (SP9), transketolase (SP28), dipeptidase (SP32), dehydropantoate 2-reductase (SP20) |
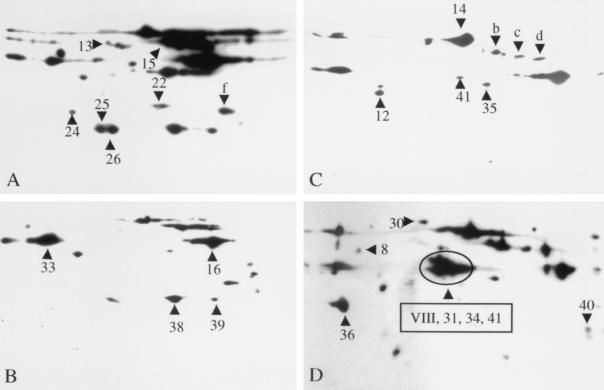
Two-dimensional Western blotting of stationary-phase culture supernatant proteins with sera from infected mice.
Culture supernatant proteins obtained from the speB mutants of MGAS 315 and MGAS 5005 grown to stationary phase were analyzed by two-dimensional Western blotting with convalescent-phase sera obtained from mice with experimental soft tissue infections caused by the wild-type parental organisms of each of these two strains. Stationary-phase cultures were used because all abundant proteins present in exponential-phase culture supernatants were present in stationary-phase preparations, but the converse was not true. The analysis was conducted with both the homologous and heterologous protein-serum pairs, and representative Western blots are shown in Fig. 3. Nineteen protein spots (SP8, SP12, SP13, SP14, SP15, SP16, SP22, SP24, SP26, SP30, SP31, SP33, SP34, SP35, SP36, SP38, SP39, SP40, and SP41) that were reactive in the Western blots matched proteins identified in Fig. 1. These 19 protein spots represent 15 distinct proteins (Table 5). SP8 (GroEL) and SP12 (phosphoglycerate kinase) are generally considered to be cytosolic proteins in bacteria. The other 13 proteins have apparent secretion signal sequences (Fig. 2) and are presumed to be proteins that are actively secreted. The identity of protein spot VIII could not be determined because it lacked homology with known proteins inferred from the available M1 genome sequence. Amino-terminal sequencing of protein spots b, c, d, and f repeatedly failed, and hence these protein spots could not be identified.
FIG. 3.
Two-dimensional Western blot analysis of stationary-phase GAS culture supernatant proteins with convalescent-phase sera obtained from mice with soft-tissue infection. Concentrated culture supernatant proteins (60 μg) were separated in the first dimension by isoelectric focusing (pH 3 to 10) and in the second dimension by SDS-PAGE with 12% (A and B) or 10% (C and D) gels. Concentrated culture supernatants from speB mutants of serotype M3 MGAS 315 (A and B) and serotype M1 MGAS 5005 (C and D) were used in the analysis. The separated proteins were transferred to nitrocellulose membranes, and Western blotting was performed with convalescent-phase sera obtained from mice infected with MGAS 315 (diluted 1:500) (A and C) and MGAS 5005 (diluted 1:1,000) (B and D). The numbered and lettered triangles indicate the proteins reactive in the Western blots that correspond to the protein spots (numbered identically) in Fig. 1.
TABLE 5.
Immunogenic proteins identified in the culture supernatants of GAS strains MGAS 315 and MGAS 5005
| Spot no. | Identical or homologous protein | Antibody ina:
|
||||
|---|---|---|---|---|---|---|
| Mouse sera
|
Human sera
|
|||||
| MGAS 315 | MGAS 5005 | Pharyngitis | ARF | STSS | ||
| 8 | GroEL | + | + | + | + | + |
| 12 | Phosphoglycerate kinase | + | + | − | − | − |
| 13 | Immunogenic secreted protein homologue | + | + | + | + | + |
| 14 | Streptolysin O | + | + | + | + | + |
| 15 | M3 protein | + | − | − | − | − |
| 16, 33 | NAD+-glycohydrolase | + | + | + | + | + |
| 18 | Glyceraldehyde-3-phosphate dehydrogenase | − | − | − | + | − |
| 22 | Human Mac-1 | + | + | + | + | + |
| 24 | Serine protease | + | + | + | + | + |
| 26 | Streptococcal pyrogenic exotoxin A | + | − | − | − | + |
| 30 | Cyclodextrin glucosyltransferase | − | + | − | − | − |
| 31, 34, 40 | M1 protein | − | + | − | + | + |
| 35 | L. lactis 45-kDa secreted protein | + | + | + | + | + |
| 36 | Streptococcal inhibitor of complement | − | + | − | − | − |
| 38, 39 | Mitogenic factor (DNase) | + | + | + | + | + |
| 41 | Streptokinase | + | + | + | + | + |
Detection of antigen-antibody reactivity for each protein was based on representative Western blot data, as represented in Fig. 3 to 5. The Western blot results were obtained with sera from mice infected subcutaneously with MGAS 315 or MGAS 5005 and from humans with pharyngitis, acute rheumatic fever (ARF), or streptococcal toxic shock syndrome (STSS). +, antibody reactivity detected; −, no antibody reactivity detected.
