Abstract
The Babesia bovis merozoite surface antigen 1 (MSA-1), a member of the variable merozoite surface antigen (VMSA) family, is an immunodominant glycoprotein which elicits antibodies that inhibit erythrocyte invasion. While antigenic polymorphism is a general feature of vmsa genes, the molecular basis and extent of msa-1 sequence polymorphism have not been well characterized. In this study we defined the msa-1 locus in the biologically cloned Mexico Mo7 strain of B. bovis and identified the sequence differences between MSA-1 antigenically dissimilar strains. We then determined whether sequences conserved between distinct msa-1 alleles would induce cross-reactive CD4+ T lymphocytes or inhibitory antibodies. The msa-1 locus in Mo7 contains a single msa-1 gene flanked by transcribed genes with no sequence homology to members of the VMSA gene family. Argentina B. bovis strains R1A and S2P have msa-1 genes with amino acid sequences that are 98.8% identical to each other, and antibodies against S2P MSA-1 cross-react with native R1A MSA-1. In contrast, identity between the Argentina and Mexico Mo7 msa-1 alleles is only 52%, with no continuous stretch of identity longer than 16 amino acids. Despite limited sequence conservation, antibodies against R1A MSA-1 were able to inhibit invasion of erythrocytes by Mo7 merozoites. The results indicate that inhibition-sensitive epitopes are conserved despite significant sequence divergence between Mexico and Argentina strain alleles and support a conserved functional role for polymorphic MSA-1 in erythrocyte invasion.
Babesia bovis and related vector-borne apicomplexan hemoprotozoa such as Plasmodium spp. have an asexual, intraerythrocytic cycle in the mammalian host. In general the erythrocyte invasion processes for babesial and plasmodial asexual stages are similar. Extracellular merozoites attach to erythrocytes through one or more surface antigens (dependent on the invasion pathway[s] utilized), reorient as necessary to bring apical organelles close to the attachment interface, and release rhoptry products at the time of membrane invagination and entry (6, 24). Based on this model, molecules on the merozoite surface and within the apical organelles are considered vaccine candidates and several have been shown to induce significant immune protection (11, 15, 16, 20, 23, 25, 27).
The merozoite surface antigen 1 (msa-1), msa-2, and babr genes of B. bovis comprise a gene family termed the variable merozoite surface antigen (VMSA) genes (3, 9, 12). Members of this gene family encode surface proteins that are proposed to mediate initial attachment of the merozoite to the host erythrocyte. Immunization with recombinant MSA-1 (rMSA-1) induces a CD4+ T-lymphocyte response and immunoglobin G antibodies that inhibit merozoite invasion of erythrocytes in vitro (2, 8).
Allelic polymorphism is a feature of VMSA genes. Extensive rearrangement has resulted in multiple genes that have 5′ or 3′ regions of sequence identity or close similarity (3). Cross-hybridization is strong between some alleles but between others occurs only when the shared 5′ or 3′ region is utilized as a probe, indicating limited overall sequence conservation (9). Antigenic cross-reactivity among strains with dissimilar alleles has not been demonstrated using monoclonal antibodies, monospecific sera, or postinfection immune sera (14, 21, 28). Additionally, cattle infected with one msa-1 allelic strain type are not as well protected against challenge with parasites containing a heterologous msa-1 type as they are against the homologous strain (8, 26), and breakthrough populations in cattle immunized with live avirulent vaccines do not express a cross-reactive MSA-1 (17).
The genetic basis for MSA-1 polymorphism has not been investigated. Therefore, we wished to determine first whether an msa-1 gene exists in MSA-1 polymorphic strains and, if so, to characterize the polymorphic msa-1 locus and determine whether the extent of sequence similarity would allow induction of cross-strain inhibitory antibodies and cross-reactive CD4+ T lymphocytes. To address these issues, we isolated, sequenced, and compared msa-1 alleles from Mexico strains of B. bovis with alleles from two MSA-1 antigenically unrelated strains from Argentina, R1A and S2P. The results indicate limited but significant sequence identity between Mexico and Argentina strain msa-1 alleles, with conservation of the inhibition-sensitive B-lymphocyte epitope(s).
MATERIALS AND METHODS
Parasites.
The Mo7 biological clone of B. bovis was derived by limiting dilution of the Mexico strain as described elsewhere (7) and maintained as a cryopreserved stabilate in liquid nitrogen (19). The Texas B. bovis strain T2B and the Argentina strains R1A and S2P (kindly provided by Ignacio Echaide) have been previously described (1, 9). Parasites were grown in long-term microaerophilus stationary-phase culture by previously described techniques (13).
DNA analysis, cloning and sequencing.
Genomic DNA was extracted from cultured merozoites by the standard phenol-chloroform procedure. For Southern blot analysis, genomic DNA was digested with restriction enzymes, electrophoresed, transferred to nylon membranes, and hybridized as previously described (28, 29). Oligonucleotide probe primer msa1-f (5′-ATGGCTACGTTTGCTCTTTTC-3′) was labeled with digoxigenin with a 3′ tail as instructed by the manufacturer (Boehringer Mannheim). Primers msa1-f and msa1-r1 (5′-TTGCGGGGATGTTCCTGATGCAG-3′) were used to PCR amplify a digoxigenin-labeled B. bovis msa-1 5′ probe. The B. bovis Sau3A genomic library was kindly provided by Doug Jasmer and was previously described (8). Amplification and cloning of a genomic fragment of R1A containing the msa-1 locus was performed by PCR with primers orf1-f (see below) and orf3-f (see below), using Taq polymerase (Boehringer Mannheim). The 3-kb product of amplification was cloned into the vector pCR II-TOPO (Invitrogen) and sequenced. Sequencing was performed with a Prism Ready Reaction DyeDeoxy Terminator cycle sequencing kit and read with an ABI PRISM 373 genetic analyzer (Applied Biosystems).The resulting sequences were assembled and analyzed with the Genetics Computer Group program (version 9) (4). Protein secondary structure analysis also was performed using the Baylor College of Medicine search launcher (http://dot.imgen.bcm.tmc.edu:9331/) (10).
