Abstract
Deletions in the CCM1, CCM2, and CCM3 genes are a common cause of familial cerebral cavernous malformations (CCMs). In current molecular genetic laboratories, targeted next-generation sequencing or multiplex ligation-dependent probe amplification are mostly used to identify copy number variants (CNVs). However, both techniques are limited in their ability to specify the breakpoints of CNVs and identify complex structural variants (SVs). To overcome these constraints, we established a targeted Cas9-mediated nanopore sequencing approach for CNV detection with single nucleotide resolution. Using a MinION device, we achieved complete coverage for the CCM genes and determined the exact size of CNVs in positive controls. Long-read sequencing for a CCM1 and CCM2 CNV revealed that the adjacent ANKIB1 and NACAD genes were also partially or completely deleted. In addition, an interchromosomal insertion and an inversion in CCM2 were reliably re-identified by long-read sequencing. The refinement of CNV breakpoints by long-read sequencing enabled fast and inexpensive PCR-based variant confirmation, which is highly desirable to reduce costs in subsequent family analyses. In conclusion, Cas9-mediated nanopore sequencing is a cost-effective and flexible tool for molecular genetic diagnostics which can be easily adapted to various target regions.
Keywords: nanopore sequencing, long-read sequencing, CRISPR/Cas9, copy number variants, cerebral cavernous malformations, structural variants
1. Introduction
Cerebral cavernous malformations (CCMs) are thin-walled vascular lesions in the central nervous system which are prone to hemorrhage. Depending on their location, they can cause a wide range of neurological complications, such as stroke-like symptoms or seizures. The familial form of CCM, which is characterized by the presence of multiple CCMs, follows an autosomal-dominant inheritance pattern with incomplete penetrance and is caused by heterozygous germline variants in the CCM1 (KRIT1), CCM2, or CCM3 (PDCD10) gene [1].
Disease-causing variants are found in more than 90% of all familial cases [1]. Frameshift mutations account for the vast majority of pathogenic variants, followed by nonsense, splice site, and copy number variants (CNVs) [1,2,3,4]. Indeed, up to 18% of pathogenic variants found in CCM patients are large deletions [5]. Standard diagnostic techniques for CNV detection are multiplex ligation-dependent probe amplification (MLPA) and short-read gene panel sequencing in combination with specific bioinformatic pipelines. However, both methods have limitations. For example, they can hardly identify the precise breakpoints of CNVs, pathogenic variants in non-coding regions, and complex structural variants (SVs), e.g., inversions or interchromosomal insertions. This is particularly relevant since all these types of genetic variation have been described for CCM patients in the recent literature [6,7,8,9]. In the context of analyses for CNVs segregating within CCM families, cost-effectiveness is also an issue.
Besides short-read-based methods, alternative approaches like Linked-Reads by 10x Genomics, Strand-Seq, or Optical Mapping have been successfully used for SV calling. Long-read based sequencing platforms like PacBio or Oxford Nanopore have also emerged as reliable tools for identifying SVs [10]. Long-read sequencing has the potential to overcome some limitations of current short-read sequencing-based molecular genetic diagnostics. It allows bridging and precise identification of SV breakpoints, making it a robust tool in CNV detection [11,12]. In particular, the highly flexible and yet affordable MinION platform from Oxford Nanopore, which enables read lengths of more than 30 kb [13], can be a valuable tool for almost any diagnostic lab. For targeted long-read sequencing, regions of interest can be enriched by using long-range PCR [14], hybridization-based approaches [15,16], Xdrop systems [17], adaptive sampling [18], or CRISPR/Cas9-mediated target selection, which enables amplification-free sequencing of genomic DNA [19] and has already been used successfully to identify SVs in clinically relevant genes like BRCA1 [20].
Depending on the specific question and the available information from previous analyses, either a single- or dual-cut excision approach should be applied for CRISPR/Cas9-mediated nanopore sequencing. In the single-cut approach, a crRNA with a binding site near one end of the target region is used to read into the unknown one. This strategy is used for genome walking approaches or when only one side of the target region is known [21]. However, the coverage often drops toward the end of the target region. The dual-cut excision approach is based on the design of crRNAs at each end of the target region and results in higher coverage [21]. However, both ends of the target region need to be known. A combination of both approaches can be applied to larger regions [22].
In this study, we show that nanopore sequencing in combination with Cas9-mediated target selection can serve as an excellent complement to diagnostic short-read sequencing of the CCM genes. Detection of CNVs at single nucleotide resolution with moderate requirements for hardware and bioinformatics skills enables cost-effective and rapid PCR approaches for subsequent cascade analyses in CCM families. Furthermore, we demonstrate that even complex SVs can be reliably detected with targeted nanopore sequencing.
2. Results
2.1. Cas9-Mediated Nanopore Sequencing Confirmed a 2552 bp Deletion in CCM1
To cover the entire genomic loci of CCM1, CCM2, and CCM3 with long reads, we first designed specific crRNAs to facilitate nanopore sequencing (Table S1). For each gene, the crRNAs were located approximately 15 kb apart from each other to avoid incomplete coverage of the target regions. Enrichment was performed in three separate reactions, allowing parallel sequencing of the three CCM genes in one sequencing run and ensuring that sequencing reads would not be terminated early by another CRISPR/Cas9-induced double-strand break. Whenever possible, crRNAs were designed to facilitate the same sequencing direction in a walking approach.
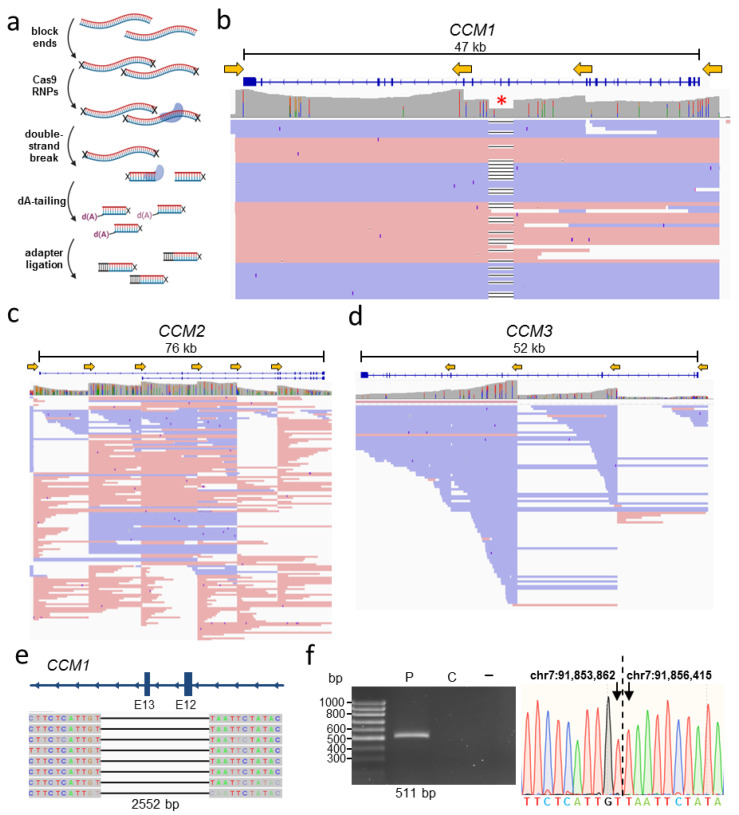
To validate our new crRNA-panel, we sequenced a sample with a known two-exon deletion in CCM1. High-molecular-weight genomic DNA was extracted, and target selection was performed by Cas9-mediated induction of double-strand breaks and ligation of sequencing adapters to cleaved ends following the Oxford nanopore protocol (Figure 1a). Long-read sequencing successfully mapped the CCM1 deletion, which started in intron 11 and ended in intron 13 with a mean sequencing coverage of 89.5× for CCM1 (SD = 19.6; Figure 1b; Table S2). In the same run, sequencing of CCM2 and CCM3 confirmed the absence of other SVs in the sample while achieving a mean sequencing coverage of 71.9× (SD = 25.3) and 31.0× (SD = 18.9), respectively (Figure 1c,d; Table S2). To refine the CNV breakpoints, we used the cuteSV tool and the Sniffles2 structural variant caller, which is implemented in the EPI2ME Labs software (Table S4). Visual inspection of our long-read data in the Integrative Genomics Viewer (IGV) finally enabled the design of deletion-specific primers (Figure 1e). The 2552 bp deletion in CCM1 and its exact breakpoints were validated by PCR and Sanger sequencing (Figure 1f; Table S5).