Although most of the protein spots identified in the analysis were present in both M1 and M3 strains, several proteins were uniquely found in the culture supernatants of one of the organisms. For example, protein spots 31, 34, 40, and VIII were identified only in the supernatant of the serotype M1 MGAS 5005 strain, whereas protein spots 15 and 25 were restricted to the supernatant of the serotype M3 MGAS 315 strain. Several of these protein spots correspond to M1 or M3 protein.
Two-dimensional Western blot analysis of culture supernatant proteins with human sera.
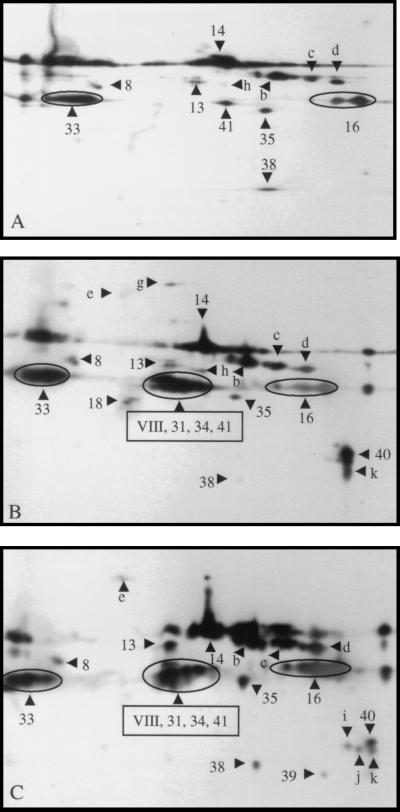
Western blot analysis was also performed for stationary-phase culture supernatant proteins of the speB mutant of serotype M1 MGAS 5005 with sera from humans with various types of GAS infection. Protein-antibody reactivity was observed with serum obtained from a pharyngitis patient, serum from a patient with acute rheumatic fever, and convalescent-phase sera from a human with streptococcal toxic shock syndrome caused by an M1 GAS strain. Representative immunologic data are shown in Fig. 4.
FIG. 4.
Two-dimensional Western blot analysis of stationary-phase GAS culture supernatant proteins with human and mouse sera. Concentrated culture supernatant proteins (60 μg) obtained from the speB mutant of MGAS 5005 (serotype M1) were separated in the first dimension by isoelectric focusing (pH 3 to 10) and in the second dimension by SDS-PAGE with 10% gels. (A) Convalescent-phase serum (diluted 1:20,000) obtained from a human with GAS pharyngitis; (B) serum obtained from a patient with acute rheumatic fever; (C) convalescent-phase serum (diluted 1:20,000) obtained from a human with streptococcal toxic shock syndrome caused by an M1 GAS strain. Control sera obtained from these patients were essentially unreactive when used at the same dilutions. The numbered and lettered triangles indicate the proteins reactive in the Western blots that correspond to the protein spots (numbered identically) in Fig. 1.
In the aggregate, the sera used had reactive antibodies against the following protein spots: SP8, SP13, SP14, SP16, SP33, SP35, SP38, a, c, d, and e. Protein spot h is absent in Fig. 4C. Protein spots 31, 34, 40, and m were present in Fig. 4B and C but absent in Fig. 4A. The occurrence of reactivity with three protein spots representing M1 protein suggests that the patients with toxic shock syndrome and acute rheumatic fever were infected by serotype M1 GAS strains, whereas the pharyngitis patient was infected with a GAS strain other than serotype M1. Antibody reactivity to SP18 was unique to the serum from the acute rheumatic fever patient (Fig. 4B). Convalescent-phase serum from the STSS patient had reactivity with protein spots i and j (Fig. 4C).
Western blot analysis of recombinant GAS proteins.
