Abstract
Haemophilus influenzae pili are surface structures that promote attachment to human epithelial cells. The five genes that encode pili, hifABCDE, are found inserted in genomes either between pmbA and hpt (hif-1) or between purE and pepN (hif-2). We determined the sequence between the ends of the pilus clusters and bordering genes in a number of H. influenzae strains. The junctions of the hif-1 cluster (limited to biogroup aegyptius isolates) are structurally simple. In contrast, hif-2 junctions are highly diverse, complex assemblies of conserved intergenic sequences (including genes hicA and hicB) with evidence of frequent recombination. Variation at hif-2 junctions seems to be tied to multiple copies of a 23-bp Haemophilus intergenic dyad sequence. The hif-1 cluster appears to have originated in biogroup aegyptius strains from invasion of the hpt-pmbA region by a DNA template containing the hif-2 genes with termini in the hairpin loop of flanking intergenic dyad sequences. The pilus gene clusters are an interesting model of a mobile “pathogenicity island” not associated with a phage, transposon, or insertion element.
Haemophilus influenzae Rd was the first free-living organism to have its genome sequenced completely (4). Significantly, this nonpathogenic laboratory strain does not contain certain DNA sequences present in disease-associated Haemophilus isolates. Virulence genes known to be missing from Rd include those encoding polysaccharide capsule (10), tryptophan catabolism determinants tnaABC (12), and hifABCDE, which encode hemagglutinating pili (23). These three groups of loci are all flanked by direct repeats, although the route for insertion of the genes into the background of the Haemophilus chromosome seems to have been different in each of the three cases (10, 12, 23). The H. influenzae serotype b capsulation genes are arranged as a compound transposon bordered by direct repeats of the 711-bp IS1016 (10). At the same genomic location in Rd, there is a single copy of IS1016 and no capsule loci. The tnaA and tnaB genes of serotype a, b, c, and f strains are located between the nlpD and mutS genes and flanked by 43-bp direct repeats of paired H. influenzae uptake signal sequences (USSs) (12). At the same nlpD-mutS map position, Rd has one copy of the paired USSs and no tryptophanase genes.
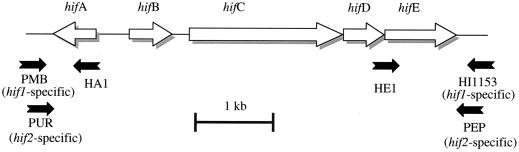
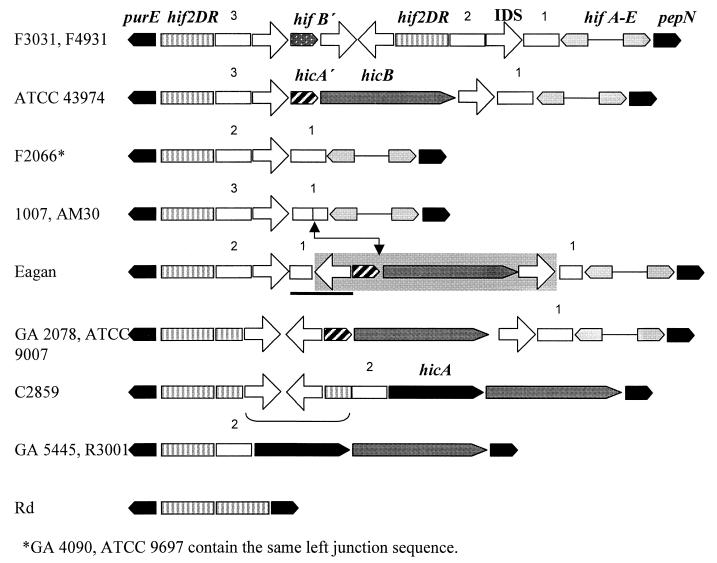
Pili elaborated by H. influenzae are thought to contribute to pathogenesis by facilitating tissue-specific attachment to host cells. The five hif genes that encode pili have been found at two distinct chromosomal locations (8, 14, 17, 23) (Fig. 1). The hif-1 location, limited to certain isolates of nonencapsulated H. influenzae biogroup aegyptius (Hae), lies between genes homologous to Escherichia coli pmbA and hpt, which are contiguous in the Rd genome. The second, more widespread location for the pilus genes (hif-2), reported in some Hae strains, nontypeable (nonencapsulated) H. influenzae (NTHI), and serotypes a, b, c, and f, is between pepN and purE homologues, also contiguous in Rd (5, 15, 19). Both hif-1 and hif-2 are flanked by direct repeats of intergenic sequences of 32 and 59 bp, respectively (named here DR-1 and DR-2, respectively). DR-1 and DR-2 show no homology to each other.
FIG. 1.
Primer binding sites in hif gene clusters. See Materials and Methods for oligonucleotide sequences.
