Abstract
Experimental findings for SARS-CoV-2 related to the glycan biochemistry of coronaviruses indicate that attachments from spike protein to glycoconjugates on the surfaces of red blood cells (RBCs), other blood cells and endothelial cells are key to the infectivity and morbidity of COVID-19. To provide further insight into these glycan attachments and their potential clinical relevance, the classic hemagglutination (HA) assay was applied using spike protein from the Wuhan, Alpha, Delta and Omicron B.1.1.529 lineages of SARS-CoV-2 mixed with human RBCs. The electrostatic potential of the central region of spike protein from these four lineages was studied through molecular modeling simulations. Inhibition of spike protein-induced HA was tested using the macrocyclic lactone ivermectin (IVM), which is indicated to bind strongly to SARS-CoV-2 spike protein glycan sites. The results of these experiments were, first, that spike protein from these four lineages of SARS-CoV-2 induced HA. Omicron induced HA at a significantly lower threshold concentration of spike protein than the three prior lineages and was much more electropositive on its central spike protein region. IVM blocked HA when added to RBCs prior to spike protein and reversed HA when added afterward. These results validate and extend prior findings on the role of glycan bindings of viral spike protein in COVID-19. They furthermore suggest therapeutic options using competitive glycan-binding agents such as IVM and may help elucidate rare serious adverse effects (AEs) associated with COVID-19 mRNA vaccines, which use spike protein as the generated antigen.
Keywords: SARS-CoV-2, COVID-19, spike protein, hemagglutination, sialic acid, CD147, electrostatic charge, glycophorin A
1. Introduction
Key to the infectivity and morbidity of SARS-CoV-2 are glycans that protrude tangentially from 22 N-linked glycosylation sites on each monomer of its spike protein [1,2,3,4,5]. These N-glycans, several of which are capped with terminal sialic acid (SA) moieties, sweep back and forth across spike protein like windshield wipers, partially shielding it from antibody binding [1,2,6,7]. For SARS-CoV-2 and several other coronaviruses, these N-glycans serve as appendages for the virus to make its initial attachments to glycoconjugates on the host cell surface [1,8,9,10,11,12,13]. These glycoconjugates are abundantly distributed on the surfaces of host cells such as red blood cells (RBCs) [14,15], platelets [16] and endothelial cells [17] and confer an associated negative electrostatic potential, additionally facilitating attachments by positively charged SARS-CoV-2 spike protein prior to viral fusion to ACE2 for replication [8,18,19,20]. For endothelial cells of blood vessel linings, for example, the disparity between 28,000 SA-tipped CD147 receptors and 175 ACE2 receptors per cell [21] provides a supporting indication of the role of glycans in widespread endothelial damage reported in COVID-19 patients [22,23].
The RBC has an especially dense surface distribution of SA, 35 million SA molecules per cell, arrayed on its sialoglycoprotein coating mainly as terminal residues of glycophorin A (GPA) [14,15]. SA in its predominant human form, Neu5Ac, is the most common terminal residue of GPA on human RBCs, with the other terminal monosaccharides of GPA matching those on SARS-CoV-2 spike N-glycans [2,5]. Through attachments to viruses via this GPA surface coating, RBCs and other blood cells can serve a host defense role [14,24,25]; however, these RBC clumps can be vascularly obstructive to the host’s detriment. Aggregates of RBCs have indeed been found in the blood of most [26,27] or a third [28] of COVID-19 patients in three clinical studies. In a study that examined the blood of hospitalized COVID-19 patients using immunofluorescence analysis, SARS-CoV-2 spike protein punctae were found on 41% of their RBCs [29]. In vitro, SARS-CoV-2 spike protein and pseudovirus attached to a nanoparticle array bearing SA derivatives [30]. Microarray detection techniques typically fail to detect these spike protein attachments to either SA [31] or CD147 [32], since they are formed through nanoscale multivalent bindings [1].
This hemagglutinating property of SARS-CoV-2 has important clinical consequences. First, with trillions of RBCs each circulating through narrow pulmonary capillaries about once per minute, even small, dynamically aggregating and disaggregating RBC clumps (as can form even in the absence of pathogens [33,34]) can impede RBC oxygenation. Peripheral ischemia, endothelial damage and vascular occlusion are indeed frequently observed in serious cases of COVID-19, as reviewed [1,23]. In COVID-19 patients, damaged endothelium of pulmonary capillaries is often observed adjoining relatively intact alveoli [35,36], while hypoxemia is manifested despite normal breathing mechanics [22,35,37,38,39]. These morbidities of COVID-19 parallel those of severe malaria, in which clumping of parasite-infected RBCs to other RBCs via SA terminal residues and endothelial cyto-adhesion also often result in fatal outcomes [1].
Although the blood cell types and processes entailed in clumping, clotting and vascular obstruction of COVID-19 are wide-ranging, virally induced hemagglutination (HA) is a central event that is amenable to in vitro study. The classic HA assay was used here to study this, using cell culture supernatants and SARS-CoV-2 trimeric spike protein. Indeed, SARS-CoV-2 spike protein mixed with human whole blood caused RBC aggregation [40], while spike protein from two other coronavirus strains also induced HA [41,42]. The HA assay can be applied further to study the HA-inhibitory effects of agents that bind to sites on spike protein, potentially shielding them from attachments to host cells. Several in silico studies [1] have found that the macrocyclic lactone ivermectin (IVM) binds with high affinity to subdomains on SARS-CoV-2 spike protein, including several glycosylated binding sites [43]. IVM achieved Nobel-prize-honored distinction for success against global parasitic scourges [44] but is of disputed efficacy in the treatment of COVID-19, as indicated, for example, by the disparity in the conclusions of this editorial [45] and its key cited meta-analysis [46].
The goals of this study were to determine whether principles of glycobiology as established for coronaviruses and in particular for SARS-CoV-2, can be validated using the classic HA assay, to test whether HA inhibition is achieved by an agent indicated to competitively bind to those glycans and to determine the comparative hemagglutinating potencies of the Wuhan virus and its Alpha, Delta and Omicron B.1.1.529 variants. The clinical relevance of testing HA induced by SARS-CoV-2 using spike protein rather than whole virus and the utility of additionally testing for HA inhibition via competitive glycan binding using IVM was suggested in an earlier study [1]. The differences in the electrostatic potential of the spike protein of these four variants were studied using molecular modeling and were related to their HA-inducing potencies as experimentally observed.
2. Results
2.1. Tests for Hemagglutination (HA) and for Its Inhibition and Reversal by IVM
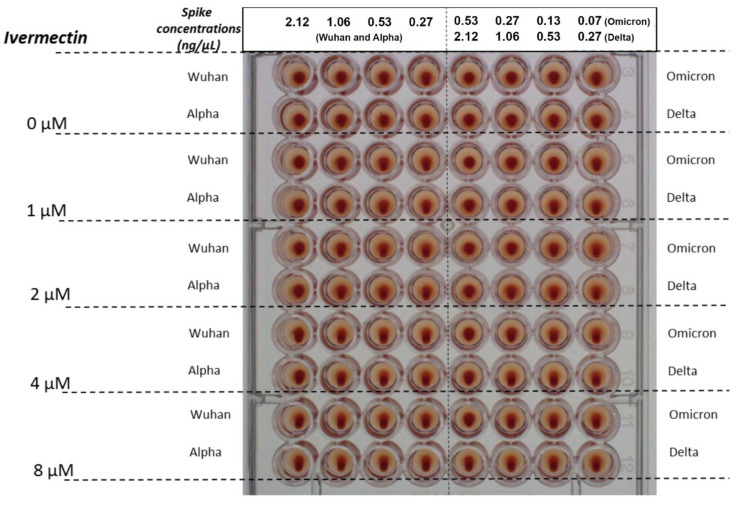
In the HA experiment, for the Wuhan, Alpha and Delta lineages, we observed HA at a spike protein concentration of 1.06 ng/µL and above, but not below. For the Omicron spike protein, we observed HA at a minimum concentration of 0.13 ng/µL and above, but not below (Figure 1 and Figure 2, Table 1).