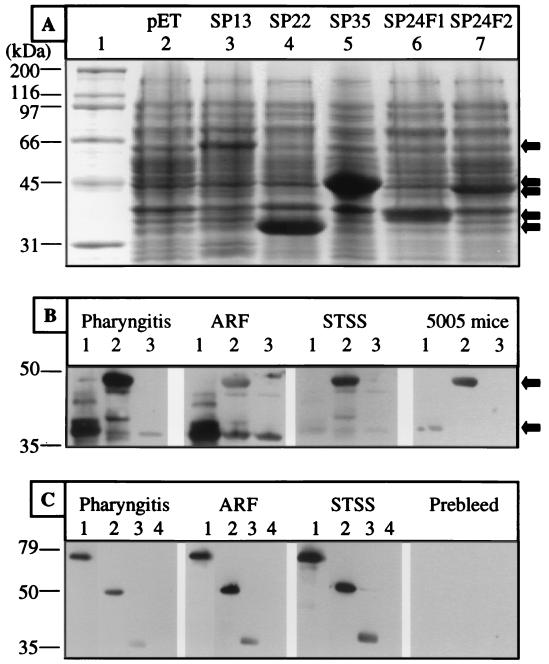
The antigenicities of SP13 (immunogenic secreted protein homologue), SP22 (Mac-1-like protein), SP24 (serine protease homologue), and SP35 (homologue of a Lactococcus lactis secreted protein) were verified by Western blot analysis with recombinant proteins expressed in E. coli (Fig. 5A). The Western blotting was conducted with pooled mouse sera obtained from animals infected subcutaneously with MGAS 5005 (serotype M1) and human sera taken from patients with pharyngitis, acute rheumatic fever, or streptococcal toxic shock syndrome. The Western blot analysis confirmed that antibodies against these proteins were present in infected hosts (Fig. 5). For example, the mouse and human sera were reactive with the two fragments of SP24 (Fig. 5). Similarly, the three human sera reacted with recombinant SP13, SP22, and SP35 (Fig. 5). Control mouse and human sera lacked reactivity with the four recombinant proteins.
FIG. 5.
Western blot analysis of recombinant GAS proteins. SP13, SP22, SP35, and two fragments of SP24 were used in the analysis. (A) SDS-PAGE of lysates of E. coli BL21 expressing the recombinant proteins. Lane 1, molecular mass markers; lane 2, empty vector; lanes 3 to 7, E. coli lysates containing recombinant proteins as indicated. (B) Western blots showing reactivity of sera with the two recombinant fragments of SP24. Sera (diluted 1:2,000) were obtained from humans with pharyngitis, acute rheumatic fever (ARF), and streptococcal toxic shock syndrome (STSS) and from mice infected subcutaneously with MGAS 5005. Lanes 1, SP24 fragment 1 (amino acid residues 57 to 282); lanes 2, SP24 fragment 2 (amino acid residues 1029 to 1366); lanes 3, vector control. (C) Western blots showing reactivity of sera with recombinant SP22 and SP35. Sera (diluted 1:10,000) were obtained from humans with pharyngitis, acute rheumatic fever, and streptococcal toxic shock syndrome and from mice prior to infection with MGAS 5005. Lanes 1, SP13; lanes 2, SP22; lanes 3, SP35; lanes 4, vector control.
DISCUSSION
In this study we conducted an initial analysis of proteins found in relative abundance in the culture supernatants of two GAS strains grown in vitro at 37°C. The study focused on speA-positive strains of serotypes M1 and M3 which are common causes of human invasive infections globally (33–35). We used isogenic speB mutants to assist the identification of proteins expressed in the stationary phase, a time when SpeB production is greatly upregulated in many strains, resulting in degradation of many GAS proteins. In the aggregate, amino-terminal amino acid sequence data were generated for 66 protein spots and 43 distinct proteins were identified. Eight of the 16 proteins with apparent signal sequences involved in active secretion have not been previously described for GAS (Table 3). These eight proteins include homologues of peptidoglycan hydrolase (SP23) (18), class B phosphatase (SPIX) (42, 47), serine protease (SP24) (50), cyclodextrin glucosyltransferase (SP30) (4), a putative secreted protein made in abundance by L. lactis (SP35) (48), an immunogenic secreted protein made by GAS (SP13) (31), a protein with regions of homology with human Mac-1 (SP22) (2, 7, 53), and a protein with no known homologue (SP27).
Previously uncharacterized and apparently secreted GAS proteins.
