Abstract
Macrocephaly frequently occurs in single-gene disorders affecting the PI3K-AKT-MTOR pathway; however, epigenetic mutations, mosaicism, and copy number variations (CNVs) are emerging relevant causative factors, revealing a higher genetic heterogeneity than previously expected. The aim of this study was to investigate the role of rare CNVs in patients with macrocephaly and review genomic loci and known genes. We retrieved from the DECIPHER database de novo <500 kb CNVs reported on patients with macrocephaly; in four cases, a candidate gene for macrocephaly could be pinpointed: a known microcephaly gene–TRAPPC9, and three genes based on their functional roles–RALGAPB, RBMS3, and ZDHHC14. From the literature review, 28 pathogenic CNV genomic loci and over 300 known genes linked to macrocephaly were gathered. Among the genomic regions, 17 CNV loci (~61%) exhibited mirror phenotypes, that is, deletions and duplications having opposite effects on head size. Identifying structural variants affecting head size can be a preeminent source of information about pathways underlying brain development. In this study, we reviewed these genes and recurrent CNV loci associated with macrocephaly, as well as suggested novel potential candidate genes deserving further studies to endorse their involvement with this phenotype.
Keywords: CNV, macrocephaly, neurodevelopmental disorders, TRAPPC9, RALGAPB, RBMS3, ZDHHC14
1. Introduction
Macrocephaly, defined as an occipitofrontal circumference (OFC) at least two standard deviations (SD) above the mean for a given age, sex, and ethnicity [1], affects about 2% of the general population and up to 5% of the children [2,3,4]. This phenotype may be driven by the expansion of the brain parenchyma, leading to a subgroup called megalencephaly, or be related to other conditions such as hydrocephalus or thickening of the frontal skull bone (cranial hyperostosis), unconnected to a primary brain development defect [5]. It can be present at birth (congenital) or be originated postnatally during the growth period (acquired), either occurring as an isolated feature (non-syndromic) or associated with other clinical signs (syndromic), including intellectual disability (ID)/neurodevelopmental delay, obesity, and overgrowth.
Because macrocephaly may indicate an underlying disorder, imaging exams, such as computerized tomography scan, head ultrasound, and magnetic resonance imaging can help to narrow the diagnosis, even in utero [6,7]. However, the etiology of most cases remains unknown, and frequently there is an absence of other significant clinical findings that could contribute to unravel the origin of this phenotype [4]. Nonetheless, it is known that several genetic syndromes have macrocephaly as a main feature, originating from de novo or inherited mutations, such as in Sotos and fragile X syndromes, respectively [2,3].
Most cases of macrocephaly with a known etiology are due to single-gene disorders affecting the PI3K-AKT-MTOR pathway, which directly acts on the process of brain development, including the maintenance, differentiation, and migration of neuronal progenitors, synaptogenesis, and regulation of protein translation. Other commonly affected pathways in macrocephaly cases are Ras/MAPK, that regulates cell cycle, development, senescence, transduces extracellular signs, and SHH, which takes part in the early development of the nervous system [8].
Several genes have already been linked to macrocephaly, amongst which we highlight two as examples. PTEN (OMIM *601728) is a tumor suppressor that interacts with several proteins within the PI3K-AKT-MTOR pathway [9], acting to restrain cell proliferation, growth, and survival [10]. PTEN haploinsufficiency during the early embryonic development leads to increased proliferation of neural progenitor cells and altered apoptosis that could result in macrocephaly [11,12]. Pathogenic variants in this gene are responsible for about 1/5 of the macrocephaly cases in autistic individuals, and extreme macrocephaly (>3 SD) frequently occurs due to PTEN de novo mutations [4,13].
The second example is the FMR1 gene (OMIM *300805), a translational regulator associated with the fragile X syndrome. Repression of this gene in the syndrome can also result in macrocephaly due to an unbalance on ribosome biogenesis, which increases the proliferation of neural progenitor cells [14,15].
Besides single nucleotide variants, epigenetic mutations, mosaicism, and copy number variations (CNVs) are emerging as important causative factors of macrocephaly, implying a greater underlying genetic heterogeneity than previously anticipated [16,17]. For example, AKT3 (OMIM *611223) is the main driver of the abnormalities in head size observed in the reciprocal CNVs at the 1q43-q44 region [18,19]. The AKT3 protein is essential for proper cell survival, proliferation, and growth, and it is mostly expressed in the brain. AKT3 duplication and other gain-of-function variants increase its catalytic-kinase activity, upregulating the signaling of the PI3K-AKT pathway [19,20].
CNVs, defined as germline deletions or duplications larger than 1 kb [21], are a remarkable source of genetic variability. They account for up to 10% of the human genomic differences and can be causative of many diseases by a variety of mechanisms, affecting gene function or dosage [22]. Rare CNVs encompassing dosage-sensitive genes are frequently associated with morbid phenotypes [23], including ID and other neurodevelopmental disorders [24,25,26]. The effects of gene dosage sensitivity can be due to various factors, the most common being haploinsufficiency, when there is a minimal threshold needed for the normal functioning of the protein, affected in the disease state by loss-of-function (LoF) mutations or heterozygous deletions [23]. Sometimes, the excess of gene product in duplications can be deleterious as well, and some genes are susceptible to any modification in dosage, resulting in reciprocal CNVs displaying most commonly “mirrored” pathogenic phenotypes [27].
Although it is the most endorsed hypothesis to explain why this happens, dosage alterations are not the single mechanism through which CNVs can lead to disease. CNVs can cause a disruption within the gene, disassociate them from their regulatory sequences, or alter chromosome three-dimensional organization. Nevertheless, it is often hard to find a single dosage-sensitive gene within a CNV that encompasses several genes, because the clinically relevant CNVs are usually rare and exhibit incomplete penetrance and variable expressivity [28].
In this study, we aim to delineate the role of CNVs in macrocephaly development through evaluation of the public database DECIPHER [29], which contains information regarding CNVs and associated phenotypes, including macrocephaly. We further elaborated a comprehensive catalogue of genes and recurrent CNV syndromes known to be associated with macrocephaly.
2. Materials and Methods
2.1. DECIPHER Patients
We searched in the public data deposited in the DECIPHER database [29] for patients carrying CNVs and presenting macrocephaly among their clinical signs (up to 3 August 2021). For further analysis, we selected only cases with de novo CNVs with a maximum size of 500 kb, excluding all variants classified as benign and patients who carried other previously identified pathogenic variants. A CNV size of 500 kb was established as threshold to reduce the number of genes to be evaluated as candidates for the macrocephaly phenotype within each variant. Genetic and clinical data of five of the recovered DECIPHER patients were previously published [30,31,32,33,34,35]. CNVs were classified following the joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the clinical genome resource (ClinGen) [36].
Genes mapped to the genomic regions of the <500 kb de novo CNVs were evaluated regarding their potential to contribute to the macrocephaly phenotype, using the database PubMed (https://pubmed.ncbi.nlm.nih.gov/), looking for previous association with macrocephaly or involvement in cellular processes whose imbalance could lead to an abnormal head size, such as cellular proliferation. The Human Protein Atlas (https://www.proteinatlas.org/) was also evaluated to confirm the protein expression in the brain.
2.2. Literature Review of Known Macrocephaly Genes and Associated CNV Syndromes
We performed an extensive review through ClinGen (https://www.clinicalgenome.org/), DECIPHER (https://www.deciphergenomics.org/), OMIM (https://www.omim.org/), and PubMed (https://pubmed.ncbi.nlm.nih.gov/) to gather updated information about the genetic mechanisms associated with macrocephaly.
Genes and CNV syndromes associated with this phenotype were filtered among the records of the OMIM database (up to 13 June 2022) according to the following criteria: (a) containing the term “macrocephaly” or “megalencephaly”, with a known molecular basis and phenotype description, or (b) containing the term “macrocephaly” or “megalencephaly”, and using the phenotype mapping key “4”, which identifies chromosomal duplication or deletion syndromes.
In the ClinGen and DECIPHER databases, we inspected all the curated recurrent CNVs recognized as dosage sensitive regions associated with clinical phenotypes [37,38], retrieving those in which macrocephaly is a typical clinical sign.
Further, we explored the PubMed repository to retrieve articles by using the terms “macrocephaly” or “megalencephaly”, aiming to complement the list of genes and CNV syndromes. This analysis was based on the evaluation of articles published in the last five years, describing genetic findings of series of patients with macrocephaly.
The collected known genes were uploaded on the WebGestalt website (http://www.webgestalt.org/) to explore the biological pathways enriched in this set (Homo sapiens; genome protein-coding genes reference list, over-representation analysis, pathway as the functional database, crossing over data with the Reactome database).