To investigate the evolution of H. influenzae pili, we sequenced the intergenic DNA between inserted hif genes and conserved chromosomal loci. In common with other recent studies (5, 15), elaborate patterns of rearrangement at the junctions of hif-2 gene clusters were found. The agent of many sequence rearrangements (and also possibly insertion of entire pilus gene complexes) seems to be a 23-bp intergenic dyad sequence (IDS) (9, 18), which is not present at the stable junctions of hif-1 clusters.
MATERIALS AND METHODS
Bacteria.
H. influenzae strains used in the DNA sequencing study are shown in Table 1. Bacteria were grown for 16 to 20 h on chocolate agar plates (Becton Dickinson, Cockeysville, Md.) at 37°C in 5% CO2.
TABLE 1.
H. influenzae strains used in this study and their pilus genotypesa
| Strain | Description | Pilus clusters |
|---|---|---|
| F3031 | Hae; BPF reference strain | Hif-1+ Hif-2+ |
| F4931 | Hae; non-BPF isolate; Brazil | Hif-1+ Hif-2+ |
| F2066 | Hae; non-BPF isolate; Brazil | Hif-1+ Hif-2+ |
| ATCC 43974 | Hae; non-BPF isolate; Texas | Hif-1+ Hif-2+ |
| ATCC 43806 | Hae; non-BPF isolate; Texas | Hif-1− Hif-2− |
| GA2078 | NTHI; Georgia | Hif-1− Hif-2+ |
| 1007 | Serotype b; Texas | Hif-1− Hif-2+ |
| ATCC 9007 | Serotype c | Hif-1− Hif-2+ |
| GA4090 | Serotype f | Hif-1− Hif-2− |
| GA5445 | NTHI; Georgia | Hif-1− Hif-2− |
Sources were described and pilus genotypes were determined in previous work (19).
PCR.
The method for preparation of PCR templates from single H. influenzae colonies and the sequences of oligonucleotides PMB, PUR, HA1, HE1, HI1153, and PEP have been described previously (19). The primer combinations used to generate specific junction regions for sequencing (Fig. 1) were as follows: for pmbA-hifA1, PMB-HA1; for hifE1-hpt, HE1-HI1153; for pmbA-hpt, PMB-HI1153; for purE-hifA2, PUR-HA1; for hifE2-pepN, HE1-PEP; and for purE-pepN, PUR-PEP. The hicA- and hicB-specific primers (with sequences in parentheses) were HICAFOR (5′-TGAAAAATTATTAGATAAGCTCG-3′) and HICAREV (5′-TTTCACTGACTTTAAAGCACCAC-3′), and HICBFOR (5′-TTAAAGGCGGTGCTTTAAAGTCAG-3′) and HICBREV (5′-TACGTTTTGTTATCTTAAACTTGG-3′). PCR was performed using a GeneAmp System 9700 (Perkin-Elmer, Norwalk, Conn.) with Taq DNA polymerase (Perkin-Elmer) and the buffer supplied. In the standard 50-μl reaction mixture, we used 2.5 U of Taq, 40 pmol of each primer, 0.8 mM deoxynucleoside triphosphates (dNTP), and 0.15 mM MgCl2. The amplification cycle consisted of an initial 3-min hold at 94°C followed by 30 s at 94°C, 60 s at 50°C, and 60 s at 72°C, repeated 30 times, and finally a 6-min 72°C hold.
DNA sequencing.
PCR fragments were purified using Qiagen PCR Clean-Up kits and sequenced by dideoxy dye terminator cycle sequencing on an ABI Prism 377 Sequencer (Perkin-Elmer). The nucleotide sequences of both strands were determined using amplification primers and primers internal to the PCR fragments. Data were analyzed using the DNAStar CLUSTALW software (7) and BLAST (1).
Nucleotide sequence accession numbers.
The left and right junctions of the F3031 hif-1 and hif-2 clusters and the left junction of the GA2078 hif-2 cluster were submitted to GenBank as accession numbers AF148694 through AF148698.
RESULTS
The strategy used in this study was to amplify DNA across the junctions of the hif gene clusters using conserved primers in pilus genes (Fig. 1) (HA1 or HE1) and in genes flanking the insertions (PMB, PUR, HI1153, and PEP). H. influenzae strains were chosen to represent a range of capsule serotypes as well as a spectrum of biogroup aegyptius strains containing the hif-1 pilus gene cluster (Table 1). The DNA sequences of the PCR products were analyzed in detail to dissect the complex structures of repeats and short sequence elements at the junctions of the pilus gene clusters with surrounding genes.
Sequence conservation in stable hif-1 pilus gene cluster junctions.