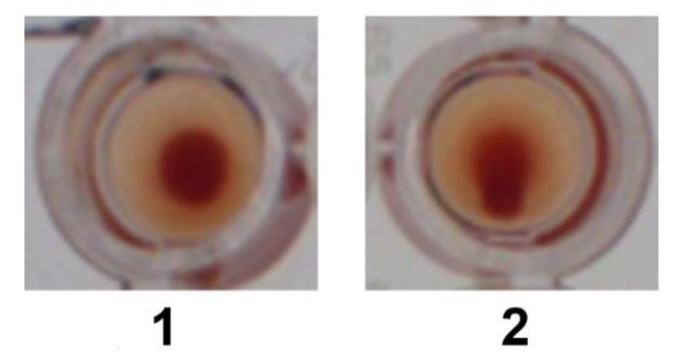
Figure 1.
Sample images of wells in which HA occurred (1; no teardrop visible) and HA did not occur (2; teardrop visible).
Figure 2.
HA as induced by spike protein concentrations of 0.27, 0.53, 1.06 and 2.12 ng/µL for the Wuhan, Alpha and Delta strains of SARS-CoV-2, and at spike protein concentrations of 0.07, 0.13, 0.27 and 0.53 ng/µL for Omicron. Effects on reversal of HA are shown for IVM at concentrations of 1, 2, 4 and 8 µM added 30 min after RBCs and spike protein. Similar results were obtained for inhibition of HA by IVM (not pictured), with differences described in Section 2.1 and summarized in Table 1.
Table 1.
Minimum concentrations of recombinant spike protein (ng/µL) needed to induce HA when added to RBC solution, and concentrations of IVM (µM) needed to inhibit or reverse this induced HA. Tests to determine these values were each done in triplicate.
| Variant | Minimum Spike Protein Concentration to Induce HA (ng/µL) | Highest Spike Protein Concentration Tested for HA (ng/µL) | Minimum IVM Concentration to Inhibit HA at Highest Spike Concentration (µM) | Minimum IVM Concentration to Release HA at Highest Spike Concentration (µM) |
|---|---|---|---|---|
| Wuhan | 1.06 | 2.12 | 1 | 1 |
| Alpha | 1.06 | 2.12 | 1 | 1 |
| Delta | 1.06 | 2.12 | 1 | 2 |
| Omicron | 0.13 | 0.53 | 1 | 2 |
In the HA inhibition experiment, IVM added to 2.5% RBC solution to attain a concentration of 1 µM at 30 min prior to spike protein partially inhibited HA, with HA observed at a spike protein concentration of 2.12 ng/µL for the Wuhan, Alpha and Delta, and of 0.27 ng/µL for the Omicron viral lineages. With IVM at 2 µM, complete inhibition of HA is observed for the Wuhan, Alpha and Delta lineages. For Omicron, IVM at 4 µM is needed to totally block HA.
In the HA reversal experiment, concentrations of IVM needed to reverse HA at the highest concentration of spike tested were 1 µM and 2 µM for the Wuhan/Alpha and Delta/Omicron viral lineages respectively.
HA was not observed with cell culture supernatants of any SARS-CoV-2 strain.
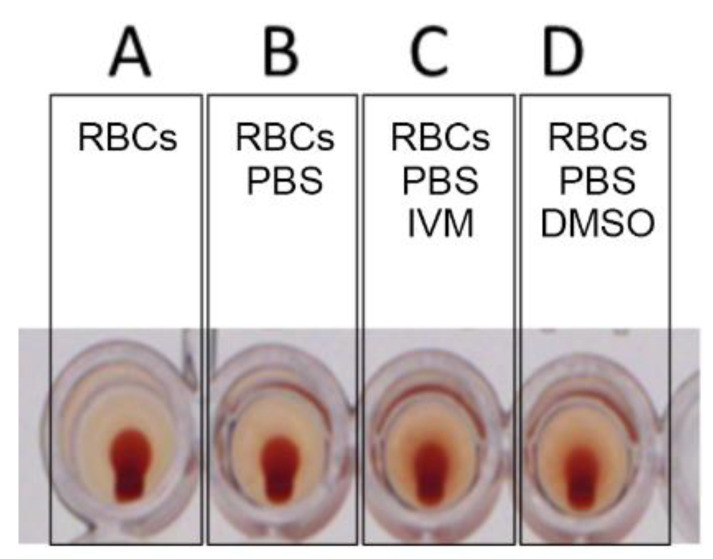
In the control experiments, RBCs alone did not exhibit HA. RBCs mixed with phosphate-buffered saline (PBS) likewise did not exhibit HA. The addition of IVM to 50 µL of 2.5% RBCs to attain a concentration of 8 µM did not cause hemolysis or induce HA. A solution of 2.5% DMSO and 97.5% water, the solvent for IVM, did not block or reverse HA (Figure 2 and Figure 3).
Figure 3.
Controls: (A) RBCs alone, (B) RBCs with PBS, (C) RBCs with PBS and IVM and (D) RBCs with PBS and DMSO. No HA is observed.
2.2. Western Blot and Quantification Analysis
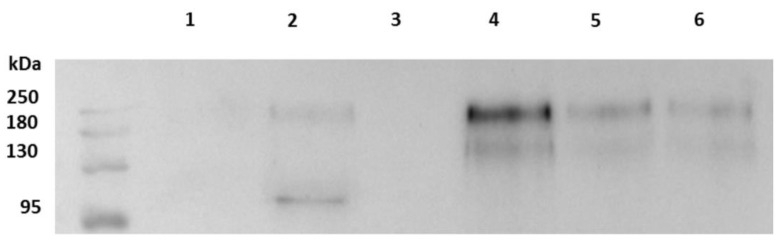
The results for quantification analysis by Western blot of spike protein of the Wuhan SARS-CoV-2 strain in cell culture supernatants are presented in Figure 4. This shows that its spike concentration is below the concentration thresholds for the induction of agglutination. Indeed, for the supernatant of the Wuhan strain in cell culture at 48 h post viral infection, the concentration of N-glycosylated spike protein is approximately 0.7 ng/µL, which dilutes to half that concentration when added to wells, approximately three times lower than the minimum concentration 1.06 ng/µL of recombinant spike found to induce HA in the HA experiment described above.
Figure 4.
Quantification by Western blot analysis of spike protein in cell culture supernatants. Lines 1 and 2: cell culture supernatants infected with SARS-CoV-2, Wuhan strain, harvested at 24 and 48 h respectively. Line 3: non-infected cell culture supernatant. Lines 4, 5 and 6: recombinant SARS-CoV-2 spike protein at 2.25, 1.25 and 0.56 ng/µL respectively.
2.3. Molecular Modeling
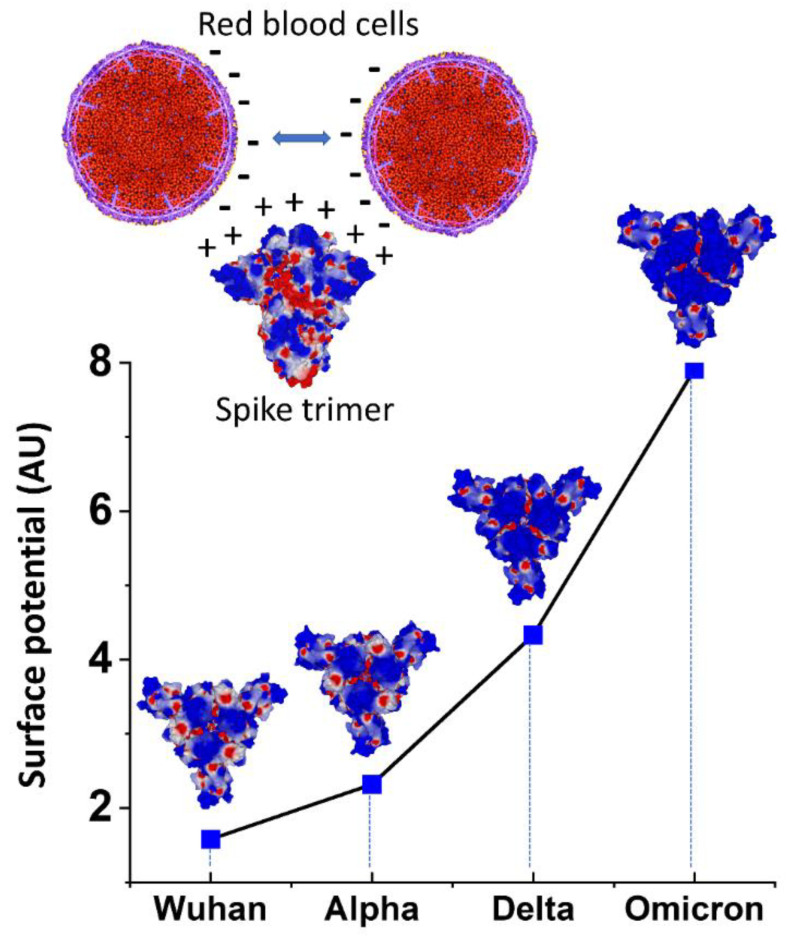
Since the surface of RBCs is electronegative due to a high expression of anionic SA in membrane proteins and gangliosides, we used molecular modeling simulations to study the electrostatic surface potential of the spike proteins used in the present study. We focused our attention on the central area of the spike trimers, which is formed by the receptor binding domain (RBD) of each monomer. The electrostatic potential of the central area of the spike trimers increased exponentially from the Wuhan initial lineage to the Omicron variant (Figure 5, lower panel). This was caused by a progressive decrease in electronegative zones (colored in red) and a concomitant increase in electropositivity (blue zones). The increase was modest between Wuhan and Alpha, larger for Delta, and reached very high levels for Omicron. As depicted in the upper panel of Figure 5, these variations in net positive electric charge between the four SARS-CoV-2 strains considered here have implications for the induction of HA, since electropositivity mitigates the electrostatic repulsion between negatively charged RBC surfaces.