3. Results
3.1. CNVs Associated with Macrocephaly in the DECIPHER Database
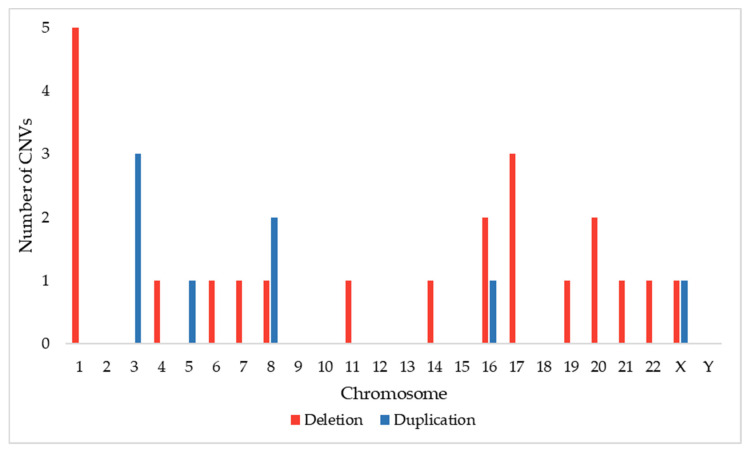
We were able to retrieve information on DECIPHER about 1033 macrocephalic patients with CNV sizes ranging from 1.72 kb to 248.96 Mb (aneuploidy), with a mean size of 7.67 Mb and a median size of 1.32 Mb. For further analysis, we selected only 29 patients with de novo <500 kb CNVs (Figure 1).
Figure 1.
Frequency of chromosomes harboring rare de novo <500 kb CNVs detected in 29 macrocephalic patients described in the DECIPHER database.
The protein-coding genes encompassed by the CNVs were evaluated to establish a possible link with macrocephaly (Table 1). In 14 patients (~48%), the CNVs included 13 genes already known to be associated with the phenotype (CHD8, GATAD2B, GPC3, KCTD13, KMT5B, KIF22, MAP2K2, MECP2, NF1, NFIA, NSD1, PUF60, and TCF20), one of them (NFIA) observed in two cases. In total, 8 patients (~53%), amid the remaining 15, harbored a CNV that encompassed, total or partially, at least one known microcephaly gene (ADNP, CACNA1G, DONSON, GRIN2A, ITSN1, MCPH1, RPL11, TBCD, and TRAPPC9), for which the reported mechanism for microcephaly is mostly LoF.
Table 1.
Description of the CNV data from the 29 macrocephalic patients with de novo <500 kb CNVs reported in the DECIPHER database and encompassed genes (in bold, known macrocephaly genes; ↓ known microcephaly genes; in red, potential candidate genes for macrocephaly).
| DECIPHER ID | Chromosomal Microarray Analysis (CMA) | Genotype-Phenotype Correlation | References | ||||
|---|---|---|---|---|---|---|---|
| Cytoband | Genomic Coordinates (GRCh38) | CNV Type | Classification | Size (kb) | Included Genes | ||
| (Protein Coding) | |||||||
| 281752 | 1q21.3 | 1:153818614-153823418 | Deletion P | Pathogenic | 5 | GATAD2B | |
| 269967 | 1p36.11 | 1:23691668-23696616 | Deletion P | Pathogenic | 5 | RPL11 ↓ | |
| 358646 | 1p31.3 | 1:61091785-61304381 | Deletion P | Pathogenic | 213 | NFIA | |
| 288170 | 1p31.3 | 1:61151026-61379931 | Deletion VUS | Pathogenic | 229 | NFIA | |
| 360889 | 1p36.33 | 1:917483-1282652 | Deletion P | VUS | 365 | AGRN/B3GALT6/C1QTNF12/C1orf159/HES4/ISG15/KLHL17/NOC2L/PERM1/PLEKHN1/RNF223/SAMD11/SCNN1D/SDF4/TNFRSF18/TNFRSF4/TTLL10/UBE2J2 | |
| 249115 | 3q25.33 | 3:159979210-160186591 | Duplication U | Likely benign | 207 | IL12A | [32] |
| Xq28 | X:154032185-154347882 | Duplication U | Pathogenic | 316 | MECP2 ↓/OPN1LW/OPN1MW/OPN1MW2/OPN1MW3/TEX28/TKTL1 | ||
| 1376 | 3q27.2 | 3:185411752-185567638 | Duplication U | VUS | 156 | LIPH/MAP3K13/TMEM41A | |
| 370137 | 3p24.1 | 3:29248723-29450441 | Duplication LP | VUS | 202 | RBMS3 | |
| 289221 | 4q22.2 | 4:93137138-93292777 | Deletion VUS | VUS | 156 | GRID2 | |
| 267309 | 5q35.3 | 5:177176839-177225635 | Duplication U | VUS | 49 | NSD1 ↓ | |
| 376373 | 6q25.3 | 6:157361157-157681369 | Deletion VUS | VUS | 320 | ZDHHC14 | |
| 288535 | 7p15.3 | 7:20953880-21052118 | Deletion VUS | Likely benign | 98 | - | |
| 339955 | 8q24.3 | 8:140361494-140527307 | Duplication VUS | VUS | 166 | AGO2/CHRAC1/TRAPPC9 ↓ | |
| 371384 | 8q24.3 | 8:143809435-143822571 | Deletion P | Pathogenic | 13 | PUF60 ↓/SCRIB ↓ | [30] |
| 314265 | 8p23.1 | 8:6518891-6801141 | Duplication P | VUS | 282 | MCPH1 ↓/ANGPT2/AGPAT5 | |
| 251808 | 11q13.2 | 11:68120554-68519565 | Deletion U | Pathogenic | 399 | LRP5/KMT5B/CHKA/C11ORF24/PPP6R3 | |
| 976 | 14q11.2 | 14:21309552-21460179 | Deletion U | Pathogenic | 151 | CHD8/RAB2B/RPGRIP1/SUPT16H | [35] |
| 253656 | 16p11.2 | 16:29837876-30179218 | Deletion U | Pathogenic | 341 | TAOK2/TLCD3B/PPP4C/CDIPT/SEZ6L2/YPEL3/MAPK3/C16orf92/INO80E/TBX6/DOC2A/ALDOA/ASPHD1/KCTD13 ↓/TMEM219/HIRIP3/MVP/GDPD3 | |
| 285342 | 16p13.2 | 16:9732634-9770161 | Deletion LP | Pathogenic | 38 | GRIN2A ↓ | |
| 301907 | 17q11.2 | 17:31334752-31340757 | Deletion P | Pathogenic | 6 | NF1 ↓ | |
| 287501 | 17q21.33 | 17:50600602-50604425 | Deletion U | Likely benign | 4 | CACNA1G ↓ | [31,33] |
| 368685 | 17q25.3 | 17:82821407-83086677 | Deletion VUS | Likely benign | 265 | ZNF750/METRNL/TBCD ↓/B3GNTL1 | |
| 300874 | 19p13.3 | 19:3979570-4131262 | Deletion P | Pathogenic | 152 | EEF2/PIAS4/MAP2K2/ZBTB7A | |
| 270687 | 20q11.23 | 20:38565558-38765539 | Deletion U | VUS | 200 | RALGAPB/SLC32A1/ADIG/ARHGAP40/ACTR5 | [34] |
| 412759 | 20q13.13 | 20:50891372-50893349 | Deletion P | Pathogenic | 2 | ADNP ↓ | |
| 249393 | 21q22.11 | 21:33581937-33883538 | Deletion U | VUS | 302 | ITSN1↓/DONSON ↓/CRYZL1 | |
| 259449 | 22q13.2 | 22:41743074-42171084 | Deletion VUS | Pathogenic | 428 | CCDC134/CENPM/CYP2D6/CYP2D7/MEI1/NAGA/NDUFA6/PHETA2/SEPTIN3/SHISA8/SMDT1/SREBF2/TCF20/TNFRSF13C/WBP2NL | |
| 270868 | Xq26.2 | X:133552850-134042983 | Deletion U | Pathogenic | 490 | GPC3 | |
| 16p11.2 | 16:29552664-30095687 | Duplication P | 543 | ALDOA/ASPHD1/C16orf54/C16orf92/CDIPT/DOC2A/HIRIP3/INO80E/KCTD13 ↓/KIF22/MAS/MVP/PAGR1/PPP4C/PRRT2/QPRT/SEZ6L2/SPN/TAOK2/TBX6/TLCD3B/TMEM219/YPEL3/ZG16 | |||
P: classified as pathogenic on DECIPHER; LP: classified as likely pathogenic on DECIPHER; VUS: classified as variant of uncertain significance on DECIPHER; U: unclassified on DECIPHER; VUS: variant of unknown significance.