Figure 1.
Cas9-targeted nanopore sequencing of the CCM genes precisely re-identified a two-exon deletion in CCM1. (a) Schematic representation of the sample preparation protocol used for Cas9-targeted nanopore sequencing. (b) CCM1 sequencing data showed a two-exon deletion spanning from intron 11 to intron 13 (*, red star). CrRNA binding sites and sequencing orientation are symbolized by yellow arrows. The coverage with a peak at 127× is displayed in gray. An excerpt of the generated reads is shown in red and blue. (c,d) CCM2 and CCM3 long-read sequencing data. The highest coverage was 119× and 87× for CCM2 and CCM3, respectively. The generated reads are shown in red and blue. (e) Breakpoints of the 2552 bp deletion in CCM1 were visually inspected in the Integrated Genomics Viewer (IGV). (f) Deletion-specific PCR detected a 511 bp band in the proband sample (P) and its absence in a healthy control (C) and negative control sample (−). Sanger sequencing of the extracted band revealed the exact breakpoints. The genomic location is based on the GRCh37 reference genome. Read data (b–e) were visualized in IGV [23]. The Locus Reference Genomic (LRG) transcripts are shown (b–e).
Because the initial sequencing depth for some areas in CCM2 and CCM3 was relatively low, we revised our CCM crRNA-panel for the following sequencing runs. The final panel combined the walking approach with the dual-cut excision approach, in which two cuts are made, one upstream and one downstream of the target region, to generate optimal coverage for CCM1, CCM2, and CCM3 (Table S1).
2.2. Targeted Nanopore Sequencing Revealed the Exact Size of Large Deletions in Familial CCM Cases
Current detection methods focus on intragenic CNVs in CCM patients. For this reason, the size of variants that are partially extragenic cannot be further characterized. Therefore, we wanted to test whether our long-read sequencing approach could specify variant breakpoints of large deletions spanning gene boundaries since this would allow the development of cost-effective PCR-based assays for familial analyses. We used a targeted approach with crRNAs located upstream of the first deleted exon that had been previously identified by NGS or MLPA analysis.
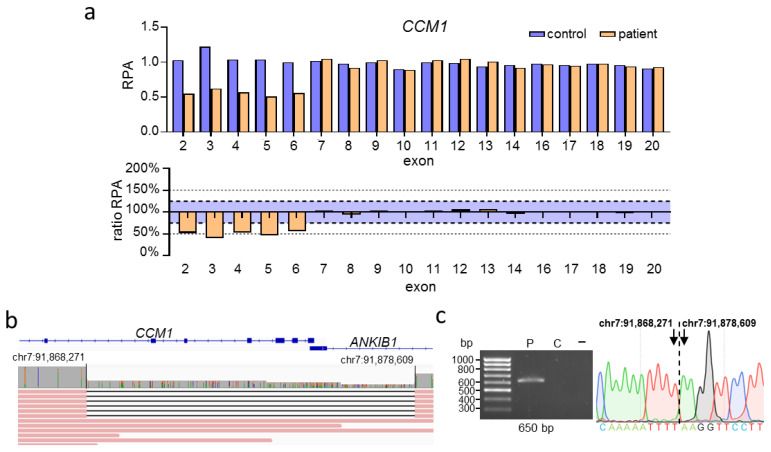
CNV analysis by MLPA for the proband presented in Figure 2 identified a heterozygous deletion of the CCM1 exons 2 to 6 (Figure 2a). No data could be generated for the noncoding exon 1 of CCM1 because the commercially available MLPA kit did not contain specific probes for this region. Over the next three years, nine relatives were tested for the presence of the variant using MLPA. We reanalyzed the DNA of the index proband by nanopore sequencing (Table S3), which revealed the deletion to be larger than anticipated. The deletion actually spans from intron 1 of the adjacent ANKIB1 gene to intron 6 of CCM1 (Figure 2b; Table S4). These results were confirmed by deletion-specific PCR and Sanger sequencing (Figure 2c; Table S5). Long-read sequencing not only enabled the design of a deletion-specific PCR assay, which can be used for further familial analyses, but also demonstrated partial deletion of ANKIB1. So far, however, little is known about the function of ANKIB1.
Figure 2.
Nanopore sequencing of a large deletion in CCM1 identified by MLPA revealed a partial deletion of ANKIB1. (a) MLPA revealed the heterozygous deletion of exons 2–6 of CCM1 (RPA = relative product area). An exon is considered to be deleted or duplicated if the ratio RPA is below 75% or above 125%, respectively (blue area). (b) Subsequent nanopore sequencing showed that the deletion comprises 10,337 base pairs, spanning from CCM1 intron 6 to ANKIB1 intron 1. Read data were inspected in IGV [23]. (c) Deletion-specific PCR detected a 650 bp band in the sample of the index proband (P) and its absence in a healthy control (C) and negative control sample (−). Sanger sequencing of the extracted band revealed the exact deletion breakpoints. The genomic location is based on the GRCh37 reference genome. The Locus Reference Genomic (LRG) transcript (CCM1) and RefSeq transcript (ANKIB1) are shown (b).