Detection of specific transcripts from the msa-1 gene.
mRNA was extracted from purified B. bovis merozoites using oligo(dT) affinity columns as instructed by the manufacturer (Ambion Inc.). MSA-1 mRNA transcripts were detected by reverse transcription (RT)-PCR analysis using primers msa1-f and msa1-r (5′- AAATGCAGAGAGAACGAAGTAG-3′) specific for the msa-1 gene. Transcripts of open reading frames 1, 3, and 4 (orf-1, -3, and -4) linked to the msa-1 gene were amplified using the following specific primers: orf-1, orf1-f (5′-GATGCTTTGGTTGACGCAAC-3′) and orf1-r (5′-GGCACTCAAACATATCGGTCAGAT-3′) orf-3, orf3-f (5′-ATGCTATTGGCTTCCAATGTC-3′) and orf3-r (5′-GTTGTTGGAGTTGCCAGCAATG-3′); and orf-4, orf4-f (5′-GATCCAGGAACTATACGCTAATAG-3′) and orf4-r (5′-CATGGAAGGCTTATGCCGTTG-3′). Products of RT-PCR were cloned into vector pCR 2.1 (Invitrogen) and sequenced.
Expression of proteins, production of antibodies, and immunoblotting.
The msa-1 ORFs were amplified from DNA extracted from the Mo7, R1A, and S2P strains by PCR using primers msa1-f and msa1-r. Amplicons were cloned into the vector pBAD/Thio-Topo (Invitrogen) for expression of rMSA-1. Primers msa1-f and msa1-r were designed to allow in-frame cloning of the inserts into the pBAD/Thio-Topo vector to produce expressed thioredoxin fusion proteins. Inclusion bodies from bacteria induced with 0.2% arabinose were prepared by sonication and high-speed centrifugation and then dissolved in 6 M urea–0.15 M NaCl–0.1 M Tris-HCl, pH 8.0. Solubilized protein was obtained after centrifugation and dialyzed against phosphate-buffered saline before immunization of mice. Relative purity of antigen was confirmed in Coomassie blue-stained sodium dodecyl sulfate-polyacrylamide gels. Antibodies against Mo7, R1A, and S2P rMSA-1 were obtained by immunization of three groups of three mice each with 10 μg of recombinant protein suspended in complete Freund's adjuvant, followed by three immunizations each of 10 μg of recombinant protein suspended in incomplete Freund's adjuvant. Murine antisera were analyzed for reactivity and specificity by immunoblotting using native antigen from parasitized erythrocytes as previously described (28).
In vitro inhibition assay.
Inhibition of B. bovis merozoites was performed as described elsewhere (9). Briefly, 5 × 105 Mo7 B. bovis merozoites were incubated with control nonimmune or anti-rMSA-1 immune bovine or murine serum, diluted 1:5 in culture medium, for 30 min at 4°C. An equal volume of 5% (vol/vol) bovine erythrocytes in culture medium was added prior to incubation in triplicate wells of 96-well plates at 37°C in a 5% CO2 atmosphere. The percentage of parasitized erythrocytes (PPE) was determined after 48 and 72 h by microscopic examination of 2,000 erythrocytes in Giemsa-stained smears prepared from each well. Results from three independent experiments were analyzed by one-tailed Student's t test.
T-cell epitope analysis.
Bovine T-lymphocyte lines specific for the parent Mexico strain of B. bovis were established with peripheral blood mononuclear cells of cow C97 (2). Cells were cultured for up to 4 weeks and stimulated weekly with irradiated (3,000 rads) peripheral blood mononuclear cells as a source of antigen-presenting cells and semiweekly with B. bovis (Mexico strain) merozoite membrane (CM) antigen (2). These cell lines are composed of predominantly CD4+ T cells (2). T lymphocytes were assayed for antigen-specific proliferation 7 days following the last subculture without antigen. Proliferation assays were carried out in replicate wells of 96-well plates (Costar, Cambridge, Mass.) as described elsewhere (2), using B. bovis CM, control uninfected red blood cell membranes (URBC), MSA-1, or control antigen consisting of Anaplasma marginale rMSP-5 (each at 1 to 25 μg/ml). The results are presented as the stimulation index (SI), calculated as mean cpm of cells cultured with B. bovis CM/mean cpm of cells cultured with URBC or as mean cpm of cells cultured with rMSA-1/mean cpm of cells cultured with rMSP-5. Responses were analyzed for statistical significance by Student's t test.
Nucleotide sequence accession numbers.
The nucleotide sequences reported here have been deposited in GenBank under accession numbers AF275908 to AF275912.
RESULTS
Genomic analysis of the msa-1 locus.