We also proposed potential candidate genes in four cases (~14%) without known macrocephaly gene within the CNV (Table 2): in one case, the detected CNV encompassed a known microcephaly gene (TRAPPC9), and in three others, the RALGAPB, RBMS3, and ZDHHC14 genes were highlighted, mainly based on an in silico analysis of their functional roles, as described in Table 2.
Table 2.
Potential candidate genes for macrocephaly and their biological functions.
| Gene | Function * | Brain Expression ꜝ | Type of Variant (Probable Effect) | Reference |
|---|---|---|---|---|
| TRAPPC9 ↓ | May function in neuronal cells differentiation. LoF associated with microcephaly r | Yes | Partial duplication (unknown) | (OMIM #613192) |
| RALGAPB | RALGAPB plays an essential role in mitosis by controlling the spatial and temporal activation of RAL GTPases in the spindle assembly checkpoint (SAC) and cytokinesis | Yes | Partial deletion (unknown) | (OMIM *618833) |
| RBMS3 | Rbms3 was shown to exhibit tumor suppressor function via regulation of c-Myc and to bind/stabilize RNA in vitro. In zebrafish, LoF disrupts craniofacial development; not previously related to human diseases | Yes | Intragenic duplication (LoF) | (OMIM *300027) [39] |
| ZDHHC14 | Overexpression of ZDHHC14 reduces cell viability and induces apoptosis by activating a classic caspase-dependent pathway, whereas heterozygous knockout of ZDHHC14 increased colony formation ability of cells. | Yes | Entire deletion (LoF) |
(OMIM *619295) |
↓: known microcephaly genes; * Described in UniProt; ꜝ According to the Human Protein Atlas.
3.2. Literature Review of Macrocephaly Genes and Associated CNV Syndromes
We assembled a list of 341 bona fide genes whose association with macrocephaly has been previously corroborated, as present in a recognizable syndrome, or when at least more than one case was reported with a gene mutation linked to macrocephaly (Table S1). Aiming to unveil the main biological processes enriched for this set of genes, we performed an analysis on WebGestalt. As expected, the set of macrocephaly genes were enriched for development of the head, skull, and central nervous system and processes related to the cell cycle. In relation to neurogenesis, enriched processes included generation of new neurons, neuron projection, and gliogenesis (Figure S1).
Furthermore, we compiled 28 genomic loci with recurrent CNV syndromes that include macrocephaly among their clinical findings: 15 deletions, 11 duplications, one triplication, and one recurrent region [del/dup], as presented on Table 3. Eighteen CNV regions had an OMIM entry, four regions were exclusively described in the ClinGen or DECIPHER databases, and five regions were retrieved from the scientific literature. Seven loci were exclusively associated with macrocephaly: 1p32p31 deletion, 3q13.31 deletion, 4q32.1q32.2 triplication, 5p13 duplication, distal 7q (7q32-qter) duplication, 14q11.2 deletion, and Xq22.3 telomeric deletion. Six loci were reported to cause macrocephaly or microcephaly with the same CNV type: 2q31.2 deletion, 10p15.3 deletion, 11q deletion, 15q11q13 deletion, 17q11.2 recurrent region (del/dup), and distal 22q11.2 duplication. A particularly interesting fact is that 17 loci (65%) exhibited mirror phenotypes: they are reciprocal deletions and duplications known to originate opposite effects on head size: 1q21.1, 4pter, 5q35, 7p22.1, 7q11.23, 8p23.1, 10q22.3q23.2, 13q31.3, 15q11q13, 15q26qter, 16p11.2, 17p13.1, 17q11.2, 17q12, 17q21.31, 19p13.13, and 22q11.2.
Table 3.
Twenty-eight recognized loci harboring deletion/duplication syndromes associated with macrocephaly (OMIM number, genomic coordinates, inheritance pattern, and ClinGen/DECIPHER data are shown, when applicable).
| Condition | OMIM # | Genomic Coordinates (hg38) | Inheritance Pattern | Association with Head Size | ClinGen/ DECIPHER |
Additional References |
|---|---|---|---|---|---|---|
| Chromosome 1p32-p31 deletion syndrome | 613735 | chr1:58193565-63125273 | Incomplete penetrance | Macrocephaly | … | [40] |
| Chromosome 1q21.1 duplication syndrome | 612475 | chr1:147105904-147917509 | Incomplete penetrance | Macrocephaly in duplication/Microcephaly in deletion (#612474) | Yes | [41] |
| Chromosome 2q31.2 deletion syndrome | 612345 | chr2:177100000-179700000 | Incomplete penetrance | Macrocephaly/Microcephaly | ... | - |
| Chromosome 3q13.31 deletion syndrome | 615433 | chr3:113700000-117600000 | de novo | Macrocephaly in deletion/Normal OFC in duplication | ... | [42,43] |
| Chromosome 4pter duplication syndrome | N/A | chr4:337779-2009235 | de novo | Macrocephaly in duplication/Microcephaly in deletion | Yes | [44] |
| Chromosome 4q32.1-q32.2 triplication syndrome | 613603 | chr4:154600000-163600000 | de novo | Macrocephaly | ... | [45] |
| Chromosome 5p13 duplication syndrome | 613174 | chr5:36845462–37231819 | de novo | Macrocephaly | ... | [46] |
| Chromosome 5q35 deletion syndrome (Sotos 1) |
117550 | chr5:176297633-177625115 | de novo | Macrocephaly in deletion/Microcephaly in duplication | Yes | [47] |
| Chromosome 7p22.1 duplication syndrome | N/A | chr7:5527147-5530600 | de novo | Relative macrocephaly in duplication/Microcephaly in deletion | ... | [48] |
| Chromosome 7q11.23 duplication syndrome | 609757 | chr7: 73330452-74728172 | Mostly de novo | Macrocephaly in duplication/Microcephaly in deletion (#194050) | Yes | - |
| Chromosome distal 7q (7(q32→qter)) duplication syndrome | N/A | chr7:128308047– 159119707 | Incomplete penetrance (can be inherited) | Macrocephaly | Yes ! | [49] |
| Chromosome 8p23.1 duplication syndrome | N/A | chr8:8242542-11908820 | de novo | Macrocephaly in duplication/Microcephaly in deletion | Yes | [28,50,51] |
| Chromosome 10p15.3 deletion syndrome | N/A | chr10:171237-2880776 | Incomplete penetrance (can be inherited) | Macrocephaly/Microcephaly | ... | [52,53,54] |
| Chromosome 10q22.3-q23.2 deletion syndrome | 612242 | chr10:80300000-95300000 | de novo | Macrocephaly in deletion/Microcephaly in duplication | Yes | - |
| Chromosome 11q deletion syndrome (Jacobsen) | 147791 | chr11:114600000-135086622 | de novo | Macrocephaly/Microcephaly | Yes | - |
| Chromosome 13q31.3 microduplication syndrome | N/A | chr13:91337007-91852603 | de novo | Macrocephaly in duplication/Microcephaly in deletion | ... | [55] |
| Chromosome 14q11.2 microdeletion syndrome | N/A | chr14:21359783-21393052 | de novo | Macrocephaly | … | [56] |
| Chromosome 15q11q13 deletion (PWS) | 176270 | chr15:22832519-28379874 | de novo | Macro-microcephaly in PWS/Microcephaly in AS | Yes | - |
| Chromosome 15q26qter duplication syndrome | 612626 | chr15:88500000-101991189 | de novo | Macrocephaly in duplication/Microcephaly in deletion | Yes | [57] |
| Chromosome 16p11.2 deletion syndrome, 593kb | 611913 | chr16:29595531-30188534 | Incomplete penetrance (can be inherited) | Macrocephaly in deletion/Microcephaly in duplication | Yes | [58] |
| Chromosome 17p13.1 duplication syndrome | N/A | chr17:7584958-8092957 | de novo | Macrocephaly in duplication/Microcephaly in deletion | ... | [45,59] |
| Chromosome 17q11.2 recurrent region 1.4Mb (del/dup) | 613675 618874 |
chr17:30780079-31937008 | Del-de novo; Dup-incomplete penetrance (can be inherited) |
Macrocephaly/Microcephaly | Yes | - |
| Chromosome 17q12 deletion syndrome | 614527 | chr17:36458167-37854616 | Mostly de novo | Macrocephaly in deletion/Microcephaly in duplication | Yes | [60] |
| Chromosome 17q21.31 deletion syndrome | 610443 | chr17:45627800-46087514 | Description of inherited cases | Macrocephaly in deletion/Microcephaly in duplication (#613533) | Yes | [61] |
| Chromosome 19p13.13 deletion syndrome | 613638 | chr19:12821186-13132186 | de novo | Macrocephaly in deletion (#613638)/Microcephaly in duplication | ... | [45] |
| Chromosome 19p13.3 microdeletion syndrome | N/A | chr19:2329321-4996917 | Description of inherited cases, although the majority are de novo | Macrocephaly in deletion/Microcephaly in duplication | … | [62,63] |
| Chromosome 22q11.2 duplication syndrome, distal | N/A | chr22:21562828-23306924 | Incomplete penetrance (can be inherited) | Macro-microcephaly in duplication/Microcephaly in deletion | Yes | [64] |
| Chromosome Xq22.3 telomeric deletion syndrome (AMME) | 300194 | chrX:104500000-109400000 | Dominant X-linked (description of inherited cases) | Macrocephaly | ... | - |
N/A: Not Available; ! includes https://dosage.clinicalgenome.org/region_search_38.cgi?loc=chr7:128,667,993-159,327,017.