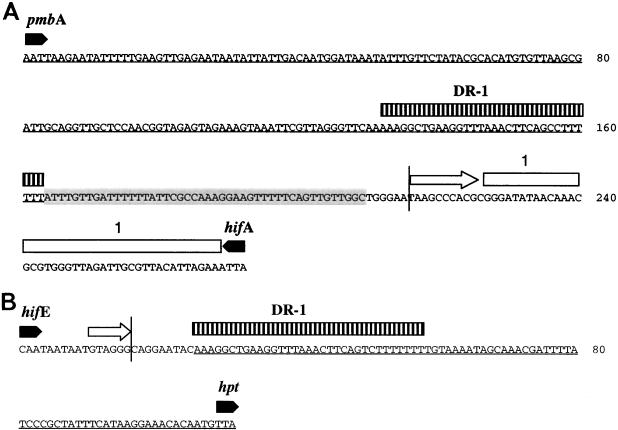
The sequences of the junctions of the hif-1 pilus cluster of Brazilan purpuric fever (BPF)-associated Hae reference strain F3031 are shown in Fig. 2. The junctions at the hif-1 locus are relatively simple and include incomplete copies of the IDS and a 32-bp direct repeat of intergenic sequence (DR-1) flanking the pilus genes hifABCDE. At the 3′ end of hifA is a 66-bp conserved junction sequence (junction sequence 1) that is also found at the 3′ end of hifA in many H. influenzae strains at both the hif-1 and hif-2 loci. Junction sequence 1 at the left end of F3031 hif-1 is 98% similar to the junction sequence in the F3031 hif-2 locus (Fig. 3A). At the left (pmbA-hifA) junction there is an AT-rich 46-bp sequence adjacent to DR-1 that is not similar to any GenBank sequence (Fig. 2A). At the right junction (Fig. 2B), the first 10 bp after the hifE1 open reading frame (ORF) are identical to the F3031 hif-2 cluster (Fig. 3B), but the subsequent 9 bases (CAGGAATAC) that lie before DR-1 are not similar to sequences associated with the pilus cluster.
FIG. 2.
Sequences of left and right ends of the hif-1 gene cluster of F3031. The 32-bp DR-1 duplications are shown as vertically striped boxes above the sequence. Open arrows represent partial IDSs. The vertical lines at the left and right ends show where flanking sequence diverges from F3031 hif-2 junctions (Fig. 3). The conserved junction sequence (junction sequence 1) is shown as an open bar. An AT-rich 46-bp sequence that is not similar to any GenBank sequence is shaded. The portion of the sequence homologous to H. influenzae Rd (Hif−) is underlined. (A) Left end: DNA sequence between the end of the hifA gene and the end of pmbA. (B) Right end: sequence between hifE and hpt.
FIG. 3.
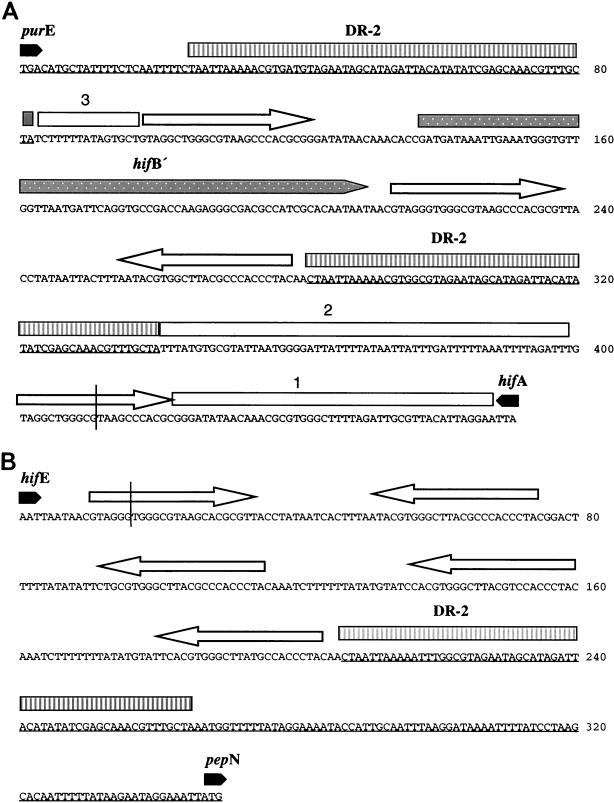
Sequences of the left and right ends of the F3031 hif-2 locus. Vertically striped boxes above the sequence show the 59-bp DR-2 duplications. Conserved junction sequences 1, 2, and 3 are shown by open bars. Open arrows depict IDSs. The vertical lines at the left and right ends show where flanking sequence diverges from F3031 hif-1 junctions (Fig. 2). Also shown is the inserted 3′-terminal portion of hifB (hifB′; stippled bar) between the leftmost DR-2s. The portion of the sequence homologous to H. influenzae Rd (Hif−) is underlined. (A) Left end: hifA-purE intergenic region. (B) Right end: hifE-pepN region.