Figure 5.
Electrostatic surface potential of SARS-CoV-2 spike trimers as denoted by color, positive in blue and negative in red. Upper panel. Under physiological conditions, RBCs maintain separation from each other due to a repulsive electric zeta potential between their negatively charged surfaces. The electropositive surface of spike protein neutralizes this zeta potential, allowing closer contacts between RBCs. Lower panel. For all variants, the electrostatic surface potential of the spike trimer is more electropositive in the central area formed by the RBD of each monomer. Quantitative analysis of the surface potential (in AU = arbitrary units) shows an exponential increase from the Wuhan to Omicron lineages.
3. Discussion
For all four SARS-CoV-2 lineages tested, Wuhan, Alpha, Delta and Omicron BA.1, spike protein mixed with human RBCs induced HA. This result provides an in vitro counterpart to the clumps of stacked RBCs (rouleaux) found, for example, in the blood of most [26,27] or a third [28] of COVID-19 patients in three clinical studies, and reinforces indications that such blood cell aggregation is key to the morbidities of this disease. HA-associated vascular obstruction in COVID-19 was further demonstrated in zebrafish embryos [47], which have capillary diameters [48] and blood cell glycosylation patterns [49] similar to those of humans. SARS-CoV-2 spike protein injected into a zebrafish embryo vein caused the formation of small RBC clumps and an associated reduction in blood flow velocity within 3–5 min after injection [47]. Also, in various in vivo or in vitro studies, SARS-CoV-2 spike protein S1 was found to cause endothelial, pulmonary and neuronal damage, as well as platelet-thrombi formation and microclots [40,50,51,52].
In addition to the glycan bindings reviewed above, electrostatic attraction could promote attachments between RBCs and SARS-CoV-2 spike protein and contribute to the observed induction of HA. As indicated here by molecular modeling simulations, the SARS-CoV-2 spike protein central region has a net positive charge, which yields an attractive force to the negatively charged RBC surface and also mitigates the repulsive electrostatic force between RBCs. This result is consistent with prior determinations of a net positive electrostatic potential of the SARS-CoV-2 spike protein [19,53]. Regardless of whether glycan bindings or electrostatic attraction is the predominant underlying force, the HA observed here suggests, more generally, that spike protein attachments to other cells such as platelets and endothelial cells may be implicated in COVID-19 morbidities, these two cells, like RBCs, having dense surface distributions of SA-tipped glycans [1] and corresponding negative surface charges [16,17]. These attachments can manifest clinically in damage to endothelial cells induced by virions or free spike protein, marked by traces of spike protein [23,35,51], and in the formation of fibrin-hardened clots that incorporate platelets, neutrophils and other blood cells [54], potentially seeded by RBC clumps.
The much stronger HA-inducing effect of the spike protein of the SARS-CoV-2 Omicron variant vs. the prior lineages tested here, with thresholds of concentration for HA induction of 0.13 ng/µL for Omicron vs. 1.06 ng/µL for the Wuhan, Alpha and Delta lineages, was an intriguing result. Under the hypothesis that RBC clumping underlies the morbidities of COVID-19, especially those related to diminished efficiency of RBC oxygenation, this finding appears paradoxical, since percentages of patients having respiratory distress or requirements for oxygen or mechanical ventilation were all sharply less for Omicron as compared with prior viral lineages [55]. Yet diminished pulmonary morbidities of Omicron can be explained by its less efficient replication compared with prior SARS-CoV-2 lineages in the lung parenchyma, encompassing the alveolar epithelium, in contrast to Omicron’s faster replication in the bronchi [56,57]. The disruption of the alveolar-capillary barrier caused by a COVID-19 infection is a prime route by which the virus enters the bloodstream [58], so limited replication of Omicron in alveolar tissue would limit systemic serum viral loads and potential for associated RBC clumping as well.
Among the attributes of the Omicron variant that are of interest for further study is that although its overall electrostatic potential is more positive than for prior lineages, its N-terminal domain (NTD) is less positive [53,59,60]. Consistent with the greater HA-inducing activity of Omicron, however, its SA-binding affinity as predicted by molecular modeling is greater than for prior lineages [61]. It has also been proposed, again based upon in silico analysis, that decreased binding affinity of mutated residues on the Omicron spike protein to a cellular receptor implicated in thromboembolic and neurological complications of COVID-19, α7nAChR, could also account for decreased morbidity of this viral variant [62].
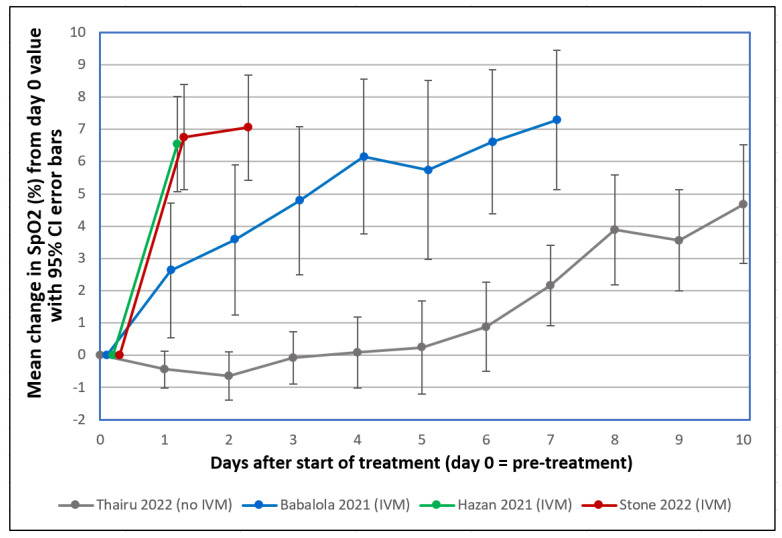
IVM, an electrically neutral molecule [63], has been found in silico to attach with a strong affinity to 10 glycan binding sites on the spike protein of the Alpha through Delta variants [43], which suggests that competitive inhibition of spike protein attachments to host cell glycoconjugates could be its means of HA inhibition. IVM at concentrations of 1–2 µM for the four different viral lineages inhibited HA when added concurrently with spike protein and also reversed HA that had been induced by the prior addition of spike protein. This HA-reversal effect could account for sharp increases observed within 24 h after administration of IVM of pre-treatment depressed SpO2 levels in severe COVID-19 patients, as summarized in Figure 6 below, which reproduces a figure from the most recent of three clinical studies reporting this effect [64]. In contrast to the sharp, rapid increases in SpO2 levels observed in these studies, moderate and severe COVID-19 patients under standard care typically manifest decreasing SpO2 values in tandem with increasing pulmonary CT abnormalities from the day of onset of disease symptoms through the second week following, as established in several studies that tracked SpO2 values, pulmonary abnormalities, or both [65,66,67,68,69,70,71].
The inhibition of HA by IVM as reported here parallels the prevention of RBC clumping in zebrafish embryos by heparan sulfate, a glycosaminoglycan likewise indicated to strongly bind to SARS-CoV-2 spike protein [49,72], when co-injected with spike protein [49]. It is noteworthy that the scattered RBC clumps observed in this zebrafish embryo study, as with those observed in the blood of COVID-19 patients [26,27,28], are much smaller than the macroscopic-scale HA—clusters of extensively interlaced RBCs—observed in this study. It is therefore likely that smaller concentrations of spike protein and IVM would be required to, respectively, induce and reverse RBC clumps of clinical relevance. Thus, the peak plasma level of IVM of approximately 412 nM, as attained about four hours after a standard oral dose of 200–350 µg/kg [1], appears to be in a range that could achieve clinical effects analogous to HA reversal observed in this study at IVM concentrations of 1–2 µM.