4. Discussion
Understanding the mechanisms of brain growth and development underlying macrocephaly can shed light to the complex process of neurodevelopment [65]. CNVs affect up to 10% of the human genome and are mostly not deleterious [66,67]. Nonetheless, in neuropsychiatric disorders, such as autism and intellectual disability—which are commonly associated with alterations in head size—there is a notable enrichment of recurrent typical CNVs, resulting from nonallelic homologous recombination of hotspots flanked by paired low copy repeats [23,68]. In fact, copy number changes often can lead to protein imbalance of the affected genes, resulting in a pathogenic phenotype in case of dosage-sensitivity [23,28,66,68]. As expected, based on theory prediction and observation in model organisms, deletions (haploinsufficiency) are more common and penetrant than duplications (triplosensitivity) for extreme developmental phenotypes [28,69]; in the investigated DECIPHER cohort, over 75% of the CNVs that met our criteria were deletions. This present study and our previous review of CNVs in microcephaly [70] generated a map of CNV loci associated with alterations in head size (Figure 2). A total of 67 loci were gathered, harboring 77 CNVs (58 deletions and 19 duplications), reinforcing the relevance of CNVs, mostly deletions, in neurodevelopmental phenotypes [28,69].
Figure 2.
Map of the loci with CNV syndromes-deletions in red, duplications in blue, or both in purple-associated with microcephaly-represented on the left side of the chromosomes-or macrocephaly-represented on the right side of the chromosomes.
The phenotypes presented by reciprocal CNVs can be allocated in four major categories: mirrored (when deletions and duplications have opposite effects), identical (both deletion and duplication result in the same phenotype spectrum), overlapping (some clinical features are present in both types of CNVs), and unique (exclusive for the deletion or duplication) [68]. Despite many cases having major driver genes responsible for the main clinical features, because of the large size of the CNVs, several genes can be affected, possibly contributing to the variability of the phenotype presented through synergistic or additive epistatic effects [66,68].
Mirror phenotypes are not universal, but frequently are present at reciprocal CNVs when head size is involved; as an example, we can cite the chromosome 5q35 region. The deletion of this region, including the NSD1 gene, results in macrocephaly, one of the phenotypes of the Sotos syndrome, while the duplication leads to a microcephalic phenotype, likely due to gene dosage effect [71]. Identical phenotypes are probably a result of a disruption in the same developmental pathways, with either LoF mutations or overexpression and enhanced gene activity leading to similar clinical features due to downstream alterations [66,68].
Revisiting the list of genomic loci linked to macrocephaly, compiled through examination of the scientific literature available at PubMed and other aforementioned public databases, three of these categories were discernible: (a) reciprocal CNVs leading to a mirror phenotype–15 out of the 28 (~53%) known recurrent CNVs identified in this study present opposite head sizes depending on the CNV type; (b) CNVs associated exclusively with macrocephaly, constituting about 25% of the syndromes identified (seven cases), and (c) the same CNV type resulting in macro and microcephaly, as presented in the seven remained cases (25%). The latter category can be illustrated by the patient 412759, who carried a pathogenic ADNP intragenic deletion, leading to LoF, which has been previously associated with the ADNP syndrome, with “large head” amongst its clinical findings, as described by Li et al. (2017) [72] and Gozes (2020) [73]. On the other side, studies using animal models and mutant embryonic stem cells were able to correlate ADNP deficiency with downregulation of the homeobox gene PAX6, which has a crucial role in neuronal progenitor cells migration and differentiation in the developing brain and has already been described in association with microcephaly [74,75].
Through examination of the DECIPHER cohort, we observed that 14 of the 29 patients (~48%) presented a CNV encompassing a known macrocephaly gene that could explain the phenotype. Particularly, the gene NFIA was found to be affected by a heterozygous intragenic deletion in two patients; its haploinsufficiency is considered a main driver to the phenotypes resulting of the chromosome 1p32-p31 deletion syndrome (OMIM #613735), especially macrocephaly and intellectual disability [76].
We found a remarkably case of a conflicting phenotype in the DECIPHER patient 269967, who presented a complete deletion of the RPL11 gene. This gene encodes a protein that is part of the large ribosomal unit, and its haploinsufficiency (amid other ribosomal proteins, such as RPL5 and RPL26, that impair the processing of pre-RNAs and the subsequent maturation of the ribosomal subunits) is the most common causative mechanism of the autosomal dominant disorder Diamond-Blackfan anemia (OMIM #612562). Almost 1/3 of the individuals with Diamond-Blackfan anemia show a degree of growth deficiency, and microcephaly is one of the present craniofacial features [77]. This is no surprise, considering that RPL11 is one of the ribosomal proteins that can interfere on the TP53-signaling pathway when in a deficiency state, leading to suppression of cell cycle progression and apoptosis due to a nucleolar stress response [78]. There is a subgroup of patients carrying chromosomal rearrangements and large deletions of other gene (RPS19) who present macrocephaly instead of microcephaly as one of the reported craniofacial abnormalities [79]; however, we found no description of RPL11 LoF and overgrowth, as presented by the patient here discussed. Considering that RPL11 is the only affected gene in the deleted segment, further genetic analysis would be needed to ensure if this is the only pathogenic variant carried by this patient and establish if this variant is indeed the cause of macrocephaly.
For the assessment of potential new candidate genes for macrocephaly, we evaluated the genomic content of the remaining 14 patients who did not carry CNVs encompassing known macrocephaly genes, but still presented protein-coding genes within the affected region. From this group, we were able to further cluster them in two categories based on similarities between them.
Eight DECIPHER patients presented a CNV that encompassed a known microcephaly gene; one of these patients (DECIPHER 339955) carried a partial duplication of TRAPPC9. Although there are duplications reported in this region in the normal population (DGV database), they do not completely overlap the distal sequence of TRAPPC9, which is duplicated in this patient, mainly exon 7. TRAPPC9 LoF is associated with a rare recessive neurodevelopmental syndrome with obesity and postnatal microcephaly as the most prominent signs [80,81], the latter likely due to the role of this protein in postmitotic neurons. It acts in the vesicular protein trafficking between the Golgi apparatus and the endoplasmic reticulum, and like several genes of this group, it is related to the proper development of the nervous system. It may also play a role in the NF-κB signaling, a crucial pathway to neuronal cell differentiation and myelin formation [82,83]. Another interesting aspect is its parent-of-origin expression bias in the brain, being predominantly expressed from the maternal allele [83]. It is important to mention that the CNV data deposited in the DECIPHER is mainly based on chromosomal microarray analysis, which hampers structural evaluation of the copy number alteration. Therefore, it is not possible to determine, in case of duplications, if an intragenic or partial duplication variant is located in tandem or elsewhere in the genome. However, it is plausible to argue that, if the duplication is in tandem, in a direct or inverted orientation, a LoF effect would be expected, though further analysis is needed to corroborate this hypothesis.