This arrangement of the hif-1 junctions was a general feature of Hae strains. Nucleotide sequences of PCR fragments containing the hifA1-pmbA and hifE1-hpt junctions (Fig. 1) of Hae isolates F4931, F2066, and ATCC 43974 (Table 1) were more than 95% similar to the F3031 sequence. To show that the pmbA-hpt intergenic region was stable in H. influenzae strains that do not have pilus genes inserted at the hif-1 locus, we determined the nucleotide sequences of Hae strain 43806 and of isolates GA5445, GA2078, and 1007 (NTHI, serotype a, and serotype b, respectively) (19). All were at least 95% similar to the corresponding pmbA-hpt of H. influenzae Rd. PCR amplification with primers PMB and HI1153 of other strains lacking hif-1 (data not shown) (19) indicate consistently identical intergenic regions.
Hypervariation and gene insertion at the ends of H. influenzae hif-2 pilus gene clusters.
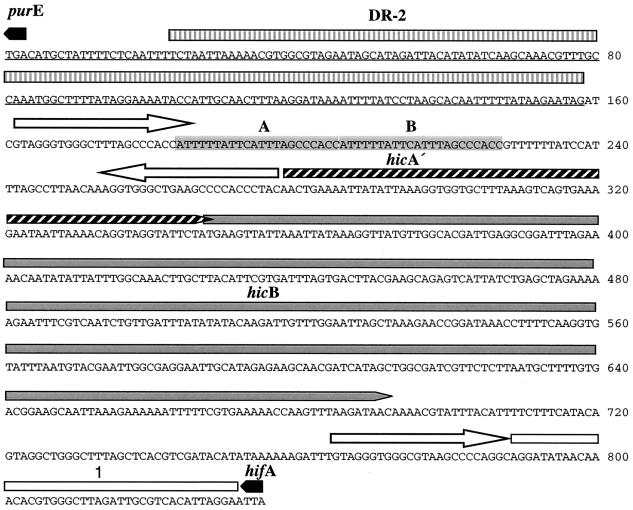
The hifA-purE and hifE-pepN regions of seven strains containing the hif-2 allele were sequenced, as well as the purE-pepN sequence of NTHI strain GA5445 lacking hif genes (Table 1; Fig. 3, 4, and 5). In contrast to the stable hif-1 cluster, hif-2 cluster ends are highly complex and variable. Sequence diversity between strains is particularly pronounced at purE-hifA2 junctions (Fig. 5), where there are conserved junction sequences (junction sequences 1, 2, and 3), two small ORFs (hicA and hicB), evidence of several short sequence insertions and deletions, and multiple copies of the 23-bp IDS.
FIG. 4.
GA2078 (NTHI) hif-2 left end nucleotide sequence. The hifA-purE sequence is shown for comparison to Fig. 3A. Note the extended (135-bp) DR-2, hicB gene, and partial hicA (hicA′; diagonally hatched bar), and also the 22-bp duplications (shaded sequences marked “A” and “B”), which are present in only one copy in ATCC 9007 and C2859.
FIG. 5.
Schematic diagram of H. influenzae hif-2 left cluster junctions. The sequences of AM30 (23), ATCC 9697, Eagan, R3001, and C2859 (15) have been reported previously. Open arrows, IDSs; open bars, conserved junction sequences. The pathway for insertion of hicAB genes into conserved junction sequence 1 of Eagan is marked with arrows and a shaded box. The portion of the Eagan left junction that is homologous to the right junction sequence of AM30 and 1007 is indicated by a solid line. Also the portion of C2859 homologous to the right junction of ATCC 9007 is bracketed. The 135-bp duplications of GA2078 and C2859 are shown as extensions of the DR-2 (hif2DR) box. Details of the right junctions are not shown. The drawing is not to scale.
BPF-associated Hae strain F3031 and Brazilian Hae strain F4931.
The left junction of the F3031 hif-2 pilus gene cluster (Fig. 3A) contains two copies of the DR-2 59-bp duplication. Sandwiched between the duplications is a 73-bp sequence homologous to the 3′-terminal portion of the hifB pilus chaperone gene. The fragment of hifB (hifB′) is flanked by copies of the IDS, GTAGGGTGGGCTTYAGCCCACCA (the underlined 13 bp is particularly strongly conserved). Between the 3′ end of the inner DR-2 and a subsequent IDS is conserved junction sequence 2, while conserved junction sequence 3 lies between the outer DR-2 and another IDS. At the hifE2-pepN junction (Fig. 3B) are strings of directly repeated IDSs and spacer sequences on opposite strands oriented to form larger inverted-repeat structures. The left and right junctions of F4931 are homologous to F3031.
Brazilian Hae strain F2066.