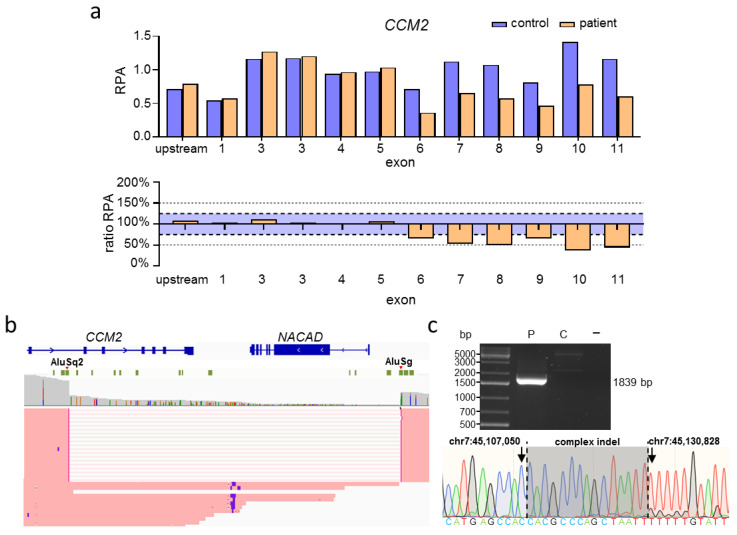
For the proband presented in Figure 3, CNV analysis by MLPA was able to identify a multi-exon deletion in CCM2, spanning from exon 6 to exon 11 (Figure 3a). Nanopore sequencing (Table S3) revealed the true size of the deletion to be 23,777 bp, covering the complete NACAD gene located downstream of CCM2 (Figure 3b). So far, the function of NACAD is unknown. Visual inspection in IGV suggested that this deletion originated from Alu-mediated recombination (Figure 3b; Table S4). Since breakpoints were localized in these highly repetitive regions, sequence alignment was not able to confidently determine the exact distal breakpoint. Nevertheless, we were able to design specific PCR primers to confirm the deletion by PCR and identify the exact breakpoints by Sanger sequencing, which also revealed an additional 15 bp indel variant between the breakpoints (Figure 3c; Table S5).
Figure 3.
Nanopore sequencing of a large CCM2 deletion suggests an Alu-mediated origin. (a) MLPA revealed a heterozygous deletion of exons 6–11 of CCM2 (RPA = relative product area). An exon is considered to be deleted or duplicated if the ratio RPA is below 75% or above 125%, respectively (blue area). (b) Subsequent nanopore sequencing showed that the deletion comprises 23,777 bp, starting in intron 5 of CCM2 and encompassing the neighboring gene NACAD. Deletion breakpoints are located inside of an AluSq2 and an AluSg repeat, respectively (green blocks). Read data were inspected in IGV [23]. (c) Deletion-specific PCR detected a 1839 bp band in the sample of the proband (P) and its absence in a healthy control (C) and negative control sample (−). Sanger sequencing of the extracted band was able to specify the deletion breakpoints and revealed a 15 bp indel variant. The genomic location is based on the GRCh37 reference genome. The Locus Reference Genomic (LRG) transcript (CCM2) and RefSeq transcript (NACAD) are shown (b).
Unfortunately, we were not able to apply our approach to re-analyze CNVs within CCM3 since no appropriate samples were available.
2.3. Targeted Nanopore Sequencing Can Be Used to Identify Complex SVs in CCM
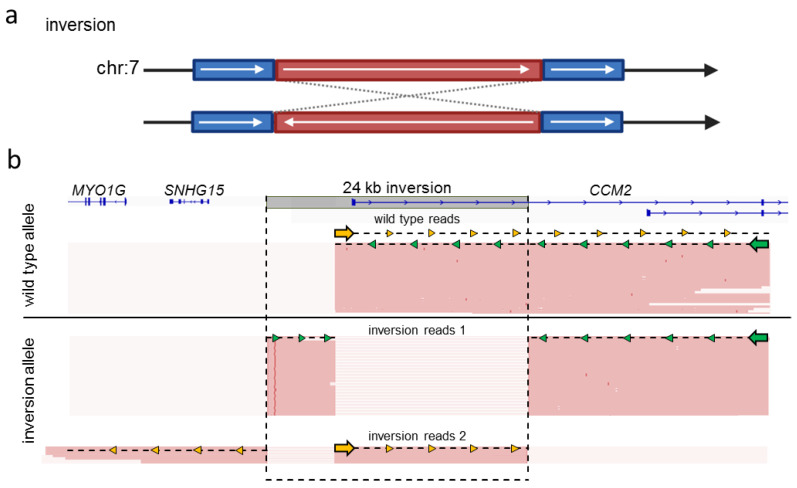
The detection of complex SVs is hard or even impossible with short-read gene panel sequencing. Since the identification of an interchromosomal insertion and an inversion in CCM2 [8,9] suggests that SVs must be considered as a possible cause of familial CCM disease, we wanted to test whether long-read sequencing could accurately detect these variants. Therefore, we reanalyzed a heterozygous 24 kb inversion on chromosome 7 (Figure 4a), which covers the first coding exon of CCM2 [8], with long-read sequencing (Figure 4b; Table S3). We used crRNAs that were part of our CCM crRNA-panel to facilitate a dual-cut excision approach covering roughly half of the coding region of CCM2. The crRNA located upstream of CCM2 was within the variant boundaries of the 24 kb inverted region, whereas the downstream crRNA was located in CCM2 intron 3. The wild-type allele was present in roughly half of all reads. Two different kinds of reads covering the inversion were present. In both cases, mapping orientation flipped after passing one of the inversion breakpoints (Figure 4b). Due to the different types of reads generated by sequencing the inversion allele, we were able to identify both breakpoints of the 24 kb inversion. This variant was discovered previously only using short-read WGS [8]. As the variant is copy number neutral and the breakpoints lie in non-coding regions, detection via targeted panel sequencing, whole-exome sequencing, or MLPA was impossible.
Figure 4.
Nanopore sequencing confidently detected a 24 kb inversion in CCM2. (a) Schematic representation of the inversion in CCM2. (b) A dual-cut approach was used to re-sequence a sample with a known heterozygous 24 kb inversion in CCM2. CrRNAs facilitated sequencing in opposing directions. One binding site was located inside of the inversion (yellow arrow), and the other one was located downstream of the variant (green arrow). Sequencing of the wild-type allele resulted in one type of reads localized between both crRNA cut sides. Two distinct read patterns visualizing the inversion could be observed depending on which crRNA initiated sequencing of the inversion allele (inversion reads 1; inversion reads 2). Read data were inspected in IGV [23]. The Locus Reference Genomic (LRG) transcripts are shown (b).
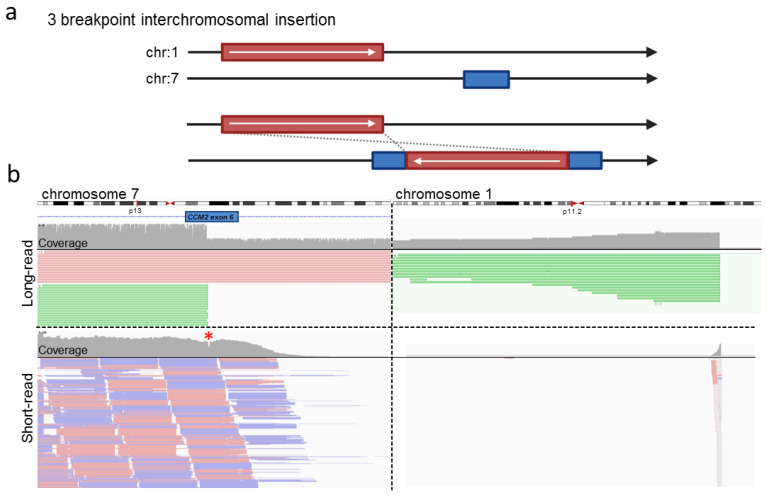
In the next step, we re-analyzed a known heterozygous interchromosomal insertion of chromosome 1 material into the coding region of CCM2 (Figure 5a). This variant which we had previously called from short read gene panel sequencing data with the SureCall software, was an ideal positive control for our approach because it already had been extensively characterized and confirmed by FISH [9]. Long-read sequencing of DNA with this variant (Table S3) generated single reads longer than 20 kb. These reads spanned the breakpoint located in CCM2 exon 6 with subsequent mapping of bases to chromosome 1 (Figure 5b). Due to the long read length, confident mapping of supplementary alignments on 1p11.2 was possible. As there were no reads covering the entire 294 kb insertion in our oxford nanopore data, an additional crRNA with a binding site downstream of the breakpoint in CCM2 would have been required in a diagnostic context to identify the second breakpoint on chromosome 1. However, due to the limited availability of DNA from this positive control, additional sequencing was unfortunately not possible in our current study.