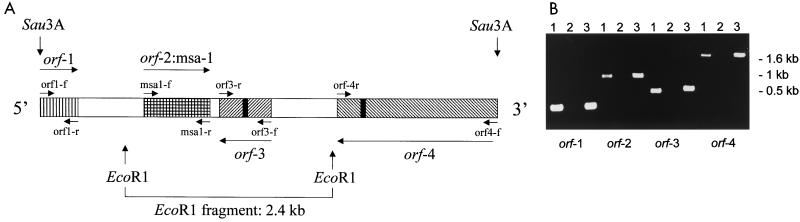
A clone designated msa-1-Sau3A was identified in a B. bovis Mo7 Sau3A genomic library by hybridization with a 5′ msa-1 probe (8, 9). The 5.52-kb genomic fragment in msa-1-Sau3A (Fig. 1A) was sequenced and determined to contain a single copy of msa-1 identical in sequence to the previously described MSA-1 cDNA clone derived from the Mexico Mo7 strain (8, 9) (GenBank accession no. AF275908). RT-PCR amplification of Mo7 mRNA using primers msa1-f and msa1-r resulted in a 901-bp fragment identical in size and sequence to genomic Mo7 DNA (Fig. 1).
FIG. 1.
Genomic analysis of the B. bovis msa-1 locus. (A) Map of the Mo7 msa-1 locus based on sequencing of a 5.520-kb fragment contained in the msa-1-Sau3A genomic clone. The four ORFs designated by patterned boxes correspond to the following genes: orf-1, low homology to polyketide synthase gene from Streptomyces spp.; orf-2, msa-1; orf-3, significant homology to N-methylaspartate receptor protein gene; orf-4, low homology to a hypothetical gene of C. elegans. Arrows indicate the orientation of the coding DNA strand. Primer sites (orf1-f, orf1-r, msa1-f, msa1-r, orf3-f, orf3-r, orf4-f, and orf4-r) are designated above and below the map of the fragment, with primer orientation indicated by arrows. EcoRI sites and the size of the corresponding EcoRI fragment (2.4 kb) are shown below the map. Intergenic regions (white boxes) and the introns in orf-3 and orf-4 (black boxes) are indicated. (B) Transcriptional analysis of the B. bovis msa-1 locus represented by an ethidium bromide-stained agarose gel of amplicons specific for each of the ORFs encoded in clone msa-1-Sau3A. For each ORF, three lanes are depicted. Lane 1 represents amplification of Mo7 mRNA with reverse transcriptase, lane 2 is a no-reverse transcriptase control, and lane 3 represents amplification of DNA. Size markers are shown at the right.
Analysis of sequence 5′ and 3′ to the Mo7 msa-1 gene identified three ORFs, none of which had significant homology to msa-1 or any other members of the vmsa gene family (Fig. 1A). An incomplete coding region termed orf-1 (GenBank accession no: AF275908), encoding a protein with low but significant homology to polyketide synthase from Streptomyces spp., is located 5′ to the msa-1 locus and is separated from the msa-1 ORF by a 0.84-kb intergenic region which does not contain other ORFs. The 420-bp incomplete orf-1 is encoded in the same orientation as msa-1 and transcribes an mRNA of 420 bp that can be amplified by RT-PCR from B. bovis mRNA using primers orf1-f and orf1-r (Fig. 1) and which is identical in sequence to orf-1 of the genomic clone msa-1-Sau3A.
The genomic fragment located 3′ to the msa-1 gene contains two additional ORFs, both in the reverse orientation relative to msa-1. The complete 612-bp coding region termed orf-3 (GenBank accession no. AF275908) is located only 160 bp immediately 3′ to msa-1 and encodes a 20-kDa protein with strong homology to N-methylaspartate receptor proteins, which are well conserved among different species (GenBank accession no. AF275908). A transcript for orf-3 detected by RT-PCR of merozoite mRNA using primers orf3-f and orf3-r (Fig. 1) was cloned and sequenced (GenBank accession no. AF275912). Comparison of sequence between the product of RT-PCR and orf3 in the msa-1-Sau3A genomic clone demonstrates the presence of a 36-bp intron with consensus intron/exon junctional splice sequences at the 5′ and 3′ ends (UG/GU… AG/GA, where “/” denotes the site of intron splicing) located between bases 2642 and 2678. Predicted secondary structure of the putative N-methylaspartate receptor protein is suggestive of a highly hydrophobic integral membrane molecule (10).
Contiguous to orf-3, and separated by an 800-bp intergenic region with no ORFs, is an incomplete and discontinuous coding region termed orf-4 (GenBank accession no. AF275908), also in the reverse orientation relative to the msa-1 gene. A database search for sequences homologous to the putative orf-4 product shows significant similarity only to a hypothetical protein (PIR accession no. T22644) from Caenorhabditis elegans. orf-4 also is transcribed, as indicated by RT-PCR analysis of mRNA using primers orf4-f and orf4-r (Fig. 1A). Comparison of orf-4 cDNA and genomic sequences reveals the presence of a 40-bp intron between bases 3926 and 3966, also with typical consensus splice sequences.
To further demonstrate that the clone msa-1-Sau3A represents the genomic region containing the msa-1 locus in the Mo7 strain, we amplified Mo7 genomic DNA using primers orf1-f and orf4-f (Fig. 1A) derived from the 5′ and 3′ regions, respectively, of the genomic clone msa-1-Sau3A. This amplification resulted in a 5.5-kb fragment identical in size and sequence to the genomic clone, demonstrating that clone msa-1-Sau3A is a faithful replica of the region containing the msa-1 locus in the Mo7 strain (data not shown).