The HA-inducing activity of SARS-CoV-2 spike protein, which is especially potent for Omicron, raises questions as to potential risks for COVID-19 mRNA vaccines, which use spike protein as the generated antigen, even though serious adverse effects (AEs) linked to spike protein, such as myocarditis [73,74,75], are rare. Detectable levels of SARS-CoV-2 spike protein and S1 in serum or plasma have been found to persist for as long as 50 days following such vaccinations [76,77,78]. The possibility that spike protein migrating into the bloodstream could in rare cases prompt such HA-associated AEs is suggested, for example, by a study of 1006 subjects experiencing AEs after receiving a Pfizer/BioNTech or Moderna mRNA vaccination, which found a significant degree of RBC aggregation in the blood of 948 of those subjects [79]. These risks may be increased for younger age groups, with 301 adolescents of 13–18 years of age who received two doses of the BNT162b2 mRNA COVID-19 vaccine in one study having a 29.2% rate of cardiac AEs, ranging from tachycardia or palpitation to myopericarditis [80]. The investigators considered chest pain, which occurred at a 4% incidence, “an alarming side effect,” however, myopericarditis cases were mostly mild and temporary. The possibility that the increased HA-inducing activity of the Omicron spike protein might result in increased HA-associated AEs from Omicron booster vaccines can be ruled out by close vaccine safety monitoring to follow up on the animal testing on these booster vaccines performed to date [81].
Additional experiments of interest for follow-up to this study include microscopic detection to check for the initial formation of RBC clumping at spike protein concentrations lower than those which induce HA. Also, the zebrafish study described above [47] could be replicated using IVM instead of heparan sulfate as the blocking agent. Finally, HA could be tested as in this experiment but using RBCs supplemented with human serum albumin (HSA) at a physiological concentration. If IVM were to bind to spike protein glycans at the same molecular region as that which binds to HSA, that could significantly limit its HA-inhibitory effect, since 93% of IVM binds to serum proteins, mainly HSA, in blood [82], and 93% of IVM would then be rendered inactive for this effect. Conversely, if, IVM were to bind to spike protein glycans and HSA each at different regions, then HSA, a large molecule (molecular mass of 66.5 kDa, vs. 875.1 Da for IVM), could considerably boost the HA-inhibitory effect of IVM through steric interference.
Figure 6.
Mean changes in oxygen saturation (SpO2) for severe COVID-19 patients following treatments including or excluding IVM. Reproduced from Stone et al., 2022 [64] (CC-BY 4.0). Patients tracked over various time periods from each regimen were those with SpO2 values all recorded on room air, having pre-treatment (day 0) values ≤ 93%. The y-axis value at day n is the mean of changes in SpO2 values from day 0 to day n, with error bars designating 95% confidence intervals. ● Thairu et al., 2022 [83,84]: 26 patients, median age 45, treated with varying combinations of lopinavir/ritonavir (Alluvia), remdesivir, azithromycin, and enoxaparin as well as zinc sulfate and vitamin C. ● Babalola et al., 2021 [84,85]: 19 patients, median age 33, treated with IVM, zinc and vitamin C, with some also given azithromycin and hydroxychloroquine. ● Hazan et al., 2021 [86]: 19 patients, median age 63, treated with IVM, doxycycline and zinc. ● Stone et al., 2022 [64]: 34 patients, median age 56.5, treated with IVM, doxycycline and zinc.
4. Materials and Methods
4.1. Source and Preparation of Red Blood Cells
Red blood cells (RBCs) were provided by the “Establishment Français du Sang” (EFS) from blood bag donors qualified as “non-therapeutic blood bag” (Convention N°7828). Twenty milliliters of whole blood (with anticoagulant EDTA) were added to a fifty-milliliter conical tube and filled with thirty milliliters of PBS at pH = 7.2 before centrifugation at 800× g for 10 min. Then, the supernatant was discarded and replaced with fresh PBS. This procedure was again repeated twice. After the last centrifugation, RBCs were diluted in PBS at a final concentration of 2.5%. RBCs were then stocked for one week at 4 °C.
4.2. Spike Proteins Preparation
Recombinant spike proteins (BioServUK, Sheffield, UK) of these four following SARS-CoV-2 strains were used for this experiment: Wuhan, Alpha, Delta and Omicron BA.1 (BSV-COV-PR-33, BSV-COV-PR-65, BSV-COV-PR-97 and BSV-COV-OM-0.1, respectively). Four concentrations of spike protein were prepared for each of these viruses by diluting stock suspension in PBS. Spike protein of each viral strain was dissolved in PBS and added to wells at final increasing concentrations of 0.27 ng/µL, 0.53 ng/µL, 1.06 ng/µL and 2.12 ng/µL for the Wuhan, Alpha and Delta lineages, and of 0.07 ng/µL, 0.13 ng/µL, 0.27 ng/µL and 0.53 ng/µL for the Omicron variant.
4.3. Cells and SARS-CoV-2 Strains Preparation
Vero E6 cells (ATCC-CRL-1586) were cultured as in previously described conditions [87,88,89] in medium (MEM, Gibco, Gaithersburg, MD, USA) with 2 mM L-glutamine and 10% fetal bovine serum (FBS) at 37 °C in a 5% CO2 incubator. We infected Vero E6 with four viral strains genotyped by whole genome next-generation sequencing (NGS) as belonging to: Pangolin lineage B.1.1 [90] (the first major lineage following the Wuhan genotype that circulated during the first epidemic period in France, designated as “Wuhan”) and three variants: Alpha (B.1.1.7), Delta (B.1.617.2) and Omicron BA.1 (B.1.1.529). Culture supernatants were harvested 24 h post-viral infection and then passed through 0.22 µM pore-sized filters (Merck Millipore, Carrigtwohill, Ireland) to remove cells and debris and obtain viral suspension for experiments.
4.4. Western Blot and Quantification Analysis
Supernatants from Wuhan SARS CoV-2 infected cells prepared as described above or uninfected cells and commercial spike were lysed with 2× Laemmli Sample Buffer (#1610737, Bio-Rad, Hercules, CA, USA) with DTT (#EU0006-B, Euromedex, Souffelweyersheim, France) added as reducing agent and then heated at 95 °C for 5 min. Proteins were separated by 10% SDS-polyacrylamide gel electrophoresis (Laemmli, 1970) and Western-blotted on a nitrocellulose membrane. After 1 h of saturation in 5% nonfat dry milk with 0.3% Tween-20 in PBS, the membrane was incubated overnight with SARS/SARS-CoV-2 Coronavirus Spike Protein (subunit 1) rabbit polyclonal antibody (Thermo Fisher Scientific, Illkirch-Graffenstaden, France) at a dilution of 1:1000 in the same buffer as for saturation. After this first incubation, the membrane was washed for 10 min three times in PBS 1X-Tween buffer and then incubated for 1 h at room temperature with peroxidase-labeled anti-rabbit donkey antibody (#NIF 824 ECL Rabbit IgG, HRP-linked whole Ab, Sigma-Aldrich Life Science, Burlington, MA, USA) diluted in saturation buffer at 1:1000. After this second incubation, the membrane was rinsed for 10 min three times in PBS 1X-Tween buffer before ECL (Western Blotting Substrate, # W1001 Promega, Madison, WI, USA) revelation by image acquisition with the Fusion Fx chemiluminescence imaging system (Vilber Lourmat, Marne-la Vallée, France). Quantification in each well was calculated by measuring band intensities using ImageQuant TL Version 7.0 analysis software (GE Healthcare, Buc, Yvelines, France). Protein markers (New England Biolabs, #P7719S) were used for molecular mass determination.
4.5. IVM Preparation
IVM was supplied by Sigma Aldrich (St Quentin Fallavier, France). Stock solution was diluted in 2.5% of DMSO and 97.5% of water. IVM, 20 µL in volume, was added in designated wells to reach final concentrations of 1, 2, 4 and 8 µM, as specified.