Figure 5.
Nanopore sequencing confidently detected an interchromosomal insertion in CCM2. (a) Schematic representation of the interchromosomal insertion in CCM2. (b) Long-read sequencing data of a previously described heterozygous interchromosomal insertion revealed about 50% of reads covering CCM2 terminate in exon 6 (green). These reads all had a supplementary alignment mapping to chromosome 1. Short-read data showed consistent coverage of CCM2 exon 6. The coverage at the position of the variant breakpoints was reduced (*, red star), and a limited number of reads had a supplementary alignment on chromosome 1. Most reads bridging to chromosome 1 had a mapping quality equal to 0 (hollow reads). Read data were inspected in IGV [23].
Taken together, our data show that even complex SVs can be detected with Cas9-based oxford nanopore sequencing.
2.4. Targeted Nanopore Sequencing on the Flongle Flow Cell Is Also Able to Determine Variant Breakpoints
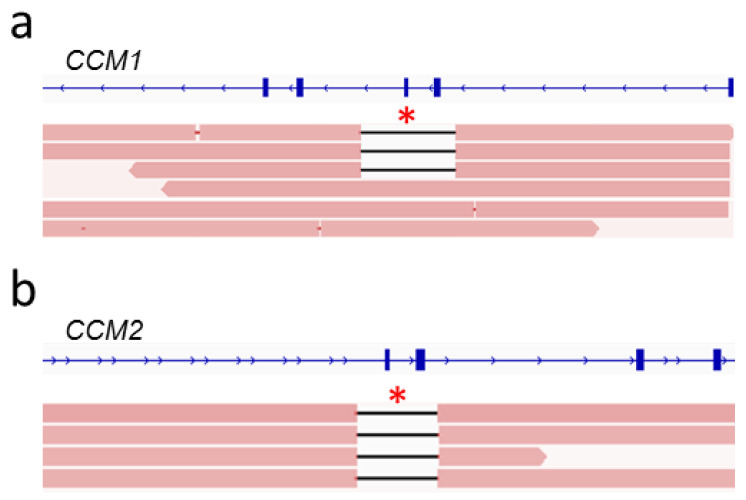
To reduce the sequencing costs, we next tested whether CNV breakpoints could also be refined by Cas9-mediated sequencing on Flongle flow cells. We first used a dual-cut approach with two crRNAs flanking the breakpoints of the intragenic CCM1 deletion described in Figure 1 and a second intragenic deletion in CCM2. Using high molecular weight DNA or medium-sized DNA treated with the Short Read Eliminator Kit, respectively, the deletion of exons 12 and 13 in CCM1 (Figure 6a) and the deletion of exons 4 and 5 in CCM2 (Figure 6b) could be re-identified with Flongle sequencing. However, the extreme bias in the variant allele read frequency of the second sample indicated limited reliability of this approach. We also tested a single-cut approach for the large CCM1 deletion described in Figure 2. Although we were able to detect the deletion, only one read covered the target region (Figure S1). Although a dual-cut approach appears to provide higher coverage, Flongle sequencing is significantly less reliable than Cas9-mediated sequencing on standard MinION flow cells.
Figure 6.
Sequencing on Flongle flow cells detected variant breakpoints in a shallow Cas9-mediated long-read approach. (a,b) Sequencing data from two dual-cut excision approaches sequenced on a Flongle flow cell. (a) Detection of the two-exon deletion (*, red star) of exons 12 and 13 in CCM1 (Figure 1) with a sequencing coverage of 6×. (b) Re-sequencing of a heterozygous two-exon deletion (*, red star) of exons 3 and 4 in CCM2 with a Flongle flow cell yielded a sequencing coverage of 4×. No wild-type reads were generated. Read data were inspected in IGV [23]. The Locus Reference Genomic (LRG) transcripts are shown (a,b).
3. Discussion
In this study, we demonstrate that the implementation of Cas9-mediated long-read sequencing can significantly improve the sensitivity and accuracy of molecular genetic diagnostics for CCM patients when used as a complementary analytical approach. It allows not only the specification of CNV breakpoints but also the detection of complex germline SVs that can be easily missed by targeted NGS approaches.
Large deletions have been reported for all three CCM genes [2,24,25,26,27,28,29,30,31,32]. Their size ranges from a few hundred kilobases to 1.9 megabases [24,30,31]. However, the actual size of CNVs in CCM1, CCM2, or CCM3 is often unknown because the identification methods do not allow accurate mapping of the breakpoints [24,26,31,33]. Indeed, we show here that the deletion of the first six exons of CCM1 was much larger than initially thought and also included parts of the neighboring ANKIB1 gene. Other studies also identified CCM1 gene deletions with a breakpoint in ANKIB1 [30]. Variants involving neighboring genes are also known for CCM2 [4,25,33] and CCM3 [28].
In molecular diagnostics, a variety of wet lab assays can be used to screen for CNVs. The MLPA technique, for example, is widely used to detect large deletions and duplications in many well-known disease genes. The resolution depends on the design of the MLPA probes, and non-coding exons might not be completely covered. Microarray-based high-density comparative genomic hybridization (aCGH) offers high-throughput analyses with a minimum resolution of 500 bp [34]. However, aCGH requires special equipment and is limited in detecting small duplications [35]. Bioinformatic advances have also made it possible to detect CNVs from high-throughput short-read panel sequencing data, allowing parallel detection of SNVs, indels, and CNVs with exon-level resolution [3,36]. Nevertheless, all techniques have disadvantages regarding targeted analyses for a familial CNV. Short-read panel sequencing is rarely used to detect known familial CNVs due to its rather high costs. However, even MLPA analysis can become time-consuming and expensive if parallel analysis of multiple samples is impossible [36]. Especially in large CCM families, several relatives often need to be screened for a familial CNV at different time points [37]. For example, in one family described here (Figure 2), nine members were sequentially tested for the identified CCM1 deletion by MLPA. Long-read sequencing is a promising option in this context. Once the breakpoints of a familial CNV have been established by long-read sequencing, subsequent analyses could be performed with inexpensive allele-specific PCR. For predictive analyses, this can reduce the hands-on time to a few hours while keeping reagent costs very low. Moreover, the MinION platform used in this study has low initial investment costs [38]. The relatively high costs of standard MinION flow cells, however, still represent a limitation of this approach. Although we could also refine the breakpoints of known CNVs with sequencing on the much cheaper Flongle flow cell, this method is not yet reproducible enough for diagnostic use. Nonetheless, since long-read sequencing with a standard MinION flow cell would be performed only once in the index patient, it can still be a cost-effective option when the need for further analyses of family members is foreseeable.