Three patients harbored a CNV affecting non-OMIM genes (RALGAPB, RBMS3 and ZDHHC14), whose functions, when disturbed, could potentially lead to abnormal head size. One of them carried a partial deletion of RALGAPB, a known tumor suppressor [84], which inhibits cell proliferation and tumor growth. Studies using animal models also emphasized the importance of the RalGAP complex for neuronal development and differentiation [85], and both knockdown and overexpression of RALGAPB in mammalian cells lead to an increase in mTORC1 activity [86]. This gene has little evidence for haploinsufficiency [87], but the inactivation of the multiprotein RalGAP complex has been proposed as a causal factor for microcephaly [88]. More studies are necessary to endorse RALGAPB as a possible candidate for macrocephaly. We observed a DECIPHER patient carrying a RBMS3 partial duplication, similar to the TRAPPC9 case previously mentioned. In vitro and in vivo studies demonstrated that rbms3 inhibits cell proliferation and promotes apoptosis due to regulation of gene transcription or RNA metabolism, and its expression is reduced in several cancers [89,90]. Defects in RNA-binding proteins, such as the aforementioned, may lead to craniofacial abnormalities. Jayasena & Bronner (2012) [39] performed a study to analyze the consequences of rbms3 LoF during zebrafish development; the mutants had a variety of abnormalities when compared to the wild type, including smaller body size and craniofacial defects due to improper cartilage formation. The reported association of rbms3 with craniofacial abnormalities in animal models and RBMS3 pHaplo of 0.89 (an ensemble machine-learning model, designed by Collins (2022) [27], that reflects the probability of haploinsufficiency for autosomal genes) indicate that this gene could be a strong candidate for the macrocephaly phenotype, even though further analysis is required to validate this assumption. ZDHHC14 expression is also reduced in several cancer types, including brain tumors; induced in vitro overexpression in gastric cancer cell lines promoted cancer cell migration and cell attachment, in addition to stimulating cell invasion [91]. In vitro studies by Yeste-Velasco et al. (2014) [92] demonstrated that whereas the overexpression promoted apoptosis through activating of the classic caspase-dependent pathway, heterozygous deletion increased colony formation. Therefore, RBMS3 and ZDHHC14 are both classified as tumor suppressor genes, and the CNVs identified in these DECIPHER patients have a probable LoF effect (intragenic duplication and entire gene deletion, respectively). Considering that the reduced expression of these two genes is reported to increase cell proliferation and/or inhibit apoptosis, they are interesting candidates and additional studies are required to provide functional support to their potential causal correlation with macrocephaly.
Finally, we found two CNVs encompassing non-coding genes that are worth mentioning. One of them was a deletion including only part of the sequence of the long intergenic non-protein coding RNA 1162 (LINC01162), mapped to 7p15.3 (patient 288535); however, despite being a validated lncRNA, there is no information regarding its function. The second CNV was an 8p23.1 duplication (patient 314265), which encompassed three protein coding genes, including part of the known microcephaly gene MCPH1, and the full sequence of its antisense lncRNA (MCPH1-AS1), a validated gene with high expression in brain tissues [93]. It is difficult to anticipate the potential impact of CNVs harboring lncRNAs; both variants were classified as likely benign according to the clinical guidelines.
Studies focusing on pathogenic variants disrupting the mechanisms that control head size are an extremely important source of information about biological pathways underlying these processes. This study reviewed the genes and CNV loci previously associated with macrocephaly in the literature as well as suggested novel potential candidate genes deserving further evaluation.
Acknowledgments
We thank the patients and their families who participated in the studies that served as the basis for this work.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes13122285/s1, Table S1: Genes with a recognizable association with macrocephaly, retrieved from OMIM and the scientific literature; Figure S1: Enriched biological processes of the gene set related to macrocephaly.
Author Contributions
Conceptualization, A.C.V.K. and G.C.B.; methodology, A.C.V.K. and G.C.B.; formal analysis, G.C.B.; investigation, G.C.B.; writing—original draft preparation, G.C.B.; writing—review and editing, A.C.V.K. and G.C.T.; supervision, A.C.V.K.; funding acquisition, A.C.V.K. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
This study makes use of data generated by the DECIPHER community. A full list of centers who contributed to the generation of the data is available from https://deciphergenomics.org/about/stats and via email from contact@deciphergenomics.org. Funding for the DECIPHER project was provided by Wellcome. Those who carried out the original analysis and collection of the data bear no responsibility for the further analysis or interpretation of them.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This research was funded by São Paulo Research Foundation [FAPESP], grant numbers 2013/080828-1 [CEPID] and 2020/15552-2; and National Council for Scientific and Technological Development [CNPq], grant numbers 157816/2018-4, 305806/2019-0 and 140271/2020-1.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Accogli A., Geraldo A.F., Piccolo G., Riva A., Scala M., Balagura G., Salpietro V., Madia F., Maghnie M., Zara F., et al. Diagnostic Approach to Macrocephaly in Children. Front. Pediatr. 2022;9:1534. doi: 10.3389/fped.2021.794069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olney A.H. Macrocephaly Syndromes. Semin. Pediatr. Neurol. 2007;14:128–135. doi: 10.1016/j.spen.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Tan A.P., Mankad K., Gonçalves F.G., Talenti G., Alexia E. Macrocephaly. Top. Magn. Reson. Imaging. 2018;27:197–217. doi: 10.1097/RMR.0000000000000170. [DOI] [PubMed] [Google Scholar]
- 4.Thomas C.N., Kolbe A.B., Binkovitz L.A., McDonald J.S., Thomas K.B. Asymptomatic macrocephaly: To scan or not to scan. Pediatr. Radiol. 2021;51:811–821. doi: 10.1007/s00247-020-04907-7. [DOI] [PubMed] [Google Scholar]
- 5.Williams C.A., Dagli A., Battaglia A. Genetic disorders associated with macrocephaly. Am. J. Med. Genet. Part A. 2008;146A:2023–2037. doi: 10.1002/ajmg.a.32434. [DOI] [PubMed] [Google Scholar]
- 6.Malinger G., Lev D., Ben-Sira L., Hoffmann C., Herrera M., Viñals F., Vinkler H., Ginath S., Biran-Gol Y., Kidron D., et al. Can syndromic macrocephaly be diagnosed in utero? Ultrasound Obstet. Gynecol. 2010;37:72–81. doi: 10.1002/uog.8799. [DOI] [PubMed] [Google Scholar]
- 7.Orru E., Calloni S.F., Tekes A., Huisman T.A.G.M., Soares B.P. The Child With Macrocephaly: Differential Diagnosis and Neuroimaging Findings. Am. J. Roentgenol. 2018;210:848–859. doi: 10.2214/AJR.17.18693. [DOI] [PubMed] [Google Scholar]
- 8.Winden K.D., Yuskaitis C.J., Poduri A. Megalencephaly and Macrocephaly. Skull Base. 2015;35:277–287. doi: 10.1055/s-0035-1552622. [DOI] [PubMed] [Google Scholar]
- 9.Li L., Ross A.H. Why is PTEN an important tumor suppressor? J. Cell. Biochem. 2007;102:1368–1374. doi: 10.1002/jcb.21593. [DOI] [PubMed] [Google Scholar]