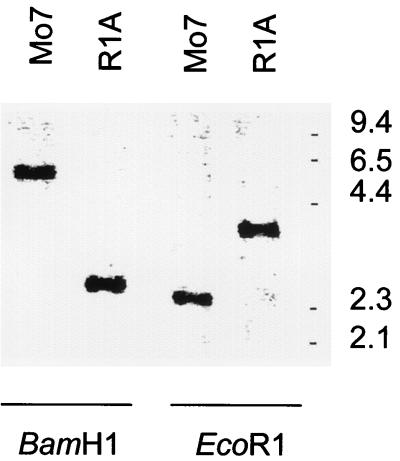
The results are consistent with the presence of a single copy of the msa-1 gene in the Mo7 strain. However, to test whether additional copies of the msa-1 gene are present in the parasite genome, Mo7 genomic DNA was probed with a digoxigenin-labeled msa1-f oligonucleotide specific for the 5′ end of msa-1 (Fig. 1A). The results are shown in Fig. 2. Digestion of Mo7 genomic DNA with BamHI, which cuts outside the 5.5-kb genomic fragment, resulted in hybridization with a single 6.0-kb band, whereas digestion with EcoRI produced a single 2.4-kb band identical in size to the predicted EcoRI fragment of clone msa-1-Sau3A (Fig. 1A and 2). The results support the presence of only a single copy of msa-1 in the Mo7 strain and are in agreement with the sequence analysis of clone msa-1-Sau3A. Similar results compatible with a single gene copy were obtained using genomic DNA from the Argentina R1A strain (Fig. 2).
FIG. 2.
Hybridization of msa-1 genes in strains Mo7 and R1A. Genomic DNA was extracted from B. bovis strains Mo7 and R1A and digested with BamHI or EcoRI. Blots were probed with the digoxigenin-labeled oligonucleotide msa1-f (Fig. 1A), specific for the 5′ end of msa-1, and hybridization was detected by chemiluminescence. Size markers (in kilobases) are shown on the right.
Characterization of msa-1 alleles in antigenically distinct strains of B. bovis.
PCR amplification of DNA from the Texas T2B and Argentina R1A and S2P strains using primers msa1-f and msa1-r, which flank the complete ORF of the msa-1 gene in the Mo7 strain (Fig. 1), resulted in bands of approximately 900 bp for each strain. Analysis of the predicted amino acid sequence of the T2B-derived DNA (GenBank accession no. AF275911) showed absolute identity with Mo7 msa-1. This result is consistent with previously detected cross-hybridization of T2B DNA in Southern blots using labeled probes derived from msa-1 of the Mo7 strain (9), as well as antibody and T-lymphocyte cross-reactivity between Mo7 and T2B MSA-1 (2, 21). Together, these data indicate that identical allelic forms of the msa-1 gene are encoded and expressed in the Texas T2B and Mo7 strains.
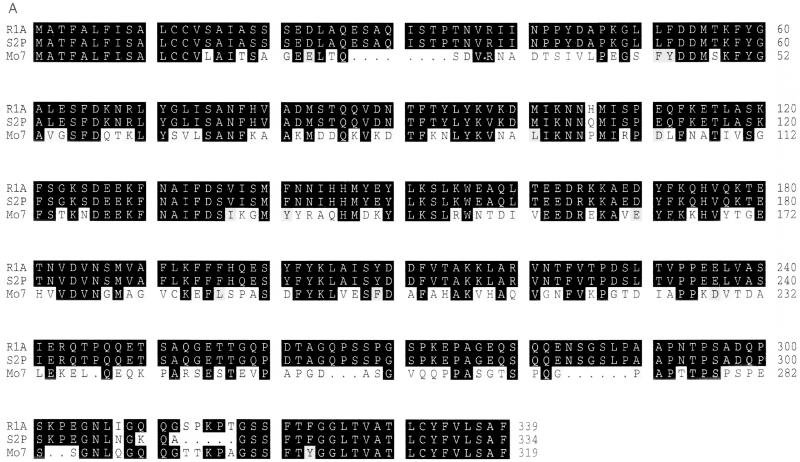
Sequence comparison of msa-1 amplified from the Argentina strains R1A (GenBank accession no. AF275910) and S2P (GenBank accession no. AF275909) demonstrates a high degree of identity (98.8%), the putative products differing by only eight amino acid changes, including three substitutions and a 5-amino-acid insertion in the S2P MSA-1 (Fig. 3A). However, sequence comparison of Mexican and Argentine MSA-1 shows extensive divergence (Fig. 3A), with overall only 52% amino acid identity between Mo7 and R1A. Predicted products of ORFs for the Argentina R1A and S2P msa-1 (334 and 339 amino acids, respectively) are longer than the predicted Mexico msa-1 ORF product (319 amino acids). The Mo7 and T2B msa-1 alleles are identical to the Argentine allele in the 5′ (first 42 nucleotides) and 3′ (last 51 nucleotides) regions, consistent with the general pattern of shared sequences among other members of the VMSA gene family (3, 9, 12). Starting at amino acid position 118 of the Mo7 sequence (amino acid position 126 of the Argentine sequence), there is a continuous stretch of 11 identical amino acids (Fig. 3A). Other identical short motifs are distributed throughout the sequence (Fig. 3A). RT-PCR analysis of R1A mRNA using primers msa1-f and msa1-r yielded a 900-bp band identical in size and sequence to the band obtained with R1A genomic DNA as a target for amplification (data not shown).
FIG. 3.