Five of these eight previously uncharacterized GAS proteins that are apparently secreted have homologues with known functions, whereas three proteins do not. SPIX is 38% identical in amino acid sequence with Morganella morganii class B acid phosphatase, and the two proteins have similar molecular masses (46). In addition, the GAS protein contains the sequence F-D-I-D-D-T-L-L-F-T-S-Q, which is very similar to a conserved structural motif, F-D-I-D-D-T-V-L-F-S-S-P, present in class B phosphatases. A second structural motif (Y-G-D-[AS]-D-[DNA]-D-[IV]) that is conserved in class B acid phosphatases also is present in SpIX (Y-G-D-S-D-E-D-I) (42, 47). Class B acid phosphatases are secreted enzymes that hydrolyze organic phosphate monoesters and also transfer the orthophosphoryl group from organic phosphoric acid esters to nucleosides and other compounds with free hydroxyl groups. The exact physiologic role of these enzymes has not been clarified, and it is not known if they participate in host-pathogen interactions.
SP23 has 46% amino acid sequence identity with Streptococcus thermophilus peptidoglycan hydrolase (18). Both of these streptococcal proteins lack the domain located in the carboxy terminus of the L. lactis major autolysin that is involved in cell wall binding.
SP30 has 30 to 50% amino acid identity and a very similar inferred molecular mass to bacterial cyclodextrin glycosyltransferases (4). These enzymes are secreted proteins and catalyze the degradation of starch into cyclodextrins that are further metabolized by the bacterium.
The inferred amino acid sequence of the protein corresponding to SP24 is homologous to those of several serine proteases. The best sequence matches were identified with serine proteases that cleave and inactivate human complement protein C5 and are made by GAS and the related pathogen group B Streptococcus (8, 15, 27). The amino acid sequences of SP24 and C5a peptidase inferred from the available M1 genome sequence are 31.4% identical in a region containing 936 amino acid residues. SP24 is also homologous to a L. lactis serine protease located on the cell surface. The L. lactis SK11 protease (50) and SP24 precursors have 1,962 and 1,647 amino acid residues, respectively, and the mature forms have 1,775 and 1,613 amino acid residues, respectively.
Importantly, SP24 has regions of homology with several serine proteases in areas containing the catalytic triad amino acid residues (aspartic acid, histidine, and serine) (Fig. 6). SP24 also has an inferred LPXTG motif located at the carboxy terminus that is characteristic of gram-positive cell surface-anchored proteins (12). Interestingly, SP24 was detected as a protein spot with an apparent molecular mass of 31 kDa, indicating that the full-length protein undergoes posttranslational cleavage. The SP24 protein fragment begins after the apparent secretion signal peptide and is composed of 280 amino acid residues that contain the aspartic acid and histidine residues of the catalytic triad, but not the serine residue. These results suggest that, unlike the L. lactis SK11 protease (50), the streptococcal serine protease homologue does not have a conventional propeptide region. It is unknown whether the 31-kDa SP24 was generated by nonspecific proteolysis or by a targeted and orchestrated GAS cleavage process that has biologic significance.
FIG. 6.
Homologous regions of several bacterial serine proteases and an inferred streptococcal serine protease. Shown are regions containing the catalytic triad amino acid residues (arrows) aspartic acid (A), serine (B), and histidine (C). The shaded amino acid residues match the consensus sequence. The numbers at the left and right sides of each sequence indicate amino acid positions. SP, serotype M1 MGAS 5005 serine protease homologue; LP, L. lactis SK11 serine protease (GenBank accession number J04962); SC, subtilisin Carlsberg (GenBank accession number X03341); PK, proteinase K (GenBank accession number X14689).
SP13 is homologous to a GAS immunogenic secreted protein of unknown function termed Isp and described by McIver et al. (31). The amino acid sequences of SP13 and Isp are 49% identical. The SP13 Isp homologue also has 43.7% amino acid sequence identity in a 113-amino-acid region with the TraG putative transfer protein encoded by a gene in staphylococcal conjugative multiresistance plasmid pSK41.
SP35 has 32.8% identity in a region of 340 amino acid residues with Usp 45, a major secreted 45-kDa protein of L. lactis with unknown function (48). Schubert et al. (43) recently reported that a homologous extracellular protein made by L. monocytogenes has peptidoglycan lytic activity.
SP22 does not have a known bacterial homologue, but it does have two features that might provide clues regarding its interaction with the host. The SP22 protein has 23% amino acid identity in a 203-residue region with the alpha subunit of human leukocyte adhesion receptor Mac-1 (1, 7, 53). Mac-1 is a complement receptor and participates in inflammation. Monoclonal antibodies directed against the Mac-1 alpha subunit inhibit neutrophil aggregation and adhesion to endothelial cells. A second feature that may provide insight regarding function is that SP22 has an arginine-glycine-aspartic acid tripeptide (RGD) which can mediate the interaction of RGD-containing proteins with human integrins (22, 45). On the basis of these structural clues, we speculate that SP22 interferes with the normal host function of Mac-1. Studies to address the role of the SP22 protein in several aspects of host-pathogen interactions are under way.