- 10.Hopkins B.D., Hodakoski C., Barrows D., Mense S.M., Parsons R.E. PTEN function: The long and the short of it. Trends Biochem. Sci. 2014;39:183–190. doi: 10.1016/j.tibs.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., Huang W.-C., Séjourné J., Clipperton-Allen A.E., Page D.T. Pten Mutations Alter Brain Growth Trajectory and Allocation of Cell Types through Elevated-Catenin Signaling. J. Neurosci. 2015;35:10252–10267. doi: 10.1523/JNEUROSCI.5272-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Junco-Clemente P., Golshani P. PTEN. Commun. Integr. Biol. 2014;7:e28358. doi: 10.4161/cib.28358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler M.G., Dasouki M.J., Zhou X.P., Talebizadeh Z., Brown M., Takahashi T.N., Miles J.H., Wang C.H., Stratton R., Pilarski R., et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J. Med. Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagerman R.J., Berry-Kravis E., Hazlett H.C., Bailey D.B., Jr., Moine H., Kooy R.F., Tassone F., Gantois I., Sonenberg N., Mandel J.L., et al. Fragile X syndrome. Nat. Rev. Dis. Prim. 2017;3:17065. doi: 10.1038/nrdp.2017.65. [DOI] [PubMed] [Google Scholar]
- 15.Li M., Shin J., Risgaard R.D., Parries M.J., Wang J., Chasman D., Liu S., Roy S., Bhattacharyya A., Zhao X. Identification of FMR1-regulated molecular networks in human neurodevelopment. Genome Res. 2020;30:361–374. doi: 10.1101/gr.251405.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pirozzi F., Nelson B., Mirzaa G. From microcephaly to megalencephaly: Determinants of brain size. Dialog Clin. Neurosci. 2018;20:267–282. doi: 10.31887/DCNS.2018.20.4/gmirzaa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tatton-Brown K., Weksberg R. Molecular Mechanisms of Childhood Overgrowth. Am. J. Med. Genet. Part C Semin. Med. Genet. 2013;163:71–75. doi: 10.1002/ajmg.c.31362. [DOI] [PubMed] [Google Scholar]
- 18.Wang D., Zeesman S., Tarnopolsky M.A., Nowaczyk M.J. Duplication of AKT3 as a cause of macrocephaly in duplication 1q43q44. Am. J. Med. Genet. Part A. 2013;161:2016–2019. doi: 10.1002/ajmg.a.35999. [DOI] [PubMed] [Google Scholar]
- 19.Lopes F., Torres F., Soares G., van Karnebeek C.D., Martins C., Antunes D., Silva J., Muttucomaroe L., Botelho L.F., Sousa S., et al. The Role of AKT3 Copy Number Changes in Brain Abnormalities and Neurodevelopmental Disorders: Four New Cases and Literature Review. Front. Genet. 2019;10:58. doi: 10.3389/fgene.2019.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alcantara D., E Timms A., Gripp K., Baker L., Park K., Collins S., Cheng C., Stewart F., Mehta S.G., Saggar A., et al. Mutations of AKT3 are associated with a wide spectrum of developmental disorders including extreme megalencephaly. Brain. 2017;140:2610–2622. doi: 10.1093/brain/awx203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeman J.L., Perry G.H., Feuk L., Redon R., McCarroll S.A., Altshuler D.M., Aburatani H., Jones K.W., Tyler-Smith C., Hurles M.E., et al. Copy number variation: New insights in genome diversity. Genome Res. 2006;16:949–961. doi: 10.1101/gr.3677206. [DOI] [PubMed] [Google Scholar]
- 22.Pös O., Radvanszky J., Styk J., Pös Z., Buglyó G., Kajsik M., Budis J., Nagy B., Szemes T. Copy Number Variation: Methods and Clinical Applications. Appl. Sci. 2021;11:819. doi: 10.3390/app11020819. [DOI] [Google Scholar]
- 23.Rice A.M., McLysaght A. Dosage sensitivity is a major determinant of human copy number variant pathogenicity. Nat. Commun. 2017;8:14366. doi: 10.1038/ncomms14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenberg C., Knijnenburg J., Bakker B., Vianna-Morgante A.M., Sloos W., A Otto P., Kriek M., Hansson K., Krepischi A., Fiegler H., et al. Array-CGH detection of micro rearrangements in mentally retarded individuals: Clinical significance of imbalances present both in affected children and normal parents. J. Med. Genet. 2005;43:180–186. doi: 10.1136/jmg.2005.032268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krepischi-Santos A., Vianna-Morgante A., Jehee F., Passos-Bueno M., Knijnenburg J., Szuhai K., Sloos W., Mazzeu J., Kok F., Cheroki C., et al. Whole-genome array-CGH screening in undiagnosed syndromic patients: Old syndromes revisited and new alterations. Cytogenet. Genome Res. 2006;115:254–261. doi: 10.1159/000095922. [DOI] [PubMed] [Google Scholar]
- 26.Martin C.L., Wain K.E., Oetjens M.T., Tolwinski K., Palen E., Hare-Harris A., Habegger L., Maxwell E.K., Reid J.G., Walsh L.K., et al. Identification of Neuropsychiatric Copy Number Variants in a Health Care System Population. JAMA Psychiatry. 2020;77:1276. doi: 10.1001/jamapsychiatry.2020.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins R.L., Glessner J.T., Porcu E., Lepamets M., Brandon R., Lauricella C., Han L., Morley T., Niestroj L.-M., Ulirsch J., et al. A cross-disorder dosage sensitivity map of the human genome. Cell. 2022;185:3041–3055.e25. doi: 10.1016/j.cell.2022.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper G.M., Coe B.P., Girirajan S., A Rosenfeld J., Vu T.H., Baker C., Williams C., Stalker H., Hamid R., Hannig V., et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdin D., Rump A., Tzschach A., Sarnow K., Schröck E., Hackmann K., Di Donato N. PUF60-SCRIB fusion transcript in a patient with 8q24.3 microdeletion and atypical Verheij syndrome. Eur. J. Med. Genet. 2018;62:103587. doi: 10.1016/j.ejmg.2018.11.021. [DOI] [PubMed] [Google Scholar]
- 31.Berecki G., Helbig K.L., Ware T.L., Grinton B., Skraban C.M., Marsh E.D., Berkovic S.F., Petrou S. Novel Missense CACNA1G Mutations Associated with Infantile-Onset Developmental and Epileptic Encephalopathy. Int. J. Mol. Sci. 2020;21:6333. doi: 10.3390/ijms21176333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bijlsma E., Collins A., Papa F., Tejada M.-I., Wheeler P., Peeters E., Gijsbers A., van de Kamp J., Kriek M., Losekoot M., et al. Xq28 duplications including MECP2 in five females: Expanding the phenotype to severe mental retardation. Eur. J. Med. Genet. 2012;55:404–413. doi: 10.1016/j.ejmg.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunii M., Doi H., Hashiguchi S., Matsuishi T., Sakai Y., Iai M., Okubo M., Nakamura H., Takahashi K., Katsumoto A., et al. De novo CACNA1G variants in developmental delay and early-onset epileptic encephalopathies. J. Neurol. Sci. 2020;416:117047. doi: 10.1016/j.jns.2020.117047. [DOI] [PubMed] [Google Scholar]
- 34.Santoro C., Malan V., Bertoli M., Boddaert N., Vidaud D., Lyonnet S. Sporadic NF1 mutation associated with a de-novo 20q11.3 deletion explains the association of unusual facies, Moyamoya vasculopathy, and developmental delay, reported by Bertoli et al. in 2009. Clin. Dysmorphol. 2013;22:42–43. doi: 10.1097/MCD.0b013e32835b8ea4. [DOI] [PubMed] [Google Scholar]
- 35.Zahir F., Firth H.V., Baross A., Delaney A.D., Eydoux P., Gibson W., Langlois S., Martin H., Willatt L., A Marra M., et al. Novel deletions of 14q11.2 associated with developmental delay, cognitive impairment and similar minor anomalies in three children. J. Med. Genet. 2007;44:556–561. doi: 10.1136/jmg.2007.050823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Riggs E.R., Andersen E.F., Cherry A.M., Kantarci S., Kearney H., Patel A., Raca G., Ritter D.I., South S.T., Thorland E.C., et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen) Genet. Med. 2020;22:245–257. doi: 10.1038/s41436-019-0686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clinical Genome Resource, Curations of Recurrent CNVs. [(accessed on 17 November 2022)]. Available online: https://search.clinicalgenome.org/kb/gene-dosage/cnv?page=1&size=All&search=
- 38.DECIPHER, CNV Syndromes. [(accessed on 17 November 2022)]. Available online: https://www.deciphergenomics.org/disorders/syndromes/list.