At the left junction of Hae strain F2066, the DR-2 was followed by junction sequences 2 and 1 separated by an IDS element (Fig. 5). The incomplete hifB sequence was not present. It is notable that the sequence of the left junction of serotype b strain Eagan (15) resembled F2066 with the addition of sequence within junction sequence 1 (Fig. 5). At the right hif-2 junction there were multiple repeats of IDS and spacer sequence in a manner analogous to F3031 (data not shown).
Texas Hae strain ATCC 43974.
In contrast to F2066, the Texas Hae strain ATCC 43974 had junction sequence 3 adjacent to the DR-2 at the left end of hif-2. Between two IDSs at the left junction was a complete ORF homologous to hicB, encoding a small acidic protein of unknown function, previously found at the left end of hif-2 in other H. influenzae strains (15). Upstream of hicB is the 3′ end of a partial copy of hicA, another gene of unknown function. Downstream of hicB was an IDS element, junction sequence 1, and hifA2. As with the other three Hae strains in this study, at the right junction there were sets of direct IDS repeats arranged as a large complex inverted repeat (data not shown).
Serotype b strain 1007.
The strain 1007 hif-2 junctions were identical to those of serotype b strain AM30 (20). Surprisingly, the entire right junction sequence of 1007 and AM30 was homologous to the left hif-2 junction of serotype b strain Eagan (15) (Fig. 5), suggesting that there may have been an independent recombination event deleting a whole set of hif genes in a common ancestor.
Serotype f strain GA4090.
The sequence of the junctions of serotype f strain GA4090 was identical to that of Hae strain F2066 (Fig. 5) and serotype f strain ATCC 9796 previously reported (15). A deletion consisting of most of the hifE gene was conserved in all of the 50 clinical serotype f isolates from the Georgia Emerging Infections Program that we tested by restriction fragment length polymorphism (RFLP)-PCR (unpublished data).
NTHI strain GA5445.
GA5445, which does not have a hif-2 pilus gene cluster insertion, has a purE-pepN sequence identical to that of NTHI strain R3001 (15) (Fig. 5). There was no duplication of the DR-2 sequence, and no IDS elements are associated with the junction. The strain contained complete copies of both hicA and hicB, arranged in tandem.
NTHI strain GA2078.
Strikingly, the length of the duplication at the left junctions of NTHI strain GA2078 and serotype c strain ATCC 9007 was 135 bp rather than the 59-bp DR-2 found in most other hif-2 clusters (Fig. 4 and 5). These two strains contain an incomplete hicA, a complete hicB, and the hifABCDE pilus gene cluster. NTHI strain C2859 (15) also contains the longer 135-bp DR-2 at the left junction, although this strain does not contain a pilus gene cluster. Instead, C2859 has complete hicAB genes, where GA2078 and 9007 have only a partial hicA sequence. Downstream of the DR-2, GA2078 contained a duplication of 22 bp (Fig. 4). The related left junction sequences of ATCC 9007 and C2859 had only one copy of this duplication. In common with all H. influenzae strains that contain pilus genes, junction sequence 1 was adjacent to the 3′ end of hifA (Fig. 5). At the GA2078 hif-2 right junction there were 3 IDS elements and the DR-2 (data not shown).
Serotype c strain ATCC 9007.
The hif-2 left junction of the ATCC 9007 isolate sequenced in our laboratory was homologous to NTHI strain GA2078, except that there was only one copy of the 22-bp duplication downstream of the DR-2 (Fig. 4). The right end of ATCC 9007 is homologous to the left end sequence immediately downstream of the first DR-2 of C2859 (Fig. 5), consisting of two IDS elements, the intervening spacer sequence, and the inner DR-2. This organization at the ATCC 9007 right end is also found in NTHI isolates AAr91 and AAr73 (GenBank accession numbers AF045064 and AF45062) (13). Comparison of hifE genes indicated further similarities between ATCC 9007 and the nontypeable strains. The sequence of hifEGA2078 is 98% similar to that of hifEAAr91, while that of hifE9007 is 98% similar to that of hifEAAr71 (13, 19). It is noteworthy that Mhlanga-Mutangadura et al. (15) sequenced the ATCC 9007 left junction and reported that there were two copies of the DR-2. The difference between the sequences was likely due to recombination between the DR-2 copies in the version of ATCC 9007 in this laboratory.
Sequence homology and host range of hicA and hicB genes.
The hicA and hicB genes and predicted gene products found in this study shared more than 95% identity with the sequences from H. influenzae strain Eagan (serotype b) and NTHI strains C2859, C2861, INT1, and R3001, described previously (15). The putative 82-amino-acid HicA and 114 amino-acid HicB gene products have predicted pI values of 10.0 and 4.8, respectively. They have no assigned function, but HicB is homologous to hypothetical proteins in the Deinococcus radiodurans genome (24) and in rickettsial species (2). Screening GenBank with the HicA and HicB protein sequences using TBLASTN revealed homology with products from predicted ORFs in an 807-bp sequence in E. coli strain ECOR32 (25). The 807-bp sequence is a replacement for a 7.6-kb DNA fragment containing an RhsB (recombination hotspot) repeat element.