SP27 has no known prokaryotic or eukaryotic homologue and, hence, is of unknown function.
Proteins generally considered to be located in the cytosol.
More than one-half of the culture supernatant proteins that we identified do not have apparent secretion signal sequences and are traditionally considered to be cytosolic proteins involved in intracellular processes. However, several of these proteins have been previously reported to be located extracellularly in streptococci, including histone-like protein (44), glyceraldehyde-3-phosphate dehydrogenase (37, 38, 51, 52), α-enolase (39), arginine deiminase (antitumor glycoprotein) (9, 23), and phosphocarrier protein (11). Glycolytic enzymes also have been detected in association with the cell surfaces of several other microbial pathogens (13, 32). We note that our analysis identified a total of eight glycolytic enzymes in the culture supernatants of the GAS strains studied. It is not clear if the proteins we identified that are generally considered to be cytosolic proteins are actively secreted by an unknown mechanism or passively released from cells undergoing autolysis. However, several of the proteins were present in abundance in the supernatants of mid-log-phase cultures, suggesting that the extracellular location of these proteins may not be solely due to passive release. Regardless of the mechanism responsible for their presence in the culture supernatant of GAS, their occurrence may influence the course of host-pathogen interactions. In this regard, we note that it has been reported that α-enolase binds to plasminogen in vitro (39), and binding of glyceraldehyde-3-phosphate dehydrogenase to human pharyngeal cells induces phosphorylation of host cell proteins (40). Hence, this enzyme appears to modulate host cell signaling events.
Immunogenicity of the culture supernatant proteins in infected hosts.
A general finding was that proteins with apparent secretion sequences were more commonly immunogenic in the few infected hosts studied than proteins lacking these sequences. Unlike the proteins without the apparent secretion signal sequences, 12 of the 16 proteins with secretion signal sequences had host antibody reactivity, and 8 of them were reactive with all five sera tested (Table 5). As expected, serotype-specific M3 protein, M1 protein, and streptococcal inhibitor of complement were not immunoreactive with some of the sera tested.
Although many antigenic proteins were identified by the two-dimensional Western blot analysis, in this study we chose to clone and overexpress only four of them, mainly to confirm their identity and immunogenicity. These four proteins were selected because they had apparent secretion signal sequences and were previously uncharacterized GAS molecules. In addition, inasmuch as two GAS extracellular proteases are known virulence factors (27, 29, 30), it is reasonable to speculate that the serine protease homologue also contributes to host-pathogen interactions. However, this issue clearly requires additional analysis. Two factors that contributed to the selection of SP22 were its regions of identity with the human Mac-1-like protein and the presence of an RGD motif. RGD motifs present in proteins made by microbial pathogens mediate binding to human integrins, thereby triggering downstream cell-signaling effects. Several microbial integrin binding proteins with the RGD motif are proven virulence factors, including the extracellular cysteine protease (SpeB) made by serotype M1 organisms (22, 45). We will report elsewhere on extensive characterization of the SP22 molecule, including cell biologic and immunologic aspects (B. Lei et al., unpublished data).
ACKNOWLEDGMENTS
We thank I. Abdi for technical assistance; J. C. Smoot and M. S. Chaussee for assistance and helpful discussions; M. Garfield for protein sequencing; H. Hill, L. G. Veasy, and D. Low for providing sera; and N. P. Hoe for critical reading of the manuscript.