- 39.Jayasena C.S., Bronner M.E. Rbms3 functions in craniofacial development by posttranscriptionally modulating TGF-β signaling. J. Cell Biol. 2012;199:453–466. doi: 10.1083/jcb.201204138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koehler U., Holinski-Feder E., Ertl-Wagner B., Kunz J., Von Moers A., Von Voss H., Schell-Apacik C. A novel 1p31.3p32.2 deletion involving the NFIA gene detected by array CGH in a patient with macrocephaly and hypoplasia of the corpus callosum. Eur. J. Pediatr. 2009;169:463–468. doi: 10.1007/s00431-009-1057-2. [DOI] [PubMed] [Google Scholar]
- 41.Brunetti-Pierri N., Berg J., Scaglia F., Belmont J., A Bacino C., Sahoo T., Lalani S.R., Graham B., Lee B., Shinawi M., et al. Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities. Nat. Genet. 2008;40:1466–1471. doi: 10.1038/ng.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molin A.-M., Andrieux J., A Koolen D., Malan V., Carella M., Colleaux L., Cormier-Daire V., David A., De Leeuw N., Delobel B., et al. A novel microdeletion syndrome at 3q13.31 characterised by developmental delay, postnatal overgrowth, hypoplastic male genitals, and characteristic facial features. J. Med. Genet. 2011;49:104–109. doi: 10.1136/jmedgenet-2011-100534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vuillaume M.-L., Delrue M.-A., Naudion S., Toutain J., Fergelot P., Arveiler B., Lacombe D., Rooryck C. Expanding the clinical phenotype at the 3q13.31 locus with a new case of microdeletion and first characterization of the reciprocal duplication. Mol. Genet. Metab. 2013;110:90–97. doi: 10.1016/j.ymgme.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 44.Bi W., Cheung S.-W., Breman A.M., Bacino C.A. 4p16.3 microdeletions and microduplications detected by chromosomal microarray analysis: New insights into mechanisms and critical regions. Am. J. Med. Genet. Part A. 2016;170:2540–2550. doi: 10.1002/ajmg.a.37796. [DOI] [PubMed] [Google Scholar]
- 45.Weise A., Mrasek K., Klein E., Mulatinho M., Llerena J., Hardekopf D., Pekova S., Bhatt S., Kosyakova N., Liehr T., et al. Microdeletion and Microduplication Syndromes. J. Histochem. Cytochem. 2012;60:346–358. doi: 10.1369/0022155412440001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan J., Zhang F., Brundage E., Scheuerle A., Lanpher B., Erickson R.P., Powis Z., Robinson H.B., Trapane P.L., Stachiw-Hietpas D., et al. Genomic duplication resulting in increased copy number of genes encoding the sister chromatid cohesion complex conveys clinical consequences distinct from Cornelia de Lange. J. Med. Genet. 2008;46:626–634. doi: 10.1136/jmg.2008.062471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Franco L.M., De Ravel T., Graham B.H., Frenkel S.M., Van Driessche J., Stankiewicz P., Lupski J.R., Vermeesch J.R., Cheung S.W. A syndrome of short stature, microcephaly and speech delay is associated with duplications reciprocal to the common Sotos syndrome deletion. Eur. J. Hum. Genet. 2009;18:258–261. doi: 10.1038/ejhg.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chui J.V., Weisfeld-Adams J.D., Tepperberg J., Mehta L. Clinical and molecular characterization of chromosome 7p22.1 microduplication detected by array CGH. Am. J. Med. Genet. Part A. 2011;155:2508–2511. doi: 10.1002/ajmg.a.34180. [DOI] [PubMed] [Google Scholar]
- 49.Bosfield K., Diaz J., Leon E. Pure Distal 7q Duplication: Describing a Macrocephalic Neurodevelopmental Syndrome, Case Report and Review of the Literature. Mol. Syndr. 2021;12:159–168. doi: 10.1159/000513453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Akcakaya N.H., Capan Y., Schulz H., Sander T., Caglayan S.H., Yapıcı Z. De novo 8p23.1 deletion in a patient with absence epilepsy. Epileptic Disord. 2017;19:217–221. doi: 10.1684/epd.2017.0906. [DOI] [PubMed] [Google Scholar]
- 51.Ballarati L., Cereda A., Caselli R., Selicorni A., Recalcati M.P., Maitz S., Finelli P., Larizza L., Giardino D. Genotype–phenotype correlations in a new case of 8p23.1 deletion and review of the literature. Eur. J. Med. Genet. 2010;54:55–59. doi: 10.1016/j.ejmg.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 52.Vargiami E., Ververi A., Kyriazi M., Papathanasiou E., Gioula G., Gerou S., Al-Mutawa H., Kambouris M., Zafeiriou D.I. Severe clinical presentation in monozygotic twins with 10p15.3 microdeletion syndrome. Am. J. Med. Genet. Part A. 2013;164:764–768. doi: 10.1002/ajmg.a.36329. [DOI] [PubMed] [Google Scholar]
- 53.DeScipio C., Conlin L., Rosenfeld J., Tepperberg J., Pasion R., Patel A., McDonald M.T., Aradhya S., Ho D., Goldstein J., et al. Subtelomeric deletion of chromosome 10p15.3: Clinical findings and molecular cytogenetic characterization. Am. J. Med. Genet. Part A. 2012;158A:2152–2161. doi: 10.1002/ajmg.a.35574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tumiene B., Čiuladaitė Ž., Preikšaitienė E., Mameniškienė R., Utkus A., Kučinskas V. Phenotype comparison confirms ZMYND11 as a critical gene for 10p15.3 microdeletion syndrome. J. Appl. Genet. 2017;58:467–474. doi: 10.1007/s13353-017-0408-3. [DOI] [PubMed] [Google Scholar]
- 55.Siavrienė E., Preikšaitienė E., Maldžienė Ž., Mikštienė V., Rančelis T., Ambrozaitytė L., Gueneau L., Reymond A., Kučinskas V. A de novo 13q31.3 microduplication encompassing the miR-17 ~ 92 cluster results in features mirroring those associated with Feingold syndrome 2. Gene. 2020;753:144816. doi: 10.1016/j.gene.2020.144816. [DOI] [PubMed] [Google Scholar]
- 56.Yasin H., Gibson W., Langlois S., Stowe R.M., Tsang E., Lee L., Poon J., Tran G., Tyson C., Wong C.K., et al. A distinct neurodevelopmental syndrome with intellectual disability, autism spectrum disorder, characteristic facies, and macrocephaly is caused by defects in CHD8. J. Hum. Genet. 2019;64:271–280. doi: 10.1038/s10038-019-0561-0. [DOI] [PubMed] [Google Scholar]
- 57.Kant S., Kriek M., Walenkamp M., Hansson K., van Rhijn A., Clayton-Smith J., Wit J., Breuning M. Tall stature and duplication of the insulin-like growth factor I receptor gene. Eur. J. Med. Genet. 2007;50:1–10. doi: 10.1016/j.ejmg.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 58.Qureshi A.Y., Mueller S., Snyder A.Z., Mukherjee P., Berman J.I., Roberts T.P., Nagarajan S.S., Spiro J.E., Chung W.K., Sherr E.H., et al. Opposing Brain Differences in 16p11.2 Deletion and Duplication Carriers. J. Neurosci. 2014;34:11199–11211. doi: 10.1523/JNEUROSCI.1366-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mooneyham K.A., Holden K.R., Cathey S., Dwivedi A., Dupont B.R., Lyons M.J. Neurodevelopmental delays and macrocephaly in 17p13.1 microduplication syndrome. Am. J. Med. Genet. Part A. 2014;164:2887–2891. doi: 10.1002/ajmg.a.36708. [DOI] [PubMed] [Google Scholar]
- 60.Mefford H., Mitchell E., Hodge J. 17q12 Recurrent Duplication. In: Adam M.P., Everman D.B., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., Amemiya A., editors. GeneReviews® [Internet] University of Washington; Seattle, WA, USA: 2016. [(accessed on 27 June 2022)]. [Updated 13 January 2022] 1993–2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK344340/ [PubMed] [Google Scholar]
- 61.Zollino M., Marangi G., Ponzi E., Orteschi D., Ricciardi S., Lattante S., Murdolo M., Battaglia D., Contaldo I., Mercuri E., et al. Intragenic KANSL1 mutations and chromosome 17q21.31 deletions: Broadening the clinical spectrum and genotype–phenotype correlations in a large cohort of patients. J. Med. Genet. 2015;52:804–814. doi: 10.1136/jmedgenet-2015-103184. [DOI] [PubMed] [Google Scholar]
- 62.Nevado J., Rosenfeld J.A., Mena R., Palomares-Bralo M., Vallespin E., Angeles Mori M., Tenorio J.A., Gripp K.W., Denenberg E., Del Campo M., et al. PIAS4 is associated with macro/microcephaly in the novel interstitial 19p13. 3 microdele-tion/microduplication syndrome. Eur. J. Hum. Genet. 2015;23:1615–1626. doi: 10.1038/ejhg.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Orellana C., Roselló M., Monfort S., Mayo S., Oltra S., Martínez F. Pure duplication of 19p13.3 in three members of a family with intellectual disability and literature review. Definition of a new microduplication syndrome. Am. J. Med. Genet. Part A. 2015;167:1614–1620. doi: 10.1002/ajmg.a.37046. [DOI] [PubMed] [Google Scholar]
- 64.Pinchefsky E., Laneuville L., Srour M. Distal 22q11.2 Microduplication. Child Neurol. Open. 2017;4:1–6. doi: 10.1177/2329048X17737651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buzanello A., Ramos A.C., Silva A.P., Ceron C., Schmitt J., Menegais K., Ribeiro N.D., Siepmann R., Nardi A. Encéfalo: Estruturas e Funções. Ação Odonto. 2013;1:10. [Google Scholar]