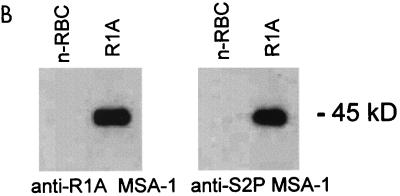
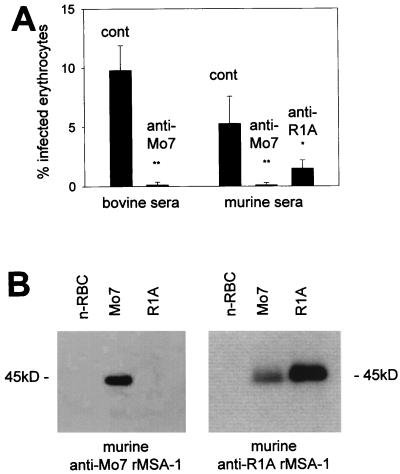
(A) Comparison of the predicted MSA-1 amino acid sequences from the Mexico Mo7 strain and the Argentina strains R1A and S2P. Amino acid substitutions, insertions, and deletions are indicated by a white background. Areas of amino acid identity have a black background, while conservative amino acid substitutions are enclosed in gray boxes. Dots indicate deletions. The number of amino acids in each sequence is shown on the right. (B) Cross-reactivity of antibodies against MSA-1 from the Argentina strains R1A and S2P. Serum raised against rMSA-1 from strain R1A or S2P was reacted in immunoblots with control URBC (n-RBC) or erythrocytes infected with the R1A strain of B. bovis. The position of the 45-kDa R1A MSA-1 molecule is indicated on the right.
To determine if R1A and S2P msa-1 transcripts are translated and expressed in merozoites, murine antisera against MSA-1 recombinant fusion proteins derived from the R1A and S2P strains were reacted with R1A merozoites in immunoblot assays. The results are shown in Fig. 3B. Murine antiserum raised against either R1A or S2P rMSA-1 bound a native protein of 45 kDa, consistent with the larger predicted size for the R1A strain MSA-1 (339 amino acids and 45 kDa) compared to MSA-1 from the Mo7 strain (319 amino acids and 42 kDa). The results indicate that R1A msa-1 is not a pseudogene and that, as expected from the high degree of sequence conservation, the Argentina strain R1A and S2P MSA-1 proteins share B-lymphocyte epitopes.
To address whether polymorphic msa-1 genes in different strains are maintained within the same locus, we amplified R1A and Mo7 genomic DNA using primers orf1-f and orf3-f, representing sequences in the 5′ and 3′ flanking ORFs of the Mo7 msa-1 locus. The expected 3.0-kb product was amplified from both the Mo7 and R1A strains. Amplification of the R1A 3-kb fragment using primers msa1-f and msa1-r produced a 900-bp fragment containing msa-1. In contrast, amplification of Mo7 and R1A genomic DNA using primers orf1-f and orf4-f resulted in a 5.5-kb product only for the Mo7 strain, suggesting that sequence in the region 3′ to orf-3 is variable between these two strains (data not shown).
In vitro inhibition.
To address whether sequences conserved between the Mexico and Argentina strain alleles encode any shared inhibition-sensitive B-lymphocyte epitopes, the homologous and heterologous inhibitory activities of antibodies raised against rMSA-1 were determined. Monospecific bovine serum antibodies against cDNA-encoded Mo7 rMSA-1 strongly neutralized Mo7 merozoite invasion (Fig. 4A), consistent with previous research showing homologous strain inhibition (8, 9). Similarly, murine antiserum against Mo7 rMSA-1 also inhibited invasion of B. bovis Mo7 merozoites (Fig. 4A). Serum antibodies against R1A rMSA-1 bound the expected 45-kDa native protein in the R1A strain and also cross-reacted with the 42-kDa native protein present in the Mo7 strain (Fig. 4B). This anti-R1A serum significantly inhibited the invasion of heterologous Mo7 merozoites (anti-R1A MSA-1 serum, 1.5% ± 0.7% PPE: control nonimmune serum, 5.3% ± 2.3% PPE, P < 0.02) (Fig. 4A). In contrast, antiserum induced by immunization of mice with Mo7 rMSA-1 did not bind R1A MSA-1 (Fig. 4B).
FIG. 4.
(A) In vitro inhibition using antiserum against Mo7 or R1A rMSA-1. B. bovis strain Mo7 merozoites were exposed to the indicated control (cont) or anti-rMSA-1 immune bovine or murine serum in vitro and incubated with bovine erythrocytes as described in Materials and Methods. The data show the mean ± standard deviation of the percentage of infected erythrocytes in three independent assays. ∗, P < 0.02; ∗∗, P < 0.001. (B) Cross-reactivity of immune sera against Mo7 and R1A rMSA-1. Murine anti-Mo7 rMSA-1 or murine anti-R1A rMSA-1 serum was reacted in immunoblots with URBC (n-RBC) or erythrocytes infected with the Mo7 or R1A strain of B. bovis. The position of the 45-kDa R1A MSA-1 molecule is indicated on the right and left.
T-lymphocyte responses.
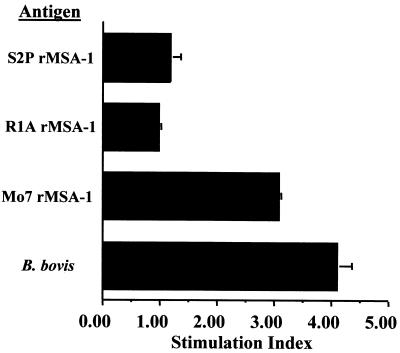
Mexico strains of B. bovis have been previously shown to induce an MSA-1-specific CD4+ T-lymphocyte response that does not cross-react with MSA-1 in Australia and Israel strains (2). To determine if the sequences conserved between R1A and Mo7 alleles of MSA-1 encode a cross-reactive T-cell epitope, we tested the effect of purified rMSA-1 on the proliferation of B. bovis-specific short-term T-lymphocyte cell lines isolated from cattle infected with the parent Mexico strain of B. bovis. The cell lines proliferated in response only to rMSA-1 derived from the Mo7 Mexico strain, not with rMSA-1 derived from the Argentina strain R1A or S2P (Fig. 5).