REFERENCES
- 1.Akesson P, Sjoholm A G, Bjorck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–1088. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 2.Arnaout M A. Structure and function of the leukocyte adhesion molecules CD11/CD18. Blood. 1990;75:1037–1050. [PubMed] [Google Scholar]
- 3.Bernhard G C, Stollerman G H. Serum inhibition of streptococcal diphosphopyridine nucleotidase in uncomplicated streptococcal pharyngitis and in rheumatic fever. J Clin Invest. 1959;38:1942–1949. doi: 10.1172/JCI103973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binder F, Huber O, Böck A. Cyclodextrin-glycosyltransferase from Klebsiella pneumoniae M5a1: cloning, nucleotide sequence and expression. Gene. 1986;47:269–277. doi: 10.1016/0378-1119(86)90070-3. [DOI] [PubMed] [Google Scholar]
- 5.Carroll J A, Garon C F, Schwan T G. Effects of environmental pH on membrane proteins in Borrelia burgdorferi. Infect Immun. 1999;67:3181–3187. doi: 10.1128/iai.67.7.3181-3187.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cash P. Proteomics in medical microbiology. Electrophoresis. 2000;21:1187–1201. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1187::AID-ELPS1187>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 7.Corbi A L, Kishimoto T K, Miller L J, Springer T A. The human leukocyte adhesion glycoprotein Mac-1 (complement receptor type 3, CD11b) a subunit. J Biol Chem. 1988;263:12403–12411. [PubMed] [Google Scholar]
- 8.Cunningham M W. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degnan B A, Fontaine M C, Doebereiner A H, Lee J J, Mastroeni P, Dougan G, Goodacre J A, Kehoe M A. Characterization of an isogenic mutant of Streptococcus pyogenes Manfredo lacking the ability to make streptococcal acid glycoprotein. Infect Immun. 2000;68:2441–2448. doi: 10.1128/iai.68.5.2441-2448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diederich L, Roth A, Messer W. A versatile plasmid vector system for the regulated expression of genes in Escherichia coli. BioTechniques. 1994;16:916–923. [PubMed] [Google Scholar]
- 11.Dubreuil J D, Jacques M, Brochu D, Frenette M, Vadeboncoeur C. Surface location of HPr, a phosphocarrier of the phosphoenolpyruvate:sugar phosphotransferase system in Streptococcus suis. Microbiology. 1996;142:837–843. doi: 10.1099/00221287-142-4-837. [DOI] [PubMed] [Google Scholar]
- 12.Fischetti V A. Surface proteins on gram-positive bacteria. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: ASM Press; 2000. pp. 11–24. [Google Scholar]
- 13.Gozalbo D, Gil-Navarro, Azorin I, Renau-Piqueras J, Martinez J P, Gil M L. The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect Immun. 1998;66:2052–2059. doi: 10.1128/iai.66.5.2052-2059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grushoff P S, Shany S, Bernheimer A W. Purification and properties of streptococcal nicotinamide adenine dinucleotide glycohydrolase. J Bacteriol. 1975;122:599–605. doi: 10.1128/jb.122.2.599-605.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill H R, Bohnsack J F, Morris E Z, Augustine N H, Parker C J, Cleary P P, Wu J T. Group B streptococci inhibit the chemotactic activity of the fifth component of complement. J Immunol. 1988;141:3551–3556. [PubMed] [Google Scholar]
- 16.Hoe N, Nakashima K, Grigsby D, Pan X, Dou S J, Naidich S, Garcia M, Kahn E, Bergmire-Sweat D, Musser J M. Rapid molecular genetic subtyping of serotype M1 Group A Streptococcus strains. Emerg Infect Dis. 1999;5:254–263. doi: 10.3201/eid0502.990210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoe N P, Nakashima K, Lukomski S, Grigsby D, Liu M, Kordari P, Dou S-J, Pan X, Vuopio-Varkila J, Salmenlinna S, McGeer A, Low D E, Schwartz B, Schuchat A, Naidich S, De Lorenzo D, Fu Y-X, Musser J M. Rapid selection of structural variants of group A Streptococcus complement-inhibiting protein in serotype M1 epidemic waves. Nat Med. 1999;5:924–929. doi: 10.1038/11369. [DOI] [PubMed] [Google Scholar]
- 18.Husson-Kao C, Mengaud J, Benbadis L, Chapot-Chartier M-P. Mur1, a Streptococcus thermophilus peptidoglycan hydrolase devoid of a specific cell wall binding domain. FEMS Microbiol Lett. 2000;187:69–76. doi: 10.1111/j.1574-6968.2000.tb09139.x. [DOI] [PubMed] [Google Scholar]
- 19.Jungblut P R, Bumann D, Haas G, Zimny-Arndt U, Holland P, Lamer S, Siejak F, Aebischer A, Meyer T F. Comparative proteome analysis of Helicobacter pylori. Mol Microbiol. 2000;36:710–725. doi: 10.1046/j.1365-2958.2000.01896.x. [DOI] [PubMed] [Google Scholar]