4.6. Tests for Hemagglutination (HA) and for Its Inhibition and Reversal by IVM
Three sets of experiments were performed to test for HA induced by SARS-CoV-2 spike protein and then for HA inhibition and reversal by IVM. To test for HA, using a 96 micro-well plate, 50 µL of 2.5% RBCs in PBS was added to wells together with 62 µL of diluted spike proteins at specified concentrations. An additional 20 µL of PBS was added to attain the same total fluid volume as used in the IVM inhibition and reversal experiments. This mixture was let sit for 30 min at room temperature under gentle agitation. Then, the plate was tilted for at least 30 s, after which, if HA had not occurred a teardrop could be observed at the bottom of the well consisting of settled RBCs (Figure 1). This teardrop was not observed if HA had occurred, i.e., if a network of linked, agglutinated RBCs had formed, as described previously [91]. To test for inhibition of HA by IVM, 50 µL of RBCs were mixed with 20 µL of IVM at specified concentrations ranging from 1–8 µM and let sit for 30 min at room temperature under gentle agitation. Then 62 µL of spike protein was added at specified concentrations, wells were let sit for an additional 30 min at room temperature under gentle agitation, and HA was determined as above. Finally, to test for reversal of HA by IVM, 50 µL of RBCs were mixed with 62 µL of spike proteins for 30 min to determine HA as above. Then 20 µL of IVM were added at specified concentrations, wells were let sit for an additional 30 min at room temperature under gentle agitation and then the plate was tilted at least 30 s and wells were rechecked for HA as described above.
The following control experiments were performed. Fifty microliters of RBCs were deposited alone in the wells to check their absence of agglutination. Twenty microliters of PBS were added to fifty microliters of RBCs to verify the absence of HA. The potential for induction of HA by IVM was tested by adding it at the highest concentration used (8 µM) to 50 µL of 2.5% RBCs. In order to test whether DMSO blocked or reversed HA, we also performed the HA inhibition and reversal experiments described above using the solvent for IVM, 2.5% DMSO and 97.5% water, but without IVM.
The HA experiment was then done with viral suspensions for each viral strain by adding to the wells the same volume of viral supernatant as for commercial spike suspensions.
Each experiment was done in triplicate.
4.7. Molecular Modeling Simulations
A complete structure of the reference spike protein was generated from the original 20B strain (pdb: 7bnm) as previously described [92]. All gaps in the pdb file were fixed by inserting the missing amino acids with the protein structure prediction service Robetta [https://robetta.bakerlab.org/, accessed on 3 November 2022] [53,93]. This source file model was used to introduce the specific mutational profiles of the indicated Alpha, Delta and Omicron variants with the MUTATE tool of Swiss-PdbViewer [53,94]. Trimeric structures in the closed pre-fusion conformation were constructed with Swiss-PdbViewer by homology with a reference model (pdb: 6VSB). All structures were then submitted to several rounds of energy minimization with the Polak–Robière algorithm [88]. The electrostatic surface potential of the spike trimers was analyzed with Molegro and quantified with ImageJ software as described previously [92].
5. Conclusions
Spike protein from four lineages of SARS-CoV-2 induced HA in human RBCs, which supports other indications that spike protein-induced RBC clumping, as well as viral attachments to other blood cells and endothelial cells, may be key to the morbidities of COVID-19. IVM, a macrocyclic lactone indicated to bind strongly to multiple glycan sites on SARS-CoV-2 spike protein, blocked HA when added to RBCs prior to spike protein and reversed HA when added afterward, which suggests therapeutic options for COVID-19 treatment using this drug or other competitive glycan-binding agents. The Omicron B.1.1.529 variant had significantly greater HA-inducing activity than the three prior lineages tested, which may relate to the findings from molecular modeling that the electrostatic charge of the central region of its spike protein was considerably more positive than for those of the prior lineages and, from a prior study, that its SA-binding affinity is greater than for prior lineages [61]. Whether increased risks of rare HA-associated serious AEs for Omicron booster vaccines might be correspondingly increased as compared with those for legacy mRNA COVID vaccines can be ruled out by close vaccine safety monitoring to follow up on the animal testing on these booster vaccines performed to date.
Acknowledgments
The authors are grateful to Barry Dancis for close review and editing of this manuscript.
Abbreviations
The following abbreviations are used in this manuscript:
| ACE2 | angiotensin converting enzyme 2 |
| CD147 | cluster of differentiation 147 protein, encoded by the BSG gene |
| COVID-19 | coronavirus disease 2019 |
| GPA | glycophorin A |
| NTD | N-terminal domain |
| PBS | phosphate-buffered saline |
| RBC | red blood cell |
| RBD | receptor binding domain |
| RCT | randomized clinical trial |
| SA | sialic acid |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
Author Contributions
Conceptualization, D.E.S., J.F. and B.L.S.; methodology, J.F. and B.L.S.; validation, C.B., P.C., J.F. and B.L.S.; formal analysis, C.B. and B.L.S.; investigation, A.B., M.M. and M.L.B.; writing—original draft preparation, D.E.S., C.B., J.F. and B.L.S.; writing—review and editing, P.C.; supervision, J.F. and B.L.S.; project administration, B.L.S. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the French Government under the “Investments for the Future” program managed by the National Agency for Research (ANR), Méditerranée-Infection 10-IAHU-03.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Scheim D.E. A Deadly Embrace: Hemagglutination Mediated by SARS-CoV-2 Spike Protein at its 22 N-Glycosylation Sites, Red Blood Cell Surface Sialoglycoproteins, and Antibody. Int. J. Mol. Sci. 2022;23:2558. doi: 10.3390/ijms23052558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen W., Hui Z., Ren X., Luo Y., Shu J., Yu H., Li Z. The N-glycosylation sites and Glycan-binding ability of S-protein in SARS-CoV-2 Coronavirus. bioRxiv. 2020 doi: 10.1101/2020.12.01.406025. [DOI] [Google Scholar]
- 3.Guo W., Lakshminarayanan H., Rodriguez-Palacios A., Salata R.A., Xu K., Draz M.S. Glycan Nanostructures of Human Coronaviruses. Int. J. Nanomed. 2021;16:4813–4830. doi: 10.2147/IJN.S302516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shajahan A., Supekar N.T., Gleinich A.S., Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30:981–988. doi: 10.1093/glycob/cwaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao C., Zeng J., Jia N., Stavenhagen K., Matsumoto Y., Zhang H., Li J., Hume A.J., Mühlberger E., van Die I., et al. SARS-CoV-2 Spike Protein Interacts with Multiple Innate Immune Receptors. bioRxiv. 2020 doi: 10.1101/2020.07.29.227462. [DOI] [Google Scholar]
- 6.Sikora M., von Bülow S., Blanc F.E.C., Gecht M., Covino R., Hummer G. Computational epitope map of SARS-CoV-2 spike protein. PLoS Comput. Biol. 2021;17:e1008790. doi: 10.1371/journal.pcbi.1008790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi Y.K., Cao Y., Frank M., Woo H., Park S.-J., Yeom M.S., Croll T.I., Seok C., Im W. Structure, Dynamics, Receptor Binding, and Antibody Binding of the Fully Glycosylated Full-Length SARS-CoV-2 Spike Protein in a Viral Membrane. J. Chem. Theory Comput. 2021;17:2479–2487. doi: 10.1021/acs.jctc.0c01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koehler M., Delguste M., Sieben C., Gillet L., Alsteens D. Initial Step of Virus Entry: Virion Binding to Cell-Surface Glycans. Annu. Rev. Virol. 2020;7:143–165. doi: 10.1146/annurev-virology-122019-070025. [DOI] [PubMed] [Google Scholar]