FIG. 5.
Proliferation of short-term T-cell lines against MSA-1. T-lymphocyte lines were established as indicated in Materials and Methods. The results are presented as the mean SI of two independent experiments performed with one cell line in culture for 2 weeks and a different cell line in culture for 4 weeks and using 5 μg of protein per ml. The SI for rMSA-1 proteins was derived by comparing the response to A. marginale rMSP-5, and the SI for B. bovis was derived by comparing the response to URBC. The mean SI against rMSA-1 Mo7 is significantly greater than the mean SI against rMSA-1 R1A or rMSA-1 S2P (P < 0.02) when analyzed by one-tailed Student's t test. For each experiment, the response to Mo7 rMSA-1 was significantly different from the response to rMSP-5 (P < 0.05), whereas there was no significant difference between the response to rMSP-5 and either rMSA-1 R1A or rMSA-1 S2P (P > 0.2).
DISCUSSION
Polymorphism among B. bovis strains has previously been reported to include a complete lack of MSA-1 B- and T-lymphocyte cross-reactivity (2, 14, 21, 26, 28). This suggested that within the population there are msa-1 alleles that encode immunologically unique MSA-1 molecules or, alternatively, that msa-1 genes in other strains do not exist. In the present study, using B. bovis strains from North and South America, which previously had been shown not to contain cross-reactive MSA-1, we identified two msa-1 alleles which differ by 48% of the deduced amino acids. Thus, while allelic variation is pronounced, msa-1 is maintained in the genome, presumably due to a necessary functional role for MSA-1 in merozoite invasion.
The VMSA family of surface proteins, of which MSA-1 is a member, also includes the MSA-2, a 44-kD glycoprotein (7, 9, 12, 22), and products of the babr genes, initially detected as a polymorphic gene family in which at least some members are tandemly arranged in the genome (3). Sequence analysis of the msa-1 locus in the Mo7 and R1A strains demonstrated that no other vmsa-related genes are present in close linkage to msa-1. Thus, not all the members of the vmsa family are tandemly arranged. Chromosomal arrangement of the vmsa family may determine the genetic mechanisms involved in msa-1 evolution and the generation of msa-1 sequence variation (5). Gene homogenization through unequal recombination or gene conversion is typically associated with evolution of genes arranged in tandem arrays and therefore is less likely to be a major mechanism for evolution of the msa-1 locus. It is more probable that homologous recombination during sexual stages in the tick vector is involved in generation of msa-1 alleles encoding polymorphic MSA-1 molecules.
Sequencing of the 5.5-kb genomic fragment obtained from the Mo7 strain, together with Southern hybridization experiments, strongly suggests that msa-1 is present as a single-copy gene in the Mo7 strain. Within the 5.5-kb genomic fragment harboring the single msa-1 gene there are at least four other genes that are transcribed in merozoites, including continuous and discontinuous ORFs located in both strands. This type of compact genomic organization resembles that of other known B. bovis loci such as rap-1 (29). Of note, the population of parasites in the uncloned R1A strain also appear to contain a single msa-1 copy with conservation of 5′ and 3′ ORFs flanking msa-1. If this feature is maintained broadly, it may allow for the direct cloning of other polymorphic msa-1 genes from B. bovis strains with immunologically unique MSA-1 glycoproteins.
Overall sequence identity between the Mexican and Argentine msa-1 alleles is much greater than between msa-1 and other members of the VMSA gene family (3, 9, 12). The longest stretches of sequence identity between the two MSA-1 proteins are in the amino (14 amino acids) and carboxy (22 amino acids with one conservative substitution) termini, consistent with the regional 5′ and 3′ pattern of sequence conservation among other vmsa genes (9). Prior to this study, the limited sequence conservation between msa-1 alleles had not been demonstrated to encode shared epitopes. Now we demonstrate that B-lymphocyte cross-reactivity can occur between MSA-1 of divergent alleles and that cross-reactive antibodies can inhibit merozoite invasion in vitro. In contrast, the absence of continuous stretches of identical amino acids long enough to constitute CD4+ T-lymphocyte epitopes is reflected in the lack of cross-reactive T-cell responses. Heterologous binding of R1A rMSA-1-specific antibodies to Mo7 MSA-1 in immunoblots is weaker than homologous binding. The reason for this reduced binding is unknown; possibly the epitope(s) recognized by cross-reactive antibodies is conformational and is not well represented in the partially denatured antigen used in immunoblots. However, we have tested the same serum in indirect immunofluorescence assays and find the same relative degree of homologous and heterologous binding (data not shown), suggesting that conformation is at least not a major cause of the differences observed. The cross-reactive epitope(s) also may be encoded by a slightly different sequence and thus not identical, resulting in a reduced affinity of heterologous antibody binding. A more likely explanation is that only a small percentage of the total antibodies generated against R1A rMSA-1 recognize a shared epitope. This explanation is consistent with the one-way cross-reactivity observed and suggests that the B-lymphocyte epitope encoded at least in part by sequences shared between alleles is not immunodominant. The limited cross-reactivity, combined with the lack of conserved CD4+ T-lymphocyte epitopes to provide help for antibody production, constitutes a severe constraint to induction of cross-protective immunity by MSA-1 immunization.