- 20.Jungblut P R, Grabher G, Stoffler G. Comprehensive detection of immunorelevant Borrelia garinii antigens by two-dimensional electrophoresis. Electrophoresis. 1999;20:3611–3622. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3611::AID-ELPS3611>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Jungblut P R, Schaible U E, Mollenkopf H J, Zimny-Arndt U, Raupach B, Mattow J, Halada P, Lamer S, Hagens K, Kaufmann S H E. Comparative proteome analysis of Mycobacterium tuberculosis and Mycobacterium bovis BCG strains: towards functional genomics of microbial pathogens. Mol Microbiol. 1999;33:1103–1117. doi: 10.1046/j.1365-2958.1999.01549.x. [DOI] [PubMed] [Google Scholar]
- 22.Kagawa T F, Cooney J C, Baker H M, McSweeney S, Liu M, Gubba S, Musser J M, Baker E N. Crystal structure of the zymogen form of the streptococcal virulence factor SpeB: an integrin-binding cysteine protease. Proc Natl Acad Sci USA. 2000;97:2235–2240. doi: 10.1073/pnas.040549997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanaoka M, Kawanaka C, Negoro T, Fukita Y, Taya K, Agui H. Cloning and expression of the antitumor glycoprotein gene of Streptococcus pyogenes Su in Escherichia coli. Agric Biol Chem. 1987;51:2641–2648. [Google Scholar]
- 24.Kellner A, Freeman E B, Carlson A S. Neutralizing antibodies to streptococcal diphosphopyridine nucleotidease in the serum of experimental animals and human beings. J Exp Med. 1958;108:299–309. doi: 10.1084/jem.108.3.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langen H, Takacs B, Evers S, Berndt P, Lahm H-W, Wipf B, Gray C, Fountoulakis M. Two-dimensional map of the proteome of Haemophilus influenzae. Electrophoresis. 2000;21:411–429. doi: 10.1002/(SICI)1522-2683(20000101)21:2<411::AID-ELPS411>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 26.LeBlanc D J, Lee L N, Inamine J M. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase Aad(9) determinant from Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35:1804–1810. doi: 10.1128/aac.35.9.1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, McLandsborough L, Kondagunta A, Cleary P P. C5a peptidase alters clearance and trafficking of group A streptococci by infected mice. Infect Immun. 1996;64:503–510. doi: 10.1128/iai.64.2.503-510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukomski S, Hoe N P, Abdi I, Rurangirwa J, Kordari P, Liu M, Shu-Jun D, Adams G G, Musser J M. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000;68:535–542. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukomski S, Montgomery C A, Rurangirwa J, Geske R S, Barrish J P, Adams G J, Musser J M. Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect Immun. 1999;67:1779–1788. doi: 10.1128/iai.67.4.1779-1788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukomski S, Sreevatsan S, Amberg C, Reichardt W, Woischnik M, Podbielski A, Musser J M. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Invest. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McIver K S, Subbarao S, Kellner E M, Heath A S, Scott J R. Identification of isp, a locus encoding an immunogenic secreted protein conserved among group A streptococci. Infect Immun. 1996;64:2548–2555. doi: 10.1128/iai.64.7.2548-2555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Modun B, Williams P. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect Immun. 1999;67:1086–1092. doi: 10.1128/iai.67.3.1086-1092.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musser J M, Hauser A R, Kim M H, Schlievert P M, Nelson K, Selander R K. Streptococcus pyogenes causing toxic shock-like-syndrome and other invasive diseases: clonal diversity and pyrogenic exotoxin expression. Proc Natl Acad Sci USA. 1991;88:2668–2672. doi: 10.1073/pnas.88.7.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Musser J M, Kapur V, Szeto J, Pan X, Swanson D, Martin D. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musser J M, Krause R M. The revival of group A streptococcal diseases with a commentary on staphylococcal toxic shock syndrome. In: Krause R M, Fauci A, editors. Emerging infections. San Diego, Calif: Academic Press; 1998. pp. 185–218. [Google Scholar]
- 36.Musser J M, Stockbauer K, Kapur V, Rudgers G W. Substitution of cysteine 192 in a highly conserved Streptococcus pyogenes extracellular cysteine protease (interleukin 1β convertase) alters proteolytic activity and ablates zymogen processing. Infect Immun. 1996;64:1913–1917. doi: 10.1128/iai.64.6.1913-1917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pancholi V, Fischetti V A. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate dehydrogenase with multiple binding activity. J Exp Med. 1992;176:415–426. doi: 10.1084/jem.176.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pancholi V, Fischetti V A. Glyceraldehyde-3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc Natl Acad Sci USA. 1993;90:8154–8158. doi: 10.1073/pnas.90.17.8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pancholi V, Fischetti V A. α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. 1998;273:14503–14515. doi: 10.1074/jbc.273.23.14503. [DOI] [PubMed] [Google Scholar]