We surveyed the incidence of homologous hicA and hicB sequences in 40 NTHI invasive disease isolates from the Georgia Emerging Infections Program by PCR using primers specific for hicA and hicB (see Materials and Methods) (data not shown). Of these strains, 72.5% (29 of 40) produced amplicons with both primer sets while only 7.5% (3 of 40) failed to produce amplicons with either the hicA or the hicB primers. The frequency of amplification of hicA only or hicB only was equal at 10% of isolates. These data show that the majority of NTHI strains in this collection contained homologous hicAB sequences, suggesting that complete or incomplete copies of the hicAB genes may be common among disease-causing NTHI strains. The failure to amplify hicAB gene fragments in a minority of strains may represent complete or partial absence of the genes or nucleotide variation in the area of the PCR primers.
DISCUSSION
The detailed sequence analysis of the junctions of the H. influenzae pilus gene clusters reveals a complex and variable organization. However, a central theme that emerges is the role of the 23-bp IDS element in the sequence rearrangements. The IDS is implicated in the frequent changes at the hypervariable junctions of the hif-2 cluster, which is the common location of the pilus genes in H. influenzae, and in the creation of the duplicated hif-1 cluster in the biogroup aegyptius strains.
Role of IDS in formation of the hif-1 cluster.
The IDS is common in H. influenzae: the Rd genome contains 14 complete IDS inverted repeats separated by a variable AT-rich spacer of 12 to 26 bases and several singular IDSs grouped tightly in five regions (18). Possibly the IDS is important in the physiology of the bacterium, forming intergenic hairpin-loop structures that influence mRNA stability or serve as targets for DNA binding proteins. The grouping of IDSs around clusters of genes in Rd and their association with sequence duplications (18) hint strongly at a role in gene movement in Haemophilus. IDS homologues are also prevalent in Neisseria gonorrhoeae and Neisseria meningitidis, where they border genes apparently inserted in the genome (16, 21).
IDSs appear to have been central to the event that produced the hif-1 gene cluster. The hif-1 and hif-2 clusters of Hae strain F3031 were shown to be more than 99% similar (17); furthermore, sequences immediately flanking the clusters are highly conserved (Fig. 2 and 3). The sequence homology abruptly ends on both sides of the clusters at a point within the IDS elements. This suggests that DNA containing hifABCDE from the hif-2 locus with IDS half-site ends invaded the pmbA-hpt region to form the hif-1 locus.
What is the mechanism for insertion of pilus genes at the hif-1 location? The generation of direct repeats at the target site, together with the presence of inverted repeats at the end of the inserted sequence, is reminiscent of transposition (11). The DR-1 is an almost perfect palindrome, suggestive of a transposase binding site. However, the direct repeats found adjacent to the inserted pilus genes are significantly longer than generally reported for transposon-mediated insertions (11). Another feature of the hif-1 duplication that differs from typical transposition is the introduction of short DNA regions of unknown origin between the inserted pilus cluster and direct repeats. There are also no sequences associated with the pilus gene cluster that show homology with a known transposase or integrase. Re-creation of the hif-1 duplication event will be essential for a complete understanding of the mechanism for insertion of the pilus genes.
IDS are central to localized variation at the hif-2 pilus junctions.
This study further documents the striking variety of localized sequence rearrangements around the H. influenzae pilus genes. Simple evolutionary relationships between strains cannot be derived from analysis of these junctions alone. At the right junction of the hif-2 cluster (Fig. 3B), direct repeats featuring IDS elements vary in copy number between strains, presumably due to slip-stranded replication (22). A similar process seems to be happening within the pilus cluster between hifA and hifB, where there are 10 directly repeated IDSs in serotype b strain AM30 (23) but only one in Hae strain F3031 (17). The left hif-2 junction is even more variable than the right (Fig. 5). Rearrangements include insertions of genes (hicA and hicB) or partial genes (hifB′ in F3031) and deletions (for example, loss of the 5′ end of hicA in ATCC 43974 and GA2078). It is also possible that pilus clusters may have been introduced and/or deleted by homologous recombination between DR-2s: for instance, between the two copies of the duplication of NTHI strain C2859. Again, IDS elements contribute to interstrain differences. IDSs flank the inserted hicAB and hifB′ and also are conspicuous as the link between the conserved junction sequences (Fig. 5). This distribution of IDS at the left hif-2 junction provides further evidence that they are recombinational hotspots. Possibly the hairpin loop is the site of frequent single- or double-stranded DNA scission, generating recombinogenic ends.