As noted above, sequence identity between the Mexican and Argentine msa-1 alleles is limited to the extreme 5′ and 3′ regions, the 11-mer peptide starting at position 118 of the Mo7 sequence (amino acid position 126 of the Argentine sequence) and small motifs of five amino acids or less throughout the sequence. The amino-terminal conserved region is typical of a hydrophobic signal sequence, with a potential cleavage site between amino acids 19 and 20 (4), and the carboxy-terminal 22 amino acids comprise a second hydrophobic domain consistent with the signal for attachment of the glycosylphosphatidylinositol anchor (4, 7) characteristic of all members of the VMSA family. BLAST and pattern searching of the database is not informative as to a potential function for the conserved 11-mer or any of the other smaller conserved motifs. Thus, how this limited sequence identity among alleles allows conservation of MSA-1 function is unknown. However, conservation of hemoprotozoan merozoite surface protein function in erythrocyte invasion can occur with significant sequence variation. For example, the primary erythrocyte binding region of Plasmodium falciparum MSP-119 can be altered beyond the retention of antibody cross-reactivity with no apparent effect on invasion or growth in vitro (18). We hypothesize that the limited sequence shared among msa-1 alleles allows merozoites expressing antigenically distinct but structurally similar molecules to invade erythrocytes via the same or a redundant pathway.
In summary, molecular characterization of msa-1 variation indicates limited but significant sequence identity between alleles in the Argentina and Mexico strains of B. bovis. While previous studies have indicated that these two alleles encode immunologically unique MSA-1 molecules, sequences conserved between them encode at least one shared B-lymphocyte epitope recognized by inhibitory antibodies against the Argentina R1A strain rMSA-1. If these same sequences are shared among other strains for which MSA-1 cross-reactivity cannot be demonstrated, it might be possible to enhance cross-inhibition of invasion by more specifically targeting the MSA-1 regions required for conservation of function. Finally, the data support the hypothesis that there are only minimal constraints on sequence variation imposed by functional conservation of molecules important for invasion (18) and that within the population, merozoites with surface molecules encoded by alternate alleles not recognized by existing antibodies or T lymphocytes may readily emerge.
ACKNOWLEDGMENTS
We thank Deb Alperin, Bev Hunter, and Carla Robertson for technical assistance, D. P. Jasmer for supplying the Sau3A genomic library, and Ignacio Echaide for providing the B. bovis strains from Argentina.
This work was supported by grants ARS CRIS-5348-32000-014-00D, USAID PCE-G-00-98-00043-00, and USDA NRI 96-35204-3667.
REFERENCES
- 1.Anziani O S, Guglielmone A A, Abdala A A, Aguirre D H, Mangold A J. Proteccion conferida por Babesia bovisvacunal en novillos Holando Argentino. Rev Med Vet (Buenos Aires) 1993;74:47–49. [Google Scholar]
- 2.Brown W C, Palmer G H, McElwain T F, Hines S A, Dobbelaere D A E. Babesia bovis: characterization of the T helper cell response against the 42-kDa merozoite surface antigen (MSA-1) in cattle. Exp Parasitol. 1993;77:97–110. doi: 10.1006/expr.1993.1065. [DOI] [PubMed] [Google Scholar]
- 3.Cowman A F, Bernard O, Stewart N, Kemp D J. Genes of the protozoan parasite Babesia bovisthat rearrange to produce RNA species with different sequences. Cell. 1984;37:653–660. doi: 10.1016/0092-8674(84)90397-0. [DOI] [PubMed] [Google Scholar]
- 4.Devereaux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graham G J. Tandem genes and clustered genes. J Theor Biol. 1995;175:71–87. doi: 10.1006/jtbi.1995.0122. [DOI] [PubMed] [Google Scholar]
- 6.Hadley T J, Klotz R W, Miller L H. Invasion of erythrocytes by malaria parasites: a cellular and molecular overview. Annu Rev Microbiol. 1986;40:451–477. doi: 10.1146/annurev.mi.40.100186.002315. [DOI] [PubMed] [Google Scholar]
- 7.Hines S A, McElwain T F, Buening G B, Palmer G H. Molecular characterization of Babesia bovismerozoite surface proteins bearing epitopes immunodominant in protected cattle. Mol Biochem Parasitol. 1989;37:1–10. doi: 10.1016/0166-6851(89)90096-0. [DOI] [PubMed] [Google Scholar]
- 8.Hines S A, Palmer G H, Jasmer D P, Goff W L, McElwain T F. Immunization of cattle with recombinant Babesia bovismerozoite antigen 1. Infect Immun. 1995;63:349–352. doi: 10.1128/iai.63.1.349-352.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hines S A, Palmer G H, Jasmer D P, McGuire T C, McElwain T F. Neutralization-sensitive merozoite surface antigens of Babesia bovisencoded by members of a polymorphic gene family. Mol Biochem Parasitol. 1992;55:85–94. doi: 10.1016/0166-6851(92)90129-8. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann K, Stoffel W. TMbase—a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- 11.Howard R J, Pasloske B L. Target antigens for asexual malaria vaccine development. Parasitol Today. 1993;9:369–372. doi: 10.1016/0169-4758(93)90085-t. [DOI] [PubMed] [Google Scholar]