- 40.Pancholi V, Fischetti V A. Regulation of the phosphorylation of human pharyngeal cell proteins by group A streptococcal surface dehydrogenase: signal transduction between streptococci and pharyngeal cells. J Exp Med. 1997;186:1633–1643. doi: 10.1084/jem.186.10.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosenkrands I, Weldingh K, Jacobsen S, Veggerby Hansen C, Florio W, Gianetri I, Andersen P. Mapping and identification of Mycobacterium tuberculosis proteins by two-dimensional gel electrophoresis, microsequencing and immunodetection. Electrophoresis. 2000;21:935–948. doi: 10.1002/(SICI)1522-2683(20000301)21:5<935::AID-ELPS935>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 42.Rossolini G K, Schippa S, Riccio M L, Berlutti F, Macaskie L E, Thaller M C. Bacterial nonspecific acid phosphohydrolases: physiology, evolution and use as tools in microbial biotechnology. Cell Mol Life Sci. 1998;54:833–850. doi: 10.1007/s000180050212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schubert K, Bichlmaier A M, Mager E, Wolff K, Ruhland G, Fiedler F. P45, an extracellular 45 kDa protein of Listeria monocytogenes with similarity to protein p60 and exhibiting peptidoglycan lytic activity. Arch Microbiol. 2000;173:21–28. doi: 10.1007/s002030050003. [DOI] [PubMed] [Google Scholar]
- 44.Stinson M W, McLaughlin R, Choi S H, Juarez Z E, Barnard J. Streptococcal histone-like protein: primary structure of hlpA and protein binding to lipoteichoic acid and epithelial cells. Infect Immun. 1998;66:259–265. doi: 10.1128/iai.66.1.259-265.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stockbauer K E, Magoun L, Liu M, Burns E H, Jr, Gubba S, Renish S, Pan X, Bodary S C, Baker E, Coburn J, Leong J M, Musser J M. A natural variant of the cysteine protease virulence factor of group A Streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins αvβ3 and αIIbβ3. Proc Natl Acad Sci USA. 1999;96:242–247. doi: 10.1073/pnas.96.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thaller M C, Lombardi G, Berlutti F, Schippa S, Rossolini G M. Cloning and characterization of the NapA acid phosphotase/phosphotransferase of Morganella morganii: identification of a new family of bacterial acid-phosphatase-encoding genes. Microbiology. 1995;141:147–154. doi: 10.1099/00221287-141-1-147. [DOI] [PubMed] [Google Scholar]
- 47.Thaller M C, Schippa S, Rossolini G M. Conserved sequence motifs among bacterial, eukaryotic, and archaeal phosphatases that define a new phosphohydrolase superfamily. Prot Sci. 1998;7:1647–1652. doi: 10.1002/pro.5560070722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Asseldonk M, Rutten G, Oteman M, Siezen R J, de Vos W M, Simons G. Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene. 1990;95:155–160. doi: 10.1016/0378-1119(90)90428-t. [DOI] [PubMed] [Google Scholar]
- 49.Veasy L G, Tani L Y, Hill H R. Persistence of acute rheumatic fever in the intermountain area of the United States. J Pediatr. 1994;124:9–16. doi: 10.1016/s0022-3476(94)70247-0. [DOI] [PubMed] [Google Scholar]
- 50.Vos P, Simons G, Siezen R J, de Vos W M. Primary structure and organization of the gene for a procaryotic, cell envelope-located serine proteinase. J Biol Chem. 1989;264:13579–13585. [PubMed] [Google Scholar]
- 51.Winram S B, Lottenberg R. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology. 1996;142:2311–2320. doi: 10.1099/13500872-142-8-2311. [DOI] [PubMed] [Google Scholar]
- 52.Winram S B, Lottenberg R. Site-directed mutagenesis of streptococcal plasmin receptor (Plr) identifies the C-terminal Lys334 as essential for plasmin binding, but mutation of the plr gene does not reduce plasmin binding to group A streptococci. Microbiology. 1998;144:2025–2035. doi: 10.1099/00221287-144-8-2025. [DOI] [PubMed] [Google Scholar]
- 53.Wright S D, Weitz J S, Huang A J, Levin S M, Silverstein S C, Leike J D. Complement receptor type three (CD11b/CD18) of human polymorphonuclear leukocytes recognizes fibrinogen. Proc Natl Acad Sci USA. 1988;85:7734–7738. doi: 10.1073/pnas.85.20.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zevering Y, Jacob L, Meyer T F. Naturally acquired human immune responses against Helicobacter pylori and implications for vaccine development. Gut. 1999;45:465–474. doi: 10.1136/gut.45.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]