The efficiencies of Cas9-mediated target selection and Oxford Nanopore sequencing reported in the literature vary within a wide range. Depending on the size of the respective target region and the individual experimental approach, on-target coverages of >500× but also <30× have been described [39,40,41,42,43,44,45]. Especially when large genes are to be covered in a Cas9-based approach with long reads, the achievable sequencing depth often becomes an issue [41]. The sequencing depths achieved in our study, which were ≥30× in all but one case (Tables S2 and S3), are within the expected range. While a higher on-target coverage would be desirable for screening approaches, these sequencing depths are more than sufficient for confirmatory analyses of known CNVs.
Breakpoint specification of known CNVs by long-read sequencing not only enables the development of efficient PCR-based assays for familial analyses but can also provide insight into the origin of CNVs. Mechanisms leading to CNV formation often involve the recombination of homologous DNA sequences. Alu-repeats, for example, belong to the short interspersed nuclear elements (SINES) that largely facilitate structural variation in the genome. They are characterized as repetitive regions about 300 bp in size and contribute to almost 11% of the human genome [46]. Because of their high homology, Alu-repeats are responsible for genomic instability. Following a double-strand break, non-allelic homologous recombination mediated by Alu-repeats can lead to deletions, duplications, and complex SVs [46]. Alu-mediated CNVs and SVs have been found in various disorders, like chronic granulomatous disease [47], limb-girdle muscular dystrophy 5 [48], and glaucoma [49]. In CCM, Alu-repeat recombination is also a known disease mechanism. Liquori and colleagues discovered a founder mutation resulting from recombination in CCM2 between an AluSx- and an AluSg-repeat region which led to the deletion of exons 2 to 10 [4]. In our second familial case, breakpoint specification with long-read sequencing also revealed Alu-mediated recombination as a possible cause of the identified deletion (Figure 3). Variant breakpoints were located in an AluSq2- and an AluSg-repeat, respectively.
Beyond accurate characterization of variant breakpoints, the utility of long-read sequencing as a complementary analysis to gene panel sequencing is also demonstrated by its ability to detect more complex SVs that are usually not captured by targeted short-read sequencing approaches [50]. Inversions or translocations, for example, might easily be missed when breakpoints are not located in coding regions. Unfortunately, short-read WGS, which has higher sensitivity for complex SVs, is not yet an option for routine diagnosis of monogenic diseases with well-established risk genes due to high investment costs and bioinformatics requirements. Targeted long-read sequencing, on the other hand, is relatively easy to implement and might confidently close the diagnostic gap for SVs [18]. Consistent with the experience of other groups in resolving complex structural rearrangements, our targeted long-read sequencing assay reliably re-identified an interchromosomal insertion and a copy number-neutral inversion in the CCM2 gene that had previously been detected in short-read approaches and elaborately validated [8,9]. However, it should be emphasized that a diagnostic SV screening approach would require higher bioinformatic skills and the use of more variant callers than an approach to refine the breakpoints of known CNVs. Despite continuous advances in sequencing chemistry and computational compensation [51,52,53], nanopore sequencing is also not yet suitable as a stand-alone diagnostic approach. In particular, the relatively high error rate of nanopore sequencing and the risk of missing whole gene deletions in Cas9-mediated targeted long-read sequencing currently limit its diagnostic power. While short-read panel sequencing remains the most cost-effective method for detecting SNVs, indels, and CNVs as a first-line approach, complementary nanopore sequencing can reduce costs and turnaround time for pathogenic familial CNVs and may also be used as a second-line approach to screen for complex SVs.
Taken together, we demonstrate that the implementation of long-read sequencing can be a great benefit to CCM diagnostics. Our experience also suggests that this approach can very easily be transferred to other diagnostic questions.
4. Materials and Methods
4.1. Design of crRNAs
CrRNAs for CCM1, CCM2, and CCM3 (Table S1) were designed and checked with the crRNA design tool from Integrated DNA Technologies (accessed between 5 May 2022 and 1 July 2022, Integrated DNA Technologies, Coralville, IA, USA). If possible, crRNAs with binding sites in exonic regions were used. If an intronic crRNA location was inevitable, we used crRNAs with binding sites outside of repetitive or poorly characterized regions. It was ensured that crRNA binding sites did not contain SNVs with a minor allele frequency greater than 0.01% listed in gnomAD v2.1.1 (accessed between 5 May 2022 and 1 July 2022).
4.2. Nanopore Sequencing
All study participants gave written informed consent for genetic analyses. High-molecular-weight genomic DNA was isolated from fresh blood or frozen blood samples using the Monarch HMW DNA Extraction Kit (New England BioLabs, Ipswich, MA, USA). The NucleoSpin Blood L Kit (Macherey-Nagel, Düren, Germany) was used to isolate medium-sized DNA fragments that are typically used in routine diagnostics. To eliminate short reads, the Short Read Eliminator Kit (Circulomics, Baltimore, MD, USA) was used. If necessary, additional purification of genomic DNA samples was performed by magnetic bead clean-up with AMPure XP beads (Beckman Coulter, Brea, CA, USA). Target selection was performed either with the Cas9 Sequencing Kit (SQK-CS9109, Oxford Nanopore Technologies, Oxford, United Kingdom) or the Ligation Sequencing Kit (SQK-CS9109, Oxford Nanopore Technologies) in combination with the following additional components: dATP Solution, NEBNext Quick Ligation Module, Quick CIP, Taq DNA Polymerase (New England BioLabs) and IDTE pH 8.0 (Integrated DNA Technologies). For Cas9 cleavage, Alt-R S.p. Cas9 Nuclease V3 (Integrated DNA Technologies), Alt-R CRISPR-Cas9 tracrRNA (Integrated DNA Technologies), and Alt-R CRISPR-Cas9 crRNAs (Integrated DNA Technologies) were used. Briefly, for targeted sequencing of a known variant, up to 5 µg of genomic DNA was used for dephosphorylation. DNA was then cleaved by RNP complexes with crRNAs located directly up- or downstream of the variant. After poly-A-tailing and AMPure XP bead purification, sequencing adapters were ligated, and sequencing was started. Long-read-sequencing was performed on a MinION device (Oxford Nanopore Technologies) equipped with R9.4.1 flow cells (Oxford Nanopore Technologies).
For combined sequencing of CCM1, CCM2, and CCM3, up to 15 µg of genomic DNA were divided and dephosphorylated in three separate tubes. The tubes were treated with RNP complexes originating from one of three crRNA-pools (Table S1). After poly-A-tailing, the three tubes were mixed, and AMPure XP bead purification, adapter ligation, and sequencing were performed as usual. Target selection for Cas9-mediated nanopore sequencing on Flongle flow cells (Oxford Nanopore Technologies) was performed with the Cas9 Sequencing Kit as mentioned above. Loading of the Flongle flow cell was done with the Flongle Flow Cell Priming Kit (EXP-FSE001, Oxford Nanopore Technologies) and according to the “Loading the Flongle flow cell” section of the SQK-LSK109 sequencing protocol.