- 9.Fantini J., Di Scala C., Chahinian H., Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents. 2020;55:105960. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ströh L.J., Stehle T. Glycan Engagement by Viruses: Receptor Switches and Specificity. Annu. Rev. Virol. 2014;1:285–306. doi: 10.1146/annurev-virology-031413-085417. [DOI] [PubMed] [Google Scholar]
- 11.Morniroli D., Giannì M.L., Consales A., Pietrasanta C., Mosca F. Human Sialome and Coronavirus Disease-2019 (COVID-19) Pandemic: An Understated Correlation? Front. Immunol. 2020;11:1480. doi: 10.3389/fimmu.2020.01480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neu U., Bauer J., Stehle T. Viruses and sialic acids: Rules of engagement. Curr. Opin. Struct. Biol. 2011;21:610–618. doi: 10.1016/j.sbi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen L., McCord K.A., Bui D.T., Bouwman K.M., Kitova E.N., Elaish M., Kumawat D., Daskhan G.C., Tomris I., Han L., et al. Sialic acid-containing glycolipids mediate binding and viral entry of SARS-CoV-2. Nat. Chem. Biol. 2022;18:81–90. doi: 10.1038/s41589-021-00924-1. [DOI] [PubMed] [Google Scholar]
- 14.Baum J., Ward R.H., Conway D.J. Natural selection on the erythrocyte surface. Mol. Biol. Evol. 2002;19:223–229. doi: 10.1093/oxfordjournals.molbev.a004075. [DOI] [PubMed] [Google Scholar]
- 15.Aoki T. A Comprehensive Review of Our Current Understanding of Red Blood Cell (RBC) Glycoproteins. Membranes. 2017;7:56. doi: 10.3390/membranes7040056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seaman G.V.F. Electrochemical features of platelet interactions. Thromb. Res. 1976;8:235–246. doi: 10.1016/0049-3848(76)90066-9. [DOI] [PubMed] [Google Scholar]
- 17.Strilić B., Eglinger J., Krieg M., Zeeb M., Axnick J., Babál P., Müller D.J., Lammert E. Electrostatic Cell-Surface Repulsion Initiates Lumen Formation in Developing Blood Vessels. Curr. Biol. 2010;20:2003–2009. doi: 10.1016/j.cub.2010.09.061. [DOI] [PubMed] [Google Scholar]
- 18.Boulant S., Stanifer M., Lozach P.-Y. Dynamics of virus-receptor interactions in virus binding, signaling, and endocytosis. Viruses. 2015;7:2794–2815. doi: 10.3390/v7062747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hassanzadeh K., Perez Pena H., Dragotto J., Buccarello L., Iorio F., Pieraccini S., Sancini G., Feligioni M. Considerations around the SARS-CoV-2 Spike Protein with Particular Attention to COVID-19 Brain Infection and Neurological Symptoms. ACS Chem. Neurosci. 2020;11:2361–2369. doi: 10.1021/acschemneuro.0c00373. [DOI] [PubMed] [Google Scholar]
- 20.Pawłowski P.H. Additional Positive Electric Residues in the Crucial Spike Glycoprotein S Regions of the New SARS-CoV-2 Variants. Infect. Drug Resist. 2021;14:5099–5105. doi: 10.2147/IDR.S342068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmetaj-Shala B., Vaja R., Atanur S.S., George P.M., Kirkby N.S., Mitchell J.A. Cardiorenal Tissues Express SARS-CoV-2 Entry Genes and Basigin (BSG/CD147) Increases With Age in Endothelial Cells. JACC Basic Transl. Sci. 2020;5:1111–1123. doi: 10.1016/j.jacbts.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huertas A., Montani D., Savale L., Pichon J., Tu L., Parent F., Guignabert C., Humbert M. Endothelial cell dysfunction: A major player in SARS-CoV-2 infection (COVID-19)? Eur. Respir. J. 2020;56:2001634. doi: 10.1183/13993003.01634-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheim D.E. From Cold to Killer: How SARS-CoV-2 Evolved without Hemagglutinin Esterase to Agglutinate, then Clot Blood Cells in Pulmonary and Systemic Microvasculature. [(accessed on 17 October 2022)]. Available online: https://osf.io/sgdj2.
- 24.De Back D.Z., Kostova E., Klei T., Beuger B., van Zwieten R., Kuijpers T., Juffermans N., van den Berg T., Korte D., van Kraaij M., et al. RBC Adhesive Capacity Is Essential for Efficient ‘Immune Adherence Clearance’ and Provide a Generic Target to Deplete Pathogens from Septic Patients. Blood. 2016;128:1031. doi: 10.1182/blood.V128.22.1031.1031. [DOI] [Google Scholar]
- 25.Varki A., Gagneux P. Multifarious roles of sialic acids in immunity. Ann. N. Y. Acad. Sci. 2012;1253:16–36. doi: 10.1111/j.1749-6632.2012.06517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berzuini A., Bianco C., Migliorini A.C., Maggioni M., Valenti L., Prati D. Red blood cell morphology in patients with COVID-19-related anaemia. Blood Transfus. 2021;19:34–36. doi: 10.2450/2020.0242-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melkumyants A., Buryachkovskaya L., Lomakin N., Antonova O., Serebruany V. Mild COVID-19 and Impaired Blood Cell–Endothelial Crosstalk: Considering Long-Term Use of Antithrombotics? Thromb. Haemost. 2022;122:123–130. doi: 10.1055/a-1551-9911. [DOI] [PubMed] [Google Scholar]
- 28.Lakhdari N., Tabet B., Boudraham L., Laoussati M., Aissanou S., Beddou L., Bensalem S., Bellik Y., Bournine L., Fatmi S., et al. Red blood cells injuries and hypersegmented neutrophils in COVID-19 peripheral blood film. medRxiv. 2020 doi: 10.1101/2020.07.24.20160101. [DOI] [Google Scholar]
- 29.Lam L.M., Murphy S.J., Kuri-Cervantes L., Weisman A.R., Ittner C.A.G., Reilly J.P., Pampena M.B., Betts M.R., Wherry E.J., Song W.-C., et al. Erythrocytes Reveal Complement Activation in Patients with COVID-19. medRxiv. 2020 doi: 10.1101/2020.05.20.20104398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker A.N., Richards S.-J., Guy C.S., Congdon T.R., Hasan M., Zwetsloot A.J., Gallo A., Lewandowski J.R., Stansfeld P.J., Straube A., et al. The SARS-COV-2 Spike Protein Binds Sialic Acids and Enables Rapid Detection in a Lateral Flow Point of Care Diagnostic Device. ACS Cent. Sci. 2020;6:2046–2052. doi: 10.1021/acscentsci.0c00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao W., Ma B., Li Z., Wang X., Gao X., Li Y., Qin B., Shang S., Cui S., Tan Z. Binding of the SARS-CoV-2 spike protein to glycans. Sci. Bull. 2021;66:1205–1214. doi: 10.1016/j.scib.2021.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shilts J., Wright G.J. No evidence for basigin/CD147 as a direct SARS-CoV-2 spike binding receptor. bioRxiv. 2020 doi: 10.1038/s41598-020-80464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maeda N., Seike M., Kon K., Shiga T. Erythrocyte Aggregation as a Determinant of Blood Flow: Effect of pH, Temperature and Osmotic Pressure. In: Mochizuki M., Honig C.R., Koyama T., Goldstick T.K., Bruley D.F., editors. Oxygen Transport to Tissue X. Springer; New York, NY, USA: 1988. pp. 563–570. [DOI] [PubMed] [Google Scholar]
- 34.Barshtein G., Wajnblum D., Yedgar S. Kinetics of linear rouleaux formation studied by visual monitoring of red cell dynamic organization. Biophys. J. 2000;78:2470–2474. doi: 10.1016/S0006-3495(00)76791-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magro C., Mulvey J.J., Berlin D., Nuovo G., Salvatore S., Harp J., Baxter-Stoltzfus A., Laurence J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: A report of five cases. Transl. Res. 2020;220:1–13. doi: 10.1016/j.trsl.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menter T., Haslbauer J.D., Nienhold R., Savic S., Hopfer H., Deigendesch N., Frank S., Turek D., Willi N., Pargger H., et al. Postmortem examination of COVID-19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology. 2020;77:198–209. doi: 10.1111/his.14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becker R.C. COVID-19 update: COVID-19-associated coagulopathy. J Thromb. Thrombolysis. 2020;50:54–67. doi: 10.1007/s11239-020-02134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gattinoni L., Coppola S., Cressoni M., Busana M., Rossi S., Chiumello D. COVID-19 Does Not Lead to a "Typical" Acute Respiratory Distress Syndrome. Am. J. Respir. Crit. Care Med. 2020;201:1299–1300. doi: 10.1164/rccm.202003-0817LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marini J.J., Gattinoni L. Management of COVID-19 Respiratory Distress. JAMA. 2020;323:2329–2330. doi: 10.1001/jama.2020.6825. [DOI] [PubMed] [Google Scholar]