- 12.Jasmer D P, Reduker D W, Hines S A, Perryman L E, McGuire T C. Surface epitope localization and gene structure of a Babesia bovis44-kilodalton variable merozoite antigen. Mol Biochem Parasitol. 1992;55:75–84. doi: 10.1016/0166-6851(92)90128-7. [DOI] [PubMed] [Google Scholar]
- 13.Levi M G, Ristic M. Babesia bovis: continuous cultivation in a microaerophilus stationary phase culture. Science. 1980;207:1218–1220. doi: 10.1126/science.7355284. [DOI] [PubMed] [Google Scholar]
- 14.Madruga C R, Suarez C E, McElwain T F, Palmer G H. Conservation of merozoite and apical complex B cell epitopes among Babesia bigemina and Babesia bovisstrains isolated in Brazil. Vet Parasitol. 1996;61:21–30. doi: 10.1016/0304-4017(95)00809-8. [DOI] [PubMed] [Google Scholar]
- 15.Marshall V M, Silva A, Foley M, Cranmer S, Wang L, McColl D J, Kemp D J, Coppel R L. A second merozoite surface protein (MSP-4) of Plasmodium falciparumthat contains an epidermal growth factor-like domain. Infect Immun. 1997;65:4460–4467. doi: 10.1128/iai.65.11.4460-4467.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall V M, Tieqiao W, Coppel R L. Close linkage of three merozoite surface protein genes on chromosome 2 of Plasmodium falciparum. Mol Biochem Parasitol. 1998;94:13–25. doi: 10.1016/s0166-6851(98)00045-0. [DOI] [PubMed] [Google Scholar]
- 17.Molloy J B, Waldron S J, Jorgensen W K. Identification of an immunodominant 40 kDa merozoite antigen common to the Australian T and Dixie vaccine strains of Babesia bovisand the development of diagnostic tests specific for these strains. Vet Parasitol. 1995;60:229–240. doi: 10.1016/0304-4017(95)00779-5. [DOI] [PubMed] [Google Scholar]
- 18.O'Donnell R A, Saul A, Cowman A F, Crabb B S. Functional conservation of the malaria vaccine antigen MSP-119 across distantly related Plasmodiumspecies. Nat Med. 2000;6:91–95. doi: 10.1038/71595. [DOI] [PubMed] [Google Scholar]
- 19.Palmer D A, Buening G M, Carson C A. Cryopreservation of Babesia bovisfor in vitro cultivation. Parasitology. 1982;84:567–571. doi: 10.1017/s0031182000052835. [DOI] [PubMed] [Google Scholar]
- 20.Palmer G H, McElwain T F. Molecular basis for vaccine development against anaplasmosis and babesiosis. Vet Parasitol. 1995;57:233–253. doi: 10.1016/0304-4017(94)03123-e. [DOI] [PubMed] [Google Scholar]
- 21.Palmer G H, McElwain T F, Perryman L E, Davis W C, Reduker D R, Jasmer D J, Shkap V, Pipano E, Goff W L, McGuire T C. Strain variation of Babesia bovismerozoite surface exposed epitopes. Infect Immun. 1991;59:3340–3342. doi: 10.1128/iai.59.9.3340-3342.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reduker D R, Jasmer D P, Goff W L, Perryman L E, Davis W C, McGuire T C. A recombinant surface protein of Babesia boviselicits antibodies that react with live merozoites. Mol Biochem Parasitol. 1989;35:239–248. doi: 10.1016/0166-6851(89)90210-7. [DOI] [PubMed] [Google Scholar]
- 23.Ridley R G, Takacs B, Etlinger H, Scaife J G. A rhoptry antigen of Plasmodium falciparum is protective in Saimirimonkeys. Parasitology. 1990;101:187–192. doi: 10.1017/s0031182000063228. [DOI] [PubMed] [Google Scholar]
- 24.Sam—Yellowe T Y. Rhoptry organelles of the apicomplexa: their role in host cell invasion and intracellular survival. Parasitol Today. 1996;12:28–32. doi: 10.1016/0169-4758(96)10030-2. [DOI] [PubMed] [Google Scholar]
- 25.Saul A, Lawrence G, Smillie A, Rzepczyk C M, Reed C, Taylor D, Anderson K, Stowers A, Kemp R, Allworth A, Anders R F, Brown G V, Pye D, Schoofs P, Irving D O, Dyer S L, Woodrow G C, Briggs W R, Reber R, Sturchler D. Human phase I vaccine trials of 3 recombinant asexual stage malaria antigens with Montanide ISA720 adjuvant. Vaccine. 1999;17:3145–3159. doi: 10.1016/s0264-410x(99)00175-9. [DOI] [PubMed] [Google Scholar]
- 26.Shkap V, Pipano E, McElwain T F, Herzberg U, Krigel Y, Fish L, Palmer GH. Cross-protective immunity induced by Babesia bovisclones with antigenically unrelated variable merozoite surface antigens. Vet Immunol Immunopathol. 1994;41:367–374. doi: 10.1016/0165-2427(94)90108-2. [DOI] [PubMed] [Google Scholar]
- 27.Siddiqui W A, Tam L Q, Kramer K J, Hui G S, Case S E, Yamaga K M, Chang S P, Chan E B, Kan S C. Merozoite surface coat precursor protein completely protects Aotus monkeys against Plasmodium falciparummalaria. Proc Natl Acad Sci USA. 1987;84:3014–3018. doi: 10.1073/pnas.84.9.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suarez C E, McElwain T F, Echaide I E, de Echaide S T, Palmer G H. Interstrain conservation of babesial RAP-1 surface exposed epitopes despite rap-1genomic polymorphism. Infect Immun. 1994;62:3576–3579. doi: 10.1128/iai.62.8.3576-3579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suarez C E, Palmer G H, Hotzel I, McElwain T F. Structure, sequence, and transcriptional analysis of the Babesia bovis rap-1multigene locus. Mol Biochem Parasitol. 1998;93:215–224. doi: 10.1016/s0166-6851(98)00032-2. [DOI] [PubMed] [Google Scholar]