Live basecalling was performed with Guppy integrated in the MinKNOW software (version 22.03.6, Oxford Nanopore Technologies). FASTQ files were combined with cygwin64 (version 1.7.35), and alignment to the GRCh37 reference genome was carried out with the MinKNOW software (version 22.03.6, Oxford Nanopore Technologies) utilizing the minimap2 aligner. Index files were generated with samtools (version 1.10) for bioinformatics analyses and igvtools [23] for visual inspection. SVs or CNVs were called with cuteSV (version 2.0.2) [54] and the human variation workflow of the EPI2ME Labs software (version 3.1.5, Oxford Nanopore Technologies) which utilizes Sniffels2 [55]. After narrowing down the SV or CNV breakpoints with the bioinformatic tools, long-read sequencing data were further visually inspected with IGV [23] (version 2.13.0, Broad Institute, Cambridge, MA, USA) to design variant-specific PCR assays.
4.3. Multiplex Ligation-Dependent Probe Amplification, PCR, and Sanger Sequencing
For copy number variation analysis by MLPA, the kits P130-A3 and P131-B1 containing probes for CCM1, CCM2, and CCM3 were used according to the manufacturer’s instructions (MRC-Holland, Amsterdam, The Netherlands). Fragment analysis was performed on a SeqStudio Genetic Analyzer (Applied Biosystems, Waltham, MA, USA) or an ABI 310 Genetic Analyzer (Applied Biosystems), and the data were analyzed with the SeqPilot software (version 5.1.0, JSI Medical Systems, Ettenheim, Germany). Visualization was performed with the GraphPad Prism software (version 8.0.1, GraphPad Software, Inc, San Diego, CA, USA).
For breakpoint confirmation, variant-specific primers (Integrated DNA Technologies) were designed, and PCR reactions were carried out with the Taq DNA-Polymerase (Thermo Fisher Scientific, Waltham, MA, USA). To amplify GC-rich regions, the OneTaq DNA Polymerase (New England BioLabs) or the GC-RICH PCR-System (Roche, Basel, Switzerland) was used with GC-Rich Buffer. PCR products were evaluated on a 1.5% agarose gel imaged on a ChemiDoc XRS+ system (BioRad, Hercules, CA, USA). Bands were cut from the gel and extracted with the Zymoclean Gel DNA Recovery Kit (Zymo Research, Irvine, CA, USA). The sequencing reaction was done with the BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific). After cleanup, the samples were sequenced on a SeqStudio Genetic Analyzer. Sequencing data were evaluated with SnapGene Viewer (Dotmaticsm, Boston, MA, USA).
4.4. Illumina Sequencing
Hybridization capture-based target enrichment of genomic DNA samples for CCM1, CCM2, and CCM3 was performed using an Agilent SureSelectQXT custom enrichment kit (Panel ID: 3152261, Agilent Technologies, Santa Clara, CA, USA). Illumina sequencing was performed on an Illumina MiSeq platform with 2 × 150 cycles (Illumina, San Diego, CA, USA). Basecalling and alignment to the GRCh37 reference genome were performed with the MiSeq Reporter Software (version 2.6.2, Illumina). Data were inspected with IGV [23] (version 2.13.0).
Acknowledgments
Sina Ramcke is thanked for her excellent technical assistance. Figure 1a, Figure 4a, and Figure 5a were created with biorender.com (accessed on 28 October 2022).
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232415639/s1.
Author Contributions
Conceptualization, M.R., C.D.M., and U.F.; formal analysis, D.S., L.B. and R.A.P.; investigation, D.S., L.B., J.L.F. and O.J.S.; genetic counseling, M.R., U.F., and S.H.; writing—original draft preparation, D.S., R.A.P. and L.B.; writing—review and editing, M.R. and U.F.; visualization, D.S. and L.B.; supervision, U.F., and M.R.; funding acquisition, C.D.M., and M.R. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Informed consent was obtained from all subjects involved in the study. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was approved by the Ethics Committee of the University Medicine Greifswald (BB 047/14a—25 January 2018).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The raw sequencing data are not publicly available because they contain additional genetic information that could compromise the privacy of research participants.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation; No. RA2876/2-2) and by the Research Network Molecular Medicine of the University Medicine Greifswald (FOVB-2022-14). MR was supported by a clinician scientist scholarship from the Gerhard Domagk program of the University of Medicine Greifswald.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Spiegler S., Rath M., Paperlein C., Felbor U. Cerebral Cavernous Malformations: An Update on Prevalence, Molecular Genetic Analyses, and Genetic Counselling. Mol. Syndromol. 2018;9:60–69. doi: 10.1159/000486292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Felbor U., Gaetzner S., Verlaan D.J., Vijzelaar R., Rouleau G.A., Siegel A.M. Large germline deletions and duplication in isolated cerebral cavernous malformation patients. Neurogenetics. 2007;8:149–153. doi: 10.1007/s10048-006-0076-7. [DOI] [PubMed] [Google Scholar]
- 3.Much C.D., Schwefel K., Skowronek D., Shoubash L., von Podewils F., Elbracht M., Spiegler S., Kurth I., Flöel A., Schroeder H.W.S., et al. Novel Pathogenic Variants in a Cassette Exon of CCM2 in Patients With Cerebral Cavernous Malformations. Front. Neurol. 2019;10:1219. doi: 10.3389/fneur.2019.01219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liquori C.L., Berg M.J., Squitieri F., Leedom T.P., Ptacek L., Johnson E.W., Marchuk D.A. Deletions in CCM2 Are a Common Cause of Cerebral Cavernous Malformations. Am. J. Hum. Genet. 2007;80:69–75. doi: 10.1086/510439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riant F., Cecillon M., Saugier-Veber P., Tournier-Lasserve E. CCM molecular screening in a diagnosis context: Novel unclassified variants leading to abnormal splicing and importance of large deletions. Neurogenetics. 2013;14:133–141. doi: 10.1007/s10048-013-0362-0. [DOI] [PubMed] [Google Scholar]
- 6.Riant F., Odent S., Cecillon M., Pasquier L., de Baracé C., Carney M.P., Tournier-Lasserve E. Deep intronic KRIT1 mutation in a family with clinically silent multiple cerebral cavernous malformations. Clin. Genet. 2014;86:585–588. doi: 10.1111/cge.12322. [DOI] [PubMed] [Google Scholar]
- 7.Pilz R.A., Skowronek D., Hamed M., Weise A., Mangold E., Radbruch A., Pietsch T., Felbor U., Rath M. Using CRISPR/Cas9 genome editing in human iPSCs for deciphering the pathogenicity of a novel CCM1 transcription start site deletion. Front. Mol. Biosci. 2022;9:953048. doi: 10.3389/fmolb.2022.953048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spiegler S., Rath M., Hoffjan S., Dammann P., Sure U., Pagenstecher A., Strom T., Felbor U. First large genomic inversion in familial cerebral cavernous malformation identified by whole genome sequencing. Neurogenetics. 2018;19:55–59. doi: 10.1007/s10048-017-0531-7. [DOI] [PubMed] [Google Scholar]