The junctions of the hif-1 cluster in Hae strains are more tightly conserved across strains than the hif-2 junctions, probably because the former possess only partial IDS sites. It is significant that the left hif-2 junctions of the Hae strains F2066 and ATCC 43974 are very different from those of F3031 and F4931 (Fig. 5), illustrating the variation that can be generated at this location in the short time since the common ancestor of the four strains acquired the hif-1 cluster. The leftmost DR-2 is strongly conserved in the Hae strains (Fig. 5), indicating that recombination events are occurring distal to the 3′ end of this sequence. The fact that the DR-2s of F2066 and F3031 abut different junction sequences (junction sequence 2 versus 3) implicates the 3′ end of the 59-bp duplication as another recombination hotspot (a similar pattern occurs with serotype b strains Eagan and AM30 [Fig. 5]).
Possible evolutionary significance of hypervariable pilus junctions.
Is there an evolutionary process driving some of the genetic rearrangement seen in this study? One indication of a role for natural selection is the observation by Mhlanga-Mutangadura et al. (15) that expression of pili and carriage of hic genes were mutually exclusive in their small sample group (nine H. influenzae strains). Although it is not certain that the ORFs even encode proteins, we can speculate that perhaps HicA and HicB perform a function antagonistic to that of pili yet necessary for invasion of certain niches. An analogous situation involving the Hia and HMW adhesins is already known to exist in NTHI strains (18). Possibly the Hic proteins may be a novel adhesin (14), or even function to block pilus-mediated attachment. Acquisition of hic or hif genes by an H. influenzae strain might profoundly influence the interaction with its human host. Evidence from sequencing this small population of pilus cluster junctions suggests that strains with different pedigrees may have picked up hic genes independently (for example, ATCC 43974, Eagan, C2859, and GA5445 [Fig. 5]). Since having both hif and hic genes would be nonproductive, selection would favor loss of one or the other cluster. The NTHI strain C2859 might be a case of a strain that has deleted its pilus genes, while strains Eagan (serotype b), ATCC 43974 (Hae), and GA2078 (NTHI) have deleted the 5′ end of hicA at least. Thus, the hypervariability of the left hif-2 junction might be an incidental result of selection for certain H. influenzae strains to oscillate between possession of either hic or hif clusters. It is also notable that some important pathogenic H. influenzae strains, such as serotype f strains, neither express complete pili nor carry hic genes.
In summary, we believe that the insertion of the hifA through hifE pilus genes as a unit into the chromosome has been a rare event in the evolution of H. influenzae. The most ancient insertion appears to have been in the hif-2 location, between purE and pepN, generating 59-bp DR-2 duplications. Another insertion of pilus genes at the hif-2 site, this time generating 135-bp duplications, may have occurred in the forerunner of some NTHI and serotype c strains. In Hae, a hif-2 pilus cluster was the template for an insertion at the hif-1 location, between pmbA and hpt, that generated the 32-bp DR-1. In addition to rare insertions, more frequent DNA rearrangements internal to the pilus genes and flanking sequence have taken place over the course of H. influenzae evolution. These rearrangements may include movement of pilus clusters by homologous recombination at DR-2 sites. There has been expansion and contraction of IDS repeat units between hifA and hifB and at the right hif-2 junction. It is possible that these repeats might influence hif gene expression. At the hypervariable left hif-2 junction there has been insertion and deletion of two small ORFs, hicA and hicB, as well as other rearrangements, also often associated with IDS elements.
The H. influenzae pilus clusters are an unusual paradigm for a pathogenicity island (6), which contain some elements suggestive of transposase or site-specific recombinase activity. Understanding the selective pressures which result in the appearance of genes and other structures (such as IDS elements) in these chromosomal locations will benefit the study of pathogenic bacteria such as H. influenzae.
ACKNOWLEDGMENTS
We acknowledge the technical assistance of Samantha Terris and Julie Turner in the production of the manuscript.