- 66.Deshpande A., Weiss L.A. Recurrent reciprocal copy number variants: Roles and rules in neurodevelopmental disorders. Dev. Neurobiol. 2018;78:519–530. doi: 10.1002/dneu.22587. [DOI] [PubMed] [Google Scholar]
- 67.Pös O., Radvanszky J., Buglyó G., Pös Z., Rusnakova D., Nagy B., Szemes T. DNA copy number variation: Main characteristics, evolutionary significance, and pathological aspects. Biomed. J. 2021;44:548–559. doi: 10.1016/j.bj.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Golzio C., Katsanis N. Genetic architecture of reciprocal CNVs. Curr. Opin. Genet. Dev. 2013;23:240–248. doi: 10.1016/j.gde.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zarrei M., MacDonald J., Merico D., Scherer S. A copy number variation map of the human genome. Nat. Rev. Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 70.Tolezano G., Bastos G., Krepischi A. Burden of rare copy number variants in microcephaly: A Brazilian cohort of 185 microcephalic patients and review of the literature. J. Autism Dev. Disord. 2022. accepted . [DOI] [PubMed]
- 71.Dikow N., Maas B., Gaspar H., Kreiss-Nachtsheim M., Engels H., Kuechler A., Garbes L., Netzer C., Neuhann T.M., Koehler U., et al. The phenotypic spectrum of duplication 5q35.2-q35.3 encompassing NSD1: Is it really a reversed Sotos syndrome? Am. J. Med. Genet. Part A. 2013;161:2158–2166. doi: 10.1002/ajmg.a.36046. [DOI] [PubMed] [Google Scholar]
- 72.Li C., Wang Y., Szybowska M. Novel features of Helsmoortel-Van der Aa/ADNP syndrome in a boy with a known pathogenic mutation in the ADNP gene detected by exome sequencing. Am. J. Med. Genet. Part A. 2017;173:1994–1995. doi: 10.1002/ajmg.a.38201. [DOI] [PubMed] [Google Scholar]
- 73.Gozes I. The ADNP Syndrome and CP201 (NAP) Potential and Hope. Front. Neurol. 2020;11:608444. doi: 10.3389/fneur.2020.608444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pinhasov A., Mandel S., Torchinsky A., Giladi E., Pittel Z., Goldsweig A.M., Servoss S.J., E Brenneman D., Gozes I. Activity-dependent neuroprotective protein: A novel gene essential for brain formation. Dev. Brain Res. 2003;144:83–90. doi: 10.1016/S0165-3806(03)00162-7. [DOI] [PubMed] [Google Scholar]
- 75.Sun X., Peng X., Cao Y., Zhou Y., Sun Y. ADNP promotes neural differentiation by modulating Wnt/β-catenin signaling. Nat. Commun. 2020;11:2984. doi: 10.1038/s41467-020-16799-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Revah-Politi A., Ganapathi M., Bier L., Cho M.T., Goldstein D.B., Hemati P., Iglesias A., Juusola J., Pappas J., Petrovski S., et al. Loss-of-function variants in NFIA provide further support that NFIA is a critical gene in 1p32-p31 deletion syndrome: A four patient series. Am. J. Med. Genet. Part A. 2017;173:3158–3164. doi: 10.1002/ajmg.a.38460. [DOI] [PubMed] [Google Scholar]
- 77.Sieff C. Diamond-Blackfan Anemia. In: Adam M.P., Everman D.B., Mirzaa G.M., Pagon R.A., Wallace S.E., Bean L.J.H., Gripp K.W., Amemiya A., editors. GeneReviews® [Internet] University of Washington; Seattle, WA, USA: 2009. [(accessed on 11 July 2022)]. [updated 17 June 2021] 1993–2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK7047/ [PubMed] [Google Scholar]
- 78.Chakraborty A., Uechi T., Nakajima Y., Gazda H.T., O’Donohue M.-F., Gleizes P.-E., Kenmochi N. Cross talk between TP53 and c-Myc in the pathophysiology of Diamond-Blackfan anemia: Evidence from RPL11-deficient in vivo and in vitro models. Biochem. Biophys. Res. Commun. 2018;495:1839–1845. doi: 10.1016/j.bbrc.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 79.Farrar J.E., Dahl N. Untangling the Phenotypic Heterogeneity of Diamond Blackfan Anemia. Semin. Hematol. 2011;48:124–135. doi: 10.1053/j.seminhematol.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mochida G.H., Mahajnah M., Hill A.D., Basel-Vanagaite L., Gleason D., Hill R.S., Bodell A., Crosier M., Straussberg R., Walsh C.A. A Truncating Mutation of TRAPPC9 Is Associated with Autosomal-Recessive Intellectual Disability and Postnatal Microcephaly. Am. J. Hum. Genet. 2009;85:897–902. doi: 10.1016/j.ajhg.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mbimba T., Hussein N.J., Najeed A., Safadi F.F. TRAPPC9: Novel insights into its trafficking and signaling pathways in health and disease (Review) Int. J. Mol. Med. 2018;42:2991–2997. doi: 10.3892/ijmm.2018.3889. [DOI] [PubMed] [Google Scholar]
- 82.Rasika S., Passemard S., Verloes A., Gressens P., El Ghouzzi V. Golgipathies in Neurodevelopment: A New View of Old Defects. Dev. Neurosci. 2018;40:396–416. doi: 10.1159/000497035. [DOI] [PubMed] [Google Scholar]
- 83.Liang Z.S., Cimino I., Yalcin B., Raghupathy N., Vancollie V.E., Ibarra-Soria X., Firth H.V., Rimmington D., Farooqi I.S., Lelliott C.J., et al. Trappc9 deficiency causes parent-of-origin dependent microcephaly and obesity. PLoS Genet. 2020;16:e1008916. doi: 10.1371/journal.pgen.1008916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Personnic N., Lakisic G., Gouin E., Rousseau A., Gautreau A., Cossart P., Bierne H. A role for Ral GTPase-activating protein subunit β in mitotic regulation. FEBS J. 2014;281:2977–2989. doi: 10.1111/febs.12836. [DOI] [PubMed] [Google Scholar]
- 85.Wagner M., Skorobogatko Y., Pode-Shakked B., Powell C.M., Alhaddad B., Seibt A., Barel O., Heimer G., Hoffmann C., Demmer L.A., et al. Bi-allelic Variants in RALGAPA1 Cause Profound Neurodevelopmental Disability, Muscular Hypotonia, Infantile Spasms, and Feeding Abnormalities. Am. J. Hum. Genet. 2020;106:246–255. doi: 10.1016/j.ajhg.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Martin T.D., Chen X.-W., Kaplan R., Saltiel A., Walker C.L., Reiner D.J., Der C.J. Ral and Rheb GTPase Activating Proteins Integrate mTOR and GTPase Signaling in Aging, Autophagy, and Tumor Cell Invasion. Mol. Cell. 2014;53:209–220. doi: 10.1016/j.molcel.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Clinical Genome Resource. RALGAPB Dosage Sensitivity. [(accessed on 22 November 2022)]. Available online: https://search.clinicalgenome.org/kb/gene-dosage/RALGAPB#report_details_haploinsufficiency.
- 88.Cloëtta D., Thomanetz V., Baranek C., Lustenberger R.M., Lin S., Oliveri F., Atanasoski S., Rüegg M.A. Inactivation of mTORC1 in the Developing Brain Causes Microcephaly and Affects Gliogenesis. J. Neurosci. 2013;33:7799–7810. doi: 10.1523/JNEUROSCI.3294-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen J., Kwong D.L.-W., Zhu C.-L., Chen L.-L., Dong S.-S., Zhang L.-Y., Tian J., Qi C.-B., Cao T.-T., Wong A.M.G., et al. RBMS3 at 3p24 Inhibits Nasopharyngeal Carcinoma Development via Inhibiting Cell Proliferation, Angiogenesis, and Inducing Apoptosis. PLOS ONE. 2012;7:e44636. doi: 10.1371/journal.pone.0044636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang C., Wu Y., Liu Y., Pan F., Zeng H., Li X., Yu L. Tumor Suppressor Effect of RBMS3 in Breast Cancer. Technol. Cancer Res. Treat. 2021;20:1–9. doi: 10.1177/15330338211004921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oo H.Z., Sentani K., Sakamoto N., Anami K., Naito Y., Uraoka N., Oshima T., Yanagihara K., Oue N., Yasui W. Overexpression of ZDHHC14 promotes migration and invasion of scirrhous type gastric cancer. Oncol. Rep. 2014;32:403–410. doi: 10.3892/or.2014.3166. [DOI] [PubMed] [Google Scholar]
- 92.Yeste-Velasco M., Mao X., Grose R., Kudahetti S.C., Lin D., Marzec J., Vasiljević N., Chaplin T., Xue L., Xu M., et al. Identification of ZDHHC14 as a novel human tumour suppressor gene. J. Pathol. 2014;232:566–577. doi: 10.1002/path.4327. [DOI] [PubMed] [Google Scholar]
- 93.Fagerberg L., Hallström B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., et al. Analysis of the Human Tissue-specific Expression by Genome-wide Integration of Transcriptomics and Antibody-based Proteomics. Mol. Cell. Proteom. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study makes use of data generated by the DECIPHER community. A full list of centers who contributed to the generation of the data is available from https://deciphergenomics.org/about/stats and via email from contact@deciphergenomics.org. Funding for the DECIPHER project was provided by Wellcome. Those who carried out the original analysis and collection of the data bear no responsibility for the further analysis or interpretation of them.