- 9.Pilz R.A., Schwefel K., Weise A., Liehr T., Demmer P., Spuler A., Spiegler S., Gilberg E., Hübner C.A., Felbor U., et al. First interchromosomal insertion in a patient with cerebral and spinal cavernous malformations. Sci. Rep. 2020;10:6306. doi: 10.1038/s41598-020-63337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahmoud M., Gobet N., Cruz-Dávalos D.I., Mounier N., Dessimoz C., Sedlazeck F.J. Structural variant calling: The long and the short of it. Genome Biol. 2019;20:246. doi: 10.1186/s13059-019-1828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mantere T., Kersten S., Hoischen A. Long-Read Sequencing Emerging in Medical Genetics. Front. Genet. 2019;10:426. doi: 10.3389/fgene.2019.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonoda K., Ishihara H., Sakazaki H., Suzuki T., Horie M., Ohno S. Long-Read Sequence Confirmed a Large Deletion Including MYH6 and MYH7 in an Infant of Atrial Septal Defect and Atrial Arrhythmias. Circ. Genom. Precis. Med. 2021;14:e003223. doi: 10.1161/CIRCGEN.120.003223. [DOI] [PubMed] [Google Scholar]
- 13.Mohammadi M.M., Bavi O. DNA sequencing: An overview of solid-state and biological nanopore-based methods. Biophys Rev. 2022;14:99–110. doi: 10.1007/s12551-021-00857-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson C.M., Holliday D.L., Crinnion L.A., Bonthron D.T. Long-read nanopore DNA sequencing can resolve complex intragenic duplication/deletion variants, providing information to enable preimplantation genetic diagnosis. Prenat. Diagn. 2022;42:226–232. doi: 10.1002/pd.6089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steiert T.A., Fuß J., Juzenas S., Wittig M., Hoeppner M.P., Vollstedt M., Varkalaite G., ElAbd H., Brockmann C., Görg S., et al. High-throughput method for the hybridisation-based targeted enrichment of long genomic fragments for PacBio third-generation sequencing. NAR Genom. Bioinform. 2022;4:lqac051. doi: 10.1093/nargab/lqac051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M., Beck C.R., English A.C., Meng Q., Buhay C., Han Y., Doddapaneni H.V., Yu F., Boerwinkle E., Lupski J.R., et al. PacBio-LITS: A large-insert targeted sequencing method for characterization of human disease-associated chromosomal structural variations. BMC Genomics. 2015;16:214. doi: 10.1186/s12864-015-1370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madsen E.B., Höijer I., Kvist T., Ameur A., Mikkelsen M.J. Xdrop: Targeted sequencing of long DNA molecules from low input samples using droplet sorting. Hum. Mutat. 2020;41:1671–1679. doi: 10.1002/humu.24063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller D.E., Sulovari A., Wang T., Loucks H., Hoekzema K., Munson K.M., Lewis A.P., Fuerte E.P.A., Paschal C.R., Walsh T., et al. Targeted long-read sequencing identifies missing disease-causing variation. Am. J. Hum. Genet. 2021;108:1436–1449. doi: 10.1016/j.ajhg.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilpatrick T., Lee I., Graham J.E., Raimondeau E., Bowen R., Heron A., Downs B., Sukumar S., Sedlazeck F.J., Timp W. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat. Biotechnol. 2020;38:433–438. doi: 10.1038/s41587-020-0407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walsh T., Casadei S., Munson K.M., Eng M., Mandell J.B., Gulsuner S., King M.C. CRISPR-Cas9/long-read sequencing approach to identify cryptic mutations in BRCA1 and other tumour suppressor genes. J. Med. Genet. 2021;58:850–852. doi: 10.1136/jmedgenet-2020-107320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.López-Girona E., Davy M.W., Albert N.W., Hilario E., Smart M.E.M., Kirk C., Thomson S.J., Chagné D. CRISPR-Cas9 enrichment and long read sequencing for fine mapping in plants. Plant Methods. 2020;16:121. doi: 10.1186/s13007-020-00661-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fiol A., Jurado-Ruiz F., López-Girona E., Aranzana M.J. An efficient CRISPR-Cas9 enrichment sequencing strategy for characterizing complex and highly duplicated genomic regions. A case study in the Prunus salicina LG3-MYB10 genes cluster. Plant Methods. 2022;18:105. doi: 10.1186/s13007-022-00937-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergametti F., Denier C., Labauge P., Arnoult M., Boetto S., Clanet M., Coubes P., Echenne B., Ibrahim R., Irthum B., et al. Mutations within the Programmed Cell Death 10 Gene Cause Cerebral Cavernous Malformations. Am. J. Hum. Genet. 2005;76:42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liquori C.L., Penco S., Gault J., Leedom T.P., Tassi L., Esposito T., Awad I.A., Frati L., Johnson E.W., Squitieri F., et al. Different spectra of genomic deletions within the CCM genes between Italian and American CCM patient cohorts. Neurogenetics. 2008;9:25–31. doi: 10.1007/s10048-007-0109-x. [DOI] [PubMed] [Google Scholar]
- 26.Choe C.U., Riant F., Gerloff C., Tournier-Lasserve E., Orth M. Multiple cerebral cavernous malformations and a novel CCM3 germline deletion in a German family. J. Neurol. 2010;257:2097–2098. doi: 10.1007/s00415-010-5648-7. [DOI] [PubMed] [Google Scholar]
- 27.Cigoli M.S., Avemaria F., De Benedetti S., Gesu G.P., Accorsi L.G., Parmigiani S., Corona M.F., Capra V., Mosca A., Giovannini S., et al. PDCD10 Gene Mutations in Multiple Cerebral Cavernous Malformations. PLoS ONE. 2014;9:e110438. doi: 10.1371/journal.pone.0110438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nardella G., Visci G., Guarnieri V., Castellana S., Biagini T., Bisceglia L., Palumbo O., Trivisano M., Vaira C., Scerrati M., et al. A single-center study on 140 patients with cerebral cavernous malformations: 28 new pathogenic variants and functional characterization of a PDCD10 large deletion. Hum. Mutat. 2018;39:1885–1900. doi: 10.1002/humu.23629. [DOI] [PubMed] [Google Scholar]
- 29.Fusco C., Copetti M., Mazza T., Amoruso L., Mastroianno S., Nardella G., Guarnieri V., Micale L., D′Agruma L., Castori M. Molecular diagnostic workflow, clinical interpretation of sequence variants, and data repository procedures in 140 individuals with familial cerebral cavernous malformations. Hum. Mutat. 2019;40:e24–e36. doi: 10.1002/humu.23851. [DOI] [PubMed] [Google Scholar]
- 30.Muscarella L.A., Guarnieri V., Coco M., Belli S., Parrella P., Pulcrano G., Catapano D., D′Angelo V.A., Zelante L., D′Agruma L. Small Deletion at the 7q21.2 Locus in a CCM Family Detected by Real-Time Quantitative PCR. J. Biomed. Biotechnol. 2010;2010:854737. doi: 10.1155/2010/854737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benedetti V., Canzoneri R., Perrelli A., Arduino C., Zonta A., Brusco A., Retta S.F. Next-Generation Sequencing Advances the Genetic Diagnosis of Cerebral Cavernous Malformation (CCM) Antioxidants. 2022;11:1294. doi: 10.3390/antiox11071294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mondéjar R., Solano F., Rubio R., Delgado M., Pérez-Sempere Á., González-Meneses A., Vendrell T., Izquierdo G., Martinez-Mir A., Lucas M. Mutation Prevalence of Cerebral Cavernous Malformation Genes in Spanish Patients. PLoS ONE. 2014;9:e86286. doi: 10.1371/journal.pone.0086286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denier C., Goutagny S., Labauge P., Krivosic V., Arnoult M., Cousin A., Benabid A.L., Comoy J., Frerebeau P., Gilbert B., et al. Mutations within the MGC4607 gene cause cerebral cavernous malformations. Am. J. Hum. Genet. 2004;74:326–337. doi: 10.1086/381718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pös O., Radvanszky J., Styk J., Pös Z., Buglyó G., Kajsik M., Budis J., Nagy B., Szemes T. Copy Number Variation: Methods and Clinical Applications. Appl. Sci. 2021;11:819. doi: 10.3390/app11020819. [DOI] [Google Scholar]