This work was supported by a Veterans Affairs Merit Grant awarded to M.M.F.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson J O, Andersson S G. Genome degradation is an ongoing process in Rickettsia. Mol Biol Evol. 1999;16:1178–1191. doi: 10.1093/oxfordjournals.molbev.a026208. [DOI] [PubMed] [Google Scholar]
- 3.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 4.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M, Utterback T, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 5.Geluk F, Eijk P P, van Ham M, Jansen H M, van Alphen L. The fimbrial gene cluster of nontypeable Haemophilus influenzae. Infect Immun. 1998;66:406–417. doi: 10.1128/iai.66.2.406-417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hacker J G, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 7.Higgins P G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignments on a computer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 8.Kar S, To S C, Brinton C C., Jr Cloning and expression in Escherichia coli of LKP pilus genes from a nontypeable Haemophilus influenzae strain. Infect Immun. 1990;58:903–908. doi: 10.1128/iai.58.4.903-908.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlin S, Mrazek J, Campbell A M. Frequent oligonucleotides and peptides of the Haemophilus influenzae genome. Nucleic Acids Res. 1996;24:2463–4272. doi: 10.1093/nar/24.21.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kroll J S, Loynds B M, Moxon E R. The Haemophilus influenzae capsulation gene cluster: a compound transposon. Mol Microbiol. 1991;5:1549–1560. doi: 10.1111/j.1365-2958.1991.tb00802.x. [DOI] [PubMed] [Google Scholar]
- 11.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin K, Morlin G, Smith A, Nordyke A, Eisenstark A, Golomb M. The tryptophanase gene cluster of Haemophilus influenzae type b: evidence for horizontal gene transfer. J Bacteriol. 1998;180:107–118. doi: 10.1128/jb.180.1.107-118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCrea K W, St. Sauver J L, Marrs C F, Clemans D L, Gilsdorf J R. Immunologic and structural relationships of the minor pilus subunits among Haemophilus influenzae isolates. Infect Immun. 1998;66:4788–4796. doi: 10.1128/iai.66.10.4788-4796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCrea K W, Watson W J, Gilsdorf J R, Marrs C F. Identification of hifD and hifE in the pilus gene cluster of Haemophilus influenzae type b strain Eagan. Infect Immun. 1994;62:4922–4928. doi: 10.1128/iai.62.11.4922-4928.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mhlanga-Mutangadura T, Morlin G, Smith A L, Eisenstark A, Golomb M. Evolution of the major pilus gene cluster of Haemophilus influenzae. J Bacteriol. 1998;180:4693–4703. doi: 10.1128/jb.180.17.4693-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkhill J, Achtman M, James K D, Bentley S D, Churcher C, Klee S R, Morelli G, Basham D, Brown D, Chillingworth T, Davies R M, Davis P, Devlin K, Feltwell T, Hamlin H, Holroyd S, Jagels K, Leather S, Moule S, Mungall K, Quail M A, Rajandream M A, Rutherford K M, Simmonds M, Skelton J, Whitehead S, Spratt B G, Barrell B G. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 17.Read T D, Dowdell M, Satola S W, Farley M M. Duplication of pilus gene complexes of Haemophilus influenzae biogroup aegyptius. J Bacteriol. 1996;178:6564–6570. doi: 10.1128/jb.178.22.6564-6570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Read T D, Farley M M. Conserved extragenic DNA elements in Haemophilus influenzae. Mol Microbiol. 1997;23:627–628. doi: 10.1046/j.1365-2958.1997.d01-1862.x. [DOI] [PubMed] [Google Scholar]
- 19.Read T D, Satola S W, Opdyke J O, Farley M M. Copy number of pilus gene clusters in Haemophilus influenzae and variation in the hifE pilin gene. Infect Immun. 1998;66:1622–1631. doi: 10.1128/iai.66.4.1622-1631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.St. Geme J W, III, Kumar V V, Cutter D, Barenkamp S J. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect Immun. 1998;66:364–368. doi: 10.1128/iai.66.1.364-368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tettelin H, Saunders N J, Heidelberg J, Jeffries A C, Nelson K F, Eisen J A, Ketchum K A, Hood D W, Peden J F, Dodson R J, Nelson W C, Gwinn M L, DeBoy R, Peterson J D, Hickey E K, Haft D H, Salzberg S L, White O, Fleischmann R D, Dougherty B A, Mason T, Ciecko A, Parksey D S, Blair E, Cittone H, Clark E B, Cotton M D, Utterback T R, Khouri H, Qin H, Vamathevan J, Gill J, Scarlato V, Masignani V, Pizza M, Grandi G, Sun L, Smith H O, Fraser C M, Moxon E R, Rappuoli R, Venter J C. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 22.van Ham S M, van Alphen L, Mooi F R, van Putten J P R. Phase variation of H. influenzae fimbriae: transcriptional control of two divergent genes through a variable combined promoter region. Cell. 1993;73:1187–1196. doi: 10.1016/0092-8674(93)90647-9. [DOI] [PubMed] [Google Scholar]
- 23.van Ham S M, van Alphen L, Mooi F R, van Putten J P M. The fimbrial gene cluster of Haemophilus influenzae. Mol Microbiol. 1994;13:673–684. doi: 10.1111/j.1365-2958.1994.tb00461.x. [DOI] [PubMed] [Google Scholar]
- 24.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, Moffat K S, Qin H, Jiang L, Pamphile W, Crosby M, Shen M, Vamathevan J J, Lam P, McDonald L, Utterback T, Zalewski C, Makarova K S, Aravind L, Daly M J, Fraser C M, et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao S, Sandt C H, Feulner G, Vlazny D A, Gray J A, Hill C W. Rhs elements of E. coli K-12: complex composites of shared and unique components that have different evolutionary histories. J Bacteriol. 1993;175:2799–2808. doi: 10.1128/jb.175.10.2799-2808.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]