- 40.Grobbelaar L.M., Venter C., Vlok M., Ngoepe M., Laubscher G.J., Lourens P.J., Steenkamp J., Kell D.B., Pretorius E. SARS-CoV-2 spike protein S1 induces fibrin(ogen) resistant to fibrinolysis: Implications for microclot formation in COVID-19. Biosci. Rep. 2021;41:BSR20210611. doi: 10.1042/BSR20210611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultze B., Gross H.J., Brossmer R., Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J. Virol. 1991;65:6232–6237. doi: 10.1128/jvi.65.11.6232-6237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Callebaut P.E., Pensaert M.B. Characterization and isolation of structural polypeptides in haemagglutinating encephalomyelitis virus. J. Gen. Virol. 1980;48:193–204. doi: 10.1099/0022-1317-48-1-193. [DOI] [PubMed] [Google Scholar]
- 43.Aminpour M., Cannariato M., Safaeeardebili M.E., Preto J., Moracchiato A., Doria D., Donato F., Zizzi E.A., Deriu M.A., Scheim D.E., et al. In Silico Analysis of the Multi-Targeted Mode of Action of Ivermectin and Related Compounds. Computation. 2022;10:51. doi: 10.3390/computation10040051. [DOI] [Google Scholar]
- 44.Santin A.D., Scheim D.E., McCullough P.A., Yagisawa M., Borody T.J. Ivermectin: A multifaceted drug of Nobel prize-honored distinction with indicated efficacy against a new global scourge, COVID-19. New Microbes New Infect. 2021;43:100924. doi: 10.1016/j.nmni.2021.100924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdool Karim S.S., Devnarain N. Time to Stop Using Ineffective COVID-19 Drugs. N. Engl. J. Med. 2022;387:654–655. doi: 10.1056/NEJMe2209017. [DOI] [PubMed] [Google Scholar]
- 46.Shafiee A., Teymouri Athar M.M., Kohandel Gargari O., Jafarabady K., Siahvoshi S., Mozhgani S.-H. Ivermectin under scrutiny: A systematic review and meta-analysis of efficacy and possible sources of controversies in COVID-19 patients. Virol. J. 2022;19:102. doi: 10.1186/s12985-022-01829-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng Y., Zhao J., Li J., Guo Z., Sheng J., Ye X., Jin G., Wang C., Chai W., Yan J., et al. SARS-CoV-2 spike protein causes blood coagulation and thrombosis by competitive binding to heparan sulfate. Int. J. Biol. Macromol. 2021;193:1124–1129. doi: 10.1016/j.ijbiomac.2021.10.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Au Sam H., Storey Brian D., Moore John C., Tang Q., Chen Y.-L., Javaid S., Sarioglu A.F., Sullivan R., Madden Marissa W., O’Keefe R., et al. Clusters of circulating tumor cells traverse capillary-sized vessels. Proc. Natl. Acad. Sci. USA. 2016;113:4947–4952. doi: 10.1073/pnas.1524448113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamakawa N., Vanbeselaere J., Chang L.-Y., Yu S.-Y., Ducrocq L., Harduin-Lepers A., Kurata J., Aoki-Kinoshita K.F., Sato C., Khoo K.-H., et al. Systems glycomics of adult zebrafish identifies organ-specific sialylation and glycosylation patterns. Nat. Commun. 2018;9:4647. doi: 10.1038/s41467-018-06950-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colunga Biancatelli R.M.L., Solopov P.A., Sharlow E.R., Lazo J.S., Marik P.E., Catravas J.D. The SARS-CoV-2 spike protein subunit S1 induces COVID-19-like acute lung injury in Κ18-hACE2 transgenic mice and barrier dysfunction in human endothelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 2021;321:L477–L484. doi: 10.1152/ajplung.00223.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nuovo G.J., Magro C., Shaffer T., Awad H., Suster D., Mikhail S., He B., Michaille J.-J., Liechty B., Tili E. Endothelial cell damage is the central part of COVID-19 and a mouse model induced by injection of the S1 subunit of the spike protein. Ann. Diagn. Pathol. 2021;51:151682. doi: 10.1016/j.anndiagpath.2020.151682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perico L., Morigi M., Galbusera M., Pezzotta A., Gastoldi S., Imberti B., Perna A., Ruggenenti P., Donadelli R., Benigni A., et al. SARS-CoV-2 Spike Protein 1 Activates Microvascular Endothelial Cells and Complement System Leading to Platelet Aggregation. Front. Immunol. 2022;13:827146. doi: 10.3389/fimmu.2022.827146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fantini J., Yahi N., Colson P., Chahinian H., La Scola B., Raoult D. The puzzling mutational landscape of the SARS-2-variant Omicron. J. Med. Virol. 2022;94:2019–2025. doi: 10.1002/jmv.27577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Price L.C., McCabe C., Garfield B., Wort S.J. Thrombosis and COVID-19 pneumonia: The clot thickens! Eur. Respir. J. 2020;56:2001608. doi: 10.1183/13993003.01608-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maslo C., Friedland R., Toubkin M., Laubscher A., Akaloo T., Kama B. Characteristics and Outcomes of Hospitalized Patients in South Africa During the COVID-19 Omicron Wave Compared With Previous Waves. JAMA. 2022;327:583–584. doi: 10.1001/jama.2021.24868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hui K.P.Y., Ho J.C.W., Cheung M.-C., Ng K.-C., Ching R.H.H., Lai K.-L., Kam T.T., Gu H., Sit K.-Y., Hsin M.K.Y., et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature. 2022;603:715–720. doi: 10.1038/s41586-022-04479-6. [DOI] [PubMed] [Google Scholar]
- 57.Peacock T.P., Brown J.C., Zhou J., Thakur N., Sukhova K., Newman J., Kugathasan R., Yan A.W.C., Furnon W., De Lorenzo G., et al. The altered entry pathway and antigenic distance of the SARS-CoV-2 Omicron variant map to separate domains of spike protein. bioRxiv. 2022 doi: 10.1101/2021.12.31.474653. [DOI] [Google Scholar]
- 58.D’Agnillo F., Walters K.-A., Xiao Y., Sheng Z.-M., Scherler K., Park J., Gygli S., Rosas L.A., Sadtler K., Kalish H., et al. Lung epithelial and endothelial damage, loss of tissue repair, inhibition of fibrinolysis, and cellular senescence in fatal COVID-19. Sci. Transl. Med. 2021;13:eabj7790. doi: 10.1126/scitranslmed.abj7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar S., Karuppanan K., Subramaniam G. Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: A comparative sequence and structural-based computational assessment. J. Med. Virol. 2022;94:4780–4791. doi: 10.1002/jmv.27927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pascarella S., Ciccozzi M., Bianchi M., Benvenuto D., Cauda R., Cassone A. The value of electrostatic potentials of the spike receptor binding and N-terminal domains in addressing transmissibility and infectivity of SARS-CoV-2 variants of concern. J. Infect. 2022;84:e62–e63. doi: 10.1016/j.jinf.2022.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lam S.D., Waman V.P., Orengo C., Lees J. Insertions in the SARS-CoV-2 Spike N-Terminal Domain May Aid COVID-19 Transmission. bioRxiv. 2021 doi: 10.1101/2021.12.06.471394. [DOI] [Google Scholar]
- 62.Doria D., Santin A.D., Tuszynski J.A., Scheim D.E., Aminpour M. Omicron SARS-CoV-2 Spike-1 Protein’s Decreased Binding Affinity to α7nAChr: Implications for Autonomic Dysregulation of the Parasympathetic Nervous System and the Cholinergic Anti-Inflammatory Pathway—An In Silico Analysis. BioMedInformatics. 2022;2:553–564. doi: 10.3390/biomedinformatics2040035. [DOI] [Google Scholar]
- 63.Yagisawa M., Omura S. (Ōmura Satoshi Memorial Institute, Kitasato University, Tokyo, Japan). Personal communication. 2022. (affiliation for both)
- 64.Stone J.C., Ndarukwa P., Scheim D.E., Dancis B.M., Dancis J., Gill M.G., Aldous C. Changes in SpO2 on Room Air for 34 Severe COVID-19 Patients after Ivermectin-Based Combination Treatment: 62% Normalization within 24 Hours. Biologics. 2022;2:196–210. doi: 10.3390/biologics2030015. [DOI] [Google Scholar]
- 65.Osman A.M., Farouk S., Osman N.M., Abdrabou A.M. Longitudinal assessment of chest computerized tomography and oxygen saturation for patients with COVID-19. Egypt. J. Radiol. Nucl. Med. 2020;51:255. doi: 10.1186/s43055-020-00376-y. [DOI] [Google Scholar]