- 35.Coughlin C.R., 2nd, Scharer G.H., Shaikh T.H. Clinical impact of copy number variation analysis using high-resolution microarray technologies: Advantages, limitations and concerns. Genome Med. 2012;4:80. doi: 10.1186/gm381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh A.K., Olsen M.F., Lavik L.A.S., Vold T., Drabløs F., Sjursen W. Detecting copy number variation in next generation sequencing data from diagnostic gene panels. BMC Med. Genomics. 2021;14:214. doi: 10.1186/s12920-021-01059-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaetzner S., Stahl S., Sürücü O., Schaafhausen A., Halliger-Keller B., Bertalanffy H., Sure U., Felbor U. CCM1 gene deletion identified by MLPA in cerebral cavernous malformation. Neurosurg. Rev. 2007;30:155–159; discussion 159–160. doi: 10.1007/s10143-006-0057-1. [DOI] [PubMed] [Google Scholar]
- 38.Brinkmann A., Ulm S.L., Uddin S., Förster S., Seifert D., Oehme R., Corty M., Schaade L., Michel J., Nitsche A. AmpliCoV: Rapid Whole-Genome Sequencing Using Multiplex PCR Amplification and Real-Time Oxford Nanopore MinION Sequencing Enables Rapid Variant Identification of SARS-CoV-2. Front. Microbiol. 2021;12:651151. doi: 10.3389/fmicb.2021.651151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alfano M., De Antoni L., Centofanti F., Visconti V.V., Maestri S., Degli Esposti C., Massa R., D′Apice M.R., Novelli G., Delledonne M., et al. Characterization of full-length CNBP expanded alleles in myotonic dystrophy type 2 patients by Cas9-mediated enrichment and nanopore sequencing. elife. 2022;11:e80229. doi: 10.7554/eLife.80229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giesselmann P., Brändl B., Raimondeau E., Bowen R., Rohrandt C., Tandon R., Kretzmer H., Assum G., Galonska C., Siebert R., et al. Analysis of short tandem repeat expansions and their methylation state with nanopore sequencing. Nat. Biotechnol. 2019;37:1478–1481. doi: 10.1038/s41587-019-0293-x. [DOI] [PubMed] [Google Scholar]
- 41.Iyer S.V., Kramer M., Goodwin S., McCombie W.R. ACME: An Affinity-based Cas9 Mediated Enrichment method for targeted nanopore sequencing. bioRxiv. 2022:preprint. [Google Scholar]
- 42.Mizuguchi T., Toyota T., Miyatake S., Mitsuhashi S., Doi H., Kudo Y., Kishida H., Hayashi N., Tsuburaya R.S., Kinoshita M., et al. Complete sequencing of expanded SAMD12 repeats by long-read sequencing and Cas9-mediated enrichment. Brain. 2021;144:1103–1117. doi: 10.1093/brain/awab021. [DOI] [PubMed] [Google Scholar]
- 43.Sone J., Mitsuhashi S., Fujita A., Mizuguchi T., Hamanaka K., Mori K., Koike H., Hashiguchi A., Takashima H., Sugiyama H., et al. Long-read sequencing identifies GGC repeat expansions in NOTCH2NLC associated with neuronal intranuclear inclusion disease. Nat. Genet. 2019;51:1215–1221. doi: 10.1038/s41588-019-0459-y. [DOI] [PubMed] [Google Scholar]
- 44.Stangl C., de Blank S., Renkens I., Westera L., Verbeek T., Valle-Inclan J.E., González R.C., Henssen A.G., van Roosmalen M.J., Stam R.W., et al. Partner independent fusion gene detection by multiplexed CRISPR-Cas9 enrichment and long read nanopore sequencing. Nat. Commun. 2020;11:2861. doi: 10.1038/s41467-020-16641-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace A.D., Sasani T.A., Swanier J., Gates B.L., Greenland J., Pedersen B.S., Varley K.E., Quinlan A.R. CaBagE: A Cas9-based Background Elimination strategy for targeted, long-read DNA sequencing. PLoS ONE. 2021;16:e0241253. doi: 10.1371/journal.pone.0241253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deininger P. Alu elements: Know the SINEs. Genome Biol. 2011;12:236. doi: 10.1186/gb-2011-12-12-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gentsch M., Kaczmarczyk A., van Leeuwen K., de Boer M., Kaus-Drobek M., Dagher M.C., Kaiser P., Arkwright P.D., Gahr M., Rösen-Wolff A., et al. Alu-repeat-induced deletions within the NCF2 gene causing p67-phox-deficient chronic granulomatous disease (CGD) Hum. Mutat. 2010;31:151–158. doi: 10.1002/humu.21156. [DOI] [PubMed] [Google Scholar]
- 48.Pluta N., Hoffjan S., Zimmer F., Köhler C., Lücke T., Mohr J., Vorgerd M., Nguyen H.H.P., Atlan D., Wolf B., et al. Homozygous Inversion on Chromosome 13 Involving SGCG Detected by Short Read Whole Genome Sequencing in a Patient Suffering from Limb-Girdle Muscular Dystrophy. Genes. 2022;13:1752. doi: 10.3390/genes13101752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schilter K.F., Reis L.M., Sorokina E.A., Semina E.V. Identification of an Alu-repeat-mediated deletion of OPTN upstream region in a patient with a complex ocular phenotype. Mol. Genet. Genomic. Med. 2015;3:490–499. doi: 10.1002/mgg3.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stephens Z., Wang C., Iyer R.K., Kocher J.P. Detection and visualization of complex structural variants from long reads. BMC Bioinformatics. 2018;19:508. doi: 10.1186/s12859-018-2539-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rang F.J., Kloosterman W.P., de Ridder J. From squiggle to basepair: Computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018;19:90. doi: 10.1186/s13059-018-1462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delahaye C., Nicolas J. Sequencing DNA with nanopores: Troubles and biases. PLoS ONE. 2021;16:e0257521. doi: 10.1371/journal.pone.0257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Z., Mai Y., Liu D., He W., Lin X., Xu C., Zhang L., Meng X., Mafofo J., Zaher W.A., et al. Fast-bonito: A faster deep learning based basecaller for nanopore sequencing. Artif. Intell. Life Sci. 2021;1:100011. doi: 10.1016/j.ailsci.2021.100011. [DOI] [Google Scholar]
- 54.Jiang T., Liu Y., Jiang Y., Li J., Gao Y., Cui Z., Liu Y., Liu B., Wang Y. Long-read-based human genomic structural variation detection with cuteSV. Genome Biol. 2020;21:189. doi: 10.1186/s13059-020-02107-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ewels P.A., Peltzer A., Fillinger S., Patel H., Alneberg J., Wilm A., Garcia M.U., Di Tommaso P., Nahnsen S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020;38:276–278. doi: 10.1038/s41587-020-0439-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The raw sequencing data are not publicly available because they contain additional genetic information that could compromise the privacy of research participants.