- 66.Metwally M.I., Basha M.A.A., Zaitoun M.M.A., Abdalla H.M., Nofal H.A.E., Hendawy H., Manajrah E., Hijazy R.f., Akbazli L., Negida A., et al. Clinical and radiological imaging as prognostic predictors in COVID-19 patients. Egypt. J. Radiol. Nucl. Med. 2021;52:100. doi: 10.1186/s43055-021-00470-9. [DOI] [Google Scholar]
- 67.Aoki R., Iwasawa T., Hagiwara E., Komatsu S., Utsunomiya D., Ogura T. Pulmonary vascular enlargement and lesion extent on computed tomography are correlated with COVID-19 disease severity. Jpn. J. Radiol. 2021;39:451–458. doi: 10.1007/s11604-020-01085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ding X., Xu J., Zhou J., Long Q. Chest CT findings of COVID-19 pneumonia by duration of symptoms. Eur. J. Radiol. 2020;127:109009. doi: 10.1016/j.ejrad.2020.109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y., Dong C., Hu Y., Li C., Ren Q., Zhang X., Shi H., Zhou M. Temporal Changes of CT Findings in 90 Patients with COVID-19 Pneumonia: A Longitudinal Study. Radiology. 2020;296:E55–E64. doi: 10.1148/radiol.2020200843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quispe-Cholan A., Anticona-De-La-Cruz Y., Cornejo-Cruz M., Quispe-Chirinos O., Moreno-Lazaro V., Chavez-Cruzado E. Tomographic findings in patients with COVID-19 according to evolution of the disease. Egypt. J. Radiol. Nucl. Med. 2020;51:215. doi: 10.1186/s43055-020-00329-5. [DOI] [Google Scholar]
- 71.Annunziata A., Coppola A., Carannante N., Simioli F., Lanza M., Di Micco P., Fiorentino G. Home Management of Patients with Moderate or Severe Respiratory Failure Secondary to COVID-19, Using Remote Monitoring and Oxygen with or without HFNC. Pathogens. 2021;10:413. doi: 10.3390/pathogens10040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gupta Y., Maciorowski D., Zak S.E., Kulkarni C.V., Herbert A.S., Durvasula R., Fareed J., Dye J.M., Kempaiah P. Heparin: A simplistic repurposing to prevent SARS-CoV-2 transmission in light of its in-vitro nanomolar efficacy. Int. J. Biol. Macromol. 2021;183:203–212. doi: 10.1016/j.ijbiomac.2021.04.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li C., Chen Y., Zhao Y., Lung D.C., Ye Z., Song W., Liu F.-F., Cai J.-P., Wong W.-M., Yip C.C.-Y., et al. Intravenous Injection of Coronavirus Disease 2019 (COVID-19) mRNA Vaccine Can Induce Acute Myopericarditis in Mouse Model. Clin. Infect. Dis. 2022;74:1933–1950. doi: 10.1093/cid/ciab707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trougakos I.P., Terpos E., Alexopoulos H., Politou M., Paraskevis D., Scorilas A., Kastritis E., Andreakos E., Dimopoulos M.A. Adverse effects of COVID-19 mRNA vaccines: The spike hypothesis. Trends Mol. Med. 2022;28:542–554. doi: 10.1016/j.molmed.2022.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cosentino M., Marino F. The spike hypothesis in vaccine-induced adverse effects: Questions and answers. Trends Mol. Med. 2022;28:797–799. doi: 10.1016/j.molmed.2022.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cognetti J.S., Miller B.L. Monitoring Serum Spike Protein with Disposable Photonic Biosensors Following SARS-CoV-2 Vaccination. Sensors. 2021;21:5857. doi: 10.3390/s21175857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ogata A.F., Cheng C.A., Desjardins M., Senussi Y., Sherman A.C., Powell M., Novack L., Von S., Li X., Baden L.R., et al. Circulating Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Vaccine Antigen Detected in the Plasma of mRNA-1273 Vaccine Recipients. Clin. Infect. Dis. 2022;74:715–718. doi: 10.1093/cid/ciab465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Röltgen K., Nielsen S.C.A., Silva O., Younes S.F., Zaslavsky M., Costales C., Yang F., Wirz O.F., Solis D., Hoh R.A., et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell. 2022;185:1025–2040. doi: 10.1016/j.cell.2022.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Giovannini F. Dark Field Microscopic Analysis on the Blood of 1,006 Symptomatic Persons After Anti-COVID mRNA Injections from Pfizer/BioNTech or Moderna. Int. J. Vaccine Theory Pract. Res. 2022;2:385–444. [Google Scholar]
- 80.Mansanguan S., Charunwatthana P., Piyaphanee W., Dechkhajorn W., Poolcharoen A., Mansanguan C. Cardiovascular Manifestation of the BNT162b2 mRNA COVID-19 Vaccine in Adolescents. Trop. Med. Infect. Dis. 2022;7:196. doi: 10.3390/tropicalmed7080196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.MacMillan C. The Omicron Booster: Your Questions Answered. Yale Medicine News, 27 October 2022. [(accessed on 3 November 2022)]. Available online: https://www.yalemedicine.org/news/omicron-booster-covid-19.
- 82.Klotz U., Ogbuokiri J.E., Okonkwo P.O. Ivermectin binds avidly to plasma proteins. Eur. J. Clin. Pharmacol. 1990;39:607–608. doi: 10.1007/BF00316107. [DOI] [PubMed] [Google Scholar]
- 83.Thairu Y., Babalola O.E., Ajayi A.A., Ndanusa Y., Ogedengbe J.O., Omede O. A comparison of Ivermectin and Non Ivermectin based regimen for COVID-19 in Abuja: Effects on virus clearance, Days-to-Discharge and Mortality. Res. Sq. 2022 doi: 10.21203/rs.3.rs-1373673/v1. [DOI] [Google Scholar]
- 84.Babalola O.E. (Bingham University, Jos/Karu, Nigeria). Personal communication. 2022. This communication provided deidentified patient level data for SpO2 changes post-treatment from two studies which he coauthored (Thairu et al., 2022 and Babalola et al., 2021 as cited), one using standard treatment excluding IVM (Thairu et al., 2022) and the other including IVM in the treatment regimen (Babalola et al., 2021), for those values obtained exclusively on room air and with pre-treatment SpO2 values ≤ 93%.
- 85.Babalola O.E., Ndanusa Y., Adesuyi A., Ogedengbe O.J., Thairu Y., Ogu O. A Randomized Controlled Trial of Ivermectin Monotherapy Versus HCQ, IVM, and AZ Combination Therapy in COVID-19 Patients in Nigeria. J. Infect. Dis. Epidemiol. 2021;7:233. doi: 10.23937/2474-3658/1510233. [DOI] [Google Scholar]
- 86.Hazan S., Dave S., Gunaratne A.W., Dolai S., Clancy R.L., McCullough P.A., Borody T.J. Effectiveness of ivermectin-based multidrug therapy in severely hypoxic, ambulatory COVID-19 patients. Future Microbiol. 2022;17:339–350. doi: 10.2217/fmb-2022-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Colson P., Levasseur A., Delerce J., Pinault L., Dudouet P., Devaux C., Fournier P.E., La Scola B., Lagier J.C., Raoult D. Spreading of a new SARS-CoV-2 N501Y spike variant in a new lineage. Clin. Microbiol. Infect. 2021;27:1352.e1–1352.e5. doi: 10.1016/j.cmi.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jaafar R., Boschi C., Aherfi S., Bancod A., Le Bideau M., Edouard S., Colson P., Chahinian H., Raoult D., Yahi N., et al. High Individual Heterogeneity of Neutralizing Activities against the Original Strain and Nine Different Variants of SARS-CoV-2. Viruses. 2021;13:2177. doi: 10.3390/v13112177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boschi C., Colson P., Bancod A., Moal V., La Scola B. Omicron Variant Escapes Therapeutic Monoclonal Antibodies (mAbs) Including Recently Released Evusheld®, Contrary to 8 Prior Main Variant of Concern (VOC) Clin. Infect. Dis. 2022;75:e534–e535. doi: 10.1093/cid/ciac143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Townsend A., Rijal P., Xiao J., Tan T.K., Huang K.A., Schimanski L., Huo J., Gupta N., Rahikainen R., Matthews P.C., et al. A haemagglutination test for rapid detection of antibodies to SARS-CoV-2. Nat. Commun. 2021;12:1951. doi: 10.1038/s41467-021-22045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fantini J., Yahi N., Azzaz F., Chahinian H. Structural dynamics of SARS-CoV-2 variants: A health monitoring strategy for anticipating COVID-19 outbreaks. J. Infect. 2021;83:197–206. doi: 10.1016/j.jinf.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kim D.E., Chivian D., Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526–